Abstract
Macrophage recruitment and pro-inflammatory differentiation are hallmarks of various diseases, including infection and sepsis. Although studies suggest that mitochondria may regulate macrophage immune responses, it remains unclear whether mitochondrial mass affects macrophage pro-inflammatory differentiation. Here, we found that lipopolysaccharide (LPS)-activated macrophages possess higher mitochondrial mass than resting cells. Therefore, this study aimed to explore the functional role and molecular mechanisms of increased mitochondrial mass in pro-inflammatory differentiated macrophages. Results show that an increase in the mitochondrial mass of macrophages positively correlates with inflammatory cytokine generation in response to LPS. RNA-seq analysis revealed that LPS promotes signal transducers and activators of transcription 2 (Stat2) and dynamin-related protein 1 (Drp1) expression, which are enriched in positive mitochondrial fission regulation. Meanwhile, knockdown or pharmacological inhibition of Drp1 blunts LPS-induced mitochondrial mass increase and pro-inflammatory differentiation. Moreover, Stat2 boosts Drp1 phosphorylation at serine 616, required for Drp1-mediated mitochondrial fission. LPS also causes Stat2-and Drp1-dependent biogenesis, which contributes to the generation of additional mitochondria. However, these mitochondria are profoundly remodeled, displaying fragmented morphology, loose cristae, reduced Δψm, and metabolic programming. Furthermore, these remodeled mitochondria shift their function from ATP synthesis to reactive oxygen species (ROS) production, which drives NFκB-dependent inflammatory cytokine transcription. Interestingly, an increase in mitochondrial mass with constitutively active phosphomimetic mutant of Drp1 (Drp1S616E) boosted pro-inflammatory response in macrophages without LPS stimulation. In vivo, we also demonstrated that Mdivi-1 administration inhibits LPS-induced macrophage pro-inflammatory differentiation. Importantly, we observed Stat2 phosphorylation and Drp1-dependent mitochondrial mass increase in macrophages isolated from LPS-challenged mice. In conclusion, we comprehensively demonstrate that a Stat2-Drp1 dependent mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages. Therefore, targeting the Stat2-Drp1 axis may provide novel therapeutic approaches for treating infection and inflammatory diseases.
Keywords: Stat2, Drp1, Mitochondrial mass, Pro-inflammatory macrophage, Lipopolysaccharide, Reactive oxygen species
Graphical abstract
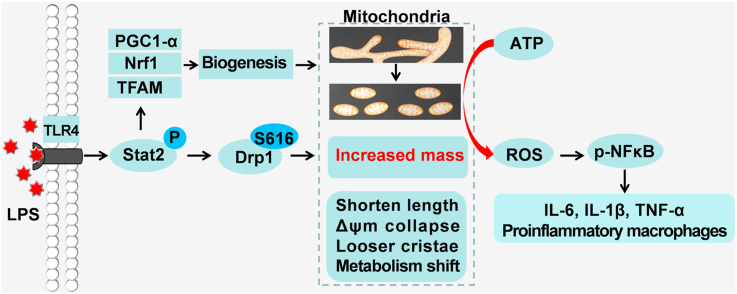
Schematic for the key role of Stat2-Drp1 induced mitochondrial mass increase in promoting pro-inflammatory differentiation of macrophages. LPS-activated macrophages possess more mitochondria by inducing Stat2-Drp1-dependent mitochondrial fission and biogenesis. However, these mitochondria undergo profound remodeling, including fragmented morphology, loose cristae, reduced Δψm, and metabolic programming, which induces a functional shift from ATP synthesis to ROS production, and further drives transcription of NFκB-dependent inflammatory cytokine production.
Highlights
-
•
Increased macrophage mitochondrial mass correlates with inflammatory cytokines.
-
•
LPS increases mitochondrial fission regulatory protein (Stat2 and Drp1) expression.
-
•
Increased Stat2 and Drp1 contribute to generation of additional mitochondria.
-
•
New mitochondria are morphologically altered and metabolically reprogrammed.
-
•
Reprogrammed mitochondria shift from ATP synthesis to ROS production.
Abbreviations
- BMDMs
bone marrow-derived macrophages
- DCFH-DA
2,7-dichlorodi-hydrofluorescein diacetate
- Dcn
decorin
- Drp1
dynamin-related protein
- ECAR
extracellular acidification rate
- ELISA
enzyme-linked immunosorbent assay
- EtBr
ethidium bromide
- ETC
electron transport chain
- FCCP
carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- Fis1
mitochondrial fission protein 1
- Fpkm
fragments per kilobase million
- FSC
forward scatter channel
- IL
interleukin
- iNOS
inducible nitric oxide;
- LPS
lipopolysaccharide;
- Mff
mitochondrial fission factor
- MFI
mean fluorescence intensity
- MIEF1
mitochondrial elongation factor 1
- Miga2
mitoguardin 2
- MitoQ
mitoquinone
- MOI
multiplicity of infection
- mtDNA
mitochondrial DNA
- MTG
MitoTracker-Green
- Mus
musculus
- NFκB
nuclear transcription factor-kappa B
- NLPR3
NLR family, pyrin domain-containing 3
- Nrf1
nuclear respiratory factor 1
- OCR
oxygen consumption rate
- PGC-1α
Peroxisome proliferator activator receptor gamma-coactivator 1α
- PMA
phorbol-12-myristate-13-acetate
- PMs
peritoneal macrophages
- Prkn
Parkin RBR E3 ubiquitin protein ligase
- Rho-123
rhodamine 123
- ROS
reactive oxygen species
- SSC
side scatter channel
- Stat2
signal transducers and activators of transcription 2
- TAK-242
Resatorvid
- TFAM
mitochondrial transcription factor A
- TFI
total fluorescence intensity
- TLR4
Toll-like receptor 4
- TMRM
Tetramethylrhodamine methyl ester
- TNF-a
tumor necrosis factor alpha
- 2DG
2-deoxy-d-glucose
- Δψm
mitochondrial membrane potential
1. Introduction
Macrophages are monocyte-derived innate immune cells that regulate tissue homeostasis and adaptive responses. They exist in most tissues of the human body and display high plasticity. In general, macrophages are differentiated into two main phenotypes: pro-inflammatory (M1), and anti-inflammatory (M2) [1]. In response to bacteria and lipopolysaccharide (LPS), resting macrophages undergo pro-inflammatory differentiation, characterized by release of large amounts of cytokines such as tumor necrosis factor alpha (TNF-α), interleukin (IL)-1β, and IL-6, as well as reactive oxygen species (ROS) [2]. These pro-inflammatory mediators are critical for the organism's immune defense and for microbial killing. However, continuous inflammatory activation in macrophages may cause collateral tissue damage and chronic inflammation. Macrophage elicited inflammation is the hallmark of various pathologies, including infection, sepsis, and radiation sickness [3]. Therefore, targeting M1 phenotype macrophages may provide novel therapeutic opportunities for inflammatory-related diseases.
Mitochondria are vital organelles in eukaryotic cells that regulate numerous biological processes ranging from ROS production, energy metabolism, and stress response to cell fate [4]. Recently, a growing body of literature has emphasized the critical role of mitochondrion as a key intracellular signaling platform modulating innate immune and inflammatory processes [5,6]. Specifically, pro-inflammatory differentiated macrophages switch their core glucose metabolic pathway from mitochondrial oxidative phosphorylation to aerobic glycolysis [7]. Hence, inhibition of glycolysis with 2-deoxyglucose (2DG) attenuates LPS-induced production of ROS and pro-inflammatory factors [8]. Mitochondrial DNA (mtDNA) release is also regarded as a danger signal for the induction of ROS-dependent inflammation in macrophages. Moreover, mitochondrial ROS (mtROS) promotes macrophages inflammatory responses by activating nuclear factor-κB (NFκB) and the NLR family pyrin domain-containing 3 (NLRP3) inflammasome [9,10]. Alternatively, scavenging mtROS reduces production of IL-1β and IL-18 in LPS-activated macrophages. These findings suggest that ROS is a key factor involved in mitochondrial homeostasis and inflammatory signaling, however, the mechanism by which mitochondria, in LPS-activated macrophages, produce ROS is not well characterized.
Mitochondria continuously remodel their mass, shape, size, and function by undergoing alternate processes of fission and fusion [11]. In general, excessive fission causes mitochondria to become fragmented into small pieces, while increased fusion supports the integration of adjacent mitochondria. Dynamin-related protein 1 (Drp1), an important cytosolic GTPase, is indispensable for mitochondrial fission. Drp1 becomes activated via phosphorylation, which is initiated at several sites, including S616 and S637. Phosphorylation at S637 limits fission by blocking Drp1 recruitment to the mitochondria, while phosphorylation at S616 boosts fission by driving Drp1 mitochondrial translocation [12]. Upon activation, Drp1 is recruited to the mitochondrial surface where it interacts with accessory proteins, such as mitochondrial fission protein 1 (Fis1), mitochondrial elongation factor 1 (Mief1) and mitochondrial fission factor (Mff) [13,14]. Previous reports also reveal that the balance in mitochondrial dynamics is associated with the biological functions of macrophages. Meanwhile, abnormal mitochondrial fusion-fission facilitates the progression of LPS-induced inflammation in microglial cells and sepsis in mice [15,16]. Furthermore, Drp1-dependent mitochondrial fission is required for clearance of apoptotic cells by macrophages [17]; and unbalanced fission and fusion events generally cause changes in mitochondrial mass and functional disorder. Many reports have emphasized the importance of mitochondrial mass in controlling cell fate, including cell death, cell survival and cell differentiation. Greater mitochondrial mass endows CD8+ memory T cells with a bioenergetic advantage and a rapid recall ability in response to reinfection [18]. Mitochondrial mass also controls skeletal muscle strength and tumor growth [19,20]. However, the role of mitochondrial mass for macrophage differentiation is not well understood.
In the current study, we systematically investigated the alteration of mitochondrial mass in LPS-activated macrophages and their functional roles in the modulation of macrophage differentiation and inflammatory response. More importantly, the underlying molecular mechanisms involved were explored in depth. Our work provides novel insights into the role of mitochondrial mass during the process of macrophage pro-inflammatory differentiation, which facilitates the treatment of various infectious, and inflammatory diseases.
2. Material and methods
2.1. Materials
The primary antibodies used in this study and their working dilutions are listed in Supplementary Table 1. The primers used for quantitative real-time reverse transcription PCR (qRT-PCR) were synthesized by AuGCT Biotechnology, and their sequences are listed in Supplementary Table 2. The information for all other reagents is described in the following sections.
