Abstract
To study thyroid hormone (TH) signaling in the human brain, we analyzed published microarray data sets of the temporal pole (Brodmann area 38) of 19 deceased donors. An index of TH signaling built on the expression of 19 well known TH-responsive genes in mouse brains (T3S+) varied from 0.92 to 1.1. After Factor analysis, T3S+ correlated independently with the expression of TH transporters (MCT8, LAT2), TH receptor (TR) beta and TR coregulators (CARM1, MED1, KAT2B, SRC2, SRC3, NCOR2a). Unexpectedly, no correlation was found between T3S+ vs DIO2, DIO3, SRC1, or TRα. An unbiased systematic analysis of the entire transcriptome identified a set of 1649 genes (set #1) with strong positive correlation with T3S+ (r > 0.75). Factor analysis of set #1 identified 2 sets of genes that correlated independently with T3S+, sets #2 (329 genes) and #3 (191 genes). When processed through the Molecular Signatures Data Base (MSigDB), both sets #2 and #3 were enriched with Gene Ontology (GO)-sets related to synaptic transmission and metabolic processes. Ranking individual human brain donors according to their T3S+ led us to identify 1262 genes (set #4) with >1.3-fold higher expression in the top half. The analysis of the overlapped genes between sets #1 and #4 resulted in 769 genes (set #5), which have a very similar MSigDB signature as sets #2 and #3. In conclusion, gene expression in the human temporal pole can be assessed through T3S+ and fluctuates with subtle variations in local TH signaling.
Keywords: thyroid hormones, gene expression profile, brain, hypothyroidism
Thyroid hormone (TH) signaling is initiated by binding of the active hormone triiodothyronine (T3) to TH nuclear receptors (TR), which modifies the transcriptional activity of T3-responsive genes [1]. TH might also work through noncanonical mechanisms that require binding to TR but are independent of transcriptional regulation of T3-responsive genes [2]. The study of gene expression profiles in cells, tissues, and organs of mice and rats with hypothyroidism or thyrotoxicosis identified sets of genes that respond either positively or negatively to T3 [3, 4]. Whether these genes are directly or indirectly regulated by T3 has only been established in a few cases but, in general, genes that respond rapidly with one or more TH responsive elements (TRE) tend to be directly controlled by T3. Notwithstanding, the collective changes in gene expression triggered by TH signaling in a given tissue or organ constitute the basis for TH-dependent biological effects.
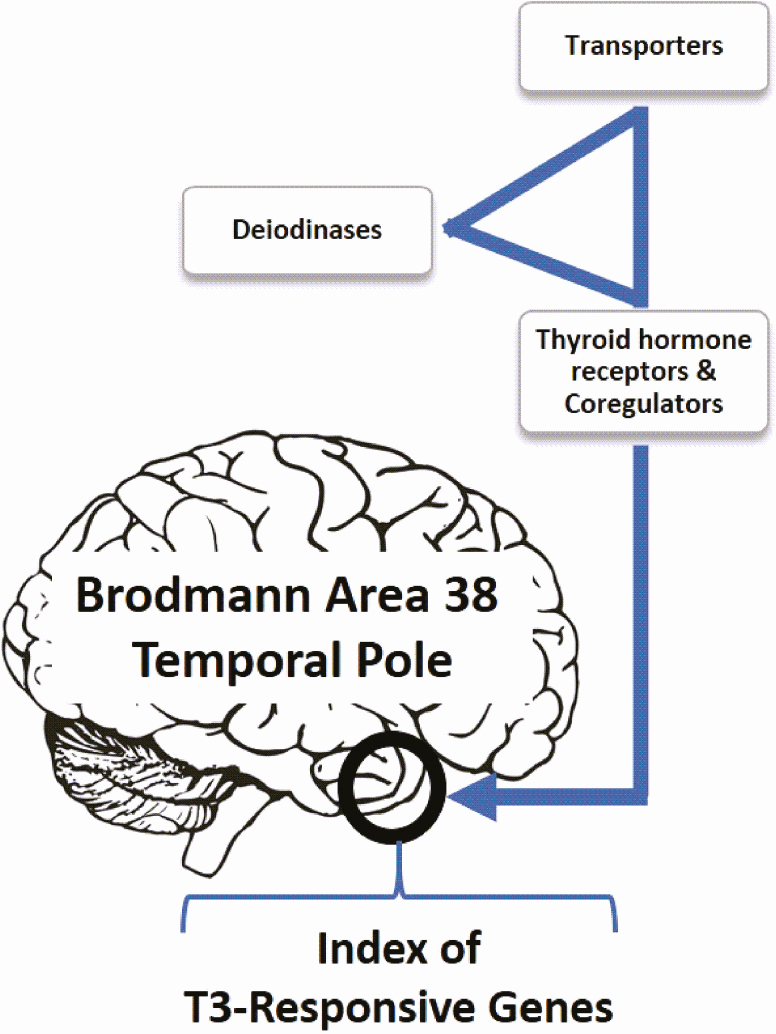
Studies in cells and animal models revealed that TH signaling in the brain depends on a triad of elements that regulate TH action: (i) transmembrane transporters, specifically, organic anion-transporting polypeptides (OATPs), monocarboxylate transporters (MCTs), and L-type amino acid transporters (LATs), that mediate passage of both T4 and T3 through the blood brain barrier and local cell membranes [5, 6]; (ii) deiodinases metabolize TH locally, specifically D2 and D3, respectively activating and inactivating TH [7]; in rats, D2-generated T3 comprises >50% of the T3 bound to TRs in the cerebral cortex [8]; (iii) TRs, mostly TRα and to a lesser extent TRβ (Fig. 1). Signaling through the triad can be initiated via circulating T3, which reaches neurons through MCT8, at the same time that circulating T4 crosses the blood-brain barrier mostly via OATPs. T4 is subsequently taken up by DIO2-expressing astrocytes, mediating T4 activation to T3; the latter acts in a paracrine fashion by entering nearby neurons via MCT8 and binding to TRs to regulate gene expression [9]. Neurons express the inactivating deiodinase D3, which converts T4 and T3 to the inactive molecules rT3 and 3,3’-T2, respectively, terminating TH signaling [10]. However, studies performed in rat brain revealed that TRs are normally almost fully occupied with T3 [8], suggesting that D3 activity in neurons does not substantially limit T3 entry or access to the neuronal nucleus. What role D3 plays in the brain remains to be determined, albeit the D3KO mouse exhibits signs of cerebral thyrotoxicosis [11]. Notwithstanding, as with all other tissues, TH signaling in the brain is likely to be unique for different areas, depending on circulating TH levels and on the exclusive blend of transporters, deiodinases, and TRs present in each area.
Figure 1.
TH signaling in the brain depends on a triad of elements that regulate TH action: (i) transmembrane transporters, ie, OATPs, MCTs, and LATs, that mediate passage of both T4 and T3 through the blood brain barrier and local cells; (ii) deiodinases metabolize TH locally, ie, DIO2 and DIO3, respectively activating and inactivating TH; (iii) TRs, mostly TRα and to a lesser extent TRβ. In the present in silico studies, microarray data previously derived from 19 human Brodmann area 38 (temporal pole) samples was used to generate a T3 positive signaling index (T3S+).
Less is known about TH signaling in the human brain. We know that the key genes involved in the triad that mediates TH signaling in the rodent brain (such as transporters, deiodinases, TRs, and co-regulators) are also expressed in the human brain (Allen Human Brain Atlas: http://human.brain-map.org/), and inactivation mutations of some of these genes have been reported to disrupt TH signaling. For example, male subjects with hemizygous mutations in MCT8 are afflicted with severe intellectual and motor disability, known as the Allan-Herndon-Dudley syndrome [12, 13], but data about the role of MCT10 and OATP1C1 in humans are limited [14, 15]. In adult humans, the presence of MCT8 is detected in neurons of the hypothalamus and in pituitary cells, but there are only few studies about the distribution of MCT8 protein in other areas of the well-developed human brains [16, 17]. The brains of 3 elderly individuals shows MCT8 protein almost exclusively in endothelial cells from different regions of the brain, including parietal, occipital, and limbic cortex, hippocampus, hypothalamus, amygdala, and cerebellum [18]. In addition, in human fetuses, MCT8 expression was detected in the blood brain barrier in all analyzed regions. Furthermore, an intense MCT8 expression was detected in other cell types, including radial glial cells in cerebral cortex and in ventricular zone [19]. Severe mental retardation (IQ < 60) is uncommon in carriers of TRβ mutations, but about 30% of affected subjects display a mild learning disability (IQ < 85), with some suggesting a relationship with attention deficit hyperactivity disorder (ADHD) [20]. At the same time, studies of patients with TRα1 mutations revealed cognitive deficits that were consistent with those seen in congenital hypothyroidism [21, 22].
