Abstract
Background
Cell-division cycle 20 (CDC20) is overexpressed in a variety of tumor cells and is negatively regulated by wild-type p53 (wtp53). Our previous study uncovered that CDC20 was upregulated and associated with poor outcome in diffuse large B-cell lymphoma (DLBCL) based on bioinformatics analysis. Dysregulation of the MDM2-p53 is a major mechanism to promote DLBCL. Thus, we hypothesized that CDC20 could be a downstream gene of the MDM2-p53 signaling pathway. However, the clinical significance and mechanistic role of a novel MDM2-p53-CDC20 signaling pathway in DLBCL have still remained unclear.
Materials and Methods
RT-qPCR was performed in MDM2 knocked down (KD) and control (Ctrl) OCI-Ly3/OCI-Ly10 cells to investigate whether CDC20 was a downstream gene of the MDM2-p53 pathway. The effects of CDC20 on cell proliferation, cell cycle and apoptosis were assessed, as well as the role of CDC20 in suppressing tumorigenicity in vivo. Furthermore, we also investigated the roles of CDC20 and MDM2 in progression of DLBCL and the underlying mechanisms.
Results
The results of RT-qPCR revealed that CDC20 was downregulated while TP53 was upregulated in MDM2 KD OCI-Ly3 and OCI-Ly10 cells. It was unveiled that the expression levels of CDC20 and MDM2 were upregulated in DLBCL tissues and cells, and high CDC20 expression was correlated with adverse clinical features and poor outcome. Functional assays showed that downregulation of CDC20 could inhibit proliferation, induce apoptosis and cell cycle arrest in vitro. In addition, inactivation of the MDM2-p53 pathway by downregulation of MDM2 restored wtp53 expression level and reduced CDC20 protein level in OCI-Ly3 and OCI-Ly10 cells. Besides, targeting CDC20 was found to suppress tumorigenesis of DLBCL in vivo.
Conclusion
CDC20 was identified as a key downstream gene of the MDM2-p53 signaling pathway in DLBCL and may be used as a novel target gene to guide therapeutic applications.
Keywords: DLBCL, MDM2-p53 signaling pathway, CDC20, tumor progression
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most frequently diagnosed subtype of hematological malignancy, with an annual incidence rate of over 100,000 cases worldwide.1–3 The pathophysiological mechanisms underlying the tumorigenesis of DLBCL are still unclear, and further developments are essential to understand the molecular mechanisms of the occurrence and progression of DLBCL.
As one of the most extensively explored tumor suppressors, wild-type p53 (wtp53) inhibits tumor growth and promotes cell cycle arrest, apoptosis and senescence in response to diverse forms of cellular stress, thereby maintaining genome stability.4 In addition, TP53 is frequently wild-type in DLBCL, and TP53 gene mutations have been observed in 22% of DLBCLs.5,6 The wtp53 is negatively regulated by murine double minute 2 (MDM2), an E3 ubiquitin ligase, and is subsequently destroyed by poly-ubiquitination.7 MDM2 is frequently overexpressed in different types of cancer, and Moller et al reported that 43% of DLBCL patients had MDM2 overexpression.6 However, the correlation between MDM2 overexpression and clinical outcomes in DLBCL is inconsistent.8,9 The MDM2 gene is trans-activated by p53, which is subsequently perturbed by MDM2-mediated ubiquitination, thereby forming a negative feedback loop.9 Malonia et al demonstrated that the F-box protein FBXO31 was responsible for promoting MDM2 degradation as an upstream tumor suppressor and caused an increase in p53 expression level.10 However, studies about downstream molecules of the MDM2-p53 singling pathway have been scarcely carried out.
Ubiquitination plays significant roles in cellular processes that regulate cell proliferation, apoptosis, cell cycle progression, DNA damage, migration, and invasion.11–13 Ubiquitin proteasome system-mediated ubiquitination is a posttranslational modification and controls the protein degradation of cellular processes.14 It has been reported that multi-subunit E3 ubiquitin ligase enzymes exert effects on the cell cycle progression and cell division.15,16 Anaphase-promoting complex (APC), the most complex E3 ubiquitin ligase, is composed of at least 14 subunits and one co-activator (CDC20 [cell-division cycle protein 20] or CDH1 [CDC20 homologue 1]).17 APCCDC20 plays a vital role in the transition from metaphase to anaphase by destructing critical cell cycle regulators.18 CDC20 exhibits its oncogenic function in a plethora of cancer cells.19,20 Meanwhile, CDC20 overexpression has been found in a variety of human tumors, such as pancreatic cancer,21 breast cancer,22 lung cancer,23,24 hepatocellular carcinoma,25 and glioblastoma,26 and it may be an independent marker for predicting clinical outcomes of pancreatic cancer.21 However, a limited number of studies concentrated on the oncogenic role of CDC20 in human DLBCL. We, for the first time, reported that CDC20 expression is elevated in DLBCL and is associated with poor overall survival (OS) based on results of previous bioinformatics analysis.27 However, the clinical role and mechanism of CDC20 in the development of DLBCL have still remained elusive.
In recent years, studies have determined the downstream substrates of CDC20, namely Cyclin B, Nek2A, and Bim, and also found that these downstream substrates were targeted by CDC20 for ubiquitination and subsequent degradation.28–31 Although Kidokoro et al pointed out that CDC20 expression is negatively regulated by p53,24 a small number of researches have concentrated on the upstream regulators of CDC20. In the present study, we found that CDC20 acted as a downstream gene of the MDM2-p53 singling pathway and played a critical role in the pathogenesis of DLBCL. Additionally, the MDM2-p53 signaling pathway was involved in the regulation of CDC20 expression, and both MDM2 and CDC20 could predict the prognosis of DLBCL.
Materials and Methods
Data Mining in Bioinformatics
High-throughput data of RNA-seq gene expression data (level 3) and clinical information for patients diagnosed with DLBCL were downloaded from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/). The Gene Expression Omnibus (GEO) datasets GSE5631532 (containing gene expression data of 89 DLBCL tissues and 33 normal tonsil tissues), GSE3201833 (containing gene expression data of 22 DLBCL tissues and 7 normal lymph nodes), GSE313129,34 (containing gene expression and survival data of 498 DLBCL patients), and GSE1084635,36 (containing gene expression and survival data of 420 DLBCL patients) were used. The limma R and edge R package were used for screening differentially expressed genes (DEGs) between DLBCL samples and non-cancerous tissues in GEO and TCGA datasets, respectively. The adjusted P<0.01 and | log2 FC | > 1 were used as the cut-off criterion for DEGs identification. Venn diagram was generated using the FunRich software.37 Survival analysis of the GEO data was performed using the R2 (http://r2.amc.nl).
