Abstract
The current study demonstrates the synthesis of coumarin-triazole hybrids 8 (a-e) in four steps starting from substituted salicylaldehyde 1 (a-e), and diethyl malonate 2. The spectroscopic studies provide the structure proofs of the new compounds, and the molecular structure of an intermediate 3a by crystallographic studies. The crystal structure analysis revealed the C–H...O, C–H... π, C–O...π and π...π molecular interactions. Further, the intermolecular interactions were quantified using Hirshfeld surface analysis and the DFT method B3LYP functional with 6–311++ G (d,p) basis set was employed to optimize the molecular geometry. The synthesized new coumarin-triazole hybrids, 8 (a–e) were screened for their α-amylase inhibitory potentials, and the results suggest that amongst the series, compounds 8c, and 8e show the promising inhibition of the enzyme, and might act as lead molecules for anti-diabetic activities. To understand the mode of action in silico molecular docking and ADME screening were performed.
Keywords: Organic chemistry, Theoretical chemistry, Amylase, Diabetes, Hydrazides, Pechmann, X-ray diffraction, In silico molecular docking
Organic chemistry; Theoretical chemistry, Amylase; Diabetes; Hydrazides; Pechmann; X-ray diffraction; In silico molecular docking.
1. Introduction
The incidence of diabetes is tremendously increasing in recent decades all over the globe, and the investigation of small molecules with potent antidiabetic activities is one of the most interesting fields of research. Diabetes mellitus (DM) is a chronic metabolic disease identified by the imbalance in glucose homeostasis leads to a rise of glucose level in blood [1, 2]. Type 2 diabetes and hypertension are very common risk factor which leads to cardiovascular disease as well as leads neurological disorders [3]. Heterocyclic chemistry has dominated the field with decades of history and prospects and plays a primordial role in the synthesis of future drugs. Amongst the heterocycles, coumarins, and triazoles have attained more attention for their natural occurrence and immense biological activities. Coumarins are prepared via Pechmann condensation, catalytic annulation phenolic acetates with acrylates [4], ZnBr2 catalyzed reaction of salicylaldehyde with ynamides [5], Amberlyst-15 catalyzed reaction of 1,3-dihydroxybenzene and ethyl acetoacetate under microwave irradiation in a solvent free conditions [6] etc. Naturally occurring coumarins in the glycoside forms are useful in medicine [7], and coumarins shows antimicrobial [8], anti-HIV [9], radical scavenging [10], and anti-viral [11] properties.
Five-membered heterocycles, specifically, 1,2,4-triazoles plays a pivotal role in medicinal chemistry for their versatile applications. 1,2,4-Triazoles were synthesized by; a cycloaddition of nitriles and amidines in the presence MCM-41/copper(I) complex as heterogeneous catalyst [12], iodine catalyzed multicomponent reaction of aryl hydrazines, paraformaldehyde, NH4OAc, and alcohols under electrochemical conditions [13], regio-selective synthesis involving Ag(I) catalyzed reaction of ethyl cyanoacetate with aryldiazoniumtetrafluoroborate in water at 0 °C [14], Iodine catalyzed environmentally benign C–N bond formation from isothiocyanates in water at room temperature [15], palladium and copper catalyzed arylation of styrene [16], triflic anhydride activated microwave-induced one-pot synthesis involving secondary amides and hydrazides [17], and by the reaction of hydrazides with isothiocyanate under optimized reaction conditions [18] 1,2,4-Triazoles were reported to show DHFR inhibitor [19], and antimicrobial [20] activities.
1,2,4-Triazoles are versatile scaffolds in the synthesis of molecular hybrids, and 1,2,4-triazole clubbed heterocycles exhibit enhanced biological activities comparable with the parent moieties, and therefore have greater applications in medicinal chemistry [21]. For instance, the fused 1,2,4-triazole-imidazolidines possess the enhanced anti-proliferative, and HDAC-tubulin inhibitor [22], isonicotinic acid tethered phenacyl triazoles show anticonvulsant [23], S,N-bis(acyclonucleoside) derivatives exhibit anti-tubercular [24], Cox-2 inhibitor [25], properties. Coumarin tethered pyrazole-oxadiazole hybrids show improved antimicrobial and antioxidant activities [26], for instance, Naproxen analogs of 1,2,4-triazole-5-thiones show potential antinociceptive and anti-inflammatory [27], triazolo [4,3-b][1,2,4] triazepine-8(9H)-ones have α-amylase and α-glucosidase inhibitory [28], and 4-amino-1,2,4-triazole-Schiff's bases exhibit good anti-diabetic [29], activities. In this context, this current study presents the synthesis of coumarin-triazole hybrids, in vitro screening of their anti-diabetic activity, in silico docking and ADME screening studies. The structure proofs were obtained by spectral analysis, one of the intermediate 3a by crystallographic, also by the computational analysis using Hirshfeld surface studies and density functional theory to substantiate the experimental results.
