Abstract
Alcohol and drug use can dysregulate neural circuit function to produce a wide range of neuropsychiatric disorders, including addiction. To understand the neural circuit computations that mediate behavior, and how substances of abuse may transform them, we must first be able to observe the activity of circuits. While many techniques have been utilized to measure activity in specific brain regions, these regions are made up of heterogeneous sub-populations, and assessing activity from neuronal populations of interest has been an ongoing challenge. To fully understand how neural circuits mediate addiction-related behavior, we must be able to reveal the cellular granularity within brain regions and circuits by overlaying functional information with the genetic and anatomical identity of the cells involved. The development of genetically encoded calcium indicators, which can be targeted to populations of interest, allows for in vivo visualization of calcium dynamics, a proxy for neuronal activity, thus providing an avenue for real-time assessment of activity in genetically and anatomically defined populations during behavior. Here, we highlight recent advances in calcium imaging technology, compare the current technology with other state-of-the-art approaches for in vivo monitoring of neural activity, and discuss the strengths, limitations, and practical concerns for observing neural circuit activity in preclinical addiction models.
Keywords: Alcohol, Ethanol, Abuse, Substance, Addiction, GCaMP, GECI, Calcium, Fluorescence, Photometry, Imaging, Indicators
Introduction
Alcohol and drugs of abuse alter the computations performed in neural circuits, which can result in maladaptive behaviors, including addiction (Dong, Taylor, Wolf, & Shaham, 2017; Lüscher, 2016; Mulholland, Chandler, & Kalivas, 2016; Stuber, Hopf, Tye, Chen, & Bonci, 2010; Volkow, Wang, Tomasi, & Baler, 2013). Our ability to understand, treat, and mitigate dysfunctional communication between neural circuits, which underlies addiction, relies first on our ability to observe and record cellular activity in the brain in vivo. Observing neural activity in vivo has been a primary and ongoing challenge in neuroscience. Currently, there are many techniques that allow for real-time assessment of neural activity; however, each of these approaches comes with advantages and limitations. Here, we will highlight the advantages of calcium imaging, introduce common methods for in vivo imaging, and discuss the limitations and practical concerns for integrating this approach with preclinical alcohol abuse models. Our goal is to provide a broad introduction and resource for preclinical addiction neuro-scientists who may not have extensive training in optics or in vivo recording techniques.
Calcium imaging leverages the fact that action potentials, a ubiquitous currency of neuronal communication, are generated via rapid ion flux, including calcium, across the cytoplasmic membrane (Baker, Hodgkin, & Ridgway, 1971; Tank, Sugimori, Connor, & Llinas, 1988). Thus, changes in intracellular calcium concentration can be used as a proxy for action potential activity (Kerr et al., 2000; Regehr, Connor, & Tank, 1989). Calcium indicators are fluorescent molecules or dyes which, when bound to calcium, increase in fluorescence intensity. By observing changes in fluorescence emitted from the indicator over time, calcium concentration can be visualized, and action potential activity can be inferred. While there are many approaches to visualizing neuronal activity, including indicators that are responsive to a wide array of factors involved in cellular activity such as chloride (Berglund et al., 2006; Verkman et al., 1989) or voltage (Hochbaum et al., 2014; Peterka et al., 2011; St-Pierre et al., 2014), as well as imaging of radiolabeled tracers (Raichle, 1983), this review will focus on a specific type of optical recording: in vivo imaging of genetically encoded calcium indicators (GECIs). We will broadly cover the advantages and limitations of the most applicable in vivo GECI imaging techniques for investigating the pathology of addiction (Fig. 1). For additional resources on this topic, as well as information regarding other activity indicators and preparations, we point the reader toward several other informative reviews (Broussard, Liang, & Tian, 2014; Germond, Fujita, Ichimura, & Watanabe, 2016; Girven & Sparta, 2017; Lin & Schnitzer, 2016; Resendez et al., 2016; Sepehri Rad et al., 2017; Storace et al., 2016; Wachowiak & Knöpfel, 2009; Yang & Yuste, 2017. For a review of insights into the circuit basis of addiction from functional magnetic resonance imaging (fMRI) and positron emission tomography (PET) literatures, see Dupuy & Chanraud, 2016; Parvaz, Alia-Klein, Woicik, Volkow, & Goldstein, 2011; Wiers, Cabrera, Skarda, Volkow, & Wang, 2016.
Fig. 1. Commonly used approaches for in vivo calcium imaging.
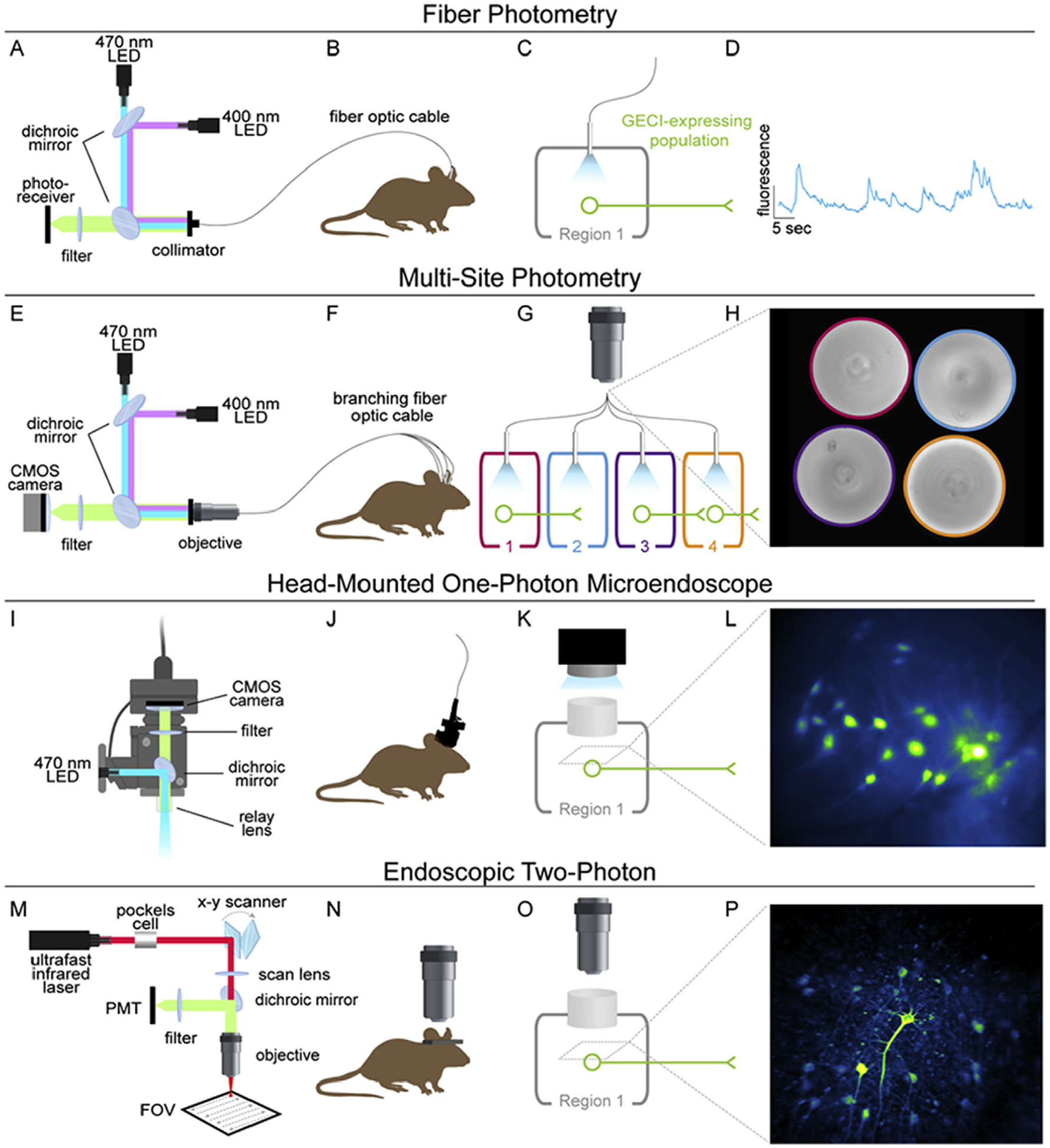
(A) Example of light path setup for typical fiber photometry recordings. Light from fiber-coupled LEDs for GECI and control isosbestic excitation (470 and 400 nm, respectively) are combined into one path via dichroic mirrors and delivered to the region of interest through a fiber optic patch cable coupled to a chronically implanted optic fiber. Emission is filtered and collected by a photoreceiver. (B) The patch cable is connected to the implanted fiber just prior to the imaging session, and allows for relative ease of movement during free behavior. (C) The optic fiber is implanted into the GECI-expressing region. This approach can be easily used to record from GECI-expressing soma or axons. (D) Example data obtained by fiber photometry recordings. Because emitted photons collected by the fiber are “scrambled” the signal is collected as “bulk fluorescence” rather than an image of the tissue. The resultant data are two-dimensional (fluorescence × time), and reflect the calcium activity of the entire GECI-expressing population, as opposed to obtaining activity resulting from single cells. (E) Example of a light path typically implemented for multi-site photometry recordings. Excitation is similar to single-site photometry, described above; however, in this case, an objective and CMOS camera are used to obtain an image of the back of a branching patch cord. This allows for simultaneous assessment of bulk fluorescence activity from multiple GECI-expressing regions within the same animal by separately assessing fluorescence within each fiber of the patch cord. (F) The branching patch cord is attached to multiple chronically implanted optic fibers allowing recordings to be performed in freely moving animals. (G) Fibers can be implanted in multiple GECI-expressing regions to record from the soma and axons of the same region (1 and 2) and/or in distinct populations (3 and 4). Note that if one GECI-expressing region receives input from another GECI-expressing region, the signal may contain activity from both regions, which cannot be distinguished in the recording. For example, the signal resulting from the fiber implanted in Region 4 may reflect somatic activity from cells in Region 4 as well as axonal activity from terminals arising from Region 3. (H) Example image of four optic fibers within the branching patch cord obtained from a multi-site photometry recording. Fluorescence within each fiber is averaged to obtain a bulk fluorescence signal arising from each of the four implanted fibers. (I) Example of a light path within a miniature head-mounted one-photon microscope. Excitation light is reflected by a dichroic mirror and passed through a relay lens. Resulting emission is filtered and imaged on a CMOS sensor. (J) The miniature microscope is attached to the animal via a magnetic baseplate just prior to the imaging session, allowing visualization of GECI-expressing cells during free behavior. The data are collected on the head, and a digitized signal is relayed through the cable to a data acquisition box. (K) Example of experimental implementation, in which a chronically implanted GRIN lens allows optical access to a deep-brain region containing GECI-expressing cells. (L) Example of an image obtained via a miniature head-mounted microscope. Fluorescence can be assessed within each GECI-expressing cell allowing for assessment of calcium activity with cellular resolution. (M) Example of a light path for two-photon microscopy. Femtosecond pulses of infrared light are delivered to the sample through a Pockels cell, x-y scanner, and scan lens (note that many setups also include a mechanism for z-plane scanning for volumetric imaging). Excitation light is passed through an objective and rapidly scanned across the sample. Emission from each excitation point is collected by a PMT and an image is constructed by attributing fluorescence to a point in x-y space based on the focal point of the laser. (N) Recordings are performed while the animal is head-fixed under the microscope. Note that head-mounted two-photon microscopes have been developed, but have not yet been widely implemented. (O) Example of experimental implementation, where a chronically implanted GRIN lens allows optical access to a deep-brain region containing GECI-expressing cells. Note that two-photon imaging can also be used to image axonal activity, but this approach is prohibitively difficult for deep-brain regions. (P) Example of an image obtained via two-photon microscopy through a GRIN lens. Fluorescence can be assessed within each GECI-expressing cell allowing for assessment of calcium activity with single cell resolution. CMOS, complementary metal-oxide-semiconductor; FOV, field of view; PMT, photomultiplier tube.
Calcium imaging has been used for decades to visualize neuronal activity (Connor, 1986; Lipscombe et al., 1988; Tank et al., 1988), but has only recently seen widespread use in freely-moving behavioral neuroscience. This is largely due to several advances that have significantly improved the feasibility and expanded the scope of possible questions that can be addressed with this approach.
First, the creation of genetically encoded calcium indicators, combined with advances in viral-mediated gene transfer technologies and transgenic animal availability, has allowed for targeted expression of GECIs in many different populations of interest (Daigle et al., 2018; DeNardo & Luo, 2017; He et al., 2016). This is in contrast to synthetic calcium indicators and calcium sensitive dyes, which require invasive and laborious loading procedures to introduce the indicator into cells of interest.
Second, GECIs have improved dramatically in temporal resolution and signal-to-noise ratio (SNR) (Chen et al., 2013; Grienberger & Konnerth, 2012; Jercog, Rogerson, & Schnitzer, 2016; Sun et al., 2013). The most commonly used indicator in recent literature is GCaMP6(s/m/f), part of the GCaMP family (Mank et al., 2008; Nakai, Ohkura, & Imoto, 2001; Tian et al., 2009), has SNR sufficiently high to detect single, isolated action potentials, and provides temporal resolution in the high millisecond range (Chen et al., 2013). Note that several variants of GCaMP6 are available, which vary in brightness, SNR, and temporal resolution (Chen et al., 2013). Most recently, further alterations have been made in GCaMP7 variants (Dana et al., 2018; Muto, Ohkura, Abe, Nakai, & Kawakami, 2013; Sato et al., 2015); for the remainder of this review, we use GCaMP as an umbrella term referring to this family of GECIs.
Third, probes have been developed that allow optical access to deep-brain regions, and imaging technology has been adapted for use in freely behaving animals, greatly expanding the application of in vivo calcium imaging (Flusberg et al., 2008; Helmchen, Fee; Tank, & Denk, 2001; Reed, Yan, & Schnitzer, 2002). Previously, in vivo calcium imaging was typically restricted to superficial brain regions accessed through thinned skulls or via cranial windows, and was seldom performed in regions deeper than 1–2 mm below the surface of the brain. Further, imaging technology typically required the sample to be immobile, precluding its use during free behavior. Gradient refractive index (GRIN) lenses have allowed for in vivo imaging in the deepest regions of the rodent brain (Reed et al., 2002), and advances in imaging technology, such as miniaturized head-mounted microscopes, now allow for assessing fluorescence activity during free movement (Flusberg et al., 2008; Helmchen, Fee, Tank, & Denk, 2001). The development of new algorithms for analysis of calcium imaging data with cellular resolution has facilitated the interpretation of such data with confidence (Mukamel, Nimmerjahn, & Schnitzer, 2009; Zhou et al., 2018). Both GRIN lenses and head-mounted microscopes have recently seen widespread use in neuroscience, and have been applied to visualize calcium dynamics in deep-brain areas of freely moving rodents (Barbera et al., 2016; Cai et al., 2016; Grewe et al., 2017; Jennings et al., 2015; Pinto & Dan, 2015; Sun et al., 2015; Ziv et al., 2013).
The advances outlined above have substantially increased the applicability of calcium imaging in neuroscience. In turn, this has driven the production of commercially produced imaging systems optimized use in behavioral neuroscience. Several commercially produced systems, as well as open-source options, are now available, making calcium-imaging technology easily accessible outside the optics field. Laboratories have readily adopted these approaches to investigate the neurobiology of addiction in rodent models (Beier et al., 2017; Calipari et al., 2017, 2016; Luo, Volkow, Heintz, Pan, & Du, 2011; Xia, Nygard, Sobczak, Hourguettes, & Bruchas, 2017).