Tissue Samples and Cell Lines
This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (No. N2020030). Clinical samples were all obtained from Sun Yat-sen University Cancer Center between 2006 and 2013. All patients or their guardians provided written consent for the use of their data. The median follow-up interval for the 97 patients was 60 months (range, 3 to 133 months). The human DLBCL cell lines OCI-Ly3 (activated B-cell-derived, wild-type TP53) and OCI-Ly10 (activated B-cell-derived, wild-type TP53) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and were all cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (gibco, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, HyClone, USA) and 1% penicillin-streptomycin solution (gibco, 15,140–122, Life Technologies, USA) at 5% CO2 and 37°C. Peripheral blood mononuclear cells (PBMCs) and normal tonsil tissues were used in this study as a control.
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from PBMCs and DLBCL cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA was quantified using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Then, total RNA was reversely transcribed into first-strand cDNA using the Prime Script RT Master Mix (TaKaRa, Kyoto, Japan). RT-qPCR was performed on the LightCycler 480 System (Roche, Basel, Switzerland) with Power SYBR Green qPCR Mix (Dongsheng Biotech Co., Ltd., Guangzhou, China). All amplification reactions were conducted using the following cycling parameters: at 94 °C for 3 min, followed by 40 cycles at 95 °C for 15 s, at 60 °C for 15 s, and at 72 °C for 20 s. The 2−ΔΔCq method was utilized for the quantification of gene expression, with β-actin as an endogenous control. Primer sequences used are displayed as follows:
MDM2 forward, 5ʹ-GGCGTGCCAAGCTTCTCTGTG-3ʹ
MDM2 reverse, 5ʹ-ACCTGAGTCCGATGATTCCTGCTG-3ʹ
TP53 forward, 5ʹ-ACCGGCGCACAGAGGAAGAG-3ʹ
TP53 reverse, 5ʹ-GCCTCATTCAGCTCTCGGAACATC-3ʹ
CDC20 forward, 5ʹ-AAGACCTGCCGTTACATTCCTTCC-3ʹ
CDC20 reverse, 5ʹ-CACCAGAGCTTGCACTCCACAG-3ʹ
β-actin forward, 5ʹ-CCTGGCACCCAGCACAAT-3ʹ
β-actin reverse, 5ʹ-GGGCCGGACTCGTCATAC-3ʹ
Cell Transfection
To knockdown genes by small-interfering RNAs (siRNAs), the control (Ctrl) and target MDM2 siRNAs were synthesized by GenePharma Co., Ltd. (Shanghai, China). DLBCL cells were transfected with MDM2 siRNAs at a final concentration of 50 nM using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. After treatment, further analyses were undertaken using transfected cells.
Cell Counting Kit (CCK-8) Assay
Cell proliferation was assessed using a CCK-8 kit (K1018; APExBIO, Houston, TX, USA) based on the manufacturer’s protocol. OCI-Ly3 and OCI-Ly10 cells were seeded into 96-well plates at a density of 5×103 cells/well in 200 μL RMP-1640 medium containing 10% (v/v) FBS. The cells were then treated with Apcin (I-444; Boston Biochem, Inc., Cambridge, MA, USA), a small molecule ligand of CDC20, at different concentrations and were incubated at 37 °C for 48 h. To measure the growth rate of the cells, 10% CCK-8 (20 μL) was added per well, and the cells were further incubated at 37 °C for 2 h. The absorbance was finally determined at 450 nm using a microplate reader (M200 PRO; Tecan Group Ltd., Männedorf, Switzerland).
Flow Cytometry Analysis
For cell apoptosis analysis, cell apoptosis was evaluated using the Annexin V-FITC Apoptosis Detection Kit (KGA108; KeyGEN, Nanjing, China) and flow cytometry analysis. According to the manufacturer’s instructions, 5×105 cells were harvested, washed with phosphate-buffered saline (PBS), and then re-suspended with 500 μL Binding Buffer. The cells were successively incubated with 5μL Annexin V-FITC and 5 μL of Propidium Iodide (PI) for 15 min in dark at room temperature. The stained cells were analyzed by flow cytometry (SP6800; Sony, Tokyo, Japan) for within 1 h.
For cell cycle, the cells were suspended in a pre-cooling 70% alcohol for fixation at 4 °C overnight. On the next day, the fixed cells were washed with PBS and stained with 500 μL PI/RNase Staining Buffer (Beyotime Institute of Biotechnology, Shanghai, China) at 37 ºC for 30 min in the dark. Cell apoptosis and cell cycle analysis were carried out using the FlowJo (Tree Star, Inc., Ashland, OR, USA) and ModFit LT (BD Biosciences, Topsham, ME, USA) software, respectively.
EdU Cell Proliferation Assay
OCI-Ly3 and OCI-Ly10 cells were seeded into 12-well plates at a density of 5×105 cells/well. Besides, 48 h after stimulation with Apcin, the cells were exposed to 50 μM EdU (C10310-3; RiboBio, Guangzhou, China) for 2 h at 37 ºC. Then, the cells were stained with 1 × Apollo and 1 × Hoechst 33,342 according to the manufacturer’s instructions. Finally, the EdU-stained cells were visualized under a confocal microscope (LSM880; Carl Zeiss, Oberkochen, Germany) with excitation and emission wavelengths of 488 and 520 nm, respectively.
Immunohistochemistry (IHC)
For IHC, sections of DLBCL were incubated with primary anti-rabbit MDM2 (1:50; ab38618; Abcam, Cambridge, UK) and anti-rabbit CDC20 (1:200; Q105; Bioworld Technology Inc., St Louis Park, MN, USA) at 4 °C overnight. On the next day, the tissues were incubated with goat anti-rabbit secondary antibody (Zhongshan Goldbridge Biotechnology Co., Ltd., Beijing, China) at 37 °C for 1 h. All images were observed under a fluorescence microscope (IX73; Olympus, Tokyo, Japan).
