Abstract
Lung cancer is one of the leading causes of all cancer-related deaths. Circulating tumor DNA (ctDNA) is released from apoptotic and necrotic tumor cells. Several sensitive techniques have been invented and adapted to quantify ctDNA genomic alterations. Applications of ctDNA in lung cancer include early diagnosis and detection, prognosis prediction, detecting mutations and structural alterations, minimal residual disease, tumor mutational burden, and tumor evolution tracking. Compared to surgical biopsy and radiographic imaging, the advantages of ctDNA are that it is a non-invasive procedure, allows real-time monitoring, and has relatively high sensitivity and specificity. Given the massive research on non-small cell lung cancer, attention should be paid to small cell lung cancer.
Keywords: Lung cancer, Circulating tumor DNA, Tumor mutational burden, Minimal residual disease, Tumor evolution
Introduction
Lung cancer causes more cancer-related deaths than breast, prostate, colorectal, and brain cancers.[1] Early-stage local lung cancer has good prognosis after curable surgery, highlighting the essence of screening and early diagnosis.[2,3] However, the dismal prognosis of metastatic advanced lung cancer demands a clearer understanding of the disease.[4] Even the wide usage of surgical tumor biopsy for identifying therapeutically targetable mutations does not confer a better survival prolongation.
Tumor DNA can be released from primary tumors, circulating tumor cells (CTCs), metastatic sites, and minimal residual disease (MRD) into the bloodstream.[5–7] Such DNA, generally called circulating tumor DNA (ctDNA), was reported over 30 years ago. Mutations identified in tumor biopsy and ctDNA are highly correlated, subsequently providing an opportunity for non-invasively characterizing mutational profiles of cancer. A series of techniques developed and modified for ctDNA, such as digital polymerase chain reaction (dPCR)[8] and next-generation sequencing (NGS),[9] empower blood examination with both high sensitivity and specificity in detecting mutations.[10] Further retrospective and prospective studies verify the utility of ctDNA in cancer diagnosis and screening, prognosis prediction, MRD identification, therapy response monitoring, and resistance mechanism characterization. ctDNA, together with CTCs and tumor exosomes, marks an era of liquid biopsy and makes a non-invasive and real-time monitoring of disease progression possible.
Evolution, a guiding principle in understanding tumor progression, metastasis, and therapeutic response, characterizes cancer hallmarks such as clonal selection and heterogeneity under selection pressure.[11] Tracking the evolutionary dynamics using multi-region exome sequencing in ctDNA helps determine subclones, which can subsequently result in relapse and metastasis. Tumor phylogenetic trees, which visually define evolutionary histories and explicit clonal and subclonal events, emphasize the clonality of driver events for drug targets and call for intervention before clinical recurrence.[12,13] The combination of ctDNA detection and longitudinal evolutionary profiling endows a new dimension in tumor research.
This review mainly focuses on the clinical application of ctDNA in lung cancer, including screening and early diagnosis, predicting prognosis and staging, profiling cancer-associated mutation and structural alterations, heterogeneity, relapse, treatment response, and resistance. A graphic illustration points out the clinical applications of ctDNA during the course of lung cancer in the setting of both early-stage and advanced metastatic disease [Figure 1]. New ctDNA detection techniques are also discussed in this review.
Figure 1.
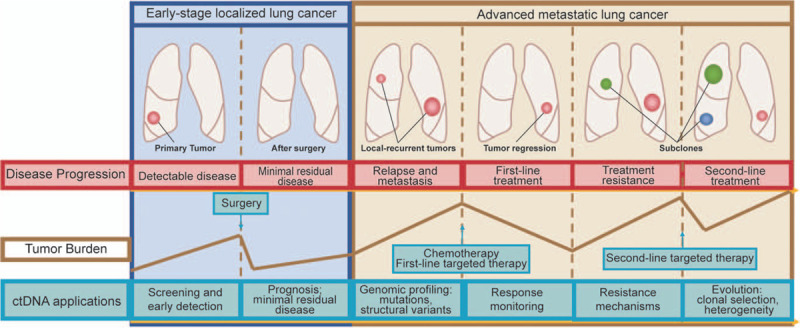
Applications of circulating tumor DNA (ctDNA) in lung cancer across the entire disease course, including screening and early diagnosis, minimal residual disease, prognosis prediction, genomic profiling, identification of resistance mechanism, and tracking of tumor evolution.
Biology of ctDNA: Release and Clearance
Despite multiple origins, most ctDNAs are released passively into the circulation as ∼166-bp double-stranded DNA fragments by apoptotic and necrotic tumor cells.[6,7] A study that used DNA electrophoresis revealed that ctDNA could also be released actively from tumor cells.[14] In addition to ctDNA, CTCs, circulating exosomes,[15] and blood platelets[16] may also become candidates for liquid biopsy. According to observation, the median half-life of ctDNA in non-small cell lung cancer (NSCLC) is 35 min,[17] providing possibilities for real-time cancer evaluation with ctDNA. ctDNA level is relatively lower compared with cell-free DNA (cfDNA), therefore, adding difficulty in sensitively detecting ctDNA. Except for plasma, cerebrospinal fluid has also been demonstrated as a good source of ctDNA and outperformed tumor biopsy tissues in detecting genomic alterations in glioblastoma.[18] Considering lung cancer, tumor DNA can also be detected in the sputum and pleural fluids. Correlation studies focusing on clinical applications of these sources of tumor DNA should be given intensive attention.