Utility of in vivo calcium imaging
There are many approaches for assessing real-time neural activity in behaving animals. Generally, these can be divided into two main categories: 1) electrical recordings, and 2) optical recordings. To highlight the experimental questions for which the features of in vivo GECI imaging are most advantageous, we will briefly contrast this approach with electrical recording techniques.
Extracellular electrophysiology has historically been the primary tool for assessing in vivo real-time neural activity, and has been implemented over the last half-century to assess the effects of alcohol and other addictive drugs in several model organisms (Aguilar-Rivera, Casanova, Gatica, Quirk, & Fuentealba, 2015; Chan, Wheeler, & Wheeler, 2016; Fanelli, Klein, Reese, & Robinson, 2013; Georges, Le Moine, & Aston-Jones, 2006; Hampson, Porrino, Opris, Stanford, & Deadwyler, 2011; Janak, Chang, & Woodward, 1999; Le Bars, Menetrey, Conseiller, & Besson, 1975; Liu, Jiang, Zhong, Wu, & Luo, 2010; Mahler et al., 2014; Margolis, Hjelmstad, Fujita, & Fields, 2014; Morra, Glick, & Cheer, 2010; Murray et al., 2015; Nicola & Deadwyler, 2000; Peoples et al., 1999; Peoples & West, 1996; Perra et al., 2005; Robinson & Carelli, 2008; Trantham, Szumlinski, McFarland, Kalivas, & Lavin, 2002; Wheeler et al., 2008) (for review see Deadwyler, 2010; Wheeler & Carelli, 2009). In vivo electrophysiology typically involves inserting one or more glass pipettes or wires (electrodes) into a region of interest to record changes in voltage during behavior or in anesthetized animals. Provided that the tip of the electrodes are sufficiently small (low micron scale), the observed changes in voltage are the result of action potentials in a single or small number of cells, allowing for single-cell, single action potential resolution (in the case of more than one cell being detected by a single wire or contact, the cells can be computationally separated based on spike waveform) (Chorev, Epsztein, Houweling, Lee, & Brecht, 2009; Gerstein & Clark, 1964; Harris, Henze, Csicsvari, Hirase, & Buzsaki, 2000). While this approach provides high temporal (microsecond domain and faster) and spatial (low micrometer) resolution, traditional in vivo electrophysiology provides no mechanism to ascertain the anatomical, morphological, or genetic identity of the recorded cells.
The power of calcium imaging lies in its ability to overlay functional activity with genetic and anatomical identity, with high yields and low false-positive cell identification. Further, spatial and morphological information are obtained in parallel, which, in addition to providing information about the anatomy of the cell, can also be used to identify and follow the same cells over many sessions/days to determine within-cell changes in activity over time (Grewe et al., 2017; Sheintuch et al., 2017). These advantages come with several sacrifices compared to electrical recordings, which must be considered in technique selection and experimental design. Foremost, calcium activity is a proxy for action potentials, and thus requires tempered interpretation when inferring spiking activity (Harris, Quiroga, Freeman, & Smith, 2016; Theis et al., 2016). Further, temporal resolution of GECIs are an order of magnitude, or more, slower than the timescale over which action potentials occur. Thus, when determining whether calcium imaging is the most appropriate approach for an experimental question, it is important to consider that many of these features can also be achieved through electrical recordings. Indeed, several combinatorial approaches have been developed which allow cell identification in electrophysiological recordings. Juxtacellular labeling (Pinault, 1996; Schreihofer & Guyenet, 1997), antidromic stimulation (Dreifuss & Kelly, 1972; Mantz, Thierry, & Glowinski, 1989), and phototagging (Cohen, Haesler, Vong, Lowell, & Uchida, 2012; Lima, Hromadka, Znamenskiy, & Zador, 2009; Nieh et al., 2015; Senn et al., 2014) are popular approaches for post hoc assessment of certain aspects of cell identity in electrophysiological recordings.
Of these approaches, phototagging (also termed photostimulation-assisted identification of neuronal populations [PINP]) most overlaps with GECI imaging in terms of the information garnered, and the experimental questions that it can address. This approach uses an optrode (combined optic fiber and electrophysiology electrode) to deliver light to a region where cells have been transduced with an excitatory opsin, such as channelrhodopsin-2 (ChR2), while simultaneously recording their activity (Cohen et al., 2012; Kim, Adhikari, & Deisseroth, 2017; Lima et al., 2009; Nieh et al., 2015; Senn et al., 2014) (Fig. 2A). Cells that display excitatory responses to light (i.e., “phototagged units”) can be assumed to belong to the population that expresses the opsin. Because opsins can be delivered to genetically or anatomically defined populations, similar to GECIs, this provides an alternative approach to overlaying functional activity with cell identity. This approach is attractive compared to GECI imaging because it circumvents precautions related to temporal resolution, and provides a direct measure of spiking activity as opposed to a proxy measure. Further, information is gathered from both the population of interest as well as non-opsin expressing cells in the area, while GECI imaging obtains information only from transgene-expressing cells.
Fig. 2. Recurrent excitation during ChR2-assisted cell identification in single-unit electrophysiology recordings (phototagging).
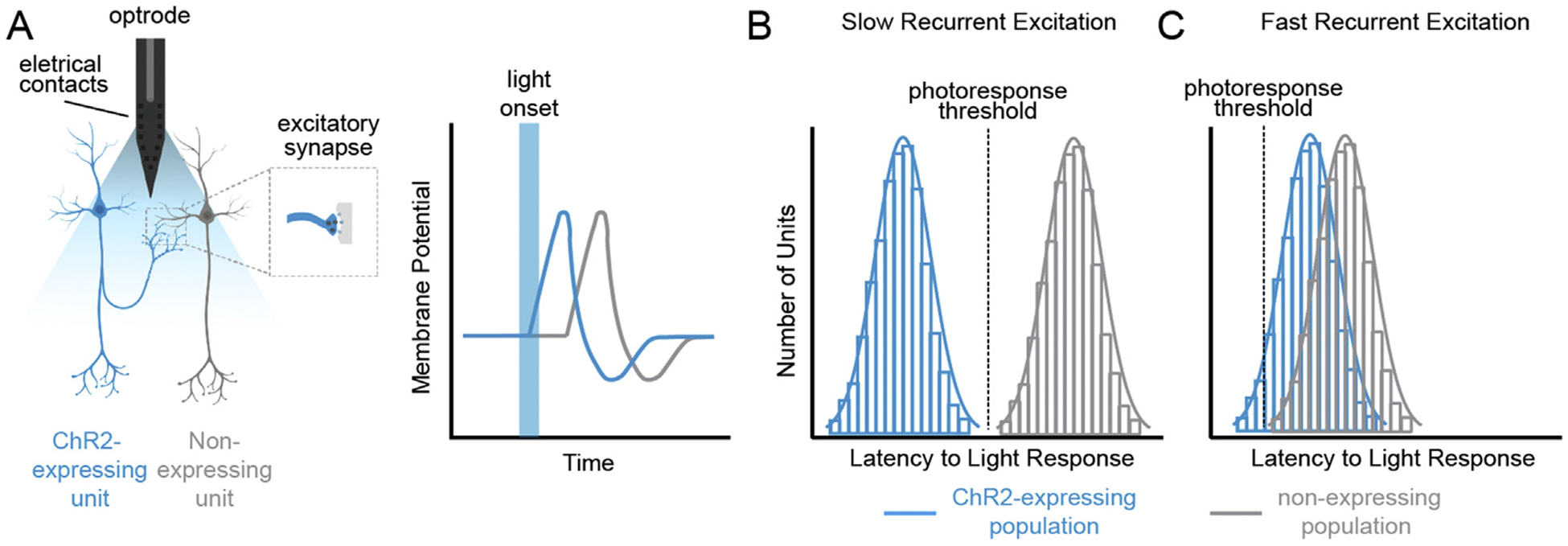
Viral transduction of ChR2 to specific sub-populations within a brain region allow for their identification during in vivo single-unit recordings by assessing each unit’s response to light. Cells that are light responsive (i.e., “phototagged”) can be assumed to be part of the population to which the opsin was delivered. (A) However, as illustrated here, activation of non-ChR2-expressing cells within the same region (non-expressing neighbors) can occur via lateral excitatory connections. By using whole-cell patch clamp in ex vivo slices to record from cells that are visually identified as expressing or non-expressing, it is possible to determine response latencies between the two populations. A photoresponse threshold is then determined and applied post hoc to the in vivo data set, and only cells that respond to light faster than non-expressing neighbors are considered to be phototagged units. (B) In regions where recurrent excitation is slow, application of the photoresponse threshold is unambiguous. (C) In regions with fast recurrent excitation, expressing and non-expressing units may have similar response latencies. In this example, ChR2-expressing cells to the right of the photoresponse threshold will incorrectly (false negative) be classified as non-expressing, and there will be a low yield of cells identified as phototagged.
However, phototagging suffers from its own set of disadvantages. Most importantly, recurrent excitation often occurs within the region of interest as a result of neurotransmitter release from the photoexcited cells. For example, photoactivation of opsin-expressing cells can result in activation of non-expressing neighbors within the region through feedforward excitation mediated by monosynaptic glutamate release from lateral connections to neighboring cells, and/or through feedforward disinhibition from light-evoked GABA release onto local inhibitory neurons, which in turn synapse onto other non-opsin expressing neighbors. Because stimulation of opsin-expressing cells can result in activation of non-expressing neighboring cells (Fig. 2A), it is necessary to qualify light-responsive units as phototagged based on their response latency. Recurrent excitation can differ between regions, populations of interest, and viral/recording strategies, thus a static photoresponse latency threshold cannot be implemented; instead, response latencies must be determined directly in the population/region of interest (Beyeler et al., 2016). Photoresponse thresholds can be determined in ex vivo slices where opsin-expressing and non-expressing cells can be identified by visualizing the fluorophore fused to the opsin. During whole-cell recording in brain slices where cells can be visually identified as opsin-expressing or non-expressing, response latencies can be determined for the two populations by photostimulating the slice, and the photoresponse threshold can be applied post hoc to the in vivo data set to exclude cells that are likely to have been activated via recurrent excitation (Beyeler et al., 2016; Nieh et al., 2015). Exclusion of light-responsive non-expressing neighbors can also be achieved in vivo through pharmacological blockade of the receptors mediating recurrent excitation, but difficulty in simultaneous recording and drug delivery has thus far limited this approach to superficial brain regions (Lima et al., 2009). However, drug delivery during electrophysiological recordings has been achieved in deep-brain areas for other experimental purposes (du Hoffmann & Nicola, 2014; Owesson-White et al., 2016). In regions that exhibit slow recurrent excitation, there is clear separation in the response latencies of opsin-expressing and non-expressing neighbors (Fig. 2B). In contrast, in regions that exhibit fast recurrent excitation, definitively identifying phototagged units can be ambiguous, resulting in a high probability of false-negative cell identification and low yields (Fig. 2C).
Another factor that can contribute to low yields in phototagging experiments is that the electrode is inserted into the brain region of interest, but cannot be directly targeted to the opsin-expressing cells, and thus obtaining recordings from the population of interest occurs stochastically. An additional limitation of in vivo extracellular recordings is that because a single wire often detects the activity of multiple neurons, units typically must be separated from each other and from background noise based on the features of the action potential waveform detected (termed “spike sorting”) (Harris et al., 2016). This process may lead to bias toward detection of cells that are more active, and comparison of extracellular spike-sorted data to ground-truth data obtained via simultaneous cell-attached recordings suggests that spike detection errors rates are often in the range of 5–10%, but can be as high as 50%, depending on biological (e.g., synchronous activity) and analytical factors (e.g., manual vs. automated spike sorting) (Harris et al., 2000, 2016).
Importantly, small movements of the tissue and the implanted wires over time produce “drift”, which can result in different cells being recorded at the electrode tip over days. While computational approaches have been developed to attempt to identify whether the same cells are recorded over time (Dickey, Suminski, Amit, & Hatsopoulos, 2009; Tolias et al., 2007), implementation of these approaches is controversial, and most electrophysiology studies refrain from making claims regarding within-cell changes in function unless the recording is performed within a single session/day. This limitation is particularly important in the context of addiction research, as addiction is a chronic disorder that can often take years to develop, and manifests itself over the course of a lifetime (Dawson, Goldstein, Chou, Ruan, & Grant, 2008; Grant & Dawson, 1998). In understanding such a protracted pathology, the use of GECI imaging to identify cells longitudinally, based on spatial and morphological information, is a powerful asset.
In summary, careful consideration should be given to selecting a technique for in vivo recordings of identified neuronal populations, as every approach has distinct pros and cons. Nonetheless, GECI imaging allows for high yield assessment of functional, spatial, and morphological information in targeted populations, and this highly paralleled approach holds great promise for unraveling the circuit-based computations that orchestrate behavior, and their dysregulation by chronic use of alcohol and abused drugs. Of course, we acknowledge that achieving a deep understanding of motivated behaviors will require integration of information garnered with diverse technologies (for review see Tye, 2018).
Approaches to calcium imaging
There are many different approaches to in vivo GECI imaging, which vary in the way the brain region of interest is optically accessed, the light excitation source, and the microscope/sensor setup that is used to excite and record the GECI. All of these approaches first require expression of a GECI, which can be achieved in a non-conditional or cell-type specific fashion through several means, such as transgenic animals or viral-mediated gene transfer (for review of transgene targeting strategies see Fenno et al., 2014; Luo, Callaway, & Svoboda, 2008; Neve & Neve, 2001). Once expression is achieved, a microscopy approach to observing the cells must be selected; all of these approaches are based on the principles of fluorescence light microscopy, but each has advantages and disadvantages, which can be leveraged depending on the experimental question.
Bulk fluorimetry/fiber photometry
Optic fiber-based observations of GECI fluorescence, such as fiber photometry, are perhaps the easiest to implement out of the currently available approaches to in vivo imaging. Fiber photometry utilizes chronically implanted optic fibers (typically 300–400 μm in diameter), similar to those that are used for optogenetics (Sparta et al., 2011), to excite the GECI and record the resulting fluorescence (Adelsberger, Garaschuk, & Konnerth, 2005; Cui et al., 2013, 2014; Gunaydin et al., 2014; Kudo et al., 1992; Kupferschmidt, Juczewski, Cui, Johnson, & Lovinger, 2017) (Fig. 1A–D). Because these fibers “scramble” the photons that are collected, spatial information is lost, and only “bulk” fluorescence arising from the entire GECI-expressing population under the fiber is collected, as opposed to an image that is obtained with traditional fluorescence microscopy. This population-based assessment thus is presumed to reflect the summed neural activity within the entire transduced population (for comparison of in vivo electrophysiology data to photometry signal, see London et al., 2018). A major advantage of this approach is the relative ease of implementation and analysis, resulting in low attrition rates and reduced need for computational approaches. For example, fiber placements that are “in the neighborhood” of the GECI-expressing population are sufficient to obtain a signal, while approaches with higher spatial resolution require that the sample be at an ideal focal distance from the probe (note that within-subject comparisons may be most appropriate due to variations in signal intensity due to fiber placement). Furthermore, excitation and photon collection are typically achieved through LEDs and a photoreceiver, respectively, and does not require an objective (Fig. 1A), making the setup inexpensive compared to traditional microscopy. Because the resultant data are 2-dimensional (fluorescence × time) (Fig. 1D), data storage requirements are low, and analysis is less computationally demanding than cellular resolution recordings, which result in high-dimensional data sets (fluorescence × space × time). This approach can also be used to record from terminal fields, where calcium activity tracks neurotransmitter release (Parker et al., 2016) with relative ease (Barker et al., 2017; Li et al., 2017; Menegas, Babayan, Uchida, & Watabe-Uchida, 2017) (single-axon recordings, as opposed to population assessments, are possible with higher resolution systems, but are prohibitively difficult in deep-brain areas, and require head fixation [Howe & Dombeck, 2016; Kuchibhotla et al., 2017]). Due to the relatively small footprint of the fiber-optic on the skull surface (the fiber is typically housed in a 1.25- or 2.5-μm diameter ferrule), several probes can be implanted in one animal, allowing for simultaneous multi-region imaging (Adelsberger, Zainos, Alvarez, Romo, & Konnerth, 2014; Calipari et al., 2017; Guo et al., 2015; Kim et al., 2016) (Fig. 1E–H). The small size of the probe and optic patch cable in fiber photometry also allows for relative ease of movement of the animal compared to other approaches, such as head-mounted microendoscopes (discussed below), which are both heavier and larger. Finally, bulk fluorescence recordings utilize sensors with fast sample rates (in the kilohertz range). While the kinetics of currently available GECIs do not necessitate fast sampling, this may become more relevant as faster indicators are developed (Marshall et al., 2016).
