Abstract
Eukaryotic mRNAs are 5′ end capped with a 7-methylguanosine, which is important for processing and translation of mRNAs. Cap methyltransferase 1 (CMTR1) catalyzes 2′-O-ribose methylation of the first transcribed nucleotide (N1 2′-O-Me) to mask mRNAs from innate immune surveillance by retinoic-acid-inducible gene-I (RIG-I). Nevertheless, whether this modification regulates gene expression for neuronal functions remains unexplored. Here, we find that knockdown of CMTR1 impairs dendrite development independent of secretory cytokines and RIG-I signaling. Using transcriptomic analyses, we identify altered gene expression related to dendrite morphogenesis instead of RIG-I-activated interferon signaling, such as decreased calcium/calmodulin-dependent protein kinase 2α (Camk2α). In line with these molecular changes, dendritic complexity in CMTR1-insufficient neurons is rescued by ectopic expression of CaMK2α but not by inactivation of RIG-I signaling. We further generate brain-specific CMTR1-knockout mice to validate these findings in vivo. Our study reveals the indispensable role of CMTR1-catalyzed N1 2′-O-Me in gene regulation for brain development.
Keywords: 2′-O-ribose methylation, CaMK2, cap1 modification, CMTR, dendrite development, innate immunity, RIG-I
Graphical Abstract

Highlights
-
•
Every mRNA molecule in neurons is N1 2′-O methylated by CMTR1
-
•
CMTR1 is essential for neuromorphogenesis and brain development
-
•
CMTR1 deficiency does not activate RIG-I and interferon signaling
-
•
CMTR1 promotes Camk2α expression to support dendrite development
Lee et al. demonstrate that CMTR1-catalyzed 2′-O-ribose methylation in mRNAs is important for dendritic morphogenesis and brain development. CMTR1 is dispensable for silencing RIG-I-activated innate immunity in neurons. Transcriptomic profiling and rescue experiments show Camk2α as the most downregulated gene in the absence of CMTR1, suggesting a role in dendrite development.
Introduction
In all eukaryotes and many viruses, the 5′ end of transcripts is modified with a 7-methylguanosine (m7G), rendering the terminal dinucleotide resistant to ribonuclease digestion (Shuman, 2002). This m7G structure, called cap0 (m7GpppNN, N: any nucleotide), is important for RNA stability, splicing, nucleocytoplasmic transport, and translation initiation (Cheng et al., 2006; Hernández et al., 2010; Ramanathan et al., 2016). Except for yeast mRNAs with the primitive m7G cap, the cap structure in higher eukaryotes has additional methylation at the 2′-O-ribose position of the first and second nucleotides by cap methyltransferase 1 (CMTR1) and CMTR2, respectively, to produce cap1 (m7GpppNmN) and cap2 (m7GpppNmNm) structures (Bélanger et al., 2010; Werner et al., 2011).
Yeast mRNAs are not cap1 modified, so the cap0 structure, which recruits the assembly of the eukaryotic initiation factor (eIF) 4F complex (i.e., eIF4E, 4G, and 4A) by direct binding of eIF4E to the m7G cap, is sufficient for “cap-dependent” translation. An early study indicated that cap1 exists in every mRNA molecule, whereas cap2 is present in ∼50% of mRNA molecules in HeLa cells (Furuichi et al., 1975). Although the cap-methyl-transferring enzyme activity was first detected in HeLa extracts (Langberg and Moss, 1981), CMTR1, the enzyme catalyzing 2′-O-ribose methylation of the first nucleotide in mRNAs (hereafter called N1 2′-O-Me), was not identified until 2010 (Bélanger et al., 2010). Dom3Z/DXO, a mammalian homolog of yeast Rai1 and Dxo1, possesses decapping, pyrophosphohydrolase, and 5′–3′ exoribonuclease activities to degrade GpppG-RNA and m7GpppG (cap0)-RNA from the 5′ end (Jiao et al., 2013), but it could not degrade cap1-oligoribonucleotide in vitro (Picard-Jean et al., 2018). Moreover, a previous study showed that cap1 modification accompanied with cytoplasmic polyadenylation in c-Mos mRNA promotes translation during oocyte maturation (Kuge et al., 1998).
The 5′ cap composition is also a determinant of self- (host) versus non-self RNA during viral infection (Leung and Amarasinghe, 2016). Viruses with inactive 2′-O methyltransferase (MTase), such as West Nile virus, Poxvirus, and Coronavirus mutants, are attenuated in vivo because interferon (IFN)-induced proteins with tetratricopeptide repeat 1 (IFIT1) bind to cap0 viral RNAs better than cap1 viral RNAs and consequently prevent the recruitment of eIF4E for viral protein synthesis (Daffis et al., 2010; Habjan et al., 2013). CMTR1 is also known as IFN-stimulated gene 95 kDa protein (ISG95), and its expression is upregulated by viral infection in various cells (Geiss et al., 2003; Guerra et al., 2003; Su et al., 2002). Moreover, the cap1 structure is suggested to prevent cellular mRNAs from being recognized as non-self molecules by the RNA sensor retinoic-acid-inducible gene-I (RIG-I) (Schuberth-Wagner et al., 2015).
The previous findings suggest that the cap1 moiety may regulate mRNA stability or translation while serving as a molecular signature for self-transcript identification. Nevertheless, none of these findings have been validated in vivo. Here, we investigated CMTR1 function in neurons, in which RNA modifications control molecular diversity to support their complex morphologies and functions (Huang and Lu, 2018; Noack and Calegari, 2018). To avoid any interference from innate immunity, we also studied CMTR1 in RIG-I-knockout (KO) and mitochondrial antiviral signaling protein (MAVS)-KO neurons. We found that impaired dendrite development in CMTR1-knockdown (KD) neurons resulted from downregulated transcription of calcium/calmodulin-dependent protein kinase 2α (Camk2α). Unexpectedly, CMTR1-KD did not trigger innate immune responses in neurons. Moreover, the findings from the KD neurons were recapitulated in CMTR1-KO cortices. Therefore, CMTR1-catalyzed methylation regulates CaMK2α expression for dendritic morphogenesis but is dispensable for masking cellular mRNAs from RIG-I detection.
Results
Nuclear MTase Activity of CMTR1 Is Required for Dendritic Development
The three cap structures showing the methylated positions with corresponding MTases are illustrated in Figure 1 A. In agreement with cap1 as the dominant cap structure in mammalian cells and tissues (Akichika et al., 2019; Furuichi et al., 1975; Wang et al., 2019), CMTR1 is expressed in various tissues (Figure S1A) and localized predominately in the nucleus of neurons and in microtubule-associated protein 2 (MAP2)-negative non-neuronal cells (Figure S1B). Notably, the expression of CMTR1 in neurons remained high during the first 6 days in vitro (DIV6), the critical period for axonal and dendritic outgrowth, and declined thereafter to a steady level (Figure 1B). Thus, we examined whether CMTR1 is involved in neuromorphogenesis by infecting DIV2 neurons with lentivirus expressing control short hairpin RNA (shRNA) (siCTL) or rat CMTR1-targeted shRNA (siCM1#1 or siCM1#2) along with mCherry red fluorescent protein (RFP) to mark infected cells. The neurons at DIV7 were harvested for immunoblotting to confirm the diminished CMTR1 level (Figure S2A) or fixed for MAP2 immunostaining. Because CMTR1-KD did not affect MAP2 expression (Figure S2B), we used an MAP2-immunostained signal to outline dendritic processes (Figure S2C). RFP-positive (i.e., lentivirus-infected) and MAP2-positive pyramidal neurons were selected for Sholl analysis (Sholl, 1953). Both siCM1#1 and siCM1#2 neurons exhibited defective dendritic arborization, as indicated by the decreased number of dendritic intersections (Figure S2D), thereby leading to reduced total dendritic number and length (Figure S2E).
