Abstract
Summary
The molecular changes induced by perturbations such as drugs and ligands are highly informative of the intracellular wiring. Our capacity to generate large datasets is increasing steadily. A useful way to extract mechanistic insight from the data is by integrating them with a prior knowledge network of signalling to obtain dynamic models. CellNOpt is a collection of Bioconductor R packages for building logic models from perturbation data and prior knowledge of signalling networks. We have recently developed new components and refined the existing ones to keep up with the computational demand of increasingly large datasets, including (i) an efficient integer linear programming, (ii) a probabilistic logic implementation for semi-quantitative datasets, (iii) the integration of a stochastic Boolean simulator, (iv) a tool to identify missing links, (v) systematic post-hoc analyses and (vi) an R-Shiny tool to run CellNOpt interactively.
Availability and implementation
R-package(s): https://github.com/saezlab/cellnopt.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
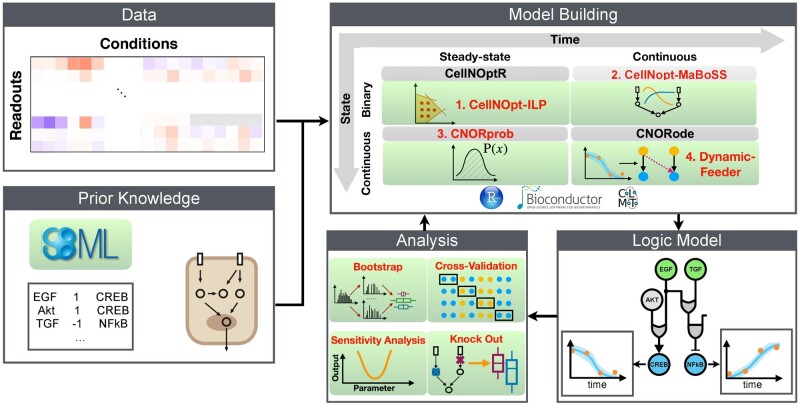
Logic networks are among the conceptually simplest modelling frameworks. They capture the mechanistic relationship between molecular entities by logic gates. Due to their simplicity, they are highly scalable and widely applied. We have previously introduced CellNetOptimiser (CellNOpt), a framework to build predictive logic models of signalling pathways by training a prior knowledge network (PKN) to biochemical data obtained from perturbation experiments (Terfve et al., 2012). The CellNOpt R packages feature different formalisms ranging from Boolean logic to logic-based ordinary differential equations(Fig. 1). The value of CellNOpt has been recently demonstrated on several applications(Supplementary Table S1).
Fig. 1.
CellNOpt pipeline with packages and features. Perturbation data are combined with prior knowledge of signalling interactions. Different modules can be used for model building: CellNOptR (Boolean logic models), CellNOpt-MaBoSS (stochastic simulations), CNORprob (probabilistic formalism), CNORode (logic-based ODEs). New packages and features are highlighted with green background and red text. (Color version of this figure is available at Bioinformatics online.)
Over the past years, we have continuously added new features and upgrades to the packages to enhance the functionality and computational efficiency of CellNOpt (Fig. 1). The current toolkit covers a unique set of features (see Supplementary Table S2). Furthermore, for alternative analyses, CellNOpt supports importing from and exporting to other tools via simple interaction files and the SBMLQual (Chaouiya et al., 2013) formats.
2 Summary of new features
We enumerate here the new features that enrich the CellNOpt framework.
2.1 CellNOpt-ILP
For the training of Boolean logic networks, we implemented an integer linear programming (ILP) formulation based on (Mitsos et al., 2009) and the IBM CPLEX optimizer. This allows us to optimize large-scale networks orders of magnitude faster than the built-in genetic algorithm. Furthermore, the method can generate pools of (near) optimal solutions, which helps identify uncertainty in the inferred network structure. We illustrate the functionality of CellNOpt-ILP on several case studies (Supplementary Text S1).
2.2 CellNOpt-MaBoSS
MaBoSS (Stoll et al., 2012) performs asynchronous stochastic simulations of logic Boolean models. It was integrated with CellNOpt to optimize Boolean networks where nodes are represented by their state probability with respect to time (Supplementary Text S2).
