Abstract
The capacity of living cells to adapt to different environmental, sometimes adverse, conditions is achieved through differential gene expression, which in turn is controlled by a highly complex transcriptional network. We recovered the full network of transcriptional regulatory associations currently known for Saccharomyces cerevisiae, as gathered in the latest release of the YEASTRACT database. We assessed topological features of this network filtered by the kind of supporting evidence and of previously published networks. It appears that in-degree distribution, as well as motif enrichment evolve as the yeast transcriptional network is being completed. Overall, our analyses challenged some results previously published and confirmed others. These analyses further pointed towards the paucity of experimental evidence to support theories and, more generally, towards the partial knowledge of the complete network.
Subject terms: Gene regulatory networks, Mathematics and computing
Introduction
Transcriptional regulation is a key mechanism for the control of genomic expression, crucial for the ability of living cells to adapt to environmental stimuli, to thrive under expectable stress and sometimes also under unexpectedly harsh conditions. Despite decades of research in the field, the actual degree of complexity underlying global transcriptional control is yet to be understood. Throughout the years, structural and biochemical studies have highlighted key features of interactions between different specific transcription factors (TFs), and of the role of promoter/enhancer structures in combinatorial regulation of eukaryotic genes1–3. There is a consensual opinion that the dominant mechanism underlying the observed complexity of gene expression patterns relies on combinatorial regulation of gene expression4,5, defined as the regulation of a single gene by two or more TFs that might either act simultaneously or independently upon different spatial or temporal conditions6,7. Nevertheless, general principles underlining the activity of TFs and the combinatorial regulation at a genomic scale are still to be established.
Few, if any, eukaryotic organisms have been as thoroughly investigated as the model yeast Saccharomyces cerevisiae. This is particularly true when referring to the study of global transcriptional control, as this organism has been among the first for which extensive analyses of transcription factor activities have been conducted. This effort is illustrated by the analysis of the effect of DNA binding activity for more than one hundred TFs, using Chromatin Immunoprecipitation (ChIP)-on-chip experiments8, or the analysis of the effect of the overexpression of 55 TFs in transcriptome remodelling9. Moreover, thousands of papers describe transcriptional associations in the model yeast S. cerevisiae, obtained through various experimental platforms and observed in many different environmental conditions. The YEASTRACT database, first launched in 200610, and regularly updated since then11–15, has provided access to all available data on transcriptional associations in S. cerevisiae, gathered from more than a thousand publications in peer-reviewed international journals, and curated by a dedicated team of yeast researchers. YEASTRACT thus provides a great platform to address transcriptional regulation at a global scale.
This accumulation of data on the yeast network has led to several computational studies of topological properties with the intent to uncover design principles of genome-wide transcriptional regulation. Most interest has been focused on in- and out-degree distributions, which correspond to the number of TFs per gene and to the number of target genes (TGs) per TF, respectively8,16–18. In 2006, Balaji et al.17 assembled and analysed the largest S. cerevisiae transcriptional network at that time, relying on the TF-DNA binding evidence data available then.
A network feature that has attracted attention is the over-representation, in the global regulatory network, of small network architectures, referred to as network motifs. In this respect, Lee et al. searched for the existence of 6 types of regulatory motifs (auto-regulation, multi-component loop, feedforward loop, single and multi-input motifs, and regulatory chain) in the yeast network8, whereas Milo et al. assessed the profiles of significant triads (motifs composed of 3 nodes), i.e., their frequencies in the yeast network compared to randomized networks19.
In this paper, we recovered the full network of transcriptional regulatory associations currently known for S. cerevisiae, as gathered in the latest YEASTRACT release15. Annotations in YEASTRACT were used to produce subnetworks that encompass associations supported by binding and/or expression evidence. In our analyses, we also included networks considered in previous studies. For all these networks, we assessed their topological characteristics aiming at uncovering properties of the transcriptional control of single genes, of specific biological functions and of the complete genome. The consideration of diverse networks allowed to observe that while some properties were verified by all of them, others clearly differed, suggesting that caution should be exercised when drawing general rules on transcriptional genome-wide networks. Furthermore, functional analyses indicated potential relationships between the connectivity (in- or out-degree) of the genes and their biological functions.
Results
Transcriptional regulatory networks of Saccharomyces cerevisiae
There are more than 195,000 regulatory associations between transcription factors (TFs) and target genes (TGs) in Saccharomyces cerevisiae, according to the data deposited in the YEASTRACT database15, which is the most comprehensive source of such interactions. In YEASTRACT, 1580 references support the stored regulatory associations. Each reference documents an average of 199.68 interactions (0.1% of the total number of interactions, with a standard deviation of 1.29). Interactions are supported by an average of 1.61 references. Table 1 provides statistics for the references that support more than 5% of the YEASTRACT interactions with the total number of documented interactions and the number of interactions supported by another reference.
Table 1.
Statistics for the five reference documents documenting more than 5% of the 195,470 interactions stored in YEASTRACT.
| Number of interactions supported by | Salin et al.20 | Reimand et al.21 | Moxley et al.22 | Chua et al.9 | Harbison et al.23 |
|---|---|---|---|---|---|
| That ref. | 74,131 | 58,089 | 25,142 | 17,756 | 10,026 |
| (% of YEASTRACT interactions) | (37.92%) | (29.71%) | (12.86%) | (9.08%) | (5.13%) |
| No other ref. | 1966 | 48,385 | 20,614 | 12,294 | 3955 |
| At least another ref. | 72,165 | 9704 | 4528 | 5462 | 6071 |
For each reference, the number of documented interactions (and percentage of the total number of interactions in YEASTRACT), the number of those interactions not documented by other references, and the number of interactions documented by at least another reference, are provided.