2.2. Animals
Wildtype (WT) C57BL/6 mice (8–10 weeks old) were obtained from the Experimental Animal Center of Fourth Military Medical University (Xi'an, China). Animals were maintained under specific pathogen-free conditions. All experimental procedures were approved by the Animal and Ethical Committee of the University, which strictly followed the National Institutes of Health (NIH) guidelines for the Use of Laboratory Animals (NIH No. 85–23, revised 2011).
2.3. Bone marrow-derived macrophages
Bone marrow-derived macrophages (BMDMs) are widely used primary macrophages that are differentiated from mammalian bone marrow (BM) cells. Briefly, 8–10-week old male C57BL/6 mice were euthanized in a 2.5% isoflurane and sacrificed by cervical dislocation. BM cells were dissociated from the bone cavity and differentiated in RPMI 1640 with 10% heat-inactivated fetal calf serum (FBS, Hyclone, SV30106.03), 1% penicillin streptomycin and 20% L929 fibroblast supernatant. After 7 days, attached BMDMs were harvested and used for further experiments.
2.4. Peritoneal macrophages isolation
To elicit large numbers of peritoneal macrophages (PMs), 8-10-week old male C57BL/6 mice were intraperitoneally injected (i.p.) with 2 mL of heat-inactivated thioglycolate media. Four days later, mice were sacrificed and injected (i.p.) with 5 mL of PBS (containing 5 mM EDTA) to wash the peritoneal cavity. Next, PMs were purified by removing non-adherent cells. Approximately 10 million PMs were collected from each mouse.
2.5. Cell culture and treatment
RAW264.7 macrophages (RAW, murine), phorbol-12-myristate-13-acetate (PMA; Sigma-Aldrich, P1585)-induced THP-1 macrophages (human), and BV-2 microglia cells (murine) were obtained from American Type Culture Collection (Manassas, VA, USA). BMDMs and PMs were harvested as described above. Cells were cultured in RPMI 1640 with 10% heat-inactivated FBS and maintained in a humidified CO2 incubator at 37 °C. To induce pro-inflammatory differentiation, macrophages were treated with 0.5 or 1 μg/mL LPS for up to 24 h. Furthermore, RAW and BMDMs were treated with 0.5 μg/mL LPS (Sigma-Aldrich, L2880) for 12 h in the presence of DMSO (Control), Drp1 inhibitor Mdivi-1 (50 μM; Selleck, S7162), mitochondrial ROS scavenger mitoquinone (MitoQ, 250 and 500 nM; Abmole, M9068) or TLR4 inhibitor resatorvid (TAK-242, 100 nM, Selleck, S7455).
2.6. Knockdown and forced expression of target genes
pSIH-H1-Puro lentiviral vectors expressing short hairpin (sh) RNA targeting murine Stat2 and Drp1, as well as pCDH-CMV-MCS-EF1-Blast-3×Flag lentiviral vectors expressing wildtype Drp1 (Flag-Drp1WT), S616 phosphorylation-defective mutant Drp1 (Flag-Drp1S616A) and (Drp1S616E, a phosphomimetic mutant) were also designed by GeneChem Co. Ltd (Shanghai, China). An empty vector (EV) served as a control. BMDMs and RAW were transfected with shDrp1 vectors or EV with polybrene (Sigma-Aldrich, H9268) according to the manufacturer's protocol. Furthermore, RAW expressed shDrp1 were rescued with either EV, Drp1WT or Drp1S616A. The multiplicity of infection (MOI) used in these experiments was 30. The transfected cells were selected with puromycin (Thermo Fisher, A1113803) and blasticidin (InvivoGen, ant-bl-05). Finally, the transfected cells were stimulated with or without 0.5 μg/mL LPS for 12 h.
2.7. RAW-ρ0 cells generation
RAW cells were long-term treated with 45 ng/mL ethidium bromide (EtBr; Invitrogen, 5585011) to inhibit mtDNA replication and transcription. After 4 weeks incubation, most of the mitochondrial DNA in RAW was deleted. Both RAW-ρ0 and RAW cells were then treated with or without 0.5 μg/mL LPS for 12 h.
2.8. Mitochondrial mass and morphology assessment
MitoTracker-Green (MTG, Molecular Probes, M7514) is a Δψm-independent dye that can monitor mitochondria. After treatment, cells were stained with MTG (100 nM) and analyzed by flow cytometry (Accuri C6, BD Biosciences, MD, USA). The mean fluorescence intensity (MFI) was quantified using the FlowJo software (Tree Star, San Jose, CA, USA). Meanwhile, laser scanning confocal microscopy (LSCM, Olympus, Tokyo, Japan) was used to observe mitochondrial morphology. The mitochondrial mass and length were analyzed by Image J software (NIH, Bethesda, MD) and averaged for 20 cells per sample.
2.9. Transmission electron microscopy (TEM)
After treatment, 2 × 106 cells were fixed in 4% glutaraldehyde (Santa Cruz, CA, USA) for 24 h at 4 °C. Samples were incubated with 1% osmium tetroxide (Electron microscopy sciences, 51007), after which they were alcohol dehydrated, and araldite embedded. Thin sections (85 nm) were stained with uranyl acetate and lead citrate. Ultrastructural analysis was conducted via TEM (FEI, Hillsboro, Oregon) at 80 kV.
2.10. ROS analysis
2,7-Dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma, D6883) is an indicator for intracellular oxidants. MitoSOX ™ Red (Molecular Probes, M36008) is a specific fluorescent probe for detection of mitochondrial ROS. Briefly, samples were stained with DCFH-DA (10 μM) and MitoSOX ™ Red (5 μM) at 37 °C for 30 min. After washing twice with PBS, cells were analyzed by flow cytometry, and their MFIs were quantified.
2.11. Δψm measurement
2.11.1. Rho-123 dye
Rhodamine 123 (Rho-123; Invitrogen, R-302) is a cell-permeant green fluorescent probe for Δψm. Briefly, samples were stained with Rho-123 (50 μM) at 37 °C for 30 min. The images were captured using LSCM, and Rho-123 MFI was assessed by Image J software. Cells were also assessed via flow cytometric assay. The ratio of Rho-123 MFI and MTG MFI was analyzed.
2.11.2. TMRM dye
Tetramethylrhodamine methyl ester (TMRM; Invitrogen, T-668) is a cell-permeant cationic lipophilic red fluorescent probe for Δψm. Briefly, samples were stained with TMRM (20 nM) and MTG TM (100 nM) at 37 °C for 30 min. Cells were analyzed using flow cytometry. The ratio of TMRM MFI and MTG MFI was assessed.
2.11.3. JC-1 dye
JC-1 (Invitrogen, T-3168) can form red fluorescent aggregates in normal mitochondria, however, it becomes transformed into green fluorescent monomers in depolarized mitochondria. Here, samples were stained with 2 μg/mL JC-1 for 30 min at 37 °C. Cells were then analyzed by flow cytometry, and the ratio of red/green was used to indicate Δψm alternation.
2.12. Enzyme-linked immunosorbent assay
Cytokine concentrations in cell supernatants and mice serum were detected using specific enzyme-linked immunosorbent assay (ELISA) kits (Cusabio, Wuhan, China) for mouse TNF-a, IL-6 and IL-1β according to the manufacturer's instructions. The optical density was monitored at 450 nm using a microplate reader (Thermo Fisher Scientific, USA). Concentrations of cytokines were calculated using the professional software Curve Expert 1.3.
2.13. ATP detection
ATP level was detected using an ATP Determination kit (Beyotime Biotechnology, S0027) according to the manufacturer's protocol. Luminescence was recorded using the luminescence plate reader (Thermo Fisher, USA).
2.14. Oxygen consumption and extracellular acidification analysis
Cells were plated at 0.5 × 106 cells/well in a 24-well Seahorse plate one day prior to measurement. The plate was incubated in glucose-supplemented Seahorse XF assay media at 37 °C for 1 h without CO2. Oxygen consumption rate (OCR) was measured with oligomycin (1 μM), carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP, 1 μM), and rotenone (0.5 μM). Meanwhile, the extracellular acidification rate (ECAR) was assessed using glucose (10 mM), oligomycin (1 μM), and 2-deoxy-d-glucose (2-DG, 0.5 μM). These plates were run on the Seahorse XF-24.
2.15. RNA-sequencing analysis
Total RNA was isolated in PBS- and LPS-treated BMDMs in triplicate. Sequencing libraries were produced using the TruSeq RNA Sample Preparation kit (Illumina, San Diego, CA, USA). RNA-sequencing (RNA-seq) analysis was conducted by Personal Biotechnology Cp. Ltd (Shanghai, China). Twenty genes enriched in mitochondrial fission and fusion regulation were analyzed. Heat maps were made based on the data shown in Supplementary Table 3; and the enrichment results for Stat2 are presented in Supplementary Table 4.
2.16. qRT-PCR
Total RNA was extracted from macrophages or tissues using the TRIzol reagent (Sigma-Aldrich, T9424). Complementary DNA (cDNA) was synthesized using the TIANScript cDNA kits (TIANGEN, KR104). Then, cDNA was amplified using a SYBR Green PCR kit (TIANGEN, FP402). qRT-PCR analysis was performed using a Cycler Sequence Detection System (Bio-Rad, CFX96, USA). The cycling parameters were as follows: 40 cycles at 95 °C for 15 s, 55 °C for 15 s and 72 °C for 15 s. Relative expression of target genes normalized to Gapdh were analyzed with CFX Manager 2.1 software (Bio-Rad).
2.17. mtDNA copy number
Total DNA in macrophages was extracted with a TIANamp Genomic DNA kit (DP304, Beijing, China) according to the manufacturer's protocol. Briefly, samples were amplified using primers for cytochrome b (mtDNA), and beta2-microglobulin (nuclear DNA) by qRT-PCR. Here, expression of cytochrome b normalized by beta2-microglobulin was used to indicate the mtDNA copy number.
2.18. Western blot
Proteins were prepared by direct lysis of cells and tissues using RIPA buffer (Beyotime Biotechnology, P0013). A total of 20 μg proteins was separated with 10–15% SDS-PAGE electrophoresis and blotted to polyvinyl difluoride membranes. The blots were blocked with PBS (pH = 7.4) containing 10% serum for 1 h, and incubated with primary antibodies overnight at 4 °C. After washing with PBS, the membranes were incubated with HRP-conjugated secondary antibodies (Sigma-Aldrich) for 2 h at a dilution of 1:3000. Finally, the images were captured with the Molecular Imager ChemiDoc XRS + System (Bio-Rad, Hercules, CA, USA). Densitometry of the bands was quantified using Quantity One software (Bio-Rad).
