Abstract
Purpose:
Treatment outcomes for childhood acute lymphoblastic leukemia (ALL) have improved steadily, but a significant proportion of patients still experience relapse due to drug resistance, which is partly explained by inherited and/or somatic genetic alternations. Recently, we and others have identified genetic variants in the ARID5B gene associated with susceptibility to ALL and also with relapse. In this study, we sought to characterize the molecular pathway by which ARID5B affects antileukemic drug response in patients with ALL.
Experimental Design:
We analyzed association of ARID5B expression in primary human ALL blasts with molecular subtypes and treatment outcome. Subsequent mechanistic studies were performed in ALL cell lines by manipulating ARID5B expression isogenically in which we evaluated drug sensitivity, metabolism, and molecular signaling events.
Results:
ARID5B expression varied substantially by ALL subtype, with the highest level being observed in hyperdiploid ALL. Lower ARID5B expression at diagnosis was associated with the risk of ALL relapse, and further reduction was noted at ALL relapse. In isogenic ALL cell models in vitro, ARID5B knockdown led to resistance specific to antimetabolite drugs (i.e., mercaptopurine and methotrexate), without affecting sensitivity to other antileukemic agents. ARID5B downregulation significantly inhibited ALL cell proliferation and caused partial cell-cycle arrest. At the molecular level, the cell-cycle checkpoint regulator p21 (encoded by CDKN1A) was most consistently modulated by ARID5B, plausibly as its direct transcription regulation target.
Conclusions:
Our data indicates that ARID5B is an important molecular determinant of antimetabolite drug sensitivity in ALL, in part through p21-mediated effects on cell-cycle progression.
Keywords: acute lymphoblastic leukemia, ARID5B, p21, relapse, antimetabolite drug resistance
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer in children and a leading cause of disease-related death in childhood (1,2). Although aggressive and fatal if not treated, ALL responds remarkably well to cytotoxic agents (e.g., glucocorticoids, methotrexate [MTX], and mercaptopurine [6-MP]) when compared to other hematological malignancies and most solid tumors (3). Contemporary combination chemotherapy can induce complete remission in nearly all children with ALL, and more than 85% of patients achieve sustained disease-free survival after ~ 2 years of risk-adapted treatment. Even with the introduction of molecularly targeted agents (e.g., imatinib), conventional chemotherapeutics remain the mainstay of ALL therapy and have proved indispensable for long-term survival (4). However, the exact mechanisms by which these cytotoxic agents exert their antileukemic effects are not completely understood. For example, a significant proportion of patients with ALL respond to initial induction therapy but nevertheless experience relapse (5,6), and many eventually succumb to their disease (7). The disparity between early response and long-term survival is a significant clinical issue for which the underlying biology is largely unknown. Identifying genetic factors that drive inherent or acquired drug resistance would not only inform more rationally designed ALL treatment regimens but could also improve fundamental understanding of the biology of leukemia drug sensitivity.
Recent genomic profiling studies have discovered a number of novel pathways involved in leukemia pathogenesis and/or treatment response. For example, genome-wide association studies by us and others have identified ARID5B as one of the top loci associated with susceptibility to ALL in children (8–18). ARID5B variants were particularly over-represented in patients with hyperdiploid ALL and also correlated with MTX metabolism in ALL cells (13,15). We have subsequently identified additional variants at this locus that influence both ALL risk and treatment outcome (16,17). ARID5B is a member of the ARID gene family characterized by a shared DNA-interacting motif (19–23). ARID genes are generally described as chromatin-remodeling factors (20,24,25), and somatic mutations in these genes have recently been described in multiple solid tumors (e.g., ARID1A) (26), signifying their importance in tumorigenesis. There is a particular paucity of functional studies of ARID5B, although the limited data available suggest that it functions as a transcription factor (19,22). In liver cells, ARID5B interacts with PHF2 as part of the chromatin-remodeling complex involved in histone methylation at genes related to chondrogenesis and glucogenesis (27). Arid5b null mice showed signs of defective lymphocyte development, especially during the first 3 months after birth (19). However, the exact role of the ARID5B gene in hematologic malignancy is largely uncharacterized, particularly in the context of antileukemic drug responses.
In this study, we systematically evaluated the expression pattern of ARID5B in pediatric ALL and its relation leukemia relapse. In vitro manipulation of ARID5B expression directly influenced the sensitivity of ALL cells to 6-MP and MTX, plausibly via p21-mediated effects on the cell cycle. Collectively, these data point to ARID5B as a potential prognostic factor in ALL and novel mechanisms related to antileukemic drug resistance.
Materials and Methods
Patients and ALL genomics
The pattern of ARID5B gene expression in newly-diagnosed ALL and its variation by molecular subtype were analyzed using two global gene expression profile datasets: 446 children from St. Jude Children’s Research Hospital (GSE33315) (28) and 106 patients from the German Cooperative Study Group for Childhood ALL and the Dutch Childhood Oncology Group (GSE13351) (29). These patients represent consecutively enrolled cases or were selected to represent the diversity of ALL subtypes (28,29).
The association of ARID5B expression with ALL relapse was examined first in 59 patients with high-risk B-ALL treated on the COG 1961 protocol (GSE7440) (30), of whom 28 experienced complete remission for at least 4 years and 31 experienced a bone marrow relapse within 3 years of diagnosis. In a second cohort of 49 patients with matched diagnosis-relapse ALL pairs (GSE28460) (31), we compared the ARID5B expression in the diagnostic bone marrow to that in relapsed leukemia in the same individual. For these analyses, patients were included primarily on the basis of sample availability (30,31).