However, there are also several drawbacks to fiber photometry as compared to approaches that provide cellular resolution. First, heterogeneity of response profiles within the population of interest are inherently lost as a consequence of reduced spatial resolution. Further, because photons reach the sensor through a fiber-optic patch cable connecting the animal to the recording system (Fig. 1B and F), noise can be introduced as a function of fiber bending during movement. Because cells are not actually visualized, it can be difficult to ascertain when fiber-bending noise, tissue autofluorescence (i.e. calcium-independent fluorescence), or other factors may be causing artifactual signal. Recently, tetherless photometry systems have been developed, where the sensor is mounted directly on the animal’s head. This precludes the need for a patch cable, allows for fully unconstrained movement, and may help to minimize movement-related artifacts introduced at the level of the patch cable (Lu et al., 2018). Wireless technology could also have significant advantages when attempting to combine calcium imaging with intravenous alcohol or drug self-administration by bypassing the need for a dual tethering system, as these behavioral approaches already require that the animal be tethered to a fluid delivery system (Oleson & Roberts, 2012; Smith & Davis, 1974).
Isosbestic control channel
As mentioned above, animal movement and tissue autofluorescence can produce noise, and because the cells are not visualized, it can be difficult or impossible to identify when noise is introduced. This is a serious concern, because transients produced by fiber bending or other noise sources can easily be attributed to neuronal activity, leading to spurious conclusions. To combat this, a control channel, where the excitation wavelength is matched to the isosbestic point of the GECI, is often implemented. An isosbestic point is a light excitation wavelength at which the photon absorbance of the sample (e.g. a GECI) does not change due to chemical or physical reactions occurring within the sample (e.g. reactions with calcium). In other words, when a GECI is excited with a wavelength close to the isosbestic point, the resulting fluorescence intensity is independent of calcium concentration present (i.e. calcium-independent fluorescence). GCaMP, for example, emits low amplitude, calcium-independent fluorescence in response to near-ultraviolet excitation (Barnett, Hughes, & Drobizhev, 2017; Tian et al., 2009). A typical approach to estimating noise is to intermittently pulse 400-nm light through the patch cable; the resulting signal is presumed to be independent of calcium activity, and is subtracted from, or compared to, the 470 nm-evoked calcium-dependent signal (Kim et al., 2016; Lerner et al., 2015).
There are several caveats that should be considered when implementing an isosbestic control channel to remove noise from fiber-based imaging systems. First, the amplitude of noise-related events is typically smaller in the 400-nm channel as compared to the 470-nm channel. This is often dealt with by scaling the isosbestic values post hoc to fit the 470-nm values (Kim et al., 2016; Lerner et al., 2015). Further, while ~400-nm excitation has been used as an isosbestic control in most photometry studies to date, determination of the precise isosbestic point can be ambiguous, and can shift depending on intracellular pH (Barnett et al., 2017). This is particularly concerning because some drugs of abuse, such as amphetamine, can dramatically alter intracellular pH (Sulzer, 2011; Sulzer & Rayport, 1990). It is also possible that 400 nm- and 470 nm-evoked signals suffer phototoxicity/photobleaching (i.e., the loss of fluorescence intensity as a function of light exposure) at different rates; if photobleaching occurs at a different rate across the imaging session it may further complicate analysis and interpretation. The complexity of the interactions between calcium concentration and GECI fluorescence argues for careful interpretation and against overreliance on an isosbestic channel as an absolute assessment of noise in fiber-based imaging systems.
In regard to reporting data obtained with these approaches, some studies have displayed activity traces where the values obtained from isosbestic excitation (often referred to as the “reference” channel) have already been scaled and subtracted or regressed from the values obtained from excitation in the calcium-sensitive range of the action spectra (often referred to as the “signal” channel). Others have displayed the activity traces from the reference and signal channels separately. We propose that the latter approach should be implemented for ease of interpretation. If only a single trace is shown, it is not possible to determine the contribution of each channel to the signal. For example, if there were a negative transient in the reference channel and no change in the signal channel, a subtracted trace would show a positive transient. Consider then, a presented trace that has had noise removed by subtraction of the reference channel. A positive transient could be a result of 1) a large decrease in the reference occurring simultaneously with a small decrease, no change, or an increase in the signal channel, or 2) a decrease, no change or a small increase in the reference channel occurring simultaneously with a large increase in the signal channel. Given the aforementioned limitations of the isosbestic control channel, it would be useful for the reader to distinguish between these scenarios.
While an isosbestic control channel should be implemented whenever possible, and can certainly help to avoid spurious conclusions, it may also be necessary in some cases to implement a calcium-insensitive fluorophore group as a negative control. This control consists of transducing the population of interest with a calcium-insensitive fluorophore with similar action spectra to that of the GECI utilized (e.g., GFP instead of GCaMP), and having the animal perform an identical behavioral task (Cui et al., 2013). If the observed signal is due to fiber bending or tissue autofluorescence, it should also be observed when only a calcium-insensitive fluorophore is present; conversely, if the signal is indeed reflective of calcium activity, it should not be observed when the GECI is not expressed. This control should be implemented if there is any ambiguity in the data obtained using the isosbestic control approach, or in cases where the imaging system does not have an isosbestic channel.
Head-mounted microendoscopes
Historically, visualizing GECIs with enough resolution to assess activity in single-cells required the use of a full-size, tabletop microscope, which precluded the use of this approach in freely moving animals. Instead, recordings were performed in anesthetized preparations or awake animals under head-restraint such that the animal was immobilized and positioned under the microscope objective. A major advance in the calcium imaging field has been the recent development of miniaturized microendoscopes, which are head-mountable and lightweight (~2 g), allowing for cellular resolution imaging during free behavior, even in small model organisms such as mice (Fig. 1I–L). These miniature microscopes typically contain both the excitation source and sensor (often a complementary metal-oxide semiconductor [CMOS] camera) within the head-mounted device, and digitized data are transmitted via a lightweight cable to a data acquisition box with a computer interface for real-time visualization and storage (Fig. 1I) (Flusberg, Jung, Cocker, Anderson, & Schnitzer, 2005; Flusberg et al., 2008). The combination of head-mountable microscopes with chronically implanted GRIN lenses (Fig. 1K) has allowed for deep-brain cellular resolution imaging of calcium dynamics in freely moving mice, rats (Helmchen, Denk, & Kerr, 2013; Sawinski et al., 2009), and songbirds (Markowitz et al., 2015; Roberts et al., 2017), and efforts are underway for imaging in non-human primates (O’Shea et al., 2017). The primary drawback of this approach is that the axial resolution of these imaging systems is poor, due to the fact that there is no mechanism for rejecting fluorescence arising from outside of the focal plane. This results in fluorescence arising from cells above and below the focal plane, as well as from the axons and dendrites of neighboring cells that may contaminate signals within the focal plane (collectively referred to as ‘neuropil contamination’); thus, to accurately determine activity within single cells, computational approaches must be implemented to denoise the data. These approaches are discussed in the Extraction of calcium signals Section, below.
Two-photon microscopy
Traditional fluorescence light microscopy, including the approaches discussed above, utilizes one-photon light sources to excite fluorophores. When one-photon excitation light is used, the sample is exposed to high-energy light whereby any single photon is capable of exciting a fluorophore; because excitation light scatters throughout the tissue, this results in fluorescence emission from areas of the sample that are outside of the focal plane, and can produce high background and out-of-focus signals, as discussed above. There are many approaches to limiting fluorescence detection to a thin focal plane of interest, including two-photon excitation. Two-photon excitation occurs when two photons excite a single molecule simultaneously. This requires generation of ultrafast pulses of excitation light, which are typically achieved through a high-powered infrared laser, pulsed into the sample on a femtosecond time scale. Photon energy is inversely proportional to wavelength; thus, at longer wavelengths, any single photon does not contain enough energy to excite a fluorophore; however, when two photons converge, they combine to excite the fluorophore and produce fluorescence. Probabilistically, convergence, and thus fluorophore excitation, only occurs at the very center of the laser focal point, thereby minimizing out-of-plane fluorescence and allowing for optical sectioning of the sample. By rapidly scanning the focal point of the laser across the sample, fluorescence is obtained point-by-point across the field of view, and because emitted fluorescence is from a known point in space, an image can be constructed. Thus, two-photon microscopy allows for high contrast imaging within a spatially restricted axial plane. Because longer wavelengths of light are also less susceptible to light scattering in tissue, spatial resolution achieved with two-photon microscopy is relatively insensitive to thickness of the sample, making this approach ideal for in vivo imaging. Further, because the focal point of the laser only dwells on, and excites, any given area of the sample for a few microseconds per collected frame, as opposed to one-photon microscopy where light exposure continuously excites large portions of the sample, photobleaching is minimized.
Until recently, the majority of in vivo calcium imaging studies utilized two-photon microscopy. Prior to the application of GRIN lenses in neuroscience, experiments largely focused on superficial cortical areas. More recently, two-photon imaging has been performed through GRIN lenses to access deep-brain regions (microendoscopic two-photon imaging) (Bocarsly et al., 2015; Calhoon et al., 2018; McHenry et al., 2017; Sato et al., 2017). The application of two-photon microscopy for GECI imaging provides great advantages, such as high resolution and contrast, which improve SNR, as well as minimal photobleaching over time. Because two-photon imaging allows for optical sectioning, the denoising algorithms that are typically required for one-photon imaging are often not necessary. An additional advantage is that two-photon excitation has high tissue and bone penetrance, allowing for more effective imaging of superficial areas of the brain with minimally invasive methods, such as transcranial imaging (Grutzendler, Yang, Pan, Parkhurst, & Gan, 2011).
However, this comes with several drawbacks as compared to the single-photon approaches. First, the equipment necessary for two-photon microscopy is much more expensive, and also much larger and heavier. This precludes head-mounted systems, and requires that the subject be immobilized under the microscope, either by anesthesia or in an awake head-fixed preparation (but see Helmchen et al., 2001, 2013). Additionally, because the laser must be scanned across the tissue, only a relatively small field of view can be assessed without significant loss of temporal resolution (but see Song et al., 2017).
A major hurdle in the application of two-photon calcium imaging to preclinical addiction models will be the adaptation of alcohol and drug self-administration procedures for use in head-fixed preparations. Given that drug reinforcement is highly context- and internal state-dependent, it is unclear what the behavioral effects of these compounds will be under such non-ethological conditions. Indeed, many abused drugs, which are canonically thought of as rewarding and reinforcing stimuli, can have aversive properties and function as punishers/stressors depending on timing, context, and dosing (Bormann & Cunningham, 1998; Risinger & Oakes, 1995; Samson & Files, 1998). It is possible that these concerns could be circumvented by fully habituating the animals to head fixation before recordings are obtained. Nonetheless, it is currently unclear how the behavioral actions of alcohol and other drugs of abuse will manifest them-selves during head restraint, and therefore extensive characterization will be required before head-fixed alcohol/drug self-administration can be considered as a viable addiction model in rodents.
Despite the possible limitations and interpretational challenges of administering alcohol and drugs in a head-fixed animal, there is still immediate and extensive utility of head-fixed two-photon imaging for exploring the pathology of addiction. A wide array of behavioral paradigms has already been adapted for use with rodents in head-fixed setups, including Pavlovian (Beyeler et al., 2016; Eshel et al., 2015; Heiney, Wohl, Chettih, Ruffolo, & Medina, 2014) and operant (Harvey, Bermejo, & Zeigler, 2001; Isomura, Harukuni, Takekawa, Aizawa, & Fukai, 2009) conditioning for non-drug stimuli, and spatial navigation tasks (Harvey, Collman, Dombeck, & Tank, 2009), among others (Guo et al., 2014; Schwarz et al., 2010). Given that drug-induced dysregulation of basic learning processes and fundamental behaviors are major sequelae of addiction, interrogation of the neural circuits mediating these processes in animals with a history of drug exposure would provide important insight into abberant circuit activity during drug-free periods.
“All-optical” simultaneous observation and manipulation of neural circuits
A long-standing promise of optical circuit dissection techniques is the ability to simultaneously manipulate and record from neural populations through a combination of indicators and opsins. This “all-optical” approach to circuit interrogation is now tractable, and holds enormous potential for not only unraveling the endogenous activity of circuits, but also assessing their causal relationship with behavioral outputs (Fig. 3). While simultaneous use of green GECIs, such as GCaMP, and a blue opsin, such as ChR2, is not possible due to the fact that they are both excited by similar wavelengths of light, red-shifted opsins allow for the possibility of both recording and manipulating cell-type specific populations within the same animal (Fig. 3C) (Klapoetke et al., 2014). For example, transduction of red-shifted opsins such as Chrimson (Klapoetke et al., 2014) or hal-orhodopsin (Zhang et al., 2007) in Region 1, combined with GECI expression in Region 2 that receives innervation from Region 1, would allow for temporally precise excitation or inhibition of local opsin-expressing terminals via application of red-shifted light through the GRIN lens or optic fiber, while simultaneously assessing the activity of downstream cells (Fig. 3A) (for review see Emiliani, Cohen, Deisseroth, & Hausser, 2015). This powerful approach to interrogating communication between circuits and the resultant effects on behavior, which could be used with any of the calcium imaging approaches discussed above, has recently been applied (Akerboom et al., 2013; Kim et al., 2016; Nieh et al., 2016; Packer, Russell, Dalgleish, & Hausser, 2015; Rickgauer & Tank, 2009). Another approach that holds great promise is the development of red-shifted calcium indicators, such as RCaMP and R-GECO, which, combined with a green GECI, allow for simultaneous assessment of activity in multiple cell-type defined populations (Dana et al., 2016; Inoue et al., 2015) (Fig. 3B).
Fig. 3. Example of approaches for all-optical circuit dissection.
