Abstract
Background:
Understanding the mechanisms that regulate hair cell (HC) differentiation in the organ of Corti (OC) is essential to designing genetic therapies for hearing loss due to HC loss or damage. We have previously identified Fibroblast Growth Factor 20 (FGF20) as having a key role in HC and supporting cell differentiation in the mouse OC. To investigate the genetic landscape regulated by FGF20 signaling in OC progenitors, we employ Translating Ribosome Affinity Purification combined with Next Generation RNA Sequencing (TRAPseq) in the Fgf20 lineage.
Results:
We show that TRAPseq targeting OC progenitors effectively enriched for RNA from this rare cell population. TRAPseq identified differentially expressed genes (DEGs) downstream of FGF20, including Etv4, Etv5, Etv1, Dusp6, Hey1, Hey2, Heyl, Tectb, Fat3, Cpxm2, Sall1, Sall3, and cell cycle regulators such as Cdc20. Analysis of Cdc20 conditional-null mice identified decreased cochlea length, while analysis of Sall1-null and Sall1-ΔZn2-10 mice, which harbor a mutation that causes Townes-Brocks syndrome, identified a decrease in outer hair cell number.
Conclusions:
We present two datasets: genes with enriched expression in OC progenitors, and DEGs downstream of FGF20 in the embryonic day 14.5 cochlea. We validate select DEGs via in situ hybridization and in vivo functional studies in mice.
Keywords: cochlea, hair cell, RNAseq, SALL1, Townes-Brocks syndrome, hearing loss
INTRODUCTION
Congenital and acquired sensorineural hearing loss are common problems, yet there are no available biologically based therapies. Congenital sensorineural hearing loss can result from defects in sensory hair cells (HCs) or specialized supporting cells (SCs) within the organ of Corti (OC).1-4 Acquired sensorineural hearing loss is commonly caused by damage to HCs.5,6 In mammals, HC loss is permanent as the mammalian OC is unable to regenerate HCs.5,7 One potential approach to treating hearing loss due to HC loss or damage is to reactivate developmental signaling pathways in latent progenitors to promote their growth and differentiation into HCs and SCs. Investigation of the developmental pathways regulating HC and SC differentiation will therefore benefit our understanding and treatment of both congenital and acquired hearing loss.
In mouse cochlea development, Fibroblast Growth Factor 20 (FGF20) signaling via FGF receptor 1 (FGFR1) is required for the differentiation of OC progenitors (prosensory cells) into HCs and SCs, specifically outer hair cells (OHCs) and outer supporting cells.8-12 Fgf20-null mice (Fgf20−/−) are deaf, with shorter cochleae and loss of OHCs and gaps of undifferentiated cells along the length of the OC interrupting the normal patterning of HCs and SCs.9 FGF20 is required during the initiation of HC and SC differentiation and Fgf20−/− mice additionally exhibit premature onset of HC differentiation, as well as delayed apical progression of HC differentiation and maturation.9,13 We do not know the mechanism by which FGF20 is required for the initiation of differentiation. We hypothesize that downstream genetic targets of FGF20 signaling in prosensory cells will be candidate effectors of HC and SC differentiation. Identifying these genes will be important for advancing therapeutics in regenerating lost or damaged HCs and will provide insight into the mechanisms underlying OC phenotypes in Fgf20−/− mice.
Here, we combined the Translating Ribosome Affinity Purification (TRAP) technology with Next Generation RNA Sequencing (TRAPseq) to study changes in gene expression patterns in prosensory cells in the presence or absence of FGF20 signaling in mice.14 TRAP allows the isolation of translating mRNA, as well as noncoding RNA bound to ribosomes, from specific cell populations without cell sorting or fine dissection.15 We used the ROSAfsTRAP allele,15 which when activated by Cre recombinase leads to the expression of a GFP-tagged ribosomal protein (L10a-eGFP). Immunoprecipitation (IP) with anti-GFP antibodies then enriches for polysomes and associated RNA. We show that by targeting the expression of L10a-eGFP to prosensory cells within the cochleae in vivo using Fgf20Cre, we were able to enrich for RNA within this relatively rare cell population. Comparing control and Fgf20−/− prosensory cell RNA, TRAPseq revealed many genes previously associated with FGF signaling, as well as genes with functional significance in cochlea development. Among these genes is Sall1, mutations of which in humans cause Townes-Brocks syndrome, a genetic condition associated with variable features that include sensorineural hearing loss.16,17
RESULTS
Fgf20Cre targets L10a-eGFP expression to the prosensory domain and Kölliker’s organ
At embryonic day 14.5 (E14.5), the floor of the cochlear duct can be divided into three sections (Fig. 1A): 1) prosensory domain (PD), which contains prosensory cells that differentiate into HCs and SCs of the OC; 2) outer sulcus (OS), epithelium that is lateral (abneural) to the prosensory domain, which develops into the lesser epithelial ridge (LER), and 3) Kölliker’s organ (KO), epithelium that is medial (neural) to the prosensory domain, which develops into the greater epithelial ridge (GER). At E14.5, Fgf20 is expressed in the prosensory domain and at postnatal day 1 (P1), the Fgf20Cre lineage includes the OC and the GER.9,10
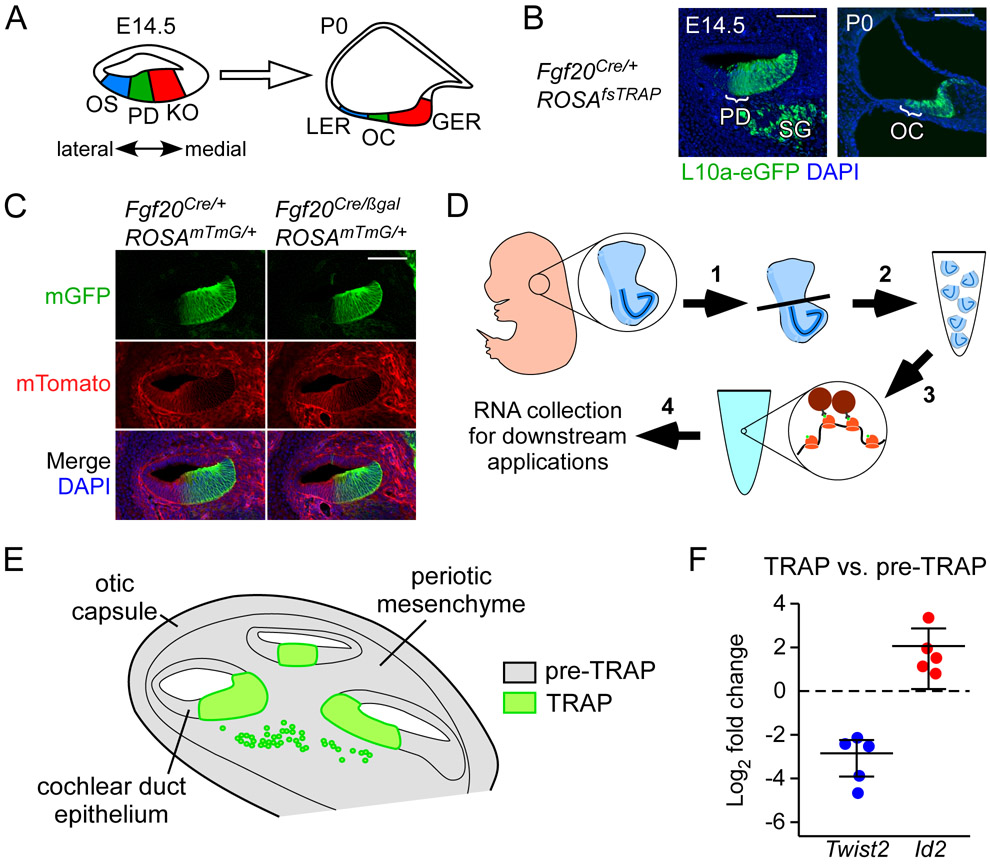
Figure 1. Fgf20Cre targets L10a-eGFP expression to the prosensory domain and Kölliker’s organ.
(A) Schematic representing cross-sectional view through the E14.5 and P0 cochlear duct. At E14.5, the epithelium at the cochlear duct floor can be divided into three regions: outer sulcus (OS), prosensory domain (PD), and Kölliker’s organ (KO). Cells from these three regions contribute to the lesser epithelial ridge (LER), organ of Corti (OC), and greater epithelial ridge (GER), respectively, at P0. Double-headed arrow indicates medial (neural) and lateral (abneural) directions. In all figures, sections through the cochlear duct are presented in this orientation.
(B) Sections through the middle turn of E14.5 and P0 Fgf20Cre/+;ROSAfsTRAP/+ cochlear ducts, showing L10a-eGFP (green) expression. At E14.5, L10a-eGFP is found in the prosensory domain (PD; bracket), Kölliker’s organ and medial wall, and spiral ganglion (SG). At P0, it is found in the organ of Corti (OC; bracket), greater epithelial ridge, and medial wall. DAPI, nuclei (blue); scale bar, 100 μm.
(C) Section through the middle turn of E14.5 cochlear ducts from Fgf20Cre/+;ROSAmTmG/+ and Fgf20Cre/βgal;ROSAmTmG/+ embryos. Cells of the Fgf20Cre lineage express mGFP (mG, green); non-lineage cells express mTomato (mT, red). DAPI, nuclei (blue); scale bar, 100 μm.
(D) Schematic showing an overview of the TRAPseq protocol (see Experimental Procedures). 1) Ventral otocysts containing the cochlea were dissected from E14.5 embryos. 2) Otocysts from each litter were pooled according to genotype to increase RNA yield. 3) Otocysts were then homogenized and centrifuged to make polysomes (pre-TRAP samples were collected at this stage). This was followed by immunoprecipitation with anti-GFP antibodies to collect L10a-eGFP labelled polysomes. 4) This produced TRAP samples, which were then purified for RNA along with pre-TRAP samples and used for downstream applications.
(E) Schematic of a cross-sectional view of the E14.5 ventral otocyst, showing three turns of the cochlear duct, surrounded by periotic mesenchyme and otic capsule. Pre-TRAP RNA (representing total input tissue) comes from the cochlear duct epithelium, periotic mesenchyme, and otic capsule (gray). TRAP RNA (representing Fgf20Cre-lineage tissue, which expresses L10a-eGFP) comes from the prosensory domain, Kölliker’s organ, medial wall of the cochlear duct, and some cells of the spiral ganglion (green).
(F) qRT-PCR showing fold change in Twist2 and Id2 expression (normalized to Gadph) in TRAP RNA samples compared to pre-TRAP samples from Fgf20Cre/+;ROSAmTmG/+ E14.5 cochleae. Each dot represents an RNA sample pooled from at least 3 embryos.
To evaluate the TRAP technique for the Fgf20 lineage, we combined the ROSAfsTRAP and Fgf20Cre alleles. The Fgf20Cre allele was made by targeted insertion of a sequence encoding a GFP-Cre fusion protein replacing exon 1 of Fgf20.10 As expected, based on prior expression and lineage tracing experiments, at E14.5, L10a-eGFP fluorescence (green) from Fgf20Cre/+; ROSAfsTRAP/+ cochleae was found in the prosensory domain, Kölliker’s organ, the cochlear duct wall more medial to the Kölliker’s organ, and some cells in the spiral ganglion (Fig. 1B). Also as expected, at P0, L10a-eGFP in Fgf20Cre/+; ROSAfsTRAP cochleae was found in the OC, the GER, and the region more medial to the GER (Fig. 1B).
Another Fgf20-null allele, Fgf20βgal, was made by targeted insertion of a sequence encoding β-Galactosidase replacing exon 1 of Fgf20.9 We combined the Fgf20Cre and Fgf20βgal alleles to generate Fgf20−/− mice (Fgf20Cre/βgal), which maintained the same dosage of Cre as control mice (Fgf20Cre/+). Importantly, based on double fluorescence expression from the ROSAmTmG Cre-reporter allele, the Fgf20Cre lineage (green) did not change in Fgf20Cre/βgal compared to Fgf20Cre/+ cochleae (Fig. 1C). Based on these results, we concluded that Fgf20Cre/+; ROSAfsTRAP/+ and Fgf20Cre/βgal; ROSAfsTRAP/+ mice will allow enrichment for prosensory cell RNA, increasing the sensitivity of RNAseq to identify changes in gene expression within these cells in the absence of FGF20 signaling.
Fgf20Cre TRAPseq enriched for prosensory domain RNA
TRAP experiments were performed at E14.5 (Fig. 1D), based on our previous findings that FGF20 signaling is required for prosensory cell differentiation at E13.5-E15.5.13 In the initial experiment, we collected pre-TRAP (pre-IP) and TRAP (post-IP) RNA from Fgf20Cre/+;ROSAfsTRAP/+ cochleae at E14.5. Pre-TRAP samples were collected prior to IP, representing whole cochlea RNA, including RNA from mesenchyme and otic capsule (Fig. 1E). Quantitative reverse transcription PCR (qRT-PCR) showed enrichment of the prosensory cell marker Id2 and depletion of the mesenchyme marker Twist2 (also called Dermo1) in TRAP RNA samples, compared to pre-TRAP RNA samples (Fig. 1F).10,18
Next, we performed TRAPseq with Fgf20−/+ (Fgf20Cre/+; ROSAfsTRAP/+) control and Fgf20−/− (Fgf20Cre/βgal; ROSAfsTRAP/+) E14.5 cochleae. Fgf20−/+ and Fgf20−/− embryos were generated at a 1:1 ratio. For each litter, cochleae from all control embryos were pooled together for RNA collection, and likewise for Fgf20−/− embryos. Each sample represents RNA from pooled tissue from a minimum of three embryos. In total, 24 libraries were sequenced, consisting of 16 TRAP samples (8 Fgf20−/+ and 8 Fgf20−/−) and 8 pre-TRAP samples (4 Fgf20−/+ and 4 Fgf20−/−). See Experimental Procedures for details.