Levels of cfDNA in the circulation are dependent on the balance between release and clearance. cfDNA clearance can occur in multiple organs such as the kidney, liver, spleen, and lymph nodes.[19] In malignant disease, the balance is broken, and the accumulation of cfDNA occurs because of a great amount of dying cells and dysfunction of the clearance system, such as in the kidney.[20] Multiple research in vitro prompted that ctDNA could enter tissue cells and, in return, affect the biological behavior of cells.[21] One interesting research showed a phenomenon that plasma from colorectal cancer patients could transform the mouse cell line NIH-3T3.[22] The fact that cell transformation and tumorigenesis are dependent on the presence of cfDNA raises the hypothesis of the active release of ctDNA into the bloodstream to enable the transformation of distant cells. Moreover, complete ctDNA clearance in the blood could serve as a prognostic marker for the efficacy of targeted therapy and chemotherapy in NSCLC patients.[23]
Technology Advances of ctDNA Detection
Detection methods for ctDNA have evolved greatly to achieve higher sensitivity and specificity and a higher correlation with tumor biopsies. Available techniques could be divided into targeted technologies and untargeted technologies. The former approach aims to detect mutations in a preset gene panel.[24] The latter approach aims to detect genomic alterations across exomes[20] or whole genome.[25] The targeted detection method shows better sensitivity while reducing the detection scope in the genome.[26] The sensitivity of dPCR ranges from 74% to 82%, and the specificity ranges from 63% to 100%.[27] The sensitivity of NGS ranges from 79% to 100%, and the specificity ranges from 94% to 100%.[28] Deeper sequencing of plasma DNA applied to selected patients with a higher tumor burden allows for higher sensitivity.
Somatic mutations and copy number alterations (CNAs) detected in ctDNA widely represent both the primary and metastatic cancer genome and overcome the limitations of a repeated invasive biopsy. PCR-based assays are also utilized to detect recurrent point mutations from a list of driver genes including EGFR and KRAS.[29–31] Usage of exome sequencing could identify mutational alterations in a series of plasma samples of NSCLC before and after treatment, which can be used for selecting a list of mutations significantly related to a specific treatment. However, the application of massively parallel sequencing encounters many limitations including low sensitivity, high cost, and need for optimization for patients. A technical report published in 2013 combined an optimized library preparation method with sophisticated bioinformatic approaches to design a personalized mutational selector to quantify genetic aberrations.[10,23] This method, which is very sensitive and economical, is called cancer personalized profiling by deep sequencing (CAPP-Seq). It achieves a sensitivity of 100% in stage II-IV NSCLC patients and 50% in stage I NSCLC patients. This method could detect cancer with a significant leading time compared to that required for traditional radiographic approaches, and the ctDNA levels quantified as mutant allele fractions (AFs) are highly correlated with tumor volume.
Applications of ctDNA in NSCLC
Screening and early diagnosis
In the early stage of NSCLC, the proven ctDNA presence in the blood qualifies ctDNA genotyping as a method for screening and early diagnosis of lung cancer. Compared to radiographic approaches and blood protein biomarkers, ctDNA is a direct measurement of the tumor based on genomic alterations. Diagnosis of cancer at the early stage of the disease allows earlier clinical intervention and improves survival. Screening in seemingly healthy individuals significantly increases false-positive rates and may cause over-diagnosis. In a study conducted on several cancer types, the sensitivity of ctDNA detection for stage IV disease was 82%; however, the sensitivity fell to 47% in stage I disease.[32] Another research recruited several female NSCLC patients, and their plasma ctDNAs, blood cell DNAs, pleural effusion supernatant DNAs, and pleural effusion pellet DNAs were collected. In order to compare different techniques including NGS techniques, droplet dPCR (ddPCR), and an amplification refractory mutation system (ARMS), the four types of samples were analyzed. This study showed that both NGS analysis and ddPCR were more sensitive and reliable over ARMS in detecting EGFR L858R and T790M mutations of early-stage NSCLC patients. This research highlighted the use of non-invasive and highly sensitive techniques such as NGS and/or ddPCR to screen cancers via ctDNA, which offers a new diagnostic and therapeutic privilege for patients.[33]
Accurate testing of selected oncogenic driver mutations at the time of initial NSCLC diagnosis is an important aspect of therapeutic management. Thus far, it is proven that ctDNA could aid in screening and early diagnosis of lung cancer with different techniques. With a novel approach called targeted error correction sequencing, somatic mutations in genes related to lung cancer could be detected in the setting of early-stage disease.[34] Nevertheless, this approach had limitations when applied to healthy individuals and asymptomatic individuals. CancerSEEK, which combined assays for genetic alterations and protein biomarkers, was tested in several early-stage cancer types including lung cancer.[35] The sensitivity of CancerSEEK in lung cancer was about 60% and the specificity was larger than 99%. It was further suggested in a prospective study that plasma ctDNA detection outperformed serum protein markers in the early diagnosis of NSCLC, through a targeted sequencing process with the Ion Personal Genome Machine and AmpliSeq cancer panel.[36] All these studies confirmed the adaptivity of ctDNA in screening and early diagnosis for lung cancer and highlighted the significance of technique improvement.