2. Experimental
2.1. Material and methods
The AR grade reagents and chemicals were procured from Sigma Aldrich, SD Fine, SRL. Thin layer chromatography (TLC) was performed with silica gel pre-coated aluminum plates (Merck, India), and visualized under UV light. 1H NMR (400 MHz) and 13C{1H} NMR (100 MHz) spectra were recorded on Agilent NMR spectrometer with DMSO-d6 and CDCl3 as solvents. Mass spectra were recorded on Lynx SCN781 spectrometer (TOF mode). Melting points were determined in open capillaries using Centex apparatus and were uncorrected.
2.2. Synthesis of coumarin hydrazides 5(a-e)
To a solution of coumarin esters 3 (a-e) (10 mmol) in ethyl alcohol (25 mL), hydrazine hydrate (10 mmol eq.) was added. Initially, the mixture was stirred at a low temperature (0–5 °C) for about an hour and then refluxed for 3 h on a water bath. The progress of the reaction was monitored by TLC, after the completion, the reaction mixture was cooled, the solvent was evaporated in vacuo to get crude coumarin hydrazides 5 (a-e), and were recrystallized from methyl alcohol.
2.2.1. 6-Chloro-2-oxo-2H-chromene-3-carbohydrazide, 5b
Obtained from 3b (10 mmol) and 4 (10 mmol) as pale yellow solid in 52% yield; 1H NMR (DMSO-d6, δ ppm): 5.20 (s, 2H, –NH2), 7.16–7.24 (m, 1H, Ar–H), 7.60–7.61 (s, 1H, Ar–H), 7.85 (s, 1H, Ar–H), 7.98 (s, 1H, 4-H), 11.01 (s, 1H, –NH); 13C{1H} NMR (DMSO-d6, δ ppm): 125.2 (1C), 127.8 (1C), 128.6 (1C), 128.7 (1C), 129.9 (1C), 130.5 (1C), 130.4 (1C), 139.2 (1C), 146.3 (1C), 151.7 (1C, COO), 163.7 (1C, CONH). MS m/z: 238.02 (M+, 35Cl, 100), 240.02 (M+2, 37Cl, 31); Anal. Calcd. for C10H7ClN2O3 (%): C, 50.33; H, 2.96; N, 11.74; Found C, 50.24; H, 2.93; N, 11.68.
2.2.2. 7-Methoxy-2-oxo-2H-chromene-3-carbohydrazide, 5d
Obtained from 3d (10 mmol) and 4 (10 mmol) as sparkle like yellow solid in 63% yield; 1H NMR (DMSO-d6, δ ppm): 3.80 (s, 3H, OCH3), 5.49 (s, 2H, –NH2), 7.36 (s, 1H, Ar–H), 7.50–7.52 (d, 1H, Ar–H), 7.92–7.94 (d, 2H, Ar–H), 11.09 (s, 1H, –NH); MS m/z: 234.74 (M+,100), Anal. Calcd. for C11H10N2O4 (%): C, 56.41; H, 4.30; N, 11.96; Found C, 56.33; H, 4.27; N, 11.90.
2.2.3. 8-Ethoxy-2-oxo-2H-chromene-3-carbohydrazide, 5e
Obtained from 3e (10 mmol) and 4 (10 mmol) as pale yellow gummy mass in 48% yield; 1H NMR (DMSO-d6, δ ppm): 1.10–1.14 (t, 3H, –CH3), 4.12–4.17 (q, 2H, –OCH2), 5.45 (s, 2H, –NH2), 7.43 (s, 1H, Ar–H), 7.51 (s, 1H, Ar–H), 7.61 (s, 1H, Ar–H), 8.04–8.08 (d, 1H, Ar–H), 11.20 (s, 1H, –NH); 13C{1H} NMR (DMSO-d6, δ ppm): 14.6 (CH3), 59.6 (OCH2), 125.3 (2C), 127.8 (1C), 128.7 (1C), 128.9 (1C), 131.1 (1C), 131.9 (1C), 132.0 (1C), 155.0 (1C, COO), 163.2 (1C, CONH). MS m/z: 248.19 (M+, 100); Anal. Calcd. for C12H12N2O4 (%): C, 58.06; H, 4.87; N, 11.29; Found C, 57.96; H, 4.84; N, 11.23.
2.3. Synthesis of intermediate compounds 7(a-e)
To a solution of coumarin hydrazide 5(a-e) (10 mmol) and alcoholic potassium hydroxide (15 mmol) in absolute ethyl alcohol (100 mL), carbon disulphide 6 (15 mmol) was added slowly at a temperature below 5 °C, and then was stirred at room temperature for 24 h. The solid formed was collected by filtration, washed with anhydrous ether (100 mL), and dried in vacuo to obtain potassium dithiocarbazinate salts 7(a-e) in quantitative yields, and were used as such without further purification.