This study was approved by the Institutional Review Board at St. Jude Children’s Research Hospital. Informed consent was obtained from parents, guardians, or patients as appropriate. This study was conducted in accordance with the U.S. Common Rule and all applicable legal regulatory requirements.
ALL cell culture and ARID5B knockdown
ALL cell lines (Nalm6, SEM, and UOC-B1 cells) were grown in culture in RPMI 1640 medium (GIBCO, Life Technologies) supplemented with 10% heat-inactivated FBS and 2 mM L-glutamine, at 37°C in 5% CO2. Stable knockdown of ARID5B was performed using lentiviral shRNAs cloned on the pLKO backbone (Sigma-Aldrich): shRNAs TRCN0000151040, TRCN0000155854, TRCN0000151281, TRCN0000152535, and TRCN0000155468 were used to target ARID5B, with shRNA SHC016 as the non-target control. Lentiviral particles containing shRNAs were produced by transient transfection of HEK293T cells with calcium phosphate (32). ALL cells were incubated with lentiviral supernatants for 48 h then subjected to selection with puromycin (2 μg/mL) for 3 days. Two hundred transduced leukemia cells were plated in methylcellulose colony-selection medium (STEMCELL Technologies) and incubated for 10 to 20 days to form single-cell colonies. Clones with shRNAs TRCN0000151040 (targeting the 3′ UTR) and TRCN0000155854 (targeting the coding region) exhibited the strongest ARID5B knockdown and remained stable for at least 10 passages (Supplementary Figure 1) and were thus selected for use in the subsequent experiments. For ARID5B re-expression, we ectopically expressed the ARID5B coding sequence in knockdown clones with shRNA TRCN0000151040 only (targeting the 3′ UTR), such that knockdown was restricted to endogenous AIRD5B with no effect on ectopic ARID5B expression. For inducible knockdown of ARID5B, shRNA TRCN0000151040 was cloned into the pLKO-Tet-On vector (Sigma-Aldrich), and transduced ALL cells were exposed to doxycycline (2 μg/mL) for ARID5B downregulation.
The efficacy of ARID5B knockdown was determined by quantitative PCR and Western blot analysis. Quantitative PCR assays were performed using the SYBR reagent (Roche Diagnostics) and an Applied Biosystems 7900 Fast Real-Time PCR System (Applied Biosystems). The primer sequences for ARID5B were AAGGTTGCCATTGGTGAAGAGTGC and GACGGCGGGCTGTTATTGTTTCAT, and the human β-actin gene was used as internal control (primer sequences: GTTGTCGACGACGAGCG and GCACAGAGCCTCGCCTT). An anti-ARID5B antibody was purchased from Sigma-Aldrich (HPA015037) and used at a 1:1000 dilution.
Orthogonally, we also performed ARID5B knockdown using the CRISPR-dCas9-KRAB system, following previously published CRISPRi method (33). Nalm6 cells were first letivirally transduced with the dCas9-KRAB-Blast plasmid (#89567, Addgene) to establish stable expression of dCas9 fusion protein. sgRNA specifically targeting ARID5B promotor (TAGAAAGAGGAGCAGCGCCC) was then introduced by a second lentiviral transduction. After selection with blasticidin and puromycin, expression of ARID5B was determined by RT-PCR described above.
Nalm6, SEM and UOC-B1 cells were obtained from the American Type Culture Collection or the German Collection of Microoganisms and Cell Cultures in 2012, respectively. Cells were authenticated by STR profiling regularly tested for mycoplasma contamination by using MycoAlert Mycoplasma Detection Kit (Lonza # LT07–118). All cell line-based experiments were completed within three weeks after thawing.
Antileukemic drug sensitivity assay and drug metabolite measurement
Antileukemic drug sensitivity was tested in ALL cells, using the MTT assay (34). Briefly, cells were seeded in 96-well plates at a density of 20,000 cells/well then treated with different concentrations of drugs (i.e., prednisone, dexamethasone, vincristine, daunorubicin, asparaginase, MTX, or 6-MP) in triplicate. After 48 h of incubation, cell viability was quantified using the MTT assay and the LC50 (concentration at which drugs kill 50% of leukemia cells) was estimated using the Prism software (GraphPad Software).
MTX and 6-MP metabolite assays were performed in accordance with our established HPLC-based procedures (35,36). Briefly, cells were plated at a density of 0.5 million cells/mL and incubated at 37°C with MTX (1 μM) in the presence of glycine (670 μM), adenosine (37 μM), and thymidine (41 μM) for 24 h. Five million cells were washed twice with ice-cold PBS, resuspended in water, and boiled for 5 min. The cell pellets were then analyzed and quantified for polyglutamated MTX (MTXPG) by using HPLC (35). For 6-MP metabolites, cells were incubated with 6-MP (10 μM) for 24 h, after which lysates were prepared by sonication. Intracellular thioguanine nucleotides were dephosphorylated, and thioguanine ribonucleosides were measured by HPLC (36).
ALL cell proliferation and cell-cycle analysis
ALL cell proliferation (for Nalm6, SEM, and UOC-B1 cells) was monitored by daily viable cell counts during a 4-day culture period. Cell growth was also examined by using the BrdU assay: cells were pulsed with 10 μM BrdU for 10 h, then BrdU uptake was quantified using flow cytometry. Cell-cycle analysis was performed by using propidium iodide staining. For each experiment, at least 10,000 events per sample were recorded by flow cytometry and data analysis was performed using FlowJo software.