Despite several methods having been applied to achieve higher sensitivity in the setting of early diagnosis and screening, mutations with low AFs might not be found owing to background noise, which could be overcome by collecting higher volumes of plasma. Additionally, it was previously found that ctDNA was slightly shorter than cfDNA, and this phenomenon encourages the application of filtering DNA fragment length by a method of experimental or in silico size selection.[7] Most importantly, broader genomic coverage and patient-specialized gene panels together produce an overall much higher sensitivity when detecting ctDNA in patients’ plasma.[10,37,38] One study adopted a method of ultra-deep sequencing in whole genome and made the most sensitive detection method possible even when patients’ plasma volume was quite low.[39]
For reliable detection of NSCLC de novo, mutation detection should be designed to reduce the false-positive rate and achieve a higher positive predictive rate. It should be noticed that some oncogenic mutations were found in healthy individuals and might interfere with the result interpretation of diagnostic tests.[38,40] Clinical outcomes in healthy individuals with elevated cfDNA mutant AFs should be prospectively traced to understand the biological and clinical interpretation of this phenomenon. Surgical tumor tissue biopsy is required for the diagnosis and mutation profiling of cancers to guide targeted therapy. However, this biopsy would be impossible in cases of locally advanced and metastatic cancer. Non-invasive ctDNA detection overcomes these shortages and can be conducted without additional harm to the patients.
Prognosis predicting, staging, and stratification of patients
In lung cancer patients, the total concentrations of cfDNA and ctDNA are higher than those in healthy individuals, and elevated concentrations of cfDNA and ctDNA are shown in correlation with tumor progression. The possible binary stratification of lung cancer patients into high- and low-ctDNA concentrations guides the prediction of prognosis. The status of EGFR mutation also serves as a prognostic biomarker, which was implied in a study evaluating the effect of ctDNA presence on prognosis among advanced-stage lung adenocarcinoma patients partially receiving epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) treatment.[41] The number of metastatic sites and the abundance of mutant EGFR ctDNA were in a strong correlation. There was a significantly shorter progression-free survival (PFS) and duration of controlled disease by EGFR-TKIs in the ctDNA-positive group than the negative group. Furthermore, there was a trend of shorter overall survival (OS) time in patients with ctDNA EGFR mutations than in patients without ctDNA EGFR mutations both in all patients and in patients receiving EGFR-TKI treatment.[41] This study supports ctDNA EGFR mutations as a biomarker for predicting distant metastasis and poor response to EGFR-TKIs. Several other studies also suggested that the presence of ctDNA EGFR mutations is associated with shorter PFS and OS.[42,43] Nevertheless, a series of studies supported the opposite correlation between ctDNA EGFR mutations and prognosis.[44,45] An open-labeled phase II study enrolling advanced NSCLC patients treated with erlotinib and pertuzumab showed that EGFR mutation detected in ctDNA suggested prolonged PFS.[46] These inconsistent results may be due to different patient inclusion criteria, different generations of EGFR-TKIs applied, different techniques adopted in ctDNA analysis, and relatively small sample sizes. These studies suggested the potential of ctDNA mutation profiling as a means for diagnosing and stratifying patients, but also reinforced significance of patient selection and therapeutic regimen choosing.
Except for recurrent mutations in NSCLC, structural alterations and epigenetic markers in ctDNA could also predict prognosis of early stage and advanced metastatic NSCLC. CNAs were identified as a prognostic factor in a series of studies on NSCLC.[47,48] A study, using a 150-gene panel and enabling CNAs detection from a very low amount of ctDNA,[49] selected patients responsive to crizotinib with MET amplification. Genome-wide hypermethylation is also frequently observed in lung cancer,[50] and hypermethylation can be evaluated in ctDNA or sputum non-invasively. Detection of cancer-specific methylation alterations opens an era for detecting epigenetic biomarkers and correlates epigenetics with prognosis and treatment response.
Non-invasive profiling of genomic characteristics
Mutation status
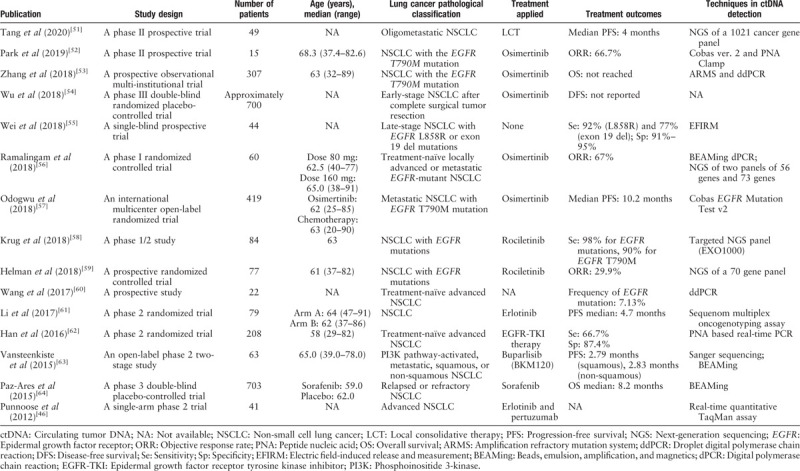
Mutation detection in ctDNA in NSCLC has been shown to present both high sensitivity and high specificity.[10] To effectively select qualified patients to provide corresponding targeted therapies, many retrospective and prospective studies have been conducted. The urge for double-blind, prospective, randomized, controlled, clinical studies has led to a plenty of studies that analyzed the EGFR mutation status of NSCLC with plasma ctDNA samples. Clinical trials focusing on ctDNA in lung cancer are summarized in Table 1.[46,51–64] A meta-analysis combining several studies published between 2007 and 2015 with various ctDNA analysis methods found the pooled sensitivity and specificity of ctDNA mutation detection as 65.7% and 99.8%, respectively.[65] In a prospective study focusing on the EGFR inhibitor gefitinib, mutation fraction differences between tumor biopsy and plasma ctDNA were compared in a cohort of over 600 patients.[66,67] The sensitivity and specificity of the applied ARMS-based EGFR detection methods in this prospective study were determined as 65.7% and 99.8%, respectively. Furthermore, this study showed that ctDNA in the plasma revealed the mutation profiles of both primary tumor sites and metastatic tumor sites, therefore, eliminating the influence of heterogeneity.