2.19. Endotoxin-induced inflammatory model in vivo
Herein, 32 WT male C57/Bl6 mice were randomly assigned to four groups (eight mice per group). Mice intraperitoneally injected (i.p.) 3% thioglycolate once to elicit macrophages. Four days later, animals were intraperitoneally injected with 20 mg/kg Mdivi-1 or DMSO for 2 h, followed by 15 mg/kg LPS or PBS stimulation (i.p.) for 6 h. Then, PMs and blood samples were harvested after mice were sacrificed by cervical dislocation.
2.20. Immunofluorescence staining
BMDMs treatment with or without LPS and MitoQ were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, X100) in PBS for 15 min. Samples were then blocked with 10% FBS for 1 h, and incubated with primary antibodies overnight at 4 °C. After washing, samples were incubated with FITC-conjugated secondary antibodies for 2 h at room temperature. DAPI was used to stain the cell nuclei. Finally, the images were captured using LSCM.
2.21. Statistical analysis
Statistical significance between two groups was analyzed using Student's t-tests with GraphPad Prism software (GraphPad, 8 San Diego, USA). Data were expressed as mean ± SEM. Correlations between two parameters were assessed by Spearman correlation analysis. p < 0.05 was considered significant.
3. Results
3.1. Pro-inflammatory differentiated macrophages have higher mitochondrial mass and lower Δψm than resting macrophages
To explore the differences in the mitochondria of resting macrophages and M1 differentiated macrophages, we measured membrane potential (Δψm) in bone marrow-derived macrophages (BMDMs) with a fluorescent probe, Rhodamine 123 (Rho-123). The accumulation of TNF-α, IL-6, and IL-1β production in supernatants indicated that LPS induced pro-inflammatory differentiation of BMDMs (Fig. S1A). Flow cytometry results showed that Rho-123 MFI was increased by approximately 2–3 fold in BMDMs stimulated with LPS for 1–24 h (Fig. S1B). However, ROS, as indicated by DCF MFI, increased by 3–11 fold in LPS-activated macrophages (Fig. S1C). Excessive ROS generally causes Δψm collapse by disrupting mitochondrial membrane homeostasis. Our results seemed to contradict previously published conclusions [21].
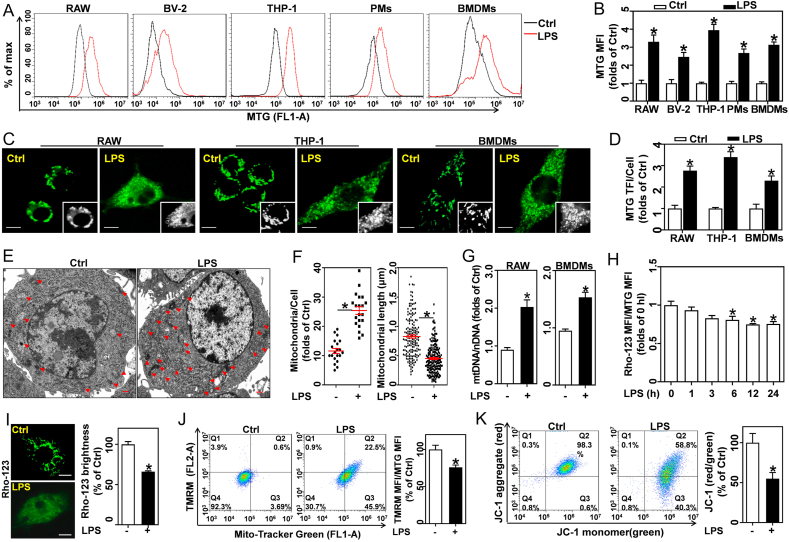
Here, we observed that LPS-treated macrophages were enlarged in size (Fig. S2A). Meanwhile, LPS-treated RAW and BMDMs showed higher forward scatter channel (FSC) signals, indicating larger cell size; and greater side scatter channel (SSC) signal, indicating higher granularity or internal complexity (Figs. S2B and S2C). MitoTracker-Green (MTG), a fluorescent probe independent of the Δψm, is widely used to measure mitochondrial mass. Interestingly, flow cytometry results showed a 2–4 folds increase in the mitochondrial mass of LPS-stimulated RAW264.7 (RAW), THP-1, BV-2, peritoneal macrophages (PMs), and BMDMs (Fig. 1A and B and S2D). In addition, mitochondrial morphology in macrophages stained with MTG was assessed by LSCM. Compared to the resting cells, LPS-treated macrophages contained more mitochondria with predominant punctate morphology (Fig. 1C and D and S3A). Although pro-inflammatory differentiated macrophages possess enlarged cell shape and increased mitochondria, their cell size did not always correlate with mitochondrial mass, particularly in primary cultured cells (Fig. S3B). Consistently, ultrastructural analysis indicated that mitochondria in LPS-treated macrophages had greater mitochondrial mass, shorter length, and looser cristae than those in resting cells (Fig. 1E and F and S3C). Moreover, the higher mtDNA/nDNA ratio confirmed that LPS-activated macrophages have increased mtDNA copy number compared to resting cells (Fig. 1G).
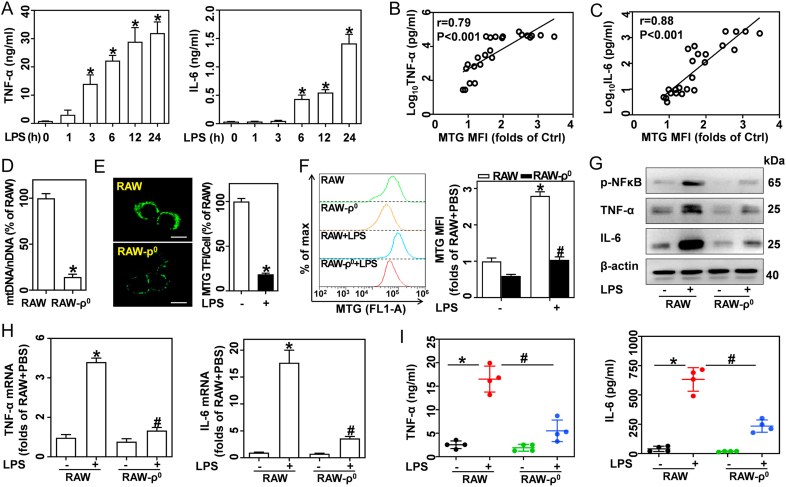
Fig. 2.
Increased mitochondrial mass contributes to inflammatory cytokine release in LPS-activated macrophages. (A–C) Cells were treated with 0.5 μg/mL LPS for indicated time points. (A) TNF-α and IL-6 in BMDM supernatants measured by ELISA, n = 4. The correlations between MTG MFI and log10TNF-α (B) or log10IL-6 (C) were evaluated by Spearman analysis, p < 0.001, n = 24. (D–I) RAW-ρ0 and wildtype RAW cells (RAW) were treated with or without 0.5 μg/mL LPS for 12 h. (D) The mtDNA/nDNA ratio was measured by qRT-PCR, n = 3. (E) MTG-stained cells captured by LSCM; scale bars, 5 μm. MTG TFI per cell was analyzed, n = 20 cells. (F) MTG-stained cells analyzed by flow cytometry, and calculated MTG MFIs, n = 3. (G) Western blotting of p-NF-κB, TNF-α, and IL-6. (H) TNF-α and IL-6 mRNA as analyzed by qRT-PCR, n = 3. (I) TNF-α and IL-6 in supernatants measured by ELISA, n = 4. Data are expressed as mean ± SEM, *p < 0.05 vs. Ctrl (A); *p < 0.05 vs. RAW (D-F, H, I), #p < 0.05 vs. RAW + LPS (F, H, I).
Fig. 1.
Pro-inflammatory differentiated macrophages show higher mitochondrial mass and lower Δψm than resting macrophages. RAW, BV-2, THP-1, PMs, and BMDMs were stimulated with 0.5 μg/mL LPS for 12 h. (A) MTG-stained cells were detected by flow cytometry. (B) Calculated MTG MFI, n = 3. (C) MTG-stained RAW, THP-1 and BMDMs were tested by LSCM; scale bars, 5 μm. (D) MTG total fluorescence intensity (TFI) per cell were analyzed, n = 20 cells. (E) Ultrastructural images of mitochondria in BMDMs captured using TEM; scale bars, 500 nm. (F) Calculated mitochondria per cell (left, n = 20 cells) and mitochondrial length (right, 165 mitochondria from 15 Ctrl cells; 203 mitochondria from 8 LPS-treated cells). (G) qRT-PCR measured mtDNA copy number in RAW and BMDMs, and expressed as mtDNA/nDNA ratio, n = 3. (H) Flow cytometry determined Rho-123 MFI/MTG MFI ratio in BMDMs treated with LPS, n = 3. (I) Rho-123 stained BMDMs captured using LSCM; scale bars, 5 μm. Rho-123 brightness was measured, n = 20 cells. (J) BMDMs co-stained with MTG and TMRM analyzed by flow cytometry, and TMRM MFI/MTG MFI ratio, n = 3. (K) JC-1 stained BMDMs analyzed by flow cytometry, and JC-1 red/green ratio, n = 3. Data are expressed as mean ± SEM, *p < 0.05, vs. Ctrl (control cells).
Given the observation that LPS-activated macrophages possess more mitochondria, we proposed that the amplification of Rho-123 MFI does not necessarily represent higher Δψm. Notably, LPS diminished the ratio of Rho-123 MFI and MTG MFI, suggesting that Rho-123 fluorescence in individual mitochondria was reduced (Figure S1B, S2D and 1H). Importantly, laser confocal analysis showed that LPS boosted Δψm loss, indicated by weakened Rho-123 brightness (Fig. 1I). Tetramethylrhodamine methyl ester (TMRM) is a cell-permeant cationic lipophilic red fluorescent probe for Δψm. Although the TMRM MFI was increased in LPS-challenged macrophages, the ratio of TMRM MFI and MTG MFI was decreased (Fig. 1J). Taken together these results indicate that pro-inflammatory differentiated macrophages experience membrane depolarization which differs from the results of previous studies that reported hyperpolarization [22]. JC-1, another probe for Δψm, can form red fluorescent aggregates in normal mitochondria, and is transformed to green fluorescent monomers in depolarized mitochondria. The ratio of JC-1 red/green is determined only be Δψm. Here, we found that LPS decreased the ratio of JC-1 red/green in BMDMs, indicating the loss of Δψm (Fig. 1K). Therefore, we demonstrated that pro-inflammatory differentiated macrophages possess more mitochondria. However, these mitochondria undergo profound remodeling, including fragmented morphology, loose cristae, and reduced Δψm.