Regulatory associations in YEASTRACT have been registered based on numerous experimental setups that may be classified into two major groups: (1) those leading to DNA binding evidence, including ChIP-on-chip, ChIP-seq, Electrophoretic Mobility Shift Assay (EMSA), or DNA footprinting; and (2) those leading to expression evidence, which is the demonstration that the deletion, mutation or overexpression of a TF affects the expression of a TG, typically including DNA microarray hybridisation, RNA-sequencing, qRT-PCR, northern blotting or the use of reporter genes. In the sequel, we will denote by:
E the set of regulatory associations supported by expression evidence;
B the set of regulatory associations supported by binding evidence;
E|B the set of regulatory associations supported by expression or binding evidence (i.e., the whole set of interactions);
E& B the set of regulatory associations supported by both expression and binding evidence.
Note that the 28 associations in YEASTRACT that still lack annotations of supporting evidence were excluded from the datasets considered in this study.
To assess whether previous observations were maintained in the updated S. cerevisiae transcriptional network, we also included in our analyses networks from two reference studies: the Costanzo et al.’s network24 that was used by Milo et al. as a source transcriptional network to uncover recurrent network motifs with the intend to disclose topological general principles19,24,25; and the Balaji et al.’s network that was assembled to analyse combinatorial regulation on a genomic scale17. In both networks, regulatory associations are supported by binding evidence only (indicated as B). Note that all the networks considered here are defined over the TFs and genes involved in transcriptional interactions, that is to say, no nodes are isolated.
Table 2 recapitulates the number of nodes and interactions for each of the six networks used in the present work, where, for each network, the nodes are those involved in interactions supported by corresponding evidence. It also indicates the proportion of actual interactions out of the possible interactions between all the identified TFs and genes, showing that the transcriptional networks are rather sparse, with a notable difference between networks containing only interactions supported by binding evidence, and those containing interactions supported by expression evidence.
Table 2.
(Top) Statistics of the six S. cerevisiae regulatory networks used in this work (TF stands for Transcription Factor, TG for Target Gene).
| Regulatory network | Evidence type | # Nodes | # Interactions | # TFs | # TGs | % Possible interactions |
|---|---|---|---|---|---|---|
| Costanzo et al.24 | B | 688 | 1079 | 131 | 592 | 1.20 |
| Balaji et al.17 | B | 4441 | 12,871 | 159 | 4408 | 1.82 |
| YEASTRACT15 | B|E | 6886 | 195,470 | 220 | 6886 | 12.90 |
| YEASTRACT15 | E | 6711 | 161,747 | 215 | 6711 | 11.21 |
| YEASTRACT15 | B | 6478 | 45,209 | 176 | 6475 | 3.97 |
| YEASTRACT15 | B&E | 3937 | 11,486 | 152 | 3912 | 1.92 |
| Network intersection | # Interactions | % Balaji | % YEASTRACT B | % YEASTRACT B&E | ||
|---|---|---|---|---|---|---|
| Balaji YEASTRACT B | 10,909 | 84.76 | 24.26 | – | ||
| Balaji YEASTRACT B&E | 3359 | 26.1 | – | 29.24 |
Each network is defined by a specific dataset, which is related to particular supporting evidence. The last column indicates the proportion of actual interactions out of the possible interactions. For example, for the YEASTRACT B&E network, there are such possible interactions, corresponding to the 152 identified TFs regulating all the 3937 TFs and TGs. (Bottom) Percentages of interaction overlaps of Balaji et al. with YEASTRACT B and with B&E networks.
Intersection assessment between Balaji et al. network on the one hand, and YEASTRACT B and B&E on the other hand shows that: YEASTRACT B contains a good proportion of Balaji’s dataset, but around 15% of it has been discarded, whereas YEASTRACT B&E only contains about 29% of Balaji’s dataset, underlining again the restricted number of associations supported by both binding and expression evidence.
Most regulatory associations based on expression evidence are associated with a sign depending on the observed effect on the TG: positive when the TF is an activator, negative when it is a repressor, more rarely dual when it has a dual effect, or possibly unknown when the observed effect has no directionality (i.e., a modified level of expression, but whereas the level increased or decreased was not clear in the supporting article).
Figure 1 displays a first assessment of the experimental basis of the data currently available in YEASTRACT. Among the documented regulatory associations, 76.87% are in the set , i.e., based on expression evidence only. Part of these associations may be indirect since there might be an intermediate TF through which the registered interaction operates. On the other hand, 17.25% of the documented regulatory associations are based on DNA binding evidence alone (in the set ). This may imply that a TF can bind to a promoter region without affecting the expression of the corresponding gene. If this is the case, it would confirm that binding evidence is not enough to establish a clear regulatory association. This is confirmed by the surprisingly restricted set of regulatory associations that are supported by both DNA binding and expression evidence (i.e., the small cardinality of the set B&E). Indeed, only 11,486 regulatory associations (5.88% of the whole interaction set B|E) can be considered reliable with evidence showing that the TF binds to the promoter region of its TG, and promotes or represses its transcription. This small number of associations in B&E may be explained by the scarcity of expression and DNA binding evidence experiments conducted in the exact same conditions26,27. In any case, as those interactions are the most reliable, this analysis suggests that our current knowledge of the transcriptional regulatory network in S. cerevisiae is still limited.
Figure 1.