Western Blot Analysis
The cells from each group were collected and washed twice with ice-cold PBS for Western blot analysis. Tumor cell lysates were generated using the KeyGEN kit (KGP2100; KeyGEN, Nanjing, China) and were re-suspended with 3× sample buffer and boiled for 10 min. The protein concentration was detected by the bicinchoninic acid (BCA) Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and were then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% skimmed milk powder for 2 h, the PVDF membranes were incubated with anti-mouse MDM2 (1:500; 556,353; BD Biosciences, Topsham, ME, USA), anti-mouse p53 (1:200; DO-1; Santa Cruz Biotechnology, Dallas, TX, USA), anti-rabbit CDC20 (1:1000; Q105; Bioworld Technology Inc., St Louis Park, MN, USA), and anti-rabbit β-actin (1:1000; P60709; Cell Signaling Technology, Inc., Danvers, MA, USA). The membranes were incubated overnight at 4°C, followed by a goat anti-rabbit or goat anti-mouse secondary antibody (1:10,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 h at 37 °C. The signals were detected using the enhanced chemiluminescence (ECL) (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Immunofluorescence
Herein, 4% paraformaldehyde-fixed cell suspensions of OCI-Ly3 and OCI-Ly10 were centrifuged in a Cytospin onto coverslips. The cells were permeabilized in 0.1% Triton X-100 for 5 min and blocked in 5% bovine serum albumin (BSA) at room temperature for 1 h, and then incubated for 1 h with primary anti-rabbit MDM2 (1:200, ab38618; Abcam, Cambridge, UK), anti-mouse p53 (1:200, DO-1, Santa Cruz) and anti-rabbit CDC20 (1:500; ab26483; Abcam, Cambridge, UK), washed thrice with PBS, and incubated with Alexa Fluor 488/555-conjugated secondary antibodies (Beyotime Institute of Biotechnology, Shanghai, China) at room temperature for 1 h in the dark. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Institute of Biotechnology, Shanghai, China) for 5 min. Images were captured using a confocal microscope (LSM880, Carl Zeiss, Oberkochen, Germany).
Xenograft Tumorigenesis in NOD/SCID Mice
The animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of the Sun Yat-sen University Cancer Center (No. 2,020,000,042). Four-week-old NOD/SCID female mice (Vital River Laboratory Animal Technology, Beijing, China) were inoculated subcutaneously with 5 × 106 OCI-Ly10 cells suspended in 100 μL PBS. Those 10 mice were randomly divided into 2 groups (n = 5 for each group), and were treated with PBS (control) or Apcin (intraperitoneal injection, 15 mg/kg, 3 times/week). The tumor size was monitored 2–3 times/week with Vernier caliper. The tumor volume was calculated using the following formula: (length × width2)/2. All mice were euthanized when the average diameter of the tumors reached 20 mm.
Statistical Analysis
Herein, SPSS 20.0 software (IBM, Armonk, NY, USA) was used to perform statistical analyses. Statistical significance between different groups was analyzed using the Student’s t-test. χ2 test was used to assess the relationship between the CDC20 expression level and clinicopathological characteristics. Kaplan-Meier survival curves were plotted to compare survival rates between different groups. P < 0.05 was considered statistically significant.
Results
MDM2 Expression Was Increased in DLBCL Cells and Tissues, and High Expression Level of MDM2 Was Associated with Poor Outcome
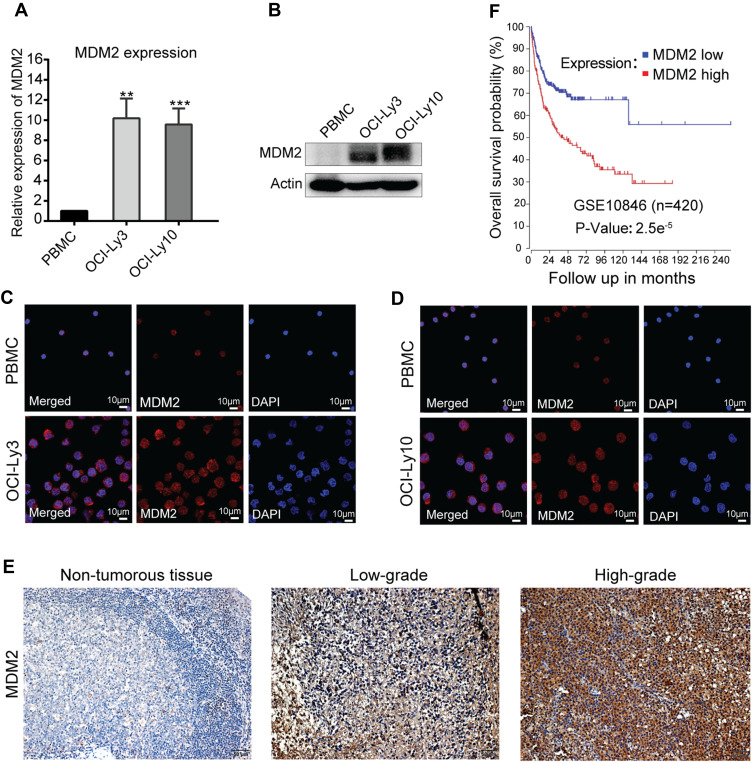
To investigate whether MDM2 was differentially expressed in DLBCL cells and PBMCs, RT-qPCR and Western blot assays were performed, respectively. The results of RT-qPCR showed that compared with PBMCs, MDM2 expression was elevated in OCI-Ly3 and OCI-Ly10 cells (Figure 1A). The results of Western blotting demonstrated the same varying tendency of mRNA levels (Figure 1B). Next, we evaluated the MDM2 expression levels in DLBCL cells and PBMCs using immunofluorescence. Our results showed an increase in the expression level of MDM2 in OCI-Ly3 and OCI-Ly10 cells (Figure 1C and D). We further evaluated the MDM2 expression level in 97 patients who were enrolled in our hospital from 2006 to 2013 by using IHC. Our results unveiled an increase in the expression level of MDM2 in DLBCL tissues (Figure 1E). Taken together, these findings indicated that the expression level of MDM2 was elevated in DLBCL cells and tissues.
Figure 1.
MDM2 expression was elevated in DLBCL cells and tissues and high expression level of MDM2 was associated with poor outcome. (A) RT-qPCR analysis of MDM2 mRNA levels in DLBCL cells and PBMCs. (B) Western blot analysis of MDM2 protein expressions in DLBCL cells and PBMCs. (C and D) Immunofluorescence staining (× 630, scale bar, 10μm) for MDM2 protein expression in PBMCs and OCI-Ly3 or OCI-Ly10 cells. (E) The expression level of MDM2 in one normal and two representative cases of total 97 samples by using IHC (× 200; scale bar, 50 μm). (F) Kaplan-Meier OS analysis of 420 DLBCL patients with different MDM2 mRNA levels based on GSE10846 (n=420). Data were analyzed through R2 (http://r2.amc.nl). **P < 0.01; ***P < 0.001.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; PBMCs, peripheral blood mononuclear cells; IHC, immunohistochemistry; OS, overall survival.