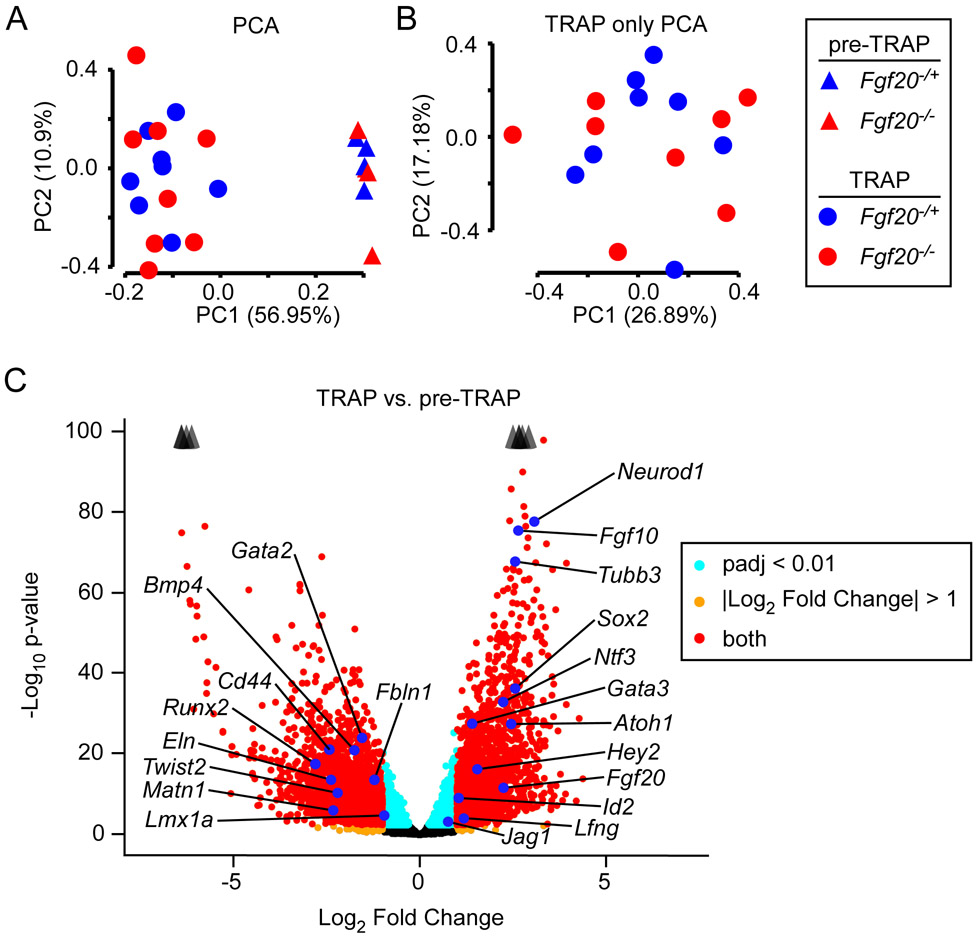
Principal component analysis (PCA) of the 24 RNAseq samples showed separation between pre-TRAP and TRAP samples along PC1 (Fig. 2A). However, there was no separation between Fgf20−/+ vs. Fgf20−/− samples along PC1 or PC2. PCA of only the 16 TRAP samples also did not show separation between Fgf20−/+ vs. Fgf20−/− samples along the first two PCs (Fig. 2B). To assess the efficiency of the TRAP technique, differentially expressed gene (DEG) analysis using DESeq2 was performed to compare pre-TRAP control samples with TRAP control samples (same genotype, Fgf20Cre/+;ROSAfsTRAP/+, for both).19 3850 DEGs were identified with adjusted p-value (padj) < 0.01 and Log2 Fold Change (LFC) < −1 or > 1 (Fig. 2C). Of these, 2017 genes had decreased expression in TRAP samples, compared to pre-TRAP (depleted by TRAP) and 1833 genes had increased expression in TRAP samples, compared to pre-TRAP (enriched by TRAP). Among the genes depleted by TRAP were mesenchymal markers Cd44 and Twist2,10,20 vasculature markers Eln and Fbln1,21,22 and chondrocyte markers Runx2 and Matn1.23,24 This was expected, since periotic mesenchyme and otic capsule were included in the input tissue but did not express L10a-eGFP. Bmp4, Lmx1a, and Gata2, markers of the outer sulcus were depleted as well.25,26 This was also expected, as the outer sulcus also did not express L10a-eGFP (Fig. 1A, B). Among the genes enriched by TRAP were prosensory domain markers Fgf20, Atoh1, Hey2, Sox2, Gata3, and Id2,9,18,27-30 Kölliker’s organ markers Lfng, Fgf10, and Jag1,26 and spiral ganglion markers Neurod1 and Tubb3,31,32 all as expected. Gene set overlap analysis with gene ontology (GO) on genes depleted by TRAP showed biological processes terms “angiogenesis” and “endochondral ossification” among the top terms (Table 1). GO analysis on genes enriched by TRAP showed biological processes terms “sensory perception of sound”, “axon guidance”, and “auditory receptor cell stereocilium organization” among the top terms (Table 2). These results strongly suggest that TRAP enriched for RNA from Fgf20Cre-lineage cells.
Figure 2. Fgf20Cre TRAPseq enriched for prosensory domain RNA.
(A) Principal Component Analysis (PCA) on 24 TRAPseq samples (8 pre-TRAP samples – 4 Fgf20−/+, 4 Fgf20−/−; 16 TRAP samples – 8 Fgf20−/+, 8 Fgf20−/−) showing separation of pre-TRAP and TRAP samples along PC1, but not of Fgf20−/+ and Fgf20−/− samples.
(B) PCA on the 16 TRAP samples (excluding the 8 pre-TRAP samples) also did not show separation between Fgf20−/+ and Fgf20−/− samples along the first two principal components.
(C) Volcano plot showing TRAP vs. pre-TRAP differentially expressed genes. Positive Log2 Fold Change value indicates enrichment by TRAP; negative Log2 Fold Change value indicates depletion by TRAP. Labeled genes represent markers of the prosensory domain, Kölliker’s organ, spiral ganglion, outer sulcus, periotic mesenchyme, and otic capsule. Padj, adjusted p-value for multiple comparisons (Benjamini-Hochberg method). The p-value plotted on y-axis is unadjusted. Arrowheads indicate genes above y-axis range.
Table 1.
Top 12 gene ontology (GO) terms from a list of 2017 differentially expressed genes depleted by TRAP, compared to pre-TRAP samples, in Fgf20Cre/+;ROSAfsTRAP/+ cochleae.
| Rank* | GO ID | GO biological processes term | p-value |
|---|---|---|---|
| 1 | GO:0006954 | inflammatory response | 3.30E-15 |
| 2 | GO:0001525 | angiogenesis | 1.60E-12 |
| 3 | GO:0030198 | extracellular matrix organization | 2.20E-11 |
| 4 | GO:0045766 | positive regulation of angiogenesis | 2.20E-10 |
| 5 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2.70E-10 |
| 6 | GO:0001974 | blood vessel remodeling | 3.20E-10 |
| 7 | GO:0007155 | cell adhesion | 8.80E-10 |
| 8 | GO:0030593 | neutrophil chemotaxis | 2.00E-09 |
| 9 | GO:0002548 | monocyte chemotaxis | 8.70E-09 |
| 10 | GO:0007186 | G-protein coupled receptor signaling pathway | 5.10E-08 |
| 11 | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2.20E-07 |
| 12 | GO:0001958 | endochondral ossification | 2.80E-07 |
Rank determined by p-value (lowest to highest).
Table 2.
Top 12 gene ontology (GO) terms from a list of 1833 differentially expressed genes enriched by TRAP, compared to pre-TRAP samples, in Fgf20Cre/+;ROSAfsTRAP/+ cochleae.
| Rank* | GO ID | GO biological processes term | p-value |
|---|---|---|---|
| 1 | GO:0007605 | sensory perception of sound | 6.30E-09 |
| 2 | GO:0007411 | axon guidance | 3.30E-08 |
| 3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 7.00E-08 |
| 4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 1.20E-06 |
| 5 | GO:0042391 | regulation of membrane potential | 2.00E-06 |
| 6 | GO:0007626 | locomotory behavior | 3.00E-06 |
| 7 | GO:0050885 | neuromuscular process controlling balance | 3.60E-06 |
| 8 | GO:0019228 | neuronal action potential | 7.20E-06 |
| 9 | GO:0014059 | regulation of dopamine secretion | 9.90E-06 |
| 10 | GO:0017158 | regulation of calcium ion-dependent exocytosis | 1.10E-05 |
| 11 | GO:0060088 | auditory receptor cell stereocilium organization | 1.70E-05 |
| 12 | GO:0045665 | negative regulation of neuron differentiation | 2.30E-05 |
Rank determined by p-value (lowest to highest).
Pre-TRAP vs. TRAP DEG analysis data are presented in Supplemental file S1. The top 50 GO terms from GO analysis on genes depleted by TRAP vs. pre-TRAP (2017 DEGs with LFC < −1 and padj < 0.01) are presented in Supplemental file S2. The top 50 GO terms from GO analysis on genes enriched by TRAP vs. pre-TRAP (1833 DEGs with LFC > 1 and padj < 0.01) are represented in Supplemental file S3. In the following text, we focus on the Fgf20−/+ vs. Fgf20−/− TRAPseq DEG analysis. We refer to the TRAP vs. pre-TRAP DEG analysis with terms “enriched by TRAP” and “depleted by TRAP.” “Enriched by TRAP” refers to transcripts with increased abundance in TRAP relative to pre-TRAP RNA from Fgf20Cre/+;ROSAfsTRAP/+ cochleae; “depleted by TRAP” refers to transcripts with decreased abundance in TRAP relative to pre-TRAP RNA from Fgf20Cre/+;ROSAfsTRAP/+ cochleae.
TRAPseq revealed known FGF target genes during organ of Corti differentiation downstream of Fgf20
DEG analysis on Fgf20−/+ vs. Fgf20−/− pre-TRAP samples resulted in, as expected, very few DEGs. In fact, there were only three DEGs with padj < 0.1: Tectb, Calb1, and Fgf20 (data not shown). DEG analysis on Fgf20−/+ vs. Fgf20−/− TRAP samples resulted in 47 DEGs with padj < 0.01 and 104 DEGs with padj < 0.1 (Fig. 3A). GO analysis with the top 362 TRAPseq DEGs (cut-off of padj < 0.5) found among the top 40 terms “sensory perception of sound,” “sensory organ morphogenesis,” “ear development,” and “inner ear receptor cell differentiation” (Table 3). Many neuronal and cell cycle biological processes terms, such as “regulation of neuron differentiation,” “forebrain neuron differentiation,” “regulation of neural precursor cell proliferation,” “cell division,” and “cell cycle arrest” were also among the top terms. Fgf20−/+ vs. Fgf20−/− DEG analysis data are presented in Supplemental file S4. The top 50 GO terms from GO analysis on 362 DEGs with padj < 0.5 are presented in Supplemental file S5.
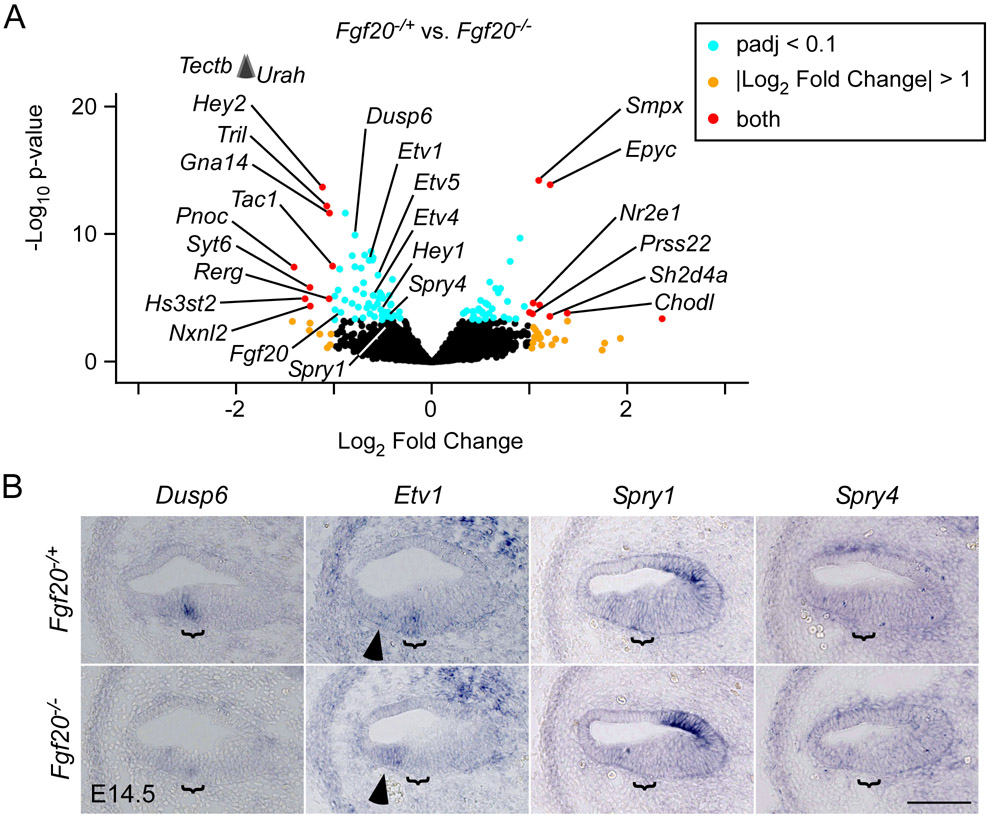
Figure 3. TRAPseq revealed known FGF target genes during organ of Corti differentiation downstream of Fgf20.
(A) Volcano plot showing Fgf20−/+ vs. Fgf20−/− differentially expressed genes. Fgf20, transcripts associated with FGF signaling, and transcripts meeting the criteria padj < 0.1 and Log2 Fold Change < −1 or > 1 are labeled, with the exception of predicted genes and unnamed transcripts. padj, adjusted p-value for multiple comparisons (Benjamini-Hochberg method). The p-value plotted on the y-axis is unadjusted. Arrowheads indicate genes above the y-axis range.
(B) RNA in situ hybridization for known FGF target genes Dusp6, Etv1, Spry1, and Spry4 on sections through the middle turn of E14.5 Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) cochlear ducts. Bracket, prosensory domain. Arrowhead, increased expression of Etv1 in the outer sulcus of Fgf20−/− cochleae. Scale bar, 100 μm.
Table 3.
Select top gene ontology (GO) terms from a list of top 362 Fgf20−/+ vs. Fgf20−/− differentially expressed genes from TRAPseq.
| Rank* | GO ID | GO biological processes term | p-value |
|---|---|---|---|
| 1 | GO:0003184 | pulmonary valve morphogenesis | 5.40E-06 |
| 2 | GO:0045664 | regulation of neuron differentiation | 6.80E-05 |
| 3 | GO:0007605 | sensory perception of sound | 1.50E-04 |
| 5 | GO:0007601 | visual perception | 3.60E-04 |
| 6 | GO:0001709 | cell fate determination | 4.10E-04 |
| 7 | GO:0046426 | negative regulation of JAK-STAT cascade | 4.10E-04 |
| 9 | GO:0021879 | forebrain neuron differentiation | 6.10E-04 |
| 12 | GO:0051301 | cell division | 1.12E-03 |
| 13 | GO:0021795 | cerebral cortex cell migration | 1.21E-03 |
| 14 | GO:0009948 | anterior/posterior axis specification | 1.23E-03 |
| 21 | GO:0090596 | sensory organ morphogenesis | 1.77E-03 |
| 28 | GO:0043583 | ear development | 2.50E-03 |
| 30 | GO:2000177 | regulation of neural precursor cell proliferation | 2.70E-03 |
| 33 | GO:0045596 | negative regulation of cell differentiation | 3.29E-03 |
| 35 | GO:0031175 | neuron projection development | 3.46E-03 |
| 38 | GO:0060113 | inner ear receptor cell differentiation | 4.33E-03 |
| 39 | GO:0007050 | cell cycle arrest | 4.76E-03 |
Rank determined by p-value (lowest to highest).
For Fgf20−/+ vs. Fgf20−/− TRAPseq DEG analysis, we considered padj < 0.1 to be statistically significant. Confirming the validity of the TRAPseq data, DEGs with padj < 0.1 included Fgf20 as well as Hey1, Hey2, Etv4, and Etv5 (Table 4), which we have previously shown by RNA in situ hybridization (ISH) are downregulated in Fgf20−/− vs. Fgf20−/+ cochleae.13 To confirm other DEGs identified by TRAPseq, we examined their expression patterns via ISH in Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) E14.5 cochleae.
Table 4.
Fgf20−/+ vs. Fgf20−/− differentially expressed genes from TRAPseq associated with FGF signaling.
| Rank* | Ensembl ID | Gene | Enrichment^ | Log2FC& | padj# |
|---|---|---|---|---|---|
| 9 | ENSMUSG00000019960 | Dusp6 | enriched | −0.79 | <0.001 |
| 21 | ENSMUSG00000004151 | Etv1 | depleted | −0.72 | <0.001 |
| 36 | ENSMUSG00000017724 | Etv4 | ENRICHED | −0.60 | <0.01 |
| 23 | ENSMUSG00000013089 | Etv5 | - | −0.55 | <0.001 |
| 74 | ENSMUSG00000031603 | Fgf20 | ENRICHED | −0.93 | 0.03 |
| 50 | ENSMUSG00000040289 | Hey1 | ENRICHED | −0.55 | 0.01 |
| 5 | ENSMUSG00000019789 | Hey2 | ENRICHED | −1.12 | <0.001 |
| 106 | ENSMUSG00000037211 | Spry1 | ENRICHED | −0.45 | 0.10 |
| 87 | ENSMUSG00000024427 | Spry4 | depleted | −0.45 | 0.06 |
Rank determined by padj (lowest to highest).
Enrichment by TRAP: results of TRAP vs. pre-TRAP comparison in Fgf20Cre/+;ROSAfsTRAP/+ cochleae. Dash (−) indicates padj > 0.05; otherwise padj < 0.05. “enriched” indicates Log2 Fold Change > 0; “ENRICHED” indicates Log2 Fold Change > 1. “depleted” indicates Log2 Fold Change < 0; “DEPLETED” indicates Log2 Fold Change < −1.
Log2 Fold Change of Fgf20−/+ vs. Fgf20−/− comparison.
Adjusted p-value of Fgf20−/+ vs. Fgf20−/− comparison.
