Abstract
The study of allosteric functional modulation in dynamic proteins is attracting increasing attention. In particular, the discovery of new allosteric sites may generate novel opportunities and strategies for drug development, overcoming the limits of classical active-site oriented drug design. In this paper, we report on the results of a novel, ab initio, fully computational approach for the discovery of allosteric inhibitors based on the physical characterization of signal propagation mechanisms in proteins and apply it to the important molecular chaperone Hsp90. We first characterize the allosteric “hot spots” involved in interdomain communication pathways from the nucleotide-binding site in the N-domain to the distal C-domain. On this basis, we develop dynamic pharmacophore models to screen drug libraries in the search for small molecules with the functional and conformational properties necessary to bind these “hot spot” allosteric sites. Experimental tests show that the selected moelcules bind the Hsp90 C-domain, exhibit antiproliferative activity in different tumor cell lines, while not affecting proliferation of normal human cells, destabilize Hsp90 client proteins, and disrupt association with several cochaperones known to bind the N- and M-domains of Hsp90. These results prove that the hits alter Hsp90 function by affecting its conformational dynamics and recognition properties through an allosteric mechanism. These findings provide us with new insights on the discovery and development of new allosteric inhibitors that are active on important cellular pathways through computational biology. Though based on the specific case of Hsp90, our approach is general and can readily be extended to other target proteins and pathways.
Introduction
The dynamic properties of proteins play key roles in all aspects of protein functions, ranging from molecular recognition and binding to enzymatic activity.1 A better knowledge of dynamics from experiments and theory makes it now feasible to model the conformational properties of several proteins at the atomic scale.2 Functional dynamics is determined by a complex interplay of covalent and noncovalent interactions that define the relative population of three-dimensional (3D) structures (determined by their free energies) and the possible interconversion kinetic pathways among them (determined by the heights of the free energy barriers between them).3,4 Binding of a ligand or substrate at an active site or of a protein partner at a certain region of the structure may select specific accessible conformations endowed with specific functional properties.5
Allosteric molecular perturbations may alter the covalent and noncovalent forces that determine the fine combination of dynamic modes at the basis of molecular recognition and function. This reverberates in a modification of the protein’s structural and/or dynamic properties, causing a response where a specific function can be switched on, fine-tuned, regulated, or blocked. Perturbation of the protein’s conformational ensemble may be achieved through several mechanisms, including ligand binding, covalent modifications, or mutations. A variation of the protein state at a certain site may thus impact on the binding affinity in a distal functional region, such as the active site or a protein contact surface.6 Information transmission between distant functional sites in proteins represents a manifestation of nonlocal interactions between residues.
The molecular mechanisms of these site-to-site communication phenomena are of great interest, especially since understanding the dynamic connectivity that favors signaling through structures may reveal new allosteric binding sites and illuminate molecular mechanisms of functional regulation.7 Moreover, achieving these goals would offer tremendous opportunities in the design of new drugs, protein engineering, and chemical genomics. Rational targeting of alternative sites may reveal new chemotypes for potential inhibitors and offer new strategies to interfere with protein–protein interactions, which are generally recognized as challenging targets.8
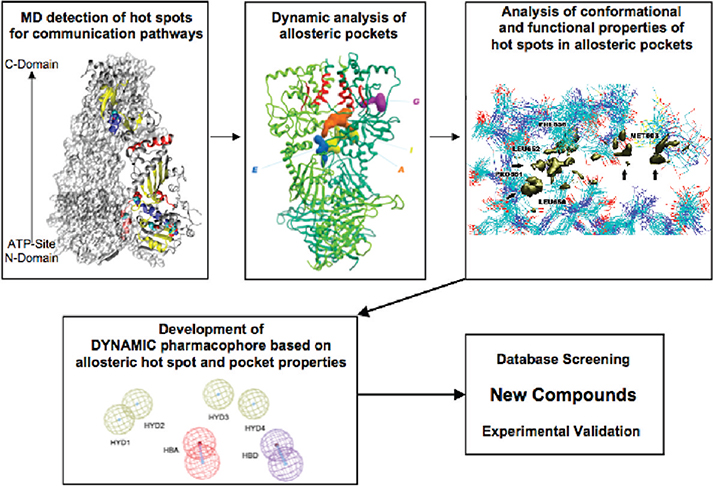
In this paper, we present a novel rational strategy for the computational-based discovery of allosteric inhibitors of molecular chaperones. In particular, we aim to perturb the functions of the activated form of the chaperone heat shock protein-90 (Hsp90), by the rational selection of antagonists with the structural and functional characteristics necessary to target “hot-spot” allosteric residues located in the C-terminal domain (CTD), which are dynamically coupled to the N-terminal ATP binding-site and may potentially affect Hsp90 chaperone function. To this end, we build on the results of a long-range coordination analysis that we developed to study Hsp90 communication pathways and dynamics at atomic resolution.9 Our approach showed that conformational changes and coordination between the N- and C-domains are responsive to specific nucleotide binding. This propagates molecular signals long-range, selectively to functionally important residues and secondary structure elements at the CTD, which define possible allosteric binding sites. The physicochemical properties of newly detected functional CTD sites are used here to build receptor-based 3D pharmacophore models. This allows us to identify novel antagonists of Hsp90 chaperone function that target a site distant from the active site, inhibit several important protein–protein interactions, and show the ability to interrupt biological pathways important for cancer cell proliferation (Scheme 1). As expected the activities of the molecules, selected from a publicly available database, do not make them immediate candidates for drug development. However, it is important to underline that our main goal is to use information on the protein dynamics to identify potential hits. Drug design and medicinal chemistry efforts can then be started on this basis to improve the activities and pharmacokinetic properties of our hits.
Scheme 1. Computational Biology Strategy for the Selection of Allosteric Inhibitors Exploiting Protein Dynamicsa.
a From the analysis of the dynamics of the activated state of the protein (Hsp90 bound to ATP), the hot spot residues active in mediating signal transduction are identified. Analysis of possible binding pockets centered on these residues identifies putative allosteric binding sites. A consensus model of functional interactions with the hot spots together with structural shape constraints is used for pharmacophore modeling. The ensemble-based approach ensures the incorporation of receptor flexibility into the pharmacophore model. Small molecule databases are then screened for leads fitting with the pharmacophoric hypothesis, and selected hits are tested experimentally.
We focus on Hsp90 as an example of a molecular system in which ligand-based activation and signal communication between physically distant domains underlies protein–protein interactions and biological function. Hsp90 is a homodimer in which each protomer is characterized by a modular architecture with three domains: an N-terminal regulatory domain (NTD), a middle domain (M-domain), and a carboxyterminal domain (CTD).10–12 The biological activity of Hsp90 depends on ATP binding and hydrolysis, which is coupled to a conformational cycle that involves the opening and closing of a dimeric molecular clamp formed by the association of the NTDs of Hsp90.13 In solution, the protein exists as a dimer owing to the stable association of highly conserved motifs in its CTD. ATP binds to the NTDs of Hsp90, stabilizes their transient dimerization,13 and sends a conformational signal to the CTD, which is responsible for the acquisition of the ATP-ase competent conformation required for chaperone activity. Moreover, the Hsp90 chaperone function is finely regulated in the cell by physical association with a number of cochaperones that regulate the ATP-ase activity or direct Hsp90 to interact with different client proteins. Different cochaperones bind to different domains of Hps90 (for a complete review see).14
Hsp90 has a well-established role in the conformational maturation, stability, and function of a wide range of “client” proteins within the cell. In cancer cells, Hsp90 is overexpressed and intersects signaling pathways essential for tumor maintenance, and its inhibition through drugs targeting the N-terminal ATP-site showed promising therapeutic perspectives.15
The results presented here open the possibility to rationally expand the chemical space of Hsp90 antagonists to effective inhibitors of allosteric communications.