We also evaluated the effects of expression levels of MDM2 on the survival of the DLBCL patients using the GEO datasets. Kaplan-Meier survival curves of GSE10846 generated by R2 showed that patients with higher expression levels of MDM2 had significantly worse OS than those with lower expression levels (Figure 1F, P=2.5e−5).
CDC20 Was Found as a Downstream Gene of the MDM2-P53 Signaling Pathway
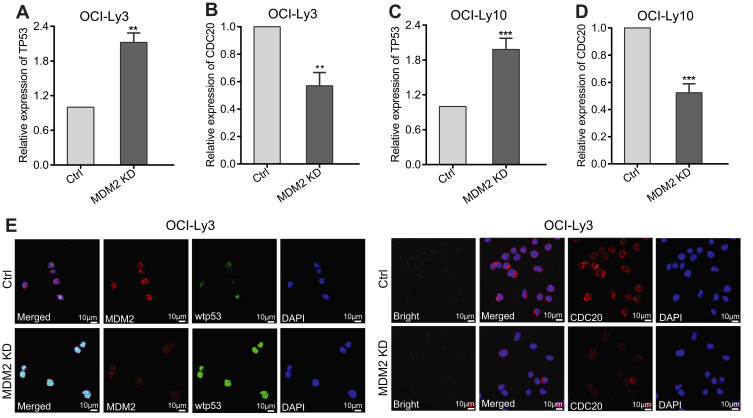
Notably, CDC20 is an oncogene and overexpressed in several types of malignancies.38 We also previously demonstrated that CDC20 was overexpressed in DLBCL and was associated with poor prognosis.27 Because of the essential role of the MDM2-p53 loop in tumorigenesis and progression of DLBCL, together with Kidokoro et al’s findings who reported that CDC20 was negatively regulated by p53 in A549 cells,25 we hypothesized that CDC20 was a downstream gene of the MDM2-p53 signaling pathway. To verify our hypothesis, we performed RT-qPCR and the results showed that CDC20 was downregulated while TP53 was upregulated in MDM2 KD OCI-Ly3 and OCI-Ly10 cells compared with Ctrl cells (Figure 2A–D). As we expected, RT-qPCR validation confirmed our hypothesis. Furthermore, immunofluorescence staining for MDM2, wtp53 and CDC20 of MDM2 KD and Ctrl OCI-Ly3 cell showed the same varying tendency as above RT-qPCR results (Figure 2E).
Figure 2.
CDC20 is a downstream gene of MDM2-P53 signaling pathway in DLBCL. (A and B) RT-qPCR detected the mRNA levels of TP53 and CDC20 in MDM2 KD and Ctrl OCI-Ly3 cells. (C and D) RT-qPCR validation of TP53 and CDC20 mRNA levels in MDM2 KD and Ctrl OCI-Ly10 cells. (E) MDM2, wtp53 and CDC20 protein expressions were analyzed by immunofluorescence staining in Ctrl and MDM2 KD OCI-Ly3 cell (× 630, scale bar, 10μm). **P < 0.01; ***P < 0.001.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; KD, knocked down; Ctrl, control; DAPI, 4ʹ,6-diamidino-2-phenylindole.
CDC20 Was Overexpressed in DLBCL and Associated with Poor Outcome
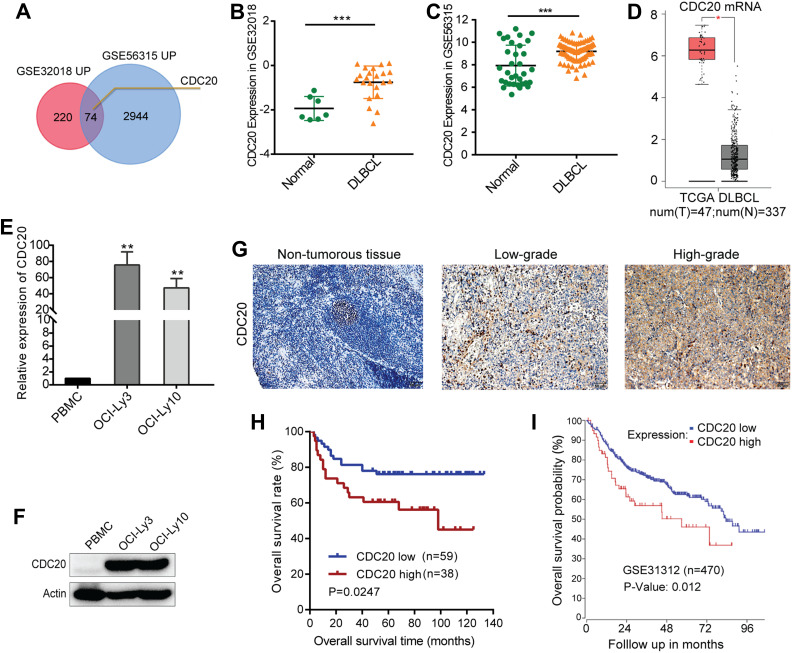
The results of bioinformatics analysis are presented in Figure 3A–D. The intersection of the GSE32018 and GSE56315 datasets for DLBCL samples and normal tissues uncovered 74 upregulated DEGs in DLBCL patients. Theses commonly upregulated genes included CDC20 (Figure 3A). The CDC20 expression level was significantly higher in DLBCL tissues compared with that in normal tissues in GSE32018 and GSE56315 datasets, respectively (Figure 3B and C). Furthermore, the TCGA DLBCL dataset confirmed that CDC20 was overexpressed in DLBCL (Figure 3D). Consistent with bioinformatics analyses, we observed that the CDC20 mRNA and protein levels were markedly increased in OCI-Ly3 and OCI-Ly10 cells compared with those in PBMCs based on results of RT-qPCR and Western blot assays (Figure 3E and F). Additionally, IHC of 97 DLBCL samples from our hospital confirmed the elevation of CDC20 expression in tumor tissues (Figure 3G). These findings indicated that CDC20 was highly expressed in DLBCL cells and tissues.
Figure 3.
CDC20 expression was elevated in DLBCL, and overexpression of CDC20 could be an unfavorable prognostic factor. (A) Venn diagram showing 74 upregulated genes in GSE32018 and GSE56315 datasets. CDC20 was up-regulated in these two GEO datasets. (B and C) Scatterplot indicated that CDC20 mRNA level was elevated in both GSE32018 and GSE56315 datasets. (D) TCGA DLBCL confirmed that CDC20 was overexpressed in DLBCL. (E and F) RT-qPCR and Western blot assays detected CDC20 expression in DLBCL cells and PBMCs. (G) IHC of CDC20 expression in one normal and two representative cases of 97 DLBCL tissues (× 200, scale bar, 50μm). (H) Kaplan–Meier OS analysis of 97 DLBCL patients with different CDC20 protein expression levels. (I) Association between CDC20 expression level and OS in DLBCL based on GSE31312 (n=470). Kaplan-Meier survival curves were generated by using R2 (http://r2.amc.nl). *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; TCGA, The Cancer Genome Atlas; num, number; T, tumor; N, normal; PBMCs, peripheral blood mononuclear cells; IHC, immunohistochemistry; OS, overall survival.
