Abstract
Proteotoxicity from insufficient clearance of misfolded/damaged proteins underlies many diseases. Carboxyl terminus of Hsc70-interacting protein (CHIP) is an important regulator of proteostasis in many cells, having E3-ligase and chaperone functions and often directing damaged proteins towards proteasome recycling. While enhancing CHIP functionality has broad therapeutic potential, prior efforts have all relied on genetic upregulation. Here we report that CHIP-mediated protein turnover is markedly post-translationally enhanced by direct protein kinase G (PKG) phosphorylation at S20 (mouse, S19 human). This increases CHIP binding affinity to Hsc70, CHIP protein half-life, and consequent clearance of stress-induced ubiquitinated-insoluble proteins. PKG-mediated CHIP-pS20 or expressing CHIP-S20E (phosphomimetic) reduces ischemic proteo- and cytotoxicity, whereas a phospho-silenced CHIP-S20A amplifies both. In vivo, depressing PKG activity lowers CHIP-S20 phosphorylation and protein, exacerbating proteotoxicity and heart dysfunction after ischemic injury. CHIP-S20E knock-in mice better clear ubiquitinated proteins and are cardio-protected. PKG activation provides post-translational enhancement of protein quality control via CHIP.
Subject terms: Mechanisms of disease, Phosphorylation, Myocardial infarction
Carboxyl terminus of Hsc70-interacting protein (CHIP) is proteostasis regulator. Here the authors show that CHIP-mediated protein turnover is enhanced by PKG-mediated phosphorylation, which results in attenuated cardiac ischemic proteotoxicity.
Introduction
The ubiquitin proteasome system (UPS) regulates post-transcriptional protein stability by identifying and then either repairing or disposing of damaged/misfolded proteins1. The degradation process employs a three-enzyme cascade (the E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases) that ultimately conjugate multiple molecules of ubiquitin, an 8 kDa protein, to selective lysine residues on a target protein. The poly-ubiquitin chain is recognized by the 26S proteasome, and the ubiquitinated substrate is then degraded and recycled back into amino acids1. In addition to having a constitutive role in protein homeostasis, the UPS is actively engaged in stress conditions where misfolded/damaged proteins accumulate, aggregate, and can cause/exacerbate disease2. Such proteotoxicity contributes to a variety of heart diseases, including myocardial ischemia/reperfusion/infarction3 and amyloidosis, and neurological disorders such as Alzheimer’s and Parkinson’s disease4.
The carboxyl terminus of Hsc70-interacting protein or CHIP (encoded by the gene known as Stub1) is an important member of the UPS system, functioning as both an E3-ligase and co-chaperone, and facilitating protein degradation5. CHIP is ubiquitously expressed, with the highest protein levels found in hypermetabolic tissues such as cardiac and skeletal muscle. The linear structure of CHIP is comprised of triple tandem tetratricopeptide repeat (TPR)-domains in the N-terminal, a central coiled-coil domain, and a C-terminal U-box domain that confers its ligase activity6. The TPR domain and the adjacent charged region of ~200 amino acids form the binding site to heat shock proteins (HSP), principally Hsc70, Hsp70, and Hsp90. HSP-CHIP interactions can modify substrate targeting and can suppress chaperone-refolding activity, ultimately transforming the machinery to one favoring protein destruction and recycling7,8. CHIP mediates protein degradation via the proteasome, as well as by autophagy-lysosome-dependent pathways9. Studies using CHIP genetic loss of function models report worsened heart responses to hemodynamic or ischemic stress10,11, accelerated aging coupled to insulin receptor dysregulation12, and exacerbated neurodegenerative disease13,14 and oncogenesis15. CHIP gene upregulation has been protective against such disorders16–18.
In contrast to artificial gene upregulation, post-translational control of CHIP functionality remains poorly understood. One effector is mono-ubiquitination that enhances CHIP substrate targeting for degradation19. CHIP phosphorylation is also detected in proteomics databases20,21, but whether this has a broad modulatory role is unknown. Protein kinase G (PKG) is the primary effector for the second messenger cyclic guanosine monophosphate (cGMP) generated by nitric oxide or natriuretic peptide stimulation. In proteotoxicity models of heart muscle22,23 and neurodegenerative disease24, PKG activation stimulates 26S proteasomes and reduces misfolded protein aggregation and associated proteotoxicity. This happens rapidly and impacts both short-lived and long-lived proteins24. We hypothesized that PKG may impact more than the proteasome by also modifying chaperone/E3-ligase functionality related to CHIP.
Here, we show that CHIP is phosphorylated by PKG at a highly conserved serine 20 (mouse; S19 human), enhancing CHIP functionality principally by increasing its post-translational half-life and protein interaction with Hsc70. Inhibiting PKG activation in myocytes and intact hearts that were then subjected to ischemic stress results in lower CHIP protein levels and its S20 phosphorylation, coupled to ubiquitinated protein accumulation with worse cytotoxicity or organ dysfunction. The opposite occurs if PKG is activated or CHIP mutated to a phosphomimetic (S20E). In vivo, mice with a CHIP S20E KI mutation are protected against myocardial infarction with improved protein quality control (PQC), smaller infarcts and enhanced function. These data reveal substantial potency to CHIP S20 (S19, human) phosphorylation as a means of protecting against damaged /misfolded protein accumulation and associated toxicity.
Results
PKG activity regulates PQC in a CHIP dependent manner
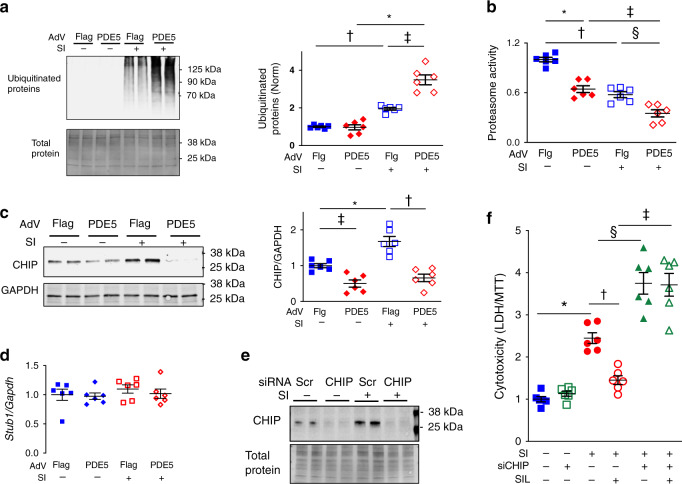
To test the intrinsic role of PKG activation on PQC, we first performed loss of function studies in cultured neonatal rat cardiomyocytes (NRCMs) transduced with an adenovirus expressing either epitope (Flag) tagged cGMP-selective phosphodiesterase 5 (PDE5Aoe) to degrade cGMP and block PKG activation, or Flag alone (Supplementary Fig. 1a, b). Exposure to 48 h of simulated ischemia (SI; deoxyglucose + acidosis + hypoxia) increased ubiquitinated proteins (UP) that further rose in cells with PDE5Aoe (Fig. 1a). PDE5oe also reduced proteasome activity in control myocytes and in those exposed to SI (Fig. 1b), the latter paralleling increased UP. Given the important role of CHIP in myocardial protein turnover, we measured steady-state levels of CHIP under these same conditions and found that while SI increased CHIP protein quantity, this was blocked by PDE5Aoe. PDE5oe reduced CHIP protein levels in controls as well, but this was amplified after SI (interaction p < 0.05, Fig. 1c). This change was post-translationally driven, as CHIP gene expression (Stub1) was unchanged in each condition (Fig. 1d).
Fig. 1. Effect of simulated ischemia (SI)+/− PKG blockade on ubiquitinated proteins and CHIP.
a Left: example immunoblot for ubiquitinated proteins from myocytes infected with adenovirus (AdV) for Flag or PDE5A-Flag (PDE5), the latter to suppress PKG activation (c.f. Supplementary Fig. 1a, b). Cells are then subjected to normal conditions or SI. Right: summary results, n = 6 biological replicates/group, 2WANOVA, Tukey multiple comparisons test (mct): *p < 10−8, †p = 0.0014, ‡p = 4 × 10−6. b Blocking PKG activation by PDE5Aoe reduces proteasome activity similarly at baseline or after SI (n = 6 biological replicates/group, 2WANOVA, Tukey mct: *p = 8 × 10−6, †8 × 10−7, ‡1 × 10−4, §0.002). c CHIP protein expression increases after SI but this is blocked by PDE5Aoe, 6 biological replicates/group, 2WANOVA, Tukey mct, *0.001, †7 × 10−6, ‡0.015. d CHIP mRNA expression is unaltered between groups, six biological replicates/group; 1WANOVA, p = 0.7. e Immunoblot showing efficacy of CHIP protein suppression in basal and SI conditions after incubation with specific but not scrambled (Scr) silencing RNA (siRNA), summary in Supplementary Fig. 1c. f LDH/MTT ratio in NRCMs exposed to control or SI conditions, +/−siRNA to CHIP (100 pM) or Scr, and with or without PDE5A inhibitor, sildenafil (SIL, 1 µM). n = 6 biological replicates/group; 1WANOVA, Sidak mct: *p = 9 × 10−6, †0.002, ‡1.2 × 10−9, §5 × 10−5. Source data are provided as a Source Data file. Individual data and mean ± SEM shown in each summary panel.