The adult human brain responds to TH as illustrated by the marked changes in mood and cognition observed during the transition from hypo- to euthyroid to thyrotoxic states [23]. However, it is not clear that the human brain responds to small changes in TH signaling. For example, mood and cognition were not affected when levothyroxine (LT4)-treated hypothyroid patients were randomized to a small increase or decrease of LT4 dose [24]. On one hand, knowledge about the several elements involved in TH signaling in the rat brain [25], including the deiodinases, suggests that homeostatic mechanisms could compensate for minimal changes in TH levels, preventing changes in T3-dependent processes in the brain [26]. On the other hand, studies of T3-responsive genes in LT4-treated hypothyroid rats with normal thyrotropin (thyroid-stimulating hormone) indicate that the minimal reduction in plasma T3 leads to brain hypothyroidism [27]. The situation in the human brain is not known but could carry significant clinical implications. For example, could the relatively lower serum T3 levels observed in LT4-treated patients [28] affect TH signaling in the brain? Or explain symptoms of impaired mood and cognition seen in some of these patients [29]?
Here we used data from a previous study of the cerebral temporal pole transcriptome of 19 deceased donors that died of trauma but were relatively young and had no history of neurological or thyroid disease, to study brain TH signaling [30]. The temporal pole corresponds to Brodmann area 38 and has strong connections with the amygdala and orbital prefrontal cortex. It is considered an association cortex involved with multimodal analysis, especially in social and emotional processing [31], all known to be affected by TH signaling [23, 32]. Furthermore, studies in humans indicate that hypo- and hyperthyroidism are associated with changes in different limbic and paralimbic structures, including the temporal gyrus [33-35]. Thus, utilizing these microarray data, we created an index of TH signaling based on the expression of known T3-responsive genes in rodents (Table 1; Supplementary Table 1 [36]). This index was further used to study whether TH signaling in these donors correlates with specific patterns of gene expression in the temporal pole.
Table 1.
Genes Positively Regulated by T3 Used to Prepare the T3S+ Index
| Gene Symbol | Function | Species | Brain Area | Main Cell Type [59] |
|---|---|---|---|---|
| ABCD2 | Import of fatty acids into organelles | Mus musculus/Rattus norvegicus | Cortex [3, 60], striatum [4, 61] | Astrocytes |
| ATP2B2 | Involved in intracellular calcium homeostasis | Mus musculus/Rattus norvegicus | Cortex [3, 60, 62] | Neurons |
| CNTNAP1 | Organization of myelinated axons/Signaling between axons and myelinating glial cells | Mus musculus/Rattus norvegicus | Cortex [3, 60, 63] hippocampus [63] | Neurons |
| DCLK1 | Involved in neuronal migration, apoptosis and neurogenesis | Mus musculus/Rattus norvegicus | Cortex [60, 64] striatum [4] | Astrocytes, neurons |
| DNM3 | Involved in vesicular transport | Mus musculus/Rattus norvegicus | Cortex [64], cerebellum [3] hippocampus [63] | Neurons, oligodendrocytes |
| HOMER1 | Acts in regulation of group 1 metabotrophic glutamate receptor function | Mus musculus/Rattus norvegicus | Cortex [62, 64] Striatum [4, 61] | Microglia |
| ITPR1 | Involved in the formation of calcium ions channel | Mus musculus/Rattus norvegicus | Cortex [60, 63] cerebellum [3, 65] striatum [4] | Neurons |
| KCNA1 | Involved in the formation of potassium channels | Mus musculus/Rattus norvegicus | Cortex [63], cerebellum [60, 65] | Neurons |
| KLF9 | Functions as a transcription factor regulating keratinocyte proliferation | Mus musculus/Rattus norvegicus | Cortex [63], cerebellum [60, 65] striatum [61] | OPC, astrocytes, oligodendrocytes |
| LNPEP | Acts in inactivation of peptide hormones | Mus musculus | Cortex [60, 64] | Astrocytes, neurons |
| NEFH | Biomarker of neuronal damage | Mus musculus/Rattus norvegicus | Cortex [60, 62, 63] cerebellum [65] hippocampus [63] striatum [4] |
Neurons |
| NEFM | Biomarker of neuronal damage | Mus musculus/Rattus norvegicus | Cortex [60, 62] striatum [4] | Neurons |
| NMT2 | Involved in regulating of signaling proteins | Mus musculus | Cortex [64] cerebellum [3] | Neurons |
| OPCML | Acts as an accessory for opioid receptor function | Mus musculus/Rattus norvegicus | Cortex [62, 64] cerebellum [3] | Neurons |
| PVALB | Involved in calcium handling | Mus musculus/Rattus norvegicus | Cortex [60, 63] cerebellum [3] hippocampus [63] striatum [4] | Neurons |
| RBBP4 | Involved in chromatin metabolism regulation | Mus musculus/Rattus norvegicus | Cortex [64] cerebellum [3, 65] | Astrocytes, neurons, oligodendrocytes |
| SEMA7A | Involved in immunomodulatory and neuronal process | Mus musculus/Rattus norvegicus | Cortex [60, 62] cerebellum [ [3] striatum [4] |
Astrocytes, oligodendrocytes |
| SORL1 | Acts in several intracellular sorting and trafficking functions/Regulation of the transformation of amyloid precursor to amyloid-β | Mus musculus/Rattus norvegicus | Cortex [62, 64] striatum [4] | Microglia |
| TBC1D30 | Involved in GTPase activity/Intracellular protein transport | Mus musculus/Rattus norvegicus | Cortex [60, 62] striatum [4, 61] | Neurons |
Abbreviations: ABCD2: ATP binding cassette subfamily D member 2; ATP2B2: ATPase, Ca++ transporting, plasma membrane 2; CNTNAP1: Contactin-associated protein 1; DCLK1: Doublecortin-like kinase 1 DNM3: Dynamin 3; HOMER1: Homer scaffold protein 1; ITPR1: Inositol 1,4,5-trisphosphate receptor type 1; KCNA1: Potassium voltage-gated channel subfamily A member 1; KLF9: Kruppel like factor 9; LNPEP: Leucyl and cystinyl aminopeptidase; NEFH: Neurofilament heavy; NEFM: Neurofilament medium; NMT2: N-myristoyltransferase 2; OPCML: Opioid binding protein/cell adhesion molecule-like PVALB: Parvalbumin; RBBP4: Retinoblastoma binding protein 4, chromatin remodeling factor; SEMA7A: Semaphorin 7A; SORL1: Sortilin related receptor 1; TBC1D30: TBC1 domain family, member 30.
1. Materials and Methods
TH signaling index
Multiple T3-responsive genes in brain tissue were previously identified in animal and cell models by contrasting gene expression between hypothyroid and thyrotoxic states. However, even if these genes are directly regulated by T3 they may also be affected by other cellular signaling pathways. It is conceivable that by looking at the behavior of a large enough group of T3-responsive genes one would maximize the reading of T3-dependent pathways while minimizing other non-T3 related influences. To study this index of TH signaling in the human brain (T3S+), we looked into a data set previously published from our laboratory [30] that contains human brain (temporal pole) microarray results and asked whether these gene sets could be used to estimate TH signaling in the human brain. To answer this question, we used a previous study [4] that looked at a series of publications in mice and rats and identified a total of 4108 T3-responsive genes in the brain. For the present analysis, we used a more stringent criteria: we only include T3-responsive genes that (i) had been reported by 3 or more publications and (ii) at least one of these reports had to have been performed in cerebral cortex. These 2 criteria led us to a much smaller list of 120 genes. Next, we selected genes that were reportedly positively regulated by T3, which reduced the list to only 30 genes—unfortunately, many studies did not specify whether the gene is positively or negatively regulated by T3. We next used these 30 genes to build a preliminary T3S+ (PT3S+) index. Subsequently, we performed several correlation rounds between each one of the 30 genes vs the PT3S+ index, removing from the PT3S+ index any gene that returned a R (Spearman) <0.5; always starting with the ones that had smaller R values. The use of this filter was justified because we wanted to create a consistent group of T3-responsive genes, knowing that we started with 30 genes identified in rodents, not humans, and that many genes were originally observed to be responsive to T3 in developing brain or cells (Table 1). Thus, it is not surprising that some of these genes might not be good surrogates of T3 action in adult humans. This filtering eliminated 11 genes, which led us to 19 consistently T3-responsive genes (positively) that were used to build the T3S+ index (Table 1; Supplementary Table 1a and 1b [36]).