We began with DEGs that have been well-linked to FGF signaling (Table 4) and were downregulated in Fgf20−/− cochleae, such as Dusp6, Etv1, Spry1, and Spry4 (Fig. 3B).33-36 By ISH, Dusp6 (Dual specificity phosphatase 6) was expressed within the prosensory domain in control cochleae, and was almost undetectable in Fgf20−/− cochleae. Etv1 (Ets variant 1) was also expressed within the prosensory domain. Interestingly, while Etv1 expression was absent in the prosensory domain in Fgf20−/− cochleae, it was increased in the outer sulcus (Fig. 3B, arrowhead). Spry1 (Sprouty homolog 1) expression was found in the prosensory domain and outer sulcus, similar to Etv1, Etv4, and Etv5 expression at this stage.13 In Fgf20−/− cochleae, Spry1 was absent from the prosensory domain, but not the outer sulcus, similar to the change in expression patterns of Etv4 and Etv5 in Fgf20−/− cochleae.13 Spry4 (Sprouty homolog 4) expression was found more diffusely in the floor of the cochlear duct, and appeared slightly decreased in the prosensory domain of Fgf20−/− cochleae, although it was difficult to tell definitively by ISH.
TRAPseq revealed many genes associated with cochlea development or hearing loss downstream of Fgf20
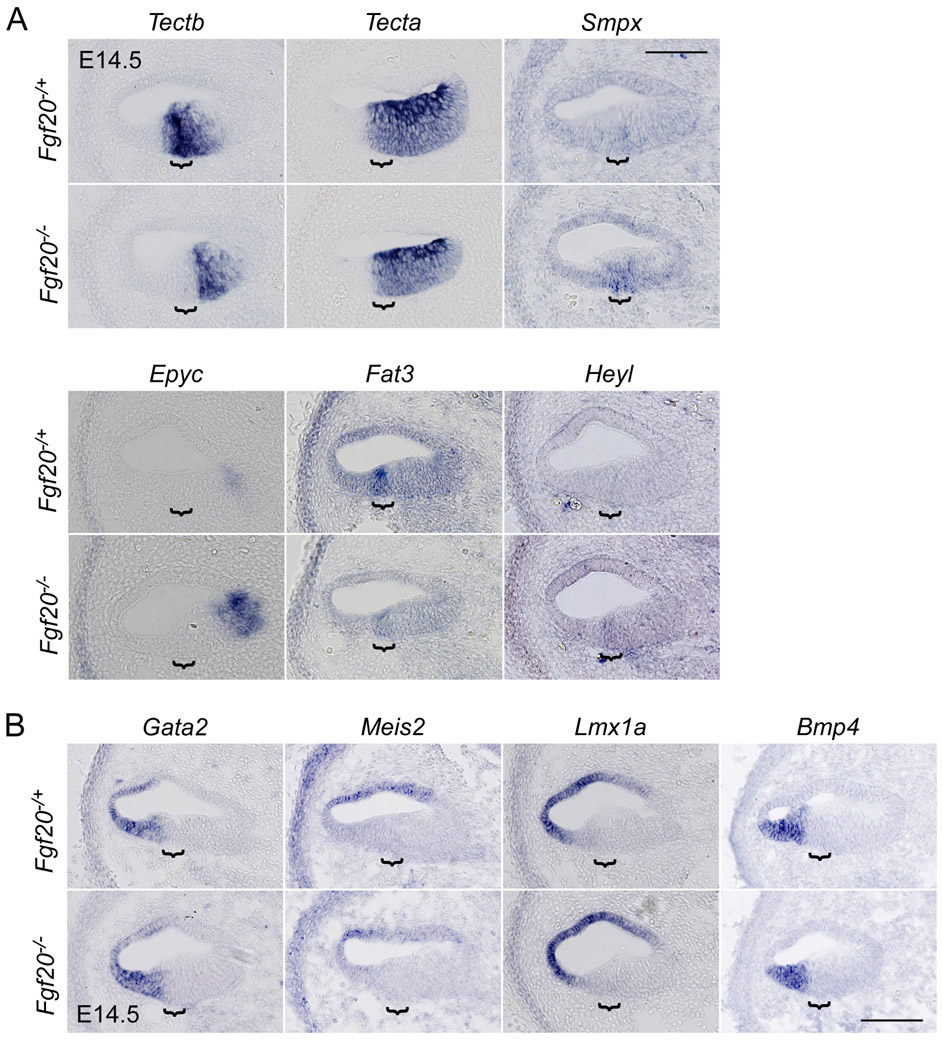
Many DEGs from Fgf20−/+ vs. Fgf20−/− TRAPseq have previously been associated with cochlea development (Table 5). We validated some interesting DEGs via ISH, including Tectb, Smpx, Epyc, Fat3, and Heyl, the results of which were all consistent with TRAPseq data (Fig. 4A). Tectb (Tectorin beta) was expressed in the prosensory domain and Kölliker’s organ and was nearly absent in the prosensory domain of Fgf20−/− cochleae. Meanwhile, Tecta (Tectorin alpha), which also trended towards lower expression per TRAPseq, but did not meet the padj cut-off (padj = 0.22), was not differentially expressed based on ISH. Smpx (Small muscle protein, X-linked) was lowly expressed in the prosensory domain and was increased in Fgf20−/− cochleae. Epyc (Epiphycan) was faintly expressed in the medial cochlear duct wall at this stage and was increased in Fgf20−/− cochleae. Fat3 (FAT atypical cadherin 3) was expressed in the prosensory domain and was decreased in Fgf20−/− cochleae. Heyl (hairy/enhancer-of-split related with YRPW motif-like), belonging to the same family as Hey1 and Hey2, was not expressed in Fgf20−/+ cochleae at E14.5, but was detected in the prosensory domain in Fgf20−/− cochleae.
Table 5.
Fgf20−/+ vs. Fgf20−/− differentially expressed genes from TRAPseq associated with hearing or cochlear development.
| Rank* | Ensembl ID | Gene | Enrichment^ | Log2FC& | padj# |
|---|---|---|---|---|---|
| 236 | ENSMUSG00000021835 | Bmp4 | DEPLETED | 0.41 | 0.38 |
| 10 | ENSMUSG00000028222 | Calb1 | ENRICHED | 0.90 | <0.001 |
| 16 | ENSMUSG00000027555 | Car13 | enriched | −0.64 | <0.001 |
| 331 | ENSMUSG00000003031 | Cdkn1b | - | −0.43 | 0.47 |
| 11 | ENSMUSG00000030862 | Cpxm2 | enriched | −0.62 | <0.001 |
| 12 | ENSMUSG00000030905 | Crym | depleted | −0.69 | <0.001 |
| 4 | ENSMUSG00000019936 | Epyc | depleted | 1.21 | <0.001 |
| 31 | ENSMUSG00000074505 | Fat3 | - | −0.96 | <0.01 |
| 47 | ENSMUSG00000015053 | Gata2 | DEPLETED | 0.55 | <0.01 |
| 28 | ENSMUSG00000032744 | Heyl | DEPLETED | 0.65 | <0.01 |
| 119 | ENSMUSG00000050100 | Hmx2 | − | 0.74 | 0.11 |
| 96 | ENSMUSG00000026686 | Lmx1a | depleted | 0.55 | 0.08 |
| 49 | ENSMUSG00000098318 | Lockd | − | −0.65 | 0.01 |
| 29 | ENSMUSG00000036446 | Lum | − | 0.70 | <0.01 |
| 61 | ENSMUSG00000027210 | Meis2 | DEPLETED | 0.48 | 0.02 |
| 168 | ENSMUSG00000030739 | Myh14 | ENRICHED | −0.41 | 0.23 |
| 97 | ENSMUSG00000004891 | Nes | DEPLETED | −0.78 | 0.08 |
| 83 | ENSMUSG00000060424 | Pantr1 | depleted | −0.51 | 0.05 |
| 53 | ENSMUSG00000045515 | Pou3f3 | depleted | −0.42 | 0.01 |
| 71 | ENSMUSG00000031665 | Sall1 | enriched | −0.49 | 0.03 |
| 177 | ENSMUSG00000049532 | Sall2 | ENRICHED | −0.39 | 0.26 |
| 89 | ENSMUSG00000024565 | Sall3 | enriched | −0.59 | 0.06 |
| 14 | ENSMUSG00000035109 | Shc4 | ENRICHED | −0.60 | <0.001 |
| 3 | ENSMUSG00000041476 | Smpx | ENRICHED | 1.09 | <0.001 |
| 225 | ENSMUSG00000074637 | Sox2 | ENRICHED | −0.45 | 0.37 |
| 18 | ENSMUSG00000061762 | Tac1 | - | −1.01 | <0.001 |
| 161 | ENSMUSG00000037705 | Tecta | ENRICHED | −0.36 | 0.22 |
| 1 | ENSMUSG00000024979 | Tectb | ENRICHED | −1.91 | <0.001 |
| 128 | ENSMUSG00000021779 | Thrb | ENRICHED | 0.45 | 0.13 |
Rank determined by padj (lowest to highest).
Enrichment by TRAP: results of TRAP vs. pre-TRAP comparison in Fgf20Cre/+;ROSAfsTRAP/+ cochleae. Dash (−) indicates padj > 0.05; otherwise padj < 0.05. “enriched” indicates Log2 Fold Change > 0; “ENRICHED” indicates Log2 Fold Change > 1. “depleted” indicates Log2 Fold Change < 0; “DEPLETED” indicates Log2 Fold Change < −1.
Log2 Fold Change of Fgf20−/+ vs. Fgf20−/− comparison.
Adjusted p-value of Fgf20−/+ vs. Fgf20−/− comparison.
Figure 4. Expression analysis of genes identified by TRAPseq associated with cochlea development or hearing loss downstream of Fgf20.
RNA in situ hybridization on sections through the middle turn of E14.5 Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) cochlear ducts. Bracket, prosensory domain. Scale bar, 100 μm.
(A) Genes expressed within the Fgf20Cre lineage: Tectb, Tecta, Smpx, Epyc, Fat3, and Heyl
(B) Genes expressed outside of the Fgf20Cre lineage: Gata2, Meis2, Lmx1a, and Bmp4
TRAPseq also identified a few transcription factors that were depleted by TRAP, but with increased expression in Fgf20−/− cochleae, including Gata2 (GATA binding protein 2), Meis2 (Meis homeobox 2), and Lmx1a (LIM homeobox transcription factor 1 alpha). Depletion by TRAP suggests that they are not highly expressed in the prosensory domain or Kölliker’s organ. By ISH, all three genes were expressed in the outer sulcus and/or roof of the cochlear duct (Fig. 4B). However, ISH did not appear to be sensitive enough to detect differences in the expression of any of these genes between Fgf20−/+ and Fgf20−/− cochleae. Bmp4 (Bone morphogenetic protein 4) was also depleted by TRAP, but not significantly changed in Fgf20−/− cochleae per TRAPseq (padj = 0.38). By ISH Bmp4 was expressed in the outer sulcus and did not show any changes in Fgf20−/− compared to Fgf20−/+ cochleae (Fig. 4B).
TRAPseq revealed decreased expression of cell cycle regulators in the absence of Fgf20
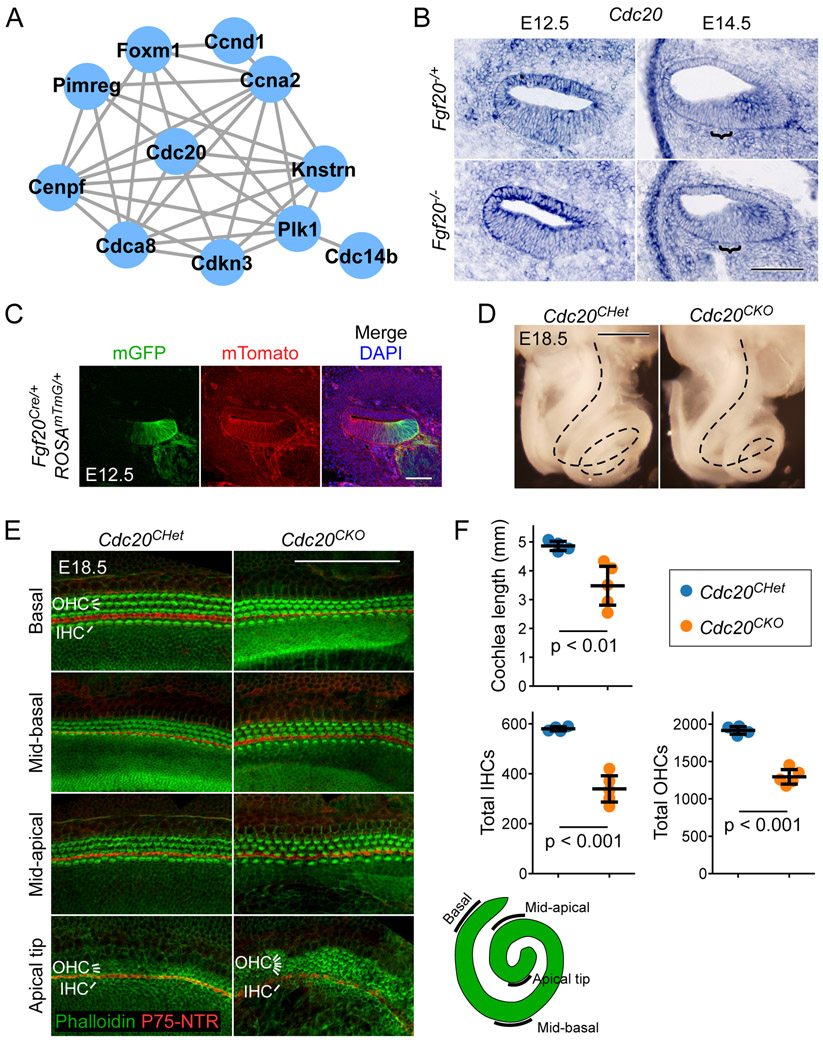
GO analysis on Fgf20−/+ vs. Fgf20−/− TRAPseq DEGs showed that the cell cycle may be affected by the loss of FGF20, with “cell division” and “cell cycle arrest” among the top terms (Table 3). This was confirmed by known and predicted protein-protein interaction (PPI) network identification via the STRING database with the top 192 DEGs, representing those with padj < 0.3.37,38 By far the largest PPI network identified consisted of cell cycle regulators (Fig. 5A). The list of top DEGs indeed showed many genes involved in cell cycle regulation, all of which were decreased in expression in Fgf20−/− cochleae (Table 6).
Figure 5. Conditional deletion of Cdc20 with Fgf20Cre resulted in a shorter cochlea.
(A) The largest protein-protein interaction network identified via the STRING database consisted of genes involved in cell cycle regulation. Lines represent known and predicted protein-protein interactions of high or very high confidence (minimum required interaction score = 0.700).
(B) RNA in situ hybridization for Cdc20 on sections through the middle turn of E12.5 and E14.5 Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) cochlear ducts. Bracket, prosensory domain. Scale bar, 100 μm.
(C) Section through the middle turn of E12.5 Fgf20Cre/+;ROSAmTmG/+ cochlear duct. Cells of the Fgf20Cre lineage express mGFP (mG, green); non-lineage cells express mTomato (mT, red). DAPI, nuclei (blue); scale bar, 100 μm.
(D) Dissected inner ears from E18.5 Cdc20CHet (Fgf20Cre/+;Cdc20flox/+) and Cdc20CKO (Fgf20Cre/+;Cdc20flox/flox) embryos with the otic capsule removed to reveal the coiled cochlea (dotted lines). Scale bar, 0.5 mm.
(E) Whole mount cochlea from E18.5 Cdc20CHet and Cdc20CKO embryos showing one row of inner hair cells (IHC) and three rows of outer hair cells (OHC) marked by phalloidin (green) and separated by inner pillar cells (p75NTR, red). Representative regions from the basal, mid-basal, and mid-apical turns, and apical tip of the cochlea are shown. See schematic showing locations of the turns of the cochlea. At the apical tip, four or more rows of OHCs were frequently observed in Cdc20CHet cochleae. Scale bar, 100 μm.
(F) Quantification of cochlea length and total number of inner and outer hair cells (IHCs and OHCs) in E18.5 Cdc20CHet (n = 4) and Cdc20CKO (n = 5) cochleae. Error bars represent mean ± std. Results were analyzed by Student’s t-test; p-values are shown.
Table 6.