2.3 CNORprob
To optimize logic networks with semi-quantitative states (between 0 and 1) at quasi-steady-state, we offer the CNORprob package. CNORprob is an R-implementation of the Matlab-based toolbox FALCON for probabilistic Boolean logic (De Landtsheer et al., 2017) (Supplementary Text S5).
2.4 Dynamic-Feeder
Dynamic-Feeder is a method to identify missing links in the PKN and provides candidates to fill knowledge gaps (Supplementary Fig. S3). It combines data-driven network inference with a protein–protein interaction network to find missing elements. For the latter, we primarily use OmniPath (Turei et al., 2016). Dynamic-Feeder generalizes our previous tool CNORfeeder (Eduati et al., 2012) to time-course data with a logic ordinary differential equations formalism (Supplementary Text S4).
2.5 Post-hoc analysis
After optimized logic models are obtained, the predictive power of the models can be assessed by cross-validation and bootstrapping. Further, the package offers parameter sensitivity analysis, and estimation of node and edge essentiality by removing them (knockout). This allows us to observe how sensitive specific proteins/interactions to perturbations are (Supplementary Text S5).
2.6 Shiny application
To build and train models with CellNOpt, CNORprob and CNORode without coding, we offer an interactive R-Shiny application (Supplementary Text S6).
In summary, the new CellNOpt features expand the options for logic modelling, in particular to analyze large datasets.
Supplementary Material
Acknowledgements
The authors thank Christian Holland for setting up the ShinyCNOR and Nicolas Palacio Escat for his help in designing Figure 1.
Funding
The work was supported by: the European Unions H2020 program (675585 Marie-Curie ITN SymBioSys); J.R.C. for Computational Biomedicine, which is partially funded by Bayer; and the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 116030 (TransQST).
Conflict of Interest: none declared.
Contributor Information
Enio Gjerga, Faculty of Medicine, Heidelberg University, Heidelberg University Hospital, Institute for Computational Biomedicine, BioQuant 69120 Heidelberg, Germany; Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE).
Panuwat Trairatphisan, Faculty of Medicine, Heidelberg University, Heidelberg University Hospital, Institute for Computational Biomedicine, BioQuant 69120 Heidelberg, Germany.
Attila Gabor, Faculty of Medicine, Heidelberg University, Heidelberg University Hospital, Institute for Computational Biomedicine, BioQuant 69120 Heidelberg, Germany.
Hermann Koch, Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE); Aachener Verfahrenstechnik, Process Systems Engineering, RWTH Aachen University, Aachen, Germany.
Celine Chevalier, Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE); University Paris-Saclay, Espace Technologique Bat. Discovery,91190 Saint-Aubin, France.
Franceco Ceccarelli, Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE); Computer Laboratory, University of Cambridge, Cambridge CB2 1TN, UK.
Aurelien Dugourd, Faculty of Medicine, Heidelberg University, Heidelberg University Hospital, Institute for Computational Biomedicine, BioQuant 69120 Heidelberg, Germany; Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE).
Alexander Mitsos, Aachener Verfahrenstechnik, Process Systems Engineering, RWTH Aachen University, Aachen, Germany.
Julio Saez-Rodriguez, Faculty of Medicine, Heidelberg University, Heidelberg University Hospital, Institute for Computational Biomedicine, BioQuant 69120 Heidelberg, Germany; Faculty of Medicine, Joint Research Centre for Computational Biomedicine (JRC-COMBINE).
References
- Chaouiya C. et al. (2013) SBML qualitative models: a model representation format and infrastructure to foster interactions between qualitative modelling formalisms and tools. BMC Sys. Biol., 7, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduati F. et al. (2012) Integrating literature-constrained and data-driven inference of signalling networks. Bioinformatics, 28, 2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsos A. et al. (2009) Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLoS Comput. Biol., 5, e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G. et al. (2012) Continuous time Boolean modeling for biological signaling: application of Gillespie algorithm. BMC Syst. Biol., 6, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terfve C. et al. (2012) CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Syst. Biol., 6, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turei D. et al. (2016) OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods, 13, 966–967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.