For clinical samples, we analyzed the correlation between CDC20 expression and clinicopathological features in 97 DLBCL patients. As shown in Table 1, the expression level of CDC20 was significantly associated with B-symptoms (χ2 = 62.505, P < 0.001), serum level of lactate dehydrogenase (LDH) (χ2 = 6.786, P = 0.009), and international prognostic index (IPI) risk (χ2 = 4.213, P = 0.040). These results indicated that CDC20 expression was correlated with adverse clinical features in DLBCL patients. We next assessed the prognostic value of CDC20 in DLBCL patients. The clinical data of 97 patients uncovered that overexpression of CDC20 was associated with an inferior OS (Figure 3H, P=0.0247). Consistent with this result, R2 was used to analyze GSE31312 dataset, and Kaplan-Meier survival curves indicated that high expression level of CDC20 had significantly worse OS than low expression level in DLBCL patients (Figure 3I, P=0.012).
Table 1.
Correlation Between CDC20 Expression and Clinicopathological Features of 97 DLBCL Patients
| Characteristics | Cases, n | CDC20 | χ2 | P-value | |
|---|---|---|---|---|---|
| Lower, n | Higher, n | ||||
| Gender | |||||
| Male | 49 | 29 | 20 | 0.112 | 0.738 |
| Female | 48 | 30 | 18 | ||
| Age | |||||
| ≤60 | 63 | 41 | 22 | 1.365 | 0.243 |
| >60 | 34 | 18 | 16 | ||
| Stage | |||||
| I–II | 39 | 25 | 14 | 0.294 | 0.588 |
| III–IV | 58 | 34 | 24 | ||
| B-symptoms | |||||
| No | 51 | 50 | 1 | 62.505 | < 0.001 |
| Yes | 46 | 9 | 37 | ||
| LDH | |||||
| Normal | 59 | 42 | 17 | 6.786 | 0.009 |
| Elevated | 38 | 17 | 21 | ||
| IPI risk group | |||||
| 0–2 | 70 | 47 | 23 | 4.213 | 0.040 |
| 3–5 | 27 | 12 | 15 | ||
| No. of extranodal sites | |||||
| 0–1 | 84 | 52 | 32 | 0.307 | 0.580 |
| ≥ 2 | 13 | 7 | 6 | ||
Abbreviations: CDC20, cell-division cycle 20; DLBCL, diffuse large B-cell lymphoma; IPI, international prognostic index; LDH, lactate dehydrogenase.
Decreased CDC20 Suppressed the Proliferation, Induced Apoptosis and G2/M Arrest in DLBCL Cells
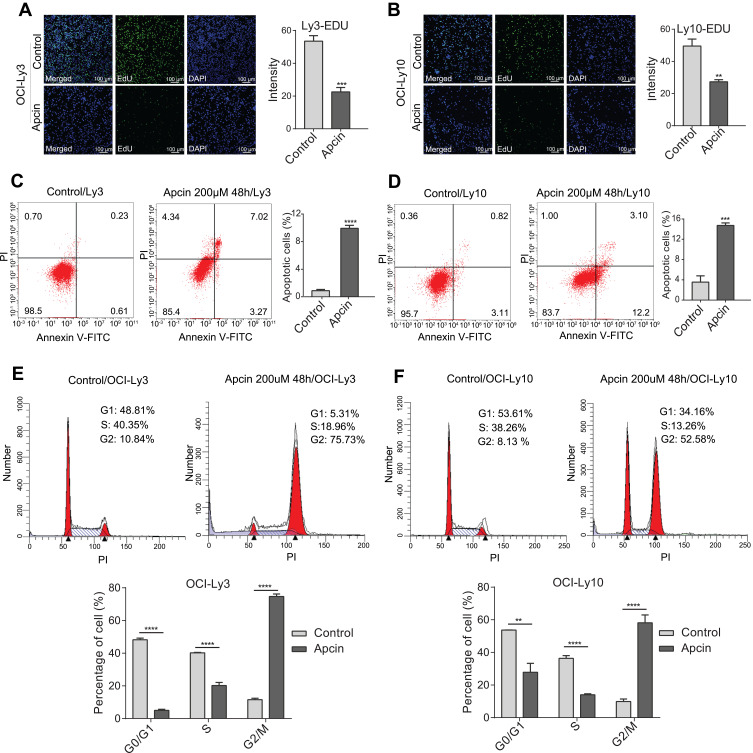
According to the above-mentioned findings, we hypothesized that CDC20 was a biologic reason for tumorigenesis of DLBCL. To assess whether CDC20 could promote cell proliferation, we decreased CDC20 protein expression using Apcin in OCI-Ly3 and OCI-Ly10 cells (Supplementary Figure 1C and D). As depicted in Supplementary Figure 1A and B, different doses of Apcin significantly suppressed the viability of OCI-Ly3 and OCI-Ly10 cells. The EdU proliferation results showed that the percentage of EdU-positive cells was noticeably decreased in OCI-Ly3 and OCI-Ly10 cells compared with that in control cells (Figure 4A and B). Collectively, these findings demonstrated the proliferative suppression effect that was induced by CDC20 inhibition in DLBCL cells.
Figure 4.
CDC20 regulated DLBCL cell proliferation, apoptosis, and cell cycle in vitro. (A and B) OCI-Ly3 and OCI-Ly10 cells were treated with Apcin (200 μM) for 48 h. The percentage of EdU-positive cells could be measured using a confocal microscope (×100; scale bar, 100 μm). (C and D) OCI-Ly3 and OCI-Ly10 cells were treated with Apcin (200 μM) and stained with Annexin V-FITC and PI, and then, the rate of apoptotic cells was analyzed by flow cytometry. (E and F) Cell cycle distribution of OCI-Ly3 and OCI-Ly10 was analyzed by using flow cytometry. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; EdU, 5-ethynyl-2ʹ-deoxyuridine; DAPI, 4ʹ,6-diamidino-2-phenylindole; Annexin V-FITC, annexin A5-fluorescein isothiocyanate; PI, propidium iodide.