Cohort of human temporal pole samples and microarray data
The present in silico studies were performed using microarray data reported previously [30]. In brief, experimental data were generated through the study of brain tissue samples collected from postmortem human donors at the University of Miami Brain Endowment Bank. Brain samples from 19 male, Caucasian, middle-aged (43.6 ± 12.2 years), adult donors without known thyroid or neurologic disease were studied as described previously. The cause of death was limited to accident or sudden cardiac death without medical intervention or prolonged agonal state. Postmortem interval at specimen collection was <24 hours, brain pH (quality measure) was >6.0. A neuroanatomist dissected homogenous samples from frozen coronal blocks based on surface and cytoarchitectural landmarks from the Brodmann area 38 (temporal cortex pole) and stored at −80 °C. Microarray analysis was performed at the Joslin Diabetes Center Genomics Core Laboratory with Genechip Human Gene 2.0 ST arrays, which utilizes a whole transcript design to assess more than 30 000 coding genes, and the Affymetrix Expression Console software [30]. Robust Multi-array Average (RMA) algorithm was used to create an expression matrix from Affymetrix data. After obtaining the expression data, a specific filter was developed by MySQL and Java software to only include unique named genes, resulting in 22 257 genes. The individual gene mRNA values of each donor were used to create the linear correlation analysis. As indicated, gene sets were analyzed using the Molecular Signatures Data Base (MSigDB) for canonical pathways enriched in the gene set. The Affymetrix data created (.CEL file) was subsequently used to analyze differentially expressed genes within the population, using Transcriptome Analysis Console (TAC, Affymetrix). These data were also processed for gene set analysis using the Gene Set Enrichment Analysis software (GSEA; Broad Institute).
THRA expression analysis.
As an additional approach to study the transcript variant 1 of the TRα gene, we performed a real-time quantitative polymerase chain reaction (RT-qPCR analysis). For this, RNA from all 19 human brain samples was extracted (RNeasy Lipid Tissue Mini Kit, Qiagen) and cDNA generated (First Strand cDNA Synthesis Kit, Roche). Genes of interest were measured by RT-qPCR (StepOnePlus Real-Time PCR Detection System, Applied Biosystems) using PowerUp SYBR Green Master Mix (Applied Biosystem) with the following conditions: 20 seconds at 95 °C, 3 seconds at 95 °C, and 30 seconds at 60 °C × 40, and 15 seconds at 95°C followed by 1 minute at 60°C and 15 seconds at 95°C. Standard curves consisting of 5 points of a serially diluted mixture of experimental and control cDNA; the amplification efficiency was 80% to 110% with a coefficient of correlation consistently >0.98. The melting curve protocol was used to verify the specificity of the amplicon generation. Cyclophilin A (CYCLOA) was used as a housekeeping internal control gene. Results were expressed as the ratio of target mRNA (THRA) to CYCLOA mRNA. The pair of primer designed are shown on Supplemental Data (Supplementary Table 2a [36]).
Homogeneity of the samples.
To test for homogeneity of all brain samples, we looked at the expression levels of 10 markers of apoptosis, having found a coefficient of variance (CV) of 2.3% to 4.8%, and also that none of the markers correlated with T3S+ (P > 0.05; Supplementary Table 3a [36]). Cerebral cortex contains multiple cell types, including astrocytes, oligodendrocytes, and neurons, and we wanted to ensure that the relative representation of these cells was not markedly different among the samples. Thus, we looked at the expression of known cell-specific markers for neurons, oligodendrocytes, and astrocytes, having found a CV from 1.7% to 9.8%. More importantly, none of these genes correlated with T3S+ (P > 0.05; Supplementary Table 3b [36]).
Statistical analysis
Nonparametric correlation (Spearman) analysis was used throughout; we qualified the correlation as strong (r > 0.7), moderate (0.5 < r < 0.7) and weak (r < 0.5); in the different gene set analyzes we also used the false discovery rate (FDR)-q value (adjusted P values according to FDR) to assess statistical significance. All data analyzes, including covariation, were using Microsoft Excel and GraphPad Prism 5 software. Unless otherwise indicated, data are the mean ± standard deviation (SD) and Spearman r correlation coefficient; P < 0.05 was used to reject the null hypothesis. We used Factor Analysis to describe variability among observed, correlated variables in terms of a potentially lower number of unobserved variables or factors. The observed variables were modelled as linear combinations of the potential factors, plus “error” terms. Factor analysis aims to find independent latent variables. We first used the Kaiser-Meyer-Olkin (KMO) analysis followed by the Bartlett test. A KMO > 0.70 was considered good and >0.85 excellent. The factor analysis used orthogonal rotation Varimax with “Kaiser” normalization; we only considered Eigenvalues > 1.0 [37].
2. Results
An index for TH signaling in the human temporal pole
The Allen Human Brain Atlas (http://human.brain-map.org/) indicates that the human temporal pole expresses key genes necessary to mediate TH signaling: (i) TH transporters at levels equivalent to the rest of the brain except for MCT8 and OATP1C that are relatively overexpressed (Z-score 1.2 to 1.5), and MCT10 that is relatively under-expressed (Z-score −0.5); (ii) TRs at levels that are below the rest of the brain (Z-score −0.7 to −1.1); (iii) deiodinases (DIO2-DIO3) at levels above the rest of the brain (Z-score 0.5-1.5).
mRNA levels for T3-responsive genes were used to create an index of TH signaling. This was done by first calculating the relative expression of each one of the 19 T3-responsive genes among the 19 donors after the level of each individual donor was divided by the average of the group (Supplementary Tables 1a and 1b [36]). Subsequently, the relative expression level of all positively regulated genes was averaged, returning an expression index (T3S+) that varied from 0.92 to 1.1, with a mean ± SD of 1.0 ± 0.042 (±2SD = 0.92-1.08) and a CV of 4%.
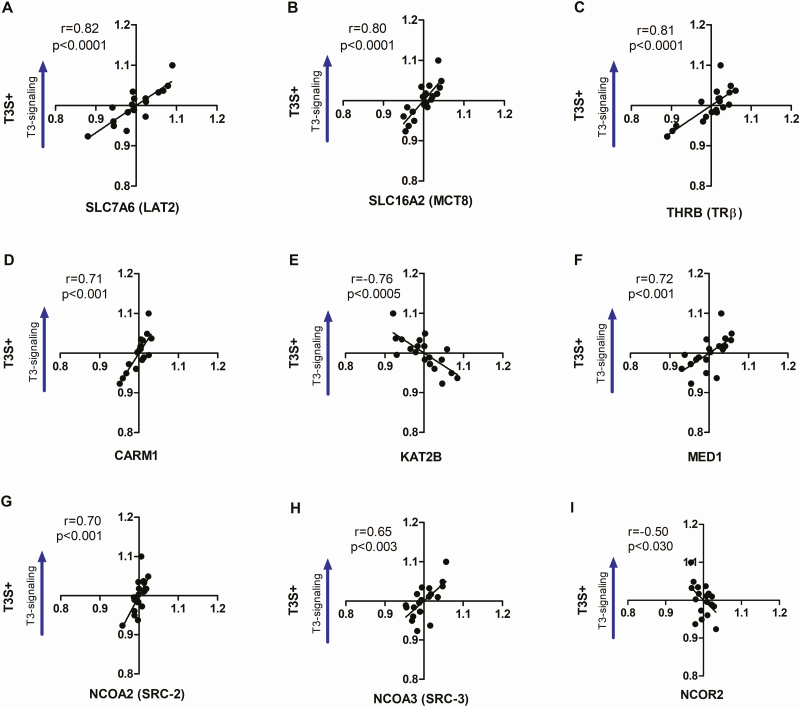
Next, we wished to validate the T3S+ and asked whether it covaries with the expression of 27 genes known to play a role in TH signaling, that is, TH transporters, deiodinases, TRs, and transcriptional modulators (Set #0). In general, we found a linear relationship between some of these variables and T3S+, with the expected positive covariation and a moderate/strong correlation with r values between 0.47 and 0.82 (Table 2). Specifically, we found that the intensity of T3S+ correlated positively with the expression of LAT1, LAT2, MCT8, and MCT10 (Table 2), whereas no correlation was found with 2 OATPs (Table 2). While it is not possible to define a causal relationship between TH signaling and the group of TH transporters, the fact that the mRNA for TH transporters are only minimally affected by TH signaling in the brain [38] suggests that the expression of TH transporters could be an important factor defining the intensity of T3S+ in this tissue. There was a moderate positive correlation between DIO2 and DIO3 mRNA levels (r = 0.52; P = 0.02), but no correlation was observed between T3S+ and the expression of DIO2 or DIO3 (Table 2). The relationship between TR expression vs T3S+ was less straightforward. TRβ covaried positively with T3S+, but TRα did not (Table 2). The latter was confirmed after TRα mRNA levels were measured using RT-qPCR and primers that were specific for TRα1 (Supplementary Table 2 [36]). There was a positive correlation between T3S+ vs the coactivators CARM1, MED1, NCOA2, NCOA3, and PPARGC1A, but not vs NCOA1 or PPARGC1B (Table 2). Furthermore, the corepressor NCOR2 covaried negatively with T3S+ (Table 2). Unexpectedly, T3S+ covaried positively with NCOR1 (Table 2) and negatively with the coactivator KAT2B (Table 2).
