Abstract
Predicting drug interactions, disposition, and side effects is central to the practice of clinical pharmacology. Until recently, the human microbiome has been an under-appreciated player in the dynamics of drug metabolism. It is now clear that humans are “superorganisms” with about 10-fold more microbial cells than human cells and harboring an immense diversity of microbial enzymes. Owing to the advent of new technologies, we are beginning to understand the human microbiome’s impact on clinical pharmacology.
The majority of the human microbiome, ~ 1 trillion cells, resides in the intestines. The intestinal microbiome is a complex community of microorganisms, typically containing hundreds of different species, many of which can be cultured in the laboratory.1 The composition of individuals’ microbiomes is remarkably resilient given that they reside in a transient environment with a continual flux of variable food sources. Microbiome composition varies considerably from person to person, and interindividual variation is often greater than day-to-day variation within the same individual. Moreover, an individual’s microbiome composition can return to its original state after extreme perturbations, such as oral antibiotics. This suggests that there are external forces selecting for a specific microbiome composition, such as an individual’s immune system.
The human microbiome is a complex system of interacting microorganisms coupled to another complex system, the human. For this reason, it is perhaps unsurprising that microbiome composition has been associated with differences in behavior, treatment outcomes, and a wide array of conditions, including arthritis, asthma, autism, behavior, and obesity. Disentangling correlation from causation is a common challenge in human microbiome studies, but new technologies are helping to elucidate the mechanistic basis underlying the microbiome’s effects. Recent work is moving toward building a framework of microbiome function within the host, with the goals of controlling and manipulating the microbiome for the benefit of human health.
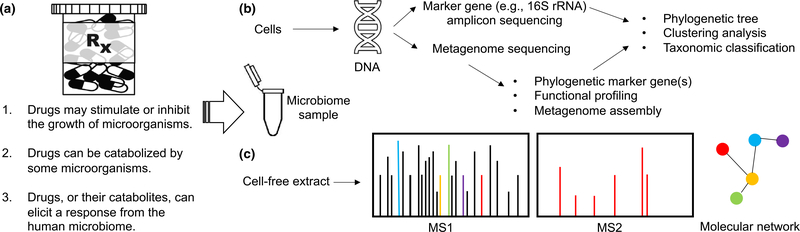
The intestinal microbiome partakes in a multi-way interaction with its host and their food or drug intake. There are three main ways in which drugs interplay with the intestinal microbiome (Figure 1a):
Drugs may stimulate or inhibit the growth of a subset of microorganisms
Drugs can be transformed or catabolized by members of the intestinal microbiome
Drugs, or their byproducts, can elicit a response from the microbiome, such as production of new compounds
Figure 1.
Schematic of experimental designs for capturing the interplay between drugs and the intestinal microbiome. (a) Drugs can change the microbiome’s composition, and the microbiome can transform or respond to drugs. (b) Microbiomics experimental pipelines typically involve extracting DNA from samples and amplifying marker gene sequences before performing reference-free (e.g., clustering) or referenced-based (e.g., classification) analyses. (c) Metabolomics studies of the exometabolome generally involve processing cell-free extracts before performing tandem mass spectrometry with or without prior separation by chromatography. The resulting MS1/MS2 data can be used to identify compounds and generate molecular networks that inform drug transformations mediated by the microbiome. For example, the red MS1 peak may result in an MS2 spectra that allows it to be related to other peaks through a molecular network. MS1, initial molecular weight; MS2, secondary fragmentation; rRNA, ribosomal RNA.
In this article, we describe modern methods for studying these three processes with microbiome samples. Although these methods are broadly applicable, we focus on green tea (GT) because it is a mixture of natural products with hallmarks of complex interactions with the intestinal microbiome.2 Similar methods could be applied to study other natural products, drugs, or drug–drug interactions.