Table 1.
Clinical trials with applications of ctDNA in lung cancer.
The mechanisms underlying EGFR-TKI treatment resistance include T790M EGFR mutations, which indicates the usage of third-generation EGFR-TKIs.[68,69] The sensitivity of detecting T790M EGFR mutation in plasma ctDNA was evaluated in multiple studies. In a study with a sample size of 54 NSCLC patients, T790M EGFR mutation was identified in plasma ctDNA. The frequency of T790M EGFR mutation was approximately 50% in patients with former response to gefitinib or erlotinib, nearly 30% in patients with prior stable diseases, and 0% in patients who were previously untreated with gefitinib or erlotinib.[70] The discrepancy among these three groups suggests that the possibility of resistance and recurrence could be identified non-invasively through the evaluation of T790M EGFR mutation status. Furthermore, the short half-life time of ctDNA enables real-time monitoring of resistance and treatment response during the entire course of treatment.
Structural variants
CNAs can be detected in ctDNA using whole-genome sequencing (WGS),[71,72] amplicon-based,[73] and hybrid-capture approaches.[69] Evaluating CNAs and mutation profiling in the same assay is very important because many clinically targetable genomic alterations in cancer are structural alterations.[47] Accurate CNA assessment in ctDNA requires a higher concentration of ctDNA in plasma, and a study emphasized that an average variant AF should be over 5% for CNA analysis. Moreover, chromosomal rearrangement in ctDNA can be identified with WGS[71,72] and hybrid-capture approaches.[28]
Application of WGS of cfDNA facilitates a non-targeted method to detect somatic CNAs, requiring low genome coverage. A study was performed in a small cohort of lung cancer patients with a relatively low sequencing depth.[74] This study suggested a significant correlation between copy number ratios in cfDNA and formalin-fixed paraffin-embedded (FFPE) tissue in advanced lung cancer patients. However, due to the relatively low sequencing depth, this method required a fraction of tumor origin in cfDNA of at least 10%. This research supported the usage of cfDNA in somatic CNAs detection and compared the efficacy of detection between the cfDNA and the FFPE tissue. Simultaneously, this research put forward the significance of cfDNA fraction in the plasma and sequencing depth.
Another study aimed to associate CNAs with clinical outcomes using a method called low-pass WGS.[75] This new method showed the efficacy of identifying focal and broad CNAs in lung cancer patients, and 0.5× coverage provided enough ability to detect CNAs. Similarly, this method required a relatively higher tumor burden and a higher fraction of ctDNA in cfDNA, and a size-based selection was suggested. Moreover, the research exhibited similarity between CNAs landscape of cfDNA and surgical tumor specimen in squamous NSCLC patients with above 25% tumor fraction in cfDNA. The discordance between cfDNA and surgical tumor specimens might be attributed to tumor heterogeneity in both space and time.
MRD and Relapse
After surgery or curative treatment, patients with possible MRD should be monitored during the entire course of treatment, generally with radiological imaging. On the one hand, radiological imaging has the advantage of a relatively low rate of damage associated with treatment. On the other hand, it has the disadvantage of limited sensitivity for the detection of micrometastases compared with overt metastases. The introduction of MRD monitoring into general clinical practice solves the listing disadvantages and facilitates the development of personalized precision medicine, considering the dimension of time. A study of lung cancer showed that post-treatment ctDNA preceded radiological imaging with a median of 5.2 months of lead time.[76] In 94% of patients undergoing recurrence, ctDNA was detected in the first post-treatment blood samples, indicating the role of reliable detection of MRD using ctDNA CAPP-Seq. Another study conducted in NSCLC patients evaluated ctDNA dynamics and the optimal timing of MRD testing.[17] The presence of MRD on day 1 after surgery could not predict clinical outcomes such as relapse-free survival (RFS) and OS. In contrast, the presence of MRD on day 3 and day 30 was predictive of an unfavorable RFS and OS. This research highlighted the proper timing for performing ctDNA detection after surgery. Post-operative ctDNA profiling is highly specific for detecting MRD and predicting relapse possibility, leading to targeted therapies on EGFR or ALK mutations as complements of surgery and chemotherapy.