Figure 1.
Nuclear CMTR1 Is Required for Dendritic Development
(A) The chemical structure of capped RNAs. RNMT (mRNA cap guanine-N7 methyltransferase), CMTR1, and CMTR2, which transfer the methyl group from S-adenosylmethionine (SAM) to the corresponding positions, are indicated. SAH, S-adenosylhomocysteine.
(B) Developmental expression of CMTR1 in neurons at the denoted days in vitro (DIV). The level of CMTR1 was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed as a relative ratio to DIV2. Data are mean ± SEM from three cultures.
(C–E) CMTR1 domain structure (C). The K239A and ΔNLS mutations render catalytic dead and cytoplasm-localized CMTR1, respectively. DIV2 rat neurons were infected with lentivirus expressing siCTL or siCM1 ± RFP or RFP-tagged human CMTR1 WT or mutants. The infected neurons at DIV7 were used for immunoblotting or immunostaining of RFP and MAP2 (D and E). Scales, 50 μm.
(F) Sholl analysis to count the dendritic intersections in every 10-μm segment away from the soma.
(G) Total dendritic number and length are expressed as mean ± SEM (n = 60 neurons from three cultures). ∗∗∗p < 0.001, two-way analysis of variance (ANOVA). See also Figures S1 and S2.
The K239-D-364-K404 triad in the catalytic domain of human CMTR1 (hCMTR1) is important for MTase activity (Smietanski et al., 2014). We used hCMTR1 for the rescue experiment because its nucleotide sequence is resistant to siCM1#1 (hereafter called siCM1)-mediated cleavage and its protein sequence shares ∼93% identity with that of the rat homolog. CMTR1 contains a nuclear localization signal (NLS) at the N terminus, followed by G patch and MTase domains. The C-terminal region contains guanylyltransferase-like and WW domains (Figure 1C). The WW domain is important for the association with RNA polymerase II for co-transcriptional N1 2′-O-Me (Galloway and Cowling, 2019). For simultaneous KD of rat CMTR1 and expression of wild-type (WT) or mutant hCMTR1, the GFP reporter in the pLL3.7-Syn plasmid (siCTL and siCM1) was replaced with an RFP or RFP-hCMTR1 construct. We generated K239A and ΔNLS (Δ3–18 amino acid [aa]) mutants, which render catalytic dead MTase and cytoplasm-localized CMTR1, respectively (Figure 1D). DIV2 cortical neurons were infected with the designated lentivirus and harvested at DIV7 for examining the KD of endogenous CMTR1 and the expression of RFP-hCMTR1 WT or mutants (Figure 1C). Similarly, the infected neurons on coverslips were processed for MAP2 immunostaining (Figure 1E) for Sholl analysis. CMTR1-KD reduced dendritic complexity (Figure 1F) and total dendritic number and length (Figure 1G), which could be rescued by ectopic expression of WT but not K239A or ΔNLS mutant. Therefore, CMTR1-mediated 2′-O-Me in the nucleus is critical to support dendritic outgrowth.
Defective Dendritic Ramification in CMTR1-KD Neurons Is Independent of Secretory Factors and RIG-I-Mediated Signaling
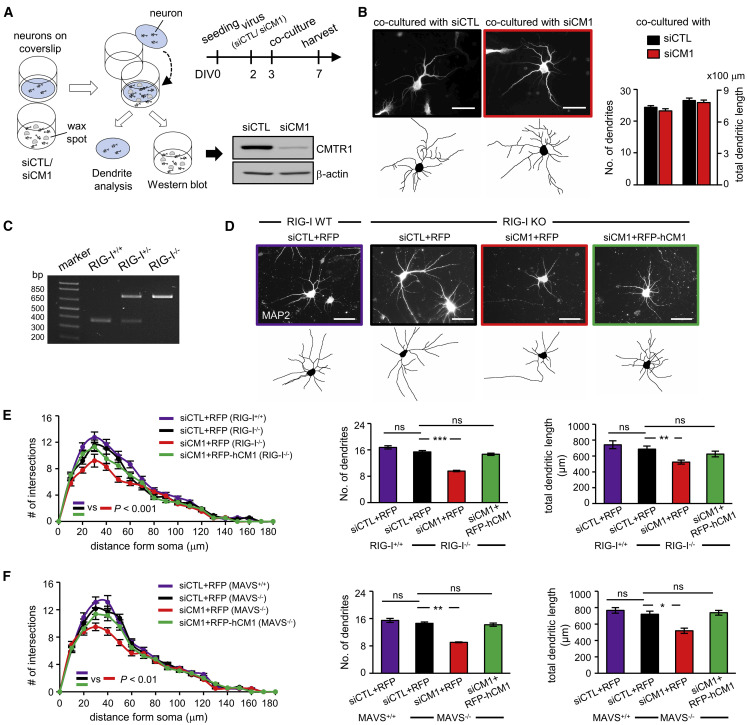
Abnormal dendritic arborization could be due to secretory factors such as a lack of brain-derived neurotrophic factor (Moya-Alvarado et al., 2018) or presence of inflammatory cytokines (O’Neill et al., 2016). KD of CMTR1 in A549 cells induces type I IFN signaling by RIG-I activation (Schuberth-Wagner et al., 2015), so abnormal dendritic development could be caused by inflammatory cytokines secreted from CMTR1-KD neurons. However, we detected no evident changes in mRNA levels of many IFN-signaling-related genes (Figure S2F). Moreover, the qRT-PCR signals of some transcripts, such as IFN-β, IFIT1, and tumor necrosis factor α (TNF-α), were too low to confidently claim the activation of innate immunity, so we performed co-culture experiments (Figure 2 A) and found that WT neurons co-cultured with siCM1 neurons exhibited normal dendritic number and length (Figure 2B). Thus, secretory factors by autocrine or paracrine action are not the culprits of defective dendritic outgrowth in CMTR1-KD neurons.
Figure 2.
CMTR1-KD Impairs Dendritic Development Independent of Secretory Factors and RIG-I/MAVS-Activated Signaling
(A) Schematic diagram of the co-culture experiment. Neurons on coverslips were co-cultured with siCTL or siCM1 neurons from DIV3 to DIV7. The neurons on dishes were harvested for western blotting.
(B) The neurons on coverslips were fixed for MAP2 immunostaining and analyzed for the number and total length of dendrites.
(C) PCR genotyping of E17.5 embryos from RIG-I heterozygous mating.
(D) RIG-I-KO neurons infected with the designated lentivirus were immunostained for MAP2. MAVS-KO neurons were processed similarly.
(E and F) The dendritic intersections, number, and length were determined. (B, E, and F) Data are mean ± SEM (n = 60 neurons from three cultures). ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, two-way ANOVA. Scales, 50 μm. See also Figure S2F.