Table 2.
Correlation Analysis of Gene Expression of Elements Involved in T3 Signaling (Set #0) and TH Positive Signaling Index (T3S+)
| Gene | T3S+ | |
|---|---|---|
| r | P | |
| TH transporters | ||
| SLC7A5 (LAT1) | 0.55 | 1.38E-02 |
| SLC7A6 (LAT2) | 0.82 | 2.07E-05 |
| SLC16A2 (MCT8) | 0.80 | 4.49E-05 |
| SLC16A10 (MCT10) | 0.59 | 7.91E-03 |
| SLCO1C1 (OATP1C1) | -0.34 | ns |
| SLCO4A1 (OATP4A1) | -0.05 | ns |
| TH receptors | ||
| THRA (TRα) | -0.04 | ns |
| THRB (TRβ) | 0.81 | 2.54E-05 |
| Deiodinases | ||
| DIO2 (D2) | -0.40 | ns |
| DIO3 (D3) | -0.28 | ns |
| Coactivators | ||
| CARM1 | 0.71 | 6.51E-04 |
| CREBBP (CBP) | 0.21 | ns |
| EP300 (P300) | -0.12 | ns |
| KAT2B | -0.76 | 1.80E-04 |
| MED1 (TRAP220) | 0.72 | 4.50E-04 |
| NCOA1 (SRC-1) | 0.38 | ns |
| NCOA2 (SRC-2) | 0.70 | 7.77E-04 |
| NCOA3 (SRC-3) | 0.65 | 2.37E-03 |
| NCOA6 | -0.07 | ns |
| PPARGC1A (PGC-1ALFA) | 0.47 | 4.22E-02 |
| PPARGC1B (PGC-1BETA) | 0.06 | ns |
| SMARCB1 (SNF5) | 0.45 | ns |
| Corepressors | ||
| HR | 0.02 | ns |
| NCOR1 | 0.56 | 1.31E-02 |
| NCOR2 | -0.50 | 2.99E-02 |
| NR0B1 (DAX-1) | -0.45 | ns |
| NR0B2 (SHP) | 0.27 | ns |
Data are the Spearman r coefficient correlation and P value.
Abbreviations: CARM1: Coactivator associated arginine methyltransferase 1; CREBBP: CREB binding protein; DIO2: Type 2 iodothyronine deiodinase; DIO3: Type 3 iodothyronine deiodinase; EP300: E1A binding protein p300; HR: lysine demethylase and nuclear receptor corepressor; KAT2B: lysine acetyltransferase 2B; MED1: mediator complex subunit 1; NCOA1: nuclear receptor coactivator 1;NCOA2: nuclear receptor coactivator 2; NCOA3: nuclear receptor coactivator 3; NCOA6: nuclear receptor coactivator 6; NCOR1: nuclear receptor corepressor 1; NCOR2: nuclear receptor corepressor 2; NR0B1: nuclear receptor subfamily 0 group B member 1; NR0B2: nuclear receptor subfamily 0 group B member 2; PPARGC1A: PPARG coactivator 1 alpha; PPARGC1B: PPARG coactivator 1 beta; SLC7A5: solute carrier family 7 member 5; SLC7A6: solute carrier family 7 member 6; SLC16A2: solute carrier family 16 member 2; SLC16A10: solute carrier family 16 member 10; SLCO1C1: solute carrier organic anion transporter family member; SLCO4A11: solute carrier organic anion transporter family member 4A1; SMARCB1: SWI/SNF-related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1; THRA: thyroid hormone receptor alpha; THRB: thyroid hormone receptor beta.
Knowing that neurons are more responsive to T3 than other cells in the central nervous system, we filtered the genes in T3S+ to include only genes typically expressed in neurons (n = 10) and created a NT3S+. We next correlated NT3S+ with the 27 genes known to play a role in TH signaling. Remarkably, the results obtained (Supplementary Table 4a and 4b [36]) were essentially similar to when T3S+ was used in the correlations (Table 2). We also used the Microsoft Excel “randbetween” function to create an additional index based on genes randomly selected from the list of 22 257 genes in the microarray (RT3S+) that functioned as a negative control. Not surprising, RT3S+ did not correlate with any of the 27 genes known to play a role in TH signaling (Supplementary Table 5a and 5b [36]).
We next reanalyzed the data in Table 2 using a Factor analysis to identify which of these genes or gene sets (factors) correlated independently with T3S+. The calculated KMO value for this dataset was 0.973, and the Bartlett test returned a P < 0.001, confirming the existence of independent factors. After only ten iterations the analysis converged on 8 independent factors (Fat 1-8) with Eigenvalues > 1.0 (Table 3); combined, these 8 factors explained 90.2% of the total variability observed (Supplementary Table 6a [36]). After the Fat 1-8 scores were tested against T3S+ through Pearson’s correlation, only Fat-1 and Fat-2 reached statistical significance, which include TRβ, 6 coregulators (CARM1, KAT2B, NCOR2, SRC2-3, MED1) and 2 transporters (LAT2, MCT8) (Fig. 2; Tables 3 and 4).
Table 3.
Factorial Charge of TH Signaling Related Genes (Set #0) in Each Factor
| TH signaling genes | Fat 1 | Fat 2 | Fat 3 | Fat 4 | Fat 5 | Fat 6 | Fat 7 | Fat 8 |
|---|---|---|---|---|---|---|---|---|
| CARM1 | 0.899 | |||||||
| THRB | 0.895 | |||||||
| KAT2B | −0.822 | |||||||
| NCOR2 | −0.852 | |||||||
| NCOA3 | 0.838 | |||||||
| MED1 | 0.756 | |||||||
| SLC7A6 | 0.702 | |||||||
| NCOA2 | 0.691 | |||||||
| SLC16A2 | 0.540 | |||||||
| CREBBP | 0.882 | |||||||
| NCOA6 | 0.863 | |||||||
| EP300 | 0.555 | |||||||
| NCOR1 | 0.547 | |||||||
| SLCO4A1 | −0.479 | |||||||
| THRA | 0.450 | |||||||
| DIO2 | 0.9 | |||||||
| DIO3 | 0.801 | |||||||
| SLCO1C1 | 0.789 | |||||||
| SMARCB1 | -0.608 | |||||||
| HR | 0.898 | |||||||
| SLC7A5 | 0.761 | |||||||
| SLC16A10 | 0.813 | |||||||
| PPARGC1A | 0.789 | |||||||
| PPARGC1B | 0.596 | |||||||
| NR0B1 | −0.919 | |||||||
| NCOA1 | 0.688 | |||||||
| NR0B2 | 0.912 |
Extraction method: Principal component analysis; Rotation method: Varimax with Kaiser normalization; Rotation converged in 10 iterations.
Figure 2.
Correlation analysis between T3S+ and the components included at the Factors 1 and 2 that reached statistical significance in Factor Analysis. TH transporters LAT2 (A), MCT8 (B), TH receptor TRβ (C), coactivators CARM1 (D), KAT2B (E), MED1 (F), SRC-2 (G), and SRC-3 (H), and corepressor NCOR2 (I). Statistical significance is shown in each graph as well as Spearman’s correlation coefficient (r).
Table 4.