Fgf20−/+ vs. Fgf20−/− differentially expressed genes from TRAPseq associated with cell cycle regulation.
| Rank* | Ensembl ID | Gene | Enrichment^ | Log2FC& | padj# |
|---|---|---|---|---|---|
| 70 | ENSMUSG00000027715 | Ccna2 | depleted | −0.33 | 0.03 |
| 32 | ENSMUSG00000070348 | Ccnd1 | - | −0.53 | <0.01 |
| 120 | ENSMUSG00000033102 | Cdc14b | - | −0.49 | 0.11 |
| 24 | ENSMUSG00000006398 | Cdc20 | depleted | −0.40 | <0.001 |
| 222 | ENSMUSG00000024791 | Cdca5 | - | −0.30 | 0.37 |
| 183 | ENSMUSG00000028873 | Cdca8 | - | −0.33 | 0.28 |
| 197 | ENSMUSG00000019942 | Cdk1 | depleted | −0.29 | 0.31 |
| 206 | ENSMUSG00000026023 | Cdk15 | ENRICHED | −0.45 | 0.32 |
| 121 | ENSMUSG00000037628 | Cdkn3 | depleted | −0.41 | 0.12 |
| 159 | ENSMUSG00000026605 | Cenpf | - | −0.92 | 0.22 |
| 158 | ENSMUSG00000001517 | Foxm1 | depleted | −0.43 | 0.22 |
| 95 | ENSMUSG00000027331 | Knstrn | - | −0.32 | 0.07 |
| 146 | ENSMUSG00000020808 | Pimreg | - | −0.37 | 0.18 |
| 147 | ENSMUSG00000030867 | Plk1 | depleted | −0.38 | 0.18 |
Rank determined by padj (lowest to highest).
Enrichment by TRAP: results of TRAP vs. pre-TRAP comparison in Fgf20Cre/+;ROSAfsTRAP/+ cochleae. Dash (−) indicates padj > 0.05; otherwise padj < 0.05. “enriched” indicates Log2 Fold Change > 0; “ENRICHED” indicates Log2 Fold Change > 1. “depleted” indicates Log2 Fold Change < 0; “DEPLETED” indicates Log2 Fold Change < −1.
Log2 Fold Change of Fgf20−/+ vs. Fgf20−/− comparison.
Adjusted p-value of Fgf20−/+ vs. Fgf20−/− comparison.
Although we have not found that Fgf20 regulates cell cycle progression by itself, Fgf20 does interact with Fgf9 and Sox2 to regulate prosensory progenitor and Kölliker’s organ proliferation, respectively.10,13 We hypothesize, therefore, that the expression changes in cell cycle regulators may reflect these functions of Fgf20. To rule out the possibility that cell cycle regulation contributes to the differentiation and patterning defect found in Fgf20−/− cochleae, we examined the largest hub of the PPI network, Cdc20 (Cell division cycle 20). Cdc20 is a coactivator of the anaphase-promoting complex (APC), the cell cycle-regulated ubiquitin ligase. Interestingly, Cdc20-APC is required for presynaptic axon differentiation in postmitotic neurons in the cerebellum.39
By ISH, Cdc20 is expressed diffusely throughout the developing cochlear duct epithelium at E12.5 (Fig. 5B). By E14.5, its expression is decreased within the prosensory domain (Fig. 5B, brackets), corresponding with prosensory cell cycle exit.40,41 There was no consistently detectable difference in Cdc20 expression between Fgf20−/+ and Fgf20−/− cochleae at E12.5 or E14.5 by ISH.
To examine whether Cdc20 may be involved in HC differentiation, we combined the Fgf20Cre and Cdc20flox alleles to conditionally delete Cdc20 from the Fgf20Cre lineage.42 Fgf20Cre targets parts of the otic epithelium as early as E10.5,10 and by E12.5 targets the medial parts of the cochlear duct floor epithelium (Fig. 5C), similar to E14.5 (Fig. 1C). Fgf20Cre/+;Cdc20flox/flox (Cdc20CKO) cochleae (length: 3.48 ± 0.75 mm) were shorter and more tightly coiled than Fgf20Cre/+;Cdc20flox/+ (Cdc20CHet) cochleae (length: 4.86 ± 0.18 mm) (Fig. 5D, F). Importantly, HCs (labeled by phalloidin, green) in Cdc20CKO cochleae exhibited the normal pattern of one row of IHCs separated from three rows of OHCs by pillar cells (inner pillar cells labeled by P75NTR, red) (Fig. 5E). Interestingly, 4 of 5 Cdc20CKO cochleae examined had 4 or more rows of OHCs at the apical tip (Fig. 5E), which may be the result of a defect in convergent extension. The tighter coil and convergent extension defect found in Cdc20CKO cochleae may be attributable to the loss of Cdc20 expression from the Kölliker’s organ and medial cochlear duct wall. Upon quantification, total number of IHCs and OHCs were decreased in Cdc20CKO (IHC: 339 ± 59; OHC: 1295 ± 108) relative to Cdc20CHet (IHC: 580 ± 9; OHC: 1916 ± 59) cochleae (Fig. 5F); however, this can likely be attributed to the shorter cochlea length.
Sall1 mutant cochleae exhibit an outer hair cell phenotype
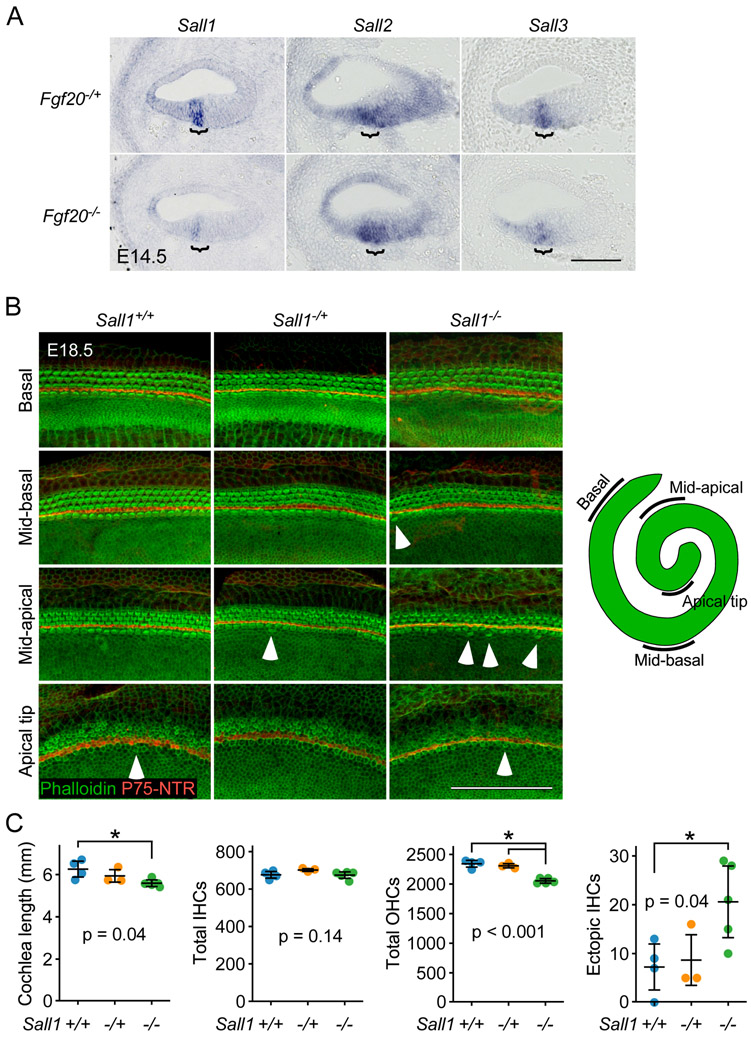
Sall1 (Sal-like 1) and Sall3 (Sal-like 3), members of a family of transcription factors, are expressed in the cochlear duct throughout development.43-46 Both were identified by TRAPseq as decreased in Fgf20−/− cochleae compared to Fgf20−/+ (Table 5). ISH showed that both Sall1 and Sall3 were expressed in the prosensory domain and were decreased in Fgf20−/− cochleae (Fig. 6A). Sall2 (Sal-like 2), another member of the same family, trended towards lower expression per TRAPseq (padj = 0.26). By ISH, Sall2 was also expressed in the prosensory domain, but was not noticeably decreased in Fgf20−/− cochleae (Fig. 6A). Sall4, the remaining member of the Sall family, was filtered out from TRAPseq DEG analysis due to insufficiently low read counts.
Figure 6. Sall1−/− cochleae exhibit an outer hair cell phenotype.
(A) RNA in situ hybridization for Sall1, Sall2, and Sall3 on sections through the middle turn of E14.5 Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) cochlear ducts. Bracket, prosensory domain. Scale bar, 100 μm.
(B) Whole mount cochlea from E18.5 Sall1+/+, Sall1−/+, and Sall1−/− embryos showing inner hair cells and outer hair cells marked by phalloidin (green) and separated by inner pillar cells (p75NTR, red). Representative regions from the basal (5% of total length from the basal tip), mid-basal (33%), and mid-apical (67%) turns, and apical tip (90%) of the cochlea are shown. See schematic showing locations of the turns of the cochlea. Numerous ectopic inner hair cells were found throughout Sall1−/− cochleae, especially towards the apex (arrowheads). Scale bar, 100 μm.
(C) Quantification of cochlea length, total number of inner hair cells (IHCs) and outer hair cells (OHCs), and total number of ectopic IHCs in E18.5 Sall1+/+ (n = 4), Sall1−/+ (n = 3), and Sall1−/− (n = 5) cochleae. Error bars represent mean ± std. Results were analyzed by one-way ANOVA. P-values shown are from the ANOVA. * indicates p < 0.05 from Tukey’s HSD (ANOVA post-hoc).
Importantly, SALL1 has been linked to Townes-Brocks syndrome (TBS) in humans, which causes sensorineural hearing loss, among other developmental defects.16 Mutations in one copy of SALL1 is responsible for TBS, although SALL1 haploinsufficiency may not be the sole causative factor, as Sall1-null mice do not recapitulate the human TBS phenotypes.43 Instead, mice expressing one copy of a Sall1 allele with mutations known to cause TBS, Sall1-ΔZn2-10 (Sall1Δ), mimic TBS defects, including hearing loss.47 This mutation results in a truncated protein encoding the N-terminus of Sall1, which has been shown to mediate transcriptional repression.48 Like wildtype Sall1, the truncated Sall1Δ protein can bind all members of the Sall family,47 and its expression alone in transgenic mice leads to derepression of Sall-regulated genes, resulting in TBS-like phenotypes.49 These results suggest that Sall1Δ may act as a dominant negative and interfere with the transcription-repressor activity of all Sall proteins.
To investigate whether Sall1 may have a role in HC differentiation, we examined Sall1-null (Sall1−/−) cochleae along with cochleae from littermate controls (Sall1+/+ and Sall1−/+) at E18.5.50 HC patterning in Sall1−/+ and Sall1−/− cochleae appeared similar to that in Sall1+/+ cochleae, with mostly one row of IHCs and three rows of OHCs throughout the entire cochlear duct (Fig. 6B). Upon quantification of cochlea length, Sall1−/− cochleae (5.59 ± 0.17 mm) were found to be slightly but statistically significantly shorter than Sall1+/+ (6.26 ± 0.43 mm) but not Sall1−/+ (5.94 ± 0.36 mm) cochleae (Fig. 6C). While the total number of IHCs did not significantly differ among the three genotypes (Sall1+/+: 676 ± 20; Sall1−/+: 701 ± 8; Sall1−/−: 674 ± 19), the total number of OHCs was significantly decreased in Sall1−/− cochleae (2053 ± 43) compared to both Sall1+/+ (2342 ± 66) and Sall1−/+ (2308 ± 43) cochleae (Fig. 6C). These results suggest that there is an OHC-specific developmental defect in Sall1−/− cochleae.
Interestingly, many ectopic IHCs (found outside of the normal row of IHCs) were present along the entire length of the Sall1−/− cochleae, especially towards the apex (Fig. 6B, arrowheads). Upon quantification, the total number of ectopic IHCs was significantly increased in Sall1−/− cochleae (21 ± 8) compared to Sall1+/+ cochleae (7 ± 5) (Fig. 6C). The number of ectopic IHCs was also increased in Sall1−/− cochleae compared to Sall1−/+ cochleae (9 ± 6), but this was not statistically significant (Fig. 6C). This finding suggests a sensory epithelium patterning defect in Sall1−/− cochleae.
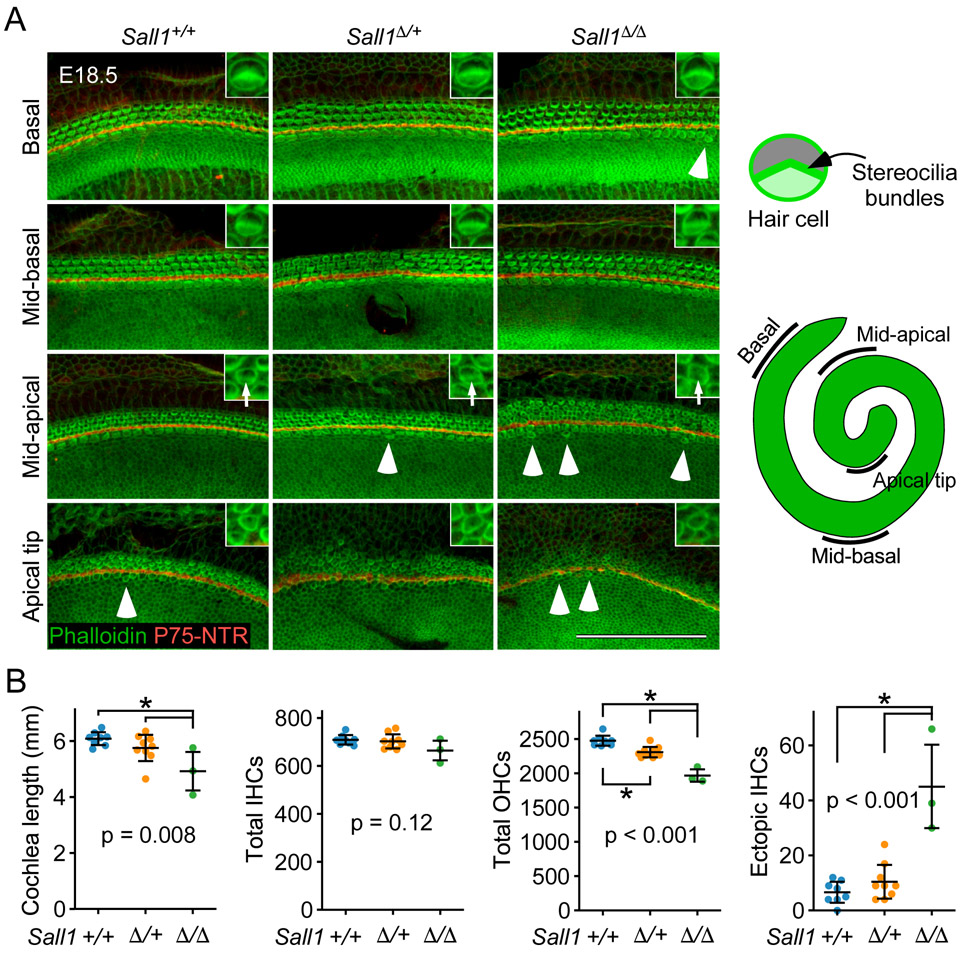
We hypothesized that the dominant negative effects of Sall1Δ may recapture the effects of decreased Sall1 and Sall3 expression found in Fgf20−/− cochleae, producing a phenotype more severe than that of Sall1−/− cochleae and more similar to that of Fgf20−/− cochleae. We examined cochleae from mice heterozygous (Sall1Δ/+) and homozygous (Sall1Δ/Δ) for the Sall1Δ mutation, as well as from wildtype littermate controls (Sall1+/+) at E18.5.47 Most Sall1Δ/Δ embryos die by E16.5.47 However, we were able to obtain three Sall1Δ/Δ embryos that survived to E18.5 (3/84 total embryos across 10 litters, with expected ratio of 1/4). Similar to Sall1−/− cochleae, the overall HC patterning appeared relatively unchanged in Sall1Δ/+ and Sall1Δ/Δ cochleae compared to Sall1+/+ cochleae (Fig. 7A).
Figure 7. Sall1-ΔZn2-10 mutant cochleae exhibit a more severe phenotype than Sall1−/− cochleae.