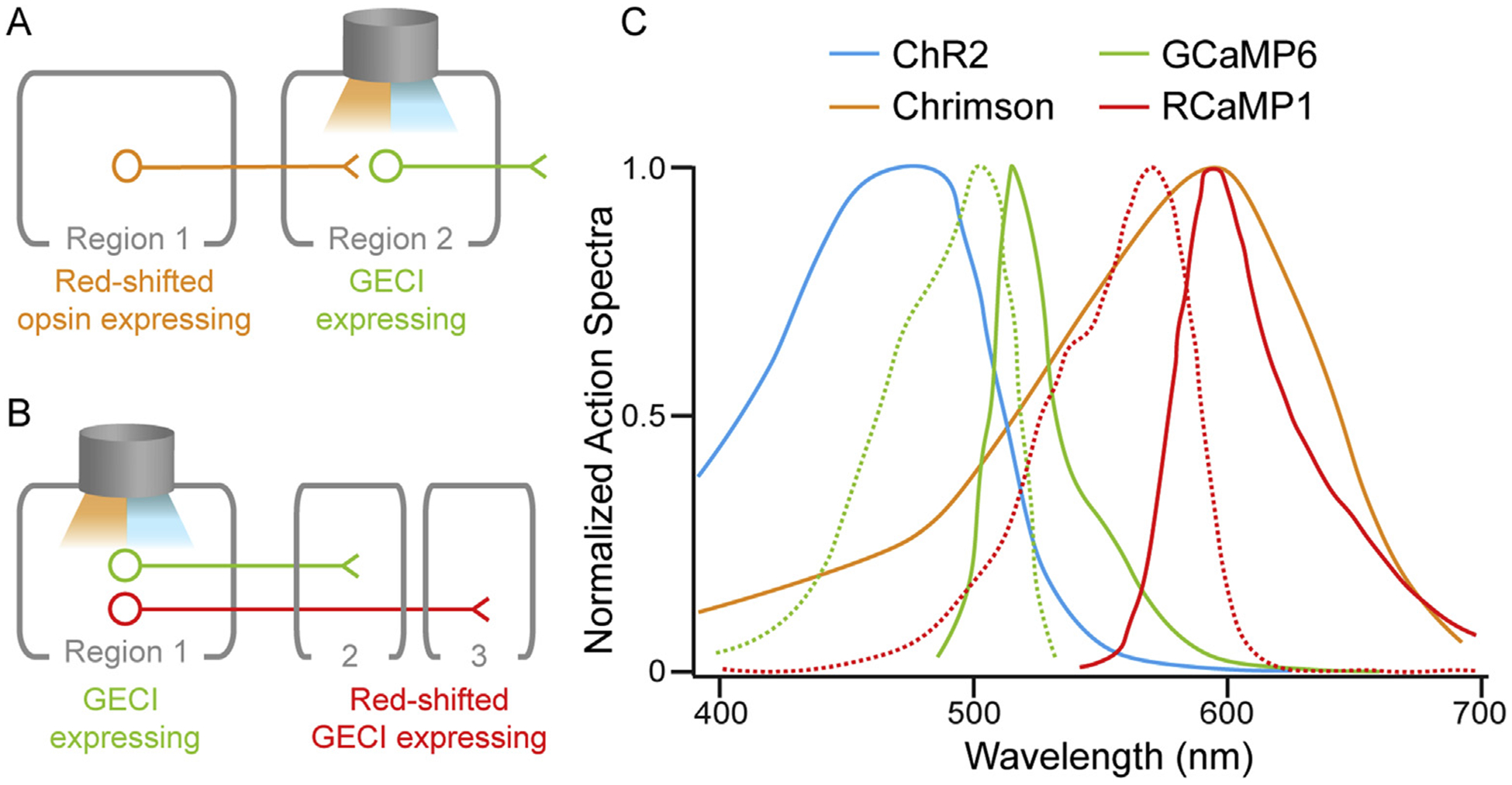
With the addition of a red excitation source, any of the microscopy approaches outlined in Fig. 1 can be used to simultaneously record from and/or activate multiple defined populations. (A) Viral transduction of a red-shifted opsin into upstream Region 1 and a GECI into downstream Region 2 allows for simultaneous recording and manipulation of defined circuits. This approach can be implemented through an optical fiber or GRIN lens to allow for simultaneous delivery of ~470 nm- (GECI excitation) and ~600-nm (red-shifted opsin excitation) light into deep-brain areas. (B) Viral-mediated delivery of green and red GECIs allows for simultaneous assessment of two anatomically distinct neuronal populations within the same brain region. Similar strategies could be used to assess genetically distinct populations. (C) Normalized action spectra as a function of wavelength for commonly used opsins and GECIs. Dotted and solid lines represent excitation and emission spectra, respectively, for GCaMP6 and RCaMP1. Note that one-photon GECI excitation is typically achieved with light powers that are approximately an order of magnitude lower than opsin excitation, which allows for simultaneous use of GCaMP6 and Chrimson with minimal cross-talk, despite somewhat overlapping spectra. Action spectra were adapted from Klapoetke et al. (2014) and Shigetomi et al. (2016).
Analysis of GECI imaging data
For the cellular-resolution approaches described above (i.e., microendoscope and two-photon imaging), data collection will result in a video, and the next step is to extract calcium signals (this step is not required for fiber photometry). Analysis of GECI imaging data can generally be divided into two categories: 1) extraction of fluorescence activity from the acquired videos, and 2) visualization/presentation and statistical analysis of the extracted values. The first category will vary greatly depending on the type of imaging system used to obtain the data, while the second is often unaffected by the acquisition method, but will vary depending on the experimental question. Here we will broadly discuss common approaches for these types of analysis.
Extraction of calcium signals
Before extraction of calcium activity, videos are often down-sampled, spatially and/or temporally, to reduce the file size and processing times for the subsequent steps. Videos are then motion-corrected to remove movement artifacts that occur during behavior. This can be achieved with a variety of software packages, many of which are freely available (Kaifosh, Zaremba, Danielson, & Losonczy, 2014; Pnevmatikakis & Giovannucci, 2017). After videos are preprocessed, there are many options for extracting single cell calcium dynamics. The most straightforward approach is to simply define a region of interest (ROI) by selecting an area encompassing a single cell and averaging the pixel intensity within the ROI to produce a “brightness over time” trace. ROIs can be manually drawn around cells, or determined using automated methods (Ozden, Lee, Sullivan, & Wang, 2008; Patel, Man, Firestein, & Meaney, 2015). Using an ROI analysis, without any correction for background noise, assumes that fluorescence within the ROI is not contaminated by fluorescence from cells above or below the focal plane, or from axons or dendrites of neighboring cells intersecting the ROI. The probability of significant neuropil contamination increases with lower-resolution imaging systems, and when GECI expressing cells are densely packed in the field of view. Depending on the degree of contamination, denoising algorithms may need to be implemented to extract accurate and reliable calcium dynamics. Contamination is particularly prevelant when imaging with head-mounted one-photon microendoscopes, because size and weight restrictions preclude the use of methods for optimizing resolution. Methods for removing noise from one-photon microendoscope recordings are rapidly changing, and the optimal approach for resolving this issue is a highly debated topic.
While head-mounted microendoscopes allow for a powerful combination of single-cell visualization and minimally restricted behavior, the lack of optical sectioning, and high levels of light scattering with one-photon excitation light result in fluorescence from cells that are out of the focal plane, emitting light, which is subsequently captured by the detector. This produces large “background” fluorescent fluctuations, as well as cells that appear overlapping in the field of view, which may result in artefactual neuropil contamination and “cross-talk” of extracted neuronal signals. To address this issue, analysis of head-mounted microscope data must employ computational methods for removing out-of-plane fluorescence (Resendez et al., 2016). There are several approaches available for denoising microendoscopic data, which we briefly describe here. ROI-based approaches, described above, have been adapted to include removal of background noise. In this approach, after raw pixel intensity is determined, contributions from background fluctuations or neighboring cells are estimated, and subtracted or regressed from the values obtained. Methods for estimating background noise vary; examples include averaging pixel intensities from the entire field of view as a proxy for background fluctuations, or in a small doughnut-shaped area around the outside of each ROI. However, these methods may also introduce artefactual transients. For example, if fluorescence resulting from in-focus cells is included in the estimation of background noise, subtraction of those values would produce a downward transient at times when the neighboring cells are active and the cell within the ROI is not; similarly, if both the cell in the ROI and a cell included in the noise estimate are simultaneously active, the extracted trace may spuriously appear flat.
To circumvent signal cross-talk that can be present in ROI analyses, principle component analysis (PCA) and independent component analysis (ICA) have been used to extract spatiotemporal profiles of neuronal activity from microendoscopic data (Mukamel et al., 2009). This method effectively identifies cell location, and isolates its activity profile; however, this approach can also result in cells being split into multiple components, because the algorithm contains no information regarding the spatial or morphological extent of a cell. Further, PCA/ICA is highly sensitive to experimenter-determined input parameters, and can suffer from poor inter-experimenter reliability. A recently developed constrained non-negative matrix factorization (CNMF) approach adapted for use with microendoscope data (CNMF-E) estimates and removes background fluctuations and noise from these data sets, and may have improved quality as compared to PCA/ICA approaches (Pnevmatikakis et al., 2016; Zhou et al., 2018). However, this approach imposes assumptions of sparse neural activity, which may not be true in all neural populations. A second assumption is that negative values are artefactual and are removed (i.e., constrained to non-negative values) (Zhou et al., 2018). GECI fluorescence faithfully tracks decreases/pauses in action potential activity, as inferred from anesthetic-induced silencing of activity in vivo (Kupferschmidt et al., 2017) and between-animal comparisons of electrophysiological and photometry data sets (London et al., 2018), or compared directly with electrical activity ex vivo via whole-cell patch-clamp recordings of GECI-expressing cells (Otis et al., 2017); thus, removal of negative transients may be erroneous.
All of the analysis methods described above have been widely implemented to extract calcium dynamics from one-photon microendoscopic imaging data (Cai et al., 2016; Cox, Pinto, & Dan, 2016; Grewe et al., 2017; Jennings et al., 2015; Murugan et al., 2017; Pinto & Dan, 2015; Yu et al., 2017), and the field has yet to reach a clear consensus as to which approach most accurately separates the activity of single cells from background noise. The lack of consensus largely results from the fact that there is no ground truth to validate the different analysis methods. A ground truth validation would require simultaneous cell-attached electrophysiological recording during imaging. This has been achieved in superficial brain regions that can be imaged without the use of GRIN lenses (Chen et al., 2013; Grewe, Langer, Kasper, Kampa, & Helmchen, 2010); however, there is currently no method for gaining simultaneous cellular resolution optical recordings and cell-attached electrical recordings from cells deep in the brain that are obstructed by the implanted lens. While simultaneous recordings under these conditions have not yet been achieved, Vander Weele et al., 2018 performed a side-by-side comparison of neural activity in projection-defined populations as measure by both in vivo cellular resolution calcium imaging and electrophysiological recordings in photo identified neurons. Ground truth validation of these analyses, and a consensus on their use, is a critical and ongoing challenge in the field. For the time being, we propose that it is best practice to report results from multiple analysis methods to 1) contrast discrepancies, and 2) limit conclusions to findings that are insensitive to methodology.
Data visualization and quantification
Following extraction of calcium activity traces from imaging videos, these data can then be analyzed to address the experimental question at hand. Values of fluorescence intensity extracted from each cell, using one of the methods described above, are often in arbitrary units; thus, these values are typically converted into a more readily interpretable unit system. The most common unit system in the extant literature is ΔF/F, where raw extracted values are converted to be expressed as a change in fluorescence over the average fluorescence of a given time period. The simplest and most common variant of this approach is to normalize the signal to the average fluorescence of the entire recording [commonly used equation: (F−F0)/F0, where F is the value of a given sample in the recording and F0 is the average of all values across the entire recording; this calculation is repeated for each sample (F) in the recording]. Thus, fluorescence intensity at each point in time is expressed as a change from the average intensity over the entire recording. Many variants of the ΔF/F conversion have been implemented. For example, the value to which activity is normalized (F0) can be a sliding window immediately prior to a behavioral event, or an average of the subset of values across the record session. A similar approach is to normalize values using a z-score transformation where values are expressed in units of standard deviations from the mean of the recording [commonly used equation: z = (x − mean)/standard deviation]. Again, with this approach values can be normalized to a pre-event window or to the entire recording. With any method of normalizing the reported units, care should be taken to ensure that results are consistent across approaches, and are insensitive to experimenter-determined parameters.
Once traces have been extracted and normalized, information related to neural activity can be evaluated. The two most common ways to assess changes in neural activity, either between or within animals, are to 1) determine the rate of transient events across a period of time, or 2) assess stimulus-evoked activity. The first approach, often referred to as event detection, typically involves setting a threshold that defines the occurrence of a calcium transient (e.g., when values are greater than 3 z scores) and applying it to each activity trace; each time the threshold is crossed, a transient event is counted, and results are reported as number of transients over time (e.g., events/minute). This approach is particularly useful for assessing changes in activity between different contexts or internal states, where activity may not be time-locked to a particular behavioral event. An example of how this type of analysis is leveraged is comparing the event rate occurring on two sides of a behavioral apparatus, where one side has a food reward, and the other does not (Jennings et al., 2015), or comparing event rate across days when animals are either food-deprived or sated (Calhoon et al., 2018).
The second approach is to align activity traces around a specific stimulus or behavioral action, such as onset of a reward-predictive tone, or initiation of a reward-paired lever (i.e., a peri-stimulus time histogram, which is commonly used in the in vivo electrophysiology literature) (Carter & Shieh, 2015). In this case, it can be determined whether there is a neural signal associated with particular behavioral events. This approach is most powerful when many trials (instances of the behavioral event of interest) can be recorded, to determine whether the signal is reliably reproduced. When this analysis is implemented with cellular resolution imaging, population responses can be assessed (e.g., x number of cells are responsive to tone A, y number of cells are responsive to tone B) (e.g., Livneh et al., 2017). An example of the way a peri-stimulus time histograms are used is comparing the neural response across trials when animals are exposed to drug-paired cues as opposed to neutral cues (Calipari et al., 2016; Xia et al., 2017). While these commonly implemented approaches are highly useful and are a great starting point for analyses, experiments often call for more specialized and complex analyses, which must be dealt with on a case-by-case basis.
Limitations and caveats
There are limitations and caveats that are, at least with the current technology, applicable to all types of GECI imaging (issues specific to certain approaches are discussed in the appropriate sections above). Importantly, changes in intracellular calcium concentration are a proxy for action potential activity rather than a direct measure. Because GECI kinetics are slower than the timescale on which action potentials are generated, baseline activity of cells cannot typically be resolved. For example, cells in prelimbic cortex spontaneously fire 1–8 times per second (Garcia, Vouimba, Baudry, & Thompson, 1999), but GECI recordings from cells in this region show transient event rates occurring only every 15–30 s (Otis et al., 2017). It is likely that calcium transients observed in vivo reflect changes in activity from baseline, such as bursting events, rather than absolute levels of spiking activity, and attempts to infer spiking activity from GECI fluorescence often have substantial error rates (Harris et al., 2016; Theis et al., 2016). Further, while it is well-established that GECI fluorescence is primarily action potential-dependent, calcium is a critically important signaling molecule, both intra- and extra-cellularly, and is released by a multitude of factors other than action potentials (Brown & MacLeod, 2001; Ghosh & Greenberg, 1995). Because calcium is important in many facets of cell function other than action potential generation, this raises concerns that GECIs, by virtue of binding calcium and essentially acting as a calcium buffer, could disrupt endogenous calcium signaling, thereby altering normal cellular function. Indeed, there are several examples of GECI expression altering cellular activity, which could be a function of GECI interference with calcium handling, as well as neurotoxicity that can be associated with viral delivery of transgenes. For example, Kupferschmidt et al. (2017) observed augmented current step elicited action potentials in GECI-expressing cells as compared to non-expressing neighbors, suggesting that GECI expression increased intrinsic excitability. Similarly, aberrant epileptiform activity has been observed in several GECI-expressing transgenic mouse lines (Steinmetz et al., 2017). It is best practice to attempt to diminish these concerns by limiting viral expression time and titer, and controls should be performed to assess potential neurotoxicity (e.g., Kupferschmidt et al., 2017). Because of the potential of GECI expression to alter cellular function, it is also critical that expression time of the GECI (i.e., time post virus injection) is matched when making between-group comparisons.