Pearson Correlation Between Factors Scores and T3S+
| Factors | Set #0 | Set #1 | ||
|---|---|---|---|---|
| Pearson r | P value | Pearson r | P value | |
| 1 | 0.67 | 0.002 | 0.42 | 0.074 |
| 2 | 0.54 | 0.016 | 0.38 | 0.112 |
| 3 | 0.07 | 0.786 | 0.54 | 0.017 |
| 4 | −0.23 | 0.35 | 0.59 | 0.007 |
| 5 | 0.15 | 0.549 | 0.04 | 0.872 |
| 6 | 0.37 | 0.124 | 0.00 | 0.992 |
| 7 | 0.10 | 0.683 | −0.03 | 0.898 |
| 8 | 0.06 | 0.816 | 0.01 | 0.959 |
| 9 | −0.08 | 0.744 | ||
| 10 | 0.12 | 0.616 | ||
| 11 | −0.07 | 0.767 | ||
| 12 | 0.06 | 0.816 | ||
| 13 | −0.02 | 0.948 | ||
| 14 | 0.00 | 0.994 | ||
| 15 | 0.05 | 0.831 | ||
| 16 | 0.01 | 0.975 | ||
| 17 | −0.05 | 0.825 | ||
| 18 | −0.01 | 0.956 | ||
Unbiased search for gene-sets and cellular pathways that correlate positively with T3S+
The fact that T3S+ correlates positively with the expression of 2 TH transporters, TRβ, and 4 coactivators, and negatively with the corepressor NCOR2 (Fig. 2) prompted us to explore the possibility that T3S+ could be used to identify T3-sensitive pathways in the human cerebral pole. To that end, we used 2 unbiased strategies: (i) correlation analysis to identify genes that correlate positively with T3S+, and (ii) TAC to contrast the transcriptome of donors with the lowest T3S+ (LT3S+) vs the highest T3S+ (HT3S+).
Correlation analysis between individual genes and T3S+
All 22 257 named genes in the microarray were tested for Spearman correlation analysis and ranked according to “r”. We found 1649 genes (set #1) that strongly correlated with T3S+ (r > 0.75; Supplementary Table 7 [36]). In contrast, when RT3S+ was used, we only identified 6 genes that correlated strongly (r > 0.75) with the 22 257 genes (Supplementary Table 5c [36]). Next, we wished to validate this analysis and tested whether similar results could be obtained by correlating the 22 257 named genes with TRα mRNA or TRβ mRNA. Whereas the correlation with TRα yielded only 7 genes with r > 0.75 and 22 genes with r > 0.70 (Supplementary Table 8 [36]), TRβ correlated strongly with 1447 genes (r > 0.75). Remarkably, 71% of these genes (1029 genes) overlapped with the genes in set #1 (Supplementary Table 9 [36]).
We next performed Factor analysis to identify which of these genes or gene sets (factors) correlated independently with T3S+. The calculated KMO value was 0.954, and the Bartlett test returned a P < 0.001, confirming the existence of independent factors. After 51 iterations the analysis converged on 18 independent factors (Fat 1-18) with Eigenvalues > 1.0 (Supplementary Table 10 [36]); combined, these 18 factors explained 100% of the total variability observed (Supplementary Table 6b [36]). After the Fat 1-18 scores were tested against T3S+ through Pearson correlation, only Fat-3 and Fat-4 reached statistical significance, which included 329 (set #2) and 191 (set #3) genes, respectively (Table 4).
The physiological context of gene sets #2 and #3 was assessed through MSigDB, that searches for canonical pathways enriched in the gene sample (Supplementary Tables 11-16 [36]). These gene sets were ranked within each category according to the enrichment scores (ES). Among the top 20 GO-sets enriched for gene set #2 under “biological processes”, “cellular component” and “molecular function” there were 14 to 15 sets related to synaptic transmission, mitochondria, and cellular transport, whereas for gene set #3 the top 20 GO-sets contained 14 to 15 sets related to ionic transport and metabolic processes.
TAC contrasting donors with the lowest T3S+ vs the highest T3S+
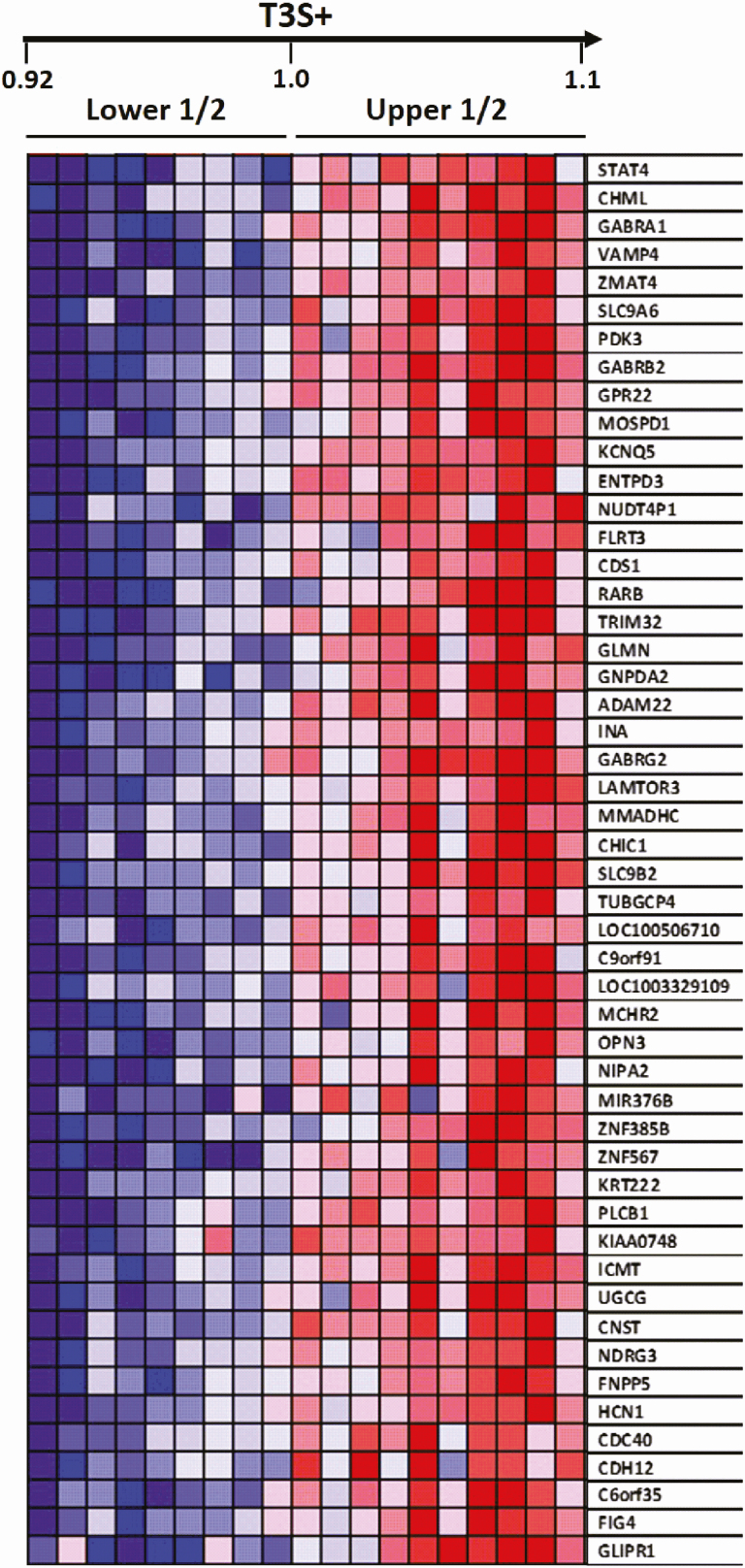
An additional approach to identify pathways that co-vary with TH signaling was to rank the group of donors according to the T3S+. This resulted in 2 subgroups of donors, one containing the lowest levels of T3S+ (LT3S+ donors # 18, 2, 17,12, 4, 9, 16, 7, 5) and the other with the highest levels of T3S+ (HT3S+ donors # 15, 8, 11, 9, 6, 11, 3, 10, 13,14) (Fig. 3; Supplementary table 1 [36]). An analysis using TAC identified 1262 genes (set #4) enriched in the HT3S+ subgroup (P < 0.05; fold change > 1.3; Supplementary Table 17 [36]).
Figure 3.
Heat map indicating the top 50 genes enriched in temporal pole samples when 2 subgroups of donors: lower (LT3S+) vs higher (HT3S+) were processed through GSEA.