2.4. Synthesis of coumarin-triazole hybrids 8(a-e)
To a suspension of the potassium salt of coumarin thiocarbazinate 7 (a-e) in water (5 mL), hydrazine hydrate (10 mmol eq.) was added. The mixture was refluxed on a water bath conditions for 18–20 h. The change of color with the liberation of H2S is observed, the reaction was continued till to get a homogeneous reaction mixture. After this, the mixture was cooled and diluted with cold water (20 mL), and neutralized with dil. hydrochloric acid to obtain the target compounds 8 (a-e), and were recrystallized from ethyl alcohol.
2.4.1. 3-(4-Amino-5-mercapto-4H-1,2,4-triazol-3-yl)-6-bromo-2H-chromen-2-one, 8a
Obtained as pale yellow gummy mass in 58% yield; 1H NMR (DMSO-d6, δ ppm): 5.61 (s, 2H, –NH2), 7.29–7.39 (m, 2H, Ar–H), 7.58–7.59 (d, 1H, Ar–H), 8.38 (s, 1H, Ar–H), 13.60 (s, 1H, –SH); 13C{1H} NMR (DMSO-d6, δ ppm): 128.7 (1C), 129.3 (1C), 131.4 (1C), 133.4 (1C), 134.0 (1C), 135.1 (1C), 135.8 (1C), 140.0 (1C), 143.9 (1C), 164.4 (COO), 179.5 (=C-SH). MS m/z: 337.06 (M+, 79Br, 100), 339.06 (M+2, 81Br, 92); Anal. Calcd. for C11H7BrN4O2S (%): C, 38.95; H, 2.08; N, 16.52; Found C, 38.88; H, 2.05; N, 16.46.
2.4.2. 3-(4-Amino-5-mercapto-4H-1,2,4-triazol-3-yl)-6-chloro-2H-chromen-2-one, 8b
Obtained as pale yellow solid in 61% yield, m.p. 234 °C; 1H NMR (DMSO-d6, δ ppm): 5.62 (s, 2H, –NH2), 7.36–7.59 (m, 3H, Ar–H), 8.38 (s, 1H, 4-H), 13.61 (s, 1H, –SH); 13C{1H} NMR (DMSO-d6, δ ppm): 107.0 (1C), 111.9 (1C), 117.9 (1C), 123.7 (1C), 124.4 (1C), 125.6 (1C), 131.6 (1C), 138.1 (1C), 140.6 (1C), 147.2 (1C), 165.3 (COO), 175.2 (=C-SH). MS m/z: 293.02 (M+, 35Cl, 100), 295.02 (M+2, 37Cl, 28); Anal. Calcd. for C11H7ClN4O2S (%): C, 44.83; H, 2.39; N, 19.01; Found C, 44.76; H, 2.36; N, 19.06.
2.4.3. 3-(4-Amino-5-mercapto-4H-1,2,4-triazol-3-yl)-8-methoxy-2H-chromen-2-one, 8c
Obtained as pale yellow gummy mass in 58% yield; 1H NMR (DMSO-d6, δ ppm): 3.77 (s, 1H, –OCH3), 5.62 (s, 2H, –NH2), 7.35–7.58 (m, 3H, Ar–H), 8.38 (s, 1H, 4-H), 13.62 (s, 1H, –SH); 13C{1H} NMR (DMSO-d6, δ ppm): 55.4 (CH3), 113.7 (1C), 119.5 (1C), 126.9 (1C), 130.6 (1C), 131.1 (1C), 131.3 (1C), 134.6 (1C), 134.7 (1C), 142.3 (1C), 166.1 (COO), 173.3 (=C-SH). MS m/z: 290.02 (M+, 100), 291.02 (M+1, 12); Anal. Calcd. for C12H10N4O3S (%): C, 49.65; H, 3.47; N, 19.30; Found C, 49.55; H, 3.45; N, 19.24.
2.4.4. 3-(4-Amino-5-mercapto-4H-1,2,4-triazol-3-yl)-7-methoxy-2H-chromen-2-one, 8d
Obtained as light yellowish solid in 62% yield, m.p. 224 °C; 1H NMR (DMSO-d6, δ ppm): 3.81 (s, 1H, –OCH3), 5.80 (s, 2H, –NH2), 7.18–7.68 (m, 3H, Ar–H), 8.40 (s, 1H, 4-H), 13.40 (s, 1H, –SH). MS m/z: 290.04 (M+, 100); Anal. Calcd. for C12H10N4O3S (%): C, 49.65; H, 3.47; N, 19.30; Found C, 49.56; H, 3.44; N, 19.33.