To test the importance of PKG-CHIP modulation on PQC after SI, myocytes were transfected with CHIP-siRNA or a scrambled control-siRNA construct, and then co-treated with (or without) the PDE5A inhibitor sildenafil (SIL, 1 μM) which stimulates PKG by suppressing cGMP hydrolysis (Fig. 1e, Supplementary Fig. 2). SIL reduced SI-induced cytotoxicity in cells expressing CHIP, however this protective effect was lost in cells lacking CHIP (Fig. 1f). Moreover, silencing CHIP worsened cytotoxicity to SI. Thus, PKG assists intrinsic protein degradation by enhancing proteasome activity and by augmenting CHIP protein expression, reducing cytotoxicity with SI.
PKG directly phosphorylates CHIP at Ser20 in cells
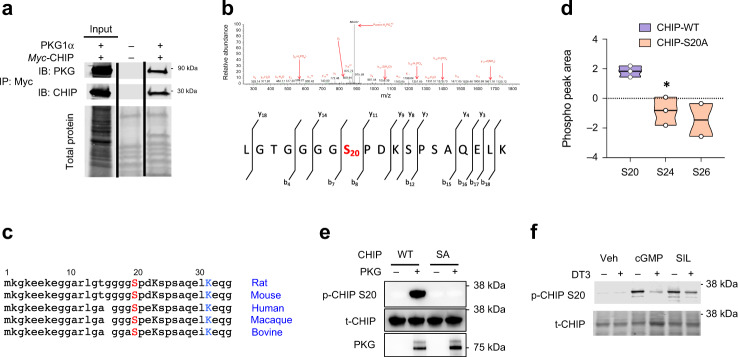
To determine if the involvement of PKG and CHIP occurs via direct protein-protein interactions, we expressed myc-tagged CHIP as bait in HEK293 cells and found PKG to be a co-immunoprecipitant (Fig. 2a). In a prior Mass Spec-derived database of proteins in adult rat myocytes subjected to 15 minutes PKG activation25, we found CHIP modified at S20 (S20 in rat and mouse, S19 human, Fig. 2b), a highly conserved residue in the TPR domain (Fig. 2c). Human CHIP peptide fragments containing amino acid residues 13–30 were incubated with cGMP/PKG and assayed by Mass Spec. We observed phosphorylation at S20 with wildtype (WT) CHIP but not a phospho-silenced CHIP-S20A mutation. Phosphorylation was also observed at neighboring S24 and S26, but only if a S20A CHIP mutant was used (Fig. 2d). This indicates the primary PKG-targeted serine on CHIP is S20. A monoclonal phospho-antibody generated against the S20 site in CHIP was tested in vitro with recombinant PKG1α and CHIP, verifying a consistent phospho-S20-CHIP signal. Use of a CHIP-S20A mutant CHIP confirmed specificity (Fig. 2e). CHIP pS20 increased in cardiomyocytes in which PKG was stimulated by addition of cGMP or PDE5 inhibition with SIL. Importantly, this was prevented by co-blockade of PKG activation by the peptide DT3 (1 μM, Fig. 2f, Supplementary Fig. 3). Thus, PKG interacts with and directly phosphorylates CHIP at serine 20.
Fig. 2. CHIP is phosphorylated by PKG at Serine 20.
a Immunoprecipitation of Myc-tagged CHIP with PKG1α. Lanes: 1-input, 2 negative control, 3-IP signal after three bead washes. Experiment replicated ×3. b Mass spectroscopy detects increased Chip-S20 (mouse sequence) phosphorylation with acute PKG activation. The lower schematic shows the sequence and fragmentation matching the spectra (above). The M2S spectra has red annotations for the assigned ion fragments, with three fragment ions localizing phosphorylation at S20 (b8 and y11 indicate it occurs on the N-terminal sequence of LGTGGGGS, b7 indicates the sequence LGTGGGG does not contain the site. Study performed with 3 biological replicates. c CHIP peptide sequence from start of N-terminus shows high level of conservation including three serines (S19, S23, S25 for human; S20, S24, S26 for mouse and rat). Lysine 30 (K30 human, K31 mouse) is also nearby, highly conserved, and known to be critical for CHIP-chaperone binding. d Mass Spec detection of serine phosphorylation of recombinant human CHIP peptide fragment containing residues 13–30 (Uniprot Q9UNE7). Only S20 phosphorylation is detected with WT CHIP. If S20A mutation is expressed, some reduced phosphorylation is observed at S24 and S26. All biological replicates shown in figure, truncated violin plot, median and 25/75%; unpaired t-test, *p = 0.01 vs CHIP-WT. e Recombinant PKG1α/CHIP assay shows pS20 signal by immunoblot for full length WT but not S20A mutant. Example of three biological replicates. f Phospho S20 antibody detects CHIP phosphorylation at S20 in rat myocytes exposed to cGMP or PDE5A inhibition, and both are blocked by concomitant PKG inhibition with DT3 (1 μM). Gel repeated ×3, with 3–4 biological replicates. Summary analysis in Supplementary Fig. 2a.
Genetic alteration of CHIP S20 alters ischemic proteotoxicity in myocytes in vitro
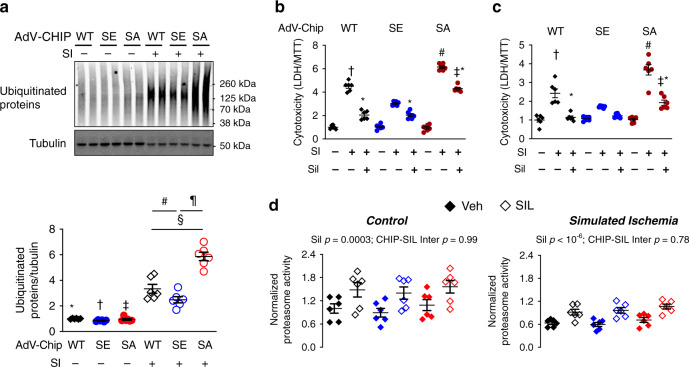
To directly probe the impact of S20 phosphorylation on proteotoxicity, we generated adenovirus expression vectors for CHIP-WT, or CHIP-S20E or CHIP-S20A. Myocytes were infected to express one of these forms, and then subjected to SI for 48 h. While each showed an increase in ubiquitinated proteins over non-stress controls, this was significantly amplified by CHIP-S20A and attenuated by CHIP-S20E expression compared to CHIP-WT (Fig. 3a). These differences were paralleled by disparities in SI-induced cytotoxicity (LDH/MTT ratio, Fig. 3b). Cells expressing CHIP-S20E mimicked the cyto-protection observed in CHIP-WT expressing myocytes co-incubated with the SIL to stimulate PKG. By contrast, cells expressing CHIP-S20A displayed greater cytotoxicity and blunted protective effects from SIL. Mutating only the neighboring serine (S24 mouse, S23 human) had negligible impact on cytotoxicity (Supplementary Fig. 4) and combining both S20 and S24 mutations yielded essentially the same response as with S20 modification alone (Fig. 3c). Thus, S20 is the primary effector of the cellular phenotype. The finding that SIL still increased proteasome activity regardless of whether WT or phospho-mutant CHIP was expressed for both control and SI conditions is consistent with PKG-stimulation of proteasome activity independent of CHIP22 (Fig. 3d).
Fig. 3. Functional regulation of UPS by Chip S20 is revealed by gain and loss of function phospho-mutants.
a Upper: representative Western blot of ubiquitinated proteins in NRCMs infected with adenovirus expressing WT, S20E (SE), or S20A (SA) CHIP, and exposed to normal or SI conditions for 48 h; Lower: summary results, data normalized for protein loading, and to control WT-CHIP; three gels, six biological replicates, 2WANOVA, Tukey mct: *p = 7 × 10−8, †3 × 10−5, ‡10−9, §4 × 10−8, ¶<10−9, #0.03. b Cytotoxicity (LDH/MTT assays) from SI is reduced by (SE) and increased by SA CHIP expression versus WT. PDE5A inhibitor sildenafil (SIL) reverses cytotoxicity in WT, but less so with SE or SA expression. 2WANOVA, 1.3 × 10−5 genotype condition interaction; Sidak mct: †p = 1.3 × 10−8 vs SE-SI, 1.2 × 10−9 vs SA-SI; #10−11 SE-SI; ‡<10−11 vs WT-SI + SIL and SE-SI+SIL; *p < 2 × 10−5 vs SI for each respective CHIP group. c Same experiment using AdV expressing combined (S20E, S24E; SE) or (S20A, S24A; SA). 2WANOVA, Sidak mct: †p = 0.01vs SE-SI, 2 × 10−5 vs SA-SI; #4.8 × 10−9 SE-SI; ‡0.03 vs SE-SI+SIL; p = 0.005 vs WT-SI + SIL; *1.4 × 10−5 vs WT-SI, 5.6 × 10−8, vs SA-SI. d Proteasome activity in resting NRCMs is augmented by PKG activation by PDE5 inhibitor sildenafil (SIL) in control myocytes and cells subjected to SI. Results of 2W-ANOVA for SIL effect and for interaction between SIL effect and gene-type of CHIP expressed are provided in the figures. N = 6 biological replicates for all conditions/groups. Source data are provided as a Source Data file. Individual data and mean ± SEM shown in each summary panel.