Next, we compared gene sets #1 and #4 having found that 769 genes (set #5) overlapped. The physiological context of gene set #5 was assessed through MSigDB, that searches for canonical pathways enriched in the gene sample (Supplementary Tables 18-20 [36]). After gene sets were ranked within each category according to enrichment score, the top 20 gene sets under “biological processes” and “cellular components” were found to contain from 14 to 15 genes sets directly related to synaptic signaling, synaptic vesicle recycling and transport, and regulation of neurotransmitter levels and release (Supplementary Tables 18 and 19 [36]). The top 20 gene sets under “molecular function” were mostly related to transmembrane transport and cellular metabolism (Supplementary Table 20 [36]).
3. Discussion
The present studies revealed that the expression of genes in the human temporal pole fluctuates with subtle changes in local TH signaling. This was concluded based on the correlation analysis of an index of TH action (T3S+) vs expression data obtained from the human cerebral temporal pole. T3S+ correlated with the expression of 2 transporters (LAT2, MCT8), TRβ, and 6 coregulators (CARM1, KAT2B, NCOR2, SRC2-3, MED1) (Fig. 2). Furthermore, unbiased analyses of expression data identified 2 groups of hundreds of genes that correlate independently with T3S+. These sets were enriched with genes involved in cellular pathways known to be regulated by TH signaling, that is, synapsis function, cellular transport, and metabolism (Supplementary Tables 11-16, 18-20 [36]). Of course, it is not expected that these hundreds of genes are directly regulated by T3. Instead, they likely reflect fine tuning of large gene networks affected by subtle variations in TH signaling. It is likely that these changes in the transcriptome will eventually be transduced into biological processes with physiological and/or clinical relevance. A remarkable finding was that the expression of slightly more than two-thirds of the genes that correlated with T3S+ also correlated with TRβ expression, but not TRα. In fact, only a handful of genes was found to correlate with TRα. While it is conceivable that the correlation indicates that T3 stimulates the expression of TRβ, this is unlikely given that neither TR is responsive to T3 in the brain [39]. These data need to be confirmed, including measurement of protein levels, but at face value, it indicates that TRβ might play a dominant role in TH signaling in the temporal pole.
The finding of a positive correlation between T3S+ and the expression of TH transporters suggests that TH transporters are limiting to TH signaling in the human temporal pole. This of course, is only if the transporters follow their mRNA levels. The available data indicate that the absence of functional transporters is limiting to TH signaling [12, 13]. However, it is accepted that as long as there is a minimum level of transporters available, additional transporter units are not expected to enhance TH signaling. This is because movement of T3 molecules across the membrane follows a concentration gradient, as T3 transport is not active. Thus, the present studies indicate that TH transporters might be limiting to TH signaling in some settings, even in the absence of inactivating mutations. These findings also beg the question of whether it is the availability of TH transporters that modulate TH signaling or vice versa. While we know that transporters facilitate TH signaling in the brain [12, 13], it remains unclear whether TH signaling affects the expression of TH transporters. Of note, changes in mRNA levels are small and there is conflicting evidence, with great interspecies variability [38]. However, the fact that the correlation between 2 transporters and T3S+ was moderate/strong, makes it likely that in the human temporal pole, TH transporters through which T3 can enter target cells are limiting and affect TH signaling. In contrast, T3S+ did not correlate with the expression of 2 OATPs (Table 2), transporters that preferentially facilitate the flux of T4 in/out of cells.
It is unexpected that the intensity of T3S+ also did not correlate with the expression of deiodinases (Table 2). Glia-specific D2KO animals exhibit a phenotype of brain hypothyroidism [40] and the global D3KO mouse exhibit a phenotype of brain thyrotoxicosis [41, 42]. At face value, this suggests that within this narrow range of TH signaling in the human brain, deiodinases play a more homeostatic role preserving TH signaling, rather than defining it. The lack of correlation could also be explained by other factors that affect deiodinase synthesis. For example, all deiodinases are selenoproteins, which require cotranslational incorporation of selencysteine into the nascent polypeptide chain [43]. Thus, mRNA levels in this case might not be the only limiting factor for the expression of the active deiodinases. In addition, there are specific mechanisms for D2 and D3 regulation. D2 is mostly regulated at posttranslational level via ubiquitination [44, 45]; D3 is not known to be ubiquitinated but has been shown to suffer posttranslation changes in subcellular localization, as it relocates to the nuclear membrane and reduces TH signaling [46].
TH coactivator and corepressors are not regulated by TH signaling in the postnatal developing cerebellum [47]. We are not aware of similar studies performed in other brain areas. Thus, the positive correlation between T3S+ and the expression of CARM1, SRC2-3, and MED1 is reassuring that these molecules positively affect TH signaling in the temporal pole (Table 2). Likewise, the negative correlation between NCOR2 and T3S+ suggests that the expression of this corepressor dampens TH signaling in the temporal pole. However, it was unexpected to see that neither TRα or SRC1 mRNA levels correlated with T3S+, and that KAT2B correlated negatively with T3S+. Both TRs are expressed in brain and mediate TH signaling, but TRα1 protein account for about 70% to 80% of total receptor content [48]. Likewise, at least in the rat brain, SRC1 is expressed ubiquitously [49]. The significance of these findings is unknown, but it could suggest that neither TRα nor SRC1 is unlikely to constitute a limiting factor for TH signaling in the temporal pole. KAT2B is an acetyl transferase previously shown to interact with TR and coactivate T3-induced gene expression [50]. However, KAT2B also coregulates other important pathways in the central nervous system, including the Hedgehog-Gli pathway [51], which inhibits TH signaling [52, 53], possibly explaining the inverse relationship with T3S+.
The analyses of the gene sets that correlated with T3S+ revealed important cellular functions in the human brain that fluctuate with TH signaling, for example, to synaptic signaling, synaptic vesicle recycling and transport, and regulation of neurotransmitter levels and release (Supplementary Tables 18-20 [36]), all carrying a high energetic cost. Thus, it is not surprising that TH signaling accelerates metabolism and energy expenditure in the human brain. In fact, this is a concept that has evolved over time. In the 1950s, while measuring cerebral blood flow and O2 consumption, investigators failed to detect differences between hyperthyroid patients and normal subjects [54, 55]. The idea of brain refractoriness to TH signaling persisted for decades until more accurate measurements could be performed. The utilization of 2-[(14)C]-deoxyglucose in mice revealed that TH signaling accelerates glucose utilization via TRα and not TRβ. While glucose utilization remained almost identical in 19 brain regions of the homozygous TRβ-PV mutant mice and their wild-type siblings, it was markedly reduced in all brain areas of the heterozygous TRα-1PV mice [56]. In humans, this was elegantly demonstrated with the use of positron emission tomography (PET) with [(18)F]- deoxyglucose to compare the relative regional cerebral glucose metabolism between untreated hypothyroid patients and healthy volunteers [57]. Hypothyroid patients exhibited lower regional activity than control subjects in the amygdala, hippocampus, and areas of the anterior cingulate cortex, and right posterior cingulate cortex. These differences were abolished with TH replacement therapy, restoring metabolic activity in brain areas that are integral to the regulation of affect and cognition [57]. In contrast, [(18)F]- deoxyglucose PET studies in hyperthyroid patients showed a decreased glucose metabolism in the limbic system (uncus and inferior temporal gyrus), but increased anxiety yielded a positive correlation with glucose metabolism in the bilateral sensory association cortex [58].
The present studies have several limitations. Although the donors were not known to have a history of neurologic or thyroid disease, we did not have serum thyroid function tests documenting biochemical euthyroidism at or before the time of death. Also, we did not measure brain tissue T3 content, but the samples were frozen, not perfused, thus were not appropriate for this measurement. In these in silico analyses, we did not measure protein levels of the TH transporters, deiodinases, or TRs. These 19 donors were relatively homogenous in demographic characteristics: all male, Caucasian, and adults. Thus, whether these results extrapolate to more diverse populations, or other brain regions, is unknown. Another limitation is the difficulty in demonstrating the homogeneity of the samples. We used the expression of genes typically expressed in different cell types of the central nervous system (Supplementary Table 3 [36]) but it is conceivable that these genes might very well be responsive to T3 as well. We know these samples were dissected by a neuroanatomist but small variations among samples might exist. Even considering all of these limitations, the strengths of the study include the relatively large number of high-quality, anatomically homogenous samples from healthy, young adult, donors that expired from traumatic accidents. The T3S+ was validated against a panel of 27 genes known to be involved in TH signaling by Factor analysis, high degree of correlation with NT3S+ and TRβ expression. A randomly generated index RT3S+ exhibited no significant correlations.