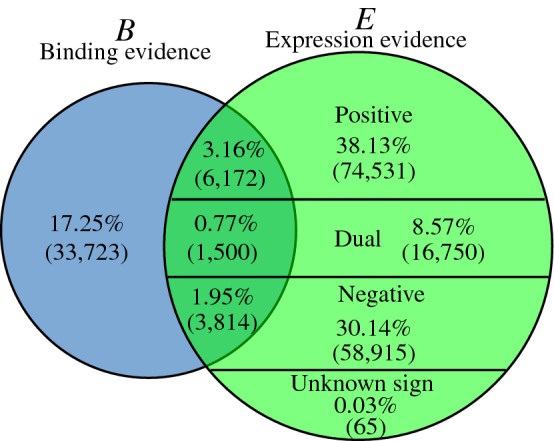
Venn diagram showing the content of YEASTRACT regulatory associations between transcription factors and target genes in S. cerevisiae according to their classification in the sets B, E and B&E, depending on the supporting experimental evidence. In the set E (and B&E), expression evidence allows to attribute a sign to the associations. Percentages refer to the fractions of the total number of associations, i.e., the cardinality of B|E.
Numbers of regulators and gene functions
In-degree analysis
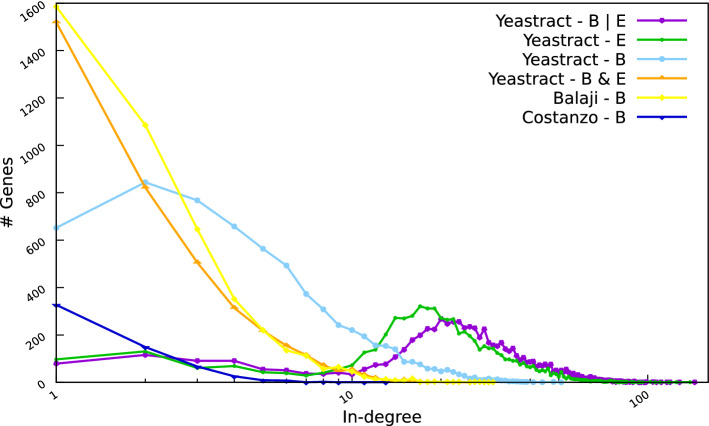
In order to assess potential trends in the transcriptional control in S. cerevisiae, we first considered the gene in-degrees, i.e., for each gene, the number of TFs controlling its transcription, regarding the different datasets. Plots in Fig. 2 indicate the in-degree distributions for the six versions of the yeast transcriptional network, considering only the regulated genes as in16,17. Hence, 3 nodes were excluded for the YEASTRACT B network, 25 for the YEASTRACT B & E network, none for the YEASTRACT B|E and E networks, 33 for the Balaji network, and 96 for the Costanzo network. Supplementary File 3 provides the in-degree distributions without excluding non-regulated nodes, where the four YEASTRACT networks now contain all the nodes present in the E|B network (i.e., the bigger set of genes). These plots further illustrate the lack of regulatory information supported by both binding and expression evidence.
Figure 2.
In-degree distributions, i.e., numbers of genes with given in-degrees. Six distinct datasets (or networks) are considered: YEASTRACT B|E (in purple), YEASTRACT E (in green), YEASTRACT B (in light blue), YEASTRACT B&E (in orange), Balaji et al.’s B17 (in yellow), Costanzo et al.’s B24 (in dark blue).
The degree distributions display similar trends for the Costanzo et al., the Balaji et al. and the YEASTRACT B&E networks, with many genes controlled by a few regulators and a few genes controlled by many regulators. For each of these networks, the degree distribution is a decreasing function. In contrast, when considering the networks from YEASTRACT B, B|E and E, the degree distributions tend to bell shaped functions, with reduced numbers of genes having small in-degrees, higher numbers of genes having intermediate in-degrees, and then again reduced numbers of genes having higher in-degrees. It is likely that these distributions follow from a high number of false positive interactions.
The in-degree analysis on the updated YEASTRACT B&E network thus confirms the previously suggested exponential in-degree distribution of the yeast transcriptional network16,17.
Relating in-degrees and biological functions
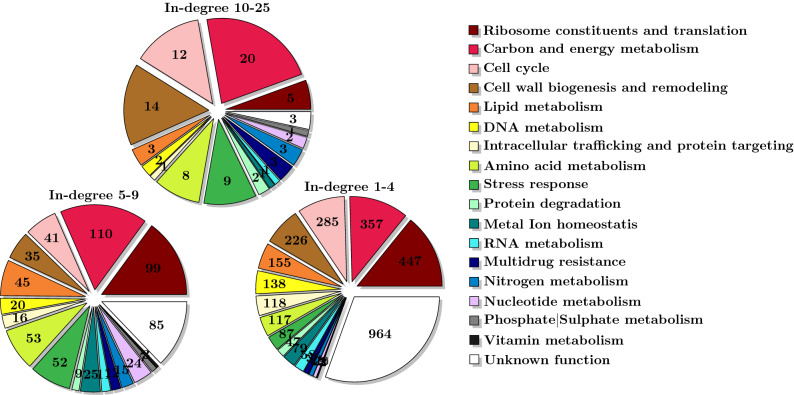
To assess whether in-degrees could relate to biological functions, we analysed the distribution of functional categories according to the in-degrees, focusing on interactions in B&E, i.e., supported by binding and expression evidence. The genes were associated to their major biological functions (see “Methods” section, and Supplementary File 4). To get a better view, we considered bins, gathering genes according to their in-degrees. These bins, appropriately defined (see “Methods” section), are as follows: degrees 1 to 4, 5 to 9, 10 to 25, including 3160, 608 and 144 genes, respectively. The results are displayed in Fig. 3.
Figure 3.
Distribution of biological functions within three in-degree bins, considering the YEASTRACT B&E network.
In this distribution of functional categories, the genes associated with the Unknown function mainly appear in the lower in-degree bin. This is most likely because these genes are less studied. More generally, the vast majority of the genes lies in the bin 1–4.
