Abstract
Deinococcus radiodurans can survive under extreme conditions, including high doses of DNA damaging agents and ionizing radiation, desiccation, and oxidative stress. Both the efficient cellular DNA repair machinery and antioxidation systems contribute to the extreme resistance of this bacterium, making it an ideal organism for studying the cellular mechanisms of environmental adaptation. The number of stress-related proteins identified in this bacterium has mushroomed in the past two decades. The newly identified proteins reveal both commonalities and diversity of structure, mechanism, and function, which impact a wide range of cellular functions. Here, we review the unique and general structural features of these proteins and discuss how these studies improve our understanding of the environmental stress adaptation mechanisms of D. radiodurans.
Keywords: DNA repair, Antioxidation, Metal ion, Deinococcus, Manganese, Zinc
1. Introduction
Since the first member, Deinococcus radiodurans R1, was discovered in X-ray sterilized canned meat in 1956, more than 35 species have been identified belonging to the genus Deinococcus to date [1]. These bacteria are widespread in the environment within hot springs [2], deserts [3], and from living organisms such as Lama glama [4]. Deinococcus is believed to be one of the toughest extremophiles in the world and is well-known for its extreme resistance to high doses of ionizing radiation. In contrast to most radiation-resistant organisms, such as spore-forming pathogens, bacteria belonging to Deinococcus show distinctly unusual characteristics. They are non-spore forming and easily genetically manipulated. For example, D. radiodurans R1, the most extensively studied Deinococcus, tolerates up to 5000 Gy of acute ionizing radiation with no loss of viability, which is 3000 times that of human cells and 250 times that of Escherichia coli [5]. It has been proposed that both the sophisticated DNA repair mechanism and antioxidation system enhance the robustness of D. radiodurans [1], [6], [7], [8].
DNA must be repaired properly upon damage to maintain life. Despite that the error-prone SOS response was not observed for D. radiodurans, it has canonical DNA repair pathways, including base/nucleotide excision repair, mismatch repair, and recombinational repair [1]. Among these pathways, homologous recombination in D. radiodurans has attracted interest over recent decades due to its astonishing efficiency for the repair of double-strand breaks (DSBs) induced by heavily ionizing radiation, which are the most lethal form of DNA damage. D. radiodurans is able to reconstruct intact genomes from shattered DNA fragments (up to 200 DSBs per haploid chromosome) within several hours following irradiation [9], [10], while a few DSBs can cause cell death in E. coli. The RecBCD pathway in E. coli is responsible for recombination initiation at DSBs, whereas the RecF pathway is primarily responsible for the repair of ssDNA gaps [11]. However, the RecF pathway is fully capable of DSB repair when RecBCD is inactive. D. radiodurans naturally lacks RecBCD recombinase, and the RecF pathway plays an essential role in recombinational DNA repair [12]. This system consists of key components conserved within its bacterial orthologs that must be activated during the recombinational repair of DSBs [12], [13]. For example, exonuclease RecJ is required in concert with helicase RecQ and/or UvrD to initiate DSB end resection [14], [15].
Previous studies have linked the radiation resistance of D. radiodurans to its prolonged desiccation tolerance phenotype, which both cause DNA damage [16], [17]. Transcriptional analysis revealed a group of genes highly induced in response to either desiccation or ionizing radiation [17]. Among these genes, five genes encoding DdrA, DdrB, DdrC, DdrD, and PprA proteins are distributed in almost all Deinococcus species. Depletion of the corresponding genes resulted in varying degrees of sensitivity to DNA damaging agents, indicating that these proteins are involved in the extreme resistance of D. radiodurans [17]. In addition, a common DNA motif called RDRM (radiation/desiccation response motif) was identified in a set of promoter regions of DNA repair genes (e.g., recA, pprA) [18]. It was recently revealed that DdrO, a self-regulated repressor specifically binding to the RDRM, tightly controls the expression of these genes under normal growth conditions [19], [20], [21], [22]. The transcription of these genes is stimulated by the metalloprotease PprI, also referred to as IrrE. DdrO is digested by the PprI protein after DNA damage, leading to the derepression of DNA repair genes. Considering the similarity with the SOS response, DdrO and PprI proteins, which are both species-specific, mediate a novel protease-based DNA damage response pathway in Deinococcus species [1].
Radiation and desiccation cause significant increases in the levels of reactive oxygen species (ROS). These small molecules are short-lived and highly reactive, and they can cause both genomic DNA and protein damage, such as 8-hydroxyguanine and protein carbonylation. It was proposed that the accumulation of oxidized protein, instead of DNA damage, is the major cause of cell death [5]. Indeed, D. radiodurans has an overall background genomic mutation rate similar to that of E. coli [23], [24]. And a proteome protection system appears to be the prevalent theory of the radiation resistance also in many other extremophiles [25]. D. radiodurans can tolerate high doses of agents that generate oxidative stress, such as hydrogen peroxide (H2O2). To cope with oxidative stress, a variety of antioxidation mechanisms of ROS detoxification have evolved, including (1) fewer respiratory chain enzymes and an effective thioredoxin reductase (TrxR) antioxidant system suppressing endogenous ROS production [26], [27]; (2) a rich pool of highly activated ROS scavenging enzymes, including catalases, superoxide dismutases (SOD), peroxidases, and Dps proteins [28], [29], [30]; (3) nonenzymatic small scavengers such as small-molecule antioxidants (e.g., manganese complexes) [31] and deinoxanthin [32], the major ketocarotenoid in D. radiodurans; And (4) Small regulatory RNAs involved in the ionizing radiation response and oxidative stress tolerance [33], [34], [35].
Manganese is a trace element ubiquitously present in nature, which is required in small amounts for the proper enzymatic activities in the cell. Compared to those in radiation-sensitive bacteria, the intracellular manganese concentration (range from 0.4 to 3 mM) and manganese-to-iron ratio (0.24) in D. radiodurans are exceptionally high, dozens of times higher than in E. coli [36]. Daly and coworkers noted that this unusual intracellular manganese-to-iron ratio correlates with the radiation resistance in D. radiodurans [36]. Given that an excessive amount of manganese causes toxicity, the intracellular manganese concentration must be tightly controlled. Multiple transporters maintain manganese homeostasis in D. radiodurans, including MntE (manganese efflux transporter), MntH (bacterial Nramp family protein) and MntABC system (ATP-binding cassette transporter). These manganese transporters have been proposed to be regulated by several regulators, such as OxyR, Fur and DtxR [1].