Tissue damage is also a major issue in calcium imaging, which is currently unavoidable for deep-brain imaging. Even optic fiber-based approaches, which are the least invasive method of accessing deep-brain areas, typically use a 300–400 μm diameter probe. GRIN lenses, which allow for cellular resolution, typically have a diameter of 500–2000 μm, and thus result in a very high volume of tissue displacement, roughly comparable to guide cannulae used for site-specific pharmacological interventions or microdialysis. For example, cellular resolution recordings from deep-brain structures in mice, such as the basolateral amygdala or medial preoptic area, have been achieved via insertion of a 600-mm diameter GRIN lens ~3.5–4.5 mm ventral from the surface of the brain (Grewe et al., 2017; McHenry et al., 2017). Gaining optical access to a region 4.5 mm below the brain surface with a 600-μm diameter GRIN lens produces a total volume of tissue displacement of 1.27 mm3. For comparison, insertion of standard 16-wire array for electrical recordings (30-μm diameter per wire) at the same depth, results in only 0.01 mm3 of total tissue displacement. In addition, it is important to consider which inputs to the region of interest may be severed by the lens or fiber. Prism lenses or mirror-tipped fibers can be used to image the region from a medial or lateral view, rather than a dorsal vantage point that is obtained with a typical probe, giving the experimenter some options as to which adjacent areas will be lesioned by the implant. While destruction of the areas that the lens passes through is certain, with such a large displacement, increased cranial pressure and tissue compaction resulting in damage to adjacent areas is also a concerning possibility. Tissue aspiration prior to lens insertion has been used in an attempt to mitigate these concerns (Resendez et al., 2016).
Another major caveat that could arise when using GECI imaging in drug self-administration studies is that many abused compounds can have direct pharmacological interactions with calcium channels and other ionotropic receptors that are calcium permeable. The premise of using calcium imaging as a proxy for neuronal activity relies on the reliable relationship between neuronal spiking and calcium influx; thus, if this relationship is altered it could produce spurious results. Abused drugs that interact directly with calcium channels or other ionotropic receptors that pass significant amounts of calcium include alcohol (Walter & Messing, 1999), nicotine (Mulle, Choquet, Korn, & Changeux, 1992), morphine (Yang, Shan, Ng, & Pang, 2000), and cocaine (Damaj, Slemmer, Carroll, & Martin, 1999; Francis, Vazquez, Papke, & Oswald, 2000). It is also possible that drugs of abuse could indirectly affect calcium handling. For example, most abused drugs increase extracellular dopamine concentrations in the striatum, and thus increase dopamine receptor activation (Di Chiara & Imperato, 1988; Young, Porrino, & Iadarola, 1991). Because dopamine receptors can alter calcium currents without producing spikes per se (Beaulieu & Gainetdinov, 2011; Hernández-López, Bargas, Surmeier, Reyes, & Galarraga, 1997; Surmeier, Bargas, Hemmings, Nairn, & Greengard, 1995), it is possible that even drugs that do not directly interact with calcium-permeable ion channels could still alter the relationship of calcium-to-spikes via indirect agonism of metabotropic receptors or interactions with signaling cascades involved in calcium handling. In situations where recordings are obtained while a drug is “on board”, it is critical that ex vivo electrophysiology validation experiments are performed to ensure that the relationship between action potentials and calcium activity is preserved in the presence of drug.
Implementation of calcium imaging in alcohol and drug abuse models
Task design
As discussed above, GECI imaging is a powerful approach that promises to provide great insight into the neurobiology of addiction. However, there are several considerations that should be taken into account when implementing this approach with alcohol and drug abuse models. One major practical concern is that all fluorescent molecules, including GECIs, are subject to photobleaching, which limits the amount of time a GECI can be imaged continuously. Doses of alcohol or drugs of abuse typically used in drug self-administration studies often engender high inter-response intervals (Lin, Pierce, Light, & Hayar, 2013; Vandaele, Cantin, Serre, Vouillac-Mendoza, & Ahmed, 2016), and can require many consumption bouts before pharmacologically relevant blood/brain concentrations of the drug are reached (Wilcox et al., 2014). Thus, tasks will need to be adapted to be compatible with the recording time limitations of GECI imaging. For example, a commonly used binge drinking model is the two-bottle choice procedure, where animals are given access to alcohol for 2 h or more (Griffin, Lopez, & Becker, 2009; Thiele & Navarro, 2014). Consumption of alcohol during the task is volitional, and the times at which the animal will choose to consume alcohol are not easily predictable. Unfortunately, due to photobleaching, recording continuously throughout a 2-h session with one-photon microscopy can be problematic. Thus, approaches must be designed where the animal will administer the drug at predictable times so that imaging can be restricted to experimenter-determined epochs.
Although it is difficult to determine exactly when animals will consume alcohol when given free access, they often exhibit “front-loading” behavior where a large portion of the total alcohol consumption occurs in the beginning of the access period (Barkley-Levenson & Crabbe, 2012; Griffin, Lopez, Yanke, Middaugh, & Becker, 2009; Rhodes et al., 2007; Wilcox et al., 2014). This front loading behavior is observed with other abused drugs as well (Ettenberg, Pettit, Bloom, & Koob, 1982; Tornatzky & Miczek, 2000), and may provide an opportunity to capture neural activity during several drug consumption bouts within a short period of time. Another solution for avoiding photobleaching is to utilize tasks that have a trial-based structure. A task that has already been used widely in the alcohol field that is readily adaptable for GECI imaging experiments is operant tasks in which the operant response results in an access period to an alcohol sipper (Hopf, Chang, Sparta, Bowers, & Bonci, 2010; Lopez & Becker, 2014; Robinson & McCool, 2015). In this case, the experimenter could simply trigger the onset of the GECI excitation light concomitantly with the operant response, with a high degree of confidence that the animal would consume alcohol within a short period of time, and terminate the excitation light after access to alcohol is removed. With this method, the experimenter could capture neural activity during many separate instances of volitional alcohol consumption over several hours while limiting the imaging time to a fraction of real-time, thereby avoiding excessive photobleaching. Similar approaches using discriminative or conditioned stimuli could be used to restrict recording times to periods in the task where alcohol or drug consumption is likely, thus minimizing photobleaching.
Timing of stimulus presentation must also be considered when designing drug self-administration tasks for calcium recordings. While timing is important in any in vivo recording, it is particularly tricky with calcium imaging due to the relatively low temporal resolution of GECIs. Indeed, calcium transients often take 1–10 s to resolve completely; thus, if multiple behavioral events occur in the window it is difficult to determine which event the transient is referencing. Take, for example, a typical operant intravenous drug self-administration procedure. Generally, in these tasks, initiation of an active operandum results in activation of a cue light or tone, concomitant with delivery of drug through an intravenous catheter (Siciliano, Ferris, & Jones, 2015; Smith & Davis, 1974). The observation of a calcium transient time-locked to the lever press would be difficult to interpret: it is not clear whether the transient is related to the motoric action of pressing the lever, the presentation of the cue light, internal expectation of drug delivery, or the delivery of the drug. Thus, alcohol and drug self-administration procedures will need to be adapted such that behavioral events are spaced sufficiently to resolve slow calcium events, and allow for readily interpretable results. This can be achieved by statically spacing the stimuli throughout the entire task; however, separating actions or cues from outcomes can also make task acquisition slow and increase attrition rates (Beylin et al., 2001). An alternative approach would be to include “probe” trials in which the events are probabilistically dissociated in time.
Practical concerns
When using in vivo GECI imaging to determine neuronal activity related to behavioral events, as with any in vivo recording, it is of the utmost importance that the events of interest are timestamped accurately within the neural data. The most common approach for timestamping behavioral events is to use transistor-transistor logic (TTL) as a means of communication between the behavioral apparatus and the imaging system. TTL signals typically consist of a binary voltage output from the behavioral apparatus; for example, when a lever is pressed the output will change from 0 V to 5 V. The most common method for transmitting TTL signals is a Bayonet Neill-Concelman (BNC) connector, which is a coaxial cable hookup featured in most behavioral systems. By connecting a BNC cable from the behavioral apparatus to the imaging system, changes in voltage can be recorded; during analysis, the times at which these changes in voltage occur can be associated with a specific frame in the imaging data. In operant self-administration studies, a typical analysis would involve quantifying the neural signal just prior to the lever press (baseline window), and after the time of the lever press (event window) to create a peri-stimulus time histogram (discussed in the Data visualization and quantification Section, above). By examining the neural activity over many of these events, it can be statistically determined whether there is a reliable neural response associated with the lever press. Similarly, by using TTL signals to communicate between the behavioral and imaging systems, the imaging system can be turned on and off to assess activity during epochs of interest over long behavioral tasks where continuous recording may not be practical (discussed in the Task design Section, above). While most behavioral and imaging systems have BNC outputs/inputs and the necessary software for this type of use, it is prudent to inquire whether a particular system has these features when determining which systems to use. Further, any system will have a finite number of BNC connections, which should also be taken into consideration if the behavioral tasks of interest have many different events that each need to be timestamped in the neural data.
As with any approach that requires the animal to be tethered, care should be taken to habituate the animal to the experimental conditions. The additional weight on the animal’s head, as well as the torque produced by the connected wire or optic fiber, can make movement difficult, especially if the task requires the animal to fit its head into a port to collect a reward, as is typical for alcohol self-administration studies. This is exacerbated by the fact that many of the currently available head-mounted microscope and photometry systems have not yet been integrated with a eletrical or optic fiber rotary joint (a device that allows the wire or patch cable to rotate with the movement of the animal, typically referred to as a ‘commutator’), which can result in tangling of the tether. In addition to habituating animals to moving with a tether attached, it is also important that the animals be habituated to being handled by the experimenter and connected to the patch cable or microscope. This requires that the animal be briefly restrained, which can be a salient stressor and which may alter alcohol-drinking or drug-taking behaviors (Chester, de Paula Barrenha, DeMaria, & Finegan, 2006; Lynch, Kushner, Rawleigh, Fiszdon, & Carroll, 1999). Finally, many behavioral apparatuses use infrared beams arrays to assess the movement of the animal, such as to determine when the animal enters the reward port as indicated by a beam break. These beams may be detected by the imaging system resulting in artefactual signals; thus, alternative approaches may be required to record the behavioral activity of the animal. For example, electrical lickometers can be used to determine when the animal is consuming alcohol (Dole, Ho, & Gentry, 1983; Robinson & McCool, 2015; Spector, Andrews-Labenski, & Letterio, 1990), and video tracking can be used to assess general locomotion (Aguiar, Mendonca, & Galhardo, 2007; Spink, Tegelenbosch, Buma, & Noldus, 2001). Similarly, care should be taken to ensure that changes in ambient light are not detected by the imaging sensor (e.g., when the animal is crossing sides in a light-dark box, or if a house light cue is used).
Technique and platform selection
As discussed in the Utility of in vivo calcium imaging Section, careful consideration should be given when determining whether GECI imaging is the most appropriate technique for an experimental question. If this determination is made, experimenters must then choose between the many available options for achieving in vivo GECI imaging. The pros and cons of each broad category of imaging techniques are discussed in the Approaches to calcium imaging Section, but within each of these categories there are also many options regarding specifics of the setup, and where to source the equipment (for in-depth review of choosing an optimal imaging approach, see Yang & Yuste, 2017).
When selecting an imaging approach for examining circuit activity in addiction models, the first criterion that should be considered is the behavioral task that will be utilized. For example, if the task requires that animals are freely moving, two-photon imaging can be eliminated from the list of options (but see Helmchen et al., 2001, 2013). If the task requires animals to be tethered to a fluid delivery system for intravenous self-administration, a wireless imaging system would be particularly attractive. The density of the population as well as expected heterogeneity of activity within the population of interest should also be considered. For particularly sparse populations, or ones where the cells are likely to have homogeneous activity profiles, fiber photometry may be the most practical approach. Additionally, for long behavioral tasks or tasks in a restricted space such as an operant box, a system that is compatible with a commutator would be advantageous, as tangling of the wire/patch cable is likely in these situations. It should also be noted that while GRIN lenses have allowed for optical access to the deep regions of the rodent brain, the difficulty of obtaining quality recordings dramatically increases with the length of the lens. When imaging with cellular-resolution through a GRIN lens, lower yields and greater movement artifacts should be expected in deeper regions; in contrast, for approaches that collect GECI fluorescence through an optic fiber, depth has little impact on recording quality, and thus may be best for deep-brain areas, especially in rats where some regions are at depths close to the length of the longest currently available GRIN lenses.
Data storage should also be taken into consideration. Using systems with cellular-resolution, it is not uncommon to generate hundreds of gigabytes, or more, of video in a single day of data collection. In contrast, for approaches that do not record an image, such as fiber photometry, hours of recording can be stored with just a few hundred megabytes or less. Big data sets can be costly in terms of processing power and time. Similarly, a major consideration for technique selection is the amount of custom programming that may be needed to analyze big data sets. Many platforms do include software that can preclude the need for custom analysis pipelines. Data downsampling, video motion correction, spatial filtering, and trimming/cropping are functions that can easily be achieved with freely available software (Kaifosh et al., 2014; Pnevmatikakis & Giovannucci, 2017; Schneider, Rasband, & Eliceiri, 2012), and are also often included in analysis packages that are included with commercially sourced imaging systems. However, the standards for analyzing GECI imaging data are rapidly changing, and there is great variability in the experimental questions that are being addressed with these techniques, making it difficult for software packages to meet everyone’s needs. Currently, data analysis for any of the GECI imaging approaches discussed here will require custom programming in most, if not all, cases. The complexity of these analyses will vary widely depending on the approach used. Even a simple analysis such as event detection (determining the time at which transients occur) will often require customization, because the characteristics of a calcium transient can vary greatly depending on the GECI, population of interest, and imaging system. Further, when using approaches with cellular resolution, such as head-mounted microendoscopes, extraction of activity traces from background noise can be highly complex and computationally demanding. There are freely available scripts for this purpose (Inan, Erdogdu, & Schnitzer, 2017; Keemink et al., 2018; Levin-Schwartz, Sparta, Cheer, & Adali, 2017; Prada et al., 2018; Zhou et al., 2018), but again, due to the wide variations in data obtained from different regions, indicators, and imaging systems, these often require extensive optimization on an experiment-by-experiment basis. Bulk fluorescence approaches (e.g., fiber photometry) are likely the best option in cases where relatively little need for custom programming is an attractive feature.
While there are far too many options and variables to provide an exhaustive list of recommendations, researchers that have not yet attempted GECI imaging can optimize their technique and platform selection by first carefully outlining the experimental questions that they hope to address. With this information in hand, researchers can weigh the usefulness of the many options available. While examining the literature and communicating with vendors will provide a great deal of information, the greatest resource available for technique selection is others in the field that have experience with these platforms. Open communication between investigators, and sharing of knowledge and expertise is essential for avoiding common pitfalls in platform selection, data acquisition, and data analysis.