RIG-I and melanoma-differentiation-associated protein 5 (MDA5) are pattern recognition receptors that sense viral RNAs with loosely defined features. Once activated by RNA molecules, both RIG-I and MDA5 convey their signaling through MAVS to induce type I IFN synthesis and innate immune responses (Reikine et al., 2014). Because RIG-I-mediated IFN responses exist in virus-infected neurons (Nazmi et al., 2011), it was unexpected that secretory factors were not involved to impair dendritic ramification. We wondered whether RIG-I, presumably activated by elevated cap1-deficient mRNAs in the CMTR1-KD condition (Schuberth-Wagner et al., 2015), may affect dendritic growth independent of IFN secretion. If so, such morphological defects should be rescued in RIG-I-KO or MAVS-KO neurons. WT and KO embryos from RIG-I heterozygous matings were used for neuronal cultures (Figure 2C). KD of CMTR1 in RIG-I-KO neurons still impaired dendritic complexity, which could be rescued by RFP-hCMTR1 expression (Figures 2D and 2E). Similar results were also found in MAVS-KO neurons (Figure 2F), so RIG-I/MAVS-mediated signaling does not contribute to dendritic maldevelopment in CMTR1-KD neurons.
CMTR1 Deficiency Alters Camk2α mRNA Expression to Affect Dendritic Morphogenesis
We used microarray and Gene Ontology (GO) analyses of genes with a corresponding mRNA level showing more than ±1.2-fold change in siCM1 neurons relative to siCTL neurons (Table S1). The GO: 0030425 dendrite category scored the highest among different biological processes (Figure 3 A). Among 48 genes in this cluster (Table S1), the top 5 to 6 genes showing the most up- or downregulation (Figure 3B) were validated by qRT-PCR (Figure 3C). Only Camk2α and Pafah1b1 levels showed a consistent reduction. We focused on Camk2α because it is the most downregulated gene and Camk2α-haploinsufficient mice also show reduced dendritic branches and length (Yamasaki et al., 2008). Indeed, the CaMK2α protein level, parallel to the change in the mRNA level, was decreased ∼2-fold in CMTR1-KD neurons and could be rescued by ectopic expression of CMTR1 WT but not the K239A or ΔNLS mutant (Figure 3D). Camk2α mRNA stability remained unchanged (Figure S3A), but the amount of 4-thiouridine (4sU)-labeled nascent Camk2α mRNA was diminished in CMTR1-KD neurons (Figures S3B and S3C). Moreover, reduced dendritic complexity in CMTR1-KD neurons could be rescued by RFP-CaMK2α expression (Figures 3E–3G), so decreased CaMK2α expression significantly accounted for dendritic maldevelopment in CMTR1-KD neurons.
Figure 3.
Knockdown of CMTR1 Decreases CaMK2α Expression to Affect Dendrite Development
(A) Gene Ontology (GO) analysis of enrichment clusters from duplicate microarrays. The statistical significance of each cluster is expressed as −log10 P.
(B) The list of genes from the dendrite GO cluster with most transcriptomic changes in siCM1 neurons. Ltbp1, latent transforming growth factor β binding protein 1; Cdkn1a, cyclin-dependent kinase inhibitor 1; Pdyn, prodynorphin; Sort1, sortilin1; Atp7a, copper-transporting P-type ATPase; Camk2α, calcium/calmodulin-dependent protein kinase 2α; Ptk2b, protein tyrosine kinase 2b; Twf1, twinfilin-1; Pafah1b1, platelet-activating factor acetylhydrolase 1b subunit 1; Prkcg, protein kinase C gamma type; Kif5a, kinesin family member 5a.
(C) The normalized RNA levels relative to Gapdh were determined by qRT-PCR.
(D) The CaMK2α level relative to GAPDH in siCTL and siCM1 neurons ± ectopic expression of denoted mutant.
(E) DIV2 neurons infected with the denoted lentivirus were immunostained with MAP2. Scales, 50 μm.
(F) Sholl analysis.
(G) Total dendritic number and length per neuron (n = 60). Data are mean ± SEM collected from ≥3 independent experiments. ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Student’s t test in (C) and (D) and two-way ANOVA in (F) and (G). See also Figure S3.
CMTR1-cKOEmx1 Mice Show Reduced Cortical Size and Abnormal Dendritic Morphology in the Absence of Innate Immunity
To assess the physiological functions of CMTR1, we generated mice carrying a targeted KO-first Cmtr1 allele (zCmtr1) and then produced mice carrying the floxed Cmtr1 allele (fCmtr1). In the zCmtr1 allele, the splicing of exon 8 to LacZ reporter resulted in a truncated transcript without a functional catalytic domain (zKO; Figure S4A). Crossing of Cmtr1 z/+ mice produced no live CMTR1-zKO (Cmtr1 z/z) mice (Figure S4B), as Cmtr1 z/z embryos at the embryonic day 8.5 (E8.5) gastrulation stage were mostly absorbed (Figure S4C). Thus, we crossed fCmtr1 mice with Emx1-Cre or Nestin-Cre mice to generate cKOEmx1 and cKONes mice (Figure 4 A). The expression of Cre in Emx1-Cre mice begins at E9.5 and is limited to the EMX1-expressing progenitors (Gorski et al., 2002). The expression of Cre in Nestin-Cre mice begins at E10.5 in pan neuron progenitors (Tronche et al., 1999), so Cmtr1 in cKONes cortices was ablated in all neurons and glia. We used cKOEmx1 cortices for anatomical and morphological analyses and cKONes cortices for molecular studies and culture. CMTR1-cKOEmx1 mice showed reduced cortical and hippocampal areas (Figure 4B). The poly(A) RNAs isolated from cKONes cortices did not contain N1 2′-O-Me (Figure 4C), and CaMK2α was also reduced in cKONes cortices (Figure 4D). Although the amounts of 4sU-incorporated transcripts in conditional wild-type (cWT) and cKONes neurons were comparable, the level of nascent Camk2α mRNA was significantly reduced in cKONes neurons (Figure 4E). In addition, DXO-KD could not restore the expression of Camk2α mRNA (Figure S5A) and protein (Figure S5B), so DXO does not degrade cap1-free Camk2α mRNA. Together with the findings in CMTR1-KD neurons (Figure S3), these results show that CMTR1 deficiency impairs Camk2α transcription rather than stability.
Figure 4.
Defective Dendritic Arborization of Cortical Pyramidal Neurons in CMTR1-cKOEmx1 Mice without Evident Inflammation
(A) The fCmtr1 mice were used to generate cKOEmx1 or cKONes mice.
(B) The dorsal view of postnatal day 22 brains with medial sagittal sections stained with hematoxylin and eosin. The outlined cortical areas are denoted.
(C) The poly(A) RNAs isolated from cortices were used to detect N1 2′-O-Me (pAm and pCm, circled by dotted line) by thin-layer chromatography.
(D) CaMK2α level in P7 cortices (n = 3 mice) were determined as mean ± SEM.
(E) 4-Thiouridine (4sU)-labeled transcripts were biotinylated, examined by dot blotting, and isolated by streptavidin beads for qRT-PCR. The nascent mRNA level of Camk2α relative to that of β-actin is expressed as mean ± SEM (n = 3 experiments).
(F) Representative z stack images of YFP-expressing cortical layer 5 neurons. Scales, 100 μm.
(G) Total dendritic intersections, number, and length from both apical and basal dendrites are mean ± SEM from 20 cWT neurons and 16 cKO neurons (three mice per group).
(H) qRT-PCR. The levels of denoted mRNAs relative to Gapdh mRNA in cortices are mean ± SEM (four mice per group). Total RNAs isolated from enterovirus-71-infected spinal cord (SC) and Japanese-encephalitis-infected cortex (brain) were positive controls of innate immunity. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Student’s t test in (D), (E), and (H) and two-way ANOVA in (G). See also Figures S4–S6.