Considering the genes associated with Stress response, 13 (8.78%) of them are in the higher in-degree bin, 87 (58.78%) in the lower 1-4 in-degree bin, and 48 (32.43%) in the intermediate 5–9 in-degree bin. Among the highly regulated stress genes, the small heat shock protein encoding genes HSP12 (19 regulators) and HSP26 (12 regulators) are both known to be required in many environmental stress conditions. HPS12 is activated upon high ethanol concentrations, glucose starvation, cell wall stress, chemical stress, oxidative stress, DNA damage, and plays a role in the protection of protein and lipid folding28. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function29. Similarly, Hsp26 is activated by a variety of triggers: heat shock, salt shock, cell cycle arrest, nitrogen starvation, carbon starvation, oxidative stress, and low pH30.
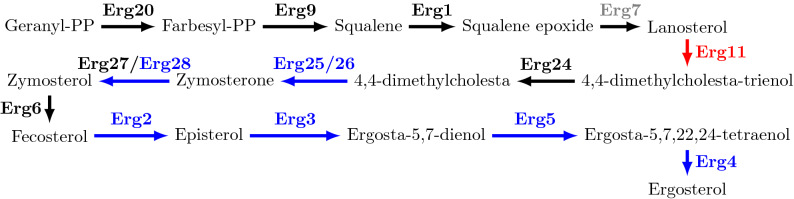
In the functional category Lipid metabolism, there are four (1.97%) genes in the higher in-degree bin: FAA1 (15 regulators), OLE1 (14 regulators), ERG11 (11 regulators), and LAC1 (10 regulators). Lac1 is a ceramide synthase subunit, that plays a crucial role in sphingolipid biosynthesis. Indeed, it catalyses the enzymatic branch point in sphingolipid biosynthesis, joining a sphingosine backbone to a very long fatty acid chain, giving rise to ceramide and controling ER-to-golgi traficking of GPI-anchored proteins. Faa1 is one of four fatty acyl-CoA synthetases, but on its own accounts for most acyl-CoA synthetase activity in yeast cells, playing a central role in both glycerolipid and sphingolipid metabolism. Ole1 is the single fatty acid desaturase in S. cerevisiae, required for mono-unsaturated fatty acid synthesis. Unsaturated fatty acids are essential for all eukaryotes as key components of cellular membranes that control the membrane organisation, fluidity and permeability, especially under stress. Finally, Erg11 is a lanosterol 14-alpha-demethylase responsible for a key step of ergosterol biosynthesis. Its inactivation not only blocks ergosterol biosynthesis, but it also leads to the accumulation of a toxic ergosterol intermediate. Other ergosterol biosynthesis encoding genes with intermediate in-degrees include ERG2, ERG3, ERG4, ERG5, ERG25, ERG26, ERG28, while the remaining steps of the pathway are regulated by a relatively low number of TFs, suggesting that they are not bottlenecks in ergosterol biosynthesis (Fig. 4).
Figure 4.
Ergosterol biosynthesis pathway, highlighting the differences in terms of individual gene regulation. Proteins in red, blue or black are encoded by genes whose transcriptional in-degree lays within the 10–25, 5–9 or bins, respectively. In grey, ERG7 is not part of the B&E network.
Finally, in the case of the Multidrug resistance (MDR) functional category, only three genes (6.67%) belong to the 10–25 in-degree bin: TPO4 with 12 regulators, and AQR1 and PDR5 with both 11 regulators. These genes are involved in multiple functions31,32. In particular, PDR5, controls resistance to a wide range of unrelated drugs, steroid transport, response to cation stress and cellular detoxification even when growing exponentially in liquid culture31. However, most MDR genes (66.67%) have less than five regulators. This is notably the case for genes encoding MDR transporters of the ATP-Binding Cassette (ABC), PDR12, PDR15 and YOR1, and of the Major Facilitator Superfamily (MFS) AZR1, DTR1, HOL1, QDR1, and QDR3, as well as the MDR transcription factors encoded by PDR1 and PDR3. Some of these genes have very specific functions. For example, although Pdr12 belongs to the ABC drug efflux pump family, it has been shown to have a distinct role in the transport of weak organic acids of intermediate lipophilicity such as sorbic and benzoic acids33. Another interesting example is the case of Pdr1, the major regulator of MDR in yeast, whose activation occurs by direct binding to drug molecules and not by increased expression in the presence of drugs34.
Numbers of targets and functions of transcription factors
Out-degree analysis
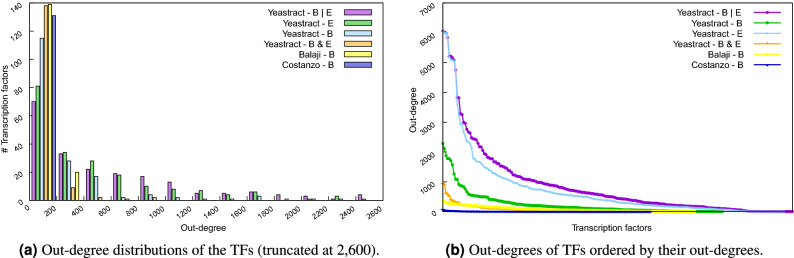
Here, we considered TF out-degrees, in order to evaluate the distribution of the number of controlled genes per TF (Fig. 5). This analysis was performed on the six networks presented in Table 2. The out-degree distribution is a decreasing function, following the same trend for all the networks, contrary to the behaviour of the in-degree distributions (Figs. 2 and 5). These results are in line with previous observations of a power law out-degree distribution in the S. cerevisiae transcriptional network16–18.
Figure 5.
Six distinct datasets (or networks) are considered: YEASTRACT B|E (purple), YEASTRACT E (green), YEASTRACT B (light blue), YEASTRACT B&E (orange), Balaji et al.’s B17 (yellow), Costanzo et al.’s B24 (in dark blue). (Left) TF out-degree distributions, i.e., number of TFs with each range of number of TGs (less than 200, between 200 and 400, etc.), truncated at 2600. (Right) TF out-degree values, in decreasing order.