Western blot analysis was performed to quantify the levels of p53, p21, p27, cyclin D, cyclin E, CDK2, and phosphorylated Rb in ALL cells under various conditions, with GAPDH as the loading control. All antibodies were purchased from Cell Signaling Technologies and were diluted at 1:1000 or according to the manufacturer’s instructions.
Gene expression profiling of ARID5B-knockdown ALL cells
Total RNA was extracted using an RNA Isolation Kit (Qiagen). Complementary DNA was synthesized from mRNA with Superscript III Reverse Transcriptase (Invitrogen, Life Technologies). Microarray experiments were performed by the Hartwell Center at St. Jude Children’s Research Hospital, using the Affymetrix Human GeneChip 1.0 ST Array (ThermoFisher). The expression of each probe-tagged gene was determined by Affymetrix Expression Console software, and pathway analyses were performed using the Gene Set Enrichment Analysis algorithm (37).
ARID5B binding to the CDKN1A cis-regulatory element
ARID5B ChIP assay was performed using the ChIP-IT High Sensitivity kit (53040; Active Motif) according to the manufacturer’s instructions. A total of 2 × 107 Nalm6 cells were fixed with formaldehyde, and sonicated chromatin was incubated with anti-ARID5B antibody (NBP1–83622; Novus Biologicals) or anti-IgG antibody (3900S; Cell Signaling Technology) overnight at 4°C. This was followed by immunoprecipitation with Protein G agarose beads for 3 h at 4°C. Precipitated DNA was analyzed by qRT-PCR with the following primers: CDKN1A forward primer 1: CACACTGCTCTATGCCAGATAC; CDKN1A reverse primer 1: CCTTAACAAGTCGGCCTGAA; CDKN1A forward primer 2: GCCTGGCTTTGAAGGTTATTG; CDKN1A reverse primer 2: AGTCATTGCTTTCCCTCAACTA; CDKN1A forward primer 3: GCACTCATGGATTCTCTCCTTTA; CDKN1A reverse primer 3: TGATCCCAGGCAAGTTGTTTA.
Additionally, we also performed CRISPRi experiments to specifically disrupt the purported interaction of ARID5B with the CDKN1A cis-regulatory element (33). Briefly, Nalm6 cells with stable dCas9-KRAB expression were lentivirally infected with a sgRNA plasmid (TTTAATCACCTTGGCCCCCG). CDKN1A expression was then quantified by RT-PCR (primer sequences: TGTCCGTCAGAACCCATGC and AAAGTCGAAGTTCCATCGCTC).
Statistical analysis
Variation in ARID5B expression across subtypes were significant in both cohorts (St. Jude and DCOG), as evaluated using the ANOVA test (38). ARID5B expression association with relapse in the COG P1961 cohort was tested using the logistic regression model, after adjusting co-variables as needed. Analysis of ARID5B level in diagnosis-relapse ALL pairs was performed using paired t-test.
Results
ARID5B expression in ALL and its association with treatment outcome
In an unselected cohort of 446 children with newly diagnosed ALL from St. Jude Children’s Research Hospital, ARID5B expression was highly variable by molecular subtype, with the highest level observed in patients with a hyperdiploid karyotype and the lowest level in patients with T-ALL (28) (Figure 1A). This subtype-dependent ARID5B expression pattern was also confirmed in an independent cohort of 106 pediatric patients with ALL from the Dutch Childhood Oncology Group (Figure 1B) (29). ARID5B expression differed by age at leukemia diagnosis (P = 0.001 for comparing age groups <10 years vs. ≥10 years; P = 5.5 × 10−6 for age treated as a continuous variable; ARID5B expression was higher in younger patients) and by gender (P = 0.02; ARID5B expression was higher in female patients), but was not related to genetic ancestry (P > 0.05). Of interest, the frequency of germline ALL risk variant in ARID5B also varies greatly by subtype, highest in the hyperdiploid subtype and least common in T-ALL (15,39,40).
Figure 1. ARID5B expression varies significantly by ALL subtype.
Gene expression was estimated by Affymetrix U133A [for the St. Jude cohort] (A) or U133-plus 2.0 [for the Dutch cohort (25)] (B) chips. ARID5B expression was consistently highest in hyperdiploid B-ALL subtypes and lowest in T-ALL than in other subtypes in both cohorts. P values were estimated by using the ANOVA test.
To examine the relation between ARID5B and ALL relapse, we first evaluated its expression in diagnostic leukemia blasts in 59 patients on the Children’s Oncology Group CCG1961 trial who had different treatment outcomes. The ARID5B transcript level in ALL cells was significantly lower in patients who experienced relapse than in those who remained in complete remission (P = 0.01), and this difference was significant even after adjusting for ALL molecular subtype, age at diagnosis, and sex (P = 0.04) (Figure 2A). Furthermore, in a cohort of 49 patients who experienced ALL relapse, we observed significant downregulation of ARID5B transcription at disease recurrence compared to the levels in matched ALL blasts at diagnosis (P = 0.0009) (Figure 2B). Taken together, these results show that ARID5B expression varied by ALL subtypes and was associated with relapse.
Figure 2. Low ARID5B expression is related to ALL relapse.
(A) ARID5B expression was significantly lower (P = 0.01 by logistic regression test) in diagnostic leukemia cells of patients who experienced relapse than in cells from relapse-free patients enrolled on the CCG1961 protocol (26). (B) ARID5B expression levels were significantly decreased (P = 0.0009 by paired t-test) in leukemia cells at relapse compared with matched diagnostic leukemia cells from the same individual.