To address whether CMTR1 affects dendritic maturation in vivo, we used Thy1-YFP-H transgenic mice, which express yellow fluorescent protein (YFP) in the subsets of cortical (layer 5) and hippocampal (CA1) pyramidal neurons (Feng et al., 2000), to produce cWT:: or cKOEmx1::Thy1-YFP mice. Immunohistochemistry confirmed the absence of CMTR1 in cortical and hippocampal pyramidal neurons (Figure S6A), so we acquired images of YFP-expressing cortical layer 5 neurons (Figure 4F) and found impaired dendritic morphology in cKOEmx1 mice (Figure 4G). Moreover, no inflammation was found in the cKOEmx1 brain because the number and morphology of microglia (Iba1-positive) appeared at the resting state (Figure S6B) and the mRNA levels of IFN-β1, TNF-α, IFIT1, and RIG-I were not upregulated in cKOEmx1 cortices (Figure 4H). In contrast, these transcripts were highly elevated in RNA-virus-infected spinal cords and brains (Figure 4H). Thus, the loss of CMTR1 did not trigger inflammatory responses to affect dendrite morphogenesis in vivo.
Discussion
This study demonstrates that CMTR1 insufficiency affects Camk2α expression to impair dendritic arborization and cortical development. Ectopic expression of CaMK2α but not depletion of RIG-I or MAVS rescued dendritic maldevelopment caused by CMTR1 deficiency, which supports that CMTR1-catalyzed N1 2′-O-Me regulates gene expression to control neuronal development.
The conserved histidine 830 in the RIG-I RNA binding pocket is important for its steric exclusion binding to cap1-methylated RNA (Devarkar et al., 2016), so cap1-deficient cellular transcripts in CMTR1-KD cells were recognized by RIG-I to activate IFN signaling (Schuberth-Wagner et al., 2015). Almost all mammalian mRNA molecules carry the cap1 structure (Akichika et al., 2019; Furuichi et al., 1975), so cap1 as a molecular signature for discriminating self and foreign transcripts seems to make perfect sense. Surprisingly, CMTR1 deficiency in neurons did not induce type-I-IFN-signaling-related gene expression (Figures 3A, 4H, and S2F) or release cytokines to affect co-cultured neurons (Figure 2A) nor resulted in any inflammatory signs in the KO brain (Figure S6B). Moreover, neurons do not appear to express other MTases to catalyze N1 2′-O-Me in the absence of CMTR1 (Figure 4C) or lack innate immunity because the expression of type I IFN genes is robustly elevated in the Japanese-encephalitis-virus-infected brain (Figure 4H), which is mediated by RIG-I activation (Kato et al., 2006). MDA5 could be another cap1-free RNA sensor (Roth-Cross et al., 2008; Züst et al., 2011). However, KD of CMTR1 in MAVS-KO neurons did not ameliorate dendritic defects (Figure 2F), so neither RIG-I nor MDA5 can detect cap1-free cellular RNA in CMTR1-deficient neurons. We reason that in the life of the mRNA after being transcribed, cap1-free mRNAs are assembled in various messenger ribonucleoproteins to escape from RIG-I surveillance. Alternatively, cap1-free mRNAs may simply not be the substrate for RIG-I. A previous study using the in vitro binding assay showed that RIG-I does not recognize cap0 single-stranded RNA even though its binding affinity for cap0 double-stranded RNA is ∼2 nM (Devarkar et al., 2016). Together with our results, N1 2′-O-Me of cellar mRNAs is not meant for escaping RIG-I surveillance, at least, in uninfected cells. Notably, CMTR1 expression was remarkably elevated in virus-infected neurons (Figure 4H). CMTR1-KO in A549 cells inhibited replication of influenza A virus (Li et al., 2020), which snatches the 5′ end of host mRNAs for priming its own transcription (Bouloy et al., 1980; Wakai et al., 2011). In contrast, CMTR1-KD enhanced replication of Zika and dengue viruses in Huh7 cells due to IFIT-1-mediated translational repression to attenuate IFN responses (Williams et al., 2020). Depending on the type of virus, CMTR1 can have proviral or antiviral effects.
The cap1 structure is implicated in posttranscriptional gene regulation (Kuge et al., 1998; Picard-Jean et al., 2018), but decreased Camk2α mRNA in CMTR1-deficient neurons resulted from reduced transcription instead of a stability change (Figures 4E and S3) or DXO-mediated degradation (Figure S5). Two possible scenarios are that Camk2α transcription is regulated by a yet-to-be-uncovered transcription factor whose posttranscriptional expression depends on cap1 modification in a DXO-independent manner or that Camk2α transcription depends on cap1 methylation because the nuclear cap-binding complex (i.e., CBP80 and CBP20) is known to promote transcription elongation (Lenasi et al., 2011). Nuclear pre-mRNA capping recruits the nuclear cap-binding complex, which then associates with various factors to regulate transcription elongation, pre-mRNA 3′-end processing, splicing, RNA decay, and nuclear export. Because these interactions are conserved from yeast to mammals, the cap0 structure alone is believed to be sufficient to regulate gene expression at these stages (Bentley, 2014; Gonatopoulos-Pournatzis and Cowling, 2014; Jiao et al., 2013; Lenasi et al., 2011; Ramanathan et al., 2016). Because cap1 is the dominant cap structure in mammals (Akichika et al., 2019; Furuichi et al., 1975; Wang et al., 2019), previous studies in mammalian cells could not exclude the contribution of cap1 in these nuclear processes to affect gene expression.
Why the ubiquitous cap1 modification affects selective gene expression is unclear. To our knowledge, only IFIT1 and DXO exhibit preferential recognition for cap0 RNA instead of cap1 RNA in in vitro binding and cleavage assays, respectively (Habjan et al., 2013; Kumar et al., 2014; Picard-Jean et al., 2018), and RIG-I specifically binds to double-stranded cap0 RNA in vitro (Devarkar et al., 2016). However, the expression of IFIT1 in our microarray data is too low in uninfected neurons (accession: GSE145223) to possibly repress translation of cap0 mRNAs. We suspect that most cap-binding proteins, such as eIF4E, with similar binding affinity to cap0 and cap1 in vitro (Haghighat and Sonenberg, 1997; Niedzwiecka et al., 2002), may exhibit a binding preference for one rather than the other cap structure, which likely depends on its associated partners and perhaps the 5′-end sequence of transcripts. What other players act in cohort with the cap1 moiety to regulate Camk2α expression requires further investigation.
CMTR1 binds RNA helicase DHX15 through its G-patch domain. One study showed that such an interaction facilitates cap1 methylation on oligoribonucleotides with structural 5′-termini in vitro (Toczydlowska-Socha et al., 2018), but the other study contradictorily suggested that DHX15 inhibits CMTR1’s MTase activity in vitro (Inesta-Vaquera et al., 2018). Moreover, overexpression of CMTR1 in HCC1806 cells enhanced translation of a small subset of mRNAs involved in proliferation and DNA damage response (Inesta-Vaquera et al., 2018). However, this study did not measure whether CMTR1 overexpression can further increase N1 2′-O-Me in poly(A) RNAs. Interestingly, the CapQuant study detected ∼10% and 3% transcripts carrying the cap0 structure in CCRF-SB cells and mouse liver, respectively (Wang et al., 2019), so the notion that mammalian mRNAs are 100% cap1 modified needs to be closely examined.