3.2. Increased mitochondrial mass contributes to inflammatory cytokine release in LPS-activated macrophages
Next, we investigated whether more mitochondrial mass contributes to the inflammatory cytokine production in LPS-activated macrophages. BMDMs were stimulated with 0.5 μg/mL LPS for 0, 1, 3, 6, 12, and 24 h. We found that LPS robustly boosted the expression (Fig. S3D) and production (Fig. 2A) of pro-inflammatory cytokines, including TNF-α and IL-6. Spearman correlation analysis showed that both TNF-α (Fig. 2B) and IL-6 (Fig. 2C) in supernatants positively correlated with mitochondrial mass in BMDMs. To further investigate the role of mitochondria mass in LPS-induced inflammatory response, we used long-term exposure to EtBr to develop RAW-ρ0 cells that lack mtDNA (Fig. 2D) as previous reports revealed that deletion of mtDNA with EtBr also reduced mitochondrial biogenesis and mitochondrial mass [23,24]. Ultrastructural analysis indicated that the mitochondria content in RAW-ρ0 cells was significantly lower than that in resting cells (Fig. S3E). Moreover, RAW-ρ0 and RAW cells were stained with MTG, and detected by LSCM (Fig. 2E) and flow cytometry (Fig. 2F). Our results show that RAW-ρ0 cells have lower mitochondrial mass than wildtype RAW cells in the absence and presence of LPS. Compared with wildtype RAW, RAW-ρ0 also exhibit decreased cell size (FSC signal) in response to LPS (Fig. S3F). Importantly, LPS-challenged RAW-ρ0 showed significantly lower TNF-α and IL-6 expression (Fig. 2G and H) and production (Fig. 2I) compared to wildtype RAW. These results indicate that increased mitochondrial mass contributes to the pro-inflammatory cytokines release in LPS-activated macrophages.
3.3. Knockdown or inhibition of Drp1 blunts LPS-induced increase of mitochondrial mass and pro-inflammatory cytokines
To elucidate the molecular mechanism of LPS-driven mitochondrial mass increase, gene expression profiles in PBS- and LPS-treated macrophages were analyzed. RNA-seq analysis results revealed that LPS caused dynamic changes in the gene transcriptome. Using a |log2-fold change|>1 and p < 0.05 as thresholds for inclusion, a total of 3919 differentially expressed genes (DEGs, 1882 up-regulated and 2037 down-regulated) were screened between the two groups. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were then conducted to analyze the DEG biological pathways and functions. Taking a false discovery rate (FDR) < 0.01 and p < 0.05 as thresholds for inclusion, a total of 1331 significant GO terms were enriched, and the top 20 terms are listed in Fig. S3G. These GO terms were associated with regulating the immune system, organelle, and metabolic processes. The KEGG enrichment analysis also revealed that 50 pathways were significantly enriched (FDR < 0.01 and p value < 0.05). As shown in Fig. S3H, the top 20 pathways were associated with the TNF, NFκB, Toll-like receptor, and JAK-STAT signaling pathways.
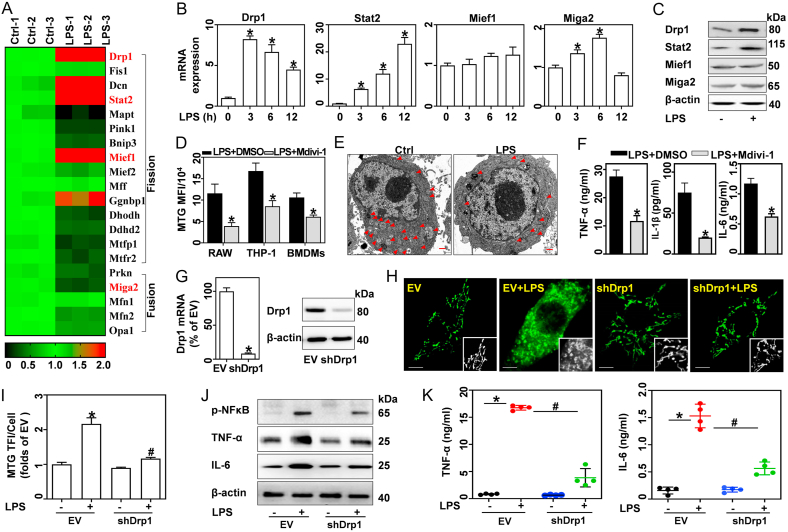
Notably, 28 GO terms (p < 0.05) were enriched in regulating mitochondria. In general, mitochondrial mass is determined by the balance of fission and fusion. Here, GO enrichment analysis indicated that 20 genes may modulate mitochondrial fission and fusion (Supplementary Table 3 and Fig. 3A). LPS caused a > 2-fold increase in positive regulators of mitochondrial fission, including, Drp1, Stat2, Mief1, and Decorin (Dcn, excluded for fragments per kilobase million (fpkm) < 1). RT-PCR and western blotting confirmed that LPS significantly enhanced the expression of Drp1 and Stat2, however, had no effect on Mief1 (Fig. 3B and C). For positive regulators of mitochondrial fusion, mitoguardin 2 (Miga2) and parkin RBR E3 ubiquitin protein ligase (Prkn, excluded for fpkm<1) were decreased by > 2-fold by LPS. However, RT-PCR and western blotting showed that LPS caused a slight increase in Miga2 mRNA and protein expression (Fig. 3B and C). We, therefore, further examined the role of Stat2 and Drp1 in modulating mitochondrial homeostasis.
Fig. 3.
Knockdown or inhibition of Drp1 blunts LPS-induced increase in mitochondrial mass and pro-inflammatory cytokine expression. (A–C) BMDMs were treated with 0.5 μg/mL LPS for 12 h or the indicated times. (A) RNA-seq analysis of gene expression profile in Ctrl and LPS-treated BMDMs. (B) qRT-PCR analysis of Drp1, Stat2, Mief1 and Miga2 mRNA expression, n = 3. (C) Proteins of Drp1, Stat2, Mief1, and Miga2 analyzed via western blotting. (D–F) BMDMs, RAW, and THP-1 cells were stimulated with LPS in the presence of DMSO or 50 μM Mdivi-1 for 12 h. (D) MTG-stained cells analyzed by flow cytometry, and calculated MFIs, n = 3. (E) Mitochondrial morphologies in BMDMs detected by TEM; scale bars, 500 nm. (F) TNF-α, IL-6, and IL-1β in supernatants tested by ELISA, n = 4. (G–K) BMDMs transfected with EV and shDrp1 were treated with or without LPS. (G) Drp1 mRNA and protein expressions detected by qRT-PCR and western blotting. (H) Confocal images of MTG-stained cells captured using LSCM, scale bars, 5 μm. (I) MTG TFI per cell, n = 20 cells. (J) p-NFκB, TNF-α, and IL-6 analyzed by western blotting. (K) TNF-α and IL-6 in supernatants analyzed by ELISA, n = 4. Data are expressed as mean ± SEM, *p < 0.05 vs. Ctrl (B); *p < 0.05 vs. LPS + DMSO (D, F); *p < 0.05 vs. EV (G, I, J), #p < 0.05 vs. EV + LPS (I, J).
As a key dynamin-related GTPase, Drp1 has a significant role in regulating mitochondrial fission [12]. We therefore first investigated the role of Drp1 in LPS-induced mitochondrial remodeling and inflammatory responses with a chemical inhibitor, Mdivi-1. Flow cytometry results showed that Mdivi-1 blunted the LPS-induced increase in mitochondrial mass (indicated by MTG MFI, Figs. S4A and 3D) and cell size (indicated by FSC, Fig. S4B) in BMDMs, RAW, and THP-1 cells. Similarly, ultrastructural analysis revealed that Mdivi-1 inhibits the LPS-induced mitochondrial division into fragments in BMDMs (Figs. 3E and S4C). Mdivi-1 also inhibited the LPS-induced production of TNF-α, IL-6, and IL-1β (Fig. 3F). Further, expression of lentiviral shRNA Drp1 (shDrp1) in BMDMs (Fig. 3G) attenuated LPS-induced increases in mitochondrial mass (Fig. 3H and I), as well as pro-inflammatory cytokine expression (Fig. 3J), and production (Fig. 3K). These findings suggest that Drp1-dependent mitochondrial fission contributes to LPS-induced mitochondrial mass increase and pro-inflammatory cytokine production.
3.4. LPS promotes mitochondrial biogenesis in a Drp1-and Stat2-dependent manner
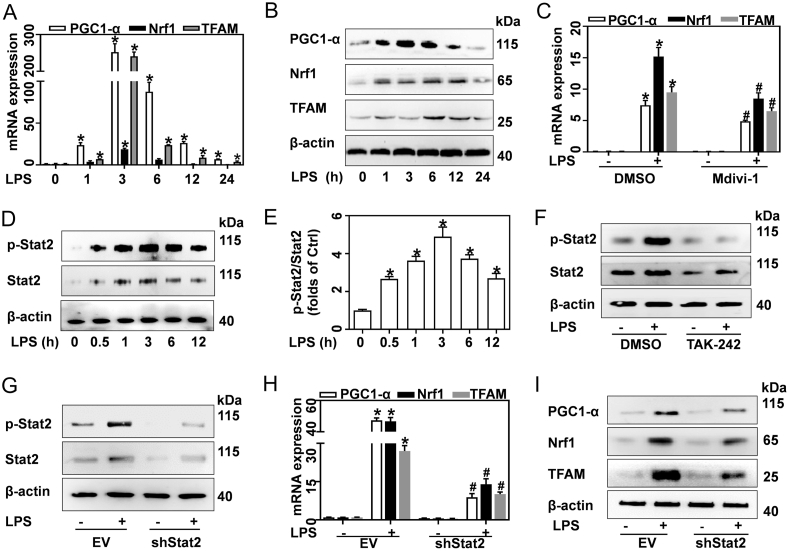
Increases in mitochondrial mass require abundant proteins encoded by mitochondrial and nuclear genomes. Mitochondrial biogenesis is the key mechanism responsible for the synthesis of these proteins, which is controlled by several transcriptional factors, including peroxisome proliferator activator receptor gamma-coactivator 1α (PGC-1α), nuclear respiratory factor 1 (Nrf1), and mitochondrial transcription factor A (TFAM). Hence, we next sought to determine whether mitochondrial biogenesis is enhanced in LPS-activated macrophages. In response to LPS, mRNA and protein expressions of PGC-1α, Nrf1, and TFAM rapidly increased within 6 h and gradually returned to normal levels at 24 h (Fig. 4A and B). Inhibition of Drp1 with Mdivi-1 abrogated the LPS-induced mitochondrial biogenesis (Fig. 4C).