(A) Whole mount cochlea from E18.5 Sall1+/+, Sall1Δ/+, and Sall1Δ/Δ embryos showing inner hair cells and outer hair cells marked by phalloidin (green) and separated by inner pillar cells (p75NTR, red). Representative regions from the basal (5% of total length from the basal tip), mid-basal (33%), and mid-apical (67%) turns, and apical tip (90%) of the cochlea are shown. See schematic to the right showing locations of the turns of the cochlea. Inset: 3.8x magnified image of a representative OHC showing stereocilia bundle formation (arrows in mid-apical region). Numerous ectopic inner hair cells were found throughout Sall1Δ/Δ cochleae, especially towards the apex (arrowheads). Scale bar, 100 μm.
(B) Quantification of cochlea length, total number of inner hair cells (IHCs) and outer hair cells (OHCs), and total number of ectopic IHCs in E18.5 Sall1+/+ (n = 8), Sall1Δ/+ (n = 9), and Sall1Δ/Δ (n = 3) cochleae. Error bars represent mean ± std. Results were analyzed by one-way ANOVA. P-values shown are from the ANOVA. * indicates p < 0.05 from Tukey’s HSD (ANOVA post-hoc).
Upon quantification of cochlea length, Sall1Δ/Δ cochleae (4.92 ± 0.84 mm) were found to be slightly but statistically significantly shorter than both Sall1+/+ (6.09 ± 0.25 mm) and Sall1Δ/+ (5.75 ± 0.50 mm, including a possible outlier at 4.65 mm) cochleae (Fig. 7B). The total number of IHCs appeared to be slightly decreased in Sall1Δ/Δ cochleae (664 ± 50) compared to Sall1+/+ (709 ± 21) and Sall1Δ/+ (702 ± 31) cochleae, but this was not statistically significant. Notably, the total number of OHCs was significantly decreased in Sall1Δ/+ cochleae (2311 ± 80) compared to Sall1+/+ cochleae (2478 ± 79), and was further significantly decreased in Sall1Δ/Δ cochleae (1969 ± 111) (Fig. 7B).
Similar to Sall1−/− cochleae, many ectopic IHCs were found throughout the length of Sall1Δ/Δ cochleae, especially towards the apex (Fig. 7A, arrowheads). Quantification of these ectopic IHCs showed a statistically significant increase in Sall1Δ/Δ (45 ± 19) compared to Sall1+/+ (7 ± 4) and Sall1Δ/+ (10 ± 7) cochleae (Fig. 7B).
In addition, the HCs in Sall1Δ/Δ cochleae appeared less mature than those found in comparative regions of Sall1+/+ and Sall1Δ/+ cochleae based on F-actin organization in phalloidin stained samples (Fig. 7A, inset). This was most apparent in the mid-apical turns, where stereocilia bundles appeared relatively well-formed in Sall1+/+ and Sall1Δ/+ cochleae (arrows in Fig. 7A, inset). In Sall1Δ/Δ cochleae, however, the stereocilia in this region appeared more immature and disorganized (arrow in Fig. 7A inset), resembling those found in the less mature apical tip [HC differentiation and maturation occur in a base-to-apex gradient,2 therefore, more apical HCs are less mature]. In the apical tip, the F-actin networks at the HC cortex in Sall1Δ/Δ cochleae appeared less dense than those found in Sall1+/+ and Sall1Δ/+ cochleae, as indicated by weaker phalloidin labeling (Fig. 7A, inset).
Overall, the Sall1Δ/Δ phenotype is similar to, but more severe than the Sall1−/− phenotype. Compared to their respective littermate Sall1+/+ cochleae, Sall1−/− cochleae were 10.7% shorter, while Sall1Δ/Δ cochleae were 19.2% shorter. Sall1−/− cochleae had a 12.3% decrease in total OHC number, while Sall1Δ/Δ cochleae had a 20.5% decrease in total OHC number. Sall1−/− cochleae had a 300% increase in the number of ectopic IHCs, while Sall1Δ/Δ cochleae had a 643% increase in the number of ectopic IHCs.
DISCUSSION
We have adapted the TRAP technique to study a relatively small population of difficult-to-isolate cells: cochlear prosensory cells. TRAP using Fgf20Cre combined with ROSAfsTRAP effectively enriched for RNA from the Fgf20Cre lineage at E14.5. We believe the pre-TRAP vs. TRAP DEG analysis provides a useful dataset for identifying genes enriched in the prosensory domain, Kölliker’s organ, and spiral ganglion of the developing cochlea.
TRAPseq DEG analysis comparing Fgf20−/+ and Fgf20−/− E14.5 cochlea samples showed decreased expression of known FGF20 signaling targets in the cochlea at this stage, as well as known FGF signaling targets in other tissues, confirming the validity of the TRAP technique. Just as importantly, TRAPseq DEGs did not include genes that we have previously shown are not downstream of FGF20, but that have been shown to be downstream of FGFR1: Cdkn1b and Sox2 (Table 5).10,11,13 Interestingly, Lockd, a non-coding RNA near the Cdkn1b locus and co-expressed with Cdkn1b,51 was significantly decreased in Fgf20−/− cochleae (Table 5). The expression of a few other genes previously shown to be downstream of FGFR1 during cochlea development, such as Fgf10, Hes5, and Ntf3,11,12 were also not significantly changed in Fgf20−/− compared to Fgf20−/+ cochleae. MAP3K4 (also called MEKK4) activity has been associated with FGF20-FGFR1 signaling during hair cell differentiation.52 However, while MAP3K4 protein levels were previously shown to be decreased in Fgf20−/− relative to Fgf20−/+ cochleae at P0, Map3k4 transcript levels were not changed in Fgf20−/− compared to Fgf20−/+ cochleae at E14.5, per TRAPseq.
As with any large data experiment, false positives and negatives are expected. Here, we used a lenient false discovery rate of 0.1 to evaluate Fgf20−/+ vs. Fgf20−/− TRAPseq results to reduce the number of false negatives at the cost of increasing false positives. While we were able to confirm many TRAPseq DEGs via ISH, as well as confirm the expression of several non-significantly changed genes as unchanged via ISH, there were discrepancies between TRAPseq and ISH results. Besides false positivity, another possible and interesting explanation for the discrepancies is that TRAPseq specifically identifies differences in ribosome-bound RNA. Such differences may not always be reflected in the whole RNA population detected by ISH.
Another caveat is that the Fgf20Cre-TRAP enrichment process is not perfect, due to technical limitations and the inclusion of Kölliker’s organ and spiral ganglion cells in the Fgf20Cre lineage. RNA from these sources dilute the RNA from the target prosensory cell population, reducing the power of TRAPseq in detecting changes within this population.
TRAPseq identified DEGs previously associated with cochlea development or hearing downstream of Fgf20
Many DEGs identified by Fgf20−/+ vs. Fgf20−/− TRAPseq have known roles in cochlea development (Table 5). Altered expression of these genes, therefore, may contribute to the Fgf20−/− phenotype. Importantly, we do not know what proportion of these DEGs are directly regulated by FGF20, and what proportion may be indirectly regulated or are markers of dysregulated differentiation. Here, we highlight some of these DEGs.
Fat3 (FAT atypical cadherin 3), encoding a mammalian homolog of the Drosophila cell adhesion molecule Fat, is required for the normal patterning of OHCs, along with Fat4.53 Fat3-null cochleae exhibits a small loss of OHCs from the base of the cochlea and a slight gain of OHCs in the mid-apex. We hypothesize that the decreased expression of Fat3 may contribute to the OHC patterning defect in Fgf20−/− cochleae.
Cpxm2 (Carboxypeptidase X 2) is one of the three genes on chromosome 7 deleted in the head bobber mouse line, which exhibits deafness and vestibular defects.54,55 However, how Cpxm2 deletion contributes to deafness in these mice has not been elucidated.
Tectb (Tectorin beta) is expressed in the prosensory domain and encodes a major glycoprotein in the tectorial membrane required for normal hearing.56,57 Interestingly, Tecta, another gene encoding a glycoprotein in the tectorial membrane, trended towards lower expression (not statistically significantly) but did not show decreased expression by ISH. The composition of the tectorial membrane in Fgf20−/− cochleae has not been studied.
Thrb (Thyroid hormone receptor beta) is expressed in the OC, GER, spiral ligament, and spiral ganglion in the neonatal cochlea. Thrb-mutant mice (both null and mutants with disrupted thyroid hormone binding) have severe hearing loss attributed to disruption of postnatal morphogenesis of the tectorial membrane.58-62 Interestingly, Thrb trended towards increased expression per TRAPseq (padj = 0.13).
Myh14 (Myosin, heavy polypeptide 14) is one of the genes encoding Myosin II. It is expressed in both developing HCs and SCs in the prenatal OC. Myosin II is required for patterning and convergent extension in the cochlea.63 A convergence and extension defect may contribute to the shortened length of Fgf20−/− cochleae. Myh14 trended towards decreased expression per TRAPseq (padj = 0.23).
Smpx (Small muscle protein, X-linked), previously shown to be expressed in HCs,64 is associated with heritable progressive hearing loss.65-67 However, Smpx-null mice have not been shown to have a hearing defect or much of an overt developmental phenotype.68 Given that Smpx is expressed in HCs, its increased expression in the prosensory domain of Fgf20−/− cochleae may reflect the premature onset of HC differentiation in these mice.
Epyc (Epiphycan), encoding a proteoglycan expressed in mature nonsensory regions of the cochlear duct, is required for normal hearing.69 Its increased expression in Fgf20−/− cochleae may also reflect premature onset of differentiation, although it has not been shown that the Kölliker’s organ and medial cochlear duct wall undergoes premature differentiation in Fgf20−/− cochleae.
Hey1 and Hey2 (hairy/enhancer-of-split related with YRPW motif 1 and 2) have been shown to be downstream of FGF20 signaling in the developing cochlea and are required to prevent premature HC differentiation.13,70 Unlike Hey1 and Hey2, Heyl is not expressed in the prosensory domain at E14.5. However, it can be detected in the developing Kölliker’s organ and differentiating supporting cells in the prosensory domain at E16.5 through at least P0.71-73 The increased expression of Heyl in Fgf20−/− cochleae may also reflect premature onset of differentiation. Additionally, Fgf20 expression in the developing OC decreases as HCs differentiate,9 corresponding with the onset of Heyl expression. This suggests that Fgf20 may inhibit Heyl expression directly. Notably, Heyl−/− mice have a normal number of organ of Corti HCs at P0.72 However, the exact role of Heyl in cochlea development has not been elucidated. It is further unclear what effect ectopic/premature expression of Heyl in the prosensory domain may have on OC differentiation.
TRAPseq identified DEGs with unknown functions in cochlea development downstream of Fgf20
Most of the DEGs identified by Fgf20−/+ and Fgf20−/− TRAPseq have no known roles in cochlea development. However, some of these are related to genes with known roles in cochlea development, suggesting possible redundancy. Here, we highlight some of the most interesting ones.
Dusp6 (Dual specificity phosphatase 6) is a known downstream target of FGF signaling and is a downstream target of FGF20 signaling in the olfactory system.36,74 Mice heterozygous for a Dusp6-null allele exhibit hearing loss, attributed to malformed otic capsule and ossicles.75 While Dusp6 is known to be expressed in the prosensory domain and the OC,76 which we confirm, its role in the development of these structures has not been investigated.
Etv4 (Ets variant 4) and Etv5 (Ets variant 5) have been shown to be downstream of FGF20/FGFR1 signaling in the developing cochlea.8,13 However, Etv1, the third member of the PEA3 group of Ets transcription factors, has not been associated with cochlea development. We show here that Etv1 expression is decreased in the prosensory domain in Fgf20−/− cochleae, while its expression is increased in the outer sulcus. This is potentially a significant pattern change, as the Fgf20−/− phenotype is more severe in the outer compartment of the OC, directly adjacent to the outer sulcus/LER. Investigating whether this increase in expression in the outer sulcus contributes to the Fgf20−/− phenotype will be addressed in future experiments.
Other DEGs with unclear functional significance but that are known to be expressed in the cochlea include the following (Table 5):
Pou3f3 (POU domain, class 3, transcription factor 3): expressed in SCs and mesenchymal cells in the cochlea.77 Based on ISH from the Eurexpress atlas, it is also expressed in the cochlear duct floor at E14.5 (http://www.eurexpress.org euxassay_019559).78 However, analysis of the Pou3f3-null mouse cochlea did not reveal any apparent developmental defects.77 Despite this, auditory and vestibular impairments have been reported in a Pou3f3 (Pou3f3L423P) mutant mouse line.79 Interestingly, the expression of Pantr1 (Pou3f3 adjacent noncoding transcript 1), a lncRNA that shares a bidirectional promoter with Pou3f3,80 was also decreased in Fgf20−/− cochleae per TRAPseq, suggesting disrupted activity at the promoter. In addition, Rorb (RAR-related orphan receptor beta), found to be increased in Fgf20−/− cochleae by TRAPseq, has an antagonistic interaction with Pou3f3 during cell fate specification in the developing neocortex.81
Calb1 (Calindin 1): expressed in mature HCs.82 Upregulation may represent premature onset of HC differentiation in Fgf20−/− cochleae.
Crym (Crystallin, mu): a thyroid hormone binding protein, highly expressed in nonsensory regions of the cochlea in adult rats.83
Shc4 (SHC family, member 4): an adaptor protein expressed in the cochlear duct floor at E14.5 and E15.5.84
Car13 (Carbonic anhydrase 13): expressed in nonsensory regions of the cochlea and the mesenchyme at E15.5 and neonatal stages.85
Tac1 (Tachykinin 1): reported to be expressed in the cochlear epithelium during development.86
Lum (Lumican): expressed in the otic capsule, some mesenchyme, and nonsensory regions of the cochlear duct.87
Nes (Nestin): expressed in the spiral ganglion and parts of the prosensory domain at E14.5 and E15.5, as well as in mature SCs.88,89
Nonsensory cell markers are upregulated in Fgf20−/− cochleae
Fgf20−/+ vs. Fgf20−/− TRAPseq also identified a few transcription factors expressed in the outer sulcus and other nonsensory cochlear epithelium: Gata2, Meis2, and Lmx1a.25,90-94 Interestingly, they were all increased in Fgf20−/− cochleae per TRAPseq suggesting that undifferentiated progenitors in Fgf20−/− cochlea may have adopted a nonsensory identity. Two other outer sulcus/nonsensory epithelial markers, Hmx2 and Bmp4,95,96 also trended toward increased expression in Fgf20−/− cochleae, albeit not significantly (padj = 0.11 and 0.38, respectively). Bmp4 is interesting because of its importance in patterning the outer sulcus, prosensory domain, and Kölliker’s organ.26
Examining the expression of these genes by ISH did not reveal noticeable changes between Fgf20−/+ and Fgf20−/− cochleae. We hypothesize that because TRAP depletes for the outer sulcus and roof of the cochlear duct, TRAPseq is highly sensitive to the expression of markers of these regions in the prosensory domain. Therefore, TRAPseq may be much more sensitive than ISH to detect small changes in the expression of genes such as Gata2, Meis2, and Lmx1a, which may represent a shift in the boundary between the prosensory domain and outer sulcus.
Cell cycle regulators are downregulated in Fgf20−/− cochleae
TRAPseq revealed many differentially expressed cell cycle regulators. We have shown before that Fgf20 by itself does not appear to regulate the cell cycle or proliferation in the developing cochlea. However, Fgf20 is redundant with Fgf9 in indirectly regulating prosensory progenitor proliferation at earlier developmental stages (E11.5-E12.5) and is redundant with Sox2 in regulating Kölliker’s organ proliferation at E14.5.10,13 Therefore, we believe the finding of differentially expressed cell cycle regulators may reflect the redundant and stage-specific functions of Fgf20 in regulating proliferation. As expected, while Cdc20 conditional-null cochleae are short, they do not exhibit the HC differentiation or patterning defect found in Fgf20−/− cochleae. We conclude that the ~10-20% decrease in length of Fgf20−/− cochleae may be attributable to decreased expression of these cell cycle regulators in sensory progenitors. It is also possible that Fgf20 has a previously unidentified role in regulating prosensory cell cycle exit, and decreased expression of these cell cycle regulator genes are reflective of premature cell cycle exit.