Blood Tumor Mutational Burden (TMB) and Immunotherapy
High TMB represents genomic instability and serves as a marker of immune checkpoint blockade therapy response, together with other biomarkers such as programmed cell death ligand-1 (PD-L1) expression.[77] Patients with NSCLC have significantly benefited from immunotherapy, particularly from immune checkpoint blockade therapies such as anti-cytotoxic T-lymphocyte-associated protein 4 inhibitors, programmed cell death-1 (PD-1) inhibitors, and PD-L1 inhibitors in the last decade. Nevertheless, despite the observed prolonged survival in patients with advanced metastatic NSCLC, the objective response rate remains relatively under expectation because of primary resistance. TMB, either in tissue (tTMB) or in plasma ctDNA (blood TMB [bTMB]), is defined as the count of total non-synonymous genomic mutations, and it has recently emerged as a powerful biomarker to select patients sensitive to immunotherapy.[78,79] bTMB could representatively reflect TMB, but larger ctDNA panels were necessary to establish a better correlation between tTMB and bTMB.[80] A recently published original investigation suggested that a well-established panel covering the whole exon of 150 selected cancer-related genes could serve as a method to identify bTMB in NSCLC patients and guide patient selection for PD-1 and PD-L1 inhibitor.[81] In the multivariable logistic analysis, patients with higher bTMB demonstrated superiorer PFS and were more likely to undergo tumor shrinkage. Meanwhile, patients responding to immune blockade therapy had significantly higher bTMB levels than those not responding. The application of bTMB detected in ctDNA served as a predictor of patients who benefited from immune blockade therapy, including atezolizumab, durvalumab, and tremelimumab.[82,83]
Evolution, Clonal Selection, Intra-tumoral, and Inter-tumoral Heterogeneity
The concept of tumor evolution not only suggests a hypothesis about the mechanism of resistance but also enlarges the perception of progression, metastasis, and therapeutic responses. Evolutionary theory in cancer research broadly covers the following aspects of tumor biological behavior: types of genomic aberrations evolution during the entire disease course, frequencies of mutations, clonal selection in tumor evolution, and high level of intra-tumoral and inter-tumoral heterogeneity.[11] Several studies adopted a bespoken patient specific multiplex-PCR NGS approach to profile the NSCLC ctDNA genome and analyzed tumor evolution, together with multi-region tumor whole-exome sequencing.[11–13] The clonal nature of driver alterations and subclone heterogeneity could be determined in these serial studies, which recruited 100 early-stage NSCLC patients prospectively and analyzed 327 tumor multi-region specimens. The Lung TRACERx (TRAcking CancER evolution through therapy) study generated phylogenetic trees tracking representative mutations from the trunk clonal population and many subclonal populations. The study addressed the clinico-pathological determinants of ctDNA, the clonal/subclonal fidelity of ctDNA, and the potential for ctDNA detection and characterization to predict relapse and targetable features of recurrent diseases. These studies showed that ctDNA presence correlated with primary tumor proliferation index (Ki-67), lymphovascular invasion, and tumor necrosis. Through longitudinal detection of ctDNA, ctDNA was detected in most relapsed cases on average 70 days before clinical confirmation by computed tomgraphy imaging. Releasing of ctDNA from different tumor regions into circulation was correlated to the volume of the tumor subclones. Data acquired were used to identify patient-specific single-nucleotide variants (SNVs) and CNAs, depicting evolutionary histories and clonal architecture. The causes of intra-tumor heterogeneity were listed as mutational processes, chromosomal instability, and genome duplication. There is a common pattern of tumor evolution with an early event of genome doubling and later events of extensive subclonal diversification. The complementation of ctDNA sequencing into routine surgical biopsies resolves difficulties in obtaining sequential tumor samples and makes dynamic observation of evolution in the process of treatment possible. These findings could assist in residual disease identification, treatment response evaluation, and targetable emerging subclones affirmation.
Applications of ctDNA in SCLC
Previous studies of large-scale sequencing revealed the genomic differences between NSCLC and SCLC.[84] SCLC patients are commonly sensitive to standard platinum and etoposide chemotherapy. However, they may have progressed into relapse and acquire resistance to standard treatment within 1 year of initial treatment, leading to a dismal 2-year survival.[85,86] Intensive studies have focused on the relationship between ctDNA and NSCLC. Nevertheless, research on applications of ctDNA in SCLC patients is relatively lacking. SCLC has a genomic landscape of high somatic tumor mutation burden because of the close relationship with carcinogens in tobacco; thus, baseline SCLC usually has high frequencies of CNAs and mutations. The most common genetic alterations are inactivation of tumor-suppressor genes, including TP53 and RB1, and point mutations in genes associated with chromatin remodeling, receptor tyrosine kinases, and the NOTCH family genes.[87] The main obstacle in applying liquid biopsy in SCLC is how to detect ctDNA in a chaotic background.[88–90] A study used NGS to detect SNVs, CNAs, insertions, and deletions with a panel of 14 genes that had frequent mutations in SCLC.[88] Eventually, the study involved testing of 140 plasma samples collected from 27 SCLC patients, and mutations associated with SCLC were found in over 80% of patients. Generally, mutant AFs of SCLC-associated gene mutations were in the range of 0.1% to 87%, and the most common mutations were in the TP53 and RB1 genes, following further observation.[87] Similar to findings in NSCLC, mutant AFs and CNAs were also in close relationship with treatment responses when considering SCLC. Additionally, ctDNA detection in SCLC patients post-operation could also provide reliable evidence of disease relapse before detectable relapse through radiographic imaging. It is advocated that more attention should be paid toward improving sequencing techniques and conducting clinical researches on SCLC.
Conclusion and Perspectives
Many studies have proven the utility of ctDNA in cancer screening and early diagnosis, genomic profiling, prognosis prediction, treatment response and resistance monitoring, MRD detection, and tumor evolution tracking. With the comprehensive characterization of the molecular landscape of NSCLC and SCLC, ctDNA provides solutions to almost all aspects of clinical considerations in a non-invasive and real-time manner. Screening and early detection will help identify early-stage lung cancer patients and indicate the need for clinical interventions. Predicting prognosis using ctDNA enables dynamic stratification of patients, thereby inferring the relative risks of relapse. Assessing treatment response and resistance makes personalized treatment possible and facilitates interventions based on resistant mechanisms. MRD detection is a sensitive approach to forecast local advances and metastases. The theory of tumor evolution will greatly shape precision medicine, considering the generality of intra-tumoral and inter-tumoral heterogeneity. Identifying driver mutations in clones and passenger mutations in subclones will lead to an era of stepwise treatment decisions.