ARID5B influences ALL sensitivity to antimetabolite drugs
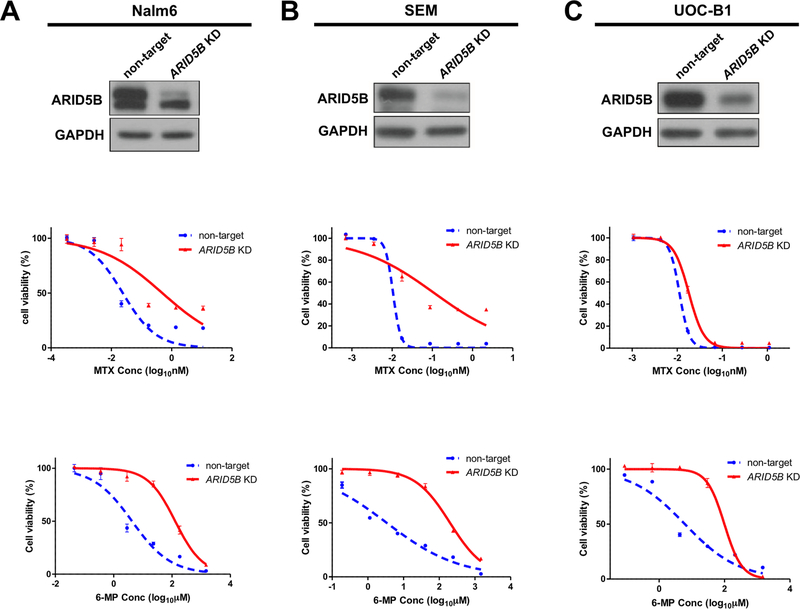
Given the observed downregulation of ARID5B expression at relapse, we hypothesized that loss of ARID5B may render ALL cells resistant to chemotherapeutic agents. To determine the effects of ARID5B on sensitivity to specific antileukemic drugs, we first established stable knockdown of this gene in a panel of human ALL cell lines, with which we tested the dose-dependent cytotoxicity of antimetabolite drugs (6-MP and MTX), glucocorticoids (prednisone, dexamethasone), daunorubicin, vincristine, and asparaginase (Figure 3 and Supplementary Table 1). In Nalm6 cells (DUX4-rearranged with ERG alternation), a 60% downregulation of ARID5B expression led to 19.4- and 30.2-fold increases in the LC50 of MTX and 6-MP, respectively (Figure 3A), with only modest effects on the sensitivity to other chemotherapeutic agents (Supplementary Table 1). In a series of Nalm6 clones with varying degrees of ARID5B knockdown, we observed a gradual increase in drug resistance as ARID5B expression decreased, with an inverse correlation between ARID5B expression and the LC50s of MTX and 6-MP (r2 = 0.5 and 0.64, respectively) (Supplementary Figure 2). Furthermore, the resistance of ARID5B-knockdown cells to MTX and 6-MP was reversed by re-expressing ARID5B (Supplementary Figure 3), indicating that the effects on drug sensitivity were specifically driven by ARID5B. Both MTX and 6-MP need to be activated intracellularly to exert cytotoxic effects. Therefore, we also examined the effects of ARID5B on drug metabolism. Upon ARID5B knockdown, the levels of MTX and 6-MP active metabolites (polyglutamated MTX and thioguanine nucleotides, respectively) were reduced significantly (Supplementary Figure 4), consistent with ALL cell resistance to these two drugs. Similar patterns of drug sensitivity and resistance were observed when ARID5B was knocked down in two other ALL cell lines (Figure 3B and C), namely SEM (MLL-rearranged) and UOC-B1 (TCF3-HLF fusion) cells. These results suggest that ARID5B selectively regulates antimetabolite drug sensitivity in ALL, with lower expression of this gene directly linked to resistance to MTX and 6-MP. The selective effects of ARID5B on 6-MP and MTX sensitivity are highly relevant because prolonged exposure to these antimetabolite drugs is indispensable to the cure of ALL, and 6-MP resistance in particular is a major cause of ALL relapse (41,42).
Figure 3. ARID5B and antimetabolite drug sensitivity in ALL Cells.
Stable knockdown of ARID5B was established using shRNA in Nalm6, SEM, and UOC-B1 cells, and its expression level was determined by Western blot analysis (top panel). GAPDH was used as a loading control. Drug sensitivity was determined using the MTT assay for MTX (middle panel) and 6-MP (bottom panel). Each experiment was performed at least three times in triplicate.
To further validate these findings, we also performed ARID5B knockdown in Nalm6 cells using the CRISPR-dCas9 system (33), in which gene transcription was inhibited by sgRNA-mediated targeting of suppressor protein KRAB to the ARID5B promoter. Again, resistance to antimetabolite drugs was observed in ALL cells with ARID5B downregulation (Supplementary Figure 5).