Over the past decade, D. radiodurans has been investigated as a model organism for studying the cellular mechanisms of environmental adaptation. Unique proteins involved in the extreme resistance have been characterized by genetic, biochemical and biophysical analyses, with their structures recently reported in literatures. Previous reviews have investigated some of these proteins as well as the mechanisms of radiation and oxidative stress resistance in D. radiodurans [1], [6], [8], [37], which will not be discussed in this review. Here, we focus on the unique and general structural properties of these stress-related proteins identified over the past few years (Table 1 and Fig. 1) and summarize unique features and mechanisms of how these proteins contribute to the extreme resistance of D. radiodurans.
Table 1.
List of stress-related proteins discussed in this review.
| Biological process | Protein | Gene locus | Metal ions in structure | Assembly | PDB code (resolution) | Reference | Comments | |
|---|---|---|---|---|---|---|---|---|
| DNA repair | DSB end resection | RecJ | DR_1126 | Mn2+ | monomer | 5F54, 5F55, 5F56 (2.3–2.7 Å) | [15] | 5′−3′ ssDNA-specific exonuclease |
| RecQ | DR_1289 | Zn2+ | monomer | 2RHF, 4Q48, 4Q47 (2.8–2.9Å) | [38], [39], [40] | ATP-dependent 3ʹ-5ʹ helicase | ||
| UvrD | DR_1775 | Mg2+ | monomer | 4C2T, 4C2U, 4C30 (2.6–4.0Å) | [41] | ATP-dependent bidirectional helicase | ||
| DNA end protection | DdrA | DR_0423 | – | large oligomer | Negative stain electron microscopy | [45] | ssDNA-binding protein | |
| Single-strand annealing | DdrB | DR_0070 | – | pentamer | 4HQB, 4NOE (2.2–2.3 Å) | [49], [50] | single-strand annealing activity | |
| Activation of DNA ligase and DNA gyrase | PprA | DR_A0346 | – | filament | 6A27, 6A28, 6A29 (1.3–2.4Å) | [55] | preventing nuclease-mediated degradation | |
| DNA damage response | PprI | DR_0167 | Zn2+ | dimer | 3DTK, 3DTE, 3DTI (2.6–3.5Å) | [57] | metalloprotease | |
| Recombinational repair | DdrO | DR_2574 | dimer | 6JQ1, 6RMQ, 6RNX (2.3–3.0Å) | [19], [20] | transcriptional repressor | ||
| Base excision repair | PolX | DR_0467 | Zn2+ | monomer | 2W9M (2.5Å) | [79] | X family DNA polymerase | |
| Recombinational repair | RecO | DR_0819 | Zn2+ | monomer | 1W3S, 1U5K, 2V1C (2.0–3.8Å) | [74], [75], [77] | promoting RecA loading on SSB-coated ssDNA | |
| RecR | DR_0198 | Zn2+ | tetramer | 1VDD, 2V1C (2.5–3.8Å) | [76], [77] | DNA binding and form complex with RecO | ||
| Antioxidation | Oxidoreductase | KatE1 | DR_1998 | tetramer | 4CAB (2.6Å) | [58] | clade 1 type of monofunctional catalase | |
| MnSOD | DR_1279 | Mn2+ | dimer | 2CDY, 2CE4, 2AW9 (2.0–2.7Å) | [59] | Mn-containing SOD | ||
| TrxR | DR_1982 | – | dimer | 2Q7V (1.9Å) | [27] | thioredoxin reductase involved in ROS scavenging | ||
| FrnE | DR_0659 | – | dimer | 5CNW 5CO3, 5COH, 5E59 (1.6–2.0Å) | [60] | cytoplasmic disulfide oxidoreductase | ||
| Metal ion homeostasis | Dps1 | DR_2263 | Zn2+ | dodecameric | 2F7N, 2C2U, 2C2F (1.1–2.0Å) | [65], [66] | ferritin family protein involved in ROS detoxification, DNA protection and iron uptake and storage. | |
| Dps2 | DR_B0092 | Fe3+ | dodecameric | 2C2J, 2C6R (2.1Å) | [67] | |||
| Mn transporter | MntH | DR_1709 | Mn2+ | monomer | 6D91, 6C3I, 6BU5, 6D9W (2.4–4.0Å) | [69] | manganese transporters belonging to bacterial Nramp family protein | |
| Quorum sensing | LuxS | DR2387 | Zn2+ | dimer | 1INN, 1VJE (1.6–1.8Å) | [80] | protein involved in quorum sensing | |
| Stress-response | DinB | DR_0053 | Zn2+ | dimer | 6IZ2 (2.1Å) | [78] | DinB/YfiT family protein | |
Fig. 1.
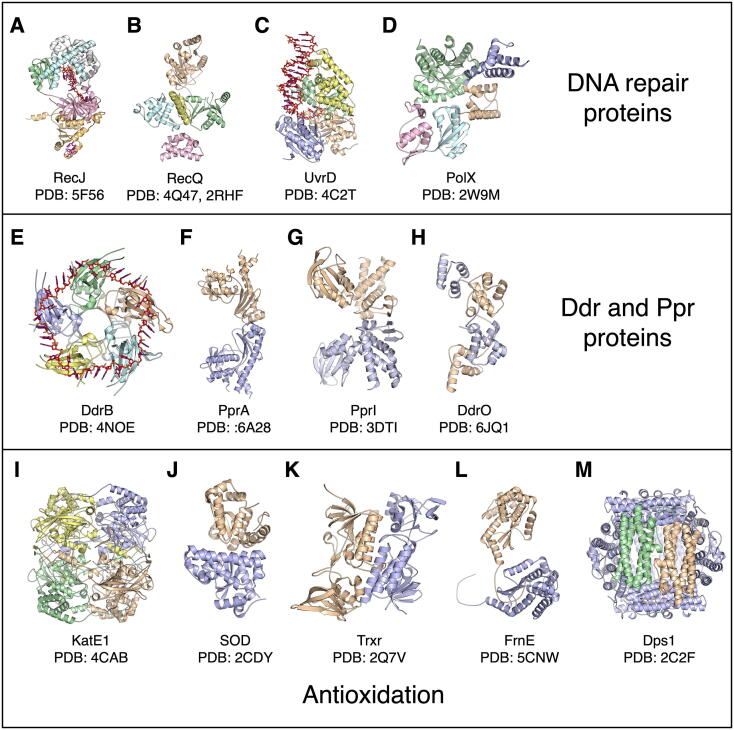
Illustration of the structures of stress-related proteins in Deinococcus species. (A–D) Protein domains from monomeric proteins are shown in distinct colors. ssDNA and the C-terminal region of drSSB protein in (A) are colored red and magenta, respectively. dsDNA in (C) is colored red. (E–M) Protomers from protein homo-oligomer structures are distinctively colored based on protein self-assembly. ssDNA in (E) is colored red. Their PDB accession codes are shown and references can be found in the Table 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Proteins in effective DNA repair processes
2.1. Proteins involved DNA end resection
DNA end resection is one of the earliest and most important steps in recombinational repair, which involves the nucleolytic digestion of DSB ends in the 5ʹ-3ʹ direction. The resultant 3ʹ single-stranded DNA (ssDNA), bound by the RecA protein, subsequently pairs with the complementary strand for DNA strand exchange reactions. In D. radiodurans, RecJ, RecQ and UvrD were reported to play roles in DSB end resection [12], [15].