In summary, our data indicate that CMTR1-catalyzed N1 2′-O-Me is an important epitranscriptomic signal for gene regulation, dendritic morphogenesis, and brain development.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-α-tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026, RRID:AB_477593 |
| Mouse monoclonal anti-β-Actin (clone AC15) | Sigma-Aldrich | Cat# A5441, RRID:AB_476744 |
| Mouse monoclonal anti-CaMKII alpha (clone 6G9) | Thermo Fisher Scientific | Cat# MA1-048, RRID:AB_325403 |
| Rabbit polyclonal anti-KIAA0082 (CMTR1) | Bethyl | Cat# A300-304A, RRID:AB_309477 |
| Mouse monoclonal anti-Glyceraldehyde-3-PDH (GAPDH) (clone 6C5) | Millipore | Cat# MAB374, RRID:AB_2107445 |
| Mouse monoclonal anti-Glial Fibrillary Acidic Protein (GFAP) (clone GA5) | Millipore | Cat# MAB360, RRID:AB_11212597 |
| Goat polyclonal anti-Histone H3 | Santa Cruz Biotechnology | Cat# sc-8654, RRID:AB_2118303 |
| Mouse monoclonal anti-Iba1 (clone GT10312) | Thermo Fisher Scientific | Cat# MA5-27726, RRID:AB_2735228 |
| Mouse monoclonal anti-MAP2A,2B (clone AP20) | Millipore | Cat# MAB378, RRID:AB_11214935 |
| Chicken polyclonal anti-MAP2 | Novus | Cat# NB300-213, RRID:AB_2138178 |
| Mouse monoclonal anti-NeuN (clone A60) | Millipore | Cat# MAB377, RRID:AB_2298772 |
| Rabbit polyclonal anti- mCherry | Abcam | Cat# ab167453, RRID:AB_2571870 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Thiouridine | Sigma-Aldrich | T4509 |
| Actinomycin D | Sigma-Aldrich | A1410 |
| Cycloheximide | Sigma-Aldrich | 01810 |
| EZ-Link HPDP-Biotin | Thermo Scientific | 21341 |
| Puromycin dihydrochloride | Sigma-Aldrich | P8833 |
| Critical Commercial Assays | ||
| KAPA mouse genotyping kit | KAPA Biosystems | KR0385 |
| ViraPower Lentiviral Expression Systems | Invitrogen | K4950-00 |
| Light Cycler 480 Probes Master | Roche | 04887301001 |
| VECTASTAIN Elite ABC-Peroxidase Kit | Vector Laboratories | Cat# PK-7100, RRID:AB_2336827 |
| PolyATtract mRNA Isolation Systems | Promega | Z5210 |
| Deposited Data | ||
| Microarray data for transcriptomic analyses | This paper | NCBI GEO: GSE145223 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse CMTR1f/f | This paper | N/A |
| Mouse RIG-I+/− | Dr. Shizuo Akira | N/A |
| Mouse MAVS+/− | Dr. Michael Gale | N/A |
| Mouse Thy1-YFP-H | Jackson Laboratory | #003782 |
| Mouse Tg-ActFLPe | Jackson Laboratory | #003800 |
| Mouse Nestin-Cre | Jackson Laboratory | #003771 |
| Mouse Emx1-IRES-Cre | Jackson Laboratory | #005628 |
| Pregnant Rat (E18.5) | Biolasco | N/A |
| Oligonucleotides | ||
| Primers for qPCR, see Table S2 | This paper | N/A |
| Primers for shRNA constructs, see Table S2 | This paper | N/A |
| Primers for genotyping, see Table S2 | This paper | N/A |
| Primers for qPCR, see Table S2 | This paper | N/A |
| mCherry-hCMTR1 cloning primer | This paper | N/A |
| RFP_F, ATGCTGGCTAGCGCCACCATGGTG AGCAAGG |
N/A | |
| RFP-hTR1_F, GGCATGGACGAGCTGTACAAGGGTACCA TGAAGAGGAGAACTGACCCAGAA |
N/A | |
| RFP-hTR1_R, TTCTGGGTCAGTTCTCCTCTTCATGGTAC CCTTGTACAGCTCGTCCATGCC |
N/A | |
| hTR1_R, ATTCAGGTTTAAACTCAGGCCCTGTGCA TCTGGA |
N/A | |
| Recombinant DNA | ||
| Human CMTR1 ORF | GenScript | OHu06786 |
| Software and Algorithms | ||
| Bonfire | Langhammer et al., 2010 | https://lifesci.rutgers.edu/∼firestein |
| MATLAB | MathWorks | https://se.mathworks.com/ |
| Prism (Version 6.0) | GraphPad | https://www.graphpad.com/ |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Metascape (online) | Zhou et al., 2019 | https://metascape.org/ |
| NeuronStudio | Rodriguez et al., 2008 | https://biii.eu/neuronstudio |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yi-Shuian Huang (yishuian@ibms.sinica.edu.tw).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The accession number for the microarray data reported in this paper is NCBI GEO: GSE145223.
Experimental Model and Subject Details
Animals and Genotyping
Experimental procedures were performed in accordance with the guidelines of the Institutional Animal Care and Utilization Committee. Mice were housed under a 12-h light-dark cycle (lights on from 8 a.m. to 8 p.m.) in a temperature- and humidity-controlled room with ad libitum access to food and water. All efforts were made to minimize the number of animals used and their suffering. To produce CMTR1-conditional KO (cKO) mice, littermates from mating Cmtr1 f/f, Emx1-Cre/+ or Cmtr1 f/+, Nestin-Cre/+ females and Cmtr1 f/f, +/+ male mice were used. RIG-I+/− mice were obtained from Dr. Fang Liao with permission from Dr. Shizuo Akira (Osaka University, Japan) (Kato et al., 2005) and MAVS+/− mice were from Dr. Sue-Jane Lin with permission from Dr. Michael Gale (University of Washington, USA) (Loo et al., 2008). Thy1-YFP-H (#003782), Tg-ActFLPe (#003800), Nestin-Cre (#003771) and Emx1-IRES-Cre (#005628) mice were obtained from the Jackson Laboratory. The genotypes were determined by PCR of tail biopsies and the KAPA mouse genotyping kit (KR0385, KAPA Biosystems) following the manufacture’s protocol. The primer sequences are in Table S2.
Generation of Mice Carrying Floxed Cmtr1 (fCmtr1) Allele
For producing chimeric mice, 4-week-old C57BL/6 female mice were super-ovulated with an intraperitoneal injection of gonadotropin from pregnant mare serum (G4877, Sigma-Aldrich), then human chorionic gonadotropin (CG1063, Sigma-Aldrich) 46 h later. Super-ovulated female mice were mated with C57BL/6 male mice to collect blastocysts. Two embryonic stem (ES) cell clones carrying KO-first with conditional potential of Cmtr1 allele were obtained from the European mouse mutant cell repository (EUMMCR) and microinjected into the cavity of blastocysts, which were recovered in M2 medium and then transferred into the uterus of pseudo-pregnant ICR female mice. Only one ES clone-derived founder produced germline-transmitted progenies. The mouse carrying the targeted Cmtr1 allele was first crossed with the Tg-ActFLPe mouse to remove the Frt-LacZ-Neo-Frt cassette and generate the floxed Cmtr1 allele (fCmtr1). The resulting line was maintained as fCmtr1 mice and then crossed with Nestin-Cre or Emx1-Cre transgenic mice to derive conditional KO (cKO) mice.