Compared with control cells, the percentage of apoptotic cells was found to be significantly increased in OCI-Ly3 and OCI-Ly10 cells (Figure 4C and D). These outcomes highlighted that downregulation of CDC20 could induce apoptosis in DLBCL cells. As shown in Figure 4E and F, DLBCL cells treated with Apcin showed a reduced number of cells in the G0/G1 and S phases, as well as a remarkably increased number of cells in the G2/M phase.
The Role of MDM2-P53-CDC20 Signaling Pathway in Human DLBCL Cells
A number of scholars pointed out that ectopically expressed p53 or DNA damage-induced endogenous p53 can downregulate CDC20 transcriptionally by direct binding to its promoter causing chromatin remodeling.39 Since the above results uncovered that CDC20 is a downstream gene of MDM2, we further studied the mechanism of MDM2-p53-CDC20 singling pathway in DLBCL.
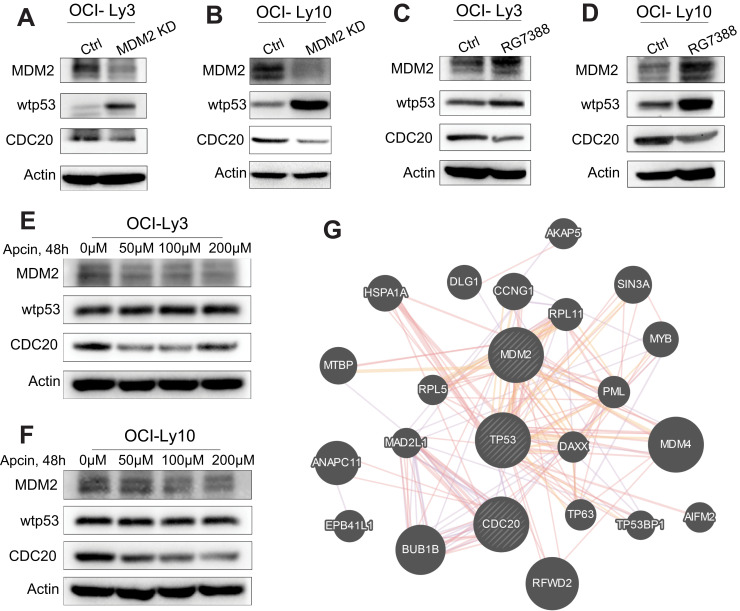
Firstly, with downregulating MDM2 by siRNA, we observed that MDM2 inhibition dramatically restored wtp53 and inhibited CDC20 protein expression in OCI-Ly3 and OCI-Ly10 cells (Figure 5A and B). RG7388 is a selective second-generation MDM2 inhibitor, which disrupts MDM2-p53 interaction and activates p53.40 We upregulated p53 using RG7388 (BioVision, 1μM, 24h) and found that protein expression of CDC20 was decreased in OCI-Ly3 and OCI-Ly10 cells (Figure 5C and D). Moreover, we noted that Apcin decreased the expression of CDC20, while this did not affect the protein expressions of MDM2 and wtp53 (Figure 5E and F). In addition, molecular network generated by online GeneMANIA (http://genemania.org/) tool indicated that both MDM2 and CDC20 interacted with TP53 (Figure 5G). Expectedly, the results suggested that CDC20 is a key downstream gene of the MDM2-p53 singling pathway, and increase of MDM2 activity upregulates CDC20 expression by inhibiting wtp53 protein expression.
Figure 5.
MDM2-p53-CDC20 signaling pathway in DLBCL cells. (A and B) OCI-Ly3 and OCI-Ly10 cells were transfected with si-MDM2, and MDM2, wtp53 and CDC20 protein expressions were detected by Western blot assay. (C and D) OCI-Ly3 and OCI-Ly10 cells were treated with RG7388, and MDM2, wtp53 and CDC20 protein expressions were identified by Western blot assay. (E and F) OCI-Ly3 and OCI-Ly10 cells were treated with Apcin, and the expression levels of MDM2, wtp53, and CDC20 were detected by Western blot assay. (G) Based on GeneMANIA (http://genemania.org/) analysis, molecular functional network of MDM2-p53-CDC20 pathway was generated.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; KD, knocked down; Ctrl, control.
Targeting CDC20 in OCI-Ly10 Cell Suppressed Tumorigenesis in NOD/SCID Mice
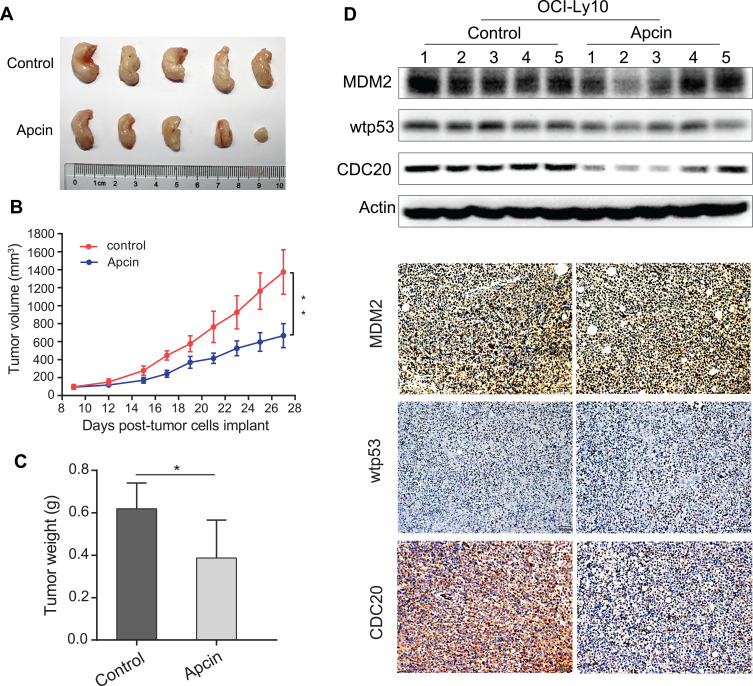
To further explore the tumor-suppressive functions of CDC20 in vivo, we used the NOD/SCID mouse model and injected 5 × 106 OCI-Ly10 cells into the flanks of the mice. After 9 days, the mice were randomly divided into two groups with similar mean tumor volumes of 100 to 120 mm3. Then, the mice were treated with PBS (control group) or Apcin (15 mg/kg, 3 times/week). Their tumor weight and size were measured for 2–3 times/week. As shown in Figure 6A–C, a more robust growth of tumor size and weight were observed in the control group than that in mice treated with Apcin. Additionally, as illustrated in the growth curve, treatment with Apcin significantly inhibited tumor growth in NOD/SCID mice bearing OCI-Ly10 xenografts (Figure 6B). The above-mentioned results were consistent with our previous in vitro findings.