Experimental Section
In this section, we describe the computational and experimental procedures in detail. In the Computational Details Section, we first describe how molecular dynamics (MD) simulations and signal communication modeling were carried out. These experimental details have already been fully described in.9 Here we are reporting them for clarity. Next we describe the development of the pharmacophore modeling, virtual ligand screening (VLS), the docking of known and new compounds to the newly discovered pockets.
In the Experimental Procedures Section, we report on the experimental procedures used to test the small molecules.
Computational Details.
MD Simulations.
The MD simulation trajectories used in this work were carried out as already described in ref 9. The details and the full description of the MD set up and runs can thus be found in the published paper dealing with the characterization of the ligand modulation of Hsp90 dynamics.9
Briefly, the crystal structure (pdb entry 2CG9)10 containing yeast Hsp90 dimer bound to ATP was employed as a starting point for the simulations. The system was solvated in a tetrahedral solvation box contains around 57 000 particles. All simulations and the analysis of the trajectories were performed using the GROMACS software package16 using the GROMOS96 force field17 and the SPC water model.18
The ATP-bound Hsp90 dimer system was first energy relaxed with 2000 steps of steepest-descent energy minimization followed by another 2000 steps of conjugate gradient energy minimization. The energy minimization was used to remove possible bad contacts from the initial structures. The system was then equilibrated by a 50 ps of MD run with position restraints on the protein and ligand to allow relaxation of the solvent molecules. The first equilibration run was followed by a second 50 ps run without position restraints on the solute. The first 5 ns of the trajectory was not used in the subsequent analysis in order to minimize convergence artifacts. Equilibration of the trajectory was checked by monitoring the equilibration of quantities, such as the root-mean-square deviation (rmsd) with respect to the initial structure, the internal protein energy, and fluctuations were calculated on different time intervals. The electrostatic term was described by using the particle mesh Ewald algorithm. The LINCS19 algorithm was used to constrain all bond lengths. For the water molecules, the SETTLE algorithm20 was used. A dielectric permittivity, ε = 1, and a time step of 2 fs were used. All atoms were given an initial velocity obtained from a Maxwellian distribution at the desired initial temperature of 300 K. The density of the system was adjusted performing the first equilibration runs at NPT condition by weak coupling to a bath of constant pressure (P0 = 1 bar, coupling time τp = 0.5 ps).21 In all simulations, the temperature was maintained close to the intended values by weak coupling to an external temperature bath21 with a coupling constant of 0.1 ps. The proteins and the rest of the system were coupled separately to the temperature bath. The structural cluster analysis was carried out using the method described by Daura and co-workers with a cutoff of 0.25 nm.22
Signal Propagation Analysis.
This approach was also already described in ref 9. It is based on the adaptation of a recent approach proposed by Bahar and co-workers to the analysis of all-atom MD simulation trajectories. The analysis of signal propagation, which was developed based on elastic network models,23 defines signal transduction events in proteins as directly related to the fluctuation dynamics of atoms, defining the communication propensities (CP) of a pair of residues as a function of the fluctuations of interresidue distances. Residues whose Cα–Cα distance fluctuates with a relatively small intensity during the trajectory are supposed to communicate more efficiently than residues whose distance fluctuations are large. In the former case, a perturbation at the one site, affecting the Cα position, is likely to be visible (reflected) at the second site, while in the latter case, the communication is less efficient due to the intrinsic amplitude of the distance fluctuations. The CP of any two residues is defined as the mean-square fluctuation of the interresidue distance defining as distance between the Cα atoms of residues i and j, respectively:
By projecting these quantities on the 3D structures of the protein bound to different ligands, it will be possible to identify possible differences in the interdomain and interprotomer long-range redistributions of interactions.
The CP was calculated for any pair of residues during the trajectory. It is worth noting that CP describes the distance fluctuation of the two residues, therefore, low CP values characterize residues that move in a highly coordinated fashion and hence may be involved in the efficient relay of conformational signals.9 The average CP value for consecutive amino acids along the sequence, calculated considering for each residue i the neighbors comprised between i − 4 and i + 4, is 0.025. The average CP value for residues distant more than 40 Å is 0.12. In the presence of ATP, around 1% of residue pairs have CP < 0.025 even if they are at distances larger than 40 Å.9 Therefore, in the presence of ligands, a number of very distant residues may have a low CP value and display high coordination despite their physical separation, and we set CP = 0.025 as a convenient threshold for discriminating high dynamic coordination at long distance. CP values at increasing distances were scanned through histogram analysis. Each bin of the histogram refers to a residue and gives the fraction of residues that have high coordination with it (CP < 0.025) at distances larger than an increasing cutoff of 40, 60, and 80 Å, respectively. Residues corresponding to histogram peaks define regions that are specifically involved in efficient long-range correlations. Results of the analysis are fully reported in ref 9.
Upon increase of the residue–residue distance in the CP scanning histograms, some peaks become progressively smaller or disappear, since the fraction of effectively coordinated residues decreases at longer physical distances. On the other hand, since the total number of possible pairs also decreases with increasing distance, for some residues the fraction of highly coordinated partners may grow at longer distances, and those residues we define to be strongly active in long-range signaling.
The residues in the C-terminal preserving the most efficient communication propensities with the ATP site were used to define the possible allosteric-binding pocket. They comprise the NTD residues 81–95 and 121–140 (Hsp90 residues numbering as in the pdb entry 2CG9) that have a long-range signaling propensity with segments 574–580 and with the two C-terminal interface helices, made of residues 645–654 (helix 4) and 661–671 (helix 5), respectively.
MD-Based Pharmacophore Modeling.
Hsp90 dimer conformations were collected at every 0.5 ns of the final 20 ns MD trajectory using the GROMACS software package and superimposed at the putative C-terminal binding pocket, i.e., residues 475–477, 591–595, 602–603, and 652–657 of one monomer and residues 502–504, 591–595, and 656–662 of another. Superimposition was performed based on backbone atoms. GREATER v1.2.2, the graphical user interface for GRID v22a, was used to calculate molecular interaction fields (MIFs) with the probes DRY (hydrophobic), O (sp2 carbonyl oxygen) and N1 (neutral flat amide NH).24 The protein was considered rigid and a 31 × 27 × 18 Å grid box was centered at the binding pocket. Grid spacing was set to 0.25 Å. Local energy minima, defined as isocontours from probes DRY (−0.8), O (−7), and N1 (−7 kcal/mol), were represented with the VMD software v1.8.6.25 Binding pocket regions with consistently favorable interactions along the MD trajectory were used to define 3D pharmacophore features of a pharmacophore hypothesis for Hsp90 C-terminal binding.