Fgf20 is upstream of Sall1, a gene implicated in human sensorineural hearing loss
We found that members of the Sall family, Sall1, Sall2, and Sall3, are expressed in the prosensory domain at E14.5, consistent with previous reports of Sall1 and Sall3 expression at around this stage.44,46 Sall1, Sall3, and potentially Sall2 showed decreased expression by TRAPseq and ISH in Fgf20−/− cochleae, suggesting that they are likely regulated by FGF20 signaling. Notably, in the kidney, Sall1 expression has been shown to be regulated by FGF signaling.97 As mentioned previously, mutations in SALL1 causes Townes-Brocks syndrome (TBS) in humans, an autosomal dominant disorder with variable presentation of phenotypes including sensorineural hearing loss.16 Sall1Δ/+ mice mimic TBS developmental defects, including hearing loss.47 Whether cochlea development is affected in Sall1−/− and Sall1Δ mice has not been reported.
We found that Sall1Δ/+ had relatively normal HC patterning, but exhibited a small decrease in the total number of OHCs. This is reminiscent of the Fgf20−/− and Fgf20;Sox2 compound mutant phenotypes, in which OHCs are the most sensitive to the loss of FGF20.9,13 We are not sure, however, how much this reduction in the number of OHCs contributes to the hearing defect found in these mice.
Sall1−/− and Sall1Δ/Δ cochleae both exhibited more severe developmental defects than Sall1Δ/+ cochleae, including shorter cochlea length, and larger decreases in the total number of OHCs. We do not know whether the decrease in HC number is solely attributable to the shorter length of these cochleae. It is possible that both the HC and cochlea length phenotypes are the result of a defect in prosensory progenitor proliferation, such as that found in Fgf20;Fgf9 double mutant mice,10 or the result of a defect in prosensory specification, such as that found in Sox2 mutant mice.28 It is also possible that the decrease in HC number is due to a defect in differentiation, similar to that found in Fgf20−/− cochleae.
Interestingly, both Sall1−/− and Sall1Δ/Δ cochleae contained numerous ectopic IHCs, found outside of the normal row of IHCs, a patterning defect that again is reminiscent of Fgf20−/− and Fgf20;Sox2 compound mutant phenotypes.9,13 Furthermore, Sall1Δ/Δ cochleae also appeared to exhibit a delay in the apical progression of HC maturation, similar to Fgf20−/− cochleae.9 The interaction between Fgf20, Sox2, and Sall1 is a topic to explore in future studies.
The Sall1Δ/Δ organ of Corti phenotype is consistently similar but more severe than that of Sall1−/− cochleae. Since the Sall1Δ truncated protein acts as a dominant negative on all members of the Sall family,47,49 this suggests that Sall2 and/or Sall3 have similar roles as Sall1 in regulating cochlea development. Based on these results, we conclude that the decreased expression of Sall1 and Sall3 likely contributes to the OHC number and IHC patterning defects found in Fgf20−/− cochleae.
Conclusions
The Fgf20−/− cochlea phenotype includes loss of two-thirds of OHCs and outer supporting cells, abnormal patterning of the remaining HCs and SCs, shorter cochlea length, premature onset of differentiation, and delayed apical progression of differentiation and maturation. Here, we did not identify one single gene that can account for the majority of the Fgf20−/− phenotype. However, we identified many FGF20-regulated genes that may contribute to parts of the phenotype. For instance, Hey1, Hey2, and possibly Heyl may account for the premature onset of differentiation phenotype; Sall1, Sall3, and Fat3 may account for the OHC differentiation, patterning, and delay in maturation phenotypes; and cell cycle regulators such as Cdc20 may account for the progenitor proliferation phenotypes in Fgf20;Fgf9 and Fgf20;Sox2 compound mutants. We conclude that FGF20 is likely not a straightforward outer HC and SC differentiation signal. Rather, the dramatic Fgf20−/− phenotype may be the result of disruptions to a combination of FGF20-regulated processes, including prosensory progenitor proliferation, differentiation, maturation, and timing of differentiation. Given the complexity of OC development, we hypothesize that small disturbances to such processes can lead to much larger defects in overall development.
EXPERIMENTAL PROCEDURES
Mice
All studies performed were in accordance with the Institutional Animal Care and Use Committee at Washington University in St. Louis (protocol #20190110 and #20170258).
Mice were group housed with littermates, in breeding pairs, or in a breeding harem (two females to one male), with food and water provided ad libitum. Mice were of mixed sexes and maintained on a mixed C57BL/6J x 129X1/SvJ genetic background, except Sall1- and Sall1Δ mice, which were maintained on an ICR genetic background. The following mouse lines were used:
Fgf20Cre: knockin allele containing a sequence encoding a GFP-Cre fusion protein replacing exon 1 of Fgf20, resulting in a null mutation.10
Fgf20βgal: knockin allele containing a sequence encoding β-galactosidase (βgal) replacing exon 1 of Fgf20, resulting in a null mutation.9
ROSAfsTRAP: knockin allele containing a loxP-Stop-loxP sequence followed by a sequence encoding L10a-eGFP, targeted to the ubiquitously expressed ROSA26 locus. Upon Cre-mediated recombination, the ribosomal protein L10a-eGFP is expressed.15
ROSAmTmG: knockin allele containing a sequence encoding a membrane-localized tdTomato (mT) flanked by loxP sequences, followed by a sequence encoding a membrane-localized eGFP (mG), targeted to the ubiquitously expressed ROSA26 locus. In the absence of Cre-mediated recombination, mT is expressed; upon Cre-mediated recombination, mG is expressed instead.98
Sall1-: knockin allele containing a sequence encoding GFP inserted in frame into exon 2 of Sall1, resulting in a null mutation.50
Sall1-ΔZn2-10 (Sall1Δ): mutant allele expressing a truncated Sall1 protein designed to mimic a mutation that causes Townes-Brocks syndrome.47
Cdc20flox: allele containing loxP sequences flanking exon 2 of Cdc20; upon Cre-mediated recombination, results in a null allele.42
Translating ribosome affinity purification (TRAP)
Affinity matrix preparation: for each immunoprecipitation (IP):
30 μl of Streptavidin MyOne T1 Dynabeads (Invitrogen, 65602) were washed in 1x PBS using an end-over-end tube rotator and a magnet, and resuspended in 88 μl of 1x PBS and conjugated to 12 μl of 1 μg/μl biotinylated protein L (Pierce 29997) in PBS for 35 min at room temperature (RT) with gentle end-over-end mixing on a tube rotator. Conjugated beads were then washed with 1x PBS + 3% IgG and protease-free BSA (Jackson ImmunoResearch, 001-000-162) x5, followed by three washes in low-salt buffer (20 mM HEPES KOH, pH 7.4, 10 mM MgCl2, 150 mM KCl, 1% NP-40 [Sigma I8896–50ML], 0.5 mM DTT [Sigma, 646563], 100 μg/ml cycloheximide [Sigma C4859-1ML]). Conjugated beads were then resuspended in low-salt buffer and mixed with 50 μg each of anti-GFP antibodies Htz-GFP-19C8 and Htz-GFP-19F7 (Memorial Sloan-Kettering Monoclonal Antibody Facility) overnight at 4°C with gentle end-over-end mixing to make the affinity matrix. Immediately before IP, the affinity matrix was washed in low-salt buffer x3.
Sample collection:
E14.5 embryos were harvested, on ice, from a mating producing a 1:1 ratio of Fgf20Cre/+;ROSAfsTRAP/+ and Fgf20Cre/βgal;ROSAfsTRAP/+ progeny. Embryos were staged based on vaginal plug (E0.5 at noon on the day plug is found) and on interdigital webbing. Embryos with too much or too little interdigital webbing were not harvested. Embryos were genotyped by LacZ staining to look for Fgf20βgal expression in back skin hair follicles (back skin from embryos were incubated in 2 mM MgCl2, 5 mM K3, 5 mM K4, 0.02% NP-40, and 1 mg/ml X-gal in N,N-dimethylformamide in 1x PBS for 30 min at 37°C, protected from light). Ventral otocysts from the embryos were dissected out in dissection buffer (1x HBSS, 2.5 mM HEPES-KOH, pH 7.4, 35 mM glucose, 4 mM NaHCO3, 100 μg/ml cycloheximide), separated from the dorsal otocyst (vestibule) without removal of the otic capsule, and pooled together by genotype. Each sample contained pooled ventral otocysts from 3-7 embryos. Pooled ventral otocysts were homogenized in lysis buffer (20 mM HEPES KOH, pH 7.4, 150 mM KCl, 10 mM MgCl2, EDTA-free protease inhibitors (Roche, 04693159001), 0.5 mM DTT, 100 μg/ml cycloheximide, 10 μl/ml rRNasin (Promega N2515), 10 μl/ml Superasin (Applied Biosystems, AM2696) using a pre-chilled Kontes homogenizer (Kontes, 885512-0020). To remove the nuclear fraction, homogenized samples were centrifuged for 10 min at 2000 g, 4°C. The supernatant (S2) was mixed with 1/8 volume of 10% NP-40 and 300 mM DHPC (reconstituted in lysis buffer; Avanti Polar Lipids 850306P) and incubated for 10 min on ice. To remove the mitochondrial fraction, samples were then centrifuged for 15 min at 20,000 g, 4°C. 60 μl of the supernatant (S20) was saved as the pre-IP (pre-TRAP) control. The pre-TRAP S20 samples were incubated at 4°C until the RNA purification step, which was performed in conjunction with TRAP samples. The rest of the S20 was used for IP.
Immunoprecipitation:
S20 was mixed with the affinity matrix for 24 hours at 4°C with end-over-end mixing. The mixture (TRAP sample) was washed in high-salt buffer (20 mM HEPES KOH, pH 7.4, 10 mM MgCl2, 350 mM KCl, 1% NP-40, 0.5 mM DTT, 100 μg/ml cycloheximide, 1 μl/ml rRNasin, 1 μl/ml Superasin) for 2 min at RT, x4.
RNA purification:
the Arcturus Picopure RNA Isolation Kit (Thermo Fisher, 12204-01) was used to isolate RNA from pre-TRAP and TRAP samples according to manufacturer’s instructions. RNA was eluted in 13 μl of elution buffer. Ventral otocysts from 3-7 embryos ranged between 4-20 ng of TRAP RNA. RNA samples were stored at −80°C until use in downstream applications.
Quantitative RT-PCR
cDNA was synthesized from pre-TRAP and TRAP RNA using the iScript Select cDNA Synthesis Kit (Bio-Rad, #170-8841) and quantified using TaqMan Fast Advanced Master Mix (Life Technologies, 4444557) with TaqMan assay probes for Twist2 and Id2. Gapdh was used as normalization control. Results were analyzed by the ΔΔCT method (normalized to Gapdh, then normalized to pre-TRAP). Each sample represents TRAP RNA from one litter.
cDNA library preparation and sequencing
cDNA library preparation and sequencing were performed at the Genome Technology Access Center (GTAC) at Washington University (gtac.wustl.edu). RNA samples were analyzed on an Agilent 2100 Bioanalyzer; all sequenced RNA samples had an RNA Integrity Number (RIN) of ≥ 8.8. Clontech SMARTer Ultra Low Input RNA for Illumina Sequencing - HV kit (Takara Bio, 634828), which utilizes both oligo(dT) and random priming, was used for cDNA library preparation and amplification. The TRAPseq results presented are from two sequencing experiments. cDNA library preparation was performed independently in the two experiments. In both experiments, 8 TRAP samples (4 Fgf20−/+ and 4 Fgf20−/−) and 4 pre-TRAP samples (2 Fgf20−/+ and 2 Fgf20−/−) were sequenced on one Illumina HiSeq 3000 lane, with single reads, 1 × 50 bps. 24 samples were sequenced in total between the two experiments (12 samples multiplexed per lane per experiment). Sequencing produced between 22 and 38 million reads per sample.
Bioinformatic analysis
Basecalling was performed with Illumina RealTimeAnalysis software. The resulting bcl files were demultiplexed with Illumina’s bclToFastq2. Both steps were performed by GTAC.
Alignment:
Reads were mapped to GRCm38.p5 (Ensemble, GCA_000001635.7) using STAR aligner,99 with the GRCm38.91 annotation file (Ensembl).100 Default parameters were used, except for the following: multi-sample 2-pass, with default settings on first pass and sjdbFileChrStartEnd (for novel splice junctions), ScoreMinOverLread=0.4, MatchNminOverLread=0.4, MismatchNmax=5 on second pass (these parameters gave the most consistent unmapped reads % across all 24 TRAPseq samples). 95-99.5% of reads were mapped per sample.
Counting and DEG analysis:
Analyses were performed in R using packages from Bioconductor (bioconductor.org). BAM files were indexed and sorted using Rsamtools.101 Gene models were defined using the GRCm38.91 annotation file (Ensembl) with GenomicFeatures.102 Reads were counted using the SummarizeOverlaps method (mode = Union) from the package GenomicAlignments.102 Genes were filtered out from downstream analysis if less than 8 of 24 samples had 25 or more reads. PC analysis showed separation between the 8 pre-TRAP samples and 16 TRAP samples along PC1, and also separation between sequencing experiment 1 and experiment 2 along PC2. Removal of Unwanted Variation from RNA-Seq Data (RUVSeq) was used to correct for this batch effect (RUVs function, k = 1).103 DESeq2 with RUVs correction factors was used for DEG analysis, with alpha = 0.1 and Benjamini-Hochberg multiple-comparisons correction.19
Pathway analysis:
gene ontology (GO) analysis was performed using the Bioconductor package topGO with the following parameters: nodeSize = 10; ontology = biological processes (BP); algorithm = elim; statistic = fisher’s exact test.104 Protein-protein interaction network analysis was performed using STRING version 11.0 with the following parameters: active interaction sources include textmining, experiments, databases, co-expression, neighborhood, gene fusion, co-occurrence; minimum required interaction score = high confidence (0.700).37,38
Sample preparation and sectioning for histology and in situ hybridization
For whole mount cochleae, inner ears were dissected out of P0 pups and fixed in 4% PFA in PBS overnight at 4°C with gentle agitation. Samples were then washed x3 in PBS. Cochleae were dissected away from the vestibule, otic capsule, and periotic mesenchyme with Dumont #55 Forceps (Roboz, RS-5010). The roof of the cochlear duct was opened by dissecting away the stria vascularis and Reissner’s membrane; tectorial membrane was removed to expose hair and supporting cells.
For sectioning, heads from E14.5 embryos were fixed in 4% PFA in PBS overnight at 4°C with gentle agitation. Samples were then washed x3 in PBS and cryoprotected in 15% sucrose in PBS overnight and then in 30% sucrose in PBS overnight. Samples were embedded in Tissue-Tek O.C.T. compound (VWR International, 4583) and frozen on dry ice. Serial horizontal sections through base of the head were cut at 12 μm with a cryostat, dried at room temperature, and stored at −80°C until use.
RNA in situ hybridization
Probe preparation:
plasmids containing the following mouse cDNA inserts were used to make digoxigenin-labeled riboprobes, and were linearized and transcribed with the indicated restriction enzyme (New England Biolabs) and RNA polymerase (New England Biolabs) according to the manufacturers’ instructions, with DIG RNA Labeling Mix (Sigma-Aldrich, 11277073910). Probes were treated with RNase-free DNase I (Sigma-Aldrich, 04716728001) for 15 min at 37°C, then hydrolyzed in 40 mM NaHCO3, 60 mM Na2CO3 at 60°C for 5-30 min, to make 300-400 bp probes. Dusp6 (412 bp, Acc65I, T7, gift of S. Mansour),76 Etv1 (2500 bp, SpeI, T7, addgene #16282, gift of S.-H. Huh), Spry1 (1500 bp, EcoRI, T7),33 Spry4 (900 bp, EcoRI, T7),33 Tectb (2746 bp, EcoRI, T7, gift of D. Wu),56 Tecta (4382 bp, NotI, T7, gift of D. Wu),56 Epyc (1522 bp, EcoRI, T7, Image clone 4037028), Heyl (1895, BamHI, T7, Image clone 40142873), Gata2 (700 bp, BamHI, T3, gift of D. Wu), Meis2 (~5000 bp, EcoRI, T3, gift of Y. Yang), Lmx1a (600 bp, SphI, Sp6, gift of D. Wu),105 and Bmp4 (AccI, T7).106
Sall1 probe was made from pBluescript KS (+)-Sall1 cDNA clone (GenBank accession no. NM_001371070.1 nucleotides 856 to 1283 inserted into the EcoRV site), which was linearized with HindIII and transcribed with T7. Sall2 probe was made from pBluescript II KS (+)-Sall2 cDNA clone (GenBank accession no. NM_015772 nucleotides 687 to 1113 inserted into the PstI/EcoRI site), which was linearized with EcoRI and transcribed with T7. Sall3 probe was made from pBluescript II KS (+)-Sall3 cDNA clone (GenBank accession no. NM_001369133 nucleotides 575 to 1124 inserted into the HincII site) which was linearized with XbaI and transcribed with T3).