PKG activity increases CHIP phosphorylation and protein expression and counters proteotoxicity after myocardial infarction in vivo
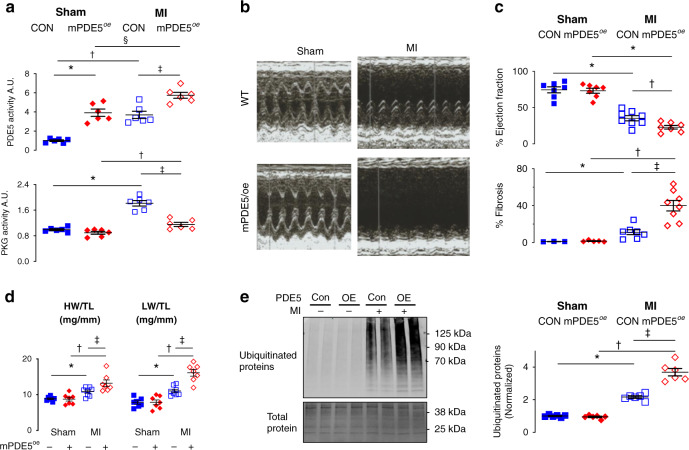
To test if intrinsic PKG activation regulates CHIP and PQC in vivo, mice expressing cardiomyocyte-restricted tetracycline-inducible phosphodiesterase 5 (mPDE5A/oe) were subjected to permanent coronary artery ligation to induce myocardial infarction (MI). In the absence of doxycycline, myocyte PDE5A activity increases 4-fold over controls in SHAM operated mice, and even somewhat more after MI (Fig. 4a, upper). However, in SHAM where PKG activity is low, this has no impact, but after MI when PKG activity normally increases, this is fully inhibited in mPDE5A/oe hearts (Fig. 4a, lower). The latter was associated with greater cardiac dilation and worse function as shown by example echocardiograms (Fig. 4b) and ejection fraction (Fig. 4c, upper). Pathological fibrosis (Fig. 4c, lower) and heart and lung weights (Fig. 4d) were also significantly greater in mPDE5A/oe hearts after MI.
Fig. 4. Suppressed myocyte PKG activity in vivo worsens infarct remodeling.
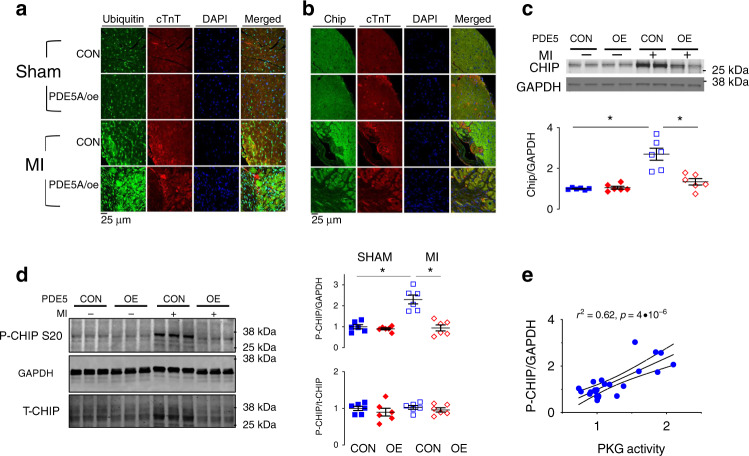
a PDE5A and PKG enzyme activity in peri-infarct region at baseline and after myocardial infarction in mice with myocyte-specific PDE5A overexpression (mPDE5oe) versus littermate controls (CON). 2WANOVA, Tukey Post mct: Upper: *10−5, †3 × 10−5, ‡8 × 10−4, §2.6 × 10−3; Lower: *2 × 10−8, †0.025, ‡9 × 10−7. b Example of M-mode echocardiograms in mPDE5/oe versus littermate controls 1-week following myocardial infarction (MI). c Summary data for left ventricular ejection fraction (EF, n = 7–8 mice/group) and for fibrosis (n = 3–8 biological replicates/group) for same study. For EF: Repeated measures mixed effects model, Holm-Sidak mct: *p < 2 × 10−8; †p = 0.03; for fibrosis: biological replicates: 3–5 – sham, 7–8 MI; Brown-Forsythe Welch ANOVA, Dunnetts mct: *p = 0.02, †0.008, ‡0.003. d Heart and Lung weight normalized to tibia length for same study (HW/TL, LW/TL respectively). N = 7–8 biological replicates; 2WANOVA, Sidaks mct: for HW/TL: *p = 0.04, †2 × 10−5, ‡0.01; LW/TL: *0.003, †8 × 10−9, ‡2 × 10−5. e Ubiquitinated protein increase after MI in the peri-infarct zone is greater in mPDE5/oe hearts. N = 6 biological replicates, 2WANOVA, Tukey mct: *8.6 × 10−6, ‡<10−8, ‡1.5 × 10−7. Source data are provided as a Source Data file. Individual data and mean ± SEM shown in each summary panel.
Global protein ubiquitination increased in the peri-infarct region after MI, and this was exacerbated in mPDE5A/oe mice (Fig. 4e). Confocal fluorescent microscopy showed that following MI, ubiquitinated proteins increased primarily in the interstitium of WT hearts. However, in mPDE5A/oe hearts, this accumulated mostly in myocytes (identified by Troponin T co-localization, Fig. 5a), consistent with the myocyte-selective decline in PKG activity in mPDE5A/oe hearts. This was paralleled by a decline in CHIP expression in myocytes as revealed by confocal microscopy (Fig. 5b). Total LV myocardial CHIP protein expression increased after MI in controls, but this was prevented in mPDE5A/oe hearts (Fig. 5c). Once again, we found CHIP mRNA expression was unaltered (Supplementary Fig. 5) so increased protein levels appeared due to post-translational modifications. We also examined proteasome activity finding it significantly reduced after MI, and further reduced in both SHAM and MI hearts with PDE5A/oe to block PKG activity (Supplementary Fig. 6).
Fig. 5. Impact of reduced in vivo myocyte PKG activity on CHIP.
a Fluorescent confocal images of transverse myocardial sections with antibody staining for ubiquitin (green), cardiac troponin T (cTnT, red), and nuclei (DAPI, blue). There is increased myocyte ubiquitin staining after MI in mPDE5/oe myocardium. b Fluorescent confocal images of same myocardium stained for CHIP (green), cTnT, and DAPI. Myocyte CHIP expression is reciprocally reduced in mPDE5/oe heart after MI. Each examination performed with biological duplicates. c Representative Western blot and summary data for total CHIP expression in both experimental groups at baseline (CON) and after MI. n = 6 biological replicates, 2WANOVA, p = 0.0011 for interaction of group and +/− MI; Tukey mct: *p < 5 × 10−6. d Example immunoblot and summary results for phospho-S20 CHIP, and ratio of pCHIP to total CHIP in peri-infarct zone before and after MI. n = 6 biological replicates; 2WANOVA (p = 0.0002 for interaction term); Tukey mct: *p = 10−5. e Myocardial PKG activity directly correlates with levels of p/t CHIP. Data are from MI study with rest and post MI data pooled in the two groups. Source data are provided as a Source Data file. Individual data and mean ± SEM shown in each summary panel.
Suppressing PKG activation in mPDE5A/oe hearts also blunted an increase in S20-CHIP phosphorylation found in the littermate controls (WT) hearts after MI (Fig. 5d). The decrease in both pS20 and total CHIP occurred in parallel, leaving the pS20/total CHIP ratio unaltered. This differs from the dissociation of pS20 from total CHIP upon acute PKG modulation (Fig. 2f), suggesting the two become linked with more chronic PKG suppression. We also found pS20-CHIP levels directly correlated with PKG activity measured in the same myocardium (Fig. 5e). Independent support for depressed pS20 CHIP in mPDE5A/oe post-MI myocardium was obtained using mass spectroscopy (Supplementary Fig. 7).
Female mice were also subjected to the same MI stress but displayed much less left ventricular dysfunction and chamber dilation as observed in males. Interestingly, enhanced female-dependent protective effects against MI were associated with both greater CHIP gene and protein levels (Supplementary Fig. 8).