RecJ is a representative member of the DHH family of proteins (defined by presence of the conserved Asp-His-His motif), which has been found in most eubacteria and archaea. Studies of D. radiodurans RecJ (drRecJ) revealed the shared and distinctive features of domain arrangement, substrate recognition and catalysis (Fig. 1A) [15]. Compared with its homologues, such as Thermus thermophilus RecJ, drRecJ is conserved within the N-terminal DHH and DHHA1 domains but has an additional C-terminal domain. Biochemical and structural studies showed that this domain interacts directly with the C-termini of the drSSB protein, which is required for the stimulation of drRecJ processing DSB ends containing 3ʹ-ssDNA overhangs. Given that drRecJ is essential for DSB end resection in D. radiodurans and that this C-terminal domain is conserved within Deinococcus species, these enhanced drRecJ-drSSB interactions ensure the drRecJ digestion of various types of DSB ends. The crystal structure of the drRecJ-DNA complex also confers its high processivity and coordination with helicases. Unlike many nucleases that bind extensively to the DNA phosphate backbone prior to cleavage, drRecJ primarily contacts DNA through stacking interactions between the side chains of the aromatic amino acids and DNA bases. Mutations of these interaction residues to Ala resulted in reduced nuclease activity and processivity. The noncanonical oligonucleotide/oligosaccharide-binding (OB) fold domain, together with the helical gateway element, is located at the entrance of the active site, which safeguards substrate DNA access. Moreover, a terminal mononucleotide-binding pocket was observed above the active site, which determines the 5ʹ to 3ʹ exonuclease activity of RecJ.
Both RecQ (drRecQ) and UrvD (drUvrD) in D. radiodurans are 3ʹ-5ʹ helicases, which unwind duplex DNA in an ATP-dependent reaction. The crystal structures of drRecQ (Fig. 1B) and drUvrD (Fig. 1C) from D. radiodurans reveal several interesting features [38], [39], [40], [41]. Unlike most RecQ proteins, drRecQ possesses a conserved helicase, RecQ-Ct, and one HRDC (helicase and RNaseD C-terminal) domain but two additional (totally three) HRDC domains [40]. Despite the similar overall fold to other HRDC domains, the C-terminal HRDC domain of drRecQ is unusually highly negatively charged with a well-ordered phosphate ion bound, which is likely to be involved in the regulation of substrate DNA binding. Similar to drRecQ, drUvrD has different characteristics from its E. coli counterparts. drUvrD is able to unwind double-stranded DNA in both the 3ʹ-5ʹ and 5ʹ-3ʹ directions in vitro, unlike other SF1A helicases, including ecUvrD [41]. Furthermore, the activity of drUvrD is modulated by additional drSSB proteins, which favors drUvrD helicase activity over translocase activity on duplex DNA containing the 5ʹ-tail. Thus, it is probably no coincidence that the direct interaction between drRecJ and drRecQ was not observed in vitro [15]. drUvrD, instead of drRecQ, seems to be the primary helicase required for coordinated activity with drRecJ for DSB end resection. These findings suggested that D. radiodurans has evolved to efficiently process DSB ends by a novel helicase-nuclease machinery.
2.2. DdrA, DdrB and PprA proteins
As mentioned above, the transcriptional levels of ddrA, ddrB and pprA are highly induced in response to ionizing radiation and desiccation in D. radiodurans. These proteins have been found only in Deinococcus species and are proposed to be involved in the DNA repair processes according to their mutants’ phenotypes: deletion of these genes sensitized D. radiodurans to ionizing radiation, DNA crosslinking reagents, or oxidative stress to varying degrees [17]. With significant differences in substrate preference, drDdrA, drDdrB and drPprA are able to bind DNA and perform unique biochemical activities [42], [43], [44].
drDdrA is evolutionarily related to the Rad52 family proteins. Despite the ability to bind ssDNA with 3ʹ-overhangs, drDdrA does not exhibit single-strand annealing activity in vitro [42], which has been observed for the human Rad52 protein. It was proposed that drDdrA is involved in the DNA end-protection system, which preserves ssDNA for recombinational repair. Using a combination of negative stain electron microscopy, genetic and biochemical techniques, the N-terminal region of DdrA from Deinococcus deserti (ddDdrA) was shown to be involved in DNA binding, likely forming a large heptameric ring-shaped complex [45]. However, no structural information is currently available for the full-length DdrA or DdrA-DNA complex. The exact role of DdrA remains unclear since the N-terminal region of ddDdrA only partially recovered the phenotype in vivo [45].
DdrB is an SSB protein that is recruited to the nucleoid in the early step of DSB repair after DNA damage [43], [46], [47]. Unlike bacterial SSBs or human RPAs, the canonical OB fold domain is absent in DdrB. Instead, DdrB possesses a unique structural topology that differs from the conserved Greek key topology observed in other OB fold-containing proteins [48]. Biochemical analysis showed that drDdrB could bind only ssDNA but not dsDNA. Moreover, drDdrB is able to perform single-strand annealing reactions similar to those of Rad52 protein, which contributes to the repair of DSBs in a RecA-independent manner [43]. Crystal structures of DdrB and DdrB-ssDNA complexes revealed an elegant mechanism for DdrB-mediated accurate annealing (Fig. 1E) [49], [50]. In contrast to dimeric drSSB [51] or ecSSB [52] proteins, drDdrB forms stacked face-to-face pentameric rings with a continuous flat DNA binding surface on each drDdrB pentamer, which is different from the intertwining of ssDNA observed in the canonical OB fold domain. Moreover, the access of free ssDNA with unpaired bases is restricted, and dissociation occurs only when two ssDNAs are fully complementary. These features ensure the high fidelity of the DdrB annealing reaction.