Primary Neuron Culture
Rat cortices isolated from embryonic day (E) 18.5 brains were cut into small pieces, washed with Hank’s balanced salt solution (HBSS) to remove debris and then digested in papain solution (0.6 mg/ml papain, 0.6 mg/ml DNase I, 0.2 mg/ml L-cysteine, 1.5 mM CaCl2 and 0.5 mM EDTA in HBSS) at 37°C for 20 min, followed by the addition of 10% horse serum in Neurobasal medium to stop the enzymatic reaction and 25-times trituration to obtain a cell suspension (Chao et al., 2012; Huang and Richter, 2007). To culture RIG-I (or MAVS) wild-type (WT) and KO neurons, cortices of E17.5 embryos from heterozygous matings were isolated and maintained individually in HBSS for ∼2-3 h on ice before determining genotypes on tail biopsies (Lu et al., 2017). The WT and KO cerebral cortices were pooled and processed similarly for neuronal cultures (Chao et al., 2012; Huang and Richter, 2007). CMTR1-cWT and -cKONes cortical neurons were prepared in the same way by using E17.5 embryos isolated from Cmtr1 f/+, Nestin-Cre/+ females crossed with Cmtr1 f/f, +/+ male mice. The cell density was 6 × 104 cells/well in a 12-well containing an 18-mm glass coverslip for immunostaining and morphological analysis and 106 cells/well in a 6-well plate for biochemical and molecular analysis. For co-culture experiments, each well in a 12-well plate was deposited with 4-5 wax spots, coated with poly-L-lysine and then seeded with 2 × 105 cells. Cortical neurons cultured in Neurobasal medium with 0.5 mM glutamine, 12.5 μM glutamate, 1X antibiotic-antimycotic and B27 supplement until the designated days in vitro (DIV) were used for experiments.
Method Details
Lentivirus Production and Lentiviral Infection
HEK293T cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. Lentivirus particles were generated by using the Virapower packaging system (Invitrogen). The mixture of 3 μg pLL3.7-Syn or pLKO plasmid and 9 μg Virapower DNA was mixed with 30 μl Lipofectamine 2000 (Invitrogen) and the DNA–liposome complex was transfected into 6 × 106 HEK293T cells overnight, then replaced with new medium, which was collected 2 days after and centrifuged at 120,000 xg for 2 h at 4°C to pellet viral particles. The viruses were resuspended in Neurobasal medium, aliquoted and stored at −80°C. For the knockdown experiment, cortical neurons at DIV2 were incubated with the designated lentiviruses overnight and harvested at DIV7 for experiments. HEK293T cells were incubated with the lentivirus in the presence of polybrene overnight, replaced with new medium for one day, then selected with puromycin to obtain stably transformed CMTR1-KD cells as described previously (Chang and Huang, 2014).
Plasmid Construction
The oligonucleotides containing short hairpin RNA (shRNA) sequences, targeted against rat CMTR1 or mouse DXO mRNA as well as a non-target control (Table S2), were cloned into HpaI and XhoI linearized pLL3.7-Syn (Huang and Richter, 2007). The TRCN0000297447 (CGTTAAGTGGTCACTCCCATT) shRNA clone against human CMTR1 was purchased from the RNAi Core Facility. To generate RFP-hCMTR1 fusion, the mCherry red fluorescent protein (RFP) plasmid (Chang and Huang, 2014) and human CMTR1 plasmid (OHu06786, GenScript) were PCR-amplified in the presence of 4 primers, RFP_F, 5′-ATGCTGGCTAGCGCCACCATGGTGAGCAAGG-3′, RFP-hTR1_R, 5′-TTCTGGGTCAGTTCTCCTCTTCATGGTACCCTTGTACAGCTCGTCCATGCC-3′, RFP-hTR1_F, 5′-GGCATGGACGAGCTGTACAAGGGTACCATGAAGAGGAGAACTGACCCAGAA-3′ and hTR1_R, 5′-ATTCAGGTTTAAACTCAGGCCCTGTGCATCTGGA-3′. The amplified fragment was digested with NheI and PmeI, then cloned into pLL3.7-Syn in which the green fluorescent protein (GFP) coding region was replaced with the RFP-hCMTR1 sequence. The K239A and deletion mutants of CMTR1 were generated by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol.
Quantitative RT-PCR (RT-qPCR)
Total RNA from cortical tissue and cells was extracted by using TRIzol reagent (Invitrogen) and analyzed by using the NanoDrop 1000 spectrophotometer (Thermo Fisher). Approximately 2 μg total RNA was incubated with RNase-free DNase for 30 min at 37°C, followed by phenol/chloroform extraction and ethanol precipitation. The purified DNA-free RNA samples were reverse-transcribed by using random primers and GoScript reverse transcriptase (Promega). The resulting cDNAs were analyzed by qPCR with the Universal Probe Library (UPL) reagent in the LightCycler 480 system (Roche). The relative expression of targets was calculated by the comparative threshold cycle value with Gapdh mRNA as the reference. The primer sequences and UPL probes designed at the Roche Assay Design Center are in Table S2.
Nucleocytoplasmic Fractionation and Western Blot Analysis
Cultured neurons were harvested in lysis buffer (10 mM HEPES, pH 7.5, 10 mM KCl, 0.1 mM EDTA and 0.5% NP-40), incubated on ice for 10 min, then centrifuged at 800 xg for 5 min at 4°C. The supernatant was collected as the cytoplasmic fraction and the pellet was washed twice with the lysis buffer, then lysed in the buffer containing 20 mM HEPES, pH 7.5 and 400 mM KCl for 10 min on ice to collect the nuclear fraction. Tissues and cells were harvested in the lysis buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5% Triton X-100, 0.1% SDS, 1 mM dithiothreitol and 1X protease inhibitor cocktail (Roche) and sonicated on ice for 20 s to break up chromosomal DNA (Misonix sonicator 3000), followed by centrifugation at 12,000 xg for 5 min at 4°C to collect supernatant. The protein concentration of collected supernatants, nuclear and cytosolic fractions was determined by use of the Pierce BCA Protein Assay Kit (Pierce), then diluted in Laemmli sample buffer. The protein samples were denatured at 95°C for 5 min, separated on 10% Tris-glycine SDS-polyacrylamide gels and transferred to 0.45 μm nitrocellulose membrane (GE Healthcare Life Science), which was incubated with the designated primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies and detection with the Immobilon Western ECL system (Millipore). The chemiluminescence signals were captured by an ECL imaging system (LAS-4000, FUJINON).
Immunofluorescence Staining, Immunohistochemistry and Image Acquisition
All solutions were prepared in 1X phosphate buffered saline (PBS) and all procedures were conducted at room temperature except for primary antibody incubation at 4°C. Cultured neurons were fixed with 4% formaldehyde for 10 min, washed with PBS for 3 times and permeabilized in 0.2% Triton X-100 for 30 min. After 3 washes of PBS, permeabilized neurons were blocked in 10% horse serum and 3% bovine serum albumin (BSA) for 60 min, then incubated with designated antibodies at 4°C for overnight. After 3 washes of PBS, proper Alexa Fluor-conjugated secondary antibodies and 4’,6-diamidino-2-phenylindole (DAPI) were added and incubated in the dark for 2 h, washed with PBS for 3 times and briefly rinsed with H2O, air-dried and mounted with ProLong Gold antifade reagent (Invitrogen). Images were acquired under a fluorescence microscope (Zeiss Imager M1). Mice were anesthetized with isoflurane inhalation and intracardially perfused with PBS and 4% formaldehyde. Brains were isolated and post-fixed in 4% formaldehyde at 4°C overnight, dehydrated in 15% (wt/vol) sucrose for 1 day and 30% (wt/vol) sucrose for another day at 4°C, embedded in Tissue-Tek OCT compound, then sectioned coronally at 25-μm thickness onto silane-coated slides by using a cryostat (Leica 3050S). Selected sections were processed for antigen retrieval in Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 9) at 90°C for 20 min, followed by 30-min permeabilization in 0.25% Triton X-100, 3 washes of PBS and 1-h blocking in 5% BSA. The remaining steps, antibody incubation and slide mounting, were as described above. Mice carrying the Thy1-YFP transgene at P18 were processed identically, but the OCT-embedded brains were sectioned coronally at 60-μm thickness onto silane-coated slides. Images of YFP-expressing pyramidal neurons in the cortical layer 5 were captured under a Zeiss inverted confocal microscope (LSM780). Each image consisted of a stack of Z series images of 1-μm spacing. For DAB (3,3′-diaminobenzidine) immunohistochemistry, horseradish peroxidase- instead of Alexa Fluor-conjugated secondary antibody was used and the immunobinding signal was developed using the Vectastain Elite ABC kit (Vector labs) following the manufacturer’s protocol. Histological staining with hematoxylin and eosin was conducted by the Pathology Core staff. Briefly, 4% formaldehyde-fixed brains were embedded in paraffin. Sections after the removal of paraffin were stained with hematoxylin and eosin. Images were acquired by using a panoramic scanner (Pannoramic 250 flash III).