CHIP S20 phosphorylation augments/stabilizes CHIP-HSC70 binding and post-translational protein stability
While CHIP has chaperone-independent activity, its binding to Hsc(Hsp70) is considered central to its functionality for promoting protein degradation7,8. S20 is located within the TPR domain of CHIP and coordinates with Hsc70 Lid-domain and the lysine residues (K597, K601) involved in protein-protein interactions (Fig. 6a). The thermodynamic impact of S20 phosphorylation on CHIP-Hsp70 binding was tested by isothermal titration calorimetry of CHIP-WT and CHIP-S20E. CHIP-S20E increased Hsp70 affinity by ~0.5 µM (59%), coupled with a 5.6 kJ mol−1 more negative enthalpy (ΔH; −26.4 ± 3.3 to −32.1 ± 0.9, p < 0.05) and a smaller change in entropy (ΔS, 25.5 ± 8.3 to 9.8 ± 3.7 J/Kmol, p < 0.05) compared to CHIP-WT. The ΔH and ΔS are consistent with our model based on the published structure of the CHIP-TPR/Hsp70-lid-tail complex26 predicting the S20E mutation (or S20 phosphorylation) would enhance the electrostatic interaction between CHIP and the Hsp70 lid domain, with an additional hydrogen bond potentially restricting motion of the lid with respect to CHIP-TPR, decreasing entropy for the CHIP/Hsp70 complex.
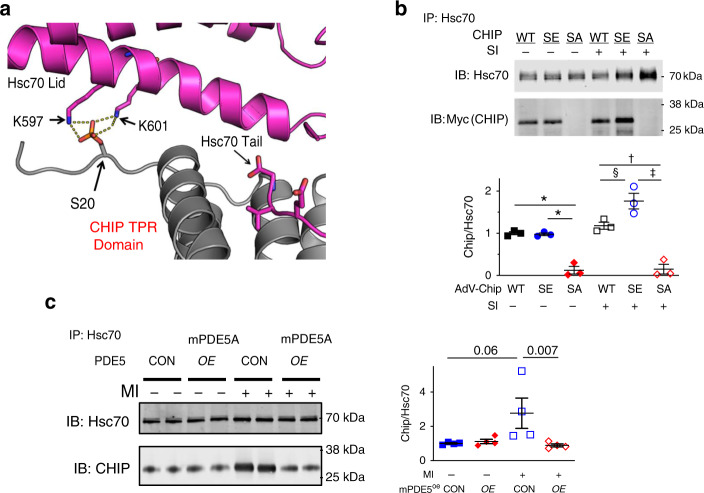
Fig. 6. PKG regulates CHIP-chaperone interaction and post-translational stability.
a Structural model of binding region between Hsc70 lid motif and the TPR region in CHIP. The location of S20 in reference to opposing lysine residues K597 and K601 in Hsc70 is shown. b Upper: example immunoprecipitation using Hsc70 as bait, and detecting Myc-tagged WT, SE, or SA CHIP in both control and SI conditions. There is minimal Hsc70-CHIP co-precipitation when CHIP-SA is expressed. Lower: summary data, n = 3 biological replicates/group, 1WANOVA for each condition, Holms-Sidaks mct: *p = 1.2 × 10−4, †0.003, ‡0.0005, §0.02. c Immunoprecipitation gels and summary data for Hsc70 and CHIP interaction in vivo with and without MI in mice with mPDE5oe and littermate controls (CON). Hsc70 is bait, and antibody detection using CHIP and Hsc70. N = 4 biological replicates/group, Kruskal-Wallis test, Dunns mct. P values in figure. Source data are provided as a Source Data file. Individual data and mean ± SEM shown in each summary panel.
To test whether thermodynamic stabilization of CHIP-Hsc70 due to S20 phosphorylation enhances protein binding, lysate from myocytes with or without exposure to SI were immunoprecipitated using Hsc70 as bait. We confirmed co-precipitation of CHIP and Hsc70 in cells expressing CHIP-WT or CHIP-S20E, but this was absent if CHIP-S20A was expressed (Fig. 6b). Reduced CHIP-Hsc70 interaction was also observed in vivo following MI in hearts with PKG activation suppressed by mPDE5A/oe, whereas this was enhanced in WT (Fig. 6c).
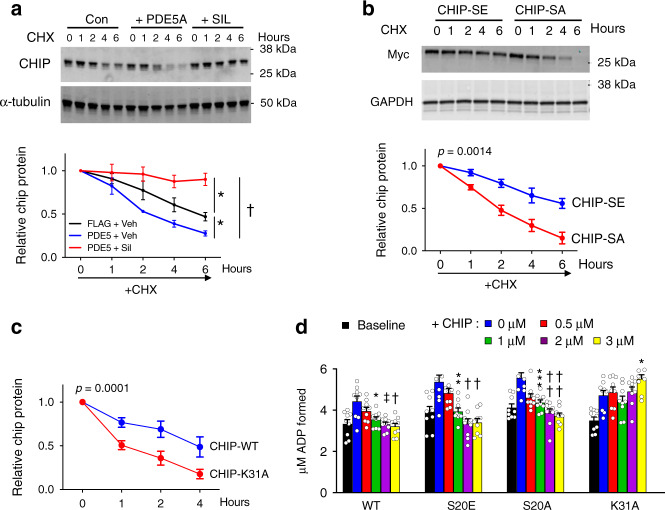
The preceding results suggested that phosphorylation of CHIP at S20 enhances its protein half-life. To test this, myocytes were treated with cycloheximide (100 µM) to block protein synthesis and then exposed to hydrogen peroxide (50 µM) as a stress stimulus. Under these conditions CHIP had a protein half-life of ~6 h (Fig. 7a), concordant with prior data27. This shortened to ~2 h if PKG activity was blocked by PDE5oe, but prolonged if PKG was stimulated by SIL. Using a similar protocol with Myc-tagged CHIP constructs, we found CHIP-S20A protein half-life was significantly shorter than CHIP-S20E (Fig. 7b).
Fig. 7. Influence of S20 modification on CHIP post-translational half-life.
a NRCMs infected with AdV-Flag or AdV-Flag-PDE5 to reduce PKG activation, and with SIL added to enhance PKG activation placed in media containing cycloheximide (CHX, 100 µM) to block protein synthesis and H2O2 (50 µM) to induce oxidative stress. N = 4–5 separate experiments with 5 independent time points per experiment. ANCOVA results for differences between paired relations: *p = 0.001; †0.000009. b Same general protocol but with NRCMs over-expressing either S20A or S20E CHIP mutants. P-value for slope difference by ANCOVA. c Same protocol but WT and K31A CHIP mutants. P-value for offset difference by ANCOVA. d Hsp70/Hsp40 dependent protein refolding ATPase assay showing capacity of CHIP to reduce activity in a dose dependent manner in wild type (WT), S20A, and S20E CHIP mutants. K31A CHIP expression prevents this negative modulation. N = 9 biological replicants, RMANOVA performed within each experiment, Dunnets mct: *p = 0.004; **0.006, ***0.002; †0.0005, ††0.0002, ‡p = 0.002 – each versus no CHIP added (blue bar). Source data are provided as a Source Data file. Regressions (a–c) and summary plot (d) each display mean ± SEM.
Mutation of CHIP Lys 31 (K30 in humans) to Ala is known to abolish CHIP-chaperone interactions, and with it, the inhibitory effect of CHIP on chaperone ATPase activity required for their protein refolding function8. As the impact of this mutation on CHIP half-life was not reported, we examined it here, and find the K30A mutation indeed shortened half-life significantly to levels observed with S20A mutants (Fig. 7c). There was, however, a striking difference between the two mutations with respect to their inhibition of Hsp70 ATPase activity. As expected, Hsp70 activity was dose-dependently inhibited by CHIP-WT, and effect that was lost with CHIP-K31A. However, both CHIP-S20E and S20A displayed similar efficacy as CHIP-WT, the latter despite its disruption of Hsc-70 interaction (Fig. 7d). This indicates that the S20A mutant destabilizes the CHIP and Hsc (Hsp) 70 interaction but does not prevent it, and chaperone activity is still modulated. However, loss of S20 phosphorylation favors CHIP protein degradation, and the reverse increased protein half-life and functionality.
