Abstract
Summary
Experimental laboratory management and data-driven science require centralized software for sharing information, such as lab collections or genomic sequencing datasets. Although database servers such as PostgreSQL can store such information with multiple-user access, they lack user-friendly graphical and programmatic interfaces for easy data access and inputting. We developed LabxDB, a versatile open-source solution for organizing and sharing structured data. We provide several out-of-the-box databases for deployment in the cloud including simple mutant or plasmid collections and purchase-tracking databases. We also developed a high-throughput sequencing (HTS) database, LabxDB seq, dedicated to storage of hierarchical sample annotations. Scientists can import their own or publicly available HTS data into LabxDB seq to manage them from production to publication. Using LabxDB’s programmatic access (REST API), annotations can be easily integrated into bioinformatics pipelines. LabxDB is modular, offering a flexible framework that scientists can leverage to build new database interfaces adapted to their needs.
Availability and implementation
LabxDB is available at https://gitlab.com/vejnar/labxdb and https://labxdb.vejnar.org for documentation. LabxDB is licensed under the terms of the Mozilla Public License 2.0.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Managing a collection of samples, plasmids or cell lines, tracking purchases and equipment and storing data annotations is common in laboratories. Within each scientific domain, laboratories manage specialized data and collections, such as fly or fish lines. For instance, high-throughput sequencing (HTS) applications have significantly expanded with many molecular biology assays (Ingolia et al., 2011; Johnson et al., 2007; Ule et al., 2003) now using HTS to address biological questions with a genome-wide point of view. As a result, current genomics projects frequently generate numerous HTS samples (e.g. 212 samples in our recent study Vejnar et al., 2019, 513 samples in White et al., 2017). The organization and sharing [e.g. on NCBI sequence read archive (SRA) (Leinonen et al., 2011)] of these datasets require not only the raw data but also detailed annotations and hierarchical structures grouping the replicates of each experiment and the sequencing runs. Tools to organize these annotations are essential for genomics and in general for data-driven science. Since the exact nature and requirements of these annotations and data structures differ across scientific fields, and since information is often provided by multiple individuals working in collaboration, it imposes strong requirements on potential solutions.
Available solutions (Anatskiy et al., 2019; Heinle et al., 2017; Hunter et al., 2017; Venco et al., 2014) to organize and share data annotations are inadequate (Supplementary Fig. S1). Some free-to-use closed-source programs such as the Google Suite offer spreadsheet-sharing solutions, but they lack structured fields essential for efficient data processing. Although commercial software, such as FileMaker Pro, handle structured data, these tools are expensive and require client installation. In addition, these available tools are not adaptable and do not provide ready-to-use solutions for lab and HTS data management.
Here, we present LabxDB, an extensible open-source solution for organizing and sharing structured data that combines open-source databases with both internet browser and programmatic interfaces. LabxDB provides several out-of-the-box databases needed in experimental laboratories such as for storing HTS sample annotations, managing laboratories purchases or collection of plasmids. The power of LabxDB is its modular implementation that facilitates organization of new data and addition of new features. LabxDB implements generic blocks that can be easily combined to create new database structures or user interfaces adapted to specific scientific fields.
2 Results
LabxDB provides two core functionalities: (i) a server-side database to store structured information and (ii) a client-side browser-based graphical user interface (GUI) and programmatic interface supporting concurrent access of multiple users (Fig. 1). A key feature of LabxDB is its ability to store hierarchically structured information with an unlimited number of nested levels. For example, a ‘project’ could include nested ‘sample’ information. LabxDB provides generic reusable and extensible blocks that allow for modular implementation of arbitrary databases. On the server, blocks are Python classes and templates handling queries from the client such as getting or removing data. On the client, blocks are (ECMAscript 6) Javascript classes used to build GUI. Each LabxDB implementation consists of a data model, a PostgreSQL database, a server handling client requests via a REST API and an optional web-based GUI described in Supplementary Material. For the GUI, we designed specific classes to display single (Supplementary Fig. S2a) or multiple nested hierarchical levels (Supplementary Fig. S2c), and to edit information (Supplementary Fig. S2b).
Fig. 1.