PprA (pleiotropic protein promoting DNA repair) is a Deinococcus species-specific protein required for DNA repair and chromosome segregation [53]. PprA is able to stimulate the biochemical activities of many proteins, such as DNA ligase [54] and DNA gyrase holoenzyme [53], by protein–protein interactions. It has also been reported that PprA has a substrate preference for DSB-containing DNA, preventing nuclease-mediated degradation [44]. However, wild type drPprA forms a wide range of oligomeric assemblies that are not suitable for crystallographic studies. By mutagenesis screening using the hydroxylamine method and error-prone PCR, crystals of two mutant drPprA proteins were obtained successfully, which contain a dimer arrangement in the asymmetric unit (Fig. 1F) [55]. The most striking feature of drPprA is that it forms a face-to-face dimeric interface and displays a novel right-handed periodic screw assembly. Such oligomeric structure may function as a platform, allowing DNA ends exposed to other DNA repair proteins. Together with the X-ray small-angle scattering results confirming the filamentous structure in solution, the drPprA structure provides valuable information for understanding the mechanism of PprA involved in DNA repair.
2.3. PprI-DdrO system
After two decades of extensive research, the PprI-DdrO system appears to be one of the most important DNA damage response systems and is conserved within Deinococcus species. While DdrO is essential for cell viability, the inactivation of PprI in Deinococcus led to a dramatic increase in sensitivity to radiation and oxidative stress [56]. In parallel to the well-documented SOS response requiring RecA-ssDNA-ATP filaments for LexA autocleavage, the cleavage of DdrO protein by PprI depends on manganese or zinc ions [21], [22]. The crystal structure of D. deserti PprI (ddPprI) has been determined as a dimer and consists of an N-terminal domain, a middle HTH motif, and a C-terminal GAF-like domain (Fig. 1G) [57]. The N-terminal domain of ddPprI is superimposable on mono-zinc metallopeptidases with one zinc ion sequestered by conserved His and Glu residues. The C-terminal GAF domain, in concert with the HTH motif, was proposed to function as the sensor domain and to undergo conformational change upon binding of the unknown ligands.
Apo structures of DdrO proteins from Deinococcus geothermalis (Fig. 1H) [20] and D. deserti [19] have been determined recently and are essentially identical. Structural analysis revealed that DdrO has an HTH-containing N-terminal domain with a pair of oppositely charged residues (RE pair) for substrate binding, which is conserved within XRE family proteins [20]. An additional C-terminal domain with a novel fold is connected by a 15 amino acid linker loop. As calculated by the PDBePISA server, DdrO has a more extensive dimeric interface largely mediated by this C-terminal domain, distinguishing DdrO from LexA and other XRE family proteins. The cleavage site region (CSR) of DdrO is located in the loop region between the last two α-helices of the C-terminal domain, which forms a stable hydrophobic core. Similar to other XRE proteins, DdrO exists as a stable dimer in solution through its NTD (Fig. 1H), which is essential for substrate DNA binding. The CSR of DdrO is involved in the crystal packing between two symmetrical protein molecules. Based on the structures of full-length ddDdrO and the CTD of ddDdrO [19], it was proposed that the CTD of DdrO can also mediate protein dimerization, adopting a much more extended conformation than in NTD dimerization. However, there is a need for more evidence to confirm DdrO dimer formation in solution. Nevertheless, given that the C-terminal domain is involved in the protein–protein interface of the DdrO dimer (both NTD-mediated dimerization and the possible CTD-mediated dimerization), cleavage of the CSR by PprI destabilizes DdrO dimer formation, further eliminating the DNA binding of DdrO. These structural features illuminate the mechanism of the DNA damage response mediated by the PprI-DdrO system.
3. Proteins involved in antioxidation systems
3.1. Oxidoreductases
Oxidoreductases play a critical role in antioxidation in D. radiodurans. At present, the structures of KatE1 [58], MnSOD [59], TrxR [27] and FrnE [60] have been determined. drKatE1 is a clade 1 type of monofunctional catalase. Mutant strain lacking the KatE1 exhibited increased sensitivity to both chronic ionizing radiation and H2O2 treatments [29], [61]. Among all the catalases characterized in D. radiodurans, KatE1 is present in most of Deinococcus species and displays the highest catalase activity, indicating that KatE1 is the major catalase in D. radiodurans [29]. The crystal structure of drKatE1 revealed a symmetric homotetramer with heme molecules in the active site (Fig. 1I) [58]. The protomer containing an alternative heme channel is very similar to previously solved monofunctional catalases, which indicates that drKatE1 is a highly conserved catalase.
D. radiodurans contains one cytoplasmic Mn-containing SOD (drSodA), which is conserved within almost all Deinococcus species. Despite the high protein sequence identity with its homologue from E. coli (ecSod), drSodA is a more efficient enzyme to eliminate superoxide radicals in vitro. Two crystal forms of drSodA were obtained with either one or two homodimers in the asymmetric unit (Fig. 1J) [59], which is almost identical to that reported previously for ecSod [62]. A positively charged loop located at the protein–protein interface of the drSodA dimer was also observed in ecSod. It has been reported that the Mn-containing SOD from E. coli is capable of binding both DNA and RNA [63]. The disruption of drSodA only moderately increased the sensitivity of D. radiodurans to ionizing radiation [1], indicating that drSodA is mainly involved in superoxide detoxification in D. radiodurans.
Thioredoxin reductase (TrxR) plays a central role in the thioredoxin system, which is involved in intracellular ROS scavenging in D. radiodurans [27]. The TrxR in D. radiodurans (drTrxR) is well conserved within Deinococcus species and has a strong cofactor preference for NADPH [27]. The crystal structure of drTrxR revealed a similar homodimeric configuration to its homologue from E. coli (ecTrxR) but showed variants of the Trx-interacting surface (Fig. 1K) [27]. Compared with ecTrxR, a conserved Phe residue from drTrxR was in a different rotamer state and caused a possible change in TrxR-Trx interaction, which may explain the much higher affinity for its own substrate drTrx1. FrnE from D. radiodurans (drFrnE) has been recently characterized as a novel cytoplasmic disulfide oxidoreductase that is also shared among Deinococcus species [60]. Crystal structures of drFrnE in different redox states have been determined (Fig. 1L) [60], representing snapshots of the catalytic steps [64]. Moreover, the structures reveal a unique C-terminal tail containing an additional dithiol motif, which is likely to facilitate the regeneration of the active site in a reduced form.
3.2. Dps proteins
DNA binding proteins from starved cells (Dps) belong to the ferritin family and play an important role in ROS detoxification, DNA protection and iron uptake and storage. D. radiodurans has two Dps proteins, Dps1 and Dps2. While Dps1 is well conserved within all Deinococcus species, Dps2 is present only in two species of Deinococcus. Compared with other bacterial Dps family members, Dps1 (drDps1) and Dps2 (drDps2) from D. radiodurans have an unusually long N-terminal extension (54 and 41 amino acids in drDps1 and drDps2, respectively) before the ferritin core structure. And these N-terminal extensions are rich in positively charged Lys and Arg residues. Crystal structures of drDps1 (Fig. 1M) and drDps2 have been determined at high atomic resolution [65], [66], [67], revealing a conserved dodecameric assembly with metal ions bound in the ferroxidase center. Notably, the N-terminal extension of drDps1 contains a novel metal ion site, which is required for DNA binding. Nevertheless, large portions of these N-terminal extension regions in drDps1 and drDps2 were disordered in solved crystal structures due to flexibility. Aided by time-resolved SAXS, it has been reported that, in contrast to drDps1, the N-terminal extension of drDps2 is essential for its dodecameric assembly and likely to be involved in membrane interactions [68]. These results suggested the distinct roles of N-terminal extension of Dps proteins in D. radiodurans.