Knock-in mice expressing CHIP-S20E are protected against MI-proteotoxicity
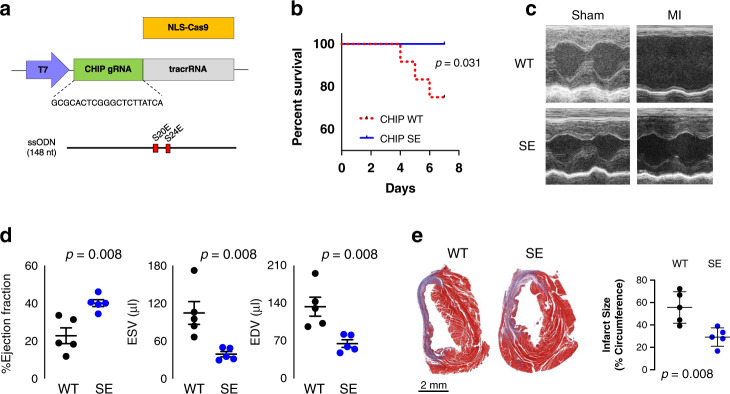
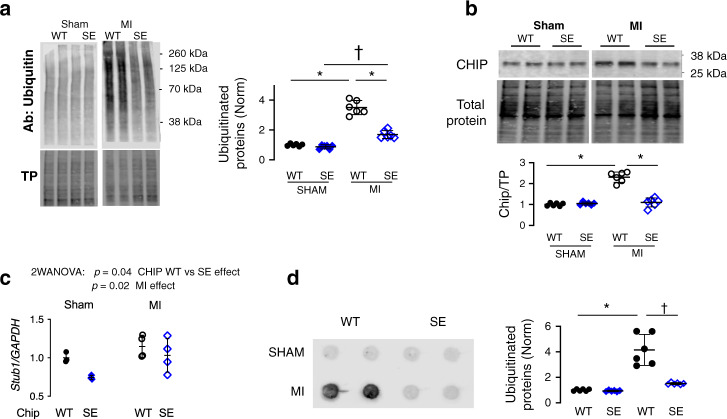
To test whether S20 phosphorylation of CHIP confers cardioprotection by enhancing PQC in vivo, we generated global CHIPS20E KI mice using CRISPR/Cas9 (Fig. 8a). Mice are born in normal Mendelian ratios and exhibit normal resting cardiac function and lifespan (Supplementary Table 1). We then tested if expression of CHIPS20E protects against cardiac damage/dysfunction following myocardial infarction, focusing on males as they displayed more severe post-MI disease. Compared to littermate controls CHIPS20E (both homozygotes and heterozygotes) subjected to MI exhibited similar lower mortality (Fig. 8b). Echocardiograms showed preserved function in CHIPS20E mice after MI (Fig. 8c, d), and reduced infarct size (Fig. 8e). These improvements were accompanied by reduced accumulation of ubiquitinated proteins (Fig. 9a). Whereas CHIP protein level rose after MI in controls, it was unaltered in KI mice (Fig. 9b), perhaps reflecting 100% of the mutant CHIP being pseudo-phosphorylated. CHIPS20E gene expression was slightly lower than WT (Fig. 9c), that might reflect longer CHIP protein half-life. To test for CHIP-S20E enhanced functionality, we determined if the mutation influenced clearance of insoluble protein aggregates. Insoluble protein aggregates were trapped by dot blot filter assay and then stained for ubiquitin. CHIP-S20E hearts had a marked clearance of ubiquitinated proteins after MI (Fig. 9d). Thus, expression of CHIP-S20E at endogenous levels is cardioprotective after MI in vivo and improves proteostasis.
Fig. 8. Mice with global S20E KI are protected against myocardial infarction.
a Guide RNA to target CHIP S20/S24 region, and the replacement RNA that includes a silent mutation of the PAM sequence to further enhance homology directed repair. b CHIPS20E (n = 10 heterozygous; n = 7 homozygous) mice have reduced mortality following myocardial infarction due to total coronary occlusion. c Example M-mode echocardiograms show improved wall motion and less heart dilation in CHIPS20E (SE) KI mice versus littermate (WT) controls. d Summary data for LV function and volumes from same experiment. N = 5 mice for each group, mean ± SEM, p-value shown for Mann–Whitney test unpaired test. e Whole heart cross-section histology shows larger infarct territory in WT versus CHIPS20E hearts. Summary quantitation (right). N = 5 biological replicates, mean ± SD, p value for Mann–Whitney unpaired test. Source data are provided as a Source Data file.
Fig. 9. Protein ubiquitination is reduced post MI in CHIPS20E mice.
a Example protein ubiquitination gel and total protein (TP) loading gel from CHIPS20E and littermate controls (WT) after Sham or MI intervention. Summary results: n = 6 biological replicates, 2WANOVA; interaction term p = 4 × 10−7, Tukey mct: *p ≤ 3 × 10−9, †3 × 10−4. b Example immunoblot for total CHIP protein expression at baseline and post-MI in the peri-infarct zone from WT and CHIPS20E mice. Summary: 6 biological replicates, 2WANOVA, Tukey mct: *p ≤ 5 × 10−10. c CHIP gene expression (Stub1/Gapdh) same study measured in Sham and post MI mice. 2WANOVA results provided in figure. d Filter trap assay for insoluble proteins probed for ubiquitin which are increased after MI in WT but maintained at low levels in CHIPS20E myocardium. N = 6 biological replicates, 2WANOVA, Tukey mct: *p = 12 × 10−7, †p = 1.9 × 10−6. Source data are provided as a Source Data file. Individual data and mean ± SD shown in each summary panel.
Discussion
The capacity of CHIP to enhance the clearance of damaged/misfolded proteins and suppress proteotoxicity is relevant to immunogenicity28, aging29, metabolic stress12, and many human diseases including cardiac and neurological10,16,30,31. Here we reveal a mechanism for post-translational augmentation of CHIP protein levels and functionality by means of PKG-mediated phosphorylation at serine 20. The result is a cascade of effects: increased interaction with co-chaperone Hsp (Hsc) 70, increased post-translational protein half-life resulting in a net rise in protein levels, and greater clearance of insoluble ubiquitinated protein aggregates in conditions of proteotoxic stress. This can provide a more rapid response to counter such stress that leverages kinase modification rather than requiring de novo gene and protein synthesis. To our knowledge, the CHIPS20E KI mouse is the first point-mutation in vivo model involving CHIP. The potency of this modification is revealed by the substantial gain-of-function it confers to counter infarction injury even in heterozygotes. Our results have therapeutic implications, given the growing role played by CHIP in cardiac, skeletal muscle, neurogenerative, and oncologic disease, and clinical armamentarium available to stimulate PKG.
CHIP pS19 (human equivalent of S20 in mouse) is found in human phospho-proteomic data bases20,21, however, we uncovered only one prior study examining any impact32. This study in mice identified CHIP-pS20 in a kinase-interactome screen for cyclin-dependent kinase 5 (CDK5), and reported that it disrupted CHIP binding to truncate apoptosis-inducing factor (AIF) leading to neuronal apoptosis. The prior study did not assess whether pS20 altered CHIP protein levels, chaperone binding, or any broader functionality, nor was there in vivo confirmation. The CHIP S20E KI mouse expresses this mutation in all cells, yet we observe no detrimental neurological or other effects. CDK5 is expressed in the heart but in contrast to PKG, is not activated by myocyte stress33. Most importantly, we find consistent protective effects from pS20 CHIP associated with increased protein levels and chaperone interaction.
The current data support a linkage between CHIP S20 phosphorylation and its enhanced binding to Hsc70 and post-translational protein level. Expression of the K31A mutant, which also prevents CHIP-chaperone binding markedly reduced post-translational half-life similar to that seen with the S20A mutant. The measured increased biophysical interaction for CHIP-S20E and associated increased protein stability further supports this. However, unlike K31A mutants that prevent chaperone interaction, the S20A mutation did not alter Hsp70/Hsp40 complex ATPase activity, a hallmark of CHIP-mediated inhibition of refolding activity. Thus, the proteins must still engage. Our cell results with CHIP-S20A indicate that while Hsc70 interactions may not be detectable by under the stringent IP conditions, they are not functionally eliminated.
While this study focused on the impact of PKG on CHIP, studies have also shown the kinase modifies many other pathways relevant to myocardial pathological signaling and remodeling34, and these could also have contributed to the net effects observed. Among these mechanisms are PKG-mediated mitochondrial protection and cytoprotection against ischemia/infarction35, activating the regulator of G-protein signaling proteins RGS2 and RGS4 to inhibit Gq-receptor coupled signaling, blocking transient receptor potential canonical channel 6 to blunt fibrosis and hypertrophy36,37, and activating tuberous sclerosis complex 2 (TSC2) to blunt the mechanistic target of rapamycin complex-1 (mTORC1) stimulating autophagy and reducing pathological growth25. Specific to the UPS, PKG activates the 26S proteasome enhancing degradation of damaged/misfolded proteins in models of heart disease and neurodegenerative22–24. The current study confirms this, but also shows PKG has a constitutive role since genetic suppression of its activity in both myocytes and intact hearts depressed proteasome activity. While an impact from these alternative PKG effectors cannot be ignored, it is intriguing that preventing S20 phosphorylation of CHIP was itself sufficient to blunt PKG-mediated protection in myocytes. Moreover, the opposite—expressing the S20E form to mimic PKG modulation was very protective by itself in cells and intact hearts against ischemic stress. This indicates that CHIP-PKG interaction is important and influential, particular in settings where PQC is sub-optimal and its enhancement beneficial.