Fig. 4.
LPS promotes mitochondrial biogenesis in a Drp1-and Stat2-dependent manner. (A–F) BMDMs were treated with 0.5 μg/mL LPS for the indicated times, or for 12 h, in the presence of DMSO, 50 μM Mdivi-1, or 100 nM TAK-242. (A) mRNA and (B) protein of PGC-1α, Nrf1, and TFAM in BMDMs treated with LPS were quantified by qRT-PCR and western blotting, respectively, n = 3. (C) PGC1A Nrf1, and TFAM mRNA in BMDMs treated with LPS and Mdivi-1 measured by qRT-PCR, n = 3. (D) Western blotting analysis of Stat2 and p-Stat2 proteins in BMDMs treated with LPS. (E) Ratio of p-Stat2/Stat2, n = 3. (F) Western blotting analysis of Stat2 and p-Stat2 proteins in BMDMs treated with LPS and TAK-242. (G–I) EV and shDrp1 transfected BMDMs were stimulated with 0.5 μg/mL LPS for 12 h. (G) Western blotting analysis of Stat2 and p-Stat2. (H) mRNA and (I) protein of PGC-1α, Nrf1 and TFAM tested by qRT-PCR and western blotting, respectively, n = 3. Data are expressed as mean ± SEM, *p < 0.05 vs. Ctrl (A, E); *p < 0.05 vs. DMSO + PBS, #p < 0.05 vs. DMSO + LPS (C); *p < 0.05 vs. EV, #p < 0.05 vs. EV + LPS (H).
As a key transcription factor, Stat2 plays an important role in regulating immune responses. In response to LPS, Stat2 phosphorylation increased in 30 min and was sustained at high levels until 12 h (Fig. 4D and E). Many reports suggest that the LPS-induced inflammatory response is a multi-step process initiated by Toll-like receptor 4 (TLR4) activation [25]. Here, we demonstrated that inhibition of TLR4 with TAK-242 abrogated LPS-induced Stat2 phosphorylation (Fig. 4F). RNA-seq analysis further suggested that Stat2 may modulate the biosynthesis of macromolecules (Supplementary Table 4). Meanwhile, knockdown of Stat2 by shRNA Stat2 (shStat2) in BMDMs (Fig. 4G) blunted LPS-induced increase of PGC-1α, Nrf1, and TFAM expression at both the mRNA and protein levels (Fig. 4H and I). Therefore, our results suggest that LPS boosts mitochondrial biogenesis in both a Stat2-and Drp1-dependent manner. Hence, enforcing biogenesis may contribute to mitochondrial mass increase by regulating cellular macromolecule biosynthesis.
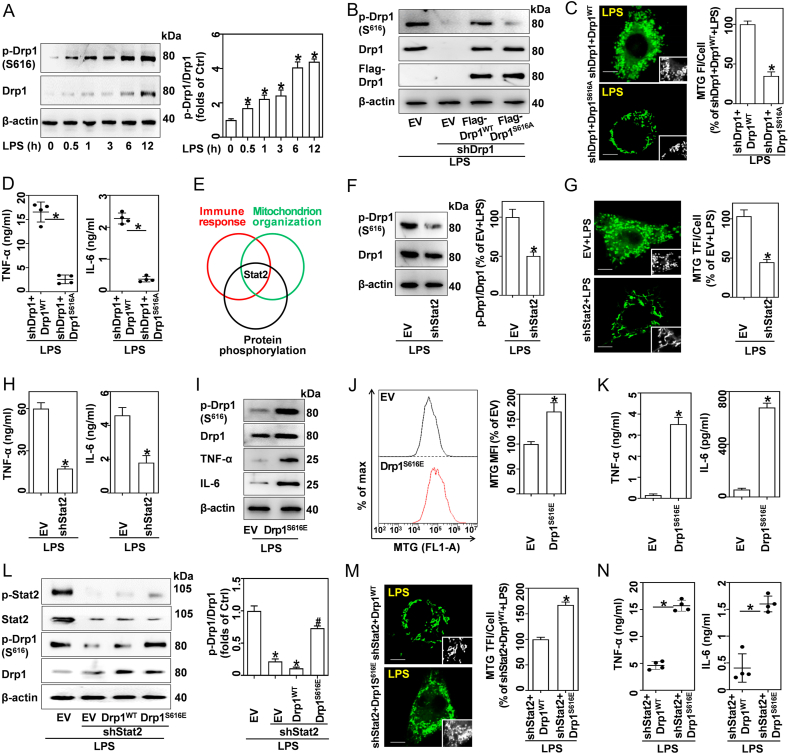
3.5. Stat2 phosphorylation of Drp1 at serine 616 is required for LPS-induced accumulation of mitochondrial mass and pro-inflammatory cytokines
Drp1 phosphorylation at the S616 site promotes its recruitment to the mitochondria, which is required for subsequent fission [26]. In this study, we found that LPS induced Drp1 S616 phosphorylation in a dose- and time-dependent manner in BMDMs (Figs. S4D and 5A). Immunofluorescence results showed that LPS induced Drp1 recruitment to the mitochondria, as indicated by increased colocalization of Drp1 and MitoTracker-Red (Fig. S4E). To further elucidate the role of p-Drp1(S616) in LPS-treated macrophages, a non-phosphorylatable mutant Drp1 (Flag-Drp1S616A), and wildtype Drp1 (Flag-Drp1WT) overexpressing vectors were designed. Here, Flag-labeled Drp1 represented exogenous Drp1. Re-expressed Drp1WT in RAW cells with stable knockdown of Drp1 restored both Drp1 and p-Drp1S616, while re-expressed Drp1S616A abrogated LPS-induced Drp1 phosphorylation at S616 (Figs. 5B and S4F). Furthermore, non-phosphorylatable mutation of Drp1 at S616 blunted the LPS-induced increase of mitochondria mass (Fig. 5C) and pro-inflammatory cytokines (Fig. 5D). Interestingly, RNA-seq analysis suggested that Stat2 was simultaneously enriched in pathways associated with regulating immune response, mitochondrion organization, and protein phosphorylation (Fig. 5E). Meanwhile, knockdown of Stat2 remarkably reversed LPS-mediated Drp1 S616 phosphorylation (Fig. 5F) and mitochondrial mass increase (Fig. 5G). In response to LPS, shStat2-transfected BMDMs exhibited decreased pro-inflammatory cytokines expression (Fig. S4G) and production (Fig. 5H) compared to the corresponding vehicle control.
Fig. 5.
Stat2 phosphorylation of Drp1 at S616 is required for LPS-induced accumulation of mitochondrial mass and inflammatory cytokines. (A) Western blotting analysis of Drp1 and p-Drp1 (S616) in BMDMs stimulated with 0.5 μg/mL LPS for indicated times, and quantified p-Drp1:Drp1 ratios, n = 3. (B–D) RAW cells were engineered to express shDrp1 and rescued with either EV, Flag-Drp1WT or Flag-Drp1S616A, prior to treatment with 0.5 μg/mL LPS for 12 h. Drp1WT, vectors expressing wildtype Drp1; Drp1S616A, vectors encoding a non-phosphorylatable mutant Drp1 at S616. (B) Western blotting analysis of p-Drp1, Drp1, and Flag-Drp1. (C) LSCM analysis of MTG-stained cells; scale bars, 5 μm. MTG TFI per cell was analyzed, n = 20 cells. (D) TNF-α and IL-6 in supernatants were quantified via ELISA, n = 4. (E) GO analysis suggests that Stat2 was enriched in pathways associated with regulation of immune response, mitochondrion organization, and protein phosphorylation. (F–H) BMDMs expressed EV and shStat2 and were treated with 0.5 μg/mL LPS for 12 h. (F) Drp1 and p-Drp1(S616) were analyzed using western blotting, and quantified p-Drp1:Drp1 ratios, n = 3. (G) LSCM analysis of MTG-stained cells; scale bars, 5 μm, and MTG TFI per cell, n = 20 cells. (H) TNF-α and IL-6 in supernatants analyzed by ELISA, n = 4. (I–K) RAW cells expressed EV and Drp1S616E. (I) Western blotting analysis of Drp1, p-Drp1, TNF-α and IL-6. (J) MTG-stained cells analyzed by flow cytometry, and calculated MFIs, n = 3. (K) TNF-α and IL-6 in supernatants analyzed by ELISA, n = 4. (L–N) RAW cells were engineered to encode shStat2 and overexpressed EV, Drp1WT and Drp1S616E (a phosphomimetic mutant), and were then treated with 0.5 μg/mL LPS for 12 h. (L) Western blotting analysis of Stat2, p-Stat2, Drp1 and p-Drp1, and quantified p-Drp1:Drp1 ratios, n = 3. (M) LSCM analysis of MTG-stained cells; scale bars, 5 μm, and MTG TFI per cell, n = 20 cells. (N) TNF-α and IL-6 in supernatants analyzed by ELISA, n = 4. Data are expressed as mean ± SEM, *p < 0.05 vs. Ctrl (A); *p < 0.05 vs. shDrp1+Drp1WT + LPS (C, D); *p < 0.05 vs. EV + LPS (F–H, L); *p < 0.05 vs. EV (J, K); #p < 0.05 vs shStat2+Drp1WT + LPS (L); *p < 0.05 vs. shStat2+Drp1WT + LPS (M, N).