Smpx probe was made from PCR product containing T7 promoter sequence (gift of J. Bok),64 and transcribed with T7. Fat3 probe was made by PCR amplification of a plasmid containing 945 bp sequence of Fat3 cDNA (gift of L. Goodrich) using primers F: 5’-ACAGCTCGCATCAGCTTCGTGT-3’ and R (containing T7 promoter sequence): 5’-GGATCCTAATACGACTCACTATAGGGAGTGCTTTGCAGGGTTCTCAGGC-3’, and transcribed with T7. Cdc20 probe was made by PCR amplification of mouse tail genomic DNA using primers F: 5’-GTTCGGGAGAGCTGAGTACG-3’ and R (containing T7 promoter sequence): 5’-GGATCCTAATACGACTCACTATAGGGAGGCTGTGTGATCTGTTGGCG-3’, and transcribed with T7.
Frozen section in situ hybridization:
frozen slides were warmed for 20 min at room temperature and then 5 min at 50°C on a slide warmer. Sections were fixed in 4% PFA in PBS for 20 min at room temperature, washed x2 in PBS and treated with pre-warmed 10 μg/ml Proteinase K (Sigma-Aldrich, 03115828001) in PBS for 1 min (E12.5 samples) or 7 min (E14.5 samples) at 37°C. Sections were then fixed in 4% PFA in PBS for 15 min at room temperature, washed x2 in PBS, acetylated in 0.25% acetic anhydrate in 0.1M Triethanolamine, pH 8.0, for 10 min, and washed again in PBS. Sections were then placed in pre-warmed hybridization buffer (50% formamide, 5x SSC buffer, 5 mM EDTA, 50 μg/ml yeast tRNA) for 3 h at 60–65°C in humidified chamber for prehybridization. Sections were then hybridized in 10 μg/ml probe/hybridization buffer overnight (12–16 h) at 60-65°C. The next day, sections were washed in 1x SSC for 10 min at 60°C, followed by 1.5x SSC for 10 min at 60°C, 2x SSC for 20 min at 37°C x2, and 0.2x SSC for 30 min at 60°C x2. Sections were then washed in KTBT (0.1 M Tris, pH 7.5, 0.15 M NaCl, 5 mM KCl, 0.1% Triton X-100) at room temperature and blocked in KTBT + 20% sheep serum + 2% Blocking Reagent (Sigma-Aldrich, 11096176001) for 4 h. Blocking Reagent was dissolved in 100 mM Maleic acid, 150 mM NaCl, pH 7.5. Sections were then incubated in sheep anti-Digoxigenin-AP, Fab fragments (1:1000, Sigma-Aldrich, 11093274910) in KTBT + 20% sheep serum + 2% Blocking Reagent overnight at 4°C. Sections were then washed x3 in KTBT for 30 min at room temperature, and then washed x2 in NTMT (0.1 M Tris, pH 9.5, 0.1 M NaCl, 50 mM MgCl2, 0.1% Tween 20) for 15 min. Sections were next incubated in NTMT + 1:200 NBT/BCIP Stock Solution (Sigma-Aldrich, 11681451001) in the dark at room temperature until color appeared. Sections were then washed in PBS, post-fixed in 4% PFA in PBS for 15 min and washed x2 in PBS. Finally, sections were dehydrated in 30% and then 70% methanol, 5 min each, followed by 100% methanol for 15 min. Sections were then rehydrated in 70% and 30% methanol and then PBS, 5 min each, and mounted in 95% glycerol.
Immunofluorescence
Whole mount:
cochleae were incubated in PBS + 0.5% Tween-20 (PBSTw) for 1 h to permeabilize. Cochleae were then blocked using PBSTw + 5% donkey serum for 1 h and then incubated in PBSTw + 1% donkey serum with the primary antibody overnight at 4°C. Cochleae were then washed x3 in PBS and incubated in PBS + 1% Tween-20 with the secondary antibody. After wash in PBS x3, cochleae were mounted in 95% glycerol or VectaShield antifade mounting medium with DAPI (Vector Labs, H-1200) with the sensory epithelium facing up.
Frozen slides were warmed for 30 min at room temperature and washed in PBS before incubating in PBS + 0.5% Triton X-100 (PBST) for 1 h to permeabilize the tissue. Sections were then blocked using in PBST + 5% donkey serum for 1 h and then incubated in PBST + 1% donkey serum with the primary antibody overnight at 4°C in a humidified chamber. Sections were then washed x3 in PBS and incubated in PBS + 1% Triton X-100 with the secondary antibody. After wash in PBS x3, slides were mounted in VectaShield antifade mounting medium with DAPI.
The following compounds and antibodies were used:
Alexa Fluor 488-conjugated Phalloidin (1:50, Invitrogen, A12379)
Rabbit anti-P75NTR (1:300, EMD Millipore, AB1554)
Alexa Fluor 555 goat anti-rabbit IgG (1:500, Invitrogen, A21428)
Imaging
Light microscopy:
slides were scanned using a Hamamatsu NanoZoomer slide scanning system with a 20x objective. Images were then processed with the NanoZoomer Digital Pathology (NDP.view2) software. 3D specimens were imaged using an Olympus SZXZ110 stereo microscope equipped with an Olympus DP70 camera.
Fluorescent microscopy was performed using a Zeiss LSM 700 confocal or Zeiss Axio Imager Z1 with Apotome 2, with z-stack step-size determined based on objective lens type (10x or 20x), as recommended by the ZEN software (around 1 μm). Fluorescent images shown are maximum projections. Images were processed with ImageJ (imagej.nih.gov).
Image analysis and quantification
Measurements and cell quantification (using the Cell Counter plugin by Kurt De Vos) were performed using Fiji.107 Total cochlear duct length was defined as the length from the very base of the cochlea to the very tip of the apex, along the tunnel of Corti, measured on whole-mount cochlea. Hair cells and stereocilia bundles were identified via Phalloidin, which binds to F-actin.108 Inner pillar cells were labeled via P75NTR.109 Inner hair cells (IHCs) were differentiated from outer hair cells (OHCs) based on their neural/abneural location, respectively, relative to P75NTR-expressing inner pillar cells. For total cell counts, IHCs and OHCs were counted along the entire length of the cochlea.
Statistical analysis and plotting
All figures were made in Canvas X (ACD systems). RNA sequencing data analysis and plotting were performed using R (r-project.org) in R studio (rstudio.com) PCA graphs were made using the plotPCA function from the Bioconductor package RUVSeq; volcano plots were made using modified code from Stephen Turner (gist.github.com/stephenturner). See Bioinformatic analysis section for more details on RNA sequencing data analysis. All other data analysis and plotting were performed using Python (python.org) in Jupyter Notebook (jupyter.org) with the following libraries: Pandas (pandas.pydata.org), Seaborn (seaborn.pydata.org), NumPy (numpy.org) and SciPy (scipy.org). Plotting was performed using the Matplotlib library (matplotlib.org). Statistics (t-test, one-way ANOVA, and two-way ANOVA) were performed using the SciPy module Stats; Tukey’s HSD was performed using the Statsmodels package (statsmodels.org). All comparisons of two means were performed using two-tailed, unpaired Student’s t-test. For comparisons of more than two means, one-way ANOVA was used. For significant ANOVA results at α = 0.05, Tukey’s HSD was performed for post-hoc pair-wise analysis. In all cases, p < 0.05 was considered statistically significant. All statistical details can be found in the figures and figure legends. In all cases, each sample (each data point in graphs) represents one animal. Based on similar previous studies, a sample size of 3-5 was determined to be appropriate. Error bars represent mean ± standard deviation. For qualitative comparisons (comparing expression via immunofluorescence or RNA in situ hybridization), at least three samples were examined per genotype. All images shown are representative. No data were excluded from analysis.
Supplementary Material
S1 – preTRAP vs. TRAP DEG analysis
S2 – GO analysis of DEGs depleted by TRAP
S3 – GO analysis of DEGs enriched by TRAP
S4 – Fgf20−/+ vs. Fgf20−/− DEG analysis
S5 – GO analysis of Fgf20−/+ vs. Fgf20−/− DEGs
Key findings:
Translating Ribosome Affinity Purification (TRAP) targeting the Fgf20-Cre lineage enriched for organ of Corti progenitor RNA
TRAP combined with RNAseq (TRAPseq) identified genes downstream of FGF20 during organ of Corti differentiation
FGF20 regulates Sall1, gene implicated in human sensorineural hearing loss
Sall1 mutant mice exhibit a decrease in the number of organ of Corti outer hair cells
ACKNOWLEDGEMENTS
We thank J. Dougherty and his laboratory for their help with the TRAP technique, B. Zhang (Department of Developmental Biology), and T. Sinnwell and M. Heinz (Genome Technology Access Center in the Department of Genetics) for their help with RNA sequencing and analysis. We thank L. Robbins for generating the Sall1, Sall2, and Sall3 plasmids used for making in situ probes. We also thank C. Ferguson for supplying us with Cdc20flox mice. This work was funded by the Department of Developmental Biology at Washington University, NIH/National Institute on Deafness and Other Communication Disorders grant DC017042 (DMO), the Washington University Institute of Clinical and Translational Sciences which is, in part, supported by the NIH/National Center for Advancing Translational Sciences, CTSA grant UL1TR002345 (JIT471 to DMO), March of Dimes grant 6-FY13-127 (MR), and the Rare Disease Foundation/BC Children’s Hospital Foundation. GTAC is partially supported by NCI Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant UL1TR000448 from the National Center for Research Resources (NCRR). HOPE Center Alafi Neuroimaging Laboratory is supported by NCRR grant 1S10RR027552, the Auditory and Vestibular Microscopy and Digital Imaging Core is supported by NIH grant P30 DC004665. The authors have no conflicts of interest to disclose.
List of abbreviations:
- DEG
differentially expressed gene
- GER
greater epithelial ridge
- GO
gene ontology
- HC
hair cell
- IHC
inner hair cell
- ISH
in situ hybridization
- IP
immunoprecipitation
- KO
Kölliker’s organ
- LER
lesser epithelial ridge
- OC
organ of Corti
- OHC
outer hair cell
- OS
outer sulcus
- padj
adjusted p-value
- PCA
principal component analysis
- PD
prosensory domain
- SC
supporting cell
- TBS
Townes-Brocks syndrome
- TRAP
translating ribosome affinity purification
- TRAPseq
TRAP combined with next generation RNA sequencing
Data availability:
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE148380 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148380).110
REFERENCES
- 1.Allen SB, Goldman J. Syndromic Sensorineural Hearing Loss (SSHL) In: StatPearls. StatPearls Publishing; 2019. Accessed April 8, 2020 http://www.ncbi.nlm.nih.gov/books/NBK526088/ [PubMed] [Google Scholar]
- 2.Basch ML, Brown RM, Jen H-I, Groves AK. Where hearing starts: the development of the mammalian cochlea. J Anat. 2016;228(2):233–254. doi: 10.1111/joa.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowl MR, Brown SDM. Genetic landscape of auditory dysfunction. Hum Mol Genet. 2018;27(R2):R130–R135. doi: 10.1093/hmg/ddy158 [DOI] [PubMed] [Google Scholar]
- 4.Wu DK, Kelley MW. Molecular Mechanisms of Inner Ear Development. Cold Spring Harb Perspect Biol. 2012;4(8):a008409. doi: 10.1101/cshperspect.a008409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong ACY, Ryan AF. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front Aging Neurosci. 2015;7:58. doi: 10.3389/fnagi.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamasoba T, Lin FR, Someya S, Kashio A, Sakamoto T, Kondo K. Current concepts in age-related hearing loss: epidemiology and mechanistic pathways. Hear Res. 2013;303:30–38. doi: 10.1016/j.heares.2013.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corwin JT, Warchol ME. Auditory hair cells: structure, function, development, and regeneration. Annu Rev Neurosci. 1991;14:301–333. doi: 10.1146/annurev.ne.14.030191.001505 [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 Is Required for Sensory Epithelial Specification in the Developing Cochlea. J Neurosci. 2008;28(23):5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huh S-H, Jones J, Warchol ME, Ornitz DM. Differentiation of the Lateral Compartment of the Cochlea Requires a Temporally Restricted FGF20 Signal. PLOS Biol. 2012;10(1):e1001231. doi: 10.1371/journal.pbio.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh S-H, Warchol ME, Ornitz DM. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife. 2015;4:e05921. doi: 10.7554/eLife.05921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono K, Kita T, Sato S, et al. FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation. PLOS Genet. 2014;10(1):e1004118. doi: 10.1371/journal.pgen.1004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirvola U, Ylikoski J, Trokovic R, Hébert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35(4):671–680. doi: 10.1016/s0896-6273(02)00824-3 [DOI] [PubMed] [Google Scholar]
- 13.Yang LM, Cheah KSE, Huh S-H, Ornitz DM. Sox2 and FGF20 interact to regulate organ of Corti hair cell and supporting cell development in a spatially-graded manner. PLOS Genet. 2019;15(7):e1008254. doi: 10.1371/journal.pgen.1008254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Zhang Y, Ma Q, et al. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci U S A. 2013;110(38):15395–15400. doi: 10.1073/pnas.1304124110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet. 1998;18(1):81–83. doi: 10.1038/ng0198-81 [DOI] [PubMed] [Google Scholar]
- 17.Rossmiller DR, Pasic TR. Hearing loss in Townes-Brocks syndrome. Otolaryngol Head Neck Surg. 1994;111(3 Pt 1):175–180. doi: 10.1177/01945998941113P103 [DOI] [PubMed] [Google Scholar]
- 18.Jones JM, Montcouquiol M, Dabdoub A, Woods C, Kelley MW. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J Neurosci. 2006;26(2):550–558. doi: 10.1523/JNEUROSCI.3859-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu H, Mitsuhashi N, Klein A, et al. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells. 2006;24(4):928–935. doi: 10.1634/stemcells.2005-0186 [DOI] [PubMed] [Google Scholar]
- 21.Cooley MA, Kern CB, Fresco VM, et al. Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev Biol. 2008;319(2):336–345. doi: 10.1016/j.ydbio.2008.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karnik SK, Brooke BS, Bayes-Genis A, et al. A critical role for elastin signaling in vascular morphogenesis and disease. Development. 2003;130(2):411–423. doi: 10.1242/dev.00223 [DOI] [PubMed] [Google Scholar]
- 23.Fujita T, Azuma Y, Fukuyama R, et al. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol. 2004;166(1):85–95. doi: 10.1083/jcb.200401138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei M, Luo J, Chen Q. Enhancing and maintaining chondrogenesis of synovial fibroblasts by cartilage extracellular matrix protein matrilins. Osteoarthritis Cartilage. 2008;16(9):1110–1117. doi: 10.1016/j.joca.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilleväli K, Matilainen T, Karis A, Salminen M. Partially overlapping expression of Gata2 and Gata3 during inner ear development. Dev Dyn. 2004;231(4):775–781. doi: 10.1002/dvdy.20185 [DOI] [PubMed] [Google Scholar]
- 26.Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP Signaling Is Necessary for Patterning the Sensory and Nonsensory Regions of the Developing Mammalian Cochlea. J Neurosci. 2010;30(45):15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch Signaling Is Not Necessary for Prosensory Induction in the Mouse Cochlea: Insights from a Conditional Mutant of RBPj. J Neurosci. 2011;31(22):8046–8058. doi: 10.1523/JNEUROSCI.6671-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiernan AE, Pelling AL, Leung KKH, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434(7036):1031–1035. doi: 10.1038/nature03487 [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Deng M, Xie X, et al. GATA3 controls the specification of prosensory domain and neuronal survival in the mouse cochlea. Hum Mol Genet. 2013;22(18):3609–3623. doi: 10.1093/hmg/ddt212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7(12):1310–1318. doi: 10.1038/nn1349 [DOI] [PubMed] [Google Scholar]
- 31.Locher H, de Groot JCMJ, van Iperen L, Huisman MA, Frijns JHM, de S Lopes SMC. Distribution and Development of Peripheral Glial Cells in the Human Fetal Cochlea. PLOS ONE. 2014;9(1):e88066. doi: 10.1371/journal.pone.0088066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 Induces Neuronal Formation in the Developing Mammalian Cochlea. J Neurosci. 2010;30(2):714–722. doi: 10.1523/JNEUROSCI.3852-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minowada G, Jarvis LA, Chi CL, et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126(20):4465–4475. [DOI] [PubMed] [Google Scholar]