Both CHIP and Hsc70 are involved in chaperone-mediated and chaperone-assisted selective autophagy38,39, and whether and how these roles are impacted by S20 modification remains to be determined. CHIP interacts with the proteasome and the lysosome for substrate degradation and is itself ubiquitinated6–9, however the regulators of CHIP protein degradation remain uncertain. Further studies are needed to determine the mechanisms regulating the turnover of CHIP, the degradation system responsible (e.g., proteasome, lysosome, or both), and how the lack of S20 phosphorylation (e.g., S20A) accelerates CHIP recycling. There are feedback loops involved in CHIP binding to heat shock element of heat shock factor-1, a transcriptional protein that, once activated by CHIP, increases transcription of Hsps such as Hsp7040. Whether pS20 modifies this also remains to be tested. Here we determined the S20 site regulates binding between CHIP and its primary cognate chaperone Hsc70 which is structurally near identical to Hsp70. Additional studies are needed to test if the interaction between CHIP and other co-chaperones (e.g., Hsp90, Bag1, Bag3) are also regulated by S20 phosphorylation. Oxidative stress reduces CHIP protein half-life41 and also depresses the functionality of myocardial PKG1α42. Whether these events are also linked remains to be tested. Lastly, the full scope of kinases that can modify CHIP S20 remains to be determined, though PKG is likely to be relevant in many tissues.
The intersection of PKG signaling with enhanced CHIP functionality is important, since from a clinical perspective, it suggests new therapeutic targets suitable for an array of existing and developing PKG activation strategies. Clinically used agents include nitric oxide donors, organic nitrates, soluble guanylate cyclase stimulators, and PDE5 inhibitors that enhance NO-dependent cGMP synthesis, and natriuretic peptides (NP), neprilysin inhibitors, and PDE9 inhibitors that increase NP-dependent cGMP stimulation43–45. All of these targets have existing small molecules or peptides in clinical use and/or under active studies in humans with cardiac or neurological disease. While PKG activation has historically been leveraged for its vasodilator capacity, the present data reveal its role in PQC by enhancing CHIP functionality. Both PKG and CHIP are widely expressed in many cell types, including the brain and cancer cells, and thus we believe the current findings will likely apply to the heart as well as other tissues, providing novel therapeutic opportunities.
Methods
Mouse models
Studies used mice (C57BL6/J background) with myocyte-specific PDE5A over-expression under control of an α-MHC tetracycline-inducible promoter46 and their littermate controls. We also generated a new global knock-mouse expressing CHIP S20E/S24E (SE) using CRISPR/Cas9 targeting/insertion methods (Transgenic Mouse Core Laboratory, Johns Hopkins University). CHIP guide RNA was designed using an algorithm developed by Ranganathan47, and subsequently cloned into [a T7 expression vector]. In vitro transcription for the guide RNA was achieved with the NEB HiScribe T7 High Yield RNA Synthesis kit, and subsequent purification with the Ambion MEGAclear Transcription Clean-up Kit. Cas9 protein was from PNA Bio, and ssODN for the S20E/S24E point mutations from Integrated DNA Technologies. C57Bl/6 blastocyst injections were performed with a mix consisting of: 30 ng/μl Cas9 protein, 12.5 ng/μl guide RNA, and 20 ng/μl ssODN. Two founders were generated, and were subsequently backcrossed into C57Bl/6J mice to eliminate potential off-target mutations from Cas9 editing. The colony was maintained by breeding heterozygous males and heterozygous females to generate knock-in homozygous, knock-in heterozygous, and wild-type littermates at a 25:50:25 ratio. Custom PCR genotyping primers are:
SE Forward: GGCGGCGAGCCTGATAAGGAG
WT Forward: GGCGGCAGCCCTGATAAGAGC
Reverse: CCTAGACTCGGGAACAGCAATCCGG
Myocardial infarction (MI)
Male and female mice (aged ~3–4 months) were subjected to transmural myocardial infarction by mid-left anterior descending artery ligation under open thoracotomy48. Controls underwent open chest surgery and LAD isolation without ligation. Data obtained 1-week after infarction included 2D echocardiography (Vevo2100, 20-MHz; Visualsonics) in conscious mice to measure LV volume and ejection fraction, with analysis blinded to study group. The protocol was approved by the Johns Hopkins Animal Care and Use Committee.
Cultured cardiomyocyte studies
Primary cultures of NRCMs were prepared as described45. After overnight culture, subsets of NRCMs were infected with adenovirus (AdV) containing a FLAG-tagged PDE5, FLAG control, Myc-tagged CHIP WT or K31A (provided by Dr. Cam Patterson), S20E or S20A (Welgen Inc., Worcester, MA) (10MOI). Forty-eight hours after infection, cells were exposed to either simulated ischemia (SI) or normal conditions. SI involved incubating cells in DMEM containing deoxyglucose (20 mM), NaCl (125 mM), KCl (8 mM), KH2PO4 (1.2 mM), MgSO4 (1.25 mM), CaCl2 (1.2 mM), NaHCO3 (6.25 mM), sodium lactate (5 mM), and HEPES (20 mM) with a pH of 6.6. Cells were then placed in an incubator chamber (Billups-Rothenberg) with 95% nitrogen, 4% carbon dioxide, and 1% oxygen. Normoxic controls were incubated in DMEM supplemented with 10% FBS in air composed of 95% carbon dioxide and 5% oxygen. RNA or protein was harvested after 48 hours of SI. Cells were also treated with sildenafil (1 µM) or vehicle, or small interference RNA (siRNA, 100 pM) specific for rat Stub1 (Qiagen, Cat. #: SI01532251) or scrambled control provided 24 h after plating and transfected with Xfect (Takara) per manufacturer protocol.
Protein analysis
Tissue and cultured cardiomyocytes were lysed in 1X lysis buffer (Cell Signaling Technology), protein concentration was measured by bicinchoninic acid assay (Pierce), and extracts were subjected to SDS-PAGE using Novex Tris-Glycine Gels (4–20% gradient gel, Life Technologies) or Mini-protean TGX Gels (4–20% gradient gel, Bio-Rad), transferred to nitrocellulose membranes, and probed with various primary antibodies from Cell Signaling Technology: CHIP (#2080S), Hsc70 (#8444S), Myc (#2276S), GAPDH (#2118S), and α-tubulin (#3878S), AbMart Inc.: p-CHIP S20 (Custom antibody, Shanghai, China; Project: 25011–1), Sigma: ubiquitin antibody (#SAB4503053), and Li-Cor: fluorescence-labeled secondary antibodies (#926-32211, 926-32210, 926-68023, or 926-68022). Gels were imaged (Odyssey, Li-Cor) and band intensity quantified (Odyssey Software 3.1).
Proteasome activity assay
Snap-frozen tissues and cultured NRCMs were homogenized on ice in cytosolic extraction buffer (50 mM Tris-HCl pH 7.5, 250 mM Sucrose, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM DTT). Protein concentrations were determined with bicinchoninic acid (BCA) reagents (Pierce) and equally concentrated in proteasome assay buffer (50 mM Tris-HCl pH 7.5, 40 mM KCl, 5 mM MgCl2, and 1 mM DTT). Chymotrypsin-like, trypsin-like, and caspase-like activities were determined in the presence of 28 μM (chymotrypsin) or 14 μM (trypsin and caspase) ATP, utilizing the following fluorogenic substrates: Suc-LLVY-AMC (18 μM, Boston Biochem #S280), Ac-RLR-AMC (45 μM, Boston Biochem #S290), and Z-Leu-Leu-Glu-AMC (40 μM, Boston Biochem #S230), respectively. The plate was read at an excitation wavelength of 380 nm and an emission wavelength of 460 nm using a Spectramax M5 (Molecular Devices). Activity was combined, using a 50:25:25 ratio for relative contribution of chymotrypsin, trypsin, and caspase-like activities49.
LDH/MTT cytotoxicity assay
Lactate dehydrogenase (LDH) leakage and (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (MTT) were assessed in cultured cardiomyocytes and media after SI from the same culture dish. LDH was assayed by commercial assay (Roche) following manufacturer’s protocol, read in 96-well microplates on a Spectramax M5 (Molecular Devices) at 490 nm absorbance. For MTT, cells were washed in phosphate-buffered saline, MTT (Sigma-Aldrich) working solution added to the culture dish, incubated at 37 °C, solubilized with acidic isopropanol, transferred to a 96 well plate, and read at 570 nm with background subtraction at 650 nm on a Spectramax M5.
Cycloheximide (CHX) chase assay
NRCMs were incubated in serum-free DMEM containing 10 μM hydrogen peroxide to induce oxidative stress and 100 μM CHX (Sigma-Aldrich) to block further protein synthesis, as described22. Cells were harvested at consecutive time points (e.g., 0, 1, 2, 4, and 6 h) after CHX and whole-cell lysates analyzed by immunoblot for CHIP or Myc (Myc-CHIP).
Tissue histology/immunostaining
The myocardium was fixed in 4% paraformaldehyde, paraffin embedded and sectioned into 4 μm slices, and stained with Masson’s Trichrome. Fibrosis was quantified by color-based pixel count and intensity in parallel sections of the ischemic and remote areas (Aperio Image Scope). Transmural infarct size was estimated as the arc angle subtended by dense scar in a mid-MI cross section divided by 360° and expressed as percent. Immunostaining for ubiquitin (Sigma) and CHIP (Cell Signaling) was performed using AlexaFluor 488 (#A-11008 Lot#1345061, used at 1:500) conjugated secondary antibodies following the manufacturer’s protocol. Counterstain was performed using Troponin-T (#A-11004 used at 1:500) (Thermo Scientific) antibodies and DAPI (Invitrogen #S36968). Image acquisition was performed on a Leica confocal microscope (TCS SPE II) at ×40 magnification.