ARID5B regulates the cell cycle and p21 signaling
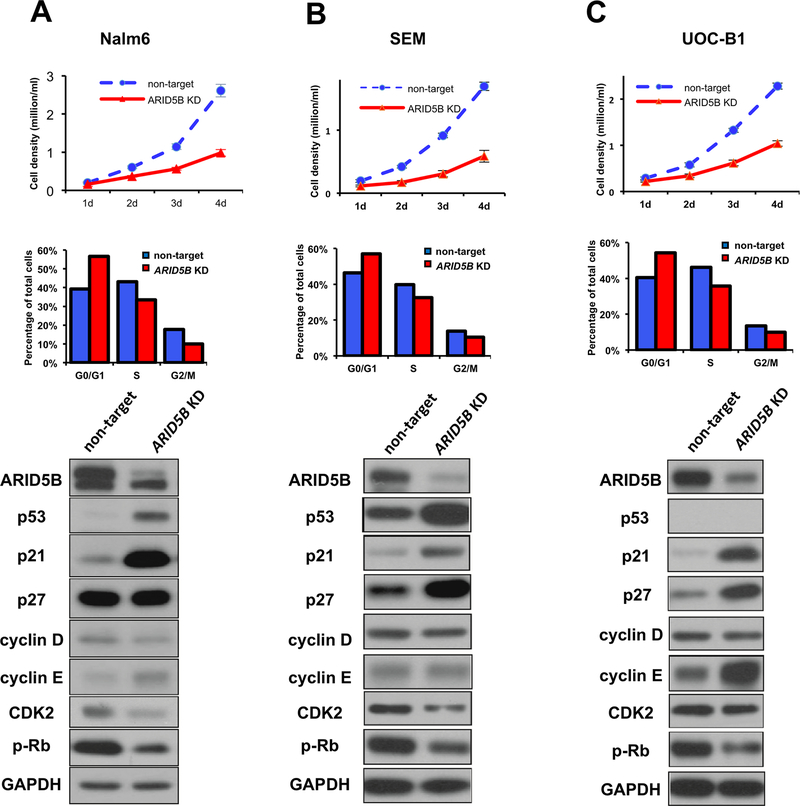
Because the cytotoxic effects of MTX and 6-MP are highly dependent on cell proliferation, we postulated that ARID5B expression affected cell-cycle. Across three ALL cell lines, cell growth was significantly impeded upon ARID5B knockdown (Figure 4, top panel). The decrease of cell growth was also confirmed by the BrdU-uptake assay (Supplementary Figure 6). When ARID5B expression was repressed, we observed partial but consistent blockade of cell cycling, with a significant increase in cells in the G0/G1 phase and a concurrent reduction of the S and G2/M population (Figure 4, middle panel). At the molecular level, the cellular machinery involved in cell-cycle checkpoint control was affected in ARID5B-knockdown cells, particularly those related to G1/S transition (Figure 4, bottom panel). For example in Nalm6 cells, ARID5B knockdown led to a dramatic upregulation of p21 and had a modest effect on p27; both of these proteins are master regulators of cyclin/CDKs during G1/S transition. Cyclin D and E levels were only modestly affected, but a reduction in CDK2 and phosphorylated Rb was readily detectable, consistent with the partial blockade of S-phase entry. The profound change in p21 and phosphorylated Rb levels following ARID5B knockdown was also confirmed in SEM and UOC-B1 cells.
Figure 4. ARID5B as a regulator of cell-cycle progression.
The proliferation of ALL cells was monitored by counting viable cells daily (top panel), and the cell-cycle distribution was determined by propidium iodide staining and flow cytometry (middle panel). Compared to non-target control cells, ARID5B knockdown cells had higher percentages of cells in the G0/G1 phases but lower percentages in the S and G2/M phases. At the molecular level, ARID5B knockdown led to consistent upregulation of p53 and p21 and downregulation of phosphorylated Rb in ALL cell lines (bottom panel). Experiments were performed in Nalm6, SEM, and UOC-B1 cells (A, B, and C, respectively).
To test the hypothesis that p21 was a direct transcription target of ARID5B, we established cell line models with inducible ARID5B knockdown. Upon the addition of doxycycline, the ARID5B level started to decline within 7 h, at which time p21 expression increased slightly. By 24 h, ARID5B was knocked down by 58% and p21 was upregulated 2.4 fold, as compared to the levels at time zero. Between 24 and 48 h, the level of p21 continued to increase and that of ARID5B remained low (Figure 5A). When doxycycline was removed from the culture medium, ARID5B expression recovered within 24 h and, consequently, the p21 level largely returned to that seen in the parental cells (Figure 5B). Subcellularly, ARID5B was restricted to the nuclei of ALL cells, but its downregulation led to an increase in both nuclear and cytoplasmic p21 (Figure 5C). When ALL cells were treated with increasing concentrations of MTX or 6-MP, the p21 level decreased gradually and became undetectable once the cells had entered apoptosis (as indicated by the cleavage of PARP) (Supplementary Figure 7). Therefore, p21 upregulation (as seen in ARID5B-knockdown cells) might directly antagonized MTX/6-MP–induced apoptosis in ALL.
Figure 5. ARID5B is a direct transcriptional regulator of p21.
To identify whether p21 was a transcription regulation target of ARID5B, we established doxycycline-dependent inducible knockdown of ARID5B in Nalm6 cells. The levels of ARID5B and p21 at different time points after doxycycline treatment were determined by Western blot analysis. The ARID5B level decreased steadily as a function of time, with a concomitant increase in p21 (A). The removal of doxycycline led to the recovery of ARID5B expression and a consequently decrease in p21 (B). Both nuclear and cytoplasmic p21 were affected by ARID5B knockdown (C). LaminB and β-actin were used as loading controls for nuclear and cytoplasmic proteins, respectively. Direct binding of ARID5B to a potential regulatory element 25 kb upstream of CDKN1A was confirmed by ARID5B ChIP-qPCR in Nalm6 cells. The ChIP assays were performed three times. The results are shown as the percentage of the input (D). To directly examine the effects of ARID5B binding, Nalm6 cells were transduced with CRISPR/dCas9-KRAB (CRISPRi) with single-guide RNA targeting ARID5B binding site. CDKN1A expression was subsequently determined by RT-PCR (E).