2.4.5. 3-(4-Amino-5-mercapto-4H-1,2,4-triazol-3-yl)-8-ethoxy-2H-chromen-2-one, 8e
Obtained as light greenish gummy mass in 56% yield; 1H NMR (DMSO-d6, δ ppm): 1.09–1.13 (t, 3H, –CH3), 3.95–4.01 (q, 2H, –OCH2), 5.92 (s, 2H, –NH2), 7.40–7.59 (m, 3H, Ar–H), 8.40 (s, 1H, 4-H), 13.62 (s, 1H, –SH); 13C{1H} NMR (DMSO-d6, δ ppm): 14.2 (CH3), 59.9 (OCH2), 116.1 (1C), 123.3 (1C), 125.3 (1C), 127.8 (1C), 128.9 (1C), 131.1 (1C), 131.9 (1C), 132.0 (1C), 145.0 (1C), 163.6 (COO), 176.2 (=C-SH). MS m/z: 304.13 (M+, 100); Anal. Calcd. for C13H12N4O3S (%): C, 51.31; H, 3.97; N, 18.41; Found C, 51.19; H, 3.94; N, 18.26.
2.5. Crystallographic studies
Single crystals were obtained by slow evaporation technique, and defect free crystal suitable for X-ray diffraction studies of dimensions 0.30 × 0.27 × 0.23 mm was chosen with the aid of polarizing microscope. X-ray intensity data were collected at 296 K using Bruker Proteum2 CCD X-ray diffractometer. The X-ray generator was operated at 45 kV and 10 mA with CuKα radiation of wavelength 1.54178 Å. Data were collected for 24 frames per set for different settings of ϕ (0° and 90°), with the scan width of 0.5° and 5 s exposure time, keeping the sample to detector distance of 45.10 mm. The complete intensity data sets were processed with SAINT PLUS [30]. The structure was solved and refined using SHELXS (direct method) and SHELXL (full-matrix least squares method on F2) programs [30, 31]. The hydrogen atoms were positioned geometrically, with C–H = 0.93 to 0.97 Å. The residual value is saturated to 0.0457 in the final cycle of refinement. The PLATON [32] program was used to calculate the geometrical parameters. ORTEP and molecular packing diagrams were drawn with the MERCURY [33] software. The parameters in CIF format are available at Cambridge crystallographic data center.
2.6. Computational studies
The theoretical studies were carried out for the 3b molecule through Hirshfeld surface analysis (HSA) and density functional theory (DFT). The HSA was carried out with Crystal Explorer 17.5 [34] to quantify the intermolecular interactions. The structural coordinates were drawn, and optimized using DFT/B3LYP hybrid functional with 6–311++G (d,p) level basis set [35, 36] in gas phase by Gaussian16 software [37]. The energies of molecular orbitals and chemical reactive descriptors were calculated. Further, the molecular electrostatic potential (MEP) map was drawn to find the chemical active regions on the three dimensional molecular surface. The optimized molecular geometry and their orbital energy states were visualized using Gauss view 6.0.8 [38] software.
2.7. α-Amylase inhibition activity (DNSA) assay
The α-amylase inhibition experiment was performed according to the reported procedure [39]. Briefly, a solution of test compounds 8 (a-e) of 25, 50, 75 and 100 mg/mL concentrations were prepared by dissolving in minimum quantity of DMSO and diluted with double distilled water. A solution of test compounds, sodium phosphate buffer (500μL/0.02M), α-amylase (0.5 mg/mL) (together of 500 μL) was incubated at 25 °C for 10 min. After this, a starch solution (500 μL/1%) in sodium phosphate buffer (0.02M, pH 6.9 with 0.006M sodium chloride) was added to each tube at an intervals of 5 s. The mixture was again incubated at 25 °C for 10 min, and DNSA (1mL) was added, and then placed in a boiling water bath for 5 min, and cooled. Finally, the mixture was diluted with distilled water (10 mL), the absorbance was recorded at 540nm. The results were calculated as;
2.8. Molecular docking and ADME predictions
Type 2 diabetes and hypertension are very common risk factor which leads to cardiovascular disease as well as leads neurological disorders [3]. The selected co-ordinates of α-amylase (PDB ID: 4W93), Angiotensin-converting enzyme (ACE, PDB ID: 1O86) are drawn from Brookhaven Protein Data Bank, these are chosen because the targeted protein was co-crystallized with inhibitor [40, 41]. The Maestro 2D sketcher was used to draw the structures of ligands, and OPLS 2005 used to compute the energy minimize. Proteins prepared by retrieving in to Maestro 9.3 platform, and protein structure corrected for the missing loops with Prime software module. H2O molecules from ACE and hetero atom were removed beyond 5 Å. Water molecules were optimized during protein pepwizard, as these are important in interaction between the receptor. OPLS 2005 force field was applied to the protein to restrained minimization, and RMSD of 0.30 Å was set to converge heavy atoms before start of docking. Grid for receptor mapping was executed by using the co-crystallized substrate or inhibitor bound to the targeted protein. Using Extra-precision (XP) docking and scoring; ligands were docked into the receptor grid of radii 20 Å and the docking calculation was done on the basis of docking score, ADME results and Glide energy. QikProp program and Maestro 9.3 were used to calculate ADME properties, and molecular visualization, respectively [42].