Conclusions
In summary, while the utility of calcium imaging for visualizing neuronal activity has been well described for several decades, its widespread use for in vivo deep-brain imaging to address questions in motivational neuroscience is still in its relative infancy. In the addiction field, this powerful tool has only just begun to be leveraged. As highlighted in this review, many questions remain regarding how best to implement and interpret this approach for investigating the neurobiology of addiction. Nonetheless, the rapid expansion in the accessibility and feasibility of calcium imaging will undoubtedly give rise to invaluable insights into the neurobiology of addiction. While the utility of calcium imaging cannot be over-stated, it is imperative that the caveats and limitations are considered and communicated such that optimal experimental design, data analysis, presentation, and interpretation can be achieved with continuity across studies.
Acknowledgments
C.A.S. is supported by NIH grants F32 MH111216 (NIMH) and K99 DA045103 (NIDA), and a NARSAD Young Investigator Award from the Brain and Behavior Research Foundation. K.M.T. is a New York Stem Cell Foundation - Robertson Investigator and McKnight Scholar, and this work was supported by funding from the JPB Foundation, the PIIF, PNDRF, JFDP, Alfred P. Sloan Foundation, New York Stem Cell Foundation, Klingenstein Foundation, McKnight Foundation, R01-MH102441 (NIMH), RF1-AG047661 (NIA), the NIH Director’s New Innovator Award DP2-DK102256 (NIDDK), and Pioneer Award DP1-AT009925 (NCCIH). We thank Dr. Caitlin M. Vander Weele for helpful input on preparing the figures.
References
- Adelsberger H, Garaschuk O, & Konnerth A (2005). Cortical calcium waves in resting newborn mice. Nature Neuroscience, 8, 988–990. 10.1038/nn1502. [DOI] [PubMed] [Google Scholar]
- Adelsberger H, Zainos A, Alvarez M, Romo R, & Konnerth A (2014). Local domains of motor cortical activity revealed by fiber-optic calcium recordings in behaving nonhuman primates. Proceedings of the National Academy of Sciences of the United States of America, 111, 463–468. 10.1073/pnas.1321612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar P, Mendonca L, & Galhardo V (2007). OpenControl: A free opensource software for video tracking and automated control of behavioral mazes. Journal of Neuroscience Methods, 166, 66–72. 10.1016/j.jneumeth.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Aguilar-Rivera MI, Casanova JP, Gatica RI, Quirk GJ, & Fuentealba JA (2015). Amphetamine sensitization is accompanied by an increase in prelimbic cortex activity. Neuroscience, 288, 1–9. 10.1016/j.neuroscience.2014.12.027. [DOI] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, et al. (2013). Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in Molecular Neuroscience, 6, 2 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PF, Hodgkin AL, & Ridgway EB (1971). Depolarization and calcium entry in squid giant axons. The Journal of Physiology, 218, 709–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera G, Liang B, Zhang L, Gerfen CR, Culurciello E, Chen R, et al. (2016). Spatially compact neural clusters in the dorsal striatum encode locomotion relevant information. Neuron, 92, 202–213. 10.1016/j.neuron.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Miranda-Barrientos J, Zhang S, Root DH, Wang HL, Liu B, et al. (2017). Lateral preoptic control of the lateral habenula through convergent glutamate and GABA transmission. Cell Reports, 21, 1757–1769. 10.1016/j.celrep.2017.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley-Levenson AM, & Crabbe JC (2012). Ethanol drinking microstructure of a high drinking in the dark selected mouse line. Alcoholism: Clinical and Experimental Research, 36, 1330–1339. 10.1111/j.1530-0277.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett LM, Hughes TE, & Drobizhev M (2017). Deciphering the molecular mechanism responsible for GCaMP6m’s Ca2+−dependent change in fluorescence. PLoS One, 12, e0170934 10.1371/journal.pone.0170934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, & Gainetdinov RR (2011). The physiology, signaling, and pharmacology of dopamine receptors. Pharmacological Reviews, 63, 182–217. 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- Beier KT, Kim CK, Hoerbelt P, Hung LW, Heifets BD, DeLoach KE, et al. (2017). Rabies screen reveals GPe control of cocaine-triggered plasticity. Nature, 549, 345–350. 10.1038/nature23888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyeler A, Namburi P, Glober GF, Simonnet C, Calhoon GG, Conyers GF, et al. (2016). Divergent routing of positive and negative information from the amygdala during memory retrieval. Neuron, 90, 348–361. 10.1016/j.neuron.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beylin AV, Gandhi CC, Wood GE, Talk AC, Matzel LD, & Shors TJ (2001). The role of the hippocampus in trace conditioning: Temporal discontinuity or task difficulty? Neurobiology of Learning and Memory, 76, 447–461. 10.1006/nlme.2001.4039. [DOI] [PubMed] [Google Scholar]
- Bocarsly ME, Jiang WC, Wang C, Dudman JT, Ji N, & Aponte Y (2015). Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomedical Optics Express, 6, 4546–4556. 10.1364/boe.6.004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann NM, & Cunningham CL (1998). Ethanol-induced conditioned place aversion in rats: Effect of interstimulus interval. Pharmacology, Biochemistry, and Behavior, 59, 427–432. [DOI] [PubMed] [Google Scholar]
- Broussard GJ, Liang R, & Tian L (2014). Monitoring activity in neural circuits with genetically encoded indicators. Frontiers in Molecular Neuroscience, 7, 97 10.3389/fnmol.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, & MacLeod RJ (2001). Extracellular calcium sensing and extracellular calcium signaling. Physiological Reviews, 81, 239–297. 10.1152/physrev.2001.81.1.239. [DOI] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, et al. (2016). A shared neural ensemble links distinct contextual memories encoded close in time. Nature, 534, 115–118. 10.1038/nature17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoon GG, Sutton AK, Chang C-J, Libster AM, Glober GF, Leveque CL, et al. (2018). Acute food deprivation rapidly modifies valence-coding microcircuits in the amygdala. Retrieved from https://www.biorxiv.org/content/early/2018/03/19/285189.
- Calipari ES, Bagot RC, Purushothaman I, Davidson TJ, Yorgason JT, Peña CJ, et al. (2016). In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proceedings of the National Academy of Sciences of the United States of America, 113, 2726–2731. 10.1073/pnas.1521238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calipari ES, Juarez B, Morel C, Walker DM, Cahill ME, Ribeiro E, et al. (2017). Dopaminergic dynamics underlying sex-specific cocaine reward. Nature Communications, 8, 13877 10.1038/ncomms13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, & Shieh J (2015). Electrophysiology Guide to research techniques in neuroscience (2nd ed.). San Diego: Academic Press. [Google Scholar]
- Chan CL, Wheeler DS, & Wheeler RA (2016). The neural encoding of cocaine-induced devaluation in the ventral pallidum. Neurobiology of Learning and Memory, 130, 177–184. 10.1016/j.nlm.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester JA, de Paula Barrenha G, DeMaria A, & Finegan A (2006). Different effects of stress on alcohol drinking behaviour in male and female mice selectively bred for high alcohol preference. Alcohol and Alcoholism, 41, 44–53. 10.1093/alcalc/agh242. [DOI] [PubMed] [Google Scholar]
- Chorev E, Epsztein J, Houweling AR, Lee AK, & Brecht M (2009). Electrophysiological recordings from behaving animals–going beyond spikes. Current Opinion in Neurobiology, 19, 513–519. 10.1016/j.conb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Cohen JY, Haesler S, Vong L, Lowell BB, & Uchida N (2012). Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature, 482, 85–88. 10.1038/nature10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JA (1986). Digital imaging of free calcium changes and of spatial gradients in growing processes in single, mammalian central nervous system cells. Proceedings of the National Academy of Sciences of the United States of America, 83, 6179–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Pinto L, & Dan Y (2016). Calcium imaging of sleep-wake related neuronal activity in the dorsal pons. Nature Communications, 7, 10763 10.1038/ncomms10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Luo G, Pham MD, Lovinger DM, et al. (2014). Deep brain optical measurements of cell type-specific neural activity in behaving mice. Nature Protocols, 9, 1213–1228. 10.1038/nprot.2014.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, et al. (2013). Concurrent activation of striatal direct and indirect pathways during action initiation. Nature, 494, 238–242. 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, et al. (2018). A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell, 174 10.1016/j.cell.2018.06.035, 465–480.e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaj MI, Slemmer JE, Carroll FI, & Martin BR (1999). Pharmacological characterization of nicotine’s interaction with cocaine and cocaine analogs. The Journal of Pharmacology and Experimental Therapeutics, 289, 1229–1236. [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, et al. (2016). Sensitive red protein calcium indicators for imaging neural activity. Elife, 5 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse B, Hasseman JP, Tsegaye G, et al. (2018). High-performance GFP-based calcium indicators for imaging activity in neuronal populations and microcompartments. bioRxiv. 10.1101/434589 [This article is a preprint and has not been peer-reviewed]. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Goldstein RB, Chou SP, Ruan WJ, & Grant BF (2008). Age at first drink and the first incidence of adult-onset DSM-IV alcohol use disorders. Alcoholism: Clinical and Experimental Research, 32, 2149–2160. 10.1111/j.1530-0277.2008.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deadwyler SA (2010). Electrophysiological correlates of abused drugs: Relation to natural rewards. Annals of the New York Academy of Sciences, 1187, 140–147. 10.1111/j.1749-6632.2009.05155.x. [DOI] [PubMed] [Google Scholar]
- DeNardo L, & Luo L (2017). Genetic strategies to access activated neurons. Current Opinion in Neurobiology, 45, 121–129. 10.1016/j.conb.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, & Imperato A (1988). Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proceedings of the National Academy of Sciences of the United States of America, 85, 5274–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey AS, Suminski A, Amit Y, & Hatsopoulos NG (2009). Single-unit stability using chronically implanted multielectrode arrays. Journal of Neurophysiology, 102, 1331–1339. 10.1152/jn.90920.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dole VP, Ho A, & Gentry RT (1983). An improved technique for monitoring the drinking behavior of mice. Physiology & Behavior, 30, 971–974. [DOI] [PubMed] [Google Scholar]
- Dong Y, Taylor JR, Wolf ME, & Shaham Y (2017). Circuit and synaptic plasticity mechanisms of drug relapse. The Journal of Neuroscience, 37, 10867–10876. 10.1523/jneurosci.1821-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreifuss JJ, & Kelly JS (1972). Recurrent inhibition of antidromically identified rat supraoptic neurones. The Journal of Physiology, 220, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy M, & Chanraud S (2016). Imaging the addicted brain: Alcohol. International Review of Neurobiology, 129, 1–31. 10.1016/bs.irn.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Emiliani V, Cohen AE, Deisseroth K, & Hausser M (2015). All-optical interrogation of neural circuits. The Journal of Neuroscience, 35, 13917–13926. 10.1523/jneurosci.2916-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel N, Bukwich M, Rao V, Hemmelder V, Tian J, & Uchida N (2015). Arithmetic and local circuitry underlying dopamine prediction errors. Nature, 525, 243–246. 10.1038/nature14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettenberg A, Pettit HO, Bloom FE, & Koob GF (1982). Heroin and cocaine intravenous self-administration in rats: Mediation by separate neural systems. Psychopharmacology (Berl), 78, 204–209. [DOI] [PubMed] [Google Scholar]
- Fanelli RR, Klein JT, Reese RM, & Robinson DL (2013). Dorsomedial and dorsolateral striatum exhibit distinct phasic neuronal activity during alcohol self-administration in rats. The European Journal of Neuroscience, 38, 2637–2648. 10.1111/ejn.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, et al. (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nature Methods, 11, 763–772. 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Jung JC, Cocker ED, Anderson EP, & Schnitzer MJ (2005). In vivo brain imaging using a portable 3.9 gram two-photon fluorescence microendoscope. Optics Letters, 30, 2272–2274. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RP, Ko TH, et al. (2008). High-speed, miniaturized fluorescence microscopy in freely moving mice. Nature Methods, 5, 935–938. 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MM, Vazquez RW, Papke RL, & Oswald RE (2000). Subtype-selective inhibition of neuronal nicotinic acetylcholine receptors by cocaine is determined by the alpha4 and beta4 subunits. Molecular Pharmacology, 58, 109–119. [DOI] [PubMed] [Google Scholar]
- Garcia R, Vouimba RM, Baudry M, & Thompson RF (1999). The amygdala modulates prefrontal cortex activity relative to conditioned fear. Nature, 402, 294–296. 10.1038/46286. [DOI] [PubMed] [Google Scholar]
- Georges F, Le Moine C, & Aston-Jones G (2006). No effect of morphine on ventral tegmental dopamine neurons during withdrawal. The Journal of Neuroscience, 26, 5720–5726. 10.1523/jneurosci.5032-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond A, Fujita H, Ichimura T, & Watanabe TM (2016). Design and development of genetically encoded fluorescent sensors to monitor intracellular chemical and physical parameters. Biophysical Reviews, 8, 121–138. 10.1007/s12551-016-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein GL, & Clark WA (1964). Simultaneous studies of firing patterns in several neurons. Science, 143, 1325–1327. 10.1126/science.143.3612.1325. [DOI] [PubMed] [Google Scholar]
- Ghosh A, & Greenberg ME (1995). Calcium signaling in neurons: Molecular mechanisms and cellular consequences. Science, 268, 239–247. [DOI] [PubMed] [Google Scholar]
- Girven KS, & Sparta DR (2017). Probing deep brain circuitry: New advances in in vivo calcium measurement strategies. ACS Chemical Neuroscience, 8, 243–251. 10.1021/acschemneuro.6b00307. [DOI] [PubMed] [Google Scholar]
- Grant BF, & Dawson DA (1998). Age of onset of drug use and its association with DSM-IV drug abuse and dependence: Results from the national longitudinal alcohol epidemiologic survey. Journal of Substance Abuse, 10, 163–173. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Grundemann J, Kitch LJ, Lecoq JA, Parker JG, Marshall JD, et al. (2017). Neural ensemble dynamics underlying a long-term associative memory. Nature, 543, 670–675. 10.1038/nature21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Langer D, Kasper H, Kampa BM, & Helmchen F (2010). High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nature Methods, 7, 399–405. 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- Grienberger C, & Konnerth A (2012). Imaging calcium in neurons. Neuron, 73, 862–885. 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Griffin WC 3rd, Lopez MF, & Becker HC (2009). Intensity and duration of chronic ethanol exposure is critical for subsequent escalation of voluntary ethanol drinking in mice. Alcoholism: Clinical and Experimental Research, 33, 1893–1900. 10.1111/j.1530-0277.2009.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin WC 3rd, Lopez MF, Yanke AB, Middaugh LD, & Becker HC (2009). Repeated cycles of chronic intermittent ethanol exposure in mice increases voluntary ethanol drinking and ethanol concentrations in the nucleus accumbens. Psychopharmacology (Berl), 201, 569–580. 10.1007/s00213-008-1324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzendler J, Yang G, Pan F, Parkhurst CN, & Gan WB (2011). Transcranial two-photon imaging of the living mouse brain. Cold Spring Harbor Protocols, 2011. 10.1101/pdb.prot065474. pii:pdb.prot065474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, et al. (2014). Natural neural projection dynamics underlying social behavior. Cell, 157, 1535–1551. 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Hires SA, Li N, O’Connor DH, Komiyama T, Ophir E, et al. (2014). Procedures for behavioral experiments in head-fixed mice. PLoS One, 9, e88678 10.1371/journal.pone.0088678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Zhou J, Feng Q, Lin R, Gong H, Luo Q, et al. (2015). Multi-channel fiber photometry for population neuronal activity recording. Biomedical Optics Express, 6, 3919–3931. 10.1364/boe.6.003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson RE, Porrino LJ, Opris I, Stanford T, & Deadwyler SA (2011). Effects of cocaine rewards on neural representations of cognitive demand in nonhuman primates. Psychopharmacology (Berl), 213, 105–118. 10.1007/s00213-010-2017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, & Buzsaki G (2000). Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. Journal of Neurophysiology, 84, 401–414. 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Harris KD, Quiroga RQ, Freeman J, & Smith SL (2016). Improving data quality in neuronal population recordings. Nature Neuroscience, 19, 1165–1174. 10.1038/nn.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey MA, Bermejo R, & Zeigler HP (2001). Discriminative whisking in the head-fixed rat: Optoelectronic monitoring during tactile detection and discrimination tasks. Somatosensory & Motor Research, 18, 211–222. [DOI] [PubMed] [Google Scholar]