Relating out-degrees and biological functions
To evaluate if out-degree values could have a functional meaning, we focused on the YEASTRACT B&E network, which includes more reliable associations, and we assessed the functions of the 25 TFs with higher out-degrees and the 25 TFs with lower out-degrees.
Identified TFs with lower and higher out-degrees were grouped according to their major functional categories (Table 3). Overall, this analysis suggests that a few functions are specific to TFs with high out-degrees and others are specific to TFs with low out-degrees. This is the case for the function Ubiquitous, which is associated to TFs playing a role in the response to, and control of, a broad range of biological processes (such TFs are impossible to assign to a specific biological process, and appear to act more like general TFs). This function associated to the TFs Cbf1, Rap1, Sfp1, Ifh1 and Fhl1, stands out as exclusive of this higher out-degree group, that is no TF with a degree lower than 3 is associated with this function. Significantly, these transcription factors are responsible for the simultaneous transcriptional control of genes involved in numerous cell functions, including cell cycle, carbon and nitrogen metabolism, or ribosome biogenesis.
Table 3.
The set of 25 TFs with higher out-degree (higher than 91) and 25 TFs with lower out-degree (lower or equal than 3) from the YEASTRACT B&E network.
| Higher out-degree | Lower out-degree | ||||
|---|---|---|---|---|---|
| TF | # Targets | Function | TF | # Targets | Function |
| Cbf1 | 622 | Ubiquitous | Stb4 | 3 | Unknown function |
| Rap1 | 573 | Yjl206c | 1 | ||
| Fhl1 | 199 | Hms2 | 1 | ||
| Sfp1 | 187 | Ppr1 | 3 | Nucleotide metabolism | |
| Ifh1 | 126 | Urc2 | 2 | ||
| Ino4 | 183 | Lipid metabolism | Gzf3 | 3 | Nitrogen metabolism |
| Ino2 | 172 | Fzf1 | 2 | Sulphite metabolism | |
| Ixr1 | 202 | Hypoxia | Cha4 | 2 | Aminoacid metabolism |
| Gcn4 | 517 | Aminoacid metabolism | Azf1 | 2 | Alternative carbon source |
| Met32 | 336 | Gal80 | 2 | ||
| Msn2 | 905 | Stress response | Rsf2 | 2 | |
| Msn4 | 395 | Mig2 | 2 | ||
| Yap1 | 297 | Aca1 | 1 | ||
| Yrr1 | 220 | Mal63 | 1 | ||
| Cin5 | 203 | Rtg2 | 1 | ||
| Rpn4 | 181 | Usv1 | 1 | ||
| Pdr1 | 172 | Rds1 | 3 | Stress response | |
| Skn7 | 152 | Yap3 | 2 | ||
| Hsf1 | 138 | Haa1 | 2 | ||
| Ste12 | 942 | Cell cycle | Rdr1 | 1 | |
| Sok2 | 364 | Ime1 | 3 | Cell cycle | |
| Tec1 | 311 | Kar4 | 3 | ||
| Fkh1 | 290 | Ndd1 | 2 | ||
| Swi4 | 175 | Tbf1 | 2 | ||
| Ndt80 | 169 | Wtm1 | 1 | ||
The functional categories Alternative carbon sources, Sulphite metabolism and Unknown function appear at the other extreme (out-degrees lower or equal than 3), and are not associated with TFs with higher out-degrees. TFs in the Unknown function thus seem to control a small number of genes that are not well characterised. The Alternative carbon sources function includes transcription factors such as Azf1, Gal80, Aca1, Mal63 or Usv1, which control the use of non-preferential carbon sources including galactose, maltose, sucrose, oleate, glycerol, acetate or ethanol. These factors control a limited number of genes as it takes only a few metabolic steps to convert the alternative carbon sources into a metabolite that can then be channelled through central carbon metabolism pathways such as glycolysis/gluconeogenesis and the TCA cycle. The function Sulphite metabolism includes a single factor, Fzf1, and involves a relatively low number of genes, acting directly on the uptake and metabolisation of sulphur containing molecules.
While the remaining functional categories include both TFs with high and low out-degrees, these TFs play different role in these processes; a factor with a higher out-degree displays a major influence on the associated processes. For example, transcriptional regulators involved in the Stress response include those required for the so-called general stress response, Msn2 and Msn4, which control the expression of many genes in response to a multitude of stress stimuli, including heat shock, osmotic shock, oxidative stress, low pH, glucose starvation, sorbic acid and high ethanol concentrations35. The major regulators of oxidative stress response in yeast, Yap1 and Skn7, are also present in the 25 TFs with higher out-degrees. The regulatory network underlying oxidative stress response is known to be highly complex, encompassing a large number of genes, as oxidative stress leads to damage in most of the cell components.
Remarkably, the Stress response category is also well represented among the 25 TFs with lower out-degrees. However, these TFs have a more restricted role in each process, as they target only a few genes involved in the response to stress agents for which the cell displays more specific and narrow-scope response mechanisms.
Of course, we cannot exclude the possibility that the low out-degree of some of these TFs (e.g. Haa1) also reflects the lack of expression and/or DNA binding experiments conducted in the precise conditions in which they are activated.