3. Results and discussion
3.1. Chemistry
In a multi-step route, a series of 3-(4-amino-5-mercapto-4H-1,2,4-triazol-3-yl)-2H-chromen-2-ones, 8 (a-e) were efficiently synthesized starting from various substituted salicylaldehyde 1 (a-e) and diethylmalonate 2. Initially, series of ethyl 2-oxo-2H-chromene-3-carboxylates 3 (a–e) [43-45] were prepared from substituted salicylaldehyde, 1 (a-e), diethylmalonate, 2 in the presence of few drops of pyridine under solvent free conditions. Then, a solution of compounds 3 (a–e) (10 mmol), hydrazine hydrate, 4 (10 mmol eq.) in ethyl alcohol (15 mL) was heated on a water bath for 2 h to obtain hydrazides 5 (a–e). The reaction of 5 (a–e) with CS2, 6 in the presence of KOH in ethyl alcohol yielded the intermediate dithiocarbazate salts, 7 (a–e), which were used in situ without being isolated and purified, the in situ reaction of 7 (a–e) with hydrazine hydrate, 4 (10 mmol eq.) under reflux conditions produced the target compounds 8 (a–e) in good yields (Figure 1).
Figure 1.
Synthetic route for coumarin-triazole hybrids, 8 (a-e).
3.2. Spectral studies
Structure proofs of the synthesized compounds 5b, 5d, 5e, and 8 (a–e) obtained by 1H NMR, 13C{1H} NMR, Mass spectroscopic studies, and elemental analysis. Compounds 5b, 5d, and 5e show singlets at δ 5.20, 5.49, 5.45 ppm for –NH2; at δ 7.98, 7.86, 8.08 ppm for C4–H; and δ 11.01, 11.09, 11.20 ppm for –NH protons. These data were in agreement and comparable with the reported molecules 5a [46], and 5c [47]. Compound 5d show singlet at δ 3.80 ppm for OCH3 protons; 5e show quartet δ 4.12–4.17 (J = 21.6 Hz) ppm, and a triplet at δ 1.10–1.14 (J = 14 Hz) ppm. All compounds show the other signals as multiplet in the aromatic region in their respective spectra. The compounds 8 (a-e) show singlets at δ 5.61–5.92 ppm for –NH2; at δ 8.38–8.40 ppm for C4–H; and at δ 13.40–13.62 ppm for –SH protons. These spectral data are in good agreement with the values of structurally similar molecule 3-(4-amino-5-mercapto-4H-1,2,4-triazolyl-3-yl)-2H-chromen-2-one reported by Raslan and Khalil [48]. Further all compounds show signals due to substituent and aromatic protons in the respective region in their spectra.
Compounds, 5b and 5e show resonance signals at δ 151.7 and 155.0 ppm for lactone-COO; at δ 163.7 and 163.2 ppm for CONH carbons. Compound 5e show signals for CH3, and OCH2 carbons at δ 14.6 and 59.6 ppm. The aromatic carbons appear in the range δ 125.2–146.3 ppm. These data were in good matching and comparable to reported molecules 5a [46], and 5c [47]. Compounds 8 (a-e) show resonance signals in the region δ 142.3–147.2, 163.6–166.1, and 173.3–179.5 ppm for C=N (triazole ring), COO, and C-SH carbons; and were in good matching with the data of structurally identical reported molecule 3-(4-amino-5-mercapto-4H-1,2,4-triazol-3-yl)-2H-chromen-2-one [48, 49, 50]. They also show the signals for aromatic and other carbons in the expected region.
Compounds, 5b show base peak at m/z 238.02 owing to its molecular mass (C10H7ClN2O3; MW = 238.01) and (M+2) peak at m/z 240.02 with abundance of 33% due to 37Cl isotopic mass; 5d show base peak at m/z 234.74 (C11H10N2O4; MW = 234.34), and 5e show base peak at m/z 248.19 (C12H12N2O4; MW = 248.08). Compounds 8 (a-e) show base peaks corresponding to their molecular mass, however, in addition to the base peaks compound 8a show (M+2) peak at m/z 339.06 with a relative abundance of 96% due to 81Br isotopic mass, and 8b show (M+2) peak at m/z 295.02 (31%) due to 37Cl isotopic mass; which confirm the structure of new compounds.