Clinical application of ctDNA for early detection faces several obstacles. The biggest obstacle is the required improvement of both sensitivity and specificity. Methods have been consistently improved to meet the clinical requirements of lower cost and higher efficacy. The rates of mutation detection in ctDNA could be improved by advanced genomic approaches such as NGS that have higher sensitivity to identify rare mutations in matched ctDNA and tumor tissue samples. Several studies have been inclined to discover the biology of early-stage lung cancer after the invention of sensitive ctDNA detecting techniques, which improve NGS library preparation and downstream bioinformatic analysis. Besides, due to high dynamic range of ctDNA concentration with current techniques, the correlation between ctDNA concentration and tumor burden is still questioned and limits the clinical application. Generally, the number of patients enrolled in ctDNA research is much smaller than the respective number for other clinical studies, and the clinical interpretation of results is hampered. Composite gene panel needs to be tested in clinical studies with well-established endpoints to demonstrate the clinical utility. Moreover, selection of appropriate time points for ctDNA screening would be crucial to detect ctDNA derived from the resistant tumor cell clones. Also, in order to control errors in the process of ctDNA extraction, quantification, analytical pipeline and reporting, great efforts have been made to standardize ctDNA analysis before integrating it into clinical practice by a variety of international consortia.
This review provides adequate information on the implications of ctNDA in lung cancer and sheds light on future research directions. With a focus on NSCLC, this review also highlights the most recent clinical advances in SCLC. ctDNA, as an important component of liquid biopsies, is bound to demonstrate its potential utility in clinical settings.
Funding
This work was supported by grants from the Intergovernmental International Science, Technology and Innovation Cooperation Key Project of the National Key R&D Programme (NKP) (No. 2017YFE0110300), the National Natural Science Foundation of China (No. 81602162), and the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (CAMS-I2M) (No. 2016-I2M-1-002).
Conflicts of interest
None.
Footnotes
How to cite this article: Li RY, Liang ZY. Circulating tumor DNA in lung cancer: real-time monitoring of disease evolution and treatment response. Chin Med J 2020;20:2476–2485. doi: 10.1097/CM9.0000000000001097
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Osarogiagbon RU, Veronesi G, Fang W, Ekman S, Suda K, Aerts JG, et al. Early-stage NSCLC: advances in thoracic oncology 2018. J Thorac Oncol 2019; 14:968–978. doi: 10.1016/j.jtho.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nesbitt JC, Putnam JB, Jr, Walsh GL, Roth JA, Mountain CF. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg 1995; 60:466–472. doi: 10.1016/0003-4975 (95)00169-l. [DOI] [PubMed] [Google Scholar]
- 4.Sandler AB, Nemunaitis J, Denham C, von Pawel J, Cormier Y, Gatzemeier U, et al. Phase III trial of gemcitabine plus cisplatin versus cisplatin alone in patients with locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol 2000; 18:122–130. doi: 10.1200/jco.2000.18.1.122. [DOI] [PubMed] [Google Scholar]
- 5.Pisetsky DS, Fairhurst AM. The origin of extracellular DNA during the clearance of dead and dying cells. Autoimmunity 2007; 40:281–284. doi: 10.1080/08916930701358826. [DOI] [PubMed] [Google Scholar]
- 6.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61:1659–1665. [PubMed] [Google Scholar]
- 7.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016; 35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999; 96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010; 2:20ra14.doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz R, Schäffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet 2017; 18:213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov 2017; doi: 10.1158/2159-8290.Cd-17-0343. [DOI] [PubMed] [Google Scholar]
- 13.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017; 545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001; 313:139–142. doi: 10.1016/s0009-8981 (01)00665-9. [DOI] [PubMed] [Google Scholar]
- 15.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014; 24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorber L, Zwaenepoel K, Deschoolmeester V, Van Schil PE, Van Meerbeeck J, Lardon F, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017; 107:100–107. doi: 10.1016/j.lungcan.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Chen K, Zhao H, Shi Y, Yang F, Wang LT, Kang G, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res 2019; 25:7058–7067. doi: 10.1158/1078-0432.Ccr-19-1213. [DOI] [PubMed] [Google Scholar]
- 18.Duan H, Hu JL, Chen ZH, Li JH, He ZQ, Wang ZN, et al. Assessment of circulating tumor DNA in cerebrospinal fluid by whole exome sequencing to detect genomic alterations of glioblastoma. Chin Med J 2020; 133:1415–1421. doi: 10.1097/cm9.0000000000000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung F, Kulasingam V, Diamandis EP, Hoon DS, Kinzler K, Pantel K, et al. Circulating tumor DNA as a cancer biomarker: fact or fiction? Clin Chem 2016; 62:1054–1060. doi: 10.1373/clinchem.2016.260331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014; 346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011; 11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 22.Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, Anker P, Herrera-Goepfert R, Medina-Velázquez LA, et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One 2012; 7:e52754.doi: 10.1371/journal.pone.0052754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Hu C, Xie Z, Wu L, Zhu Z, Rao C, et al. Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl Lung Cancer Res 2020; 9:269–279. doi: 10.21037/tlcr.2020.03.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 25.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol 2011; 136:527–539. doi: 10.1309/ajcpr1svt1vhugxw. [DOI] [PubMed] [Google Scholar]