Querying the CDKN1A locus for an ARID motif (43), we identified a potential ARID5B binding site 25kb distal to the transcription start site of CDKN1A. Interestingly, this locus overlaps with an open chromatin segment in a number of hematopoietic cells (lymphoid-primed multipotential progenitors, granulocyte-monocyte progenitors, monocytes, and B cells) on the basis of ATAC-seq signal (44) and histone modification marks (H3K27Ac, H3K4me1, and H3K4me3) (Supplementary Figure 8). Using ARID5B ChIP-qPCR, we experimentally verified ARID5B binding to this putative cis-regulatory element (Figure 5D). More importantly, targeting dCas9-KRAB to interrupt ARID5B binding at this site, using the CRISPRi method (33), resulted in a 1.58-fold change in CDKN1A expression (Figure 5E). Collectively, our results suggest that ARID5B is a putative transcription suppressor of CDKN1A.
Discussion
ARID5B was first linked to ALL in a series of reports in which germline intronic variants of the ARID5B gene were identified as modifying the risk of developing leukemia (13,15). Despite repeated validation in independent studies across diverse populations (8,9,11,12,14,16–18,45–47), the exact effects of these genetic polymorphisms on ARID5B activity and more importantly the biological processes by which this protein regulates normal and malignant hematopoiesis remain largely unknown. Our current study, therefore, represents one of the first attempts to characterize ARID5B signaling in ALL. ARID5B appeared to be abundantly expressed in ALLs, with the exception of T-ALL. Its particularly elevated transcription in hyperdiploid ALL is consistent with the observation that children with ARID5B risk alleles were most likely to develop this specific subtype of ALL.
When we downregulated ARID5B in a panel of ALL cells of diverse subtypes, there was a consistent decrease in proliferation, partly due to cell-cycle blockade at the G1/S checkpoint. This is relevant because multiple therapeutic agents for ALL (e.g., MTX and 6-MP) target DNA synthesis and nucleotide metabolism with cell cycle–dependent cytotoxic effects. For example, reduced cell cycling would decrease the incorporation of 6-MP metabolites into DNA, leading to lower levels of DNA damage and subsequent apoptosis (48). Indeed, it has long been postulated that the remarkable response of ALL to MTX/6-MP–based therapy is at least partly due to the rapid cycling of ALL leukemia cells. Because prolonged exposure to these chemotherapeutic agents is essential for long-term remission in children with ALL, genetic factors modulating their sensitivity are likely to influence ALL relapse, as seen in the case of ARID5B. Interestingly, low expression of ARID5B was associated with high rates of relapse, and ARID5B expression was also downregulated at relapse, suggesting that ARID5B suppression may confer inherent drug resistance at ALL diagnosis and may also be responsible for acquired drug resistance at ALL relapse. It should be noted, however, that the exact degree to which ARID5B contributes to MTX and 6-MP resistance in ALL remains unclear, and it is plausible that this gene represents a component of a larger network or pathway of genes governing ALL sensitivity to the antimetabolite drugs.
Our results also shed important light on possible functions of ARID5B in hematopoietic tissue, particularly the effects of this gene on cell-cycle regulation. Although the expression of several cell-cycle and checkpoint genes changed significantly upon ARID5B knockdown, the most consistent alteration across ALL cell lines of different subtypes was the upregulation of p21. As a master regulator of both G1/S and G2/M checkpoints, p21 expression is itself tightly regulated via p53-dependent mechanisms and also via pathways that do not involve p53 (49–51). Upregulation of p21 occurs rapidly upon exposure to chemotherapeutics as a means of delaying the initiation of apoptosis, and it may be involved in the decision between cell-cycle arrest and apoptosis (52). Inducible ARID5B knockdown in ALL cell lines led to p21 suppression in a time-dependent fashion within 24 h, suggesting a possible direct link. It is, therefore, reasonable to hypothesize that the downregulation of ARID5B activates p21 expression, which in turn blocks cyclin and CDK signaling and eventually triggers cell-cycle arrest. There is some evidence that the AT-rich interactive (ARID) domain in ARID5B possesses sequence-specific DNA-binding affinity and can interact with and guide epigenetic regulators (e.g., PHF2) (19,21–23,27). Combining motif analysis, histone modification, ATAC-seq data, and ChIP-qPCR, we indeed identified an ARID5B binding site within a putative cis-regulatory element upstream of CDKN1A (Supplemental Figure 8 and Figure 5D), and the disruption of which altered CDKN1A transcription.
It is entirely plausible that p21 is one of many ARID5B target genes and thus is only partially responsible for the effects of ARID5B knockdown on cell-cycle and drug resistance. In fact, our global expression profiling studies in ALL cell lines identified other genes that were differentially expressed upon ARID5B knockdown, with an enrichment of components of the p53 signaling pathway (Supplemental Figure 9). These results provide interesting leads for further mechanistic studies in the future. Leung et al. recently reported that ARID5B is a direct target of TAL1 and functions as an activator of MYC transcription in T-ALL (53). Interestingly, ARID5B knockdown also led to significant delay in cell growth in T-ALL, but this was primarily the result of increased apoptosis, with only modest effects on the cell-cycle distribution.
There is still much to learn about the functions of ARID5B in blood cell development and diseases, especially the functional consequences of disease-related genetic variants in this gene. In addition to ALL, ARID5B variants have also been implicated in genetic susceptibility to autoimmune diseases (e.g., lupus, rheumatoid arthritis, type 2 diabetes, and Graves disease) (54–57). Elucidating ARID5B biology may therefore have significance in improving our understanding of immune regulation in general.
Supplementary Material
Translational Relevance.