Sholl Analysis
Dendrite tracking and process connection were performed by using Neuro J (a plugin of ImageJ) (Schindelin et al., 2012) and NeuronStudio (Rodriguez et al., 2008), respectively. The data were then compiled and analyzed by using Bonfire (Langhammer et al., 2010) to count the number of dendritic intersections in every 10-μm segment away from the soma in concentric circles.
Microarray and Gene Ontology Analysis
Total RNA isolated from control and CMTR1-KD neurons at DIV7 (2 samples per group) were submitted to a commercial service (Welgene, Taiwan). Briefly, total RNA (0.2 μg per sample) was amplified and labeled with Cy3, and 0.6 μg Cy3-labled cRNA was fragmented to ∼50-100 nt, which were used for array (Agilent, whole rat genome 4x44K microarray) hybridization at 65°C for 17 h. The images of hybridized Cy3 signals were analyzed and quantified by using Feature extraction 10.7.3.1 software (Agilent). From the normalized data, we eliminated the genes with unknown EntrezGene ID and with signal/noise ratio of expression < 1. The genes with expression (averaged from duplicate datasets) showing more than ± 1.2-fold change in KD versus control neurons were selected for Metascape gene ontology analysis (Zhou et al., 2019). The term “neuron” was applied to all selected ontologies, including KEGG Functional Sets, KEGG Pathway, KEGG Structural Complexes, GO Biological Processes/Cellular Components/Molecular Functions, Reactome Gene Sets and CORUM database.
Thin Layer Chromatography (TLC) Detection of N1 2′-O-Me
Total RNA extracted from HEK293T cells and cortical tissues was purified by using the PolyATtract mRNA Isolation System (Promega) to isolate poly(A) RNA, which was decapped and dephosphorylated by using Cap-Clip acid pyrophosphatase (CELLSCRIPT) and FastAP alkaline phosphatase (Thermo Fisher), respectively. The resulting 5′-OH RNA was radiolabeled with T4 polynucleotide kinase (Thermo Fisher) in the presence of [γ-32P] ATP, digested to mononucleotides with Nuclease P1 (Sigma Aldrich), then separated on a PEI cellulose F-coated TLC plate (Millipore) with isobutyric acid: 0.5M ammonium hydroxide (67:33) in the first dimension and isopropanol: HCl: H2O (68:18:14) in the second dimension as modified from the previous protocol (Kruse et al., 2011). The radioactive image was acquired by using Typhoon 9410 Imager (GE Healthcare Life Science).
4-Thiouridine (4sU)-Labeling and Isolation of Nascent RNAs
Cultured neurons at DIV7 were incubated with 50 μM 4sU (T4509, Sigma-Aldrich) for 12 h prior to isolating total RNA with TRIzol (Invitrogen). The 4sU-incorporated nascent transcripts were biotinylated with 20 fmol EZ-LINK HPDP-Biotin (Thermo Fisher) per μg total RNA in 10 mM Tris pH 7.5 and 1 mM EDTA at 60°C for 3 h. The biotinylated samples were denatured at 65°C for 10 min and chilled on ice for 5 min. A part of reaction containing 1 μg total RNA was applied to nitrocellulose membrane. The membrane was UV (120,000 J/cm2)-crosslinked for 2 min, blocked with 3% BSA in PBS for 30 min and washed with PBS for 3 times, followed by 1-h incubation of horse peroxidase-conjugated streptavidin (PK6102, Vestastain) at room temperature. After 3 washes of PBS, the membrane was developed with the Immobilon Western ECL system (Millipore). The remained biotinylated samples were incubated with streptavidin paramagnetic beads (Z5481, Promega), which were pre-treated with 1% polyvinylpyrrolidone (P5288, Sigma-Aldrich) for 10 min to block non-specific binding. After 30-min incubation at room temperature, the beads were washed 5 times with 10 mM Tris pH 7.5, 1 mM EDTA and 1 M NaCl and then eluted with 100 mM dithiothreitol. The eluted RNAs were extracted with phenol/chloroform and precipitated with ethanol, followed by reverse transcription with oligo-dT primers and GoScript reverse transcriptase (Promega) and qPCR.
Quantification and Statistical Analysis
Data are expressed as mean ± SEM. GraphPad Prism software was used to evaluate statistical differences between groups. Single-factor comparisons were determined by two-tailed Student’s t test. Multivariate data were analyzed by two-way ANOVA with Tukey post hoc comparisons. Sample sizes and statistical methods are described in figure legends.
Acknowledgments
The authors thank Fang Liao for RIG-I mice and EV71-infected spinal cord cDNA, Sue-Jane Lin for MAVS mice, Yi-Ling Lin for JEV-infected brain cDNA, Guey-Shin Wang for Histone3 antibody, and Yi-Ping Hsueh for advice on dendritic morphology analysis. We thank the National RNAi Core Facility at Academia Sinica for the shRNA clones, DNA sequencing support under the Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-115), institutional pathology core for H&E staining, and light microscopy core for technical assistance in image acquisition. This work was supported by Ministry of Science and Technology (MoST108-2320-B-001-020-MY3 and AS-KPQ-109-BioMed), National Health Research Institutes (NHRI-EX109-10719SI), and Academia Sinica (AS-SUMMIT-109) in Taiwan.
Author Contributions
Y.-L.L. designed and performed the experiments, analyzed data, and wrote the manuscript. F.-C.K. conducted tissue immunoblotting, and C.-H.L. isolated E8.5 embryos for genotyping. Y.-S.H. designed and supervised the study, co-wrote the manuscript, and is responsible for its content.
Declaration of Interests
The authors declare no competing interests.
Published: October 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108269.