The experimental IR spectrum, the compound 3a show characteristic stretching bands for C–H, C=O (ester and lactone), C=C, and C–Br functions (Figure S1b). Whereas, the theoretical DFT-IR spectrum show absorption bands with little higher absorption frequencies (Figure S1a). The stretching absorption bands appear in the experimental FTIR spectrum at 2983, 1831, 1606, 1318, 1291, and 769 cm−1 were assigned to C–H, COO-lactone, COO-ester, C=C, C–O, and C–Br vibrations. The theoretical FTIR spectrum from DFT analysis shows the absorption bands at 3041, 1834, 1589, 1306, 1286, and 606 cm−1 were assigned to C–H, COO-lactone, COO-ester, C=C, C–O, and C–Br stretching vibrations.
In experimental 1H NMR spectrum, compound 3a (Figure S2b), shows a triplet at δ 1.12 (J = 14Hz) ppm due to ester CH3 protons, a quartet at δ 4.16 (J = 21.2Hz) ppm due to OCH2 protons, one doublet each at δ 7.34, and δ 7.51 ppm due to aromatic protons. A singlet observed at δ 8.83 ppm was due to C-4 of coumarin ring. The theoretical 1H NMR spectrum obtained from NMR-DB (Figure S2a) shows triplet at δ 1.29 (J = 7.1Hz) ppm, quartet at δ 4.10 (J = 7.1Hz) ppm due to ester CH3, OCH2 protons. two doublets at δ 7.50, and δ 7.79 ppm due to aromatic protons. A singlet observed at δ 8.83 ppm was due to C-4 of coumarin ring. The theoretical 1H NMR (chemical shifts and splitting) and the reported 1H NMR data of the compound [43, 44, 45] are in good agreement with each other as seen in Figure S2.
3.3. Crystallographic studies
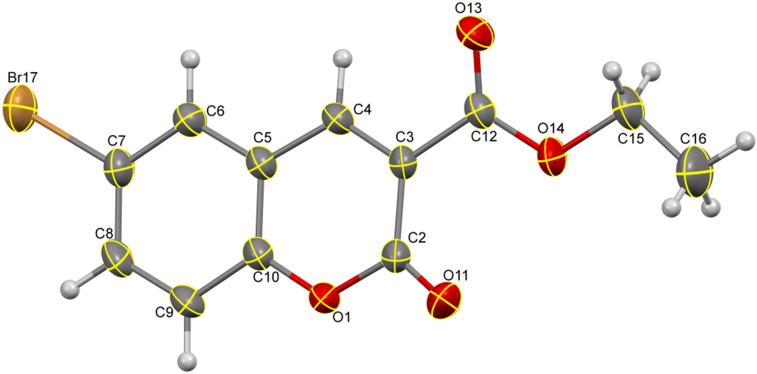
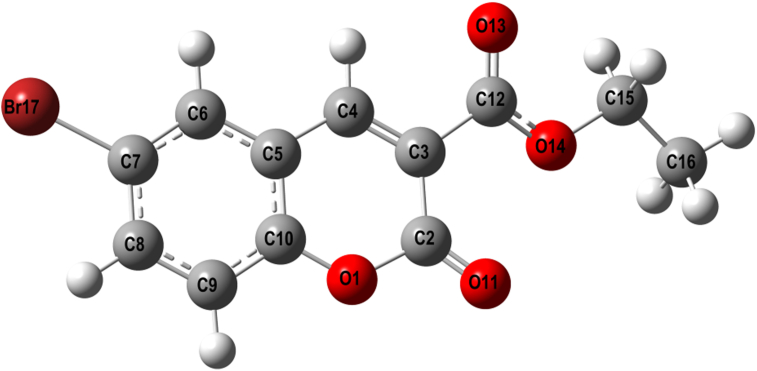
The compound 3a (CCDC#: 2005362) was crystallized from ethanol, shows following crystallographic parameters: a = 5.8503 (6) Å, b = 13.2759 (14) Å, c = 14.8674 (16) Å, α = 90o, β = 92.641 (3)o, γ = 90° and V = 1153.3 (2) Å3. The compound crystallized in the monoclinic crystal system with P21/n space group, whereas, the reported crystal structure is monoclinic with P21/c space group [51]. The ORTEP of molecule 3a is depicted in Figure 2, and crystal structure refinement data were summarized in Table 1. The packing of the molecules viewed along the a-axis (Figure S3), down the b-axis (Figure S4), and along the c-axis (Figure S5). The H-bond interactions are shown by dotted lines, which lead to the 2-dimentional layered stacking. The bond length, bond and torsion angles are given in Table S1, Table S2 and Table S3.