- 26.Komatsubara KM, Sacher AG. Circulating tumor DNA as a liquid biopsy: current clinical applications and future directions. Oncology (Williston Park) 2017; 31:618–627. [PubMed] [Google Scholar]
- 27.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2:1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paweletz CP, Sacher AG, Raymond CK, Alden RS, O’Connell A, Mach SL, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res 2016; 22:915–922. doi: 10.1158/1078-0432.Ccr-15-1627-t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011; 17:7808–7815. doi: 10.1158/1078-0432.Ccr-11-1712. [DOI] [PubMed] [Google Scholar]
- 30.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 31.Gautschi O, Huegli B, Ziegler A, Gugger M, Heighway J, Ratschiller D, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett 2007; 254:265–273. doi: 10.1016/j.canlet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6:224ra224.doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Lv J, Wan S, Xin J, Xie T, Li T, et al. High sensitive and non-invasive ctDNAs sequencing facilitate clinical diagnosis and clinical guidance of non-small cell lung cancer patient: a time course study. Front Oncol 2018; 8:491.doi: 10.3389/fonc.2018.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017; 9:eaan2415.doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018; 359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen KZ, Lou F, Yang F, Zhang JB, Ye H, Chen W, et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep 2016; 6:31985.doi: 10.1038/srep31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 2015; 21:4586–4596. doi: 10.1158/1078-0432.Ccr-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016; 34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Wang Y, Shi W, Zhu M, Liu Z, Luo N, et al. Serial ultra-deep sequencing of circulating tumor DNA reveals the clonal evolution in non-small cell lung cancer patients treated with anti-PD1 immunotherapy. Cancer Med 2019; 8:7669–7678. doi: 10.1002/cam4.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016; 7:9707–9717. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim T, Kim EY, Lee SH, Kwon DS, Kim A, Chang YS. Presence of mEGFR ctDNA predicts a poor clinical outcome in lung adenocarcinoma. Thorac Cancer 2019; 10:2267–2273. doi: 10.1111/1759-7714.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21:3196–3203. doi: 10.1158/1078-0432.Ccr-14-2594. [DOI] [PubMed] [Google Scholar]
- 43.Lee JY, Qing X, Xiumin W, Yali B, Chi S, Bak SH, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016; 7:6984–6993. doi: 10.18632/oncotarget.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan G, Zhang K, Ding J, Li J. Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2017; 8:33922–33932. doi: 10.18632/oncotarget.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao C, Yuan JQ, Yang ZY, Fu XH, Wu XY, Tang JL. Blood as a substitute for tumor tissue in detecting EGFR mutations for guiding EGFR TKIs treatment of nonsmall cell lung cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2015; 94:e775.doi: 10.1097/md.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012; 18:2391–2401. doi: 10.1158/1078-0432.Ccr-11-3148. [DOI] [PubMed] [Google Scholar]
- 47.Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med 2019; 25:738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 48.Kutilin DS, Airapetova TG, Anistratov PA, Pyltsin SP, Leiman IA, Karnaukhov NS, et al. Copy number variation in tumor cells and extracellular DNA in patients with lung adenocarcinoma. Bull Exp Biol Med 2019; 167:771–778. doi: 10.1007/s10517-019-04620-y. [DOI] [PubMed] [Google Scholar]
- 49.Peng H, Lu L, Zhou Z, Liu J, Zhang D, Nan K, et al. CNV detection from circulating tumor DNA in late stage non-small cell lung cancer patients. Genes (Basel) 2019; 10:926.doi: 10.3390/genes10110926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki M, Yoshino I. Aberrant methylation in non-small cell lung cancer. Surg Today 2010; 40:602–607. doi: 10.1007/s00595-009-4094-6. [DOI] [PubMed] [Google Scholar]
- 51.Tang C, Lee WC, Reuben A, Chang L, Tran H, Little L, et al. Immune and circulating tumor DNA profiling after radiation treatment for oligometastatic non-small cell lung cancer: translational correlatives from a mature randomized phase II trial. Int J Radiat Oncol Biol Phys 2020; 106:349–357. doi: 10.1016/j.ijrobp.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 52.Park CK, Cho HJ, Choi YD, Oh IJ, Kim YC. A phase II trial of osimertinib in the second-line treatment of non-small cell lung cancer with the EGFR T790M mutation, detected from circulating tumor DNA: LiquidLung-O-Cohort 2. Cancer Res Treat 2019; 51:777–787. doi: 10.4143/crt.2018.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Zhu L, Xia B, Chen E, Zhao Q, Zhang X, et al. Epidermal growth factor receptor (EGFR) T790M mutation identified in plasma indicates failure sites and predicts clinical prognosis in non-small cell lung cancer progression during first-generation tyrosine kinase inhibitor therapy: a prospective observational study. Cancer Commun (Lond) 2018; 38:28.doi: 10.1186/s40880-018-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu YL, Herbst RS, Mann H, Rukazenkov Y, Marotti M, Tsuboi M. ADAURA: phase III, double-blind, randomized study of osimertinib versus placebo in EGFR mutation-positive early-stage NSCLC after complete surgical resection. Clin Lung Cancer 2018; 19:e533–e536. doi: 10.1016/j.cllc.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Wei F, Strom CM, Cheng J, Lin CC, Hsu CY, Soo Hoo GW, et al. Electric field-induced release and measurement liquid biopsy for noninvasive early lung cancer assessment. J Mol Diagn 2018; 20:738–742. doi: 10.1016/j.jmoldx.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramalingam SS, Yang JC, Lee CK, Kurata T, Kim DW, John T, et al. Osimertinib as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer. J Clin Oncol 2018; 36:841–849. doi: 10.1200/jco.2017.74.7576. [DOI] [PubMed] [Google Scholar]