Supplemental Information
References
- Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O., Suzuki T. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:eaav0080. doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- Bélanger F., Stepinski J., Darzynkiewicz E., Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 2010;285:33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M., Plotch S.J., Krug R.M. Both the 7-methyl and the 2′-O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc. Natl. Acad. Sci. USA. 1980;77:3952–3956. doi: 10.1073/pnas.77.7.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.W., Huang Y.S. Arsenite-activated JNK signaling enhances CPEB4-Vinexin interaction to facilitate stress granule assembly and cell survival. PLoS One. 2014;9:e107961. doi: 10.1371/journal.pone.0107961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao H.W., Lai Y.T., Lu Y.L., Lin C.L., Mai W., Huang Y.S. NMDAR signaling facilitates the IPO5-mediated nuclear import of CPEB3. Nucleic Acids Res. 2012;40:8484–8498. doi: 10.1093/nar/gks598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. USA. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G., Mellor R.H., Bernstein M., Keller-Peck C., Nguyen Q.T., Wallace M., Nerbonne J.M., Lichtman J.W., Sanes J.R. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Morgan M., Shatkin A.J., Jelinek W., Salditt-Georgieff M., Darnell J.E. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway A., Cowling V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:270–279. doi: 10.1016/j.bbagrm.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss G.K., Carter V.S., He Y., Kwieciszewski B.K., Holzman T., Korth M.J., Lazaro C.A., Fausto N., Bumgarner R.E., Katze M.G. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 2003;77:6367–6375. doi: 10.1128/JVI.77.11.6367-6375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T., Cowling V.H. Cap-binding complex (CBC) Biochem. J. 2014;457:231–242. doi: 10.1042/BJ20131214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L., Jones K.R. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra S., López-Fernández L.A., Pascual-Montano A., Muñoz M., Harshman K., Esteban M. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 2003;77:6493–6506. doi: 10.1128/JVI.77.11.6493-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M., Hubel P., Lacerda L., Benda C., Holze C., Eberl C.H., Mann A., Kindler E., Gil-Cruz C., Ziebuhr J. Sequestration by IFIT1 impairs translation of 2'O-unmethylated capped RNA. PLoS Pathog. 2013;9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A., Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- Hernández G., Altmann M., Lasko P. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem. Sci. 2010;35:63–73. doi: 10.1016/j.tibs.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Huang Y.S., Lu W.H. Decoding hidden messages in neurons: insights from epitranscriptome-controlled and specialized ribosome-controlled translation. Curr. Opin. Neurobiol. 2018;48:64–70. doi: 10.1016/j.conb.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Huang Y.S., Richter J.D. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol. 2007;431:143–162. doi: 10.1016/S0076-6879(07)31008-2. [DOI] [PubMed] [Google Scholar]
- Inesta-Vaquera F., Chaugule V.K., Galloway A., Chandler L., Rojas-Fernandez A., Weidlich S., Peggie M., Cowling V.H. DHX15 regulates CMTR1-dependent gene expression and cell proliferation. Life Sci. Alliance. 2018;1:e201800092. doi: 10.26508/lsa.201800092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Chang J.H., Kilic T., Tong L., Kiledjian M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kruse S., Zhong S., Bodi Z., Button J., Alcocer M.J., Hayes C.J., Fray R. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 2011;1:126. doi: 10.1038/srep00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge H., Brownlee G.G., Gershon P.D., Richter J.D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5’ppp- mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langberg S.R., Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methyltransferases from HeLa cells. J. Biol. Chem. 1981;256:10054–10060. [PubMed] [Google Scholar]
- Langhammer C.G., Previtera M.L., Sweet E.S., Sran S.S., Chen M., Firestein B.L. Automated Sholl analysis of digitized neuronal morphology at multiple scales: Whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry A. 2010;77:1160–1168. doi: 10.1002/cyto.a.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T., Peterlin B.M., Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Amarasinghe G.K. When your cap matters: structural insights into self vs non-self recognition of 5′ RNA by immunomodulatory host proteins. Curr. Opin. Struct. Biol. 2016;36:133–141. doi: 10.1016/j.sbi.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Clohisey S.M., Chia B.S., Wang B., Cui A., Eisenhaure T., Schweitzer L.D., Hoover P., Parkinson N.J., Nachshon A. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat. Commun. 2020;11:164. doi: 10.1038/s41467-019-13965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.H., Yeh N.H., Huang Y.S. CPEB2 Activates GRASP1 mRNA Translation and Promotes AMPA Receptor Surface Expression, Long-Term Potentiation, and Memory. Cell Rep. 2017;21:1783–1794. doi: 10.1016/j.celrep.2017.10.073. [DOI] [PubMed] [Google Scholar]
- Moya-Alvarado G., Gonzalez A., Stuardo N., Bronfman F.C. Brain-Derived Neurotrophic Factor (BDNF) Regulates Rab5-Positive Early Endosomes in Hippocampal Neurons to Induce Dendritic Branching. Front. Cell. Neurosci. 2018;12:493. doi: 10.3389/fncel.2018.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi A., Dutta K., Basu A. RIG-I mediates innate immune response in mouse neurons following Japanese encephalitis virus infection. PLoS One. 2011;6:e21761. doi: 10.1371/journal.pone.0021761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A., Marcotrigiano J., Stepinski J., Jankowska-Anyszka M., Wyslouch-Cieszynska A., Dadlez M., Gingras A.C., Mak P., Darzynkiewicz E., Sonenberg N. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- Noack F., Calegari F. Epitranscriptomics: A New Regulatory Mechanism of Brain Development and Function. Front. Neurosci. 2018;12:85. doi: 10.3389/fnins.2018.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E., Kwok B., Day J.S., Connor T.J., Harkin A. Amitriptyline protects against TNF-α-induced atrophy and reduction in synaptic markers via a Trk-dependent mechanism. Pharmacol. Res. Perspect. 2016;4:e00195. doi: 10.1002/prp2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Jean F., Brand C., Tremblay-Létourneau M., Allaire A., Beaudoin M.C., Boudreault S., Duval C., Rainville-Sirois J., Robert F., Pelletier J. 2′-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS One. 2018;13:e0193804. doi: 10.1371/journal.pone.0193804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A., Robb G.B., Chan S.H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikine S., Nguyen J.B., Modis Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Ehlenberger D.B., Dickstein D.L., Hof P.R., Wearne S.L. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S. A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1-2'O-Methylated Self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- Smietanski M., Werner M., Purta E., Kaminska K.H., Stepinski J., Darzynkiewicz E., Nowotny M., Bujnicki J.M. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 2014;5:3004. doi: 10.1038/ncomms4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Pezacki J.P., Wodicka L., Brideau A.D., Supekova L., Thimme R., Wieland S., Bukh J., Purcell R.H., Schultz P.G., Chisari F.V. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczydlowska-Socha D., Zielinska M.M., Kurkowska M., Astha, Almeida C.F., Stefaniak F., Purta E., Bujnicki J.M. Human RNA cap1 methyltransferase CMTr1 cooperates with RNA helicase DHX15 to modify RNAs with highly structured 5′ termini. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20180161. doi: 10.1098/rstb.2018.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Wakai C., Iwama M., Mizumoto K., Nagata K. Recognition of cap structure by influenza B virus RNA polymerase is less dependent on the methyl residue than recognition by influenza A virus polymerase. J. Virol. 2011;85:7504–7512. doi: 10.1128/JVI.02375-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Alvin Chew B.L., Lai Y., Dong H., Xu L., Balamkundu S., Cai W.M., Cui L., Liu C.F., Fu X.Y. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019;47:e130. doi: 10.1093/nar/gkz751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Purta E., Kaminska K.H., Cymerman I.A., Campbell D.A., Mittra B., Zamudio J.R., Sturm N.R., Jaworski J., Bujnicki J.M. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011;39:4756–4768. doi: 10.1093/nar/gkr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Gokhale N.S., Snider D.L., Horner S.M. The mRNA Cap 2′-O-Methyltransferase CMTR1 Regulates the Expression of Certain Interferon-Stimulated Genes. MSphere. 2020;5:e00202-20. doi: 10.1128/mSphere.00202-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki N., Maekawa M., Kobayashi K., Kajii Y., Maeda J., Soma M., Takao K., Tanda K., Ohira K., Toyama K. Alpha-CaMKII deficiency causes immature dentate gyrus, a novel candidate endophenotype of psychiatric disorders. Mol. Brain. 2008;1:6. doi: 10.1186/1756-6606-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the microarray data reported in this paper is NCBI GEO: GSE145223.