In conclusion, the transcriptome of the human temporal pole can be assessed through an index of TH signaling T3S+ and was found to correlate with subtle variations in TH signaling. The expression of several hundred genes related to synaptic signaling and cellular metabolism as well as other multiple cellular processes, correlated positively with T3S+, within a narrow ~10% window. It is fascinating to see that TH signaling in the temporal pole exhibits subtle fluctuation that is likely to have significant physiological/pathophysiological consequences. At this time, we do not know the source of this fluctuation, which could due to normal physiological variations around an optimal level, or just reflective of individual-specific variations in the brain setup of TH signaling parameters, or both.
Acknowledgments
We would like to thank the University of Miami Brain Endowment Bank for their collaboration.
Financial Support: This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP Grant 2013/09148-0) and National Institute of Diabetes and Digestive and Kidney Diseases (DK65055) (to A.C.B.).
Glossary
Abbreviations
- CV
coefficient of variance
- D2/DIO2
type 2 deiodinase
- D3/DIO3
type 3 deiodinase
- GO
Gene Ontology
- GSEA
Gene Set Enrichment Analysis
- KMO
Kaiser-Meyer-Olkin
- LT4
levothyroxine
- MSigDB
Molecular Signatures Data Base
- PET
positron emission tomography
- RT-qPCR
real-time quantitative polymerase chain reaction
- T3
triiodothyronine
- T4
thyroxine
- TAC
Transcriptome Analysis Console
- TH
thyroid hormone
- TR
thyroid hormone nuclear receptors
- TRE
TH responsive element
Additional Information
Disclosures: A.B. is a consultant for Allergan Inc, Synthonics Inc and BLA Technology LLC; E.A.M. is co-founder of Equilibrate Therapeutics LLC; the other authors have nothing to disclose. The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References
- 1. Liu YY, Brent GA. Thyroid hormone and the brain: mechanisms of action in development and role in protection and promotion of recovery after brain injury. Pharmacol Ther. 2018;186:176-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hönes GS, Rakov H, Logan J, et al. Noncanonical thyroid hormone signaling mediates cardiometabolic effects in vivo. Proc Natl Acad Sci U S A. 2017;114(52):E11323-E11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chatonnet F, Guyot R, Picou F, Bondesson M, Flamant F. Genome-wide search reveals the existence of a limited number of thyroid hormone receptor alpha target genes in cerebellar neurons. Plos One. 2012;7(5):e30703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chatonnet F, Flamant F, Morte B. A temporary compendium of thyroid hormone target genes in brain. Biochim Biophys Acta. 2015;1849(2):122-129. [DOI] [PubMed] [Google Scholar]
- 5. Bernal J, Guadaño-Ferraz A, Morte B. Thyroid hormone transporters-functions and clinical implications. Nat Rev Endocrinol. 2015;11(12):690. [DOI] [PubMed] [Google Scholar]
- 6. Morte B, Bernal J. Thyroid hormone action: astrocyte-neuron communication. Front Endocrinol (Lausanne). 2014;5:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hernandez A, Morte B, Belinchón MM, Ceballos A, Bernal J. Critical role of types 2 and 3 deiodinases in the negative regulation of gene expression by Tɜin the mouse cerebral cortex. Endocrinology. 2012;153(6):2919-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crantz FR, Silva JE, Larsen PR. An analysis of the sources and quantity of 3,5,3’-triiodothyronine specifically bound to nuclear receptors in rat cerebral cortex and cerebellum. Endocrinology. 1982;110(2):367-375. [DOI] [PubMed] [Google Scholar]
- 9. Freitas BC, Gereben B, Castillo M, et al. Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest. 2010;120(6):2206-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tu HM, Legradi G, Bartha T, Salvatore D, Lechan RM, Larsen PR. Regional expression of the type 3 iodothyronine deiodinase messenger ribonucleic acid in the rat central nervous system and its regulation by thyroid hormone. Endocrinology. 1999;140(2):784-790. [DOI] [PubMed] [Google Scholar]
- 11. Hernandez A, Martinez ME, Fiering S, Galton VA, St Germain D. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest. 2006;116(2):476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dumitrescu AM, Liao XH, Best TB, Brockmann K, Refetoff S. A novel syndrome combining thyroid and neurological abnormalities is associated with mutations in a monocarboxylate transporter gene. Am J Hum Genet. 2004;74(1):168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friesema EC, Grueters A, Biebermann H, et al. Association between mutations in a thyroid hormone transporter and severe X-linked psychomotor retardation. Lancet. 2004;364(9443):1435-1437. [DOI] [PubMed] [Google Scholar]
- 14. Visser WE, Friesema EC, Visser TJ. Thyroid hormone transporters: the knowns and the unknowns. Mol Endocrinol. 2011; 25:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schroeder AC, Privalsky ML. Thyroid hormones, t3 and t4, in the brain. Front Endocrinol (Lausanne). 2014;5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alkemade A, Friesema EC, Unmehopa UA, et al. Neuroanatomical pathways for thyroid hormone feedback in the human hypothalamus. J Clin Endocrinol Metab. 2005;90(7):4322-4334. [DOI] [PubMed] [Google Scholar]
- 17. Alkemade A, Friesema EC, Kuiper GG, et al. Novel neuroanatomical pathways for thyroid hormone action in the human anterior pituitary. Eur J Endocrinol. 2006;154(3):491-500. [DOI] [PubMed] [Google Scholar]
- 18. Wilpert NM, Krueger M, Opitz R, et al. Spatiotemporal changes of cerebral monocarboxylate transporter 8 expression. Thyroid. 2020;30(9):1366-1383. [DOI] [PubMed] [Google Scholar]
- 19. López-Espíndola D, García-Aldea Á, Gómez de la Riva I, et al. Thyroid hormone availability in the human fetal brain: novel entry pathways and role of radial glia. Brain Struct Funct. 2019;224(6):2103-2119. [DOI] [PubMed] [Google Scholar]
- 20. Persani L, Campi I. Syndromes of resistance to thyroid hormone action. Exp Suppl. 2019;111:55-84. [DOI] [PubMed] [Google Scholar]
- 21. Moran C, Schoenmakers N, Agostini M, et al. An adult female with resistance to thyroid hormone mediated by defective thyroid hormone receptor alpha. J Clin Endocrinol Metabol. 2013;98(11):4254-4261. [DOI] [PubMed] [Google Scholar]
- 22. Bochukova E, Schoenmakers N, Agostini M, et al. A mutation in the thyroid hormone receptor alpha gene. N Engl J Med. 2012;366(3):243-249. [DOI] [PubMed] [Google Scholar]
- 23. Noda M. Possible role of glial cells in the relationship between thyroid dysfunction and mental disorders. Front Cell Neurosci. 2015;9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samuels MH, Schuff KG, Carlson NE, Carello P, Janowsky JS. Health status, psychological symptoms, mood, and cognition in L-thyroxine-treated hypothyroid subjects. Thyroid. 2007;17(3):249-258. [DOI] [PubMed] [Google Scholar]
- 25. Silva JE, Larsen PR. Regulation of thyroid hormone expression at the prereceptor and receptor levels. In: Hennemann G, ed. Thyroid Hormone Metabolism. 1st ed. New York and Basel: Marcel Dekker, Inc.; 1986:441-500. [Google Scholar]
- 26. Peeters R, Fekete C, Goncalves C, et al. Regional physiological adaptation of the central nervous system deiodinases to iodine deficiency. Am J Physiol Endocrinol Metab. 2001;281(1):E54-E61. [DOI] [PubMed] [Google Scholar]
- 27. Werneck de Castro JP, Fonseca TL, Ueta CB, et al. Differences in hypothalamic type 2 deiodinase ubiquitination explain localized sensitivity to thyroxine. J Clin Invest. 2015;125(2):769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdalla SM, Bianco AC. Defending plasma T3 is a biological priority. Clin Endocrinol (Oxf). 2014;81(5):633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saravanan P, Chau WF, Roberts N, Vedhara K, Greenwood R, Dayan CM. Psychological well-being in patients on ‘adequate’ doses of l-thyroxine: results of a large, controlled community-based questionnaire study. Clin Endocrinol (Oxf). 2002;57(5):577-585. [DOI] [PubMed] [Google Scholar]