Quantitative real-time PCR
Total RNA was extracted with Trizol Reagent (Invitrogen) from myocardium or isolated myocytes per manufacturer’s instructions, then reverse transcribed into cDNA using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, Life Technologies). cDNA was subjected to PCR amplification using either TaqMan specific primers (Applied Biosystems) for CHIP (Stub1, mouse #Mm00490634_m1, rat #Rn01751757_g1) or gylceraldehyde-3-phosphate dehydrogenase (GAPDH, mouse #99999915_g1, rat #Rn01775763_g1) (Applied Biosystems). The Biorad CFX384 qPCR machine was used to determine the threshold cycle (Ct) values determined by crossing point method, were normalized to the respective housekeeping gene GAPDH (Applied Biosystems) values for each run.
PKG and PDE5A activity assay
In vitro PKG activity was measured by colorimetric assay (Cyclex), and PDE5A activity by fluorescence polarization assay (Molecular Probes) following manufacture protocols of cell lysates from either from heart tissues or NRCMs. PKG activity is assessed by detecting phosphorylation of a pseudo-substrate, while PDE5 activity is measured by fluorescence polarization assay under linear conditions with or without PDE5A inhibitor (sildenafil 1 μM).
Recombinant proteins for in vitro assays
Human HSP40 (DnaJB1), human CHIP and CHIP point mutants in pET30a with a C-terminal His-tag and hHsc70 (HSPA8) wild type with an N-terminal His-tag in pCOLA-Duet1 were expressed in BL21(DE3) and purified on Ni-NTA (Qiagen). Hsc70 and CHIP proteins were induced and expressed for 22 h at 18 °C. Hsp40 protein was induced and expressed for 5 h at 25 °C. Required stock solutions of proteins were prepared by diluting high concentrated stocks in protein storage buffer into protein storage buffer, 20 mM HEPES (pH 7.4), 150 mM NaCl. Human, active PKG1α purified from Sf9 cells was purchased (SRP0371, Sigma).
PKG and CHIP/CHIP-S20A in vitro kinase assays
Aliquots of recombinant CHIP or PKG1α were diluted to either 12.5 μM or 0.25 μM in Kinase Dilution Buffer II (K23-09, SignalChem: 5 mM MOPS, pH 7.2, 2.5 mM β-glycerol-phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 50 ng/μl BSA, 0.5 mM DTT). A 250 µM stock solution (5X) of ATP was made in Kinase Assay Buffer I (K01-09 SignalChem: 25 mM MOPS, pH 7.2, 12.5 mM β-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT). A 100 μM cGMP solution was purchased (#G47-09, SignalChem). The final buffer composition of complete reactions was 10.5 mM MOPS, 4 mM Beta-glycerol-phosphate, 8 mM MgCl2, 1.6 mM EGTA, 0.64 mM EDTA, 0.08 mM DTT, 10 µM cGMP, 50 µM ATP. CHIP or PKG1α were used at final concentrations of 2.5 μM or 0.1 μM, respectively (25:1 molar ratio). Reactions were started with the addition of the 5X ATP solution, and incubated for 30 °C for 1 h. Aliquots were either flash frozen for phospho-proteomic analysis or used for reducing SDS-PAGE/immunoblot analysis. Controls included no substrate (CHIP) or no kinase (PKG1α). Experimental triplicates were run on multiple days to confirm findings.
Mass spectrometry was performed at the UNC-CH Proteomics Center (Chapel Hill, NC, USA) as described50. Samples were reduced, alkylated and digested with trypsin overnight. After digestion, peptides were desalted using C18 spin columns and dried down via vacuum centrifugation. Each sample was analyzed in technical duplicate by LC/MS/MS using the Easy nLC 1200-QExactive HF (Thermo). Data were searched against a Uniprot E.Coli database (Uniprot E.Coli proteome ID (UP000000625) available at), appended with human CHIP-WT, CHIP-S19A, and PKG1α sequences, using Sequest within Proteome Discoverer (v2.1). Data were filtered using an FDR of 1% and peak areas were mean normalized.
Isothermal titration calorimetry
Isothermal titration calorimetry was carried out using a TA Instruments NanoITC equilibrated at 25 °C and utilizing a stir speed of 600 rpm. Experiments were conducted in triplicate and data represent the average ± one standard deviation. Titrations utilized 350 µM WT or S20E CHIP titrated into 50 µM Hsp70 (385–641) as a series of twenty-four 2 µL injections.
Chaperone refolding ATPase activity assay
Human HSP40 (DnaJB1), human CHIP and CHIP point mutants in pET30a with a C-terminal His-tag and hHsc70 (HSPA8) wild type with an N-terminal His-tag in pCOLA-Duet1 were expressed in BL21(DE3) and purified on Ni-NTA (Qiagen). Hsc70 and CHIP proteins were induced and expressed for 22 h at 18 °C. Hsp40 protein was induced and expressed for 5 h at 25 °C. Required stock solutions of proteins were prepared by diluting high concentrated stocks in protein storage buffer into protein storage buffer, 20 mM HEPES (pH7.4), 150 mM NaCl. Hsc70 and Hsp40 were used at a concentration of 0.5 µM and 0.25 µM, respectively, with a gradient of CHIP protein concentrations (0.5–6 µM). Hsp40-stimulated ATPase activity of Hsc70 was measured using fluorescent polarization with the Transcreener® ADP2 FP Assay (BellBrook Labs, 3010-1 K) on a Clariostar plate reader (BMG) in 384 well black, non-binding plates (Greiner, 784900). Optimized instrument settings were used: Excitation wavelength: 590 nm; Emission wavelength: 675 nm; Required value: 5%; Target mP: 20; Gain A: 1824; Gain B: 1854; Focal Height: 12.4 mm. Values of µM ADP formed in the reactions were interpolated from an ATP/ADP standard curve run in parallel.
Statistical analysis
Data are presented by the individual experimental results along with mean ± SEM. Data were processed with Microsoft Excel Version 16. Group comparisons were performed by 1- or 2-way ANOVA. Multiple comparisons testing was performed by Holm-Sidak or Tukey’s test using Prism 8 software. Data that were not normally distributed were assessed by non-parametric analysis (Kruskal Wallis) test and Dunn’s test for multiple comparisons. Sample sizes and individual statistical results for all analyses are provided in the figures or legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was supported by NIH-NHLBI (HL-135827, HL-119012, HL-093432, HHSN268201000032C), Fondation Leducq TransAtlantic Network of Excellence, Peter Belfour Laboratory (D.A.K.), American Heart Association Mid-Atlantic Fellowship Grant (M.J.R., D.L.) and SDG 16SDG26960000 (R.C.P.) and CDA 18CDA34110140 (M.J.R.), NHLBI-T32-HL-07227 (M.J.R.), Amyloidosis Foundation Donald C. Brockman Memorial Research Grant (M.J.R.), Max Kade Fellowship by the Austrian Academy of Sciences (P.P.R.), NIH F31-HL134196 (K.M.K.), NIH F31-HL HL143905 (B.D.E.), NIH-NIA-AG-061188 (J.C.S.), NIH-NIGMS GM-R35128595 (R.C.P.), NSF MCB 1552113 (R.C.P.), and Miami University through the Robert H. and Nancy J. Blayney Professorship (R.C.P.). Special thanks to Lee Graves and Laura Herring at The University of North Carolina Michael Hooker Proteomics Center.
Source data
Author contributions
M.J.R. first identified CHIP modification and the concept of the study, performed the majority of experiments, and organized and analyzed most of the data. C.U.O. performed mouse echocardiography and analysis. R.S.H. analyzed the direct regulation of CHIP S20 by PKG, performed CHIP refolding assay, and validated the phospho-S20 CHIP antibody. K.M.K.-S. played a key role in designing the knock-in CHIP-SE mouse and generate genotyping primers. D.D. determined the half-life of CHIP full length and site mutant proteins during oxidative stress. M.I.A. assessed cardiac protein quality control in the newly generated CHIP S20 knock-in mice. P.P.R. performed the myocardial infarction (and sham) surgeries on phosphodiesterase 5 overexpression mice and immunostaining for CHIP and ubiquitin. S.M. performed a number of cellular/molecular assays for the study. B.L.D.-E. performed PKG-Chip interaction studies and analyzed data. R.J.H. performed the PKG-kinome proteomics analysis. C.V. contributed molecular signaling data and helped generate constructs. H.Z. and M.M.M. performed isothermal titration calorimetry and analysis. V.A. performed immunoprecipitation of CHIP and HSC70 in phosphodiesterase 5 overexpression mice and CHIP S20 mutant cell models. V.S.H. performed PKG activity assays and contributed molecular signaling data. D.I.L. provided PKG and PDE5 assay assistance to many in the study. M.S. generated numerous plasmids and viral vectors used for the study. J.E.V.-E. supervised the proteomics study, and provided grant support, assistance with project development and the preparation of the manuscript. M.S.W. contributed insights regarding CHIP function and regulation. R.C.P. supervised H.Z. and M.M.M. during isothermal titration calorimetry, modeled CHIP S20 phosphorylation-HSC70 tail interaction, and helped with the preparation of the manuscript. J.C.S. supervised R.S.-H. and C.V., provided plasmid constructs, cell lines, and mouse heart tissue, contributed insights of CHIP function and regulation, and assisted with preparation of the manuscript. D.A.K. conceived and directed the overall study, provided the majority of grant support for the research, provided extensive scientific input throughout its development, substantially contributed to data presentation and analysis, and was responsible for the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Source data are provided with this paper. Any remaining data that support the results of the study will be available from the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021531.