3.3. MntH
As mentioned above, D. radiodurans has an exceptionally high intracellular Mn concentration, which was proposed to play a critical role in antioxidation. To our knowledge, the structural information currently available for manganese transporters in D. radiodurans comes from the study of MntH (drMntH) [69]. drMntH belongs to the Nramp family transporters, which are involved in the high-affinity system for Mn uptake. The crystal structure of drMntH was originally solved in an inward-facing conformation in the presence of a monoclonal antibody fragment, which revealed a pseudosymmetric LeuT-fold-like conformation, as observed in the structures of other bacterial Nramp proteins. However, through the transport cycle, drMntH undergoes a more agile conformational change distinct from the rigid body movement of canonical LeuT-fold transporters. Given that these are the first solved complementary structures of the same Nramp protein, it remains to be investigated whether this metal transport mechanism is conserved among MntH proteins.
4. Metalloenzymes
Divalent metal ions, including Mg2+, Mn2+, Zn2+ and Ca2+, are essential for the enzymatic activities of many metalloproteins and are frequently used to promote or improve protein crystallization. For nucleic-acid-processing enzymes, these divalent metal ions serve as protein cofactors involved in substrate DNA/RNA binding (in DNA binding proteins), nucleophilic attack (in nucleases and polymerases) and ATP hydrolysis (in helicases). Given their highly specific role in cellular processes, these divalent metal ions must be carefully selected to avoid possible enzymatic abnormalities. For example, different divalent cations cause alterations in the kinetics and fidelity of DNA polymerases [70]. DNA nucleases exhibit altered substrate binding preferences in catalytic activities depending on the identity of the divalent cations in the reaction [71].
D. radiodurans has an unusual metal ion content with a high intracellular Mn concentration (~millimolar level), which is different from other radiation-sensitive bacteria. Therefore, it is interesting to investigate the possible role of these metal ions in proteins that function in the extreme resistance of D. radiodurans. Mn and Zn metal ions, which come from either protein copurification or the reservoir solution for crystallization, are two divalent metal ions frequently observed in the currently solved structures of Deinococcus proteins. Mn ions are found in the structures of RecJ [15], RNase J [72], MazG [73], MntH [69], and SodA [59], and the structures of PprI [57], RecO [74], [75], RecR [76], [77], RecQ [40], DinB [78], PolX [79], Dps1 [66], LuxS [80], and several peptidases [81], [82] contain Zn ions.
However, the Mn and Zn ions present in these structures are not fully consistent with their biochemical activities. Mn ions are preferred for the enzymatic activities of RecJ [15], MazG [83], and PolX [79], while Zn ions are involved in protein folding and substrate processing/binding by RNase J [72], PprI [21], RecOR [77], DinB [78], and LuxS [80]. Compared with E. coli homologues that mainly use Mg ions as cofactors, RecJ [15], MazG [83], MutS2 [84] and PolX [79] in D. radiodurans exhibited a strong preference for Mn over Mg, which might be due to the intrinsic properties of these two metal ions, e.g., the coordination geometry required for catalysis. Moreover, Mn was essential for the protein dimerization of RNase J from D. radiodurans [72], which is likely to be involved in the balance of its exo- and endonucleolytic activity. These structures suggest a broad role for metal ions in protein-function regulation in D. radiodurans.
5. Summary and outlook
To date, more than 900 protein structures from Deinococcus species have been deposited in the Protein Data Bank, providing valuable information on the mechanisms of the extreme resistance of D. radiodurans. And structural studies of large protein complexes including ribosomal proteins and ribonucleoprotein complexes shed new light on the mechanisms of antibiotic inhibition and RNA degradation [85], [86]. These proteins, with both commonalities and diversity in structural features, are involved in the efficient repair of DNA damage and strong antioxidation systems. While several studies revealed both protein conformations and biochemical activities shared with homologous proteins, some of these proteins contain novel protein folds, protein–protein interaction interfaces and altered metal ion binding sites, which give rise to their distinct functions in vivo. For example, many nucleic-acid binding proteins (e.g., RecJ and DdrO) have unique domain arrangement and substrate binding capabilities, which shed light on the prompt DNA damage repair in in D. radiodurans.
D. radiodurans can survive under extreme conditions, and the first crystal structure of a D. radiodurans protein was determined in 2001 [80]. However, after 20 years of probing, our knowledge of how these proteins contribute to the robustness of D. radiodurans remains vague. Almost all the Deinococcus proteins solved by X-ray crystallography come from D. radiodurans, the most extensively studied Deinococcus species, and most structural information of protein–protein or protein-substrate complex listed in Table 1 are currently not available from other bacteria. DdrB and DdrO (both species-specific) are the only two proteins with cross-species structural information available. Although these two proteins from two different Deinococcus species are structurally conserved, it is still unclear whether the structures and functions of other species-specific proteins from D. radiodurans, such as PprA, PprM and DdrR, are universal among Deinococcus species. Additional structural information (e.g., DdrO in complex with DNA or PprI) is needed to elucidate the detailed mechanism of the PprI-DdrO system in Deinococcus species.
There are still a large number of questions that need to be addressed. What is the structural basis of nucleic acid-processing enzymes involved in DNA repair processes in D. radiodurans? Structural information of these proteins (e.g. RecQ, RecOR and RecA) in complex with substrate DNA is required to elucidate the structure–function relationships. Given that species-specific Ddr and Ppr proteins were highly induced after environmental stresses and their DNA-binding capabilities, biochemical and structural studies may shed light on their shared and unique molecular mechanisms underlying diverse DNA repair pathways. Compared with DNA repair proteins exhibiting characteristic structural features, two major ROS scavenging enzymes from D. radiodurans, KatE1 and SodA, show striking similarities to their bacterial counterparts, indicating possible shared reductase-based antioxidation mechanisms. In view of the antioxidation role of elevated intracellular Mn concentrations in D. radiodurans, it is thus important to evaluate the protein structures of Mn transporters to better understand the mechanisms and regulation of the intracellular Mn homeostasis in D. radiodurans. Remarkably, structural studies revealed novel OB domains (with distinctive protein folding [15], [50], assembly [51] and DNA recognition [15]) in DNA binding/processing proteins in D. radiodurans, which are critical for the DNA repair process. More work is needed to identify the molecular and structural features of these proteins and their complex with DNA, which could potentially illuminate their function in DNA repair. In addition, protein posttranslational modifications (PTMs) including the phosphorylation [87], PARylation [88], and succinylation [89] of several stress-response-related proteins from D. radiodurans, have been characterized and are likely to affect the regulation of protein activities, structures, and subcellular localization. Further studies are needed to obtain more detailed insights into the structural properties resulting from PTMs. In general, structural studies of stress-related proteins that contribute to the robustness of D. radiodurans remain important topics for further research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFA0503900), and the grants from National Natural Science Foundation of China (31500656, 31670819).