Pocket analysis was also carried out with the PocketFinder module of the ICM suite.26 Probe atoms (carbon, oxygen, and nitrogen atoms) were placed at the center of higher density areas and converted into a pharmacophore hypothesis using the Catalyst ViewHypothesis workbench of the Catalyst v4.1.1 software. Local energy minima identified with the DRY, O, and N1 probes were converted into hydrophobic and hydrogen-bond acceptor and donor features, respectively. Flexibility was taken into account with 1.6 Å radium tolerances around each pharmacophore feature, i.e., spherical volumes where matching chemical groups should be located. Projection points from which the extended hydrogen-bond partner participates, i.e., Arg591 and/or Ser657 hydrogen bonding an acceptor group and Asp503 and/or Ser602 hydrogen bonding a donor group, were created in order to mimic the location of their side chains during the MD simulation. Shape filtering was done by filling the common binding cavity along the last 10 ns MD trajectory with chemical probes (carbon atoms) and converting them into inclusion volumes using the convert molecule to shape tool of the Catalyst ViewHypothesis workbench. The minimum similarity tolerance was set to 0.5.
The final pharmacophore hypothesis consisted of a 3D arrangement of six features (i.e., four hydrophobic regions and one each hydrogen-bond acceptor and donor) located at defined positions. These were surrounded by 1.6 Å radium tolerance spheres, assessing the area in space that should be matched by corresponding chemical functions of the virtual screening molecules. The hydrogen-bond acceptor and donor features additionally include a vector indicating the direction of the interaction. The desirable shape of the new virtual screening hits was delimited by a series of inclusion volumes. Table 1 reports on the distance constraints for the pharmacophore model generated.
Table 1.
Pharmacophore Conformational Propertiesa
| HYD1 | HYD2 | HYD3 | HYD4 | HBA | |
|---|---|---|---|---|---|
| HYD2 | 2.6 | ||||
| HYD3 | 9.9 | 7.9 | |||
| HYD4 | 13.4 | 11.7 | 4.5 | ||
| HBA | 9.8 | 8.1 | 5.3 | 5.6 | |
| HBD | 15.8 | 14.1 | 7.0 | 2.6 | 7.1 |
Distance constraints in Å.
Virtual Screening.
The NCI database was downloaded from the 2007 release of the ZINC library27 and converted into a multiconformer Catalyst database. A maximum of 100 conformations, within a 20 kcal/mol energy range above the calculated global minimum, were generated for each molecule using the “FAST” conformational analysis model of catDB utility program. The pharmacophore hypothesis was screened using the “fast flexible database search” settings.
Docking of Known and Newly Discovered Compounds.
Initial models for novobiocin, its derivatives, and the selected small molecules described in the paper were generated using the standard building blocks of MAESTRO v8.5 and minimized with MACROMODEL v8.1,28,29 using the Merck molecular force field (MMFF),30 the Polak–Ribiere conjugate gradient (PRCG) minimization method with an energy convergence criterion of 0.05 kJ/mol and the generalized Born equation/surface area (GB/SA)31 continuum solvation model with parameters for water (dielectric constant ε = 78). Five thousand steps of the systematic unbounded multiple minimum (SUMM) method implemented in MACROMODEL were used in order to allow a full exploration of the conformational space. Autodock Tools v1.5 was used to prepare ligands and receptors for docking, namely, to remove water molecules, add hydrogens, compute Gasteiger charges,32 and merge nonpolar hydrogens. Side chain charges were assigned according to their pKa. Blind docking experiments on the whole Hsp90 C-terminus domain were performed with the novobiocin derivatives using AutoDock v4.0.33 Grid maps were generated with AutoGrid v4.0 using a 0.375 Å grid spacing. The Lamarckian genetic algorithm was employed for all docking runs. An initial population of 150 individuals randomly placed on the Hsp90 C-terminus domain was created. Random orientations and torsions were used. The number of generations was set to 25 million, and the maximum number of energy calculations was set to 27 thousands. A mutation rate of 0.02 and a crossover rate of 0.8 were used, and the local search frequency was set up at 0.06. Two hundred independent runs were performed for each compound with the parameters described above. Results differing by less than 2 Å in positional rmds were clustered together and represented by the result with the most favorable free energy of binding.
Initial geometries for the virtual screening hit compounds were collected from the ZINC database.27 Docking runs were limited to the allosteric binding pocket at the dimer interface.
Experimental Procedures.
Cell Viability, Elisa Tests, and Akt Folding. Cells and Cell Cultures.
Human prostate adenocarcinoma PC3 and lung adenocarcinoma H460 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in cultures as recommended by the supplier. Human umbilical vein endothelial cells (HUVEC) were obtained from Clonetics. Rat A10 smooth muscle cells were the generous gift of Dr. Michael Conte, University of California, San Francisco.
Antibodies.
Antibodies to b-actin (Sigma-Aldrich) and Akt (CST, Inc., Danvers, MA) were used.
Binding Assays.
Plastic microtiter wells were coated with increasing concentrations (0–150 μM) of the various compounds, blocked in 3% gelatin, and further mixed with recombinant full length Hsp90 or Hsp90 C-domain (residues 629–732, 1 mg/mL) produced in BL-21 E.Coli as a GST fusion protein, and further isolated from the GST frame by thrombin cleavage. After a 2 h incubation at 22 °C, compound binding under the various conditions tested was detected with an antibody to Hsp90, followed by a peroxidase-conjugated secondary reagent and quantification of absorbance at 405 nm.
Cell Viability Analysis.
The various normal or tumor cell types (2 × 105/ml, 50 mL) were seeded in triplicates in 96-well plates and incubated with increasing concentrations of the various Hsp90-C terminus compounds (0–150 mM) for 16 h at 22 °C. At the end of the incubation, cultures were analyzed for cell viability by an 3(4,5-dimethyl-thyazoyl-2-yl)2,5 diphenyl-tetrazolium bromide (MTT) colorimetric assay with absorbance at 405 nm. In other experiments, tumor cell types were incubated with various concentrations of Hsp90 C-terminus compounds, and whole cell extracts were analyzed by Western blotting.
Statistical Analysis.
Data were analyzed using the two-sided unpaired t test on a GraphPad software package for Windows (Prism). A p value of 0.05 was considered as statistically significant.
Cochaperone and Client Protein Interactions with Coimmunoprecipitation Assays. Cell Culture, Transfection, and Immunoprecipitation.
COS7 cells (American Type Culture Collection) were cultured in a temperature-controlled incubator (37 °C and 5% CO2) in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% (v/v) fetal bovine serum (FBS), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.0), 2 mM glutamine, 1 mM of sodium pyruvate, and nonessential amino acids (Biosource/Invitrogen). Cells were transiently transfected with pcDNA3 empty vector or pcDNA3 containing flag-tagged wild-type human Hsp90alpha by using FuGene6 (Roche Applied Science), following the manufacturer’s instructions. Twenty-four hours after transfection, cells were treated with 100 μM of indicated compounds for 1 h. Then, cells were lysed (20 mM HEPES, 100 mM NaCl, 1 mM MgCl2, 0.1% NP-40, 20 mM Na2MoO4, phosphatase inhibitor (Roche), and protease inhibitors(Roche)), and incubated with anti-flag antibody-conjugated beads (Sigma) for 2 h at 4 °C. Coimmunoprecipitated proteins were identified by immunoblotting with indicated antibodies recognizing Flag (Affinity Bioreagents), ERBB2 (Santa Cruz), CDK4 (Santa Cruz), p60Hop (Cell Signaling), p50Cdc37 (Neomarkers), p23 (Affinity Bioreagents), or AHA1 (Rockland). See also ref 34.