Enriched regulatory motifs in the S. cerevisiae transcriptional network
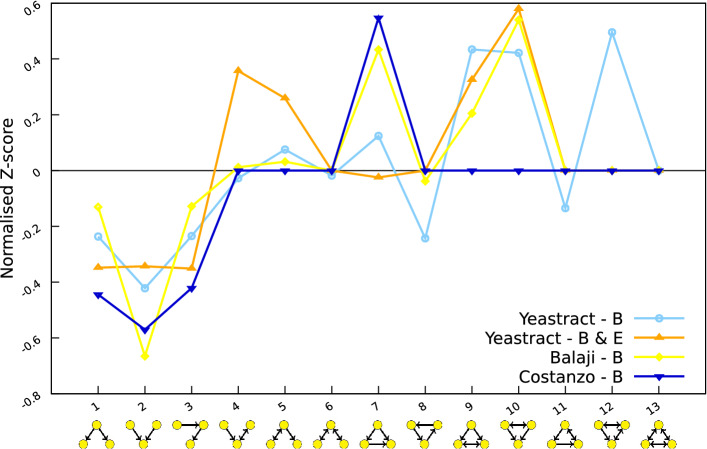
In 2004, Milo et al. presented an approach to assess network local structures in terms of recurring transcriptional motifs. In particular, they considered the triad significance profile, which relates to the enrichment significance of the numbers of directly connected triads (defined as 3 node subgraphs) in a given network, compared to these numbers in random networks. Here, we repeated this analysis to assess the motif profile of the YEASTRACT database content, restricted to triadic motifs. We compared the enrichment of the 13 triadic motifs for the datasets from Costanzo et al.24 (used by Milo et al.19 in their analysis), from Balaji et al.17 and from the YEASTRACT binding (B) and binding and expression (B&E) evidence (Fig. 6).
Figure 6.
Motif profiles of four S. cerevisiae transcriptional networks.
According to the analysis performed by Milo et al.19, biological networks would exhibit a motif profile that is preserved across organisms, with a clear under-representation of the triads 1, 2 and 3, and over-representation of the motif 7, known as feed-forward loop (dark blue profile in Fig. 6). Interestingly, this profile is not conserved when considering other versions of the yeast transcriptional network (Fig. 6). In particular, the feed-forward loop is less over-represented in the YEASTRACT networks, and the motif 10 is over-represented in all but Costanzo et al. network. In this triad, two TFs cross-regulate themselves and target the same gene. Altogether, these results suggest that the over-representation of specific motifs depends on the data underlying the considered transcriptional network.
The regulatory associations documented in the YEASTRACT datasets relate to all experimental conditions, whereas the networks considered by Milo et al. and that of Balaji et al. are based on data obtained for cells growing under Control conditions. In order to analyse comparable datasets, we isolated the YEASTRACT interactions seen to occur only in Control conditions, confirming the different motif profiles (Supplementary File 5), with the exception of the motif 12, which is not over-represented anymore. Indeed, different environmental conditions may lead to the activation of different TFs and potentially to different motif profiles. To test this hypothesis, the YEASTRACT network of transcriptional associations supported by binding and expression evidence (B& E) was filtered according to the specific environmental conditions in which the associations were found to take place. We performed this analysis for the two transcriptional networks as stored in YEASTRACT releases of 201714 and 201915 (the Supplementary File 6 provides an overview of the differences between these two networks). We observed that the motif profiles for the Control environmental condition of the 2017 and 2019 networks are similar, whereas they differ for the Stress environmental condition (see Supplementary File 7). This highlights again that motif profiles may vary as new data become available (even with small variations as it is the case here). It also supports the fact that there is no immediate functional significance of the over-representation of a network motif36, and that function does not follow motif structure neither structure follows function, as proposed by Payne and Wagner37.
Altogether these results show that, unless having the “full network”, one should not draw definite conclusions on motif profiles, as these vary significantly between different versions of the transcriptional network.
Discussion
Networks have proved to be a convenient unifying representation for a wide range of biological processes involving genes, proteins, metabolites, etc. Indeed, cellular responses to environmental signals are governed by complex networks encompassing protein–protein, protein-to-gene and metabolic interactions. In this work, the transcriptional regulatory network of the model yeast Saccharomyces cerevisiae was globally evaluated in terms of topological features, using previous data as well as the most recent data available in the YEASTRACT database.
Investigation of the regulatory associations gathered into YEASTRACT database highlighted the lack of associations supported by binding evidence. This suggests that the current network of regulatory associations in S. cerevisiae is far from being complete (Fig. 1). Further systematic assessment of transcriptional regulation in yeast is thus still needed, combining different technical approaches.
An exponential distribution of the in-degree in the yeast network was previously suggested, where a few genes would be regulated by a high number of regulators, whereas many genes would be regulated by a few regulators16,17. Our analyses showed that, while this distribution seems to be maintained in the YEASTRACT B&E network, this is not the case when considering larger, yet less reliable YEASTRACT networks (Fig. 2).
By relating functions and in-degrees, it appeared that, given specific biological processes, some involved genes are more tightly regulated than others, operating in broader scopes of conditions. This is the case for the stress responsive genes HSP12 and HSP26 that have over 12 regulators, and that are involved in a variety of stress conditions. In contrast, a good proportion (58.78%) of the stress genes have less than 5 regulators, and seem to be activated under the control of a lower number of signaling pathways. Furthermore, we found that for the Lipid metabolism functional category, most genes have less than 10 regulators, except a few genes that seem to require a tighter regulation because of their critical role. For example, the crucial nature of the step catalysed by Erg11 in ergosterol biosynthesis might explain why ERG11 has a higher in-degree, when compared to other players in lipid metabolism. Furthermore, the importance of ergosterol in the organisation of the plasma membrane may explain the intermediate in-degree of a good part of the remaining genes controlling its production. Finally, in the case of the Multidrug resistance genes, most of them have a low in-degree. A possible explanation for this may be that yeast does not often resort to the MDR network since natural habitats are normally devoided of chemical stress agents such as drugs. Nevertheless, conclusions about genes with lower in-degrees should be taken with caution. Indeed, a low number of regulators may be due to the lack of knowledge, as suggested by the high number of such genes associated with Unknown function.