Moreover, we developed a vector that encodes a constitutively active phosphomimetic mutant of Drp1, Drp1S616E (Fig. 5I). Interestingly, RAW cells overexpressing Drp1S616E showed higher mitochondrial mass (5J) and proinflammatory cytokine levels (Fig. 5I and K) compared to RAW cells transfected with EV. Moreover, RAW cells with Stat2 knockdown were overexpressed with either Drp1WT or Drp1S616E. As shown in Fig. 5L, transfection with Drp1WT failed to increase Drp1 S616 phosphorylation in shStat2-transfected BMDMs, while overexpression of Drp1S616E induced increased phosphorylation. In response to LPS, shStat2+Drp1S616E cells exhibited an apparent increase in mitochondrial fragmentation and inflammatory cytokines, while shStat2+Drp1WT cells blunted these processes (Fig. 5M and N). Moreover, shStat2+Drp1S616E cells displayed slightly higher expression of PGC-1α, Nrf1, and TFAM compared to shStat2+Drp1WT cells (Fig. S4H). Therefore, our findings demonstrate that Stat2 phosphorylation of Drp1 at serine 616 is crucial for LPS-induced increase of mitochondrial mass and pro-inflammatory cytokines.
3.6. The Stat2-Drp1 axis induces mitochondria in LPS-activated macrophages to generate ROS rather than ATP which drives inflammation
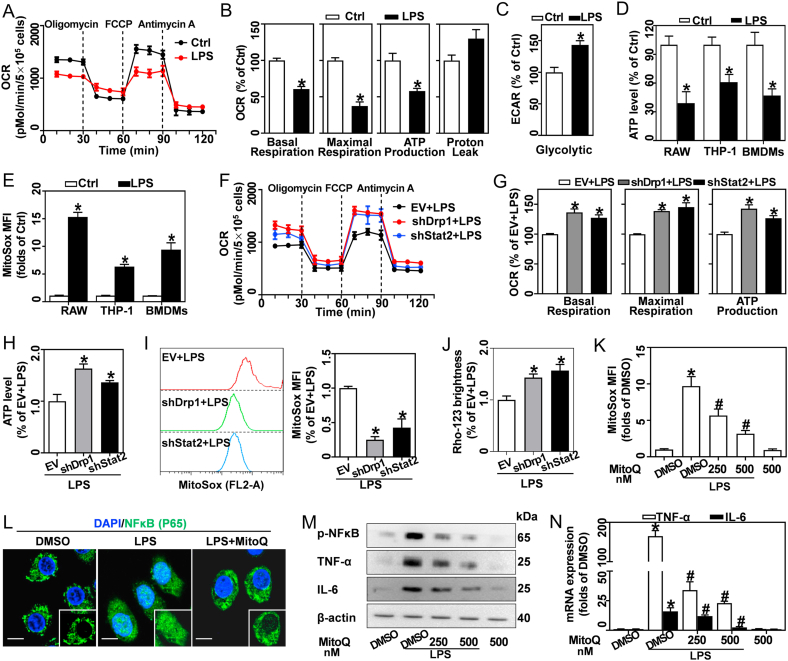
Pro-inflammatory differentiated macrophages undergo metabolic programming from oxidative phosphorylation to aerobic glycolysis [27]. Here, we evaluated the OCR and extracellular acidification rate (ECAR) by Seahorse assay. In response to LPS, basal respiration, maximal respiration, and ATP production were remarkably reduced in BMDMs (Fig. 6A and B). In contrast, LPS-activated BMDMs showed higher glycolytic activity than resting macrophages (Figs. S5A and 6C). ATP synthesis efficiency via glycolysis is much lower than mitochondrial oxidative phosphorylation; and thus, ATP content was decreased in LPS-treated RAW, THP-1, and BMDMs (Fig. 6D). In contrast, we found that LPS-induced a 7–15 folds increase in mitochondrial ROS, as indicated by MitoSOX (Figs. S5B and 6E). Moreover, mitochondrial ROS was increased in parallel with the mitochondrial mass in LPS-activated macrophages (Figs. S5C and S5D). Meanwhile, RAW-ρ0 cells lacking mitochondrial DNA displayed lower ROS generation compared to normal RAW cells (Fig. S5E). Cumulatively, these results indicate that mitochondrial fragmentation drives ROS generation rather than ATP production.
Fig. 6.
Stat2-Drp1 induces mitochondria in LPS-activated macrophages to generate ROS rather than ATP, which then drives inflammation. (A–E) RAW, THP-1, and BMDMs were treated with 0.5 μg/mL LPS for 12 h. (A) Oxygen consumption rate (OCR) in BMDMs tested by Seahorse assay. (B) Quantitative levels of basal respiration, maximal respiration, ATP production, and proton leakage, n = 3. (C) Glycolysis in BMDMs determined from extracellular acidification rate (ECAR) by Seahorse assay, n = 3. (D) ATP levels in LPS-treated RAW, THP-1 and BMDMs measured using a specific ATP Determination kit, n = 3. (E) Mitochondrial ROS in LPS-treated RAW, THP-1, and BMDMs detected using MitoSOX by flow cytometry, n = 3. (F–J) BMDMs expressing EV, shDrp1 or shStat2 were stimulated with 0.5 μg/mL LPS for 12 h. (F) OCR evaluated by Seahorse assay. (G) Basal respiration, maximal respiration, and ATP production quantified, n = 3. (H) ATP levels measured, n = 3. (I) MitoSOX stained cells tested by flow cytometry, and analysis of MFIs, n = 3. (J) LSCM analyzed Rho-123 brightness, n = 20 cells. (K–N) BMDMs stimulated with 0.5 μg/mL LPS for 12 h in the presence of DMSO or MitoQ (250 and 500 nM). (K) MitoSOX stained cells analyzed by flow cytometry, and calculated MFIs, n = 3. (L) NFκB expression (FITC-labeled, green), analyzed by immunofluorescence; DAPI (blue) stains nuclei. (M) Protein bands of p-NFκB, TNF-α, and IL-6 measured by western blotting. (N) TNF-α and IL6 mRNA analyzed by qRT-PCR. Data are expressed as mean ± SEM, *p < 0.05 vs. Ctrl (B–E); *p < 0.05 vs. EV + LPS (G–J); *p < 0.05 vs. DMSO + PBS, #p < 0.05 vs. DMSO + LPS (K, N). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, we investigated whether the Stat2-Drp1 pathway is associated with mitochondrial functional switching of LPS-treated macrophages. Interestingly, knockdown of Drp1 and Stat2 restored LPS-induced reduction of OCR parameters, including basal respiration, maximal respiration, and ATP production (Fig. 6F and G). Meanwhile, the LPS-induced ATP decrease (Fig. 6H), ROS increase (Fig. 6I) and Δψm loss (Figs. S5F and 6J) were reversed by shDrp1 and shStat2.
Previous studies demonstrated that ROS may act as a mitochondrial redox signal to drive inflammation [28,29]. Hence, MitoQ, a mitochondria-targeted ROS scavenger, was used to limit LPS-induced ROS production (Fig. 6K). As a key transcription factor of pro-inflammatory cytokines, NF-κB phosphorylation and nuclear recruitment are required for its activation. Here we demonstrated that LPS promoted NF-κB phosphorylation and nuclear translocation in BMDMs, while MitoQ administration blunted these processes (Fig. 6L and M). Furthermore, inhibition of mitochondrial ROS production with MitoQ abrogated LPS-induced TNF-α and IL-6 expression (Fig. 6M and N).
Together, these findings suggest that Stat2-Drp1 signaling activation induces the mitochondrial functional shift from ATP synthesis to ROS generation. The ROS emanating from the mitochondria might then play a key role in regulating inflammatory cytokine generation.
3.7. Inhibition of Drp1 reduces mitochondrial mass and inflammatory cytokines in peritoneal macrophages derived from LPS-challenged mice
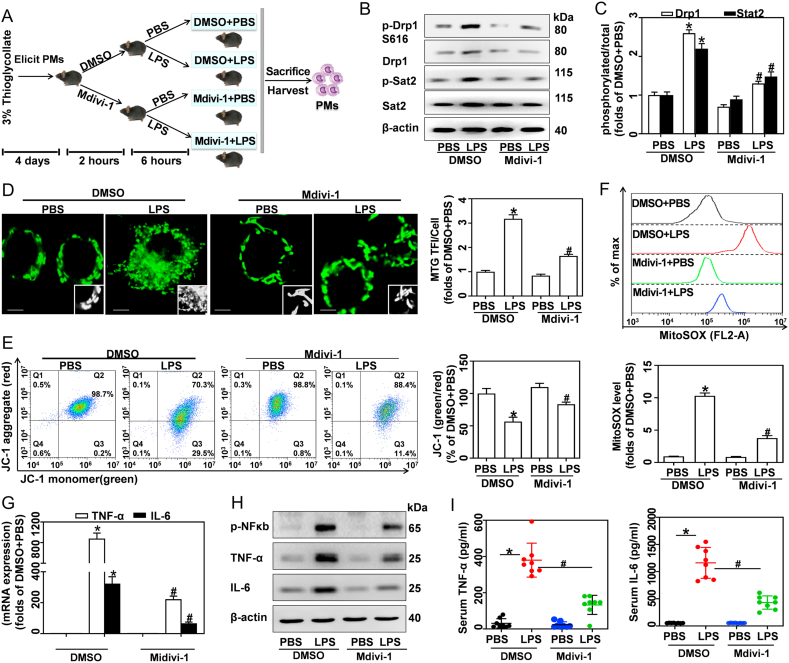
Lastly, we assessed the role of Drp1 in LPS-induced accumulation of mitochondrial mass and inflammatory cytokines in vivo (Fig. 7A). Here, peritoneal macrophages (PMs) isolated from DMSO + LPS mice exhibited higher Drp1 and Stat2 phosphorylation compared to control mice, while Mdivi-1 treatment reduced Drp1 expression and phosphorylation (Fig. 7B and C). As expected, MTG staining results showed that LPS promoted increases in mitochondria mass in PMs, while Mdivi-1 inhibited this effect (Fig. 7D). LPS-challenged mice showed reduced JC-1 aggregates and increased JC-1 monomers in their PM cells, indicating Δψm loss. Mdivi-1 treatment restored LPS-induced Δψm depolarization (Fig. 7E). Inhibition of Drp1 with Mdivi-1 blunted LPS-induced mitochondrial ROS production (Fig. 7F), NFκB phosphorylation and pro-inflammatory cytokine expression in PMs (Fig. 7G and H). Furthermore, DMSO + LPS mice exhibited higher serum TNF-α and IL-6 levels than DMSO + PBS mice, indicating that LPS causes obvious inflammation in vivo. Promisingly, Mdivi-1 treatment alleviated the inflammation in LPS-challenged mice (Fig. 7I). These results clearly demonstrate that the Drp1-dependent mitochondrial mass increase in macrophages is required for the pro-inflammatory effects of LPS in vivo.
Fig. 7.