- 34.Münchberg SR, Steinbeisser H. The Xenopus Ets transcription factor XER81 is a target of the FGF signaling pathway. Mech Dev. 1999;80(1):53–65. doi: 10.1016/s0925-4773(98)00193-2 [DOI] [PubMed] [Google Scholar]
- 35.Willardsen M, Hutcheson DA, Moore KB, Vetter ML. The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech Dev. 2014;131:57–67. doi: 10.1016/j.mod.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang LM, Huh S-H, Ornitz DM. FGF20-Expressing, Wnt-Responsive Olfactory Epithelial Progenitors Regulate Underlying Turbinate Growth to Optimize Surface Area. Dev Cell. 2018;46(5):564–580.e5. doi: 10.1016/j.devcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Kim AH, Yamada T, et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326(5952):575–578. doi: 10.1126/science.1177087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126(8):1581–1590. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y-S, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133(15):2817–2826. doi: 10.1242/dev.02453 [DOI] [PubMed] [Google Scholar]
- 42.Manchado E, Guillamot M, de Cárcer G, et al. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell. 2010;18(6):641–654. doi: 10.1016/j.ccr.2010.10.028 [DOI] [PubMed] [Google Scholar]
- 43.Nishinakamura R, Matsumoto Y, Nakao K, et al. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128(16):3105–3115. [DOI] [PubMed] [Google Scholar]
- 44.Ott T, Kaestner KH, Monaghan AP, Schütz G. The mouse homolog of the region specific homeotic gene spalt of Drosophila is expressed in the developing nervous system and in mesoderm-derived structures. Mech Dev. 1996;56(1):117–128. doi: 10.1016/0925-4773(96)00516-3 [DOI] [PubMed] [Google Scholar]
- 45.Ott T, Parrish M, Bond K, Schwaeger-Nickolenko A, Monaghan AP. A new member of the spalt like zinc finger protein family, Msal-3, is expressed in the CNS and sites of epithelial/mesenchymal interaction. Mech Dev. 2001;101(1–2):203–207. doi: 10.1016/s0925-4773(00)00552-9 [DOI] [PubMed] [Google Scholar]
- 46.Parrish M, Ott T, Lance-Jones C, Schuetz G, Schwaeger-Nickolenko A, Monaghan AP. Loss of the Sall3 Gene Leads to Palate Deficiency, Abnormalities in Cranial Nerves, and Perinatal Lethality. Mol Cell Biol. 2004;24(16):7102–7112. doi: 10.1128/MCB.24.16.7102-7112.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiefer SM, Ohlemiller KK, Yang J, McDill BW, Kohlhase J, Rauchman M. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes–Brocks syndrome birth defects. Hum Mol Genet. 2003;12(17):2221–2227. doi: 10.1093/hmg/ddg233 [DOI] [PubMed] [Google Scholar]
- 48.Kiefer SM, McDill BW, Yang J, Rauchman M. Murine Sall1 represses transcription by recruiting a histone deacetylase complex. J Biol Chem. 2002;277(17):14869–14876. doi: 10.1074/jbc.M200052200 [DOI] [PubMed] [Google Scholar]
- 49.Kiefer SM, Robbins L, Barina A, Zhang Z, Rauchman M. SALL1 truncated protein expression in Townes-Brocks syndrome leads to ectopic expression of downstream genes. Hum Mutat. 2008;29(9):1133–1140. doi: 10.1002/humu.20759 [DOI] [PubMed] [Google Scholar]
- 50.Takasato M, Osafune K, Matsumoto Y, et al. Identification of kidney mesenchymal genes by a combination of microarray analysis and Sall1-GFP knockin mice. Mech Dev. 2004;121(6):547–557. doi: 10.1016/j.mod.2004.04.007 [DOI] [PubMed] [Google Scholar]
- 51.Paralkar VR, Taborda CC, Huang P, et al. Unlinking an lncRNA from Its Associated cis Element. Mol Cell. 2016;62(1):104–110. doi: 10.1016/j.molcel.2016.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haque K, Pandey AK, Zheng H-W, Riazuddin S, Sha S-H, Puligilla C. MEKK4 Signaling Regulates Sensory Cell Development and Function in the Mouse Inner Ear. J Neurosci. 2016;36(4):1347–1361. doi: 10.1523/JNEUROSCI.1853-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139(10):1806–1820. doi: 10.1242/dev.077461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buniello A, Hardisty-Hughes RE, Pass JC, Bober E, Smith RJ, Steel KP. Headbobber: a combined morphogenetic and cochleosaccular mouse model to study 10qter deletions in human deafness. PLOS One. 2013;8(2):e56274. doi: 10.1371/journal.pone.0056274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Somma G, Alger HM, McGuire RM, et al. Head bobber: an insertional mutation causes inner ear defects, hyperactive circling, and deafness. J Assoc Res Otolaryngol. 2012;13(3):335–349. doi: 10.1007/s10162-012-0316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rau A, Legan PK, Richardson GP. Tectorin mRNA expression is spatially and temporally restricted during mouse inner ear development. J Comp Neurol. 1999;405(2):271–280. doi: [DOI] [PubMed] [Google Scholar]
- 57.Russell IJ, Legan PK, Lukashkina VA, Lukashkin AN, Goodyear RJ, Richardson GP. Sharpened cochlear tuning in a mouse with a genetically modified tectorial membrane. Nat Neurosci. 2007;10(2):215–223. doi: 10.1038/nn1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forrest D, Erway LC, Ng L, Altschuler R, Curran T. Thyroid hormone receptor beta is essential for development of auditory function. Nat Genet. 1996;13(3):354–357. doi: 10.1038/ng0796-354 [DOI] [PubMed] [Google Scholar]
- 59.Griffith AJ, Szymko YM, Kaneshige M, et al. Knock-in mouse model for resistance to thyroid hormone (RTH): an RTH mutation in the thyroid hormone receptor beta gene disrupts cochlear morphogenesis. J Assoc Res Otolaryngol. 2002;3(3):279–288. doi: 10.1007/s101620010092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaukua N, Shahidi MK, Konstantinidou C, et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature. 2014;513(7519):551–554. doi: 10.1038/nature13536 [DOI] [PubMed] [Google Scholar]
- 61.Ng L, Cordas E, Wu X, Vella KR, Hollenberg AN, Forrest D. Age-related hearing loss and degeneration of cochlear hair cells in mice lacking thyroid hormone receptor β1. Endocrinology. 2015;156(10):3853–3865. doi: 10.1210/en.2015-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharlin DS, Visser TJ, Forrest D. Developmental and cell-specific expression of thyroid hormone transporters in the mouse cochlea. Endocrinology. 2011;152(12):5053–5064. doi: 10.1210/en.2011-1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto N, Okano T, Ma X, Adelstein RS, Kelley MW. Myosin II regulates extension, growth and patterning in the mammalian cochlear duct. Development. 2009;136(12):1977–1986. doi: 10.1242/dev.030718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon H, Lee DJ, Kim MH, Bok J. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol. 2011;44(1):69–78. doi: 10.5115/acb.2011.44.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdelfatah N, Merner N, Houston J, et al. A Novel Deletion in SMPX Causes a Rare form of X-Linked Progressive Hearing Loss in Two Families Due to a Founder Effect. Hum Mutat. 2013;34(1):66–69. doi: 10.1002/humu.22205 [DOI] [PubMed] [Google Scholar]
- 66.Huebner AK, Gandia M, Frommolt P, et al. Nonsense mutations in SMPX, encoding a protein responsive to physical force, result in X-chromosomal hearing loss. Am J Hum Genet. 2011;88(5):621–627. doi: 10.1016/j.ajhg.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schraders M, Haas SA, Weegerink NJD, et al. Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am J Hum Genet. 2011;88(5):628–634. doi: 10.1016/j.ajhg.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palmer S, Groves N, Schindeler A, et al. The Small Muscle-Specific Protein Csl Modifies Cell Shape and Promotes Myocyte Fusion in an Insulin-like Growth Factor 1–Dependent Manner. J Cell Biol. 2001;153(5):985–998. doi: 10.1083/jcb.153.5.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanada Y, Nakamura Y, Ishida Y, et al. Epiphycan is specifically expressed in cochlear supporting cells and is necessary for normal hearing. Biochem Biophys Res Commun. 2017;492(3):379–385. doi: 10.1016/j.bbrc.2017.08.092 [DOI] [PubMed] [Google Scholar]
- 70.Benito-Gonzalez A, Doetzlhofer A. Hey1 and Hey2 Control the Spatial and Temporal Pattern of Mammalian Auditory Hair Cell Differentiation Downstream of Hedgehog Signaling. J Neurosci. 2014;34(38):12865–12876. doi: 10.1523/JNEUROSCI.1494-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham-McDonogh O. Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev Biol. 2008;316(1):87–99. doi: 10.1016/j.ydbio.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 Regulation by FGF Provides a Notch-Independent Mechanism for Maintaining Pillar Cell Fate in the Organ of Corti. Dev Cell. 2009;16(1):58–69. doi: 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol. 2011;352(2):329–340. doi: 10.1016/j.ydbio.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 74.Dickinson RJ, Keyse SM. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci. 2006;119(22):4607–4615. doi: 10.1242/jcs.03266 [DOI] [PubMed] [Google Scholar]
- 75.Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134(1):167–176. doi: 10.1242/dev.02701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urness LD, Li C, Wang X, Mansour SL. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev Dyn. 2008;237(1):163–169. doi: 10.1002/dvdy.21380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mutai H, Nagashima R, Sugitani Y, Noda T, Fujii M, Matsunaga T. Expression of Pou3f3/Brn-1 and its genomic methylation in developing auditory epithelium. Dev Neurobiol. 2009;69(14):913–930. doi: 10.1002/dneu.20746 [DOI] [PubMed] [Google Scholar]
- 78.Diez-Roux G, Banfi S, Sultan M, et al. A High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo. PLOS Biol. 2011;9(1):e1000582. doi: 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar S, Rathkolb B, Kemter E, et al. Generation and Standardized, Systemic Phenotypic Analysis of Pou3f3L423P Mutant Mice. PLOS ONE. 2016;11(3):e0150472. doi: 10.1371/journal.pone.0150472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goff LA, Groff AF, Sauvageau M, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2015;112(22):6855–6862. doi: 10.1073/pnas.1411263112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oishi K, Aramaki M, Nakajima K. Mutually repressive interaction between Brn1/2 and Rorb contributes to the establishment of neocortical layer 2/3 and layer 4. Proc Natl Acad Sci U S A. 2016;113(12):3371–3376. doi: 10.1073/pnas.1515949113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waldhaus J, Durruthy-Durruthy R, Heller S. Quantitative High-Resolution Cellular Map of the Organ of Corti. Cell Rep. 2015;11(9):1385–1399. doi: 10.1016/j.celrep.2015.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Usami S, Takumi Y, Suzuki N, et al. The localization of proteins encoded by CRYM, KIAA1199, UBA52, COL9A3, and COL9A1, genes highly expressed in the cochlea. Neuroscience. 2008;154(1):22–28. doi: 10.1016/j.neuroscience.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 84.Hawley SP, Wills MKB, Rabalski AJ, Bendall AJ, Jones N. Expression patterns of ShcD and Shc family adaptor proteins during mouse embryonic development. Dev Dyn. 2011;240(1):221–231. doi: 10.1002/dvdy.22506 [DOI] [PubMed] [Google Scholar]
- 85.Wu L, Sagong B, Choi JY, Kim U-K, Bok J. A systematic survey of carbonic anhydrase mRNA expression during mammalian inner ear development. Dev Dyn. 2013;242(3):269–280. doi: 10.1002/dvdy.23917 [DOI] [PubMed] [Google Scholar]
- 86.Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477(4):412–421. doi: 10.1002/cne.20257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ficker M, Powles N, Warr N, Pirvola U, Maconochie M. Analysis of genes from inner ear developmental-stage cDNA subtraction reveals molecular regionalization of the otic capsule. Dev Biol. 2004;268(1):7–23. doi: 10.1016/j.ydbio.2003.11.023 [DOI] [PubMed] [Google Scholar]
- 88.Chow C, Trivedi P, Pyle M, Matulle J, Fettiplace R, Gubbels SP. Evaluation of Nestin Expression in the Developing and Adult Mouse Inner Ear. Stem Cells Dev. 2016;25(19):1419–1432. doi: 10.1089/scd.2016.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chow CL, Guo W, Trivedi P, Zhao X, Gubbels SP. Characterization of a unique cell population marked by transgene expression in the adult cochlea of nestin-CreER(T2)/tdTomato-reporter mice. J Comp Neurol. 2015;523(10):1474–1487. doi: 10.1002/cne.23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haugas M, Lilleväli K, Hakanen J, Salminen M. Gata2 is required for the development of inner ear semicircular ducts and the surrounding perilymphatic space. Dev Dyn. 2010;239(9):2452–2469. doi: 10.1002/dvdy.22373 [DOI] [PubMed] [Google Scholar]
- 91.Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, Wu DK. Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol. 2009;333(1):14–25. doi: 10.1016/j.ydbio.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mann ZF, Gálvez H, Pedreno D, et al. Shaping of inner ear sensory organs through antagonistic interactions between Notch signalling and Lmx1a. eLife. 2017;6:e33323. doi: 10.7554/eLife.33323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, Fritzsch B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008;334(3):339–358. doi: 10.1007/s00441-008-0709-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sánchez-Guardado LÓ, Ferran JL, Rodríguez-Gallardo L, Puelles L, Hidalgo-Sánchez M. Meis gene expression patterns in the developing chicken inner ear. J Comp Neurol. 2011;519(1):125–147. doi: 10.1002/cne.22508 [DOI] [PubMed] [Google Scholar]
- 95.Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18(9):3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W, Chan EK, Baron S, Van De Water TR, Lufkin T. Hmx2 homeobox gene control of murine vestibular morphogenesis. Development. 2001;128(24):5017–5029. [DOI] [PubMed] [Google Scholar]
- 97.Poladia DP, Kish K, Kutay B, et al. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291(2):325–339. doi: 10.1016/j.ydbio.2005.12.034 [DOI] [PubMed] [Google Scholar]
- 98.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 99.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Howe KL, Contreras-Moreira B, De Silva N, et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020;48(D1):D689–D695. doi: 10.1093/nar/gkz890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morgan M, Pagès H, Obenchain V, Hayden N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. Published online 2018. R package version 1.32.0. http://bioconductor.org/packages/Rsamtools. doi: 10.18129/B9.bioc.Rsamtools [DOI] [Google Scholar]
- 102.Lawrence M, Huber W, Pagès H, et al. Software for Computing and Annotating Genomic Ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32(9):896. doi: 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. Published online 2016. R package version 2.32.0. https://bioconductor.org/packages/topGO. doi: 10.18129/B9.bioc.topGO [DOI] [Google Scholar]
- 105.Huang M, Sage C, Li H, Xiang M, Heller S, Chen Z-Y. Diverse expression patterns of LIM-homeodomain transcription factors (LIM-HDs) in mammalian inner ear development. Dev Dyn. 2008;237(11):3305–3312. doi: 10.1002/dvdy.21735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9(17):2105–2116. doi: 10.1101/gad.9.17.2105 [DOI] [PubMed] [Google Scholar]
- 107.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Avinash GB, Nuttall AL, Raphael Y. 3-D analysis of F-actin in stereocilia of cochlear hair cells after loud noise exposure. Hear Res. 1993;67(1):139–146. doi: 10.1016/0378-5955(93)90241-R [DOI] [PubMed] [Google Scholar]
- 109.Mueller KL, Jacques BE, Kelley MW. Fibroblast Growth Factor Signaling Regulates Pillar Cell Development in the Organ of Corti. J Neurosci. 2002;22(21):9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 – preTRAP vs. TRAP DEG analysis
S2 – GO analysis of DEGs depleted by TRAP
S3 – GO analysis of DEGs enriched by TRAP
S4 – Fgf20−/+ vs. Fgf20−/− DEG analysis
S5 – GO analysis of Fgf20−/+ vs. Fgf20−/− DEGs
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE148380 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148380).110