- Harvey CD, Collman F, Dombeck DA, & Tank DW (2009). Intracellular dynamics of hippocampal place cells during virtual navigation. Nature, 461, 941–946. 10.1038/nature08499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiney SA, Wohl MP, Chettih SN, Ruffolo LI, & Medina JF (2014). Cerebellar-dependent expression of motor learning during eyeblink conditioning in head-fixed mice. The Journal of Neuroscience, 34, 14845–14853. 10.1523/jneurosci.2820-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W, & Kerr JN (2013). Miniaturization of two-photon microscopy for imaging in freely moving animals. Cold Spring Harbor Protocols, 2013, 904–913. 10.1101/pdb.top078147. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Fee MS, Tank DW, & Denk W (2001). A miniature head-mounted two-photon microscope. High-resolution brain imaging in freely moving animals. Neuron, 31, 903–912. [DOI] [PubMed] [Google Scholar]
- Hernández-López S, Bargas J, Surmeier DJ, Reyes A, & Galarraga E (1997). D1 receptor activation enhances evoked discharge in neostriatal medium spiny neurons by modulating an L-type Ca2+ conductance. The Journal of Neuroscience, 17, 3334–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, et al. (2016). Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron, 91, 1228–1243. 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Hoffmann J, & Nicola SM (2014). Dopamine invigorates reward seeking by promoting cue-evoked excitation in the nucleus accumbens. The Journal of Neuroscience, 34, 14349–14364. 10.1523/jneurosci.3492-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf FW, Chang SJ, Sparta DR, Bowers MS, & Bonci A (2010). Motivation for alcohol becomes resistant to quinine adulteration after 3 to 4 months of intermittent alcohol self-administration. Alcoholism: Clinical and Experimental Research, 34, 1565–1573. 10.1111/j.1530-0277.2010.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe MW, & Dombeck DA (2016). Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature, 535, 505–510. 10.1038/nature18942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan H, Erdogdu MA, & Schnitzer M (2017). Robust estimation of neural signals in calcium imaging In Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, et al. (Eds.), Advances in neural information processing systems (Vol. 30, pp. 2901–2910). Curran Associates, Inc. [Google Scholar]
- Inoue M, Takeuchi A, Horigane S, Ohkura M, Gengyo-Ando K, Fujii H, et al. (2015). Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nature Methods, 12, 64–70. 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- Isomura Y, Harukuni R, Takekawa T, Aizawa H, & Fukai T (2009). Microcircuitry coordination of cortical motor information in self-initiation of voluntary movements. Nature Neuroscience, 12, 1586–1593. 10.1038/nn.2431. [DOI] [PubMed] [Google Scholar]
- Janak PH, Chang JY, & Woodward DJ (1999). Neuronal spike activity in the nucleus accumbens of behaving rats during ethanol self-administration. Brain Research, 817, 172–184. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J, et al. (2015). Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell, 160, 516–527. 10.1016/j.cell.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jercog P, Rogerson T, & Schnitzer MJ (2016). Large-scale fluorescence calcium-imaging methods for studies of long-term memory in behaving mammals. Cold Spring Harbor Perspectives in Biology, 8 10.1101/cshperspect.a021824. pii:a021824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, & Losonczy A (2014). SIMA: Python software for analysis of dynamic fluorescence imaging data. Frontiers in Neuroinformatics, 8, 80 10.3389/fninf.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keemink SW, Lowe SC, Pakan JMP, Dylda E, van Rossum MCW, & Rochefort NL (2018). FISSA: A neuropil decontamination toolbox for calcium imaging signals. Scientific Reports, 8, 3493 10.1038/s41598-018-21640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, & Schafer WR (2000). Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron, 26, 583–594. [DOI] [PubMed] [Google Scholar]
- Kim CK, Adhikari A, & Deisseroth K (2017). Integration of optogenetics with complementary methodologies in systems neuroscience. Nature Reviews Neuroscience, 18, 222–235. 10.1038/nrn.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Yang SJ, Pichamoorthy N, Young NP, Kauvar I, Jennings JH, et al. (2016). Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nature Methods, 13, 325–328. 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, et al. (2014). Independent optical excitation of distinct neural populations. Nature Methods, 11, 338–346. 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchibhotla KV, Gill JV, Lindsay GW, Papadoyannis ES, Field RE, Sten TA, et al. (2017). Parallel processing by cortical inhibition enables context-dependent behavior. Nature Neuroscience, 20, 62–71. 10.1038/nn.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Akita K, Nakamura T, Ogura A, Makino T, Tamagawa A, et al. (1992). A single optical fiber fluorometric device for measurement of intracellular Ca2+ concentration: Its application to hippocampal neurons in vitro and in vivo. Neuroscience, 50, 619–625. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt DA, Juczewski K, Cui G, Johnson KA, & Lovinger DM (2017). Parallel, but dissociable, processing in discrete corticostriatal inputs encodes skill learning. Neuron, 96 10.1016/j.neuron.2017.09.040, 476–489.e475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bars D, Menetrey D, Conseiller C, & Besson JM (1975). Depressive effects of morphine upon lamina V cells activities in the dorsal horn of the spinal cat. Brain Research, 98, 261–277. [DOI] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, et al. (2015). Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell, 162, 635–647. 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Schwartz Y, Sparta DR, Cheer JF, & Adali T (2017). Parameter-free automated extraction of neuronal signals from calcium imaging data. In Paper presented at the 2017 IEEE international conference on acoustics, speech and signal processing (ICASSP). [Google Scholar]
- Lima SQ, Hromadka T, Znamenskiy P, & Zador AM (2009). PINP: A new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One, 4, e6099 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XB, Pierce DR, Light KE, & Hayar A (2013). The fine temporal structure of the rat licking pattern: What causes the variability in the interlick intervals and how is it affected by the drinking solution? Chemical Senses, 38, 685–704. 10.1093/chemse/bjt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, & Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nature Neuroscience, 19, 1142–1153. 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Madison DV, Poenie M, Reuter H, Tsien RW, & Tsien RY (1988). Imaging of cytosolic Ca2+ transients arising from Ca2+ stores and Ca2+ channels in sympathetic neurons. Neuron, 1, 355–365. [DOI] [PubMed] [Google Scholar]
- Li L, Tang Y, Sun L, Rahman K, Huang K, Xu W, et al. (2017). In vivo fiber photometry of neural activity in response to optogenetically manipulated inputs in freely moving mice. Journal of Innovative Optical Health Sciences, 10, 1743001 10.1142/S1793545817430015. [DOI] [Google Scholar]
- Liu F, Jiang H, Zhong W, Wu X, & Luo J (2010). Changes in ensemble activity of hippocampus CA1 neurons induced by chronic morphine administration in freely behaving mice. Neuroscience, 171, 747–759. 10.1016/j.neuroscience.2010.09.052. [DOI] [PubMed] [Google Scholar]
- Livneh Y, Ramesh RN, Burgess CR, Levandowski KM, Madara JC, Fenselau H, et al. (2017). Homeostatic circuits selectively gate food cue responses in insular cortex. Nature, 546, 611–616. 10.1038/nature22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London TD, Licholai JA, Szczot I, Ali MA, LeBlanc KH, Fobbs WC, et al. (2018). Coordinated ramping of dorsal striatal pathways preceding food approach and consumption. The Journal of Neuroscience, 38, 3547–3558. 10.1523/jneurosci.2693-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MF, & Becker HC (2014). Operant ethanol self-administration in ethanol dependent mice. Alcohol, 48, 295–299. 10.1016/j.alcohol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Gutruf P, Xia L, Bhatti DL, Wang X, Vazquez-Guardado A, et al. (2018). Wireless optoelectronic photometers for monitoring neuronal dynamics in the deep brain. Proceedings of the National Academy of Sciences of the United States of America, 115, E1374–E1383. 10.1073/pnas.1718721115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, & Svoboda K (2008). Genetic dissection of neural circuits. Neuron, 57, 634–660. 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Volkow ND, Heintz N, Pan Y, & Du C (2011). Acute cocaine induces fast activation of D1 receptor and progressive deactivation of D2 receptor striatal neurons: In vivo optical microprobe [Ca2+]i imaging. The Journal of Neuroscience, 31, 13180–13190. 10.1523/jneurosci.2369-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C (2016). The emergence of a circuit model for addiction. Annual Review of Neuroscience, 39, 257–276. 10.1146/annurev-neuro-070815-013920. [DOI] [PubMed] [Google Scholar]
- Lynch WJ, Kushner MG, Rawleigh JM, Fiszdon J, & Carroll ME (1999). The effects of restraint stress on voluntary ethanol consumption in rats. Experimental and Clinical Psychopharmacology, 7, 318–323. [DOI] [PubMed] [Google Scholar]
- Mahler SV, Vazey EM, Beckley JT, Keistler CR, McGlinchey EM, Kaufling J, et al. (2014). Designer receptors show role for ventral pallidum input to ventral tegmental area in cocaine seeking. Nature Neuroscience, 17, 577–585. 10.1038/nn.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Santos AF, Direnberger S, Mrsic-Flogel TD, Hofer SB, Stein V, et al. (2008). A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nature Methods, 5, 805–811. 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantz J, Thierry AM, & Glowinski J (1989). Effect of noxious tail pinch on the discharge rate of mesocortical and mesolimbic dopamine neurons: Selective activation of the mesocortical system. Brain Research, 476, 377–381. [DOI] [PubMed] [Google Scholar]
- Margolis EB, Hjelmstad GO, Fujita W, & Fields HL (2014). Direct bidirectional mu-opioid control of midbrain dopamine neurons. The Journal of Neuroscience, 34, 14707–14716. 10.1523/jneurosci.2144-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz JE, Liberti WA 3rd, Guitchounts G, Velho T, Lois C, & Gardner TJ (2015). Mesoscopic patterns of neural activity support songbird cortical sequences. PLoS Biology, 13, e1002158 10.1371/journal.pbio.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Li JZ, Zhang Y, Gong Y, St-Pierre F, Lin MZ, et al. (2016). Cell-type-specific optical recording of membrane voltage dynamics in freely moving mice. Cell, 167 10.1016/j.cell.2016.11.021, 1650–1662.e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry JA, Otis JM, Rossi MA, Robinson JE, Kosyk O, Miller NW, et al. (2017). Hormonal gain control of a medial preoptic area social reward circuit. Nature Neuroscience, 20, 449–458. 10.1038/nn.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Babayan BM, Uchida N, & Watabe-Uchida M (2017). Opposite initialization to novel cues in dopamine signaling in ventral and posterior striatum in mice. Elife, 6 10.7554/eLife.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JT, Glick SD, & Cheer JF (2010). Neural encoding of psychomotor activation in the nucleus accumbens core, but not the shell, requires cannabinoid receptor signaling. The Journal of Neuroscience, 30, 5102–5107. 10.1523/jneurosci.5335-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, & Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron, 63, 747–760. 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Chandler LJ, & Kalivas PW (2016). Signals from the fourth dimension regulate drug relapse. Trends in Neurosciences, 39, 472–485. 10.1016/j.tins.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle C, Choquet D, Korn H, & Changeux JP (1992). Calcium influx through nicotinic receptor in rat central neurons: Its relevance to cellular regulation. Neuron, 8, 135–143. [DOI] [PubMed] [Google Scholar]
- Murray JE, Belin-Rauscent A, Simon M, Giuliano C, Benoit-Marand M, Everitt BJ, et al. (2015). Basolateral and central amygdala differentially recruit and maintain dorsolateral striatum-dependent cocaine-seeking habits. Nature Communications, 6, 10088 10.1038/ncomms10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan M, Jang HJ, Park M, Miller EM, Cox J, Taliaferro JP, et al. (2017). Combined social and spatial coding in a descending projection from the prefrontal cortex. Cell, 171 10.1016/j.cell.2017.11.002, 1663–1677.e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Ohkura M, Abe G, Nakai J, & Kawakami K (2013). Real-time visualization of neuronal activity during perception. Current Biology, 23, 307–311. 10.1016/j.cub.2012.12.040. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, & Imoto K (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nature Biotechnology, 19, 137–141. 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Neve RL, & Neve KA (2001). Overview of neural gene expression. Current Protocols in Neuroscience. 10.1002/0471142301.ns0405s00. [DOI] [PubMed] [Google Scholar]
- Nicola SM, & Deadwyler SA (2000). Firing rate of nucleus accumbens neurons is dopamine-dependent and reflects the timing of cocaine-seeking behavior in rats on a progressive ratio schedule of reinforcement. The Journal of Neuroscience, 20, 5526–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieh EH, Matthews GA, Allsop SA, Presbrey KN, Leppla CA, Wichmann R, et al. (2015). Decoding neural circuits that control compulsive sucrose seeking. Cell, 160, 528–541. 10.1016/j.cell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieh EH, Vander Weele CM, Matthews GA, Presbrey KN, Wichmann R, Leppla CA, et al. (2016). Inhibitory input from the lateral hypothalamus to the ventral tegmental area disinhibits dopamine neurons and promotes behavioral activation. Neuron, 90, 1286–1298. 10.1016/j.neuron.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea DJ, Trautmann E, Chandrasekaran C, Stavisky S, Kao JC, Sahani M, et al. (2017). The need for calcium imaging in nonhuman primates: New motor neuroscience and brain-machine interfaces. Experimental Neurology, 287, 437–451. 10.1016/j.expneurol.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleson EB, & Roberts DC (2012). Cocaine self-administration in rats: Threshold procedures In Kobeissey F (Ed.), Psychiatric disorders. Methods in molecular biology (methods and protocols) (pp. 303–319). Humana Press. [DOI] [PubMed] [Google Scholar]