- 30. McAninch EA, Jo S, Preite NZ, et al. Prevalent polymorphism in thyroid hormone-activating enzyme leaves a genetic fingerprint that underlies associated clinical syndromes. J Clin Endocrinol Metab. 2015;100(3):920-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olson IR, Plotzker A, Ezzyat Y. The Enigmatic temporal pole: a review of findings on social and emotional processing. Brain. 2007;130(Pt 7):1718-1731. [DOI] [PubMed] [Google Scholar]
- 32. Bauer M, London ED, Silverman DH, Rasgon N, Kirchheiner J, Whybrow PC. Thyroid, brain and mood modulation in affective disorder: insights from molecular research and functional brain imaging. Pharmacopsychiatry. 2003;36(Suppl 3):S215-S221. [DOI] [PubMed] [Google Scholar]
- 33. Singh S, Modi S, Bagga D, Kaur P, Shankar LR, Khushu S. Voxel-based morphometric analysis in hypothyroidism using diffeomorphic anatomic registration via an exponentiated lie algebra algorithm approach. J Neuroendocrinol. 2013;25(3):229-234. [DOI] [PubMed] [Google Scholar]
- 34. Zhang W, Song L, Yin X, et al. Grey matter abnormalities in untreated hyperthyroidism: a voxel-based morphometry study using the DARTEL approach. Eur J Radiol. 2014;83(1):e43-e48. [DOI] [PubMed] [Google Scholar]
- 35. Göttlich M, Heldmann M, Göbel A, Dirk AL, Brabant G, Münte TF. Experimentally induced thyrotoxicosis leads to increased connectivity in temporal lobe structures: a resting state fMRI study. Psychoneuroendocrinology. 2015;56:100-109. [DOI] [PubMed] [Google Scholar]
- 36. Marcelino CP, McAninch EA, Fernandes GW, Bocco B, Ribeiro MO, Bianco AC Data from: Temporal Pole Responds to Subtle Changes in Local Thyroid Hormone Signaling. Dryad Digital Repository. Deposited 18 August 2020. 10.5061/dryad.bg79cnp7w. [DOI] [PMC free article] [PubMed]
- 37. Tabachnick BG, Fidell LS. Using Multivariate Statistics. 5th ed. Boston: Pearson/Allyn & Bacon; 2007. [Google Scholar]
- 38. Muzzio AM, Noyes PD, Stapleton HM, Lema SC. Tissue distribution and thyroid hormone effects on mRNA abundance for membrane transporters Mct8, Mct10, and organic anion-transporting polypeptides (OATPs) in a teleost fish. Comp Biochem Physiol A Mol Integr Physiol. 2014;167:77-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hodin RA, Lazar MA, Chin WW. Differential and tissue-specific regulation of the multiple rat c-erbA messenger RNA species by thyroid hormone. J Clin Invest. 1990;85(1):101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bocco BM, Werneck-de-Castro JP, Oliveira KC, et al. Type 2 deiodinase disruption in astrocytes results in anxiety-depressive-like behavior in male mice. Endocrinology. 2016;157(9):3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peeters RP, Hernandez A, Ng L, et al. Cerebellar abnormalities in mice lacking type 3 deiodinase and partial reversal of phenotype by deletion of thyroid hormone receptor α1. Endocrinology. 2013;154(1):550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hernandez A, Quignodon L, Martinez ME, Flamant F, St Germain DL. Type 3 deiodinase deficiency causes spatial and temporal alterations in brain T3 signaling that are dissociated from serum thyroid hormone levels. Endocrinology. 2010;151(11):5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23(1):38-89. [DOI] [PubMed] [Google Scholar]
- 44. Zavacki AM, Arrojo E Drigo R, Freitas BC, et al. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol. 2009;29(19):5339-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arrojo E Drigo R, Egri P, Jo S, Gereben B, Bianco AC. The type II deiodinase is retrotranslocated to the cytoplasm and proteasomes via p97/Atx3 complex. Mol Endocrinol. 2013;27(12):2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jo S, Kalló I, Bardóczi Z, et al. Neuronal hypoxia induces Hsp40-mediated nuclear import of type 3 deiodinase as an adaptive mechanism to reduce cellular metabolism. J Neurosci. 2012;32(25):8491-8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinez de Arrieta C, Koibuchi N, Chin WW. Coactivator and corepressor gene expression in rat cerebellum during postnatal development and the effect of altered thyroid status. Endocrinology 2000;141:1693–1698. [DOI] [PubMed] [Google Scholar]
- 48. Manzano J, Morte B, Scanlan TS, Bernal J. Differential effects of triiodothyronine and the thyroid hormone receptor beta-specific agonist GC-1 on thyroid hormone target genes in the b ain. Endocrinology. 2003;144(12):5480-5487. [DOI] [PubMed] [Google Scholar]
- 49. Charlier TD, Balthazart J. Modulation of hormonal signaling in the brain by steroid receptor coactivators. Rev Neurosci. 2005;16(4):339-357. [DOI] [PubMed] [Google Scholar]
- 50. Sharma D, Fondell JD. Temporal formation of distinct thyroid hormone receptor coactivator complexes in HeLa cells. Mol Endocrinol. 2000;14(12):2001-2009. [DOI] [PubMed] [Google Scholar]
- 51. Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M, Helin K. Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73(20):6323-6333. [DOI] [PubMed] [Google Scholar]
- 52. Dentice M, Luongo C, Huang S, et al. Sonic hedgehog-induced type 3 deiodinase blocks thyroid hormone action enhancing proliferation of normal and malignant keratinocytes. Proc Natl Acad Sci U S A. 2007;104(36):14466-14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dentice M, Bandyopadhyay A, Gereben B, et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7(7):698-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sokoloff L, Wechsler RL, Mangold R, Balls K, Kety SS. Cerebral blood flow and oxygen consumption in hyperthyroidism before and after treatment. J Clin Invest. 1953;32(3):202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sokoloff L, Wechsler RL, Balls K, Kety S. The relation of the cerebral O2 consumption to the total body metabolism in hyperthyroidism. J Clin Invest. 1950;29(6):847. [PubMed] [Google Scholar]
- 56. Itoh Y, Esaki T, Kaneshige M, et al. Brain glucose utilization in mice with a targeted mutation in the thyroid hormone alpha or beta receptor gene. Proc Natl Acad Sci U S A. 2001;98(17):9913-9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bauer M, Silverman DH, Schlagenhauf F, et al. Brain glucose metabolism in hypothyroidism: a positron emission tomography study before and after thyroid hormone replacement therapy. J Clin Endocrinol Metab. 2009;94(8):2922-2929. [DOI] [PubMed] [Google Scholar]
- 58. Schreckenberger MF, Egle UT, Drecker S, et al. Positron emission tomography reveals correlations between brain metabolism and mood changes in hyperthyroidism. J Clin Endocrinol Metab. 2006;91(12):4786-4791. [DOI] [PubMed] [Google Scholar]
- 59. McKenzie AT, Wang M, Hauberg ME, et al. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci Rep. 2018;8(1):8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morte B, Ceballos A, Diez D, et al. Thyroid hormone-regulated mouse cerebral cortex genes are differentially dependent on the source of the hormone: a study in monocarboxylate transporter-8- and deiodinase-2-deficient mice. Endocrinology. 2010;151(5):2381-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chatonnet F, Picou F, Fauquier T, Flamant F. Thyroid hormone action in cerebellum and cerebral cortex development. J Thyroid Res. 2011;2011:145762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morte B, Díez D, Ausó E, et al. Thyroid hormone regulation of gene expression in the developing rat fetal cerebral cortex: prominent role of the Ca2+/calmodulin-dependent protein kinase IV pathway. Endocrinology. 2010;151(2):810-820. [DOI] [PubMed] [Google Scholar]
- 63. Royland JE, Parker JS, Gilbert ME. A genomic analysis of subclinical hypothyroidism in hippocampus and neocortex of the developing rat brain. J Neuroendocrinol. 2008;20(12):1319-1338. [DOI] [PubMed] [Google Scholar]
- 64. Cuevas E, Ausó E, Telefont M, Morreale de Escobar G, Sotelo C, Berbel P. Transient maternal hypothyroxinemia at onset of corticogenesis alters tangential migration of medial ganglionic eminence-derived neurons. Eur J Neurosci. 2005;22(3):541-551. [DOI] [PubMed] [Google Scholar]
- 65. Dong H, Wade M, Williams A, Lee A, Douglas GR, Yauk C. Molecular insight into the effects of hypothyroidism on the developing cerebellum. Biochem Biophys Res Commun. 2005;330(4):1182-1193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.