Figure 2.
ORTEP diagram of molecule 3a with thermal ellipsoids drawn at 50% probability.
Table 1.
Crystal structure refinement statistics of ethyl 6-bromo-2-oxo-2H-chromene-3-carboxylate, 3a.
| Parameter | Value |
|---|---|
| CCDC No. | 2005362 |
| Empirical formula | C12H9BrO4 |
| Formula weight | 218.20 |
| Temperature | 296 K |
| Wavelength | 1.54178 Å |
| Crystal system, space group | Monoclinic, P21/n |
| Unit cell dimensions | a = 5.8503 (6) Å, b = 13,2759 (14) Å, c = 14.8674 (16) Å, α = 90o, β = 92.641 (3)o, γ = 90° |
| Volume | 1047.1 (2) Å3 |
| Z | 4 |
| Density (calculated) | 1.711 Mg m −3 |
| Absorption coefficient | 4.891 mm −1 |
| F000 | 592 |
| Crystal size | 0.30 × 0.27 × 0.23 mm |
| Θ range for data collection | 5.96o to 65.36o |
| Index ranges | −6 ≤ h ≤ 6, −15 ≤ k ≤ 15 |
| −17 ≤ l ≤ 17 | |
| Reflections collected | 16788 |
| Independent reflections | 1900 [Rint = 0.0566] |
| Absorption correction | multi-scan |
| Refinement method | Full matrix least-squares on F2 |
| Data/restraints/parameters | 1900/0/155 |
| Goodness-of-fit on F2 | 1.083 |
| Final [I > 2σ(I)] | R1 = 0.0457, wR2 = 0.1215 |
| R indices (all data) | R1 = 0.0459, wR2 = 0.1217 |
| Largest diff. peak and hole | 0.644 and -0.810 e Å−3 |
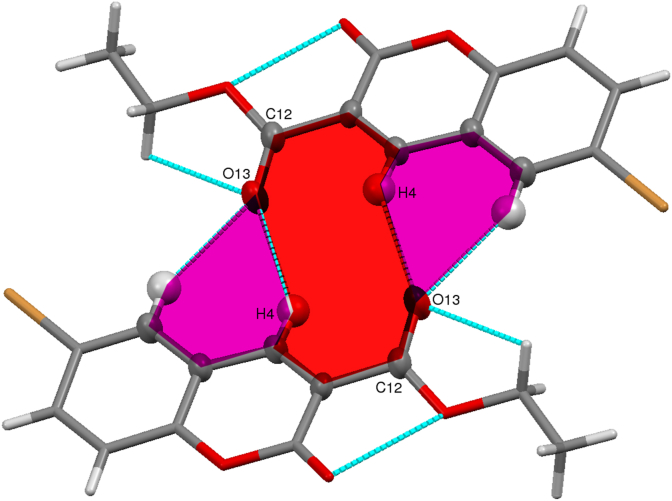
The parameters such as bond lengths, bond angles, and torsion angles of the molecule, 3a were compared with the reported data [51], and most of the values are in good agreement. The slight changes observed in the bond lengths for the O13–C12, Br17–C7, O11–C2, O1–C2, O14–C15, and C2–C3 bonds were 1.204(4), 1.897(3), 1.198(4), 1.387(3), 1.459(4), and 1.461 (4) Å, respectively; for the same bonds the reported bond lengths are 1.198(4), 1.888(3), 1.189(4), 1.379(3), 1.455(4), and 1.465 (4) Å. The bond angles for the C10–O1–C2, O1–C10–C5, O11–C2–O1, C4–C3–C12, C10–C5–C6 are 122.7(2), 120.7(2), 116.1(3), 117.4(2), 118.7(3)o; whereas the reported values for the same bond angles are 123.1(2), 121.0 (2), 116.8(3), 117.7(3), and 119.0 (17)o, respectively. The slight changes in the torsion angles observed for C2–C3–C12–O13, O11–C2–C3–C12 are -154.6 (3)o, 5.5 (5)o; but the reported torsion angles values for the same are -155.0 (3)o, 6.1 (3)o, respectively. Further, the molecular structure exhibits the similar kind of molecular interactions as possessed by the compound reported earlier. The C–H...O intermolecular interactions stabilizes the crystal structure (Table S4). The interactions such as C–H...π, C–O...π and π…π also stabilizes the structure, and leads to the R22 (14), R22 (10) and R21 (6) supramolecular synthons [51, 52, 53] as shown in Figure 3.
Figure 3.
Intermolecular interactions leading to the supramolecular ring motif. The colored region indicates R22 (14) [pink colored region indicates the R21 (6) and the red color indicates the R22 (10)].