Our analysis of TFs out-degrees confirmed the power law distribution previously proposed by different authors. It also corroborated the property established by Ouma et al.18, basically stating that subnetworks resulting from a sampling of the associations (i.e., the edges) would follow a power law out-degree distribution. According to what was observed for the in-degrees, the TFs with high out-degrees are known to be key regulators involved in a range of biological processes, whereas the TFs with low out-degrees would play a role in very specific functions or have yet no known function.
To uncover potential structural design principles of the yeast transcriptional network, we performed an analysis of motif profiles, focusing on triads. We showed that these motif profiles vary as the network is enriched with novel regulatory associations, and that they also depend on the environmental conditions in which the regulatory associations take place.
Altogether, our study provides a detailed analysis of the most recent version of the transcriptional network, as stored in the YEASTRACT database. Moreover, it highlights the complexity of the transcription regulatory processes that control gene expression, as well as our limited knowledge of the complete S. cerevisiae regulatory network. While previously observed properties were confirmed, other could not be retrieved when considering the up-to-date S. cerevisiae network. Hence, it seems to be hard to find general principles concerning the yeast transcriptional network as long as the complete network is not known. Still, while being cautious in trying to get definite conclusions, assessment of current genome wide transcriptional networks from topological and functional view points can still be informative. Promises of network biology probably rely on disclosing biological mechanisms rather than general principles.
Methods
All networks considered (Table 2), as well as processing scripts and intermediate results mentioned below are provided as Supplementary Files.
Data extraction from the YEASTRACT database
The YEASTRACT database contains all regulatory associations between transcription factors and target genes documented in Saccharomyces cerevisiae, up to July 2019. These regulatory associations were obtained from thousands of publications and curated by experts15.
The full network (B|E) considered is provided in Supplementary File 1. Regulatory associations from the full network are annotated with information extracted from one or more supporting papers, whenever such information is available. Regulatory associations can be classified according to the set of employed experiments as: Binding, when having at least one publication based on TF–DNA binding assays; or Expression, when having at least one publication based on expression evidence and no publication based on TF–DNA binding assays. Then, regulatory associations having at least one publication based on expression evidence, are further classified as: Positive, when all publications supported a positive effect; Negative, when all publications supported a negative effect; or Dual, otherwise.
Additionally, each regulatory association is annotated with the environmental condition in which it was found to take place, such as: Biofilm formation, Carbon source quality/availability, Cell cycle/morphology, Human niche conditions, In vitro, Lipid supplementation, Nitrogen source quality/availability, Oxygen availability, Stress, Unstressed log-phase growth (control).
Supplementary File 2, provides a script with trimming capabilities to generate all the sub-networks and associated network measures from Table 2 derived from the original full network. For example, networks used for the motif profile analyses displayed in Supplementary Files 5 and 7 include regulatory associations supported by both Binding and Expression evidence, where the full network is further filtered by a given environmental condition.
Node degree analysis
Supplementary File 2, provides the degree.py Python script to compute, for each network in Table 2, the in- and out-degrees of all the network’s nodes, saving these results in intermediate files. The script then uses these intermediate files to call GNUplot in order to generate the in- and out-degree distributions comparing all networks. In particular, it generates Figs. 2 (for the in-degree) and 5 (for the out-degree). Additionally, it selects the 25 TFs with higher and the 25 TFs with lower out-degrees used in Fig. 3.
Functional analysis
In order to evaluate a possible link between gene in-degree or TF out-degree and the functional categories associated to these genes and TFs, we performed a classification of the biological function of all the genes/TFs in the YEASTRACT B & E network, based on the description available in the Saccharomyces Genome Database (http://www.yeastgenome.org), as well as the associated Gene Ontology terms and relevant literature. This classification was manually performed for genes/TFs with at least 5 regulators (752 genes), which were then used to train a Support Vector Machine text classifier based on the Python scikit-learn (https://scikit-learn.org/) library. We then used this text classifier to classify the remaining 3,160 genes. The code is available in Supplementary File 2.
To obtain statistically significant groups, we performed Chi-squared tests for association between consecutive in-degrees, which led to the definition of three bins: in-degrees 1–4, 5–9 and 10–25 (see Supplementary File 4).
The number of genes associated with each biological function in each of the three bins is presented in Fig. 3. Additionally, the 25 TFs with higher out-degree and the 25 TFs with lower out-degree are presented in Table 3 together with their functional classifications. Supplementary File 4 includes the Spreadsheets for both the in-degree and out-degree functional classifications.
Motif analysis
In order to assess the network recurring transcriptional motifs, we considered the triad significance profile presented by Milo et al.19, where the number of connected triads in the network is computed and compared with random networks of the same dimensions in order to assess each triad enrichment significance (Z-score). To compute the triad enrichment, we considered the motif discovery software tool gtrieScanner (http://www.dcc.fc.up.pt/gtries/) by Pedro Ribeiro38.
Supplementary File 2 provides the motifs.py Python script which, for each network from Table 2, starts by converting the supplied network of genes into an equivalent network of numerical nodes. It then computes the triad enrichment, using gtrieScanner with the following parameters: -s 3 for motifs of size 3, -m subgraphs dir3.str for the subgraph list of size 3 to be considered, -d to consider directed graphs, -f simple for non-weighted graphs, -r 10000 to generate 10,000 random networks and -g network to supply the numerical network previously created. It produces the result file containing the corresponding Z-score for each motif, and a file listing all occurrences of the motifs in the supplied numerical network. Then, the motifs.py Python script parses the results and automatically performs the necessary corrections considered by Milo et al.19: (a) the Z-score of a given motif is set to 0 if the motif has less than 4 occurrences; (b) the triad significance profile is computed as normalisation of the Z-score to length 1 considering the following formula: . The normalised triad significance profiles of all networks are saved in corresponding files which are used by GNUplot to produce the motif profiles in Fig. 6 and Supplementary Files 5 and 7.