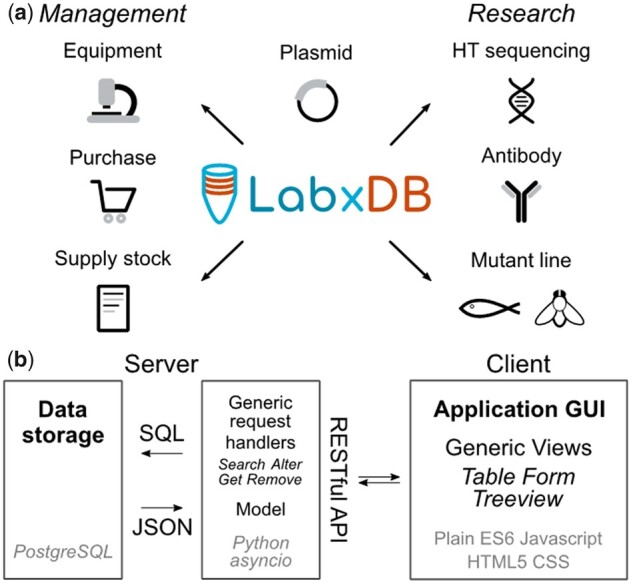
(a) Example applications of LabxDB for lab management and research data annotation. (b) Diagram describing the client–server structure of LabxDB. Server stores the data in PostgreSQL, hosts the data model and responds to client requests. Communications between the client and the server are using a RESTful API, while the results of SQL queries addressed to PostgreSQL are returned as JSON data. To implementLabxDB databases (such as LabxDB seq), generic request handlers (Search, Alter etc.) and generic views (Table, Form etc.) are used to implement the server and client sides respectively. Technologies used are indicated in light gray. (Color version of this figure is available at Bioinformatics online.)
LabxDB includes several preconfigured implementations, including a simple purchase-tracking database designed to track order requests from multiple users. We also developed a more complex database, LabxDB seq, that stores genomic HTS sample annotations. LabxDB seq leverages the hierarchical capabilities of LabxDB and is composed of four nested levels: project, sample, replicate and run. In this structure, within a project, the same experiment is performed in replicates for each sample, and each replicate is composed of one or more sequencing runs. To implement the GUI, we designed specific editing forms by extending generic LabxDB classes that allow users to create their structure of replicates, samples and projects and annotate all levels at the same time. All requests handled by the server use generic LabxDB server classes. We implemented procedures written in PL/pgSQL to add automatic unique identifiers (similarly to gene IDs) to projects, samples, replicates and runs. Finally, we developed scripts to import HTS data from local sequencing facilities or public resources, such as SRA and to export data to SRA, greatly facilitating these tasks (Supplementary Fig. S3). LabxDB seq, therefore, allows scientists to manage HTS data from production to publication.
3 Conclusion
We developed LabxDB to quickly create databases, which will help scientists to organize, edit and search their data. Using the default client configuration, a new database implementation only requires the administrator to define a data model; within a few minutes, LabxDB can be deployed in the cloud. Because databases help organize and share information, we hope that LabxDB will facilitate data exchange within laboratories as well as between them. We released LabxDB under an open-source license to encourage adaptation and sharing of database structures. Finally, LabxDB can be used to automatically configure pipelines. Thus, LabxDB provides an essential link between scientist manual input of information and automatic treatment of data with bioinformatics pipelines.
Supplementary Material
Acknowledgements
We thank all members of the Giraldez laboratory for their support, their suggestions and for reporting bugs during development. We thank Daniel Burkhardt, Cassandra Kontur and Dr Mina Kojima for manuscript editing. NIH [R21 HD073768, R01 HD074078, R01 GM102251, R35 GM122580] supported this work.
Conflict of Interest: none declared.
Contributor Information
Charles E Vejnar, Department of Genetics.
Antonio J Giraldez, Department of Genetics; Yale Stem Cell Center; Yale Cancer Center, Yale University School of Medicine, New Haven, CT 06510, USA.
References
- Anatskiy E. et al. (2019) Parkour LIMS: high-quality sample preparation in next generation sequencing. Bioinformatics, 35, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Heinle C.E. et al. (2017) MetaLIMS, a simple open-source laboratory information management system for small metagenomic labs. Gigascience, 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A. et al. (2017) MASTR-MS: a web-based collaborative laboratory information management system (LIMS) for metabolomics. Metabolomics, 13, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T. et al. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S. et al. (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science, 316, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Leinonen R. et al. (2011) The sequence read archive. Nucleic Acids Res., 39, D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J. et al. (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science, 302, 1212–1215. [DOI] [PubMed] [Google Scholar]
- Vejnar C.E. et al. (2019) Genome wide analysis of 3′ UTR sequence elements and proteins regulating mRNA stability during maternal-to-zygotic transition in zebrafish. Genome Res., 29, 1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venco F. et al. (2014) SMITH: a LIMS for handling next-generation sequencing workflows. BMC Bioinformatics, 15, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.J. et al. (2017) A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife, 6, 30860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