Contributor Information
Yuejin Hua, Email: yjhua@zju.edu.cn.
Ye Zhao, Email: yezhao@zju.edu.cn.
References
- 1.Lim S., Jung J.H., Blanchard L., de Groot A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol Rev. 2019;43:19–52. doi: 10.1093/femsre/fuy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira A.C., Nobre M.F., Rainey F.A., Silva M.T., Wait R., Burghardt J. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int J Syst Bacteriol. 1997;47:939–947. doi: 10.1099/00207713-47-4-939. [DOI] [PubMed] [Google Scholar]
- 3.de Groot A., Chapon V., Servant P., Christen R., Saux M.F., Sommer S. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int J Syst Evol Microbiol. 2005;55:2441–2446. doi: 10.1099/ijs.0.63717-0. [DOI] [PubMed] [Google Scholar]
- 4.Copeland A., Zeytun A., Yassawong M., Nolan M., Lucas S., Hammon N. Complete genome sequence of the orange-red pigmented, radioresistant Deinococcus proteolyticus type strain MRP(T) Stand Genomic Sci. 2012;6:240–250. doi: 10.4056/sigs.2756060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly M.J. Death by protein damage in irradiated cells. DNA Repair (Amst) 2012;11:12–21. doi: 10.1016/j.dnarep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Timmins J., Moe E. A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: major findings, functional and mechanistic insight and challenges. Comput Struct Biotechnol J. 2016;14:168–176. doi: 10.1016/j.csbj.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin M., Xiao A., Zhu L., Zhang Z., Huang H., Jiang L. The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express. 2019;9:138. doi: 10.1186/s13568-019-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi H.Z., Wang W.Z., He J.Y., Ma Y., Xiao F.Z., He S.Y. Antioxidative system of Deinococcus radiodurans. Res Microbiol. 2020;171:45–54. doi: 10.1016/j.resmic.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Daly M.J., Ouyang L., Fuchs P., Minton K.W. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slade D., Lindner A.B., Paul G., Radman M. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell. 2009;136:1044–1055. doi: 10.1016/j.cell.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Pages V. Single-strand gap repair involves both RecF and RecBCD pathways. Curr Genet. 2016;62:519–521. doi: 10.1007/s00294-016-0575-5. [DOI] [PubMed] [Google Scholar]
- 12.Bentchikou E., Servant P., Coste G., Sommer S. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu G., Wang L., Chen H., Lu H., Ying N., Tian B. RecO is essential for DNA damage repair in Deinococcus radiodurans. J Bacteriol. 2008;190:2624–2628. doi: 10.1128/JB.01851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morimatsu K., Kowalczykowski S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc Natl Acad Sci U S A. 2014;111:E5133–E5142. doi: 10.1073/pnas.1420009111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng K., Xu H., Chen X., Wang L., Tian B., Zhao Y. Structural basis for DNA 5 -end resection by RecJ. Elife. 2016;5 doi: 10.7554/eLife.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattimore V., Battista J.R. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka M., Earl A.M., Howell H.A., Park M.J., Eisen J.A., Peterson S.N. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics. 2004;168:21–33. doi: 10.1534/genetics.104.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova K.S., Omelchenko M.V., Gaidamakova E.K., Matrosova V.Y., Vasilenko A., Zhai M. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Groot A., Siponen M.I., Magerand R., Eugenie N., Martin-Arevalillo R., Doloy J. Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus. Nucleic Acids Res. 2019;47:11403–11417. doi: 10.1093/nar/gkz883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu H., Wang L., Li S., Pan C., Cheng K., Luo Y. Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO. Nucleic Acids Res. 2019;47:9925–9933. doi: 10.1093/nar/gkz720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludanyi M., Blanchard L., Dulermo R., Brandelet G., Bellanger L., Pignol D. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol Microbiol. 2014;94:434–449. doi: 10.1111/mmi.12774. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Xu Q., Lu H., Lin L., Wang L., Xu H. Protease activity of PprI facilitates DNA damage response: Mn2+-dependence and substrate sequence-specificity of the proteolytic reaction. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long H.G., Kucukyildirim S., Sung W., Williams E., Lee H., Ackerman M. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol Biol Evol. 2015;32:2383–2392. doi: 10.1093/molbev/msv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long H.A., Miller S.F., Williams E., Lynch M. Specificity of the DNA Mismatch Repair System (MMR) and mutagenesis bias in Bacteria. Mol Biol Evol. 2018;35:2414–2421. doi: 10.1093/molbev/msy134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krisko A., Radman M. Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Csh Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slade D., Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obiero J., Pittet V., Bonderoff S.A., Sanders D.A. Thioredoxin system from Deinococcus radiodurans. J Bacteriol. 2010;192:494–501. doi: 10.1128/JB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reon B.J., Nguyen K.H., Bhattacharyya G., Grove A. Functional comparison of Deinococcus radiodurans Dps proteins suggests distinct in vivo roles. Biochem J. 2012;447:381–391. doi: 10.1042/BJ20120902. [DOI] [PubMed] [Google Scholar]
- 29.Jeong S.W., Jung J.H., Kim M.K., Seo H.S., Lim H.M., Lim S. The three catalases in Deinococcus radiodurans: only two show catalase activity. Biochem Biophys Res Commun. 2016;469:443–448. doi: 10.1016/j.bbrc.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Basu B., Apte S.K. Gamma radiation-induced proteome of Deinococcus radiodurans primarily targets DNA repair and oxidative stress alleviation. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daly M.J., Gaidamakova E.K., Matrosova V.Y., Kiang J.G., Fukumoto R., Lee D.Y. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B., Hua Y. Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 2010;18:512–520. doi: 10.1016/j.tim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Gao L.H., Chen X.N., Tian Y., Yan Y.L., Zhan Y.H., Zhou Z.F. The novel ncRNA OsiR positively regulates expression of katE2 and is required for oxidative stress tolerance in Deinococcus radiodurans. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai C.H., Liao R., Chou B., Contreras L.M. Transcriptional analysis of Deinococcus radiodurans reveals novel small RNAs that are differentially expressed under ionizing radiation. Appl Environ Microb. 2015;81:1745–1755. doi: 10.1128/AEM.03709-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villa J.K., Amador P., Janovsky J., Bhuyan A., Saldanha R., Lamkin T.J. A genome-wide search for ionizing-radiation-responsive elements in Deinococcus radiodurans reveals a regulatory role for the DNA gyrase subunit A gene's 5' untranslated region in the radiation and desiccation response. Appl Environ Microb. 2017;83 doi: 10.1128/AEM.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daly M.J., Gaidamakova E.K., Matrosova V.Y., Vasilenko A., Zhai M., Venkateswaran A. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 37.Blasius M., Sommer S., Hubscher U. Deinococcus radiodurans: what belongs to the survival kit? Crit Rev Biochem Mol Biol. 2008;43:221–238. doi: 10.1080/10409230802122274. [DOI] [PubMed] [Google Scholar]