Supplementary information
Acknowledgements
This work was supported by “Fundação para a Ciência e a Tecnologia” (FCT) contracts PTDC/BBB-BIO/4004/2014 and PTDC/BII-BIO/28216/2017, sabbatical grant to PTM (SFRH/BSAB/143643/2019), and PhD scholarship to MG (PD/BD/143027/ 2018). INESC-ID acknowledges support by FCT (contract UIDB/50021/2020). iBB - Institute for Bioengineering and Biosciences acknowledges support from FCT (UIDB/04565/2020). CC acknowledges support from Instituto Gulbenkian de Ciência.
Author contributions
P.T.M. and T.P. wrote the scripts and performed the network analyses. M.G. conducted the functional classification. M.T., C.C. and P.T.M. designed and supervised the project. All the authors discussed the results, contributed to the final manuscript and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pedro T. Monteiro and Tiago Pedreira.
Contributor Information
Miguel C. Teixeira, Email: mnpct@tecnico.ulisboa.pt
Claudine Chaouiya, Email: claudine.chaouiya@univ-amu.fr.
Supplementary information
is available for this paper at 10.1038/s41598-020-74043-7.
References
- 1.Klinkenberg LG, Mennella TA, Luetkenhaus K, Zitomer RS. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot. Cell. 2005;4:649–660. doi: 10.1128/ec.4.4.649-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mennella TA, Klinkenberg LG, Zitomer RS. Recruitment of tup1-ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot. Cell. 2003;2:1288–1303. doi: 10.1128/ec.2.6.1288-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sertil O, Kapoor R, Cohen BD, Abramova N, Lowry CV. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:5831–5837. doi: 10.1093/nar/gkg792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee N. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res. 2003;31:7024–7031. doi: 10.1093/nar/gkg894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britten RJ, Davidson EH. Gene regulation for higher cells: A theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 7.Davidson EH. Genomic Regulatory Systems. Amsterdam: Elsevier; 2001. [Google Scholar]
- 8.Lee TI. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 9.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira M, et al. The YEASTRACT database: A tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteiro PT, et al. YEASTRACT-DISCOVERER: New tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;36:D132–D136. doi: 10.1093/nar/gkm976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulrehman D, et al. YEASTRACT: Providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2010;39:D136–D140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeira MC, et al. The YEASTRACT database: An upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;42:D161–D166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teixeira MC, et al. YEASTRACT: An upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;46:D348–D353. doi: 10.1093/nar/gkx842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteiro PT, et al. YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guelzim N, Bottani S, Bourgine P, Képès F. Topological and causal structure of the yeast transcriptional regulatory network. Nat. Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- 17.Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J. Mol. Biol. 2006;360:213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 18.Ouma WZ, Pogacar K, Grotewold E. Topological and statistical analyses of gene regulatory networks reveal unifying yet quantitatively different emergent properties. PLoS Comput. Biol. 2018;14:e1006098. doi: 10.1371/journal.pcbi.1006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milo R, et al. Superfamilies of evolved and designed networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- 20.Salin H, et al. Structure and properties of transcriptional networks driving selenite stress response in yeasts. BMC Genomics. 2008;9:333. doi: 10.1186/1471-2164-9-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimand J, Vaquerizas JM, Todd AE, Vilo J, Luscombe NM. Comprehensive reanalysis of transcription factor knockout expression data in Saccharomyces cerevisiae reveals many new targets. Nucleic Acids Res. 2010;38:4768–4777. doi: 10.1093/nar/gkq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moxley JF, et al. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator gcn4p. Proc. Natl. Acad. Sci. 2009;106:6477–6482. doi: 10.1073/pnas.0811091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costanzo MC, et al. YPD PombePD and WormPD: Model organism volumes of the BioKnowledge Library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–7. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira MC, et al. Refining current knowledge on the yeast FLR1 regulatory network by combined experimental and computational approaches. Mol. BioSyst. 2010;6:2471. doi: 10.1039/c004881j. [DOI] [PubMed] [Google Scholar]
- 27.Lucau-Danila A, et al. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 2005;25:1860–1868. doi: 10.1128/mcb.25.5.1860-1868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert AP, et al. NMR structure of hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. PLoS ONE. 2012;7:e41975. doi: 10.1371/journal.pone.0041975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welker S, et al. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell. 2010;39:507–520. doi: 10.1016/j.molcel.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Susek RE, Lindquist S. Transcriptional derepression of the saccharomyces cerevisiae HSP26 gene during heat shock. Mol. Cell. Biol. 1990;10:6362–6373. doi: 10.1128/mcb.10.12.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golin J, Ambudkar SV. The multidrug transporter pdr5 on the 25th anniversary of its discovery: An important model for the study of asymmetric ABC transporters. Biochem. J. 2015;467:353–363. doi: 10.1042/bj20150042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.dos Santos SC, Teixeira MC, Dias PJ, Sá-Correia I. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: Understanding their physiological function through post-genomic approaches. Front. Physiol. 2014 doi: 10.3389/fphys.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mira NP, Teixeira MC, Sá-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: A genome-wide view. OMICS. 2010;14:525–40. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakur JK, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 35.Causton HC, et al. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazurie A, Bottani S, Vergassola M. An evolutionary and functional assessment of regulatory network motifs. Genome Biol. 2005;6:R35. doi: 10.1186/gb-2005-6-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne JL, Wagner A. Function does not follow form in gene regulatory circuits. Sci. Rep. 2015;5:13015. doi: 10.1038/srep13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribeiro, P. Efficient and Scalable Algorithms for Network Motifs Discovery. Ph.D. thesis, Faculty of Science, University of Porto (2011).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.