MICROBIOME SURVEYS TO SCREEN FOR DRUG EFFECTS
Many drugs have effects on microbiome composition even though they are not widely assumed to have antimicrobial properties. The activity of nonantibiotic drugs is often species specific3 and may be difficult to determine given the large number of microorganisms that inhabit human intestine. Culture-based assays serve as models for confirming a drug’s ability to modulate membership in the intestinal microbiome. For example, GT is well known for its antimicrobial properties and health benefits, which are attributed to the catabolization of GT polyphenols by the gut microbiome.2 Different concentrations of GT can inhibit the growth of microorganisms that are well known for their prevalence in the gut. Using in vitro techniques, we have found that Escherichia coli (E. coli) is inhibited on media containing a concentration equivalent to tea brewed with 1 tea bag (~ 0.5 g/L catechins), whereas 1.5 tea bags are required to inhibit the growth of Enterococcus faecium. Nevertheless, culture-based assays are laborious and limited by the fact that we do not yet know how to culture some microorganisms from the human intestine.1
The most common approach for studying the intestinal microbiome is to perform sequencing of the 16S ribosomal RNA (rRNA) gene of fecal samples before and after treatment with a drug (Figure 1b). Sequencing the 16S rRNA has become popular due to the gene’s ubiquity across all prokaryotes, ease of selective amplification with polymerase chain reaction primers targeting conserved regions, and variable regions that may provide genus-level resolution. Many companies now offer services to amplify and sequence the 16S rRNA gene from thousands of samples in microbiome studies. This allows comparison of microbiome samples originating from large cohorts across many timepoints. The resulting sequences can be used to identify many of the organisms that are present, compare diversity across samples, or construct a phylogenetic tree depicting the evolutionary relationships among microorganisms. For example, 16S rRNA sequencing has been applied to study (i) the shift in intestinal microbiomes before and after drinking GT4 and (ii) the impact of polybrominated diphenyl ethers, persistent environmental contaminants, on the regulation of primary bile acids metabolized by the mouse gut microbiome.5
Although 16S rRNA sequencing has a relatively low cost per sample, there are many challenges associated with processing and interpreting the immense number of resulting DNA sequences. As is common for observational studies, it is often impossible to distinguish correlation from causation in microbiome surveys. Furthermore, experimental issues (e.g., polymerase chain reaction artifacts and sequencing errors) can confound the analysis of 16S rRNA amplicon sequences. The 16S rRNA gene is also an imperfect marker of microbiome composition, as it does not offer species-level resolution and ignores many microorganisms (e.g., eukaryotes and viruses). For this reason, metagenome sequencing has become increasingly popular (Figure 1b). Although it is more costly per sample, metagenomics provides the ability to investigate microbial functions and even assemble some genomes without culturing.
METABOLOMICS TO PINPOINT DRUG–MICROBIOME INTERACTIONS
Although microbiome surveys have implicated the microbiome in human health, metabolomics holds the potential to shed further light on its role in the host. Metabolomics involves the use of a mass spectrometer to identify compounds according to their mass-to-charge ratio (m/z) in cell-free supernatant. Untargeted metabolomics can detect compounds at very small abundances, making it possible to screen for transformations that are mediated by members of the intestinal microbiome (Figure 1c). These transformations may cause off-target effects or alter the potency of drugs in individuals with specific microbiomes. For example, the bioactive compounds in GT are known to undergo a variety of transformations into metabolic derivatives with largely unknown effects.2 In this manner, the intestinal microbiome may play a role in promoting or disrupting the beneficial effects of drugs and natural products.
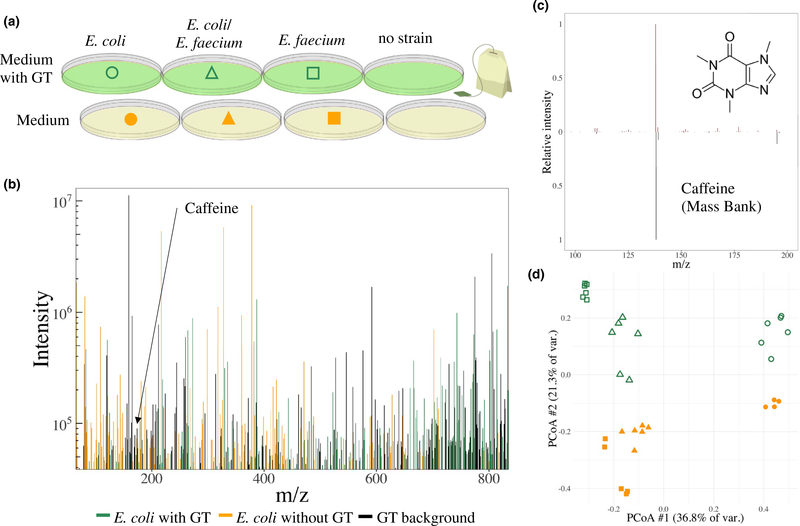
Untargeted metabolomic studies generally collect mass spectra on a variety of samples from different conditions. For example, we have applied metabolomics to study the exometabolome of common intestinal microorganisms in monoculture and coculture during growth in the presence or absence of GT (Figure 2a). Using this approach, we have observed the secretion of a number of compounds by E. coli that are induced only in the presence of GT (Figure 2b). Tandem mass spectrometry can be used to fragment specific compounds of interest within the sample. In conjunction with the initial molecular weight, the secondary fragmentation spectrum can be compared with extensive databases of known secondary fragmentation spectra to identify molecules of interest, such as caffeine in GT (Figure 2c). This data can be used in conjunction with molecular networking to identify microbially mediated transformations to drugs or natural products.