The past decades have seen increasing survival in children with ALL, but relapse still occurs in approximately 20% of patients, primarily as a result of de novo or acquired drug resistance. ARID5B has been identified by us and others to be associated with susceptibility to ALL and treatment outcomes, although the biological mechanisms by which ARID5B contributes to normal hematopoiesis and leukemogenesis are largely unknown. We found that downregulation of ARID5B led to cell proliferation inhibition, cell-cycle arrest, and resistance to antimetabolite drugs (mercaptopurine and methotrexate). At the molecular level, CDKN1A is a plausible direct transcription regulation target of ARID5B and might be an important mediator of antimetabolite drug sensitivity in ALL. Taken together, our findings point to a new mechanism of drug resistance in ALL and may aid the development of new therapies for this group of patients.
Acknowledgements
Financial support:
The authors thank the patients and parents who participated in the clinical trials included in this study. The authors thank Keith A. Laycock, PhD, ELS, for scientific editing of the manuscript. This work was supported by the National Institutes of Health (GM118578, CA021765 and GM115279), the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, and the Specialized Center of Research of Leukemia and Lymphoma Society (7010-14).
Footnotes
Disclosures of Conflicts of Interest
The authors declare no competing financial interests.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet 2008;371(9617):1030–43 doi 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Infection Greaves M., immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer 2006;6(3):193–203 doi 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Evans WE. Drug therapy - Treatment of acute lymphoblastic leukemia. New Engl J Med 2006;354(2):166–78 doi DOI 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 4.Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, et al. Improved Early Event-Free Survival With Imatinib in Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia: A Children’s Oncology Group Study. J Clin Oncol 2009;27(31):5175–81 doi 10.1200/Jco.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowitz MJ, Wood BL, Devidas M, Loh ML, Raetz EA, Salzer WL, et al. Prognostic significance of minimal residual disease in high risk B-ALL: a report from Children’s Oncology Group study AALL0232. Blood 2015;126(8):964–71 doi 10.1182/blood-2015-03-633685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowitz MJ, Devidas M, Hunger SP, Bowman WP, Carroll AJ, Carroll WL, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood 2008;111(12):5477–85 doi 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children’s Oncology Group study. Leukemia 2008;22(12):2142–50 doi 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhandari P, Ahmad F, Mandava S, Das BR. Association of Genetic Variants in ARID5B, IKZF1 and CEBPE with Risk of Childhood de novo B-Lineage Acute Lymphoblastic Leukemia in India. Asian Pacific journal of cancer prevention : APJCP 2016;17(8):3989–95. [PubMed] [Google Scholar]
- 9.Chokkalingam AP, Hsu LI, Metayer C, Hansen HM, Month SR, Barcellos LF, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Cause Control 2013;24(10):1789–95 doi 10.1007/s10552-013-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharbi H, Ben Hassine I, Soltani I, Safra I, Ouerhani S, Othmen HBH, et al. Association of genetic variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the risk of acute lymphoblastic leukemia in Tunisian children and their contribution to racial differences in leukemia incidence. Pediatric hematology and oncology 2016;33(3):157–67 doi 10.3109/08880018.2016.1161685. [DOI] [PubMed] [Google Scholar]
- 11.Hsu LI, Chokkalingam AP, Briggs FB, Walsh K, Crouse V, Fu C, et al. Association of genetic variation in IKZF1, ARID5B, and CEBPE and surrogates for early-life infections with the risk of acute lymphoblastic leukemia in Hispanic children. Cancer causes & control : CCC 2015;26(4):609–19 doi 10.1007/s10552-015-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood 2013;122(19):3298–307 doi 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nature genetics 2009;41(9):1006–10 doi 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood 2010;115(9):1765–7 doi 10.1182/blood-2009-09-241513. [DOI] [PubMed] [Google Scholar]
- 15.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nature genetics 2009;41(9):1001–5 doi 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Cheng C, Devidas M, Pei DQ, Fan YP, Yang WJ, et al. ARID5B Genetic Polymorphisms Contribute to Racial Disparities in the Incidence and Treatment Outcome of Childhood Acute Lymphoblastic Leukemia. J Clin Oncol 2012;30(7):751–7 doi 10.1200/Jco.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Yang WJ, Perez-Andreu V, Devidas M, Fan YP, Cheng C, et al. Novel Susceptibility Variants at 10p12.31–12.2 for Childhood Acute Lymphoblastic Leukemia in Ethnically Diverse Populations. Jnci-J Natl Cancer I 2013;105(10):733–42 doi 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, Trevino LR, Yang JJ, Scheet P, Pui CH, Evans WE, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia 2010;24(4):894–6 doi 10.1038/leu.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahoud MH, Ristevski S, Venter DJ, Jermiin LS, Bertoncello I, Zavarsek S, et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome research 2001;11(8):1327–34 doi 10.1101/gr.168801. [DOI] [PubMed] [Google Scholar]
- 20.Lin C, Song W, Bi X, Zhao J, Huang Z, Li Z, et al. Recent advances in the ARID family: focusing on roles in human cancer. OncoTargets and therapy 2014;7:315–24 doi 10.2147/OTT.S57023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic acids research 2005;33(1):66–80 doi 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitson RH, Huang T, Itakura K. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochemical and biophysical research communications 1999;258(2):326–31 doi DOI 10.1006/bbrc.1999.0643. [DOI] [PubMed] [Google Scholar]
- 23.Wilsker D, Patsialou A, Dallas PB, Moran E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research 2002;13(3):95–106. [PubMed] [Google Scholar]