Inhibition of Drp1 reduces mitochondrial mass and inflammatory cytokines in PMs derived from LPS-challenged mice. (A) 32 C57BL/6 mice were given 3% thioglycolate (i.p.) for 4 days to elicit macrophages. Animals were then injected (i.p.) with DMSO or Mdivi-1 (20 mg/kg) for 2 h, followed by PBS or LPS (15 mg/kg) for 6 h. PMs were isolated after mice were sacrificed. (B) Drp1, p-Drp1, p-Stat2 and Stat2 proteins in PMs analyzed by western blotting. (C) Quantification of p-Drp1:Drp1 and p-Stat2:Stat2 ratios. (D) MTG in PMs measured by LSCM; scale bars, 3 μm. Assessment of MTG TFI per cell, n = 20 cells. (E) JC-1 stained PMs analyzed by flow cytometry, and calculation of the JC-1 red:green ratio, n = 8. (F) MitoSOX stained PMs analyzed using flow cytometry, with calculated MFIs, n = 8. (G) TNF-α and IL6 mRNA in PMs analyzed by qRT-PCR, n = 3. (H) p-NFκB, TNF-α and IL-6 protein expressions analyzed by western blotting. (I) TNF-α and IL-6 in serum measured by ELISA, n = 8. Data are expressed as mean ± SEM, *p < 0.05 vs. DMSO + PBS, #p < 0.05 vs. DMSO + LPS. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Emerging evidence has revealed that mitochondria may act as a key intracellular signaling platform for regulating immune responses [30]. In this study, we demonstrated that pro-inflammatory differentiated macrophages possess greater mitochondrial mass than resting cells. Consistent with previous reports, we found that LPS induces significant macrophage spreading and enlargement. In contrast, restriction of macrophage spreading by spatial confinement blunts the LPS-induced inflammatory response [31]. Our findings also revealed that the mitochondrial mass increase correlates with cell size enlargement in LPS-treated macrophages. Meanwhile, deletion of mtDNA and inhibition of Drp1 effectively reduced mitochondrial mass and cell size in LPS-treated macrophages. Moreover, previous studies have shown that mitochondrial mass may be critical for the control of cell fate and immune responses; while higher mitochondrial mass contributes to T cell senescence and cancer cell chemo-resistance [32,33]. Increased mitochondrial mass in platelets, specifically, drives platelet hyperreactivity and inflammation in aged mice [34]. Also, PGC-1α deficient mice show lower mitochondrial mass, which causes spontaneous kidney inflammation and injury [35]. Here, we found that mitochondrial mass in LPS-activated macrophages was positively correlated with pro-inflammatory cytokine production. Meanwhile, reduced mitochondrial mass blunted the inflammatory response in LPS-activated macrophages. Importantly, an increase in mitochondrial mass with Drp1S616E boosted macrophage pro-inflammatory differentiation, although without LPS stimulation. Thus, it is possible that increased mitochondrial mass may serve as one of the most important events for macrophage elongation and pro-inflammatory differentiation.
Furthermore, our findings reveal that new mitochondria undergo profound reorganization, including fragmented morphology, loose cristae, and reduced Δψm. Δψm is generated by proton pumping complexes of the respiratory chain, which are critical for modulating mitochondrial homeostasis [36]. In fact, Mills et al. reported that LPS induces mitochondrial hyperpolarization, as indicated by higher TMRM analyzed by flow cytometry [22]. However, Rho-123 and TMRM signals in flow cytometry analysis are affected by mitochondrial mass and size, which may cause spurious results. In contrast, the ratio of JC-1 red/green is determined exclusively by Δψm, rather than by other factors such as mitochondrial mass, size, and density [37]. We therefore, proposed that it may be prudent to simultaneously detect Δψm with Rho-123 and TMRM using flow cytometry, particularly in cases involving increased mitochondrial mass. Herein, our work reveals that pro-inflammatory differentiated macrophages possess increased mitochondrial mass, however their Δψm is reduced.
Mitochondrial mass is determined by the balance between fission and fusion. Increased fusion promotes adjacent mitochondrial integration, while enforcing fission facilitates mitochondria fragmentation. Fission is controlled by several dynamin family GTPases, especially Drp1 [38]. Recently, many reports have emphasized the key role of Drp1 in various physiological and pathological processes [39]. Here, RNA-seq analysis showed that LPS significantly increased Drp1 expression, while slightly decreasing Fis1. Furthermore, abrogation of Drp1 remarkably attenuates LPS-induced mitochondrial mass increase and macrophage M1 differentiation in vivo and in vitro. It has been reported that Drp1 interaction with Fis1 is required for mitochondrial fission. Hence, selective inhibition of the Drp1/Fis1 interaction with P110 abrogates pathologic mitochondrial fragmentation [14]. However, emerging evidence indicates that Fis1 may be dispensable in mitochondrial recruitment of Drp1, and subsequent mitochondrial fission [13,40]. Moreover, mitochondrial growth requires synthesis of ETC proteins, which is modulated by mitochondrial biogenesis [41]. However, the impact of mitochondrial biogenesis on regulating the inflammatory response continues to be a controversial matter. For instance, previous reports show that increased mitochondrial biogenesis contributes to inflammation resolution [42]. Moreover, survival of critical illnesses may be associated with activation of mitochondrial biogenesis by enabling ATP demands [43]. Conversely, we speculate that early biogenesis activation contributes to mitochondrial mass increase and pro-inflammatory differentiation of macrophages.
Similar to the results reported in previous studies [31,44], our RNA-seq data revealed that LPS induced dramatic changes in the gene expression profile, and these DEGs were determined to be involved in multiple functions including, regulating the inflammatory response, cellular organelles, NF-κB pathway, and JAK-STAT pathway, etc. As a key signal transducer and transcriptional activator, Stat2 is indispensable for type I interferon (IFN)-induced antiviral and anti-tumor responses [45]. Although IFNs are the only cytokines known to directly activate Stat2, evidence also appears to support TLR-driven Stat2 signaling pathway. Stat2 −/− mice are defective in regulating TLR-dependent dendritic cell activation and CD8+ T cell proliferation [46]. Interestingly, we found that LPS boosted Stat2 expression and phosphorylation in a TLR4-dependent manner. However, the precise role of Stat2 in regulating inflammation remains unclear. For instance, Stefania et al. have reported that Stat2 deficiency does not affect the production of TNF-α and IL-6 in response to LPS [46]. Meanwhile, Radha et al. demonstrated that influenza-infected Stat2−/− mice show higher viral burden, inflammation, and morbidity than wildtype mice [47]. Meanwhile, George et al. reported that high expression of Stat2 is sufficient for inducing IL-6 expression [48]; and Alazawi et al. found that Stat2−/− mice show lower inflammatory cytokine production in response to LPS [49]. Similarly, our current findings indicate that Stat2 facilitates the inflammatory response of LPS-activated macrophages.
Recently, many studies have underscored the prominent role of Stat2 in regulation of mitochondrial homeostasis [50,51]. Moreover, Stat2 has been identified within mitochondria and has been implicated as an orchestrator of mitochondrial function and energy production [50]. In this study, we uncovered the mechanism involved in Stat2-mediated mitochondrial mass increase. Specifically, Stat2 promotes Drp1 phosphorylation at S616, which is necessary for fission by impelling Drp1 mitochondrial translocation [12]. Previously, extracellular regulated protein kinases and protein kinase A were found to activate Drp1 S616 phosphorylation [26]. In agreement with our results, Rojeen et al. report that skin fibroblasts from Stat2 mutation patients show decreased Drp1 S616 phosphorylation and mitochondrial fission [51]. However, little is known about whether Stat2 directly phosphorylates Drp1 at serine 616. Meanwhile, here we demonstrated that Stat2 favors LPS-induced mitochondrial mass increase via transcriptional activation of mitochondrial biogenesis. Similarly, previous reports have emphasized that Stat1 and Stat3 may modulate mitochondrial biogenesis and respiration, however the precise role of Stat2 is not well explored [50]. Thus, our work reveals that Stat2 is necessary for LPS-induced mitochondrial mass increase through modulation of mitochondrial fission and biogenesis.
The function of mitochondria may be determined by its mass, size, density, morphology, and distribution. Cells undergo mitochondrial remodeling and functional shift upon exposure to adverse stimuli [6]. The primary purpose of mitochondria in resting macrophages and memory T cells is ATP production through the oxidative phosphorylation pathway [52]. Alternatively, pro-inflammatory differentiated macrophages and activated effector T cells generate their ATP by nonmitochondrial sources, namely aerobic glycolysis [7]. However, there is a large discrepancy in ATP synthesis efficiency between oxidative phosphorylation (32–36 ATPs), and glycolysis (2 ATPs). Hence, we propose that increased mitochondrial mass does not necessarily equate to more ATP production. Moreover, loose cristae and reduced Δψm in mitochondria generally impair mitochondrial ETC efficiency, which may result in lower ATP synthesis and increased electron leakage, which serves as the major source of mitochondrial ROS [53]. Our work showed that pro-inflammatory differentiated macrophages repurpose their mitochondria to generate ROS rather than ATP in a Stat2-Drp1 dependent manner. Oxidative stress and inflammation are interconnected pathophysiological processes associated with various inflammatory diseases [54]. Pro-inflammatory differentiated macrophages release large amounts of ROS. In turn, ROS promotes pro-inflammatory cytokine production by regulating NF-κB, iNOS, and the NLPR3 inflammasome [9,10]. A recent report has also reported that IL-1β promotes mitochondria and ROS production in epithelial cells through an autocrine effect [55]. Thus, we demonstrated that ROS emanating from mitochondria modulate NF-κB-dependent cytokine production in pro-inflammatory differentiated macrophages.
Tissue-resident and monocyte-derived macrophage infiltration, and activation, are hallmarks of infection and sepsis. It is becoming increasingly clear that regulating pro-inflammatory macrophage differentiation into the anti-inflammatory phenotype, or suppressing the differentiation of macrophages back toward a pro-inflammatory phenotype, has an important impact on the progression and resolution of many diseases [3]. In this study, we demonstrated that pro-inflammatory differentiated macrophages possess more mitochondria by inducing Stat2-Drp1-dependent mitochondrial fission and biogenesis. Interestingly, these remodeled mitochondria displayed fragmented morphology, loose cristae, reduced Δψm, and metabolic programming, which boosted their functional shift from ATP synthesis to ROS production, and further drove transcription of NFκB-dependent inflammatory cytokines. Therefore, our study provides novel insights into the differentiation of pro-inflammatory macrophages, as well as new strategies for treatment of infectious diseases.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
The authors thank the Shanxi Provincial Key Lab of Free radical biology and medicine, Ministry of Education Key Lab of Hazard Assessment and Control in Special Operational Environment, School of Public Health, Fourth Military Medical University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101761.