Results
Background: Hot Spots in Signal Transduction from the ATP-Site to the CTD.
Different dynamic states of Hsp90 can be switched on/off in response to the presence of a specific ligand at the ATP-binding site. In this context, we generated a computational model aimed to identify the substructures (subdomains, secondary structure elements, single residues) that play a relevant role in the dynamic communication between a certain binding site and distal regions of the protein implied in function. The results showed, at atomic resolution, that the identity of mediators of the cross-talk between N- and C-domains was dependent on the specific nucleotide activating differential functional motions. Briefly, in our approach, which builds on the work of Chennubotla and Bahar,23 the CP between any two residues, as a function of fluctuation of their distance components, is evaluated. CP describes a communication time; therefore, low CP values are related to efficiently communicating residues. The threshold for high communication efficiency is the CP value calculated for four consecutive residues along the sequence. Hot spots for signal transduction are identified by calculating for each residue the fraction of all other protein residues that have high communication efficiency with it (CP lower than the threshold) at distances larger than an increasing cutoff of 40, 60, and 80 Å. Distant, physically separated residues that have a more efficient communication than that defined by the “local” threshold define the regions specifically involved in efficient long-range signal transduction.9
This analysis illuminated different pathways of signal transduction that selectively depend on the ligand identity. In particular, specific clusters of residues participate in the signal transduction from the N-terminal nucleotide-binding site to the CTD. In the ATP-bound, active form of the chaperone, long-range communication from the binding site is mainly directed to residues at the CTD interface. In particular, NTD residues 81–95 and 121–140 involved in ATP recognition (residue numbering from 2CG9.pdb) show a consistently high long-range coordination with segments 574–580 and with the C-terminal interface helices, made of residues 645–654 (helix 4) and 661–671 (helix 5), respectively (see Supporting Information, Figure S1).
Identification of Allosteric Pockets.
The C-terminal interface region with higher communication propensity with the distal ATP-binding site was then subjected to structural investigation to detect potential binding sites centered on the communication hot spots. Cluster analysis of the trajectories was used to identify the most representative conformations of the CTD. Individual frames were grouped into 21 clusters, with the most populated five accounting for 84% of the structural diversity.
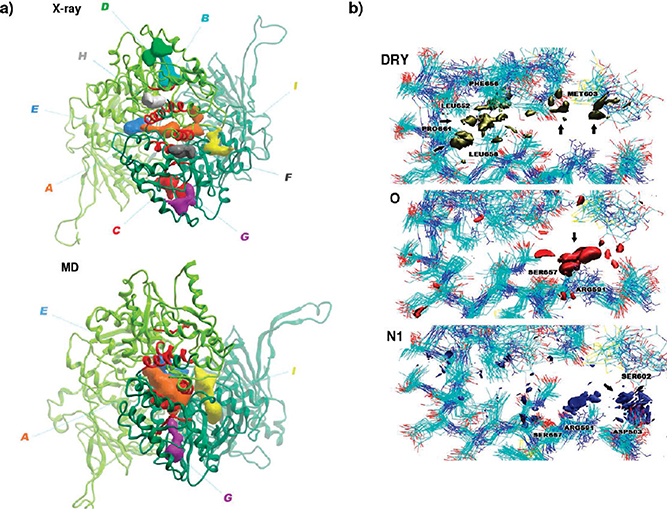
These representative structures and the original crystal structure (2CG9.pdb) were subjected to analysis with the pocketFinder module of the ICM software,26 complemented by visual inspection. Nine potential binding pockets with volume and area suitable for interaction with drug-like compounds were identified in the X-ray crystal structure (Scheme 1, Table 2, and Figure S1b,c of the Supporting Information). Interestingly, only pocket A is consistently detected in all representative MD conformations, increasing in volume and area and defining a binding tunnel at the dimer interface suitable to accommodate small compounds able to interact directly with the hot spot residues involved in efficient long-range coordination (Figure 1a, Table 2, and Figure S1c of the Supporting Information).
Table 2.
Drug-Like Binding Sites Identified with the ICM PocketFinder Modulea
| pockets |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| structure cluster time (ns) | A |
B |
C |
D |
E |
F |
G |
H |
I |
|||||||||
| volume | area | volume | area | volume | area | volume | area | volume | area | volume | area | volume | area | volume | area | volume | area | |
| X-ray t = 0 | 453.9 | 497.8 | 325.2 | 312.7 | 324.8 | 330.2 | 249.5 | 242.2 | 230.5 | 214.2 | 227.7 | 279.3 | 213.9 | 256.3 | 212.4 | 226.5 | 184.0 | 231.9 |
| 1 t = 27.9 | 243.2 | 225.6 | 409.3 | 469.2 | 151.5 | 209.0 | 455.7 | 501.3 | ||||||||||
| 2 t = 65.2 | 670.4 | 543.8 | 236.8 | 304.9 | 250.4 | 262.4 | 484.8 | 465.8 | ||||||||||
| 3 t = 46.3 | 551.6 | 549.3 | 192.5 | 207.5 | 158.9 | 204.0 | 266.9 | 290.4 | ||||||||||
| 4t = 9.5 | 331.2 | 298.7 | 453.8 | 441.2 | 192.3 | 233.9 | ||||||||||||
| 5 t = 51.1 | 532.7 | 558.2 | 375.7 | 359.3 | 157.1 | 200.4 | ||||||||||||
| residues | chain a: 331, 423, 476, 477, 502–505, 591, 592, 594, 595, 657–661 | chain b: 470, 472, 473, 507, 511, 514, 520–525, 527, 528, 582–586, 589 | chain a: 470, 472, 473, 491, 514, 521–527, 581–586, 589 | chain b: 472, 490, 491, 494, 496, 527–530, 532, 537, 540, 544, 568–570, 579 | chain a: 598–601 | chain a: 486, 573–576, 620, 621, 624, 646, 649, 650, 653 | chain a: 488, 490, 491, 494, 496, 527–530, 532, 537, 540, 568–570, 579 | chain a: 662, 665, 666, 669, 670 | chain a: 415, 418, 422, 426, 444–446, 454, 500–503, 506 | |||||||||
| chain b: 477, 478, 590, 591, 593–595, 597, 599, 606, 607, 657, 658 | chain b: 327–329, 331–333, 360, 361, 363, 364, 420, 421, 423, 424, 427 | chain b: 662, 665, 666, 669, 670, 673, 676, 677 | chain b: 486, 487, 573–576, 620, 621, 646, 649, 650, 653 | chain b: 599, 601,602 | ||||||||||||||
Volume (Å3) and area (Å2) are provided for each of these pockets. Blank spaces indicate either that no binding pocket was found or that the volume and area were relatively small for binding of drug-like compounds (volume <150 and/or area <200). Residues surrounding each of the binding pockets at the pdb entry 2CG9 are shown in the last row.
Figure 1.
Pharmacophore model and resulting small molecules. (a) 3D representations of potential ligand binding pockets identified with the ICM pocketFinder module on the CTD of the Hsp90 dimer from X-ray crystal structure and from the representative structure corresponding to 65.2 ns of the MD simulation (cluster 2). Pocket A located at the dimer interface increases in area, volume, and number of contacts with the communication hot spot residues represented with a red ribbon. (b) Isosurfaces for the DRY, O, and N1 probes from the GRID force field in the putative allosteric pocket. DRY probe highlights four hydrophobic regions related to favorable interactions with apolar residues, such as Met 603, Leu 652, Phe 656, Leu 658, and Pro 661. O and N1 probes identify two regions prone to hydrogen bonding, one of these acting mainly as acceptor from Arg 591 and Ser 657 and the other as donor to Asp 503 and Ser 602, respectively.
Allosteric Inhibitor Discovery: Pharmacophore Modeling Based on Signal Transduction Information.
Next, we used the information on signal transduction, conformational states spanned by hot spot residues, and conformational properties of pocket A together with the analysis of their chemical properties to develop pharmacophore models for virtual screening of small molecule databases. The pharmacophores are designed to recapitulate the complementary interactions necessary to guarantee productive binding with the putative allosteric site.
Structures from the final 20 ns of the MD simulations were used. Local molecular interaction fields (MIF) minima were calculated at the allosteric site with the GRID force field and probes accounting for hydrophobic (DRY) and hydrogen-bond acceptor (O) and donor (N1) interactions.35 Isosurfaces at −0.8 kcal/mol derived from the DRY probe highlight four hydrophobic regions related to favorable interactions with apolar residues, such as Met 603, Leu 652, Phe 656, Leu 658, and Pro 661 (Figure 1b). Surfaces defined at an energy level of −7 kcal/mol with the probes O and N1 identified two regions prone to hydrogen bonding, one of these acting mainly as acceptor from Arg 591 and Ser 657 (Figure 1b) and the other as donor to Asp 503 and Ser 602 (Figure 1b). The fluctuations in the positions, distances, and dihedral angles among the side chain functionalities of these critical residues were used to define the average and upper and lower boundaries in the positioning of the hydrogen-bond donor functions of the pharmacophore.
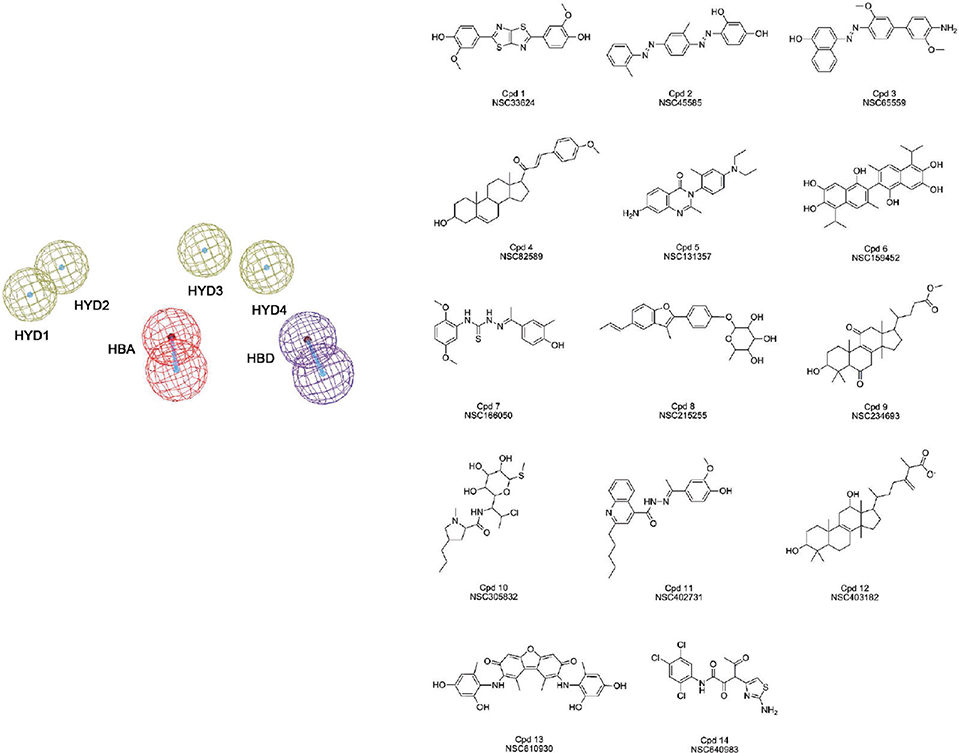
Taken together, these interactions defined a six-feature pharmacophore model for the virtual screening of new C-terminus targeted inhibitors of Hsp90 (Figure 2). The size and shape features of the new compounds were filtered with a set of inclusion volumes defined based on the radius and shape of pocket A at 65.2 ns of the MD simulation.
Figure 2.
The pharmacophore and selected hits. The resulting six-feature pharmacophore and the molecular structures of the compounds selected from virtual screening with the pharmacophore model of the NCI database.
Allosteric Inhibitors: New Hits through Pharmacophore Guided Virtual Screening.
The new allosteric pharmacophore model was used to perform a screening search of the NCI repository. The database contains a library of more than 290 000 compounds. Filtering of the database with the pharmacophore returned 36 hits (Figure 2), corresponding to 0.01% of the database.
Experimental Tests on Newly Discovered Hsp90 Inhibitors Targeting the C-terminal.
Fourteen of the selected compounds resulting from the virtual screening could be obtained from the NCI and tested for affinity for the Hsp90 full-length protein, the CTD, for their effects on cancer and normal cell viability, for the induction of degradation of specific Hsp90 client proteins, and for their activity in disrupting Hsp90 association with cochaperones.
Molecular Interactions between Selected Molecules and Hsp90.
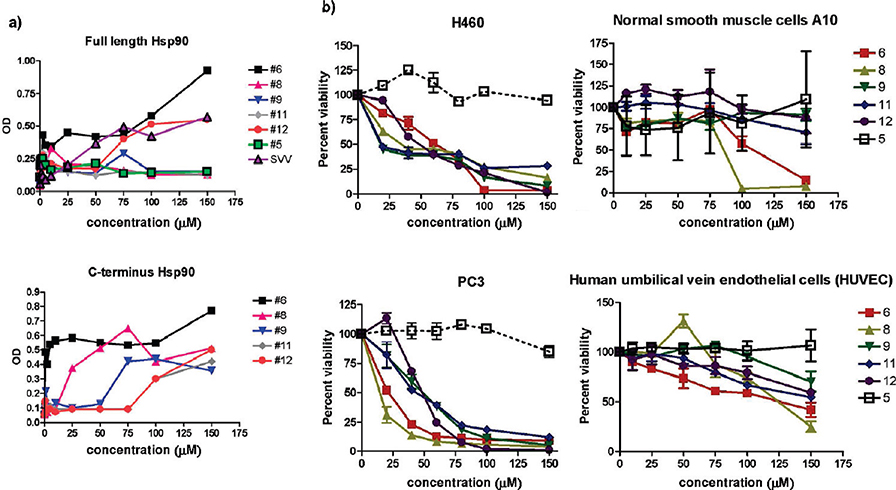
By ELISA tests several of the discovered lead compounds (namely 6, 8, 9, 11, 12) bound the recombinant isolated Hsp90 C-domain in a specific and saturable manner (Figure 3a). Functionally, treatment of lung adenocarcinoma H460 or prostate adenocarcinoma PC3 cells with the selected compounds resulted in a concentration-dependent loss of cell viability (Figure 3b). This response was specific for inhibition of cancer-related signaling, as the implicated compounds did not reduce the viability of normal A10 smooth muscle cells or human umbilical vein endothelial cells (Figure 3b).
Figure 3.
Small molecules bind to Hsp90 CTD and affect cancer cell viability. (a) ELISA. Microtiter wells were coated with the indicated increasing concentrations of small molecules and incubated with recombinant full-length or C-domain of Hsp90. Binding was determined using domain-specific antibodies to Hsp90 and quantified by absorbance at OD405. Data are the mean ±SEM of three independent experiments. (b) Inhibition of cell viability in H460 and PC3 cancer cell lines, and normal smooth muscle cells A10 and HUVEC cells, as evaluated by cell counting after a 24 h exposure to the selected small molecules. Values represent the mean (±SD) of three independent experiments.
Selected Hits Inhibit Hsp90 Chaperone Function and Impact on Hsp90 Association with Cochaperones.
We next asked whether the cytotoxic effect exerted by compounds 6, 8, 9, 11, and 12 was due to loss of Hsp90 client proteins resulting from inhibition of chaperone function. Consistent with this model, a preliminary analysis of compounds 6, 8, and 9 induced a concentration-dependent loss of the Hsp90 client protein, the kinase Akt, in tumor cells. Selected compounds were active in a concentration range between 25 and 100 μM, with activities comparable to those of known C-terminal inhibitors. As control, compound 5, which showed no effect on tumor cell viability, did not reduce Akt levels in tumor cells (Supporting Information, Figure S2).
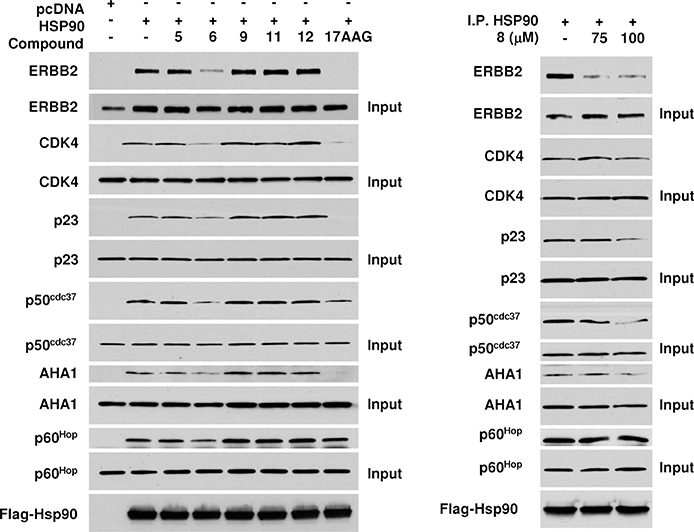
Selected compounds were also tested in a different experimental setting (see Materials and Methods in Supporting Information) using coimmunoprecipitation assays to probe the interaction of Hsp90 with client and cochaperone proteins. In this test, compound 6 clearly disrupted interactions with two kinase client proteins ERBB2 and CDK4. Moreover, compound 6 was shown to impair the binding of cochaperones p23, p50, and Aha1. While Aha1 interacts with the middle segment of Hsp90, p23 and p50 bind Hsp90 at the NTD. Since compound 6 is selected to interact with the CTD, these data suggest that it likely alters Hsp90 conformational equilibrium affecting client and cochaperone binding via an allosteric mechanism. Interestingly, compound 6 emerged as the only one able to slightly reduce Hsp90 association with p60, a cochaperone known to bind to the CTD. Binding of the inhibitor at the CTD may directly interfere with the physical binding of p60 at the same region of the protein (Figure 4).
Figure 4.
Inhibition of Hsp90 chaperone function. Client and cochaperone binding to Hsp90 is inhibited by small molecules 6 and 8. COS7 cells were transfected with wild-type Flag–Hsp90. After incubation, cells were treated with 100 μM of the indicated Hsp90 inhibitor for 1 h. Then, cells were lysed, and proteins were immunoprecipitated (IP) by a Flag antibody-conjugated agarose. Indicated coprecipitating proteins were detected by immunoblotting.
The coimmunoprecipitation experiments also showed that compound 8 could dramatically disrupt ERBB2/Hsp90 association at 75 and 100 μM doses, supporting the validity of the computational design approach (Figure 4).
Overall, these results confirm the validity of the computational approach taking the full dynamics of the protein into account to discover new allosteric sites.
Binding Poses of the Hits in Hsp90 CTD.
The molecular interactions of the compounds identified through the allosteric dynamic pharmacophore with the Hsp90 CTD were characterized via computational docking and analysis. The allosteric binding pocket is a small tunnel located at the dimer interface, delimited by residues 474–487, 502–503, 591–599, 602–603, and 652–657 from one monomer and residues 502–504, 591–595, and 656–662 from another (pocket A, Figure 1, and Figure S1, Supporting Information). Although already present in the Hsp90 crystal structure, the shape of the newly found putative site increases its binding complementarity to C-terminus inhibitors during the MD simulation, in the absence of the inhibitors. Multiple structures from the MD simulation of the ATP-complex were used as targets. This is equivalent to describing relevant representatives of the ensemble of conformational states, taking flexibility of the whole protein into account.
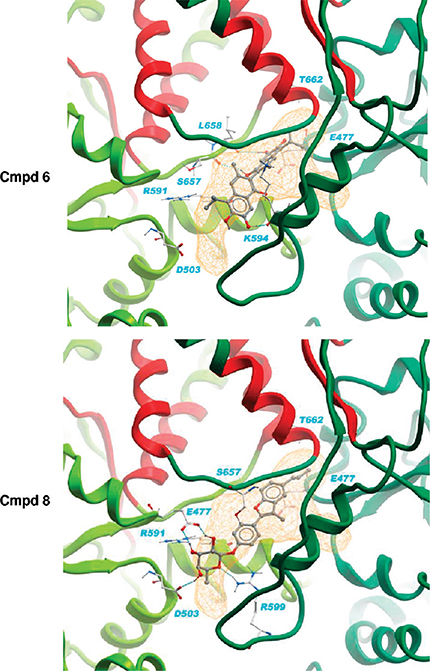
Binding poses and theoretical affinities were calculated and the results are reported in Table S1 of the Supporting Information. The whole CTD surface was scanned, and the active compounds were observed to dock selectively and favorably in the proposed allosteric pocket, consistent with MD simulations and pharmacophore analysis (Figure 5). These molecules show a good shape complementarity to pocket A at the dimer interface, establishing hydrogen bonds and hydrophobic contacts with the proposed allosteric hot spot residues.
Figure 5.
Binding of new hits (compounds 6 and 8) into the Hsp90 dimer. Lowest binding energies were obtained with a MD representative conformation (cluster 2, time = 65 200 ps). The protein complex is shown in a ribbon representation colored by a chain with the putative communication hot spot residues colored in red. The Hsp90 C-terminal binding pocket (pocket A) is shown as an orange line mesh and protein–ligand hydrogen bonds are represented with spheres.
Finally, we docked Hsp90 inhibitors targeting the CTD derived from the literature to the newly discovered allosteric pocket to further validate our approach through structure–activity relationships. No experimental structural information is available on complexes between CTD and these inhibitors. Novobiocin (IC50 700 μM) and the more potent related derivatives ND-1 (active at 100 μM) and ND-2 (active at 40 μM)36,37 (Supporting Information, Figure S3) were thus docked to the full CTD. The calculated affinities are reported in Table S2 of the Supporting Information, along with the contacts established by the docked drugs with signal transduction hot spots. Interestingly, estimated binding energies with novobiocin and related derivatives resulted in good agreement with their relative inhibition potencies. The strongest protein–small molecule interactions with novobiocin (−6.02 kcal/mol) and compounds ND-1 and ND-2 (−6.62 and −8.14 kcal/mol, respectively) were found with the representative structure of cluster 2 (65.2 ns frame of the MD simulation). The three compounds make contact with residues belonging to the communication hot spot structures. Of critical importance is the disruption of a salt bridge between Glu 477 and Arg 591 after approximately 60 ns that increases the size and the volume of the binding site and improves the calculated binding affinity for compounds ND-1 and ND-2.
The qualitative good correlation between the calculated affinities and the experimental activities of the small molecules constitutes an encouraging validation of the target and of the use of information from signal transduction analysis in the detection of putative allosteric binding sites.
Discussion
The discovery of new allosteric sites may offer novel opportunities in the identification of new drugs and in the understanding of fundamental biological processes. While consensus is increasing on the importance of allosteric motions in the context of protein functional control and regulation, the relevance of using these concepts in drug design has not been fully exploited.8,38,39 Discovering and targeting allosteric sites can in fact lead to the expansion of the chemical space of leads and to new classes of drugs. Most importantly, the discovery of new molecules targeted to allosteric sites may represent a viable strategy in the search for new protein–protein interactions inhibitors.40
Protein conformational plasticity and dynamics appear to be critical for allosteric events. In the current view of allostery, a protein populates a certain ensemble of dynamic conformational states at equilibrium, and perturbations induce a shift in the relative populations of states. Signals coded by covalent or noncovalent modifications can be transmitted long-range through pre-existing pathways4,6 that depend on the inherent topological architecture of the protein. At a more refined level, the selection of specific communication pathways between physically separate sites depends on the fine chemical properties of the modification or the binding partner. Results from several research groups have revealed the existence of alternative interaction networks with a link to dynamic motions,41–44 showing that preferred relatively small, local fluctuations in proteins lead to functionally active states.
In this paper, we have built on our previous results on the atomic level characterization of the correlations between dynamics, long-range coordination, and allosteric communication between the physically distant N- and C-domains in full-length Hsp90 to develop a new strategy for computational discovery of allosteric inhibitors. Hsp90 dynamic and functional properties appeared to be highly responsive to the presence of a specific nucleotide in the ATP-site at the N-domain. Once the principal signal transduction pathways and the correlated hot spot residues that act as communication mediators between the ATP-site and the C-terminal interface have been revealed,9 the translation of this structural dynamical information into 3D receptor-based pharmacophore models allowed us to rationally discover new C-terminal ligands able to interfere with the chaperone function.
In this framework, we have focused on the activated form of Hsp90, which represents an important target for cancer drug discovery. Our signal transduction model revealed that the most efficient long-range communication (over >60 Å) from the binding site is mainly directed to a specific subset of residues at the CTD interface. Conformational analysis of the whole simulation trajectory showed that this site could populate a set of conformations apt to optimally accommodate known CTD targeted inhibitors. Docking of novobiocin and related derivatives to different representative protein conformations actually provided semiquantitative correlations between the activities of the compounds and their calculated binding energies.
We set out to search for new molecules targeting the newly discovered putative C-terminal allosteric site. The aim was to cause a disruptive interference in the network of interactions coding for the collective motions related to the chaperone functional activity. Our rationale was that the new hits should perturb the dynamics of the CTD substructures important for signal transduction with the NTD and interfere with the chaperone molecular recognition properties, thus disrupting association with cochaperones necessary for function and ultimately blocking client folding. To this end, we developed a pharmacophore model with complementary functionalities for the C-terminal allosteric site, using multiple protein target structures to take the flexibility of the whole protein into account. The dynamic pharmacophore was then used to screen the NCI small molecule database. Strikingly, experimental tests proved that selected molecules bind the CTD of Hsp90. Moreover, they had important effects on the viability of two independent cancer cell lines (H460, lung e PC3, prostate), while affecting to a significantly lower degree the two normal cell types (endothelial cells and vascular smooth muscle). Compounds 6, 8, and 9 were demonstrated to inhibit Hsp90 chaperone function, as shown by the effects on the levels of Akt, an established Hsp90 client protein that requires a fully functional chaperone activity for folding and stability. Using a different experimental approach, compound 6 and 8 were confirmed to disrupt association with two more client proteins. Most importantly compound 6 was shown to affect binding to a specific subset of cochaperones, reducing the association of Hsp90 with p23, p50, Aha1, and p60.
Interestingly, p23 and p50 bind to Hsp90 NTD, while the activity of Aha1 and Akt depends on interactions with the M-domain.45 Since the selected molecules interact directly with the C-domain, they likely alter Hsp90 molecular recognition properties by influencing its dynamics through an allosteric mechanism.
Consequently, these hits represent new leads for the development of allosteric drugs that act by tweaking the functional dynamics of the protein toward an inactive state.
The fact that molecule 6 and 8 resulted the only active hits in this second series of experiments does not exclude that the other derivatives may show similar effects under different experimental conditions. The coimmunoprecipitation assays are in fact based on Hsp90 overexpression with a drug incubation time of 1 h. Different compounds may have different binding kinetics and affinities for the chaperone, determined by specific on/off rates or different diffusion properties within the cell. The incubation time allowed for these first control experiments thus may not be sufficient to break client/cochaperone interactions.
It is worth noting at this point that compound 6 reduces the interaction between Hsp90 and the cochaperone p60, which is known to bind the CTD. Consequently, this lead also appears to perturb the molecular recognition properties of the CTD.
Importantly, the selected hits induce the disruption of Hsp90 complexes with important kinase client proteins and with cochaperones that are fundamental for Hsp90 functional activity through both the allosteric mechanisms and the abrogation of direct interactions with the CTD. This indicates that our hits act simultaneously on different biological pathways important for cancer development. It is important to underline here that the activities of our hits are still far from the ones required for efficient pharmacological applications. However, the scope of our endeavor was to identify active hits using information on an allosteric pocket obtained directly from the study of the dynamics of a complex molecular machine. Optimization of the structures through medicinal chemistry design and synthesis are currently underway.
From the applicative point of view, the possibility to rationally discover molecules that are active via allosteric and/or direct effects may facilitate the design of experiments aimed to disrupt specific interactions and to report on the behavior of the system/pathway in which the interaction is involved. All of these aspects may be important in the development of new cancer chemotherapeutics and in increasing our understanding of fundamental biochemical processes.
Moreover, we think that strategies similar to the one presented here, in which the dynamics of the target is explicitly taken into account, may be applied to the discovery of inhibitors of protein–protein interactions or of possible drug-binding sites for targets that are not easily druggable. In the former case, by carrying out an atomic resolution analysis of the protein’s internal dynamics and coordination, it may be possible to isolate the interaction surfaces that are endowed with specific flexibility properties and that need specific remodeling for the molecular recognition and binding of a second protein partner. In the latter case, the knowledge of internal coordination may be exploited to identify sites where binding of a small molecule can induce the perturbation of important functional motions, resulting in the inhibition of the function of the protein or enzyme under exam.