Figure 2.
Studying the exometabolome of gut bacterial strains and how their interactions change in response to green tea (GT). (a) Monocultures and cocultures of Escherichia coli (E. coli) and Enterococcus faecium (E. faecium) grown in the presence and absence of GT. A negative control is used for background subtraction in subsequent steps. (b) Mass spectra of the exometabolome of E. coli grown in the presence of GT with removal of mass-to-charge ratio (m/z) features found in the medium. Peaks are colored based on whether they were found in the GT extract alone (black), E. coli grown on the medium without GT (orange), or found only when grown in the presence of GT (green). (c) Molecular structure of caffeine and secondary fragmentation spectrum of the precursor ion (top) compared with a caffeine standard (bottom). (d) Principle coordinates analysis (PCoA) of captured exometabolomes of two gut microbiota strains (E. coli and E. faecium) in replicate monocultures (circles and squares) and cocultures (triangles) grown on the medium alone (orange) or medium supplemented with GT extract (green). All samples have peaks in the medium and GT background subtracted before analysis. Samples cluster according to the growth medium and strains, revealing differences in the exometabolome elicited by growth in the presence of GT.
However, untargeted metabolomics of the bacterial exometabolome is not without drawbacks. In addition to the inability to quantify many compounds, mass spectra are rife with low-intensity peaks from unknown compounds (i.e., noise) and may be missing peaks from many present compounds for a wide variety of reasons. Notwithstanding these limitations, it is possible to use the entire initial molecular weight spectrum to compare samples with ordination techniques, such as principle coordinates analysis. Many replicates are typically used to verify that intersample differences are greater than variability from experimental sources. This approach allows the reduction of complex mass spectra with many unknowns into a manageable framework for comparing samples across treatment conditions (Figure 2d), and it may reveal the underlying compounds that are important for further investigation. Ordination techniques can be used in conjunction with dose gradients to uncover changes in the exometabolome that are elicited in response to increasing levels of drugs or natural products.
CONCLUSIONS
The impact of the human microbiome on clinical pharmacology has only recently been widely recognized. It is now commonplace for researchers to investigate the microbiome’s influence on drug interactions, disposition, and side effects. Yet, it remains challenging to determine the functional basis behind the microbiome’s role due to the abundance of interactions among members of the microbiome, their human host, and a person’s food or drug intake. Although the microbiome is indirectly considered during clinical trials, they are unlikely to capture the full variability among human microbiomes or the breadth of potential drug interactions. Experiments in laboratory (e.g., in vitro) systems offer a means of studying these interactions in a controlled setting, albeit one removed from the human host. New technologies continue to emerge that enable clinical pharmacologists to probe these interactions in high throughput. Although few well-characterized examples currently exist of the complex interplay between drugs and the intestinal microbiome, we are optimistic that many will emerge in the future as the importance of the microbiome becomes increasingly clear.
ACKNOWLEDGMENTS
The authors wish to acknowledge the support of Christopher Anderton and Rosalie Chu at Pacific Northwest National Laboratory in acquiring the mass spectrometry data shown here. We are grateful for feedback from Richard Boyce and the Center for Excellence of Natural Product-Drug Interaction Research.
FUNDING
This work was funded by the National Center for Complementary & Integrative Health at the National Institutes of Health (grant 7U54AT008909–03). Mass spectrometry was performed by the Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory (proposal 50137).
Footnotes
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
References
- 1.Forster SC et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat. Biotechnol. 37, 186–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H & Sang S Biotransformation of tea polyphenols by gut microbiota. J. Funct. Foods 7, 26–42 (2014). [Google Scholar]
- 3.Maier L et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan X et al. Green tea liquid consumption alters the human intestinal and oral microbiome. Mol. Nutr. Food Res. 62, e1800178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li CY et al. PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice. Drug Metab. Dispos. 46, 1226–1240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]