- 57.Odogwu L, Mathieu L, Goldberg KB, Blumenthal GM, Larkins E, Fiero MH, et al. FDA benefit-risk assessment of osimertinib for the treatment of metastatic non-small cell lung cancer harboring epidermal growth factor receptor T790M mutation. Oncologist 2018; 23:353–359. doi: 10.1634/theoncologist.2017-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol 2018; 29:700–706. doi: 10.1093/annonc/mdx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helman E, Nguyen M, Karlovich CA, Despain D, Choquette AK, Spira AI, et al. Cell-free DNA next-generation sequencing prediction of response and resistance to third-generation EGFR inhibitor. Clin Lung Cancer 2018; 19:518–530.e7. doi: 10.1016/j.cllc.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Bai H, Hong C, Wang J, Mei TH. Analyzing epidermal growth factor receptor mutation status changes in advanced non-small-cell lung cancer at different sampling time-points of blood within one day. Thorac Cancer 2017; 8:312–319. doi: 10.1111/1759-7714.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li T, Piperdi B, Walsh WV, Kim M, Beckett LA, Gucalp R, et al. Randomized phase 2 trial of pharmacodynamic separation of pemetrexed and intercalated erlotinib versus pemetrexed alone for advanced nonsquamous, non-small-cell lung cancer. Clin Lung Cancer 2017; 18:60–67. doi: 10.1016/j.cllc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han JY, Choi JJ, Kim JY, Han YL, Lee GK. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer 2016; 16:627.doi: 10.1186/s12885-016-2678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vansteenkiste JF, Canon JL, De Braud F, Grossi F, De Pas T, Gray JE, et al. Safety and efficacy of buparlisib (BKM120) in patients with PI3K pathway-activated non-small cell lung cancer: results from the phase II BASALT-1 study. J Thorac Oncol 2015; 10:1319–1327. doi: 10.1097/jto.0000000000000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paz-Ares L, Hirsh V, Zhang L, de Marinis F, Yang JC, Wakelee HA, et al. Monotherapy administration of sorafenib in patients with non-small cell lung cancer (MISSION) trial: a phase III, multicenter, placebo-controlled trial of sorafenib in patients with relapsed or refractory predominantly nonsquamous non-small-cell lung cancer after 2 or 3 previous treatment regimens. J Thorac Oncol 2015; 10:1745–1753. doi: 10.1097/jto.0000000000000693. [DOI] [PubMed] [Google Scholar]
- 65.Qian X, Liu J, Sun Y, Wang M, Lei H, Luo G, et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016; 7:29154–29165. doi: 10.18632/oncotarget.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, McCormack R, Webster A, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer 2014; 110:55–62. doi: 10.1038/bjc.2013.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014; 9:1345–1353. doi: 10.1097/jto.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3:75ra26.doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016; 7:11815.doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 2009; 15:2630–2636. doi: 10.1158/1078-0432.Ccr-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013; 5:30.doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012; 4:162ra154.doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015; 10:e0140712.doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor F, Bradford J, Woll PJ, Teare D, Cox A. Unbiased detection of somatic copy number aberrations in cfDNA of lung cancer cases and high-risk controls with low coverage whole genome sequencing. Adv Exp Med Biol 2016; 924:29–32. doi: 10.1007/978-3-319-42044-8_6. [DOI] [PubMed] [Google Scholar]
- 75.Chen X, Chang CW, Spoerke JM, Yoh KE, Kapoor V, Baudo C, et al. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin Cancer Res 2019; 25:2254–2263. doi: 10.1158/1078-0432.Ccr-18-1593. [DOI] [PubMed] [Google Scholar]
- 76.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017; 7:1394–1403. doi: 10.1158/2159-8290.Cd-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costantini A, Takam Kamga P, Dumenil C, Chinet T, Emile JF, Giroux Leprieur E. Plasma biomarkers and immune checkpoint inhibitors in non-small cell lung cancer: new tools for better patient selection? Cancers (Basel) 2019; 11:1269.doi: 10.3390/cancers11091269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis 2019; 11:S71–S80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greillier L, Tomasini P, Barlesi F. The clinical utility of tumor mutational burden in non-small cell lung cancer. Transl Lung Cancer Res 2018; 7:639–646. doi: 10.21037/tlcr.2018.10.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hendriks LE, Rouleau E, Besse B. Clinical utility of tumor mutational burden in patients with non-small cell lung cancer treated with immunotherapy. Transl Lung Cancer Res 2018; 7:647–660. doi: 10.21037/tlcr.2018.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019; 5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018; 24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 83.Peters S, Cho BC, Reinmuth N, Lee KH, Luft A, Ahn M-J, et al. Abstract CT074: tumor mutational burden (TMB) as a biomarker of survival in metastatic non-small cell lung cancer (mNSCLC): blood and tissue TMB analysis from MYSTIC, a phase III study of first-line durvalumab ± tremelimumab vs chemotherapy. Cancer Res 2019; 79:CT074.doi: 10.1158/1538-7445.Am2019-ct074. [Google Scholar]
- 84.Blackhall F, Frese KK, Simpson K, Kilgour E, Brady G, Dive C. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol 2018; 19:e470–e481. doi: 10.1016/s1470-2045 (18)30455-8. [DOI] [PubMed] [Google Scholar]
- 85.Osterlind K, Andersen PK. Prognostic factors in small cell lung cancer: multivariate model based on 778 patients treated with chemotherapy with or without irradiation. Cancer Res 1986; 46:4189–4194. [PubMed] [Google Scholar]
- 86.Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017; 14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015; 524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Almodovar K, Iams WT, Meador CB, Zhao Z, York S, Horn L, et al. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol 2018; 13:112–123. doi: 10.1016/j.jtho.2017.09.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet 2016; 32:360–371. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 90.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]