Figure 6.
Targeting CDC20 inhibits tumorigenesis of DLBCL in vivo. (A) OCI-Ly10 cells (5×106 cells per mouse) were subcutaneously injected into the posterior flank of NOD/SCID mice. After 9 days, the mice were randomly divided into two groups and treated with PBS or Apcin. Photographed tumors of these two groups on day 27 are shown. (B and C) The primary tumor volume and weight are also illustrated. (D) Western blotting and IHC (× 200; scale bar, 50 μm) were performed to estimate the protein expressions of MDM2, wtp53, and CDC20 in two groups. *P < 0.05; **P < 0.01.
Abbreviations: DLBCL, diffuse large B-cell lymphoma; IHC, immunohistochemistry.
Furthermore, Western blotting and IHC were used to evaluate the expression levels of CDC20, MDM2, and wtp53. It was disclosed that treatment with Apcin remarkably reduced the expression level of CDC20 in mice bearing OCI-Ly10 xenografts, while expression levels of MDM2 and wtp53 in both Apcin-treated group and control group were similar (Figure 6D). These results demonstrated that targeting CDC20 in OCI-Ly10 cells might play a carcinogenesis suppression effect in vivo.
Collectively, CDC20 was observed to act as a key downstream gene of the MDM2-p53 signaling pathway to regulate cell proliferation, cell apoptosis, and cell cycle. MDM2 overexpression has been shown to inactivate wild-type p53 function and upregulate CDC20 expression.
Discussion
In the present study, we identified CDC20 as a downstream gene of the MDM2-p53 signaling pathway in DLBCL. Our results also demonstrated that the expression levels of both MDM2 and CDC20 were elevated in DLBCL cells and tissues and were markedly associated with disease progression and poor outcomes of DLBCL. Furthermore, targeting CDC20 using small-molecule inhibitor Apcin is likely to benefit wtp53 DLBCL cells in in vitro experiments and xenograft lymphoma animal model.
Recently, studies about the correlation between MDM2 expression and clinical outcomes in DLBCL has been contradictory.8,9 To illustrate this, we studied how MDM2 affected OS in DLBCL patients from GSE10846 (n=420) using Kaplan-Meier survival analysis. The data was analyzed through R2. We pointed out that high expression level of MDM2 was correlated with poor OS in DLBCL patients.
As mentioned above, the overexpression of CDC20 has been observed and considered as a poor clinical outcome in a wide range of tumors,18,21-25 except for DLBCL. We further attempted to clarify whether CDC20 was overexpressed in DLBCL and its role and molecular mechanism in carcinogenesis and tumor progression. We found that CDC20 was overexpressed in DLBCL cells and tissues, and its overexpression predicted a poor OS compared with low CDC20 expression. Furthermore, high expression level of CDC20 was correlated with adverse clinical features of B-symptoms, LDH level and IPI risk in 97 DLBCL patients. A study reported that CDC20 is inhibited by the overexpression of p53,24 as well as the essential role of the MDM2-p53 loop in tumorigenesis and progression of DLBCL, we selected CDC20 for further in-depth exploration and hypothesized that CDC20 was a downstream gene of the MDM2-p53 signaling pathway. The RT-qPCR results showed that CDC20 was downregulated while TP53 was upregulated in MDM2 KD OCI-Ly3 and OCI-Ly10 cells compared with Ctrl cells and confirmed our hypothesis.
It has been reported that targeting CDC20 is a novel and effective cancer therapeutic strategy in several types of cancer,38 however, its clinical role and molecular mechanism in DLBCL has still remained unclear. Recently, we concluded that CDC20 may be a potential novel therapeutic target for DLBCL,27 which is consistent with Maes et al’s results.41 However, in Maes et al’s research, there was a lack of survival analysis regarding CDC20 protein expression, as well as the mechanistic role of CDC20 and its upstream regulators of MDM2-p53 in the pathogenesis of DLBCL. Hence, we used Apcin, a small molecule ligand of CDC20,42 to study the anti-tumor effects of CDC20 on DLBCL cell lines and NOD/SCID mice. In agreement with previous studies regarding cell cycle and apoptosis in other tumors,43,44 we found that the knockdown of CDC20 could inhibit proliferation and growth of DLBCL cells. In the present study, silencing of CDC20 resulted in G2/M phase arrest and induced apoptosis. We further confirmed that the inhibition of CDC20 using Apcin could suppress tumorigenicity in vivo. We also demonstrated that MDM2-p53 could regulate CDC20 expression in DLBCL cells at protein level. With inhibiting MDM2 expression via siRNA, this restored wtp53 and lowered CDC20 expression in OCI-Ly3 and OCI-Ly10 cells. Meanwhile, we upregulated wtp53 using RG7388 and observed that CDC20 protein expression was decreased in OCI-Ly3 and OCI-Ly10 cells. At last, CDC20 inhibitor Apcin knocked down CDC20 expression while no significant differences in MDM2 and wtp53 expression between Apcin treated and control groups were found. Consistent with this, IHC staining for MDM2, wtp53 and CDC20 of xenograft tumor showed the same varying tendency as the results of Western blotting. Therefore, CDC20 may be considered as a downstream gene of the MDM2-p53 signaling pathway. The above-mentioned in vitro and in vivo data indicated that downregulation of CDC20 could change the biological functions and progression of DLBCL.
Conclusion
In summary, we demonstrated that the expression levels of MDM2 and CDC20 were elevated in DLBCL and were significantly associated with poor survival outcome. Our results also revealed that CDC20 could act as a downstream gene of MDM2-p53 singling pathway. Furthermore, the MDM2-p53-CDC20 signaling pathway exhibited to play a critical role in the pathogenesis of DLBCL. Downregulation of CDC20 in DLBCL cells could suppress the proliferation, induce apoptosis and G2/M cell cycle arrest. Thus, targeting CDC20 may be a promising therapeutic strategy for treating DLBCL.
Acknowledgments
We thank Sun Yat-sen University Cancer Center for the support of DLBCL samples and a good platform with excellent experimental conditions.
Author Contributions
Chengtao Sun, Mengzhen Li and Yanfen Feng are co-first authors. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interest.