3.4. Hirshfeld surface analysis
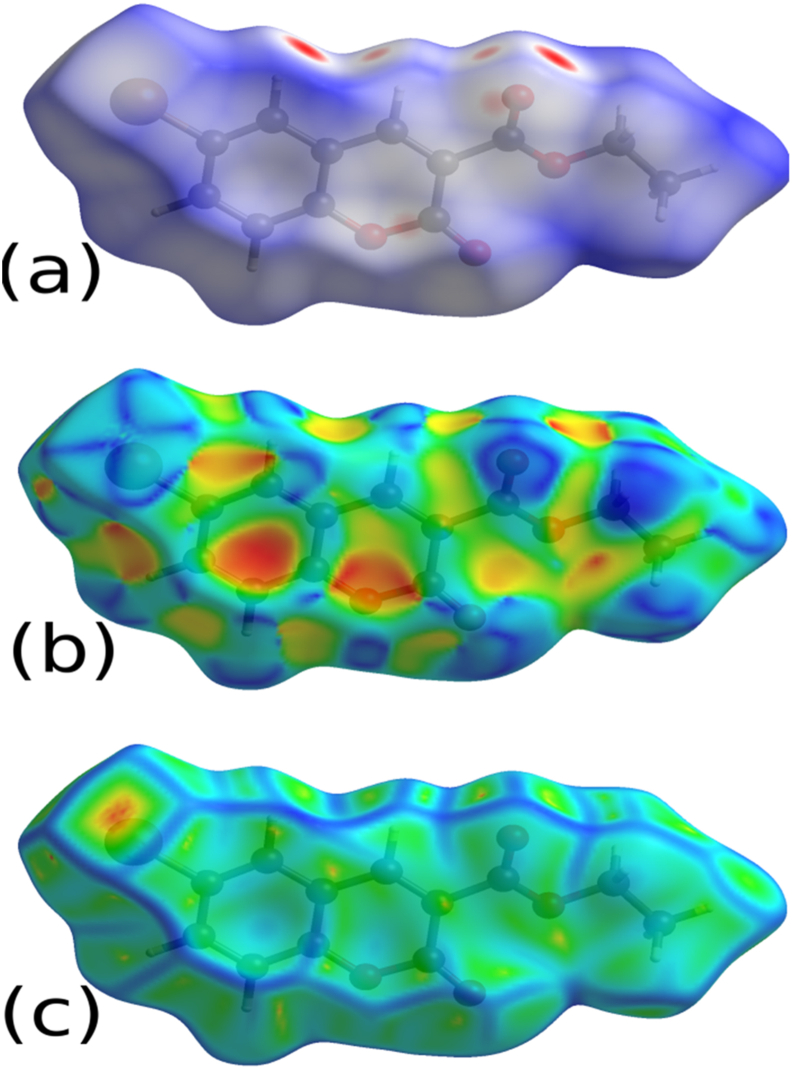
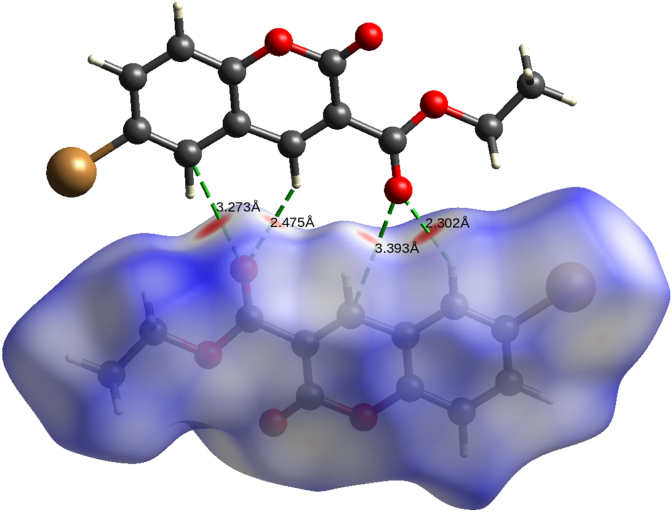
The CrystalExplorer 17.5 [34] software was used generate the 3D dnorm map and 2D fingerprint plots on molecular Hirshfeld surfaces (HS). The 3D dnorm (volume = 282.07 Å3) surfaces were generated in the color range of −0.226 au (blue) to +1.271 au (red). The dnorm (Figure 4a) on HS with color codes represents the different molecular contacts. Red region on dnorm surface represents the short intercontacts, white color represents the intercontacts with distance equal to van der Waals radii, and the blue region represent the longer intercontacts [54, 55]. The 2D fingerprint maps in expanded form [56, 57] for all contacts were drawn in the scale of 0.6–2.8 Å (Figure S6). The 2D fