Abstract
Many environmental carcinogens cause DNA damage, which can result in mutations and other alterations in genomic DNA if not repaired promptly. Because of the bulkiness of the lesions, DNA-protein crosslinks (DPCs) are one of the types of toxic DNA damage with potentially deleterious consequences. Despite the importance of DPCs, how cells remove these complex DNA adducts has been incompletely understood. However, major progress in the DPC repair field over the past five years now supports the view that cells are equipped with multiple mechanisms to cope with DPCs. Here, we first provide an overview of environmental substances that induce DPCs, describing the sources of exposure and mechanisms of DPC formation. We then review current models of DPC repair and discuss their significance for environmental carcinogens.
Keywords: DNA damage, DNA repair, cancer, carcinogen, DPC
Introduction
According to International Agency for Research on Cancer (IARC), approximately 9.6 million deaths were estimated to be caused by cancer in the world in 2018 (Ferlay et al. 2019), making cancer the second leading cause of global deaths. Roughly 20% of these cancers are estimated to be attributable to environmental sources, including household air pollution, ambient air pollution and occupational exposures (Prüss-Üstün et al. 2016). Because carcinogenesis is strongly linked to DNA damage and mutagenesis, a better understanding of how carcinogens generate DNA damage and how cells deal with those DNA lesions is critical for assessing and mitigating health risk.
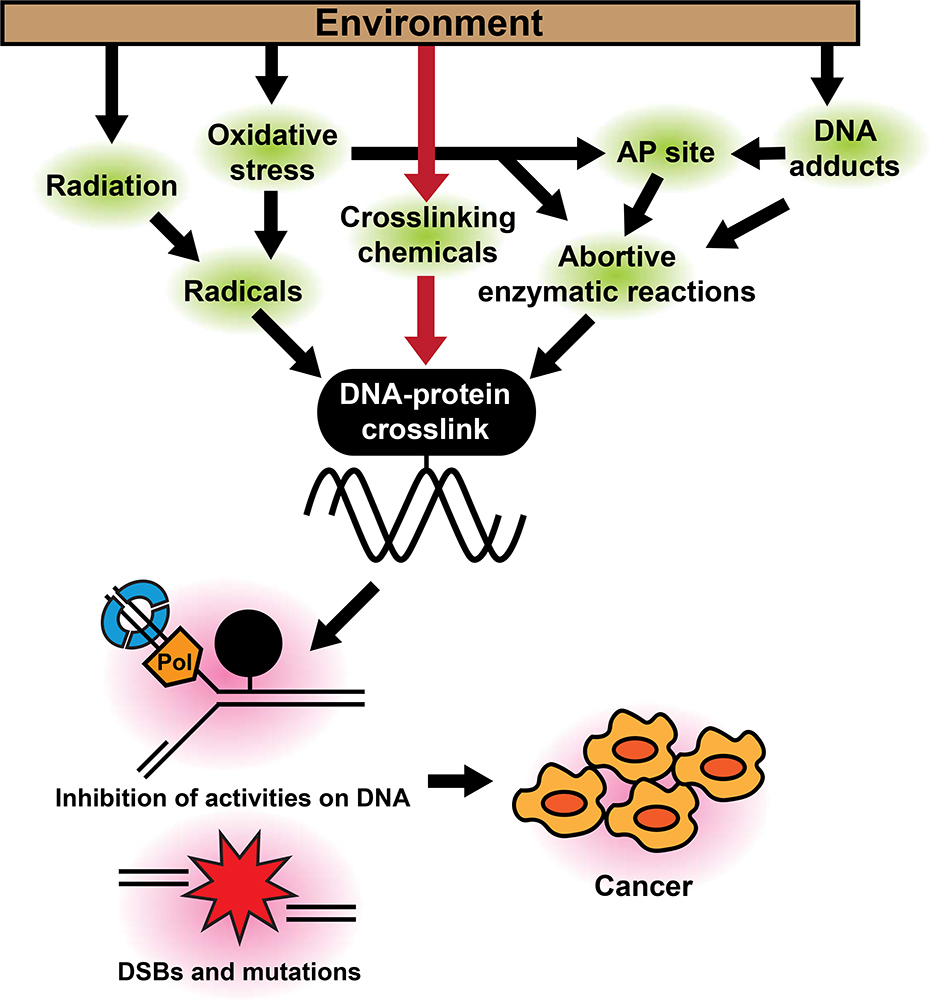
DNA-protein crosslinks (DPC) are a common type of DNA damage. During fundamental cellular activities such as DNA replication, transcription and DNA repair, numerous proteins bind to DNA. These DNA-protein interactions are mostly noncovalent and precisely regulated to prevent unnecessary binding and conflicts among DNA-binding proteins. However, because of the proximity to DNA, these DNA-binding proteins can also be crosslinked and trapped on DNA when exposed to reactive substances from the environment (Fig. 1). Compared with other DNA adducts, DPCs are extremely bulky and, therefore, difficult to repair because access of repair proteins can be hampered by steric interactions. As a consequence, burdensome DPCs persist on DNA and cause conflicts with the replication and transcription machineries (Nakano et al. 2012; Duxin et al. 2014; Ji et al. 2019; Larsen et al. 2019). Collisions of replication forks with DPCs cause fork stalling and eventually lead to double strand breaks (DSBs), which are one of the most toxic lesions for cells (Zeman and Cimprich 2014). Moreover, collisions of the transcription machinery can block transcription and cause transcriptional errors such as deletions and substitutions around DPC sites (Nakano et al. 2012). Thus, persistent DPCs impact cellular functions and challenge genomic integrity, possibly mediating carcinogenic effects of environmental carcinogens (Fig. 1).
Figure 1. Possible pathways for DPC induction by environmental substances.
Some environmental carcinogens (e.g., formaldehyde, 1,3-butadiene, and hexavalent chromium) can generate DPCs through direct crosslinking (red arrows). Others can cause DPCs via indirect mechanisms such as radical production or induction of abortive enzymatic reactions (black arrows). Persistent DPCs can inhibit important cellular activities on chromatin such as DNA replication, and induce mutations, DSBs, and genomic alterations, all of which could contribute to carcinogenesis.
In this review, we evaluate DPC formation induced by environmental substances, introduce current models of DPC repair pathways, and discuss potential contributions of DPCs to environmental risks.
I. DNA-protein-crosslinking agents in the environment
Potential carcinogenic effects of environmental agents are assessed by epidemiologic studies, bioassays using experimental animals or cells, and molecular studies. Based on these results, IARC and other organizations such as U.S. Environmental Protection Agency (EPA) categorize potential carcinogens. In the IARC categories, substances are classified from 1 to 3 (1: carcinogenic, 2A: probably carcinogenic, 2B: possibly carcinogenic, 3: not classifiable) regarding the weight of evidence for carcinogenicity in humans, i.e., how much the carcinogenicity is reliably supported by scientific data, but not the degree of toxicity (IARC 2019). In this list, there are well-known DPC-inducing substances such as formaldehyde, 1,3-butadiene and hexavalent chromium, all of which are assigned to category 1. Here, we describe these DPC-inducing substances and summarize how those were identified as carcinogens.
Formaldehyde
Formaldehyde, which is widely used in building materials such as composite wood products, glues and coatings, can be released into indoor air from these products. Formaldehyde is also produced and emitted into the air by tobacco smoking and incomplete combustion in automobile engines and stoves. In addition, workers at factories that produce formaldehyde-containing products, as well as professionals such as anatomists, pathologist and funeral industry workers who directly handle formaldehyde to preserve biological specimens, are at higher risk of occupational exposure to formaldehyde.
Carcinogenicity of formaldehyde was already suspected in 1970s because of its chemical reactivity to cause a crosslink between molecules. In addition, mutagenic effects of formaldehyde had been demonstrated in bacteria, fungi, yeasts and flies (Auerbach et al. 1977). Later, carcinogenicity of formaldehyde was shown in animal experiments in which rats exposed to high concentrations of formaldehyde by inhalation developed squamous cell carcinoma in nasal passages (Swenberg et al. 1980; Kerns et al. 1983). Increased cell proliferation following formaldehyde exposure was also reported in rat nasal epithelia in a concentration-dependent manner (Monticello et al. 1990; Monticello et al. 1996). Consistent with these animal studies, extensive epidemiologic studies revealed an increased incidence of nasopharyngeal carcinoma in formaldehyde-related workers (Hauptmann et al. 2004; Swenberg et al. 2013). In addition to cancers in nasopharyngeal region, lymphohematopoietic and brain cancers are also suspected to be caused by formaldehyde inhalation (Beane Freeman et al. 2009; Hauptmann et al. 2009).
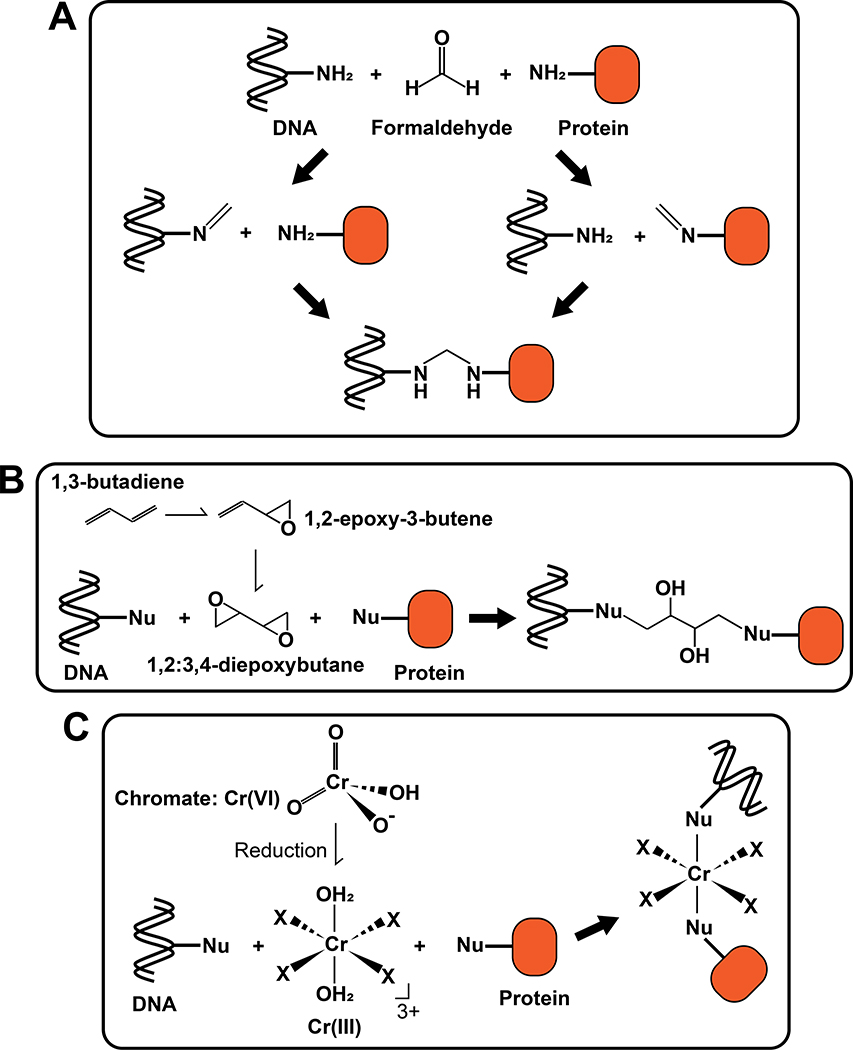
The carcinogenic effects of formaldehyde are due to its chemical reactivity with biomolecules in cells. In general, an aldehyde group can react with nucleophiles such as amine and thiol groups, generating formaldehyde adducts on DNA and protein. In addition to these simple adducts, formation of a Schiff base intermediate after the first reaction with an amine allows the second reaction with another amine (or thiol), resulting in formation of various crosslinks, including DPCs, DNA inter-strand crosslinks (ICLs) and intra-strand crosslinks (Fig. 2A) (Tretyakova et al. 2015). In the case of DPC formation, in vitro experiments showed that crosslinks are generated most frequently between lysine residues in protein and guanosine in DNA (Lu et al. 2010).
Figure 2. Formation of DPCs by environmental crosslinking substances.
(A) DPC formation induced by formaldehyde. A reaction of formaldehyde with an amine group of DNA forms a Schiff base (Left), which can react with protein to form a DPC. Alternatively, a reaction of formaldehyde with an amine group of protein forms a Schiff base (Right), which can react with DNA to form a DPC. (B) DPC formation induced by 1,3-butadiene. 1,3-butadiene is metabolized to 1,2-epoxy-3-butene and then to 1,2:3,4-diepoxybutane by cytochrome P450 (CYP450) isozymes in cells. Two epoxy groups in 1,2:3,4-diepoxybutane individually react with nucleophilic residues in DNA and protein to form a crosslink. Nu: nucleophilic residue. (C) DPC formation induced by Cr(VI). Cr(VI) is reduced to Cr(III) in cells and reactive Cr(III) generates crosslinks through reactions with nucleophilic residues on DNA and protein. Nu: nucleophilic residue. X: ligand.
Ultra-sensitive mass spectrometry has been used to quantitate DPCs induced by formaldehyde exposure in the tissues of rats and monkeys (Lai et al. 2016). A major advantage of this method is that, by using isotope-labeled formaldehyde for the exposure, DPCs generated by exogenous formaldehyde can be distinguished from those caused by endogenous formaldehyde, which can be produced as a metabolic byproduct. In this study, DPCs caused by exogenous formaldehyde were detectable in nasal epithelium but not in other distal tissues such as peripheral blood mononuclear cells, bone marrow and the liver. This is consistent with the results of animal studies, which showed nasopharyngeal carcinoma formation in exposed rats (Swenberg et al. 1980; Kerns et al. 1983).
Because formaldehyde also causes types of DNA damage other than DPCs, including ICLs and intra-strand crosslinks, it is difficult to estimate the contribution of DPCs to carcinogenicity of formaldehyde. However, one study showed that formaldehyde predominantly induced DPCs compared to ICLs using modified comet assays, which distinguished DPCs from ICLs by a proteinase K treatment (Merk and Speit 1999). This finding suggests that formaldehyde causes DPCs with high prevalence and, therefore, warrants further investigation into the role of DPCs in carcinogenic effects.
1,3-butadiene
1,3-butadiene can be produced and emitted from incomplete combustion of fuels. In addition, because 1,3-butadiene is used to manufacture synthetic rubbers, plastics and resins, workers in these industries are at higher risk of 1,3-butadiene exposure. Epidemiologic studies of U.S. workers exposed to 1,3-butadiene showed an increased incidence of lymphohematopoietic cancers, including leukemia and non-Hodgkin’s lymphoma (Acquavella and Leonard 2001). Bioassays using rats and mice with chronic 1,3-butadiene exposure demonstrated much stronger carcinogenicity than in humans. In these studies, cancers were induced in multiple sites such as blood, blood vessels, forestomach, ovary, mammary gland, liver and lung (NTP 1993). Differences in 1,3-butadiene sensitivity between humans and rodents may be explained by differences in metabolic activation of 1,3-butadiene among species and other factors such as breathing rates.
Inhaled 1,3-butadiene is metabolized by cytochrome P450 (CYP450) isozymes to 1,2-epoxy-3-butene and then to 1,2:3,4-diepoxybutane. The epoxy group reacts with nucleophilic residues such as thiols and amines. Accordingly, with two reactive moieties, 1,2:3,4-diepoxybutane can generate crosslinks, including DPCs and ICLs, in addition to DNA adducts (Fig. 2B). Mass spectrometry-based proteomics detected many crosslinked proteins on DNA, including β-actin, GAPDH, PARP1 and AGT, in cells exposed to 1,2:3,4-diepoxybutane (Michaelson-Richie et al. 2010; Gherezghiher et al. 2013). It was also shown that 1,2:3,4-diepoxybutane preferentially crosslinks the N-7 position of guanine and a cysteine or lysine side chain. Mutagenic analyses using tissue culture cells showed that diepoxide is more mutagenic than monoepoxide (Steen et al. 1997a; Steen et al. 1997b), raising the possibility that DPCs and/or DNA-DNA crosslinks underlie the higher mutagenicity of 1,2:3,4-diepoxybutane. While crosslinks induced by 1,2:3,4-diepoxybutane were shown to be predominantly ICL rather than DPCs by the modified comet assays (Wen et al. 2011), it remains unclear which lesion is responsible for the carcinogenicity of 1,2:3,4-diepoxybutane.
Hexavalent chromium: Cr(VI)
Chromium, a common metal in nature, is widely used in industries for plating, painting and alloys. As a result of these uses, workers can be exposed to chromium through inhalation. Among oxidative states of chromium (0-VI), the metallic Cr(0), trivalent Cr(III) and hexavalent states Cr(VI) are common. Cr(VI) is especially known to be water soluble. Thus, chromium is also a common contaminant in drinking water.
Epidemiologic studies of workers in chromium-related industries revealed that chromium inhalation results in an increased lung cancer risk (Mancuso 1997a; Mancuso 1997b; Proctor et al. 2014; Yatera et al. 2018). Gastrointestinal cancer risk is also increased by chromium inhalation (Welling et al. 2015). In addition, gastrointestinal cancer may be associated with chromium exposure through ingestion of chromium, although only a few studies are available and the results varied among the studies (Sun et al. 2015). In animal studies, tissue injury, inflammation and increased markers for cell proliferation and survival (Ki67 and phosphorylation of Akt) were observed in lungs of mice after inhalation exposure to chromium, supporting the increased lung cancer incidence in epidemiologic studies (Beaver et al. 2009a; Beaver et al. 2009b). For oral exposure, carcinogenic effects were observed in oral cavity of rats and small intestine of mice after 2-year exposure to sodium dichromate dihydrate, although the exposure did not affect survival (Stout et al. 2009).
Toxicity of chromium depends on its valence state, with Cr(VI) having the strongest toxic effect. Cr(VI), such as chromate (CrO42−), can be actively transported into cells by anion transporters and then reduced intracellularly to reactive Cr(III). On the other hand, extracellular Cr(III) is less toxic to cells compared to Cr(VI). This is because Cr(III) can enter cells only by passive diffusion, which is less efficient than active Cr(VI) transportation by anion transporters (Yatera et al. 2018). Therefore, extracellular reduction of Cr(VI) to Cr(III) by biologic fluids and other mechanisms is an important detoxification mechanism for Cr(VI). However, inhaled chromium-containing particles accumulate in bifurcations of the lung (Ishikawa et al. 1994), where high chromium concentrations might overwhelm local reduction capacity to reduce Cr(VI).
Cr(III) inside cells reacts with DNA and proteins to induce various types of DNA damage, including Cr(III)-DNA adducts, DPCs and ICLs (Fig. 2C) (Mattagajasingh and Misra 1996; Proctor et al. 2014; Tretyakova et al. 2015). Particularly germane to this review, Cr(III) reacts with DNA during or immediately after the reduction of Cr(VI) to Cr(III) and subsequently captures a protein to form a DPC (Macfie et al. 2010). In epidemiologic studies, detection of elevated DPC levels in lymphocytes was proposed as an indicator of chromium exposure (Zhitkovich et al. 1998). In rats, analysis of chromium-induced lesions in various organs at different times after intraperitoneal chromium injection revealed that chromium-induced DPCs persist longer than ICLs, suggesting the burdensome nature of chromium induced-DPC in vivo (Tsapakos et al. 1983).
In addition to DNA damage, epigenetic and transcriptional changes triggered by chromium-induced DPCs have also been proposed as mechanisms of carcinogenic effects (Schnekenburger et al. 2007). In particular, chromium-induced crosslinking of the HDAC1-DNMT1 complex to the promoter region of Cyp1a1 has been shown to inhibit activation of the gene. Because Cyp1a1is important for chemical detoxification in the response to benzo[a]pyrene exposure, its failed activation results in increased benzo[a]pyrene-DNA adducts in cells.
Taken together, these findings support the carcinogenicity of chromium exposure and the role of DPC formation in the process.
II. Potential DPC induction through indirect mechanisms
Environmental factors cause DPCs not only through direct crosslinking of proteins to DNA (Section I), but also through indirect mechanisms (Fig. 1). In this section, we will describe mechanisms in which DPCs can be induced indirectly by environmental carcinogens, such as generation of DNA and protein radicals and induction of abortive enzymatic reactions.
DPC formation through generation of radicals on DNA and protein
Some environmental substances can induce DPC formation indirectly via generation of radicals on DNA and/or protein. DNA and protein radicals can be produced by reactive oxygen species (ROS) and reactive nitrogen species (RNS). While these species are endogenously generated in cells through biological processes in mitochondria, peroxisomes, and the endoplasmic reticulum and other sites, many environmental factors are also known to induce these reactive species exogenously. For example, metals such as Cr, Ni, Cd, As, Pb and Be (Klein et al. 1991), quinones, (Bolton and Dunlap 2017), polycyclic aromatic hydrocarbons (PAHs) (Wilk et al. 2013), nitrogen oxides (NOx), sulfur oxides (SOx) and asbestos (Poljsak and Fink 2014) all produce ROS in cells. In highly polluted air, these environmental pollutants derived from motor and industrial exhaust, tobacco smoke and other sources are found in fine particulate matter (PM) (Adams et al. 2015; Lakey et al. 2016), inhalation of which might cause ROS/RNS generation and free radical formation on DNA and protein. Other sources of radical formation include ionizing and ultraviolet irradiation. These types of radiation generate radicals on DNA and protein by transferring its energy to a water molecule, generating hydroxyl radical (OH˙) (indirect mechanism), or directly to nucleotides (direct mechanism) (Nakano et al. 2017).
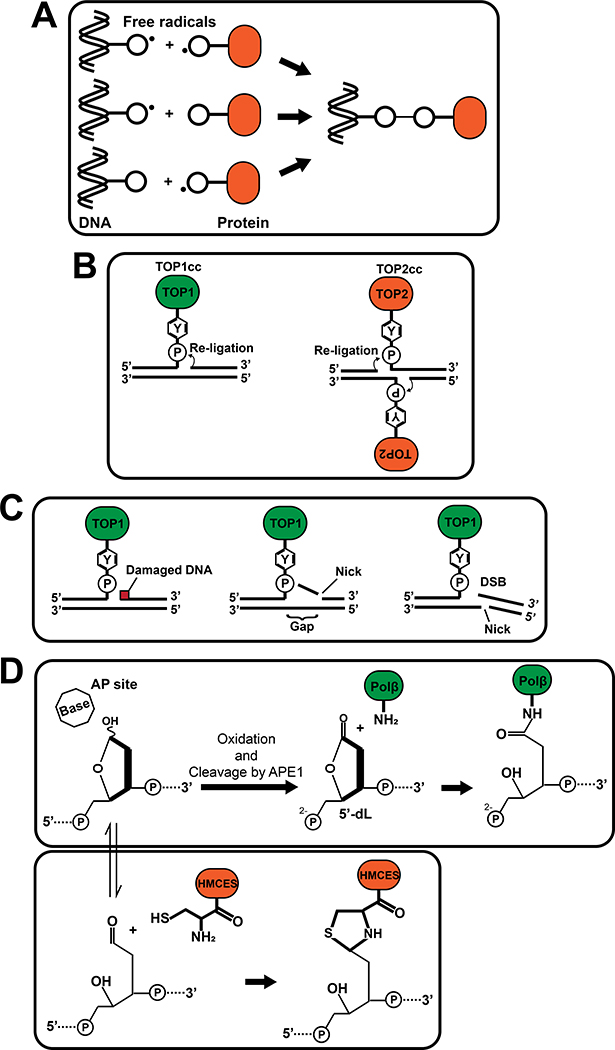
Free radicals, which have one or more unpaired electrons in a molecule, are unstable and highly reactive. When reactive radicals are generated on DNA or side chains of amino acids, those moieties react with protein and DNA, respectively, to produce DPCs (Fig. 3A) (Dizdaroglu and Gajewski 1989; Tretyakova et al. 2015) containing Thy-Lys, Thy-Tyr, Cyt-Tyr and Gua-Lys crosslinks (Cadet and Wagner 2013; Tretyakova et al. 2015; Nakano et al. 2017). Of note, oxidative DNA damages caused by ROS/RNS can further induce DPCs by distinct mechanisms involving abortive enzymatic reactions (see below). Several studies demonstrated DPC formation in cultured cells and animals after exposure to ROS/RNS-generating environmental substances, such as NiCl2 (Zhuang et al. 1994), arsenite (Ramirez et al. 2000; Bau et al. 2002), benzo[a]pyrene (a PAH) (Park et al. 2002), and SO2 (Sang et al. 2009). Taken together, DPCs can be induced by environmental pollutants through DNA and protein radical formation.
Figure 3. Indirect mechanisms of DPC formation by environmental substances.
(A) DPC formations through free radicals on DNA and/or proteins. (B) Structures of TOP1cc (left) and TOP2cc (right). A covalent bond between the topoisomerase active site tyrosine and the phosphate in DNA is formed at 3’-end (TOP1cc) or 5’-end (TOP2cc) of DNA. (C) Examples of stable TOP1cc formation through DNA damage caused by environmental carcinogens. DNA damage, such as base damages and an AP site, near TOP1-mediated incision inhibit re-ligation of TOP1 (left). A SSB downstream of the TOP1-mediated incision in the same strand (middle) or in the opposite strand (right) also prevents re-ligation by creating a gap or DSB, respectively. (D) DPC formation at AP sites. 5’-dL (5’-deoxyribonolactone) is generated when an AP site is spontaneously oxidized and cleaved by APE1, and Polβ is covalently trapped through reaction with the 5’-dL (Top). HMCES reacts with the open-ring form of an AP site to form a DPC (Bottom). Circled “P”: phosphate. “Y” in hexagon: tyrosine side chain.
DPC formation through abortive enzymatic reactions of topoisomerases
DNA damage caused by environmental factors can indirectly cause DPC formation by covalent trapping of DNA processing enzymes such as topoisomerase I (TOP1) and topoisomerase II (TOP2) on DNA. TOP1 creates a nick and releases torsional stress of DNA while a TOP2 homodimer generates a double strand break and resolves DNA catenanes (Pommier et al. 2016). To cleave a DNA phosphodiester bond, DNA topoisomerases catalyze a transesterification reaction through their active site tyrosine, which results in formation of a covalent bond with DNA (TOP1: at the 3’-end; TOP2: at the 5’-end, Fig. 3B). These covalent bonds normally occur transiently and are reversed when the enzymes re-ligate DNA. However, failures in the re-ligation step result in covalent trapping of these enzymes on DNA in a reaction intermediate state called a TOP1 or TOP2 cleavage complex (TOP1cc or TOP2cc, respectively). Importantly, DNA damage flanking the DNA incision inhibits re-ligation, resulting in formation of stable TOP1ccs and TOP2ccs. For example, in the case of TOP1, re-ligation can be blocked by base oxidation, base alkylation, an apurinic/aprymidinic (AP) site, or a DNA adduct caused by PAHs, mostly when they are at −1 to +2 position from the DNA incision. (Pourquier and Pommier 2001; Pommier et al. 2002; Pommier et al. 2016) (Fig. 3C). In addition, TOP1ccs are stabilized by nearby single strand breaks (SSBs), which are a common DNA lesion formed during base excision repair (BER, see below) of damages generated by environmental substances, or by ROS reacting with DNA backbone sugars. These SSBs generate DNA gaps or DSBs and prevent re-ligation (Fig. 3C) (Pourquier and Pommier 2001; Pommier et al. 2016). Therefore, environmental substances that induce these types of DNA damage could indirectly cause DPCs as a consequence of abortive enzymatic reactions of topoisomerases.
DPC formation through generation of AP sites
When environmental carcinogens induce DNA base damage such as oxidation and alkylation, AP sites can be generated through the action of DNA glycosylases, which remove damaged bases during the process of BER (Jacobs and Schar 2012). In BER, the AP endonuclease APE1 creates a DNA nick at the 5’-end of an AP site, resulting in formation of 5’-deoxyribose phosphate (5’-dRp). In the case of short-patch BER pathway, Polβ performs 1-bp displacing DNA synthesis from the nick and removes the excluded 5’-dRp through its 5’-dRp lyase activity, which involves β-elimination. However, spontaneous oxidization of an AP site inhibits the β-elimination reaction by Polβ, resulting in covalent trapping of Polβ (Fig. 3D) (DeMott et al. 2002; Quinones et al. 2015; Quinones and Demple 2016). Another enzyme, poly(ADP-ribose) polymerase 1 (PARP1), displays AP lyase activity at AP sites and can be trapped at 3’-end of AP site (Prasad et al. 2014). In addition, a recent study demonstrated that a protein called HMCES (5-Hydroxymethylcytosine Binding, ES Cell Specific) forms a DPC at exposed AP sites in single-stranded DNA to shield the sites from error-prone polymerases and endonucleases (Mohni et al. 2019). In this reaction, HMCES forms a covalent bond with an AP site through the α-amino group of the N-terminal cysteine, leading to formation of a stable thiazolidine DPC (Fig. 3D) (Halabelian et al. 2019; Thompson et al. 2019). In summary, environmental factors that cause base damage could indirectly induce DPCs through conversion of damaged nucleotides to AP sites and subsequent formation of DNA-protein crosslinks.
III. Mechanisms of DPC Repair
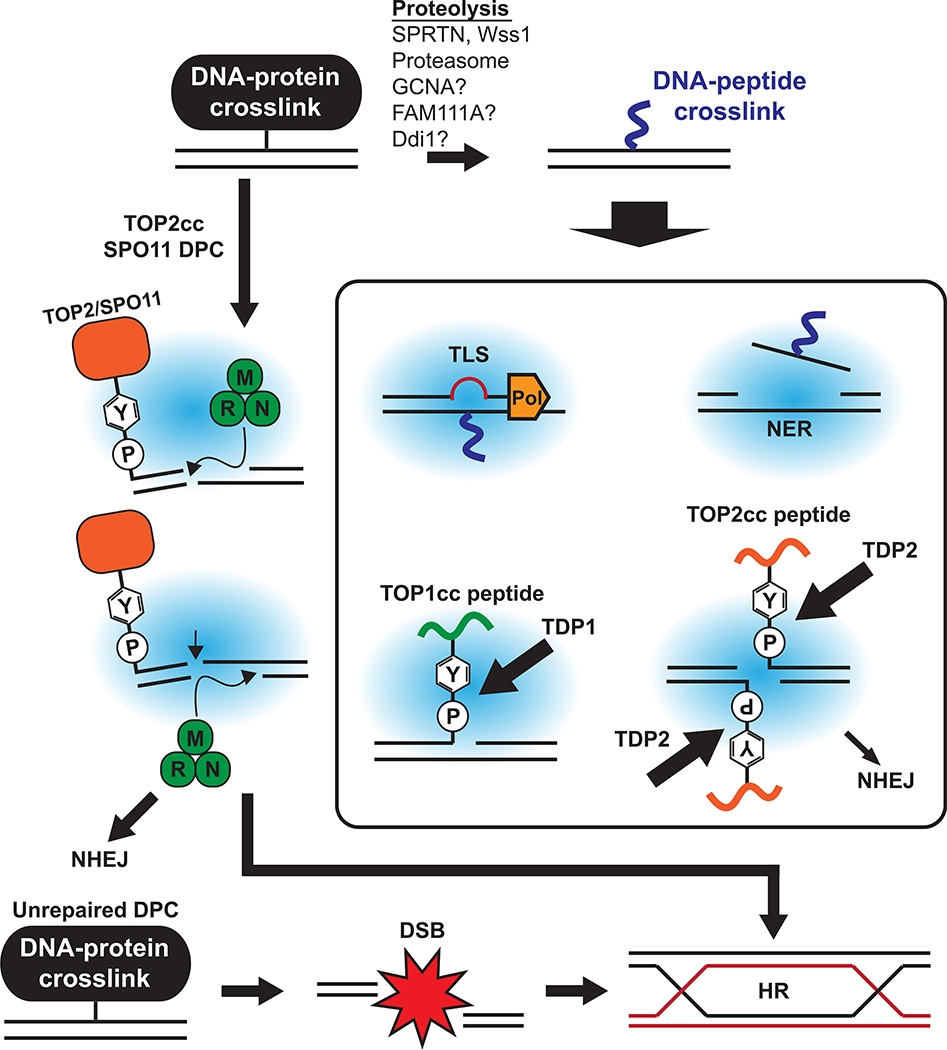
Because of the diversity of DPCs in structure, size and context, cells employ a variety of repair mechanisms to resolve different DPCs. DPCs can be classified based on the structures of their DNA components. The most common type of DPC is a protein crosslinked to an intact DNA backbone. DPCs caused by crosslinking chemicals and radicals belong to this type. DPCs at a DNA nick can be divided into two types based on the position of the attached protein. Polβ DPCs and TOP2ccs (if one TOP2 subunit is trapped) are formed at the 5’-end of a DNA nick while TOP1ccs and PARP1 DPCs are attached at the 3’-end of a DNA nick. TOP2ccs can be stabilized at the 5’-end of DSBs if both of TOP2 subunits are trapped. In addition to these structural differences, other conditions such as cell-cycle phases and other processes such as DNA replication significantly affect the choice of DPC repair pathways. Here, we describe current knowledge of the mechanisms by which DPCs are processed and repaired depending on the situation (Fig. 4).
Figure 4. Crosstalk of DNA repair pathways for DPC resolution.
DPC proteins can be degraded via replication-dependent or -independent proteolysis to a peptide level. The resulting DNA-peptide crosslinks are bypassed by TLS during DNA replication and repaired later by NER. Phosphotyrosyl bonds of TOP1cc and TOP2cc are cleaved by TDP1 and TDP2, respectively. DPCs at 5’-end of DSBs can be removed together with a flanking DNA fragment by nuclease activity of the MRN complex. HR is required to repair DSBs that are generated as a DPC repair intermediate or those that result from replication collisions with DPCs. NHEJ is also employed during TOP2cc processing.
Degradation of DPC proteins by proteases
DPCs, which are generally bulky, limit the access of repair proteins due to steric hindrance. To solve this problem, cells employ various proteases to reduce the size of the protein component of DPCs, facilitate the downstream repair processes, and allow other cellular processes such as DNA replication to proceed.
(1). Replication-coupled proteolysis by SPRTN
A recent breakthrough in the DPC repair field is the discovery of the DNA-dependent metalloproteases Wss1 in yeasts and its counterpart SPRTN in higher eukaryotes (Stingele et al. 2014; Lopez-Mosqueda et al. 2016; Stingele et al. 2016; Vaz et al. 2016; Maskey et al. 2017; Morocz et al. 2017). SPRTN contains a metalloprotease domain SprT at its N-terminus and degrades DPC proteins in a replication-coupled manner to mitigate the effect of DPCs on DNA replication forks. SPRTN depletion impairs removal of formaldehyde-induced DPCs and etoposide-induced TOP2ccs (Lopez-Mosqueda et al. 2016; Stingele et al. 2016; Morocz et al. 2017; Hu et al. 2020). In addition, SPRTN depletion in human and mouse cells causes elevation of total levels of DPCs and accumulation of spontaneously stabilized TOP1ccs (Vaz et al. 2016; Maskey et al. 2017). Accordingly, cells lacking SPRTN are more sensitive to DPC-inducing chemicals such as formaldehyde as well as to camptothecin and etoposide, which stabilize TOP1ccs and TOP2ccs, respectively (Davis et al. 2012; Stingele et al. 2016; Vaz et al. 2016; Maskey et al. 2017; Morocz et al. 2017). These findings suggest that SPRTN plays a major role in the repair of DPCs produced endogenously and exogenously.
SPRTN appears to be particularly important for removing DPCs during replication. SPRTN levels are high in S and G2 phases (Mosbech et al. 2012). Moreover, SPRTN-mediated DPC repair requires S phase entry (Vaz et al. 2016). In agreement with these findings, cell-free DNA replication experiments using Xenopus egg extracts demonstrated that SPRTN promotes degradation of DPCs at DNA replication forks (Larsen et al. 2019).
SPRTN is regulated at several levels. The first known regulatory step is its chromatin recruitment, which is mediated by SPRTN mono-ubiquitination and is dependent on the ubiquitin-binding UBZ4 domain. Roughly a half of SPRTN is present in cells as a monoubiquitinated form, but chromatin-loaded SPRTN is enriched in the non-ubiquitinated form (Stingele et al. 2016). Thus, it appears that mono-ubiquitination blocks chromatin association of SPRTN. Formaldehyde treatment, but not other types of replication stress, leads to a reduction in monoubiquitinated SPRTN levels concomitant with an increase in SPRTN loading on chromatin. Thus, DPCs might signal for SPRTN deubiquitination, possibly through replication fork collisions, to increase SPRTN chromatin loading. However, it is currently unclear whether the chromatin loading of SPRTN represents its recruitment to replication forks stalled at DPCs. Indeed, how SPRTN is recruited to replication forks stalled at DPCs is not fully understood. The C-terminal region of SPRTN contains a PCNA interacting peptide (PIP) box and a UBZ4 domain; however, their requirement for SPRTN recruitment to replication forks is not clear (Maskey et al. 2017; Morocz et al. 2017; Larsen et al. 2019).
Another level of SPRTN regulation is at the level of enzyme activity. SPRTN is a DNA-dependent metalloprotease that contains multiple DNA binding domains (Stingele et al. 2016; Vaz et al. 2016; Morocz et al. 2017; Toth et al. 2017; Li et al. 2019). In biochemical experiments, double-strand DNA (dsDNA) partially activates SPRTN, causing degradation of only SPRTN itself, while single-strand DNA (ssDNA) can fully activate SPRTN and stimulate degradation of other proteins on DNA (Lopez-Mosqueda et al. 2016; Stingele et al. 2016; Vaz et al. 2016; Morocz et al. 2017). In vitro studies using a cell-free replication system suggest that ssDNA is generated when a replicative helicase bypasses a DPC and this ssDNA exposure is required for SPRTN-mediated DPC degradation (Larsen et al. 2019; Sparks et al. 2019). This ssDNA exposure is assisted by an accessory DNA helicase, RTEL1, which unwinds dsDNA beyond the DPC. Thus, ssDNA exposure at DPCs might be a key signal for enzymatic activation of SPRTN.
SPRTN has been shown to cleave only unstructured protein region in vitro (Vaz et al. 2016). Recent structural study of the SPRTN SprT protease domain further supported this notion by revealing its narrow substrate-binding groove (Li et al. 2019), suggesting that unfolding of substrates might be required for efficient degradation. Indeed, it was demonstrated that the p97 ATPase, an interactor of SPRTN that possesses protein unfolding activity, is important for SPRTN to process TOP1cc. In the case of TOP1cc degradation, p97 recruitment to TOP1ccs is assisted by a p97 co-factor, TEX264, which bridges SUMO-1-modified TOP1ccs and p97 (Fielden et al. 2020).
From these findings, an emerging model for SPRTN-mediated DPC repair is as follows: (i) the replication machinery encounters a DPC and ssDNA is exposed around DPCs, (ii) a possible deubiquitination step stimulates SPRTN-loading on chromatin, (iii) exposed ssDNA around DPCs fully activates the protease domain of SPRTN to degrade DPC proteins with possible involvement of cofactors, and (iv) SPRTN autocleavage potentially terminates its function.
(2). DPC proteolysis by the ubiquitin-proteasome pathway
The ubiquitin-proteasome system, which plays a central role in destruction of unfolded proteins and proteins with short half-lives, is also involved in degradation of DPC proteins. Many known DPCs such as TOP1ccs, TOP2ccs as well as DPCs containing Polβ, PARP1 or HMCES have been reported to be degraded by the proteasome (Desai et al. 1997; Mao et al. 2001; Quinones et al. 2015; Mohni et al. 2019; Prasad et al. 2019). DPCs induced by environmental carcinogens such as formaldehyde and Cr(VI) have also been shown to be degraded by a proteasome-dependent mechanism (Quievryn and Zhitkovich 2000; Zecevic et al. 2010). A recent in vitro study using Xenopus egg extracts revealed a detailed mechanism of poly-ubiquitination and proteasome-dependent degradation of DPC proteins (Larsen et al. 2019). Collision of a replisome with a DPC triggers stepwise poly-ubiquitination of the DPC protein, first by the ubiquitin E3 ligase TRAIP, which promotes helicase bypass of the DPC and subsequent ssDNA exposure, and then by a second currently unknown E3 ligase, which is thought to facilitate proteasomal degradation of DPC proteins. Through these mechanisms, the ubiquitin-proteasome pathway degrades DPC proteins independently of SPRTN but in a replication-coupled manner.
SUMOylation of DPCs is another mechanism of tagging DPCs for proteasomal degradation. In a Xenopus cell free system, replication-independent SUMOylation of DPC proteins has been reported (Larsen et al. 2019). In human cells, formaldehyde exposure induces global SUMOylation of chromatin proteins (Borgermann et al. 2019). In a seemingly analogous manner, treatment with 5-aza-dC, which induces DPCs containing the DNA methyltransferase DNMT1, results in SUMOylation of DNMT1 in a replication-independent manner, and this SUMOylation is required for subsequent ubiquitination and proteasomal degradation of DNMT1 DPCs. A similar role of DPC SUMOylation in inducing proteasomal degradation has also been reported with TOP1ccs and TOP2ccs (Sun et al. 2019). In particular, SUMOylation of topoisomerase DPCs by the SUMO ligase PIAS4 and subsequent poly-ubiquitination by the SUMO-targeted ubiquitin ligase RNF4 induces proteasomal degradation of TOP1ccs and TOP2ccs. Although it remains to be seen whether the SUMO-ubiquitin cascade applies to other DPCs, DPC SUMOylation is emerging as a mechanism for initiating replication-independent degradation of DPCs by the proteasome.
(3). Other potential proteases for DPC repair
GCNA (also known as ACRC) is a germline- and stem cell-specific putative metalloprotease in the SprT family, the same protein family that contains SPRTN. GCNA has recently been implicated in resolving DPCs in germ cells and early embryos (Bhargava et al. 2019; Borgermann et al. 2019; Dokshin et al. 2019). These studies suggest that GCNA works in G2/M phases and mainly targets TOP2ccs. GCNA is targeted to SUMOylated DPC proteins through its SUMO-interacting motifs (SIMs) (Borgermann et al. 2019), reinforcing the importance of SUMOylation in DPC repair.
Non-SprT proteases are also reported to be involved in DPC repair. FAM111A, a chymotrypsin-like serine protease that localizes at replication forks through PCNA, protects replication forks from DPCs via its protease activity (Kojima et al. 2020). Knockout of FAM111A sensitizes cells to various DPC-inducing agents including formaldehyde, TOP1 and TOP2 inhibitors. Spontaneous accumulation of TOP1cc was also observed in FAM111A knockout cells, suggesting non-redundant roles of FAM111A and SPRTN in DPC repair. FAM111A contains a ssDNA-binding domain, and a point mutation that disrupts ssDNA binding diminishes FAM111A autocleavage activity in vivo. These findings suggest that FAM111A might undergo DNA-dependent protease activation, an aspect that might be shared with SPRTN regulation.
A putative aspartic protease, Ddi1, was also recently implicated in DPC repair in yeasts (Serbyn et al. 2019). This study showed that a deletion of ddi1 sensitized cells to DPC-inducing agents, including formaldehyde in the absence of wss1. Replication-coupled Ddi1 recruitment to TOP1ccs was also shown in the absence of Wss1 and Tdp1 (see below). These findings suggest redundant roles of Wss1 and Ddi1 in DPC repair in yeasts. Given that the ddi1 gene is conserved in humans (DDI1 and DDI2), it is possible that the human homologs, DDI1 and DDI2, play a similar role. The presence of multiple DPC proteases underscores the importance of DPC repair as well as the diverse cellular contexts in which cells need to deal with these toxic DNA lesions.
Hydrolysis of phosphotyrosyl bonds by TDP1 and TDP2
Topoisomerases are trapped on DNA through covalent bonds between the active site tyrosines of the enzymes and phosphate groups of the cleaved DNA backbone. Tyrosyl-DNA phosphodiesterases are enzymes that directly hydrolyze these phosphotyrosyl bonds (TDP1 for TOP1ccs and TDP2 for TOP2ccs) (Yang et al. 1996; Pouliot et al. 1999; Cortes Ledesma et al. 2009). Because intact DPCs potentially hinder the access of TDP enzymes, crosslinked TOP1 and TOP2 proteins might be proteolyzed to peptides, presumably by the proteasome or SPRTN, before being processed by TDP enzymes (Debethune et al. 2002; Interthal and Champoux 2011; Gao et al. 2014). In the case of TOP1ccs, cleavage of the phosphotyrosyl bond by TDP1 leaves a DNA nick with 3’-phosphate and 5’-OH, which is converted to a ligatable nick with 3’-OH and 5’-phosphate by bifunctional polynucleotide kinase 3’-phosphatase (PNKP) before ligation (El-Khamisy 2011). Similarly, a TOP2cc-containing DNA end becomes, after hydrolysis by TDP2, a ligatable single strand break (if one TOP2 subunit is trapped) or a ligatable DSB, which is preferentially repaired by non-homologous end joining (NHEJ) in an error-free manner (Gomez-Herreros et al. 2013). Interestingly, a recent study demonstrated that the SUMO ligase ZATT promotes proteasome-independent resolution of TOP2ccs by enhancing TDP2 recruitment through SUMOylation of TOP2ccs and by inducing a conformational change of TOP2ccs through a direct interaction (Schellenberg et al. 2017). Because TOP1 and TOP2 are abundant enzymes, these specialized repair mechanisms are important to protect the genome form TOP1ccs and TOP2ccs.
Translesion synthesis (TLS) over DNA-peptide crosslinks (DpCs)
Even if DPC proteins are degraded to small peptides by proteolysis, replicative polymerases are not able to replicate over DNA-peptide crosslink (DpC) lesions because the high fidelity DNA polymerases cannot insert nucleotides across the peptide adducts. Under these circumstances, cells employ TLS polymerases, which have larger active sites that can accommodate lesions, to bypass these adducts (Sale et al. 2012; Vaisman and Woodgate 2017). Y-family polymerases (Polη, Polι, Polκ and Rev1) insert a nucleotide across the lesion, and the B-family polymerase Polζ performs the extension from the unpaired DNA terminus. Switching to Y-family polymerases is promoted by RAD6/RAD18-mediated mono-ubiquitination of PCNA at K164, which facilitates recruitment of Y-family polymerases through interactions with their ubiquitin-binding domains. After lesion bypass is complete, a replicative polymerase continues DNA synthesis. DPC protein degradation followed by TLS over DpC has been demonstrated in an in vitro study using Xenopus egg extracts (Duxin et al. 2014). The remaining DpCs can be repaired by the nucleotide excision repair (NER) mechanism (see below).
While TLS promotes uninterrupted DNA replication at a DpC, it is an error prone process because TLS polymerases do not always insert the correct nucleotide across peptide adducts. Indeed, TLS over DpCs was reported to be mutagenic (Minko et al. 2008; Pande et al. 2017) although the mutagenic potential seems to vary depending on the site of the DpC. Interestingly, SPRTN was shown to have a function in the regulation of TLS in part through recruitment of the p97 segregase to evict TLS polymerases (Centore et al. 2012; Ghosal et al. 2012; Juhasz et al. 2012; Machida et al. 2012; Mosbech et al. 2012; Kim et al. 2013). This may imply that SPRTN coordinates proteolysis of DPC proteins and subsequent TLS over DpC lesions to minimize mutations.
Nucleotide excision repair (NER)
Nucleotide excision repair (NER) removes damaged nucleotides by “cut-and-patch” mechanism (Marteijn et al. 2014). This pathway starts with recognition of DNA helix distortions by the XPC-RAD23B-CETN2 complex followed by recruitment of the transcription initiation factor IIH (TFIIH) complex. Subsequently, helicase activity of TFIIH unwinds the damaged region, and the strand is excised by the XPF-ERCC1 complex and XPG at 5’- and 3’-ends, respectively. As a consequence, a 22–30 bp gap is generated, which will be filled by DNA polymerases. In general, intact DPCs are too bulky to be repaired by NER (Reardon and Sancar 2006). Indeed, NER-deficient cells displayed accumulation of DpCs with a peptide less than 7.4 kDa following exposure to formaldehyde, suggesting that NER is responsible for removal of DpCs containing a short peptide (Nakano et al. 2007; Nakano et al. 2009). Furthermore, in vitro incision assays using cell extracts also showed consistent results, in which DNA with a DPC was incised by the NER pathway only when the crosslinked polypeptides were less than 6.8 kDa. These findings indicate that preprocessing of DPC proteins by proteolysis (presumably by SPRTN and the proteasome) might be necessary before NER.
Excision of terminal DPCs by MRE11-RAD50-NBS1 (MRN)
The MRE11-RAD50-NBS1 (MRN) complex is a multifunctional nuclease complex involved in multiple aspects of the DNA damage response (Syed and Tainer 2018). When DPCs are present at the 5’-end of DSBs as is the case for TOP2ccs, those DPCs can be removed together with a flanking DNA fragment by nuclease activity of the MRN complex (Aparicio et al. 2016; Deshpande et al. 2016; Hoa et al. 2016). In the presence of DPCs at the 5’-end of DSB, MRN creates a nick at 15–20 bp away from the DPC by its endonuclease activity and then utilizes its 3’−5’ exonuclease activity to digest a DNA strand from the nick, which generates an exposed ssDNA region. The last step is another endonucleolytic cleavage of DNA opposite the initial nick so that the attached protein can be released together with the franking DNA fragment. In addition to TOP2ccs, a meiotic recombination protein SPO11 (subunit A of topoisomerase VI), which has similar enzymatic activity to TOP2, is also repaired by this mechanism (Neale et al. 2005; Milman et al. 2009). After removal of DPCs, the DSB ends enable subsequent repair by NHEJ or homologous recombination (HR) (Fig. 4).
IV. Crosstalk among DNA repair pathways
An emerging theme is that multiple repair pathways are involved in DPC repair and often process DPCs step by step. For example, when a replication fork encounters a DPC, replication-coupled proteolysis pathways involving SPRTN or the proteasome degrade the DPC protein. Subsequently, the DpC is bypassed by TLS to allow DNA replication to proceed, and the bypassed DpCs are eventually repaired by NER.
In addition to DPC repair pathways, HR, a DSB repair mechanism, is also important for cell survival after exposure to DPC-inducing agents such as formaldehyde (Nakano et al. 2007; Ridpath et al. 2007; de Graaf et al. 2009; Nakano et al. 2009), chromium (Bryant et al. 2006), and TOP1/2 poisons (Maede et al. 2014). It is possible that HR is crucial after these exposures because collisions of the DNA replication machinery with DPCs and other DNA crosslinks generate DSBs. In addition, DPC repair processes produce DSBs as an intermediate, which requires HR to complete the repair. Another DSB repair mechanism, NHEJ, might also be employed in the case of TOP2cc repair to deal with DSBs generated after hydrolysis of phosphotyrosyl bonds by TDP2 or after DNA end resection by MRN (see above) (Gomez-Herreros et al. 2013; Hoa et al. 2016). Consistent with these notions, genetic studies showed that knockout of NHEJ components such as KU70 sensitized cells to TOP2 inhibitors, but not to TOP1 inhibitors (Maede et al. 2014).
Another mechanism that might be important for the cellular response to DPC lesions is the replication stress response pathway, which stabilizes and resolves stalled replication forks. This pathway is important because DPCs cause conflicts with replication forks, and persistent fork stalling and fork collapse are detrimental to cells. Many HR components such as BRCA1/2, RAD51 and RAD51 paralogs (RAD51C, XRCC2 and XRCC3), are also involved in this pathway (Liao et al. 2018). In addition, genetic screening identified components of the Fanconi anemia pathway to be important for protecting cells from formaldehyde and TOP1/2 poisons (Ridpath et al. 2007; Maede et al. 2014). Given that FANCD2 protects stalled forks from nuclease-mediated degradation (Schlacher et al. 2012), this pathway may contribute to fork protection when replication forks collide with DPCs. Consistent with this possibility, an in vitro study using Xenopus egg extracts showed that FANCI-FANCD2, a key component in Fanconi anemia pathway, did not directly participate in DPC repair (Duxin et al. 2014). Taken together, in addition to the mechanisms that directly process DPC lesions, HR, NHEJ and replication stress response pathways also influence the response to DPC lesions and all of these pathways contribute to cell survival (Fig. 4).
V. Conclusions and perspectives
Recent studies have revealed that cells are equipped with various mechanisms to deal with endogenously produced DPCs, and that defects in DPC repair have severe consequences. Individuals with the Ruijs-Aalfs syndrome, a human genetic disease caused by mutations in SPRTN gene, exhibit genomic instability, premature aging and early onset of hepatocellular carcinomas (Ruijs et al. 2003; Lessel et al. 2014). Similar phenotypes were also seen in a mouse model carrying Sprtn hypomorphic alleles, accompanied by TOP1cc accumulation and tumor formation in the liver (Maskey et al. 2014; Maskey et al. 2017). These facts highlight the negative effects of DPCs and their association with carcinogenesis. However, cells deal with high loads of endogenous DPCs by deploying amazing repair capacities as demonstrated by extensive DPC accumulation in SPRTN deficient cells and mice (Vaz et al. 2016; Maskey et al. 2017). This high DPC repair capacity, however, might obscure biological assessment of DPC-inducing agents, potentially leading to underestimation of abilities of environmental substances to induce DPC adducts. Moreover, the capacity of repair pathways for certain types of DPCs might vary among cell types. For example, formaldehyde-induced DPCs are repaired much more slowly in human peripheral blood lymphocytes than other cell types (Quievryn and Zhitkovich 2000). Therefore, it will be important to use relevant cell types and models in assessing DPC induction by environmental factors and also in studying repair mechanisms. Currently, DPC repair mechanisms are often studied using model substrates and treatments with high doses of DPC-inducing drugs. However, environmentally induced DPCs might be different from endogenously generated DPCs in their types of crosslinks and in their difficulty of repair. Thus, it will be important to investigate whether and how cells can resolve environmentally induced DPCs through the repair mechanisms that are designed for repairing endogenous DPCs.
Acknowledgements
We thank Scott H. Kaufmann for critical reading of the manuscript. This work was supported by the National Institutes of Health (R01 CA233700 to Y. J. M.).
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- Acquavella JF, Leonard RC. 2001. A review of the epidemiology of 1,3-butadiene and chloroprene. Chem Biol Interact 135–136:43–52. [DOI] [PubMed] [Google Scholar]
- Adams K, Greenbaum DS, Shaikh R, van Erp AM, Russell AG. 2015. Particulate matter components, sources, and health: Systematic approaches to testing effects. J Air Waste Manag Assoc 65:544–558. [DOI] [PubMed] [Google Scholar]
- Aparicio T, Baer R, Gottesman M, Gautier J. 2016. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J Cell Biol 212:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach C, Moutschen-Dahmen M, Moutschen J. 1977. Genetic and cytogenetical effects of formaldehyde and related compounds. Mutat Res 39:317–361. [DOI] [PubMed] [Google Scholar]
- Bau DT, Wang TS, Chung CH, Wang AS, Wang AS, Jan KY. 2002. Oxidative DNA adducts and DNA-protein cross-links are the major DNA lesions induced by arsenite. Environ Health Perspect 110 Suppl 5:753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane Freeman LE, Blair A, Lubin JH, Stewart PA, Hayes RB, Hoover RN, Hauptmann M. 2009. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute Cohort. J Natl Cancer Inst 101:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver LM, Stemmy EJ, Constant SL, Schwartz A, Little LG, Gigley JP, Chun G, Sugden KD, Ceryak SM, Patierno SR. 2009a. Lung injury, inflammation and Akt signaling following inhalation of particulate hexavalent chromium. Toxicol Appl Pharmacol 235:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver LM, Stemmy EJ, Schwartz AM, Damsker JM, Constant SL, Ceryak SM, Patierno SR. 2009b. Lung inflammation, injury, and proliferative response after repetitive particulate hexavalent chromium exposure. Environ Health Perspect 117:1896–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava V, Goldstein CD, Russell L, Xu L, Ahmed M, Li W, Casey A, Servage K, Kollipara R, Picciarelli Z, Kittler R, Yatsenko A, Carmell M, Orth K, Amatruda JF, Yanowitz JL, Buszczak M. 2019. GCNA Preserves Genome Integrity and Fertility Across Species. Dev Cell 52:38–52 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Dunlap T. 2017. Formation and Biological Targets of Quinones: Cytotoxic versus Cytoprotective Effects. Chem Res Toxicol 30:13–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgermann N, Ackermann L, Schwertman P, Hendriks IA, Thijssen K, Liu JC, Lans H, Nielsen ML, Mailand N. 2019. SUMOylation promotes protective responses to DNA-protein crosslinks. EMBO J 38:e101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Ying S, Helleday T. 2006. Homologous recombination is involved in repair of chromium-induced DNA damage in mammalian cells. Mutat Res 599:116–123. [DOI] [PubMed] [Google Scholar]
- Cadet J, Wagner JR. 2013. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol 5:a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Yazinski SA, Tse A, Zou L. 2012. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell 46:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. 2009. A human 5’-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461:674–678. [DOI] [PubMed] [Google Scholar]
- Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. 2012. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol 19:1093–1100. [DOI] [PubMed] [Google Scholar]
- de Graaf B, Clore A, McCullough AK. 2009. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst) 8:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debethune L, Kohlhagen G, Grandas A, Pommier Y. 2002. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res 30:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. 2002. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J Biol Chem 277:7637–7640. [DOI] [PubMed] [Google Scholar]
- Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. 1997. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem 272:24159–24164. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Lee JH, Arora S, Paull TT. 2016. Nbs1 Converts the Human Mre11/Rad50 Nuclease Complex into an Endo/Exonuclease Machine Specific for Protein-DNA Adducts. Mol Cell 64:593–606. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M, Gajewski E. 1989. Structure and mechanism of hydroxyl radical-induced formation of a DNA-protein cross-link involving thymine and lysine in nucleohistone. Cancer Res 49:3463–3467. [PubMed] [Google Scholar]
- Dokshin GA, Davis GM, Sawle AD, Eldridge MD, Nicholls PK, Gourley TE, Romer KA, Molesworth LW, Tatnell HR, Ozturk AR, de Rooij DG, Hannon GJ, Page DC, Mello CC, Carmell MA. 2019. GCNA Interacts with Spartan and Topoisomerase II to Regulate Genome Stability. Dev Cell 52:53–68 e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, Walter JC. 2014. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF. 2011. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol Med 3:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, Bray F. 2019. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144:1941–1953. [DOI] [PubMed] [Google Scholar]
- Fielden J, Wiseman K, Torrecilla I, Li S, Hume S, Chiang SC, Ruggiano A, Narayan Singh A, Freire R, Hassanieh S, Domingo E, Vendrell I, Fischer R, Kessler BM, Maughan TS, El-Khamisy SF, Ramadan K. 2020. TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts. Nat Commun 11:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, Pommier Y. 2014. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2). J Biol Chem 289:17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherezghiher TB, Ming X, Villalta PW, Campbell C, Tretyakova NY. 2013. 1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res 12:2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. 2012. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J Biol Chem 287:34225–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilon A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, Cortes-Ledesma F. 2013. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet 9:e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halabelian L, Ravichandran M, Li Y, Zeng H, Rao A, Aravind L, Arrowsmith CH. 2019. Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition. Nat Struct Mol Biol 26:607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann M, Lubin JH, Stewart PA, Hayes RB, Blair A. 2004. Mortality from solid cancers among workers in formaldehyde industries. Am J Epidemiol 159:1117–1130. [DOI] [PubMed] [Google Scholar]
- Hauptmann M, Stewart PA, Lubin JH, Beane Freeman LE, Hornung RW, Herrick RF, Hoover RN, Fraumeni JF Jr., Blair A, Hayes RB. 2009. Mortality from lymphohematopoietic malignancies and brain cancer among embalmers exposed to formaldehyde. J Natl Cancer Inst 101:1696–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsutsui K, Takeda S, Sasanuma H. 2016. Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes . Mol Cell 64:580–592. [DOI] [PubMed] [Google Scholar]
- Hu Q, Klages-Mundt N, Wang R, Lynn E, Kuma Saha L, Zhang H, Srivastava M, Shen X, Tian Y, Kim H, Ye Y, Paull T, Takeda S, Chen J, Li L. 2020. The ARK Assay Is a Sensitive and Versatile Method for the Global Detection of DNA-Protein Crosslinks. Cell Rep 30:1235–1245 e1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. 2019. Agents Classified by the IARC Monographs, Volume 1–125 https://monographs.iarc.fr/agents-classified-by-the-iarc/. Accessed 3 February 2020. [Google Scholar]
- Interthal H, Champoux JJ. 2011. Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1. Biochem J 436:559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Nakagawa K, Satoh Y, Kitagawa T, Sugano H, Hirano T, Tsuchiya E. 1994. “Hot spots” of chromium accumulation at bifurcations of chromate workers’ bronchi. Cancer Res 54:2342–2346. [PubMed] [Google Scholar]
- Jacobs AL, Schar P. 2012. DNA glycosylases: in DNA repair and beyond. Chromosoma 121:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Park D, Kropachev K, Kolbanovskiy M, Fu I, Broyde S, Essawy M, Geacintov NE, Tretyakova NY. 2019. 5-Formylcytosine-induced DNA-peptide cross-links reduce transcription efficiency, but do not cause transcription errors in human cells. J Biol Chem 294:18387–18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. 2012. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res 40:10795–10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. 1983. Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res 43:4382–4392. [PubMed] [Google Scholar]
- Kim MS, Machida Y, Vashisht AA, Wohlschlegel JA, Pang YP, Machida YJ. 2013. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res 41:1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CB, Frenkel K, Costa M. 1991. The role of oxidative processes in metal carcinogenesis. Chem Res Toxicol 4:592–604. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Machida Y, Palani S, Caulfield TR, Radisky ES, Kaufmann SH, Machida YJ. 2020. FAM111A protects replication forks from protein obstacles via its trypsin-like domain. Nat Commun 11:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Yu R, Hartwell HJ, Moeller BC, Bodnar WM, Swenberg JA. 2016. Measurement of Endogenous versus Exogenous Formaldehyde-Induced DNA-Protein Crosslinks in Animal Tissues by Stable Isotope Labeling and Ultrasensitive Mass Spectrometry. Cancer Res 76:2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey PS, Berkemeier T, Tong H, Arangio AM, Lucas K, Poschl U, Shiraiwa M. 2016. Chemical exposure-response relationship between air pollutants and reactive oxygen species in the human respiratory tract. Sci Rep 6:32916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Gao AO, Sparks JL, Gallina I, Wu RA, Mann M, Raschle M, Walter JC, Duxin JP. 2019. Replication-Coupled DNA-Protein Crosslink Repair by SPRTN and the Proteasome in Xenopus Egg Extracts. Mol Cell 73:574–588 e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, Cabrera E, Freire R, Pope K, Nahid A, Norris F, Leventer RJ, Delatycki MB, Barbi G, von Ameln S, Hogel J, Degoricija M, Fertig R, Burkhalter MD, Hofmann K, Thiele H, Altmuller J, Nurnberg G, Nurnberg P, Bahlo M, Martin GM, Aalfs CM, Oshima J, Terzic J, Amor DJ, Dikic I, Ramadan K, Kubisch C. 2014. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet 46:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Raczynska JE, Chen Z, Yu H. 2019. Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan. Cell Rep 26:3336–3346 e3334. [DOI] [PubMed] [Google Scholar]
- Liao H, Ji F, Helleday T, Ying S. 2018. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep 19:e46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I. 2016. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. Elife 5:e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg JA. 2010. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc 132:3388–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfie A, Hagan E, Zhitkovich A. 2010. Mechanism of DNA-protein cross-linking by chromium. Chem Res Toxicol 23:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Kim MS, Machida YJ. 2012. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle 11:3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maede Y, Shimizu H, Fukushima T, Kogame T, Nakamura T, Miki T, Takeda S, Pommier Y, Murai J. 2014. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol Cancer Ther 13:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso TF. 1997a. Chromium as an industrial carcinogen: Part I . Am J Ind Med 31:129–139. [DOI] [PubMed] [Google Scholar]
- Mancuso TF. 1997b. Chromium as an industrial carcinogen: Part II. Chromium in human tissues. Am J Ind Med 31:140–147. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 2001. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem 276:40652–40658. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. 2014. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15:465–481. [DOI] [PubMed] [Google Scholar]
- Maskey RS, Flatten KS, Sieben CJ, Peterson KL, Baker DJ, Nam HJ, Kim MS, Smyrk TC, Kojima Y, Machida Y, Santiago A, van Deursen JM, Kaufmann SH, Machida YJ. 2017. Spartan deficiency causes accumulation of Topoisomerase 1 cleavage complexes and tumorigenesis. Nucleic Acids Res 45:4564–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. 2014. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat Commun 5:5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattagajasingh SN, Misra HP. 1996. Mechanisms of the carcinogenic chromium(VI)-induced DNA-protein cross-linking and their characterization in cultured intact human cells. J Biol Chem 271:33550–33560. [DOI] [PubMed] [Google Scholar]
- Merk O, Speit G. 1999. Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ Mol Mutagen 33:167–172. [DOI] [PubMed] [Google Scholar]
- Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. 2010. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J Proteome Res 9:4356–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. 2009. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol 29:5998–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. 2008. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat Res 637:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohni KN, Wessel SR, Zhao R, Wojciechowski AC, Luzwick JW, Layden H, Eichman BF, Thompson PS, Mehta KPM, Cortez D. 2019. HMCES Maintains Genome Integrity by Shielding Abasic Sites in Single-Strand DNA. Cell 176:144–153 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticello TM, Morgan KT, Hurtt ME. 1990. Unit length as the denominator for quantitation of cell proliferation in nasal epithelia. Toxicol Pathol 18:24–31. [DOI] [PubMed] [Google Scholar]
- Monticello TM, Swenberg JA, Gross EA, Leininger JR, Kimbell JS, Seilkop S, Starr TB, Gibson JE, Morgan KT. 1996. Correlation of regional and nonlinear formaldehyde-induced nasal cancer with proliferating populations of cells. Cancer Res 56:1012–1022. [PubMed] [Google Scholar]
- Morocz M, Zsigmond E, Toth R, Enyedi MZ, Pinter L, Haracska L. 2017. DNA-dependent protease activity of human Spartan facilitates replication of DNA-protein crosslink-containing DNA. Nucleic Acids Res 45:3172–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, Hartmann-Petersen R, Lukas J, Choudhary C, Pocock R, Bekker-Jensen S, Mailand N. 2012. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol 19:1084–1092. [DOI] [PubMed] [Google Scholar]
- Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. 2009. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem 284:27065–27076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H. 2007. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell 28:147–158. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ouchi R, Kawazoe J, Pack SP, Makino K, Ide H. 2012. T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription. J Biol Chem 287:6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Xu X, Salem AMH, Shoulkamy MI, Ide H. 2017. Radiation-induced DNA-protein cross-links: Mechanisms and biological significance. Free Radic Biol Med 107:136–145. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436:1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. 1993. NTP Toxicology and Carcinogenesis Studies of 1,3-Butadiene (CAS No. 106–99-0) in B6C3F1 Mice (Inhalation Studies). Natl Toxicol Program Tech Rep Ser 434:1–389. [PubMed] [Google Scholar]
- Pande P, Ji S, Mukherjee S, Scharer OD, Tretyakova NY, Basu AK. 2017. Mutagenicity of a Model DNA-Peptide Cross-Link in Human Cells: Roles of Translesion Synthesis DNA Polymerases. Chem Res Toxicol 30:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Ha EH, Lee KH, Hong YC. 2002. Benzo[a]pyrene-induced DNA-protein crosslinks in cultured human lymphocytes and the role of the GSTM1 and GSTT1 genotypes. J Korean Med Sci 17:316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljsak B, Fink R. 2014. The protective role of antioxidants in the defence against ROS/RNS-mediated environmental pollution. Oxid Med Cell Longev 2014:671539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Kohlhagen G, Laco GS, Kroth H, Sayer JM, Jerina DM. 2002. Different effects on human topoisomerase I by minor groove and intercalated deoxyguanosine adducts derived from two polycyclic aromatic hydrocarbon diol epoxides at or near a normal cleavage site. J Biol Chem 277:13666–13672. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL. 2016. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286:552–555. [DOI] [PubMed] [Google Scholar]
- Pourquier P, Pommier Y. 2001. Topoisomerase I-mediated DNA damage. Adv Cancer Res 80:189–216. [DOI] [PubMed] [Google Scholar]
- Prasad R, Horton JK, Chastain PD 2nd, Gassman NR, Freudenthal BD, Hou EW, Wilson SH. 2014. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res 42:6337–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Horton JK, Dai DP, Wilson SH. 2019. Repair pathway for PARP-1 DNA-protein crosslinks. DNA Repair (Amst) 73:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, Suh M, Campleman SL, Thompson CM. 2014. Assessment of the mode of action for hexavalent chromium-induced lung cancer following inhalation exposures. Toxicology 325:160–179. [DOI] [PubMed] [Google Scholar]
- Prüss-Üstün A, Wolf J, Corvalán C, Bos R, Neira M. 2016. Preventing disease through healthy environments : a global assessment of the burden of disease from environmental risks. Geneva: World Health Organization; 147 p. [Google Scholar]
- Quievryn G, Zhitkovich A. 2000. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21:1573–1580. [PubMed] [Google Scholar]
- Quinones JL, Demple B. 2016. When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions. DNA Repair (Amst) 44:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones JL, Thapar U, Yu K, Fang Q, Sobol RW, Demple B. 2015. Enzyme mechanism-based, oxidative DNA-protein cross-links formed with DNA polymerase beta in vivo. Proc Natl Acad Sci U S A 112:8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez P, Del Razo LM, Gutierrez-Ruiz MC, Gonsebatt ME. 2000. Arsenite induces DNA-protein crosslinks and cytokeratin expression in the WRL-68 human hepatic cell line. Carcinogenesis 21:701–706. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Sancar A. 2006. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci U S A 103:4056–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. 2007. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res 67:11117–11122. [DOI] [PubMed] [Google Scholar]
- Ruijs MW, van Andel RN, Oshima J, Madan K, Nieuwint AW, Aalfs CM. 2003. Atypical progeroid syndrome: an unknown helicase gene defect? Am J Med Genet A 116A:295–299. [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang N, Hou L, Yun Y, Li G. 2009. SO(2) inhalation induces protein oxidation, DNA-protein crosslinks and apoptosis in rat hippocampus. Ecotoxicol Environ Saf 72:879–884. [DOI] [PubMed] [Google Scholar]
- Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, Butler LR, Williams JG, Munoz-Cabello AM, Mueller GA, London RE, Cortes-Ledesma F, Williams RS. 2017. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 357:1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Talaska G, Puga A. 2007. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol 27:7089–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbyn N, Noireterre A, Bagdiul I, Plank M, Michel AH, Loewith R, Kornmann B, Stutz F. 2019. The Aspartic Protease Ddi1 Contributes to DNA-Protein Crosslink Repair in Yeast. Mol Cell 77:1066–1079. [DOI] [PubMed] [Google Scholar]
- Sparks JL, Chistol G, Gao AO, Raschle M, Larsen NB, Mann M, Duxin JP, Walter JC. 2019. The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell 176:167–181 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen AM, Meyer KG, Recio L. 1997a. Analysis of hprt mutations occurring in human TK6 lymphoblastoid cells following exposure to 1,2,3,4-diepoxybutane. Mutagenesis 12:61–67. [DOI] [PubMed] [Google Scholar]
- Steen AM, Meyer KG, Recio L. 1997b. Characterization of hprt mutations following 1,2-epoxy-3-butene exposure of human TK6 cells. Mutagenesis 12:359–364. [DOI] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, Tsutakawa SE, Borg A, Kjaer S, Tainer JA, Skehel JM, Groll M, Boulton SJ. 2016. Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Mol Cell 64:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. 2014. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158:327–338. [DOI] [PubMed] [Google Scholar]
- Stout MD, Herbert RA, Kissling GE, Collins BJ, Travlos GS, Witt KL, Melnick RL, Abdo KM, Malarkey DE, Hooth MJ. 2009. Hexavalent chromium is carcinogenic to F344/N rats and B6C3F1 mice after chronic oral exposure. Environ Health Perspect 117:716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Brocato J, Costa M. 2015. Oral Chromium Exposure and Toxicity. Curr Environ Health Rep 2:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jenkins LM, Su YP, Nitiss KC, Nitiss JL, Pommier Y. 2019. A conserved SUMO-Ubiquitin pathway directed by RNF4/SLX5-SLX8 and PIAS4/SIZ1 drives proteasomal degradation of topoisomerase DNA-protein crosslinks. BioRxiv 10.1101/707661. [DOI] [Google Scholar]
- Swenberg JA, Kerns WD, Mitchell RI, Gralla EJ, Pavkov KL. 1980. Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res 40:3398–3402. [PubMed] [Google Scholar]
- Swenberg JA, Moeller BC, Lu K, Rager JE, Fry RC, Starr TB. 2013. Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicol Pathol 41:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A, Tainer JA. 2018. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem 87:263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PS, Amidon KM, Mohni KN, Cortez D, Eichman BF. 2019. Protection of abasic sites during DNA replication by a stable thiazolidine protein-DNA cross-link. Nat Struct Mol Biol 26:613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Hegedus L, Juhasz S, Haracska L, Burkovics P. 2017. The DNA-binding box of human SPARTAN contributes to the targeting of Poleta to DNA damage sites. DNA Repair (Amst) 49:33–42. [DOI] [PubMed] [Google Scholar]
- Tretyakova NY, Groehler At, Ji S. 2015. DNA-Protein Cross-Links: Formation, Structural Identities, and Biological Outcomes. Acc Chem Res 48:1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapakos MJ, Hampton TH, Wetterhahn KE. 1983. Chromium(VI)-induced DNA lesions and chromium distribution in rat kidney, liver, and lung. Cancer Res 43:5662–5667. [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. 2017. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52:274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, Drobnitzky N, Freire R, Amor DJ, Lockhart PJ, Kessler BM, McKenna GW, Gileadi O, Ramadan K. 2016. Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Mol Cell 64:704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling R, Beaumont JJ, Petersen SJ, Alexeeff GV, Steinmaus C. 2015. Chromium VI and stomach cancer: a meta-analysis of the current epidemiological evidence. Occup Environ Med 72:151–159. [DOI] [PubMed] [Google Scholar]
- Wen Y, Zhang PP, An J, Yu YX, Wu MH, Sheng GY, Fu JM, Zhang XY. 2011. Diepoxybutane induces the formation of DNA-DNA rather than DNA-protein cross-links, and single-strand breaks and alkali-labile sites in human hepatocyte L02 cells. Mutat Res 716:84–91. [DOI] [PubMed] [Google Scholar]
- Wilk A, Waligorski P, Lassak A, Vashistha H, Lirette D, Tate D, Zea AH, Koochekpour S, Rodriguez P, Meggs LG, Estrada JJ, Ochoa A, Reiss K. 2013. Polycyclic aromatic hydrocarbons-induced ROS accumulation enhances mutagenic potential of T-antigen from human polyomavirus JC. J Cell Physiol 228:2127–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Burgin AB Jr., Huizenga BN, Robertson CA, Yao KC, Nash HA. 1996. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci U S A 93:11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatera K, Morimoto Y, Ueno S, Noguchi S, Kawaguchi T, Tanaka F, Suzuki H, Higashi T. 2018. Cancer Risks of Hexavalent Chromium in the Respiratory Tract. J UOEH 40:157–172. [DOI] [PubMed] [Google Scholar]
- Zecevic A, Hagan E, Reynolds M, Poage G, Johnston T, Zhitkovich A. 2010. XPA impacts formation but not proteasome-sensitive repair of DNA-protein cross-links induced by chromate. Mutagenesis 25:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitkovich A, Voitkun V, Kluz T, Costa M. 1998. Utilization of DNA-protein cross-links as a biomarker of chromium exposure. Environ Health Perspect 106 Suppl 4:969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Huang X, Costa M. 1994. Protein oxidation and amino acid-DNA crosslinking by nickel compounds in intact cultured cells. Toxicol Appl Pharmacol 126:319–325. [DOI] [PubMed] [Google Scholar]