- 38.Killoran M.P., Keck J.L. Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic Acids Res. 2008;36:3139–3149. doi: 10.1093/nar/gkn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Hou H., Du Q., Zhang W., Liu G., Shtykova E.V. Solution small angle X-ray scattering (SAXS) studies of RecQ from Deinococcus radiodurans and its complexes with junction DNA substrates. J Biol Chem. 2013;288:32414–32423. doi: 10.1074/jbc.M113.502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S.C., Huang C.H., Yang C.S., Way T.D., Chang M.C., Chen Y. Crystal structure of Deinococcus radiodurans RecQ helicase catalytic core domain: the interdomain flexibility. Biomed Res Int. 2014;2014 doi: 10.1155/2014/342725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stelter M., Acajjaoui S., McSweeney S., Timmins J. Structural and mechanistic insight into DNA unwinding by Deinococcus radiodurans UvrD. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0077364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris D.R., Tanaka M., Saveliev S.V., Jolivet E., Earl A.M., Cox M.M. Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu G., Lu H., Wang L., Chen H., Xu Z., Hu Y. DdrB stimulates single-stranded DNA annealing and facilitates RecA-independent DNA repair in Deinococcus radiodurans. DNA Repair (Amst) 2010;9:805–812. doi: 10.1016/j.dnarep.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Narumi I., Satoh K., Cui S., Funayama T., Kitayama S., Watanabe H. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol Microbiol. 2004;54:278–285. doi: 10.1111/j.1365-2958.2004.04272.x. [DOI] [PubMed] [Google Scholar]
- 45.Gutsche I., Vujicic-Zagar A., Siebert X., Servant P., Vannier F., Castaing B. Complex oligomeric structure of a truncated form of DdrA: a protein required for the extreme radiotolerance of Deinococcus. Biochim Biophys Acta. 2008;1784:1050–1058. doi: 10.1016/j.bbapap.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Norais C.A., Chitteni-Pattu S., Wood E.A., Inman R.B., Cox M.M. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem. 2009;284:21402–21411. doi: 10.1074/jbc.M109.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouthier de la Tour C., Boisnard S., Norais C., Toueille M., Bentchikou E., Vannier F. The deinococcal DdrB protein is involved in an early step of DNA double strand break repair and in plasmid transformation through its single-strand annealing activity. DNA Repair (Amst) 2011;10:1223–1231. doi: 10.1016/j.dnarep.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiman-Marangos S., Junop M.S. The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Res. 2010;38:3432–3440. doi: 10.1093/nar/gkq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiman-Marangos S.N., Peel J.K., Weiss Y.M., Ghirlando R., Junop M.S. Crystal structure of the DdrB/ssDNA complex from Deinococcus radiodurans reveals a DNA binding surface involving higher-order oligomeric states. Nucleic Acids Res. 2013;41:9934–9944. doi: 10.1093/nar/gkt759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugiman-Marangos S.N., Weiss Y.M., Junop M.S. Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB. Proc Natl Acad Sci U S A. 2016;113:4308–4313. doi: 10.1073/pnas.1520847113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernstein D.A., Eggington J.M., Killoran M.P., Misic A.M., Cox M.M., Keck J.L. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci U S A. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webster G., Genschel J., Curth U., Urbanke C., Kang C., Hilgenfeld R. A common core for binding single-stranded DNA: structural comparison of the single-stranded DNA-binding proteins (SSB) from E. coli and human mitochondria. FEBS Lett. 1997;411:313–316. doi: 10.1016/s0014-5793(97)00747-3. [DOI] [PubMed] [Google Scholar]
- 53.Devigne A., Guerin P., Lisboa J., Quevillon-Cheruel S., Armengaud J., Sommer S., Bouthier-de-la-Tour C., Servant P. PprA protein is involved in chromosome segregation via its physical and functional interaction with DNA gyrase in irradiated Deinococcus radiodurans bacteria. mSphere. 2016;1 doi: 10.1128/mSphere.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kota S., Kamble V.A., Rajpurohit Y.S., Misra H.S. ATP-type DNA ligase requires other proteins for its activity in vitro and its operon components for radiation resistance in Deinococcus radiodurans in vivo. Biochem Cell Biol. 2010;88:783–790. doi: 10.1139/o10-075. [DOI] [PubMed] [Google Scholar]
- 55.Adachi M., Shimizu R., Shibazaki C., Satoh K., Fujiwara S., Arai S. Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans. FASEB J. 2019;33:3647–3658. doi: 10.1096/fj.201801506R. [DOI] [PubMed] [Google Scholar]
- 56.Hua Y., Narumi I., Gao G., Tian B., Satoh K., Kitayama S. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem Biophys Res Commun. 2003;306:354–360. doi: 10.1016/s0006-291x(03)00965-3. [DOI] [PubMed] [Google Scholar]
- 57.Vujicic-Zagar A., Dulermo R., Le Gorrec M., Vannier F., Servant P., Sommer S. Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae. J Mol Biol. 2009;386:704–716. doi: 10.1016/j.jmb.2008.12.062. [DOI] [PubMed] [Google Scholar]
- 58.Borges P.T., Frazao C., Miranda C.S., Carrondo M.A., Romao C.V. Structure of the monofunctional heme catalase DR1998 from Deinococcus radiodurans. FEBS J. 2014;281:4138–4150. doi: 10.1111/febs.12895. [DOI] [PubMed] [Google Scholar]
- 59.Dennis R.J., Micossi E., McCarthy J., Moe E., Gordon E.J., Kozielski-Stuhrmann S. Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:325–329. doi: 10.1107/S1744309106008402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bihani S.C., Panicker L., Rajpurohit Y.S., Misra H.S., Kumar V. drFrnE represents a hitherto unknown class of eubacterial cytoplasmic disulfide oxido-reductases. Antioxid Redox Signal. 2018;28:296–310. doi: 10.1089/ars.2016.6960. [DOI] [PubMed] [Google Scholar]