Contributor Information
Wenli Li, Email: liwenli@fmmu.edu.cn.
Chunxu Hai, Email: cx-hai@fmmu.edu.cn.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 31800706, 21677176, 31900892, 31770915].
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sica A., Erreni M., Allavena P. Macrophage polarization in pathology. Cell. Mol. Life Sci. 2015;72(21):4111–4126. doi: 10.1007/s00018-015-1995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuter S., Gupta S.C., Chaturvedi M.M. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashir S., Sharma Y., Elahi A. Macrophage polarization: the link between inflammation and related diseases. Inflamm. Res. 2016;65(1):1–11. doi: 10.1007/s00011-015-0874-1. [DOI] [PubMed] [Google Scholar]
- 4.Folmes C.D.L., Dzeja P.P., Nelson T.J. Mitochondria in control of cell fate. Circ. Res. 2012;110(4):526–529. doi: 10.1161/RES.0b013e31824ae5c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin H.S., Suh H.W., Kim S.J. Mitochondrial control of innate immunity and inflammation. Immune Netw. 2017;17(2):77–88. doi: 10.4110/in.2017.17.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tannahill G.M., Curtis A.M., Adamik J. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong W.J., Yang H.H., Guan X.X. Inhibition of glycolysis alleviates lipopolysaccharide-induced acute lung injury in a mouse model. J. Cell. Physiol. 2019;234(4):4641–4654. doi: 10.1002/jcp.27261. [DOI] [PubMed] [Google Scholar]
- 9.Zhou R., Yazdi A.S., Menu P. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 10.Brune B., Dehne N., Grossmann N. Redox control of inflammation in macrophages. Antioxidants Redox Signal. 2013;19(6):595–637. doi: 10.1089/ars.2012.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni H.M., Williams J.A., Ding W.X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015;4:6–13. doi: 10.1016/j.redox.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalska B., Duszynski J., Szymanski J. [Mechanism of mitochondrial fission - structure and function of Drp1 protein] Postepy Biochem. 2016;62(2):127–137. [PubMed] [Google Scholar]
- 13.Otera H., Wang C., Cleland M.M. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haileselassie B., Mukherjee R., Joshi A.U. Drp1/Fis1 interaction mediates mitochondrial dysfunction in septic cardiomyopathy. J. Mol. Cell. Cardiol. 2019;130:160–169. doi: 10.1016/j.yjmcc.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J., Choi H., Min J.S. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 2013;127(2):221–232. doi: 10.1111/jnc.12361. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez A.S., Elguero M.E., Finocchietto P. Abnormal mitochondrial fusion-fission balance contributes to the progression of experimental sepsis. Free Radic. Res. 2014;48(7):769–783. doi: 10.3109/10715762.2014.906592. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Subramanian M., Yurdagul A.J. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell. 2017;171(2):331–345. doi: 10.1016/j.cell.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Windt G.J., O'Sullivan D., Everts B. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. U. S. A. 2013;110(35):14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien L.C., Chen Q., Savas J. Skeletal muscle mitochondrial mass is linked to lipid and metabolic profile in individuals with spinal cord injury. Eur. J. Appl. Physiol. 2017;117(11):2137–2147. doi: 10.1007/s00421-017-3687-9. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y., Pan C., Yu L. SSBP1 upregulation in colorectal cancer regulates mitochondrial mass. Canc. Manag. Res. 2019;11:10093–10106. doi: 10.2147/CMAR.S211292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suski J.M., Lebiedzinska M., Bonora M. Relation between mitochondrial membrane potential and ROS formation. Methods Mol. Biol. 2012;810:183–205. doi: 10.1007/978-1-61779-382-0_12. [DOI] [PubMed] [Google Scholar]
- 22.Mills E.L., Kelly B., Logan A. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. J. Cell. 2016;167(2):457–470. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao L.P., Ovchinnikov D., Wolvetang E. The effect of ethidium bromide and chloramphenicol on mitochondrial biogenesis in primary human fibroblasts. Toxicol. Appl. Pharmacol. 2012;261(1):42–49. doi: 10.1016/j.taap.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Lee S.R., Heo H.J., Jeong S.H. Low abundance of mitochondrial DNA changes mitochondrial status and renders cells resistant to serum starvation and sodium nitroprusside insult. Cell Biol. Int. 2015;39(7):865–872. doi: 10.1002/cbin.10473. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y.C., Yeh W.C., Ohashi P.S. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Ko A.R., Hyun H.W., Min S.J. The differential DRP1 phosphorylation and mitochondrial dynamics in the regional specific astroglial death induced by status epilepticus. Front. Cell. Neurosci. 2016;10:124. doi: 10.3389/fncel.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo S.J., Garg N.J. Metabolic programming of macrophage functions and pathogens control. Redox Biol. 2019;24:101198. doi: 10.1016/j.redox.2019.101198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda Y., Inoguchi T. [Oxidative stress and chronic inflammation][J] Nihon Rinsho. 2016;74(Suppl 2):73–76. [PubMed] [Google Scholar]
- 29.Griffiths H.R., Gao D., Pararasa C. Redox regulation in metabolic programming and inflammation. Redox Biol. 2017;12:50–57. doi: 10.1016/j.redox.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer A., Laverny G., Bernardi L. Mitochondria: an organelle of bacterial origin controlling inflammation. Front. Immunol. 2018;9:536. doi: 10.3389/fimmu.2018.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain N., Vogel V. Spatial confinement downsizes the inflammatory response of macrophages. Nat. Mater. 2018;17(12):1134–1144. doi: 10.1038/s41563-018-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callender L.A., Carroll E.C., Bober E.A. Mitochondrial mass governs the extent of human T cell senescence. Aging Cell. 2020;19(2) doi: 10.1111/acel.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnie G., Sotgia F., Lisanti M.P. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget. 2015;6(31):30472–30486. doi: 10.18632/oncotarget.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davizon-Castillo P., Mcmahon B., Aguila S. TNF-alpha-driven inflammation and mitochondrial dysfunction define the platelet hyperreactivity of aging. Blood. 2019;134(9):727–740. doi: 10.1182/blood.2019000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontecha-Barriuso M., Martin-Sanchez D., Martinez-Moreno J.M. PGC-1alpha deficiency causes spontaneous kidney inflammation and increases the severity of nephrotoxic AKI. J. Pathol. 2019;249(1):65–78. doi: 10.1002/path.5282. [DOI] [PubMed] [Google Scholar]
- 36.Zorova L.D., Popkov V.A., Plotnikov E.Y. Mitochondrial membrane potential. Anal. Biochem. 2018;552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reers M., Smiley S.T., Mottola-Hartshorn C. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol. 1995;260:406–417. doi: 10.1016/0076-6879(95)60154-6. [DOI] [PubMed] [Google Scholar]
- 38.Fonseca T.B., Sanchez-Guerrero A., Milosevic I. Mitochondrial fission requires DRP1 but not dynamins. Nature. 2019;570(7761):E34–E42. doi: 10.1038/s41586-019-1296-y. [DOI] [PubMed] [Google Scholar]
- 39.Hu C., Huang Y., Li L. Drp1-Dependent mitochondrial fission plays critical roles in physiological and pathological progresses in mammals. Int. J. Mol. Sci. 2017;18(1) doi: 10.3390/ijms18010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y.J., Jeong S.Y., Karbowski M. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15(11):5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu W., Zhang X., Wu H. HO-1 is essential for tetrahydroxystilbene glucoside mediated mitochondrial biogenesis and anti-inflammation process in LPS-treated RAW264.7 macrophages. Oxid. Med. Cell Longev. 2017;2017:1818575. doi: 10.1155/2017/1818575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piantadosi C.A., Suliman H.B. Transcriptional control of mitochondrial biogenesis and its interface with inflammatory processes. Biochim. Biophys. Acta. 2012;1820(4):532–541. doi: 10.1016/j.bbagen.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carre J.E., Orban J.C., Re L. Survival in critical illness is associated with early activation of mitochondrial biogenesis. Am. J. Respir. Crit. Care Med. 2010;182(6):745–751. doi: 10.1164/rccm.201003-0326OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jablonski K.A., Amici S.A., Webb L.M. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens L., Van Eyck L., Jochmans D. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 2017;139(6):1995–1997. doi: 10.1016/j.jaci.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 46.Xu J., Lee M.H., Chakhtoura M. STAT2 is required for TLR-induced murine dendritic cell activation and cross-presentation. J. Immunol. 2016;197(1):326–336. doi: 10.4049/jimmunol.1500152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gopal R., Lee B., Mchugh K.J. STAT2 signaling regulates macrophage phenotype during influenza and bacterial super-infection. Front. Immunol. 2018;9:2151. doi: 10.3389/fimmu.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nan J., Wang Y., Yang J. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):3906–3911. doi: 10.1073/pnas.1714102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alazawi W., Heath H., Waters J.A. Stat2 loss leads to cytokine-independent, cell-mediated lethality in LPS-induced sepsis. Proc. Natl. Acad. Sci. U. S. A. 2013;110(21):8656–8661. doi: 10.1073/pnas.1221652110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dasgupta A., Chen K.H., Tian L. Gone fission: an asymptomatic STAT2 mutation elongates mitochondria and causes human disease following viral infection. J. Brain. 2015;138(Pt 10):2802–2806. doi: 10.1093/brain/awv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shahni R., Cale C.M., Anderson G. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 2015;138(Pt 10):2834–2846. doi: 10.1093/brain/awv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambold A.S., Pearce E.L. Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol. 2018;39(1):6–18. doi: 10.1016/j.it.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Indo H.P., Davidson M., Yen H.C. Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion. 2007;7(1–2):106–118. doi: 10.1016/j.mito.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 54.Khansari N., Shakiba Y., Mahmoudi M. Chronic inflammation, and oxidative stress as a major cause of age-related diseases and cancer. Recent Pat. Inflamm. Allergy Drug Discov. 2009;3(1):73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- 55.Clauzure M., Valdivieso A.G., Massip C.M. Disruption of interleukin-1beta autocrine signaling rescues complex I activity and improves ROS levels in immortalized epithelial cells with impaired cystic fibrosis transmembrane conductance regulator (CFTR) function. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0099257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.