Competing interests
Dr. Ranek and Kass are co-inventors of a provisional patent related to the use of CHIP phosphomutants for gene therapy of protein quality control-aggregation disorders. The remaining authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Rakesh Kukreja and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-18980-x.
References
- 1.Ciechanover A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 2.Sandri M, Robbins J. Proteotoxicity: an underappreciated pathology in cardiac disease. J. Mol. Cell Cardiol. 2014;71:3–10. doi: 10.1016/j.yjmcc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu C, et al. Inadequate ubiquitination-proteasome coupling contributes to myocardial ischemia-reperfusion injury. J. Clin. Invest. 2018;128:5294–5306. doi: 10.1172/JCI98287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Q, et al. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front Aging Neurosci. 2016;8:303. doi: 10.3389/fnagi.2016.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranek MJ, Stachowski MJ, Kirk JA, Willis MS. The role of heat shock proteins and co-chaperones in heart failure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20160530. doi: 10.1098/rstb.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 8.Narayan V, et al. Protein-protein interactions modulate the docking-dependent E3-ubiquitin ligase activity of carboxy-terminus of Hsc70-interacting protein (CHIP) Mol. Cell Proteom. 2015;14:2973–2987. doi: 10.1074/mcp.M115.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB1 regulates TFEB-induced autophagy-lysosome pathway. Embo J. 2017;36:2544–2552. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schisler JC, et al. CHIP protects against cardiac pressure overload through regulation of AMPK. J. Clin. Invest. 2013;123:3588–3599. doi: 10.1172/JCI69080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Xu Z, He XR, Michael LH, Patterson C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2836–H2842. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 12.Tawo R, et al. The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell. 2017;169:470–482.e413. doi: 10.1016/j.cell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi CH, et al. Disrupted structure and aberrant function of CHIP mediates the loss of motor and cognitive function in preclinical models of SCAR16. PLoS Genet. 2018;14:e1007664. doi: 10.1371/journal.pgen.1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu. Rev. Pharm. Toxicol. 2015;55:353–371. doi: 10.1146/annurev-pharmtox-010814-124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajiro M, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat. Cell Biol. 2009;11:312–319. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 16.Singh AK, Pati U. CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of beta-secretase. Aging Cell. 2015;14:595–604. doi: 10.1111/acel.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu CW, Zhang TP, Wang HX, Yang H, Li HH. CHIP enhances angiogenesis and restores cardiac function after infarction in transgenic mice. Cell Physiol. Biochem. 2013;31:199–208. doi: 10.1159/000343361. [DOI] [PubMed] [Google Scholar]
- 18.Cabral Miranda F, et al. CHIP, a carboxy terminus HSP-70 interacting protein, prevents cell death induced by endoplasmic reticulum stress in the central nervous system. Front. Cell Neurosci. 2014;8:438. doi: 10.3389/fncel.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaglione KM, et al. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 21.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranek MJ, Kost CK, Jr., Hu C, Martin DS, Wang X. Muscarinic 2 receptors modulate cardiac proteasome function in a protein kinase G-dependent manner. J. Mol. Cell Cardiol. 2014;69:43–51. doi: 10.1016/j.yjmcc.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VerPlank JJS, Tyrkalska SD, Fleming A, Rubinsztein DC, Goldberg AL. cGMP via PKG activates 26S proteasomes and enhances degradation of proteins, including ones that cause neurodegenerative diseases. Proc. Natl Acad. Sci. USA. 2020;117:14220–14230. doi: 10.1073/pnas.2003277117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranek MJ, et al. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature. 2019;566:264–269. doi: 10.1038/s41586-019-0895-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, et al. A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure. 2015;23:472–482. doi: 10.1016/j.str.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito AT, et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circulation Res. 2010;106:1692–1702. doi: 10.1161/CIRCRESAHA.109.214346. [DOI] [PubMed] [Google Scholar]
- 28.Brehm A, Kruger E. Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases. Semin. Immunopathol. 2015;37:323–333. doi: 10.1007/s00281-015-0486-4. [DOI] [PubMed] [Google Scholar]
- 29.Branicky R, Hekimi S. Proteostasis or aging: let the CHIPs fall where they may. Dev. Cell. 2017;41:126–128. doi: 10.1016/j.devcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Cao Z, et al. CHIP: A new modulator of human malignant disorders. Oncotarget. 2016;7:29864–29874. doi: 10.18632/oncotarget.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao M, Yan M, Zhang J, Xu Q, Chen W. Carboxy-terminus Hsc70 interacting protein exerts a tumor inhibition function in head and neck cancer. Oncol. Rep. 2017;38:1629–1636. doi: 10.3892/or.2017.5827. [DOI] [PubMed] [Google Scholar]
- 32.Kim C, et al. Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 2016;23:333–346. doi: 10.1038/cdd.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JM, Poolman RA, Brooks G. Role of G1 phase cyclins and cyclin-dependent kinases during cardiomyocyte hypertrophic growth in rats. Am. J. Physiol. 1998;275:H814–H822. doi: 10.1152/ajpheart.1998.275.3.H814. [DOI] [PubMed] [Google Scholar]
- 34.Adler J, Kuret A, Längst N, Lukowski R. Targets of cGMP/cGKI in cardiac myocytes. J. Cardiovasc. Pharm. 2020 doi: 10.1097/fjc.0000000000000817. [DOI] [PubMed] [Google Scholar]
- 35.Inserte J, Garcia-Dorado D. The cGMP/PKG pathway as a common mediator of cardioprotection: translatability and mechanism. Br. J. Pharm. 2015;172:1996–2009. doi: 10.1111/bph.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koitabashi N, et al. Cyclic GMP/PKG-dependent inhibition of TRPC6 channel activity and expression negatively regulates cardiomyocyte NFAT activation Novel mechanism of cardiac stress modulation by PDE5 inhibition. J. Mol. Cell Cardiol. 2010;48:713–724. doi: 10.1016/j.yjmcc.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takimoto E, et al. Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J. Clin. Invest. 2009;119:408–420. doi: 10.1172/JCI35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Dai Q, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dikshit P, Jana NR. The co-chaperone CHIP is induced in various stresses and confers protection to cells. Biochem. Biophys. Res. Commun. 2007;357:761–765. doi: 10.1016/j.bbrc.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura T, et al. Prevention of PKG1alpha oxidation augments cardioprotection in the stressed heart. J. Clin. Investig. 2015;125:2468–2472. doi: 10.1172/JCI80275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breitenstein S, Roessig L, Sandner P, Lewis KS. Novel sGC stimulators and sGC activators for the treatment of heart failure. Handb. Exp. Pharm. 2017;243:225–247. doi: 10.1007/164_2016_100. [DOI] [PubMed] [Google Scholar]
- 44.McMurray JJ, et al. Angiotensin-neprilysin inhibition versus enalapril in heart failure. N. Engl. J. Med. 2014;371:993–1004. doi: 10.1056/NEJMoa1409077. [DOI] [PubMed] [Google Scholar]
- 45.Lee DI, et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519:472–476. doi: 10.1038/nature14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M, et al. Myocardial remodeling is controlled by myocyte-targeted gene regulation of phosphodiesterase type 5. J. Am. Coll. Cardiol. 2010;56:2021–2030. doi: 10.1016/j.jacc.2010.08.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranganathan V, Wahlin K, Maruotti J, Zack DJ. Expansion of the CRISPR-Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat. Commun. 2014;5:4516. doi: 10.1038/ncomms5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rainer PP, et al. Cardiomyocyte-specific transforming growth factor beta suppression blocks neutrophil infiltration, augments multiple cytoprotective cascades, and reduces early mortality after myocardial infarction. Circulation Res. 2014;114:1246–1257. doi: 10.1161/CIRCRESAHA.114.302653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majetschak M. Regulation of the proteasome by ATP: implications for ischemic myocardial injury and donor heart preservation. Am. J. Physiol. Heart Circulatory Physiol. 2013;305:H267–H278. doi: 10.1152/ajpheart.00206.2012. [DOI] [PubMed] [Google Scholar]
- 50.Ehe BK, et al. Data on peptides identified by mass spectrometry analysis of in vitro DYRK1A-mediated phosphorylation sites on GLI1. Data Brief. 2017;15:577–583. doi: 10.1016/j.dib.2017.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Source data are provided with this paper. Any remaining data that support the results of the study will be available from the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021531.