- 61.Shuryak I., Matrosova V.Y., Gaidamakova E.K., Tkavc R., Grichenko O., Klimenkova P. Microbial cells can cooperate to resist high-level chronic ionizing radiation. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0189261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borgstahl G.E., Pokross M., Chehab R., Sekher A., Snell E.H. Cryo-trapping the six-coordinate, distorted-octahedral active site of manganese superoxide dismutase. J Mol Biol. 2000;296:951–959. doi: 10.1006/jmbi.1999.3506. [DOI] [PubMed] [Google Scholar]
- 63.Smolik A.C., Bengez-Pudja L., Cheng I., Mascotti D.P. Characterization of E. coli manganese superoxide dismutase binding to RNA and DNA. Bba-Proteins Proteom. 2014;1844:2251–2256. doi: 10.1016/j.bbapap.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 64.Collet J.F., Messens J. Structure, function, and mechanism of thioredoxin proteins, antioxid redox. Sign. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 65.Kim S.G., Bhattacharyya G., Grove A., Lee Y.H. Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans. J Mol Biol. 2006;361:105–114. doi: 10.1016/j.jmb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 66.Romao C.V., Mitchell E.P., McSweeney S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J Biol Inorg Chem. 2006;11:891–902. doi: 10.1007/s00775-006-0142-5. [DOI] [PubMed] [Google Scholar]
- 67.Cuypers M.G., Mitchell E.P., Romao C.V., McSweeney S.M. The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J Mol Biol. 2007;371:787–799. doi: 10.1016/j.jmb.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 68.Santos S.P., Cuypers M.G., Round A., Finet S., Narayanan T., Mitchell E.P. SAXS structural studies of Dps from Deinococcus radiodurans highlights the conformation of the mobile N-terminal extensions. J Mol Biol. 2017;429:667–687. doi: 10.1016/j.jmb.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Bozzi A.T., Zimanyi C.M., Nicoludis J.M., Lee B.K., Zhang C.H., Gaudet R. Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter. Elife. 2019;8 doi: 10.7554/eLife.41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vashishtha A.K., Wang J., Konigsberg W.H. Different divalent cations alter the kinetics and fidelity of DNA polymerases. J Biol Chem. 2016;291:20869–20875. doi: 10.1074/jbc.R116.742494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng M., Patel D., Dervan J.J., Ceska T., Suck D., Haq I. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat Struct Mol Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y., Lu M., Zhang H., Hu J., Zhou C., Xu Q. Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucleic Acids Res. 2015;43:5550–5559. doi: 10.1093/nar/gkv444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goncalves A.M., de Sanctis D., McSweeney S.M. Structural and functional insights into DR2231 protein, the MazG-like nucleoside triphosphate pyrophosphohydrolase from Deinococcus radiodurans. J Biol Chem. 2011;286:30691–30705. doi: 10.1074/jbc.M111.247999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makharashvili N., Koroleva O., Bera S., Grandgenett D.P., Korolev S. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure. 2004;12:1881–1889. doi: 10.1016/j.str.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Leiros I., Timmins J., Hall D.R., McSweeney S. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 2005;24:906–918. doi: 10.1038/sj.emboj.7600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee B.I., Kim K.H., Park S.J., Eom S.H., Song H.K., Suh S.W. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Timmins J., Leiros I., McSweeney S. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 2007;26:3260–3271. doi: 10.1038/sj.emboj.7601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J., Zhao L., Seo H.S., Jung J.H., Choi J.I., Kim M.K. Crystal structure of the highly radiation-inducible DinB/YfiT superfamily protein DR0053 from Deinococcus radiodurans R1. Biochem Biophys Res Commun. 2019;513:354–359. doi: 10.1016/j.bbrc.2019.03.209. [DOI] [PubMed] [Google Scholar]
- 79.Leulliot N., Cladiere L., Lecointe F., Durand D., Hubscher U., van Tilbeurgh H. The family X DNA polymerase from Deinococcus radiodurans adopts a non-standard extended conformation. J Biol Chem. 2009;284:11992–11999. doi: 10.1074/jbc.M809342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lewis H.A., Furlong E.B., Laubert B., Eroshkina G.A., Batiyenko Y., Adams J.M. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure. 2001;9:527–537. doi: 10.1016/s0969-2126(01)00613-x. [DOI] [PubMed] [Google Scholar]
- 81.Agrawal R., Goyal V.D., Kumar A., Gaur N.K., Jamdar S.N., Kumar A. Two-domain aminopeptidase of M1 family: structural features for substrate binding and gating in absence of C-terminal domain. J Struct Biol. 2019;208:51–60. doi: 10.1016/j.jsb.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 82.Are V.N., Kumar A., Goyal V.D., Gotad S.S., Ghosh B., Gadre R. Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases. Proteins. 2019;87:212–225. doi: 10.1002/prot.25641. [DOI] [PubMed] [Google Scholar]
- 83.Mota C.S., Goncalves A.M., de Sanctis D. Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP. FEBS J. 2016;283:4274–4290. doi: 10.1111/febs.13923. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H., Xu Q., Lu M., Xu X., Wang Y., Wang L. Structural and functional studies of MutS2 from Deinococcus radiodurans. DNA Repair (Amst) 2014;21:111–119. doi: 10.1016/j.dnarep.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 85.Wekselman I., Zimmerman E., Davidovich C., Belousoff M., Matzov D., Krupkin M. The Ribosomal protein uL22 modulates the shape of the protein exit tunnel. Structure. 2017;25:1233–1241. doi: 10.1016/j.str.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 86.Chen X.G., Taylor D.W., Fowler C.C., Galan J.E., Wang H.W., Wolin S.L. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–177. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajpurohit Y.S., Misra H.S. Structure-function study of deinococcal serine/threonine protein kinase implicates its kinase activity and DNA repair protein phosphorylation roles in radioresistance of Deinococcus radiodurans. Int J Biochem Cell Biol. 2013;45:2541–2552. doi: 10.1016/j.biocel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 88.Cho C.C., Chien C.Y., Chiu Y.C., Lin M.H., Hsu C.H. Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans. Nat Commun. 2019;10:1491. doi: 10.1038/s41467-019-09153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou C., Dai J., Lu H., Chen Z., Guo M., He Y. Succinylome analysis reveals the involvement of lysine succinylation in the extreme resistance of Deinococcus radiodurans. Proteomics. 2019;19 doi: 10.1002/pmic.201900158. [DOI] [PubMed] [Google Scholar]