- Otis JM, Namboodiri VM, Matan AM, Voets ES, Mohorn EP, Kosyk O, et al. (2017). Prefrontal cortex output circuits guide reward seeking through divergent cue encoding. Nature, 543, 103–107. 10.1038/nature21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owesson-White C, Belle AM, Herr NR, Peele JL, Gowrishankar P, Carelli RM, et al. (2016). Cue-evoked dopamine release rapidly modulates D2 neurons in the nucleus accumbens during motivated behavior. The Journal of Neuroscience, 36, 6011–6021. 10.1523/jneurosci.0393-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden I, Lee HM, Sullivan MR, & Wang SS (2008). Identification and clustering of event patterns from in vivo multiphoton optical recordings of neuronal ensembles. Journal of Neurophysiology, 100, 495–503. 10.1152/jn.01310.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HW, & Hausser M (2015). Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nature Methods, 12, 140–146. 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker NF, Cameron CM, Taliaferro JP, Lee J, Choi JY, Davidson TJ, et al. (2016). Reward and choice encoding in terminals of midbrain dopamine neurons depends on striatal target. Nature Neuroscience, 19, 845–854. 10.1038/nn.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvaz MA, Alia-Klein N, Woicik PA, Volkow ND, & Goldstein RZ (2011). Neuroimaging for drug addiction and related behaviors. Reviews in the Neurosciences, 22, 609–624. 10.1515/rns.2011.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel TP, Man K, Firestein BL, & Meaney DF (2015). Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. Journal of Neuroscience Methods, 243, 26–38. 10.1016/j.jneumeth.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples LL, Uzwiak AJ, Gee F, Fabbricatore AT, Muccino KJ, Mohta BD, et al. (1999). Phasic accumbal firing may contribute to the regulation of drug taking during intravenous cocaine self-administration sessions. Annals of the New York Academy of Sciences, 877, 781–787. [DOI] [PubMed] [Google Scholar]
- Peoples LL, & West MO (1996). Phasic firing of single neurons in the rat nucleus accumbens correlated with the timing of intravenous cocaine self-administration. The Journal of Neuroscience, 16, 3459–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perra S, Pillolla G, Melis M, Muntoni AL, Gessa GL, & Pistis M (2005). Involvement of the endogenous cannabinoid system in the effects of alcohol in the mesolimbic reward circuit: Electrophysiological evidence in vivo. Psychopharmacology (Berl), 183, 368–377. 10.1007/s00213-005-0195-0. [DOI] [PubMed] [Google Scholar]
- Pinault D (1996). A novel single-cell staining procedure performed in vivo under electrophysiological control: Morpho-functional features of juxtacellularly labeled thalamic cells and other central neurons with biocytin or neurobiotin. Journal of Neuroscience Methods, 65, 113–136. [DOI] [PubMed] [Google Scholar]
- Pinto L, & Dan Y (2015). Cell-type-specific activity in prefrontal cortex during goal-directed behavior. Neuron, 87, 437–450. 10.1016/j.neuron.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, & Giovannucci A (2017). NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. Journal of Neuroscience Methods, 291, 83–94. 10.1016/j.jneumeth.2017.07.031. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, et al. (2016). Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron, 89, 285–299. 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada J, Sasi M, Martin C, Jablonka S, Dandekar T, & Blum R (2018). An open source tool for automatic spatiotemporal assessment of calcium transients and local ‘signal-close-to-noise’ activity in calcium imaging data. PLoS Computational Biology, 14, e1006054 10.1371/journal.pcbi.1006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed WA, Yan MF, & Schnitzer MJ (2002). Gradient-index fiber-optic microprobes for minimally invasive in vivo low-coherence interferometry. Optics Letters, 27, 1794–1796. [DOI] [PubMed] [Google Scholar]
- Regehr WG, Connor JA, & Tank DW (1989). Optical imaging of calcium accumulation in hippocampal pyramidal cells during synaptic activation. Nature, 341, 533–536. 10.1038/341533a0. [DOI] [PubMed] [Google Scholar]
- Resendez SL, Jennings JH, Ung RL, Namboodiri VM, Zhou ZC, Otis JM, et al. (2016). Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nature Protocols, 11, 566–597. 10.1038/nprot.2016.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JS, Ford MM, Yu CH, Brown LL, Finn DA, Garland T Jr., et al. (2007). Mouse inbred strain differences in ethanol drinking to intoxication. Genes, Brain and Behavior, 6, 1–18. 10.1111/j.1601-183X.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Rickgauer JP, & Tank DW (2009). Two-photon excitation of channelrhodopsin-2 at saturation. Proceedings of the National Academy of Sciences of the United States of America, 106, 15025–15030. 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger FO, & Oakes RA (1995). Nicotine-induced conditioned place preference and conditioned place aversion in mice. Pharmacology, Biochemistry, and Behavior, 51, 457–461. [DOI] [PubMed] [Google Scholar]
- Roberts TF, Hisey E, Tanaka M, Kearney MG, Chattree G, Yang CF, et al. (2017). Identification of a motor-to-auditory pathway important for vocal learning. Nature Neuroscience, 20, 978–986. 10.1038/nn.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DL, & Carelli RM (2008). Distinct subsets of nucleus accumbens neurons encode operant responding for ethanol versus water. The European Journal of Neuroscience, 28, 1887–1894. 10.1111/j.1460-9568.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SL, & McCool BA (2015). Microstructural analysis of rat ethanol and water drinking patterns using a modified operant self-administration model. Physiology & Behavior, 149, 119–130. 10.1016/j.physbeh.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson HH, & Files FJ (1998). Behavioral pharmacology of alcohol In Tartar RE, Ammerman RT, & Ott PJ (Eds.), Handbook of substance abuse (pp. 23–32). Boston, MA: Springer. [Google Scholar]
- Sato M, Kawano M, Ohkura M, Gengyo-Ando K, Nakai J, & Hayashi Y (2015). Generation and imaging of transgenic mice that express G-CaMP7 under a tetracycline response element. PLoS One, 10, e0125354 10.1371/journal.pone.0125354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Motegi Y, Yagi S, Gengyo-Ando K, Ohkura M, & Nakai J (2017). Fast varifocal two-photon microendoscope for imaging neuronal activity in the deep brain. Biomedical Optics Express, 8, 4049–4060. 10.1364/boe.8.004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawinski J, Wallace DJ, Greenberg DS, Grossmann S, Denk W, & Kerr JN (2009). Visually evoked activity in cortical cells imaged in freely moving animals. Proceedings of the National Academy of Sciences of the United States of America, 106, 19557–19562. 10.1073/pnas.0903680106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreihofer AM, & Guyenet PG (1997). Identification of C1 presympathetic neurons in rat rostral ventrolateral medulla by juxtacellular labeling in vivo. The Journal of Comparative Neurology, 387, 524–536. [DOI] [PubMed] [Google Scholar]
- Schwarz C, Hentschke H, Butovas S, Haiss F, Stuttgen MC, Gerdjikov TV, et al. (2010). The head-fixed behaving rateprocedures and pitfalls. Somatosensory & Motor Research, 27, 131–148. 10.3109/08990220.2010.513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn V, Wolff SB, Herry C, Grenier F, Ehrlich I, Grundemann J, et al. (2014). Long-range connectivity defines behavioral specificity of amygdala neurons. Neuron, 81, 428–437. 10.1016/j.neuron.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Sepehri Rad M, Choi Y, Cohen LB, Baker BJ, Zhong S, Storace DA, et al. (2017). Voltage and calcium imaging of brain activity. Biophysical Journal, 113, 2160–2167. 10.1016/j.bpj.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheintuch L, Rubin A, Brande-Eilat N, Geva N, Sadeh N, Pinchasof O, et al. (2017). Tracking the same neurons across multiple days in Ca(2+) imaging data. Cell Reports, 21, 1102–1115. 10.1016/j.celrep.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Patel S, & Khakh BS (2016). Probing the complexities of astrocyte calcium signaling. Trends in Cell Biology, 26, 300–312. 10.1016/j.tcb.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano CA, Ferris MJ, & Jones SR (2015). Cocaine self-administration disrupts mesolimbic dopamine circuit function and attenuates dopaminergic responsiveness to cocaine. The European Journal of Neuroscience, 42, 2091–2096. 10.1111/ejn.12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SG, & Davis WM (1974). Intravenous alcohol self-administration in the rat. Pharmacological Research Communications, 6, 379–402. [DOI] [PubMed] [Google Scholar]
- Song A, Charles AS, Koay SA, Gauthier JL, Thiberge SY, Pillow JW, et al. (2017). Volumetric two-photon imaging of neurons using stereoscopy (vTwINS). Nature Methods, 14, 420–426. 10.1038/nmeth.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparta DR, Stamatakis AM, Phillips JL, Hovelso N, van Zessen R, & Stuber GD (2011). Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nature Protocols, 7, 12–23. 10.1038/nprot.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AC, Andrews-Labenski J, & Letterio FC (1990). A new gustometer for psychophysical taste testing in the rat. Physiology & Behavior, 47, 795–803. [DOI] [PubMed] [Google Scholar]
- Spink AJ, Tegelenbosch RA, Buma MO, & Noldus LP (2001). The EthoVision video tracking system–a tool for behavioral phenotyping of transgenic mice. Physiology & Behavior, 73, 731–744. [DOI] [PubMed] [Google Scholar]
- Steinmetz NA, Buetfering C, Lecoq J, Lee CR, Peters AJ, Jacobs EAK, et al. (2017). Aberrant cortical activity in multiple GCaMP6-expressing transgenic mouse lines. eNeuro, 4 10.1523/eneuro.0207-17.2017pii: ENEURO.0207-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storace D, Sepehri Rad M, Kang B, Cohen LB, Hughes T, & Baker BJ (2016). Toward better genetically encoded sensors of membrane potential. Trends in Neurosciences, 39, 277–289. 10.1016/j.tins.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Hopf FW, Tye KM, Chen BT, & Bonci A (2010). Neuroplastic alterations in the limbic system following cocaine or alcohol exposure In Self JKGDW (Ed.), Behavioral neuroscience of drug addiction (pp. 3–27). Berlin, Heidelberg: Springer. [DOI] [PubMed] [Google Scholar]
- Sulzer D (2011). How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron, 69, 628–649. 10.1016/j.neuron.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, & Rayport S (1990). Amphetamine and other psychostimulants reduce pH gradients in midbrain dopaminergic neurons and chromaffin granules: A mechanism of action. Neuron, 5, 797–808. [DOI] [PubMed] [Google Scholar]
- Sun XR, Badura A, Pacheco DA, Lynch LA, Schneider ER, Taylor MP, et al. (2013). Fast GCaMPs for improved tracking of neuronal activity. Nature Communications, 4, 2170 10.1038/ncomms3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Kitamura T, Yamamoto J, Martin J, Pignatelli M, Kitch LJ, et al. (2015). Distinct speed dependence of entorhinal island and ocean cells, including respective grid cells. Proceedings of the National Academy of Sciences of the United States of America, 112, 9466–9471. 10.1073/pnas.1511668112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Bargas J, Hemmings HC Jr., Nairn AC, & Greengard P (1995). Modulation of calcium currents by a D1 dopaminergic protein kinase/phosphatase cascade in rat neostriatal neurons. Neuron, 14, 385–397. [DOI] [PubMed] [Google Scholar]
- Tank DW, Sugimori M, Connor JA, & Llinas RR (1988). Spatially resolved calcium dynamics of mammalian Purkinje cells in cerebellar slice. Science, 242, 773–777. [DOI] [PubMed] [Google Scholar]
- Theis L, Berens P, Froudarakis E, Reimer J, Roman Roson M, Baden T, et al. (2016). Benchmarking spike rate inference in population calcium imaging. Neuron, 90, 471–482. 10.1016/j.neuron.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele TE, & Navarro M (2014). Drinking in the dark” (DID) procedures: A model of binge-like ethanol drinking in non-dependent mice. Alcohol, 48, 235–241. 10.1016/j.alcohol.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods, 6, 875–881. 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias AS, Ecker AS, Siapas AG, Hoenselaar A, Keliris GA, & Logothetis NK (2007). Recording chronically from the same neurons in awake, behaving primates. Journal of Neurophysiology, 98, 3780–3790. 10.1152/jn.00260.2007. [DOI] [PubMed] [Google Scholar]
- Tornatzky W, & Miczek KA (2000). Cocaine self-administration “binges”: Transition from behavioral and autonomic regulation toward homeostatic dysregulation in rats. Psychopharmacology (Berl), 148, 289–298. [DOI] [PubMed] [Google Scholar]
- Trantham H, Szumlinski KK, McFarland K, Kalivas PW, & Lavin A (2002). Repeated cocaine administration alters the electrophysiological properties of prefrontal cortical neurons. Neuroscience, 113, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM (2018). Neural circuit motifs in valence processing. Neuron, 100, 436–452. 10.1016/j.neuron.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandaele Y, Cantin L, Serre F, Vouillac-Mendoza C, & Ahmed SH (2016). Choosing under the influence: A drug-specific mechanism by which the setting controls drug choices in rats. Neuropsychopharmacology, 41, 646–657. 10.1038/npp.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Weele CM, Siciliano CA, Matthews GA, Namburi P, Izadmehr EM, Espinal I, et al. (2018). Dopamine enhances signal-to-noise ratio in cortical-brainstem encoding of aversive stimuli. Nature, 2018. 10.1038/s41586-018-0682-1 (Published online November 7, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Tomasi D, & Baler RD (2013). Unbalanced neuronal circuits in addiction. Current Opinion in Neurobiology, 23, 639–648. 10.1016/j.conb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachowiak M, & Knöpfel T (2009). Optical imaging of brain activity in vivo using genetically encoded probes In Frostig RD (Ed.), In vivo optical imaging of brain function (2nd ed., pp. 2–35). Boca Raton, FL: CRC Press/Taylor & Francis. [PubMed] [Google Scholar]
- Walter HJ, & Messing RO (1999). Regulation of neuronal voltage-gated calcium channels by ethanol. Neurochemistry International, 35, 95–101. [DOI] [PubMed] [Google Scholar]
- Wheeler RA, & Carelli RM (2009). Dissecting motivational circuitry to understand substance abuse. Neuropharmacology, 56(Suppl 1), 149–159. 10.1016/j.neuropharm.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RA, Twining RC, Jones JL, Slater JM, Grigson PS, & Carelli RM (2008). Behavioral and electrophysiological indices of negative affect predict cocaine self-administration. Neuron, 57, 774–785. 10.1016/j.neuron.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Wiers CE, Cabrera E, Skarda E, Volkow ND, & Wang GJ (2016). PET imaging for addiction medicine: From neural mechanisms to clinical considerations. Progress in Brain Research, 224, 175–201. 10.1016/bs.pbr.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Wilcox MV, Cuzon Carlson VC, Sherazee N, Sprow GM, Bock R, Thiele TE, et al. (2014). Repeated binge-like ethanol drinking alters ethanol drinking patterns and depresses striatal GABAergic transmission. Neuropsychopharmacology, 39, 579–594. 10.1038/npp.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Nygard SK, Sobczak GG, Hourguettes NJ, & Bruchas MR (2017). Dorsal-CA1 hippocampal neuronal ensembles encode nicotine-reward contextual associations. Cell Reports, 19, 2143–2156. 10.1016/j.celrep.2017.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Shan J, Ng KF, & Pang P (2000). Morphine and methadone have different effects on calcium channel currents in neuroblastoma cells. Brain Research, 870, 199–203. [DOI] [PubMed] [Google Scholar]
- Yang W, & Yuste R (2017). In vivo imaging of neural activity. Nature Methods, 14, 349–359. 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ST, Porrino LJ, & Iadarola MJ (1991). Cocaine induces striatal c-fos-immunoreactive proteins via dopaminergic D1 receptors. Proceedings of the National Academy of Sciences of the United States of America, 88, 1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Ahrens S, Zhang X, Schiff H, Ramakrishnan C, Fenno L, et al. (2017). The central amygdala controls learning in the lateral amygdala. Nature Neuroscience, 20, 1680–1685. 10.1038/s41593-017-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, et al. (2018). Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife, 7 10.7554/eLife.28728 pii:e28728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, et al. (2013). Long-term dynamics of CA1 hippocampal place codes. Nature Neuroscience, 16, 264–266. 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]