Our findings point to several features that make approaches, such as the one presented here, attractive for the discovery and development of allosteric inhibitors of protein functions and interactions. The concept of using a combination of structural, dynamic, and long-range correlation information led us to rationally discover a new and diverse set of chemical structures with drug-like properties able to target allosteric sites very distant from the active site. In this context, we could expand the molecular diversity space of Hsp90 antagonists, selecting molecules with promising anticancer activities.
Incorporating information on functional dynamics, internal residue–residue coordination, and protein flexibility can help unveil possible binding states of the receptor that are available on the protein’s energy landscape but may not be immediately evident in a single-structure representation. The discovery of alternative states can thus unveil possible allosteric binding sites, allow structurally different ligands to occupy the same site, or guide design efforts aimed at the functional and structural modification of existing leads to target-specific receptor geometries.
Overall, the use of biophysical and computational models taking dynamic and communication into account combined with pharmacophore development and screening may be useful to find new chemotypes for specific functions, to increase the yields of drug screening, and to help design new allosteric leads with important therapeutic opportunities.
Supplementary Material
Acknowledgment.
This work was supported by a grant from Associazione Italiana Ricerca sul Cancro (AIRC) to G.C. G.M. gratefully acknowledges support from a “L’Oréal-Unesco for Women in Science” grant.
Footnotes
Supporting Information Available: Contains all the description of the materials and methods used for the calculations and experimental procedures and description of additional tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Henzler-Wildman K; Kern D Nature 2007, 450, 964–972. [DOI] [PubMed] [Google Scholar]
- (2).Smock RG; Gierasch LM Science 2009, 324, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schrank TP; Bolen DW; Hilser VJ Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 16984–16989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hilser VJ Science 2010, 327, 653–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Boehr DD; Nussinov R; Wright PE Nat. Chem. Biol. 2009, 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).del Sol A; Tsai C-J; Ma B; Nussinov R Structure 2009, 17, 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hardy JA; Wells JA Curr. Opin. Struct. Biol. 2004, 14, 706–715. [DOI] [PubMed] [Google Scholar]
- (8).Wells JA; McCLendon CL Nature 2007, 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- (9).Morra G; Verkhivker GM; Colombo G PLoS Comput. Biol. 2009, 5, e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ali MMU; Roe SM; Vaughan CK; Meyer P; Panaretou B; Piper PW; Prodromou C; Pearl LH Nature 2006, 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shiau AK; Harris SF; Southworth DR; Agard DA Cell 2006, 127, 329–340. [DOI] [PubMed] [Google Scholar]
- (12).Dollins DE; Warren JJ; Immormino RM; Gewirth DT Mol. Cell 2007, 28, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pearl LH; Prodromou C; Workman P Biochem. J. 2008, 410, 439–453. [DOI] [PubMed] [Google Scholar]
- (14).Zuehlke A; Johnson JL Biopolymers 2010, 93, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Biamonte MA; Van de Water R; Arndt JW; Scannevin RH; Perret D; Lee WC J. Med. Chem. 2010, 53, 3–17. [DOI] [PubMed] [Google Scholar]
- (16).Van der Spoel D; Lindahl E; Hess B; Groenhof G; Mark AE; Berendsen HJ C. J. Comput. Chem. 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- (17).Scott WRP; Hunenberger PH; Tironi IG; Mark AE; Billeter SR; Fennen J; Torda AE; Huber T; Kruger P; Gunsteren WFV. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar]
- (18).Berendsen HJC; Grigera JR; Straatsma PR J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar]
- (19).Hess B; Bekker H; Fraaije JGEM; Berendsen HJ C. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar]
- (20).Miyamoto S; Kollman PA J. Comput. Chem. 1992, 13, 952–962. [Google Scholar]
- (21).Berendsen HJC; Postma JPM; van Gunsteren WF; Di Nola A; Haak JR J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar]
- (22).Daura X; Jaun B; Seebach D; Gunsteren W. F. v.; Mark AE. J. Mol. Biol. 1998, 280, 925–932. [DOI] [PubMed] [Google Scholar]
- (23).Chennubhotla C; Bahar I PLoS Comput. Biol. 2007, 3, 1716–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Goodford PJ J. Med. Chem. 1985, 28, 849–857. [DOI] [PubMed] [Google Scholar]
- (25).Humphrey W; Dalke A; Schulten KJ Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (26).Abagyan RA; Totrov MM; Kuznetsov DA J. Comput. Chem. 1994, 15, 488–506. [Google Scholar]
- (27).Irwin JJ; Shoichet BK J. Chem. Inf. Model. 2005, 45, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).MacroModel, version 8.1; Schrodinger: New York, NY, 2008. [Google Scholar]
- (29).Mohamadi F; Richards NGJ; Guida WC; Liskamp R; Lipton M; Caufield C; Chang G; Hendrickson T; Still WC J. Comput. Chem. 1990, 11, 440–467. [Google Scholar]
- (30).Halgren TA J. Comput. Chem. 1996, 490–519. [Google Scholar]
- (31).Still WC; Tempczyk A; Hawley RC; Hendrickson TJ Am. Chem. Soc. 1990, 112, 6127–6129. [Google Scholar]
- (32).Gasteiger J; Marsili M Tetrahedron 1980, 3219–3228. [Google Scholar]
- (33).Morris GM; Goodsell DS; Halliday RS; Huey R; Hart WE; Belew RK; Olson AJ J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar]
- (34).Tsutsumi S; Mollapour M; Graf C; Lee CT; Scroggins BT; Xu WP; Haslerova L; Hessling M; Konstantinova AA; Trepel JB; Panaretou B; Buchner J; Mayer MP; Prodromou C; Neckers L Nat. Struct. Mol. Biol. 2009, 16, 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Neves MAC; Dinis TCP; Colombo G; Melo ML S. J. Med. Chem. 2009, 52, 143–150. [DOI] [PubMed] [Google Scholar]
- (36).Le Bras G; Radanyi C; Peyrat JF; Brion JD; Alami M; Marsaud V; Stella B; Renoir JM J. Med. Chem. 2007, 50, 6189–6200. [DOI] [PubMed] [Google Scholar]
- (37).Yu XM; Shen G; Neckers L; Blake H; Holzbeierlein J; Cronk B; Blagg BS J. J. Am. Chem. Soc. 2005, 127, 12778–12779. [DOI] [PubMed] [Google Scholar]
- (38).Hardy JA; Wells JA Curr. Opin. Struct. Biol. 2004, 14, 706–715. [DOI] [PubMed] [Google Scholar]
- (39).Swain JF; Gierasch LM Curr. Opin. Struct. Biol. 2006, 16, 102–108. [DOI] [PubMed] [Google Scholar]
- (40).Zorn JA; Wells JA Nat. Chem. Biol. 2010, 6, 179–188. [DOI] [PubMed] [Google Scholar]
- (41).Suel GM; Lockless SW; Wall MA; Ranganathan R Nat. Struct. Biol. 2003, 10, 59–69. [DOI] [PubMed] [Google Scholar]
- (42).Lee J; Natarajan M; Nashine VC; Socolich M; Vo T; Russ WP; Benkovic SJ; Ranganathan R Science 2008, 322, 438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Swain JF; Dinler G; Sivendran R; Montgomery DL; Stotz M; Gierasch LM Mol. Cell 2007, 26, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Peng JW Structure 2009, 17, 310–320. [Google Scholar]
- (45).Zuehlke A; Johnson JL Biopolymers 2010, 93, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.