References
- 1.Huang W, Xue X, Shan L, Qiu T, Guo L, Ying J. Clinical significance of PCDH10 promoter methylation in diffuse large B-cell lymphoma. BMC Cancer. 2017;17(1):815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–494.e415. doi: 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogai H, Dorken B, Lenz G. Pathogenesis of non-Hodgkin’s lymphoma. J Clin Oncol. 2011;29(14):1803–1811. doi: 10.1200/JCO.2010.33.3252 [DOI] [PubMed] [Google Scholar]
- 4.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107. doi: 10.1038/nrc2584 [DOI] [PubMed] [Google Scholar]
- 5.Drakos E, Singh RR, Rassidakis GZ, et al. Activation of the p53 pathway by the MDM2 inhibitor nutlin-3a overcomes BCL2 overexpression in a preclinical model of diffuse large B-cell lymphoma associated with t(14;18)(q32;q21). Leukemia. 2011;25(5):856–867. doi: 10.1038/leu.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moller MB, Ino Y, Gerdes AM, Skjodt K, Louis DN, Pedersen NT. Aberrations of the p53 pathway components p53, MDM2 and CDKN2A appear independent in diffuse large B cell lymphoma. Leukemia. 1999;13(3):453–459. doi: 10.1038/sj.leu.2401315 [DOI] [PubMed] [Google Scholar]
- 7.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocque GL, Møller MB, Colleoni GWB, et al. Poor survival predicted by MDM2 oncoprotein expression in diffuse large B-cell lymphoma (DLBCL) with wild-type TP53 gene. Blood. 2008;112(11):5269. doi: 10.1182/blood.V112.11.5269.5269 [DOI] [Google Scholar]
- 9.Xu-Monette ZY, Moller MB, Tzankov A, et al. MDM2 phenotypic and genotypic profiling, respective to TP53 genetic status, in diffuse large B-cell lymphoma patients treated with rituximab-CHOP immunochemotherapy: a report from the International DLBCL Rituximab-CHOP consortium program. Blood. 2013;122(15):2630–2640. doi: 10.1182/blood-2012-12-473702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci. 2015;112(28):8632–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994 [DOI] [PubMed] [Google Scholar]
- 12.Ding D, Ao X, Liu Y, et al. Post-translational modification of Parkin and its research progress in cancer. Cancer Commun (Lond). 2019;39(1):77. doi: 10.1186/s40880-019-0421-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14(4):233–247. doi: 10.1038/nrc3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau AW, Fukushima H, Wei W. The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front Biosci. 2012;17:2197–2212. doi: 10.2741/4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Zhang C, Hu W, Feng Z. Parkinson’s disease-associated protein Parkin: an unusual player in cancer. Cancer Commun (Lond). 2018;38(1):40. doi: 10.1186/s40880-018-0314-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang L, Barford D. Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr Opin Struct Biol. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Yu H. Mutual regulation between the spindle checkpoint and APC/C. Semin Cell Dev Biol. 2011;22(6):551–558. doi: 10.1016/j.semcdb.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, Roychoudhury S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28(1):81–92. doi: 10.1093/carcin/bgl100 [DOI] [PubMed] [Google Scholar]
- 20.Mao DD, Gujar AD, Mahlokozera T, et al. A CDC20-APC/SOX2 signaling axis regulates human glioblastoma stem-like cells. Cell Rep. 2015;11(11):1809–1821. doi: 10.1016/j.celrep.2015.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang DZ, Ma Y, Ji B, et al. Increased CDC20 expression is associated with pancreatic ductal adenocarcinoma differentiation and progression. J Hematol Oncol. 2012;5:15. doi: 10.1186/1756-8722-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karra H, Repo H, Ahonen I, et al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer. 2014;110(12):2905–2913. doi: 10.1038/bjc.2014.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol. 2012;106(4):423–430. doi: 10.1002/jso.23109 [DOI] [PubMed] [Google Scholar]
- 24.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. 2008;27(11):1562–1571. doi: 10.1038/sj.onc.1210799 [DOI] [PubMed] [Google Scholar]
- 25.Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji P, Smith SM, Wang Y, et al. Inhibition of gliomagenesis and attenuation of mitotic transition by MIIP. Oncogene. 2010;29(24):3501–3508. doi: 10.1038/onc.2010.114 [DOI] [PubMed] [Google Scholar]
- 27.Sun C, Cheng X, Wang C. Gene expression profiles analysis identifies a novel two-gene signature to predict overall survival in diffuse large B-cell lymphoma. Biosci Rep. 2019;39(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1(2):82–87. doi: 10.1038/10049 [DOI] [PubMed] [Google Scholar]
- 29.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27(3):462–473. doi: 10.1016/j.molcel.2007.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes MJ, Kimata Y, Wattam SL, et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol. 2006;8(6):607–614. doi: 10.1038/ncb1410 [DOI] [PubMed] [Google Scholar]
- 31.Wan L, Tan M, Yang J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell. 2014;29(4):377–391. doi: 10.1016/j.devcel.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dybkaer K, Bogsted M, Falgreen S, et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J Clin Oncol. 2015;33(12):1379–1388. doi: 10.1200/JCO.2014.57.7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Abad C, Pisonero H, Blanco-Aparicio C, et al. PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood. 2011;118(20):5517–5527. doi: 10.1182/blood-2011-03-344374 [DOI] [PubMed] [Google Scholar]
- 34.Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP consortium program study. Leukemia. 2012;26(9):2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardesa-Salzmann TM, Colomo L, Gutierrez G, et al. High microvessel density determines a poor outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus chemotherapy. Haematologica. 2011;96(7):996–1001. doi: 10.3324/haematol.2010.037408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515 [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther. 2015;151:141–151. doi: 10.1016/j.pharmthera.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee T, Nath S, Roychoudhury S. DNA damage induced p53 downregulates Cdc20 by direct binding to its promoter causing chromatin remodeling. Nucleic Acids Res. 2009;37(8):2688–2698. doi: 10.1093/nar/gkp110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Q, Zhang Z, Liu JJ, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56(14):5979–5983. doi: 10.1021/jm400487c [DOI] [PubMed] [Google Scholar]
- 41.Maes A, Maes K, De Raeve H, et al. The anaphase-promoting complex/cyclosome: a new promising target in diffuse large B-cell lymphoma and mantle cell lymphoma. Br J Cancer. 2019;120(12):1137–1146. doi: 10.1038/s41416-019-0471-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sackton KL, Dimova N, Zeng X, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514(7524):646–649. doi: 10.1038/nature13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manchado E, Guillamot M, de Carcer G, et al. Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell. 2010;18(6):641–654. doi: 10.1016/j.ccr.2010.10.028 [DOI] [PubMed] [Google Scholar]
- 44.Hadjihannas MV, Bernkopf DB, Bruckner M, Behrens J. Cell cycle control of Wnt/beta-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012;13(4):347–354. doi: 10.1038/embor.2012.12 [DOI] [PMC free article] [PubMed] [Google Scholar]