- 24.Guan B, Wang TL, Shih IM. ARID1A, a Factor That Promotes Formation of SWI/SNF-Mediated Chromatin Remodeling, Is a Tumor Suppressor in Gynecologic Cancers (vol 71, pg 6718, 2011). Cancer research 2012;72(12):3116– doi 10.1158/0008-5472.CAN-12-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. The Biochemical journal 2004;383(Pt 2):319–25 doi 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bitler BG, Fatkhutdinov N, Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin Ther Targets 2015;19(11):1419–22 doi 10.1517/14728222.2015.1062879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nature cell biology 2011;13(6):668–75 doi 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JH, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481(7380):157–63 doi 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JGCAM, Peters STCJM, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 2009;10(2):125–34 doi 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhojwani D, Kang H, Menezes RX, Yang W, Sather H, Moskowitz NP, et al. Gene expression signatures predictive of early response and outcome in high-risk childhood acute lymphoblastic leukemia: A Children’s Oncology Group Study [corrected]. J Clin Oncol 2008;26(27):4376–84 doi 10.1200/JCO.2007.14.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood 2011;118(19):5218–26 doi 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Zhang H, Yang WJ, Yadav R, Morrison AC, Qian MX, et al. Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nature communications 2015;6 doi UNSP 7553 10.1038/ncomms8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert LA, Larson MH, Morsut L, Liu ZR, Brar GA, Torres SE, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013;154(2):442–51 doi 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans WE, Relling MV, Rodman JH, Crom WR, Boyett JM, Pui CH. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. New Engl J Med 1998;338(8):499–505 doi Doi 10.1056/Nejm199802193380803. [DOI] [PubMed] [Google Scholar]
- 35.French D, Yang W, Cheng C, Raimondi SC, Mullighan CG, Downing JR, et al. Acquired variation outweighs inherited variation in whole genome analysis of methotrexate polyglutamate accumulation in leukemia. Blood 2009;113(19):4512–20 doi 10.1182/blood-2008-07-172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dervieux T, Chu Y, Su Y, Pui CH, Evans WE, Relling MV. HPLC determination of thiopurine nucleosides and nucleotides in vivo in lymphoblasts following mercaptopurine therapy. Clinical chemistry 2002;48(1):61–8. [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Team RC. R: A language and environment for statistical computing. 2013.
- 39.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31–12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. Journal of the National Cancer Institute 2013;105(10):733–42 doi 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Andreu V, Roberts KG, Xu H, Smith C, Zhang H, Yang W, et al. A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood 2015;125(4):680–6 doi 10.1182/blood-2014-09-595744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li BS, Li H, Bai Y, Kirschner-Schwabe R, Yang JJ, Chen Y, et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nature medicine 2015;21(6):563–71 doi 10.1038/nm.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer JA, Wang JH, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nature genetics 2013;45(3):290–4 doi 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 1998;14(1):48–54 doi DOI 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 44.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature genetics 2016;48(10):1193–203 doi 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross JA, Linabery AM, Blommer CN, Langer EK, Spector LG, Hilden JM, et al. Genetic variants modify susceptibility to leukemia in infants: a Children’s Oncology Group report. Pediatric blood & cancer 2013;60(1):31–4 doi 10.1002/pbc.24131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vijayakrishnan J, Kumar R, Henrion MY, Moorman AV, Rachakonda PS, Hosen I, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia 2017;31(3):573–9 doi 10.1038/leu.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Healy J, Richer C, Bourgey M, Kritikou EA, Sinnett D. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica 2010;95(9):1608–11 doi 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panetta JC, Evans WW, Cheok MH. Mechanistic mathematical modelling of mercaptopurine effects on cell cycle of human acute lymphoblastic leukaemia cells. Brit J Cancer 2006;94(1):93–100 doi 10.1038/sj.bjc.6602893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aliouat-Denis CM, Dendouga N, Van den Wyngaert I, Goehlmann H, Steller U, van de Weyer I, et al. p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol Cancer Res 2005;3(11):627–34 doi 10.1158/1541-7786.MCR-05-0121. [DOI] [PubMed] [Google Scholar]
- 50.Kim WH, Kang KH, Kim MY, Choi KH. Induction of p53-independent p21 during ceramide-induced G1 arrest in human hepatocarcinoma cells. Biochem Cell Biol 2000;78(2):127–35. [PubMed] [Google Scholar]
- 51.Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes & development 1995;9(8):935–44. [DOI] [PubMed] [Google Scholar]
- 52.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71 doi 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Leong WZ, Tan SH, Ngoc PCT, Amanda S, Yam AWY, Liau WS, et al. ARID5B as a critical downstream target of the TAL1 complex that activates the oncogenic transcriptional program and promotes T-cell leukemogenesis. Genes & development 2017;31(23–24):2343–60 doi 10.1101/gad.302646.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G, Watanabe M, Imai Y, Hara K, Manabe I, Maemura K, et al. Associations of variations in the MRF2/ARID5B gene with susceptibility to type 2 diabetes in the Japanese population. Journal of human genetics 2012;57(11):727–33 doi 10.1038/jhg.2012.101. [DOI] [PubMed] [Google Scholar]
- 55.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nature genetics 2012;44(5):511–6 doi 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 56.Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. American journal of human genetics 2013;92(1):41–51 doi 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomer Y, Hasham A, Davies TF, Stefan M, Concepcion E, Keddache M, et al. Fine mapping of loci linked to autoimmune thyroid disease identifies novel susceptibility genes. J Clin Endocrinol Metab 2013;98(1):E144–52 doi 10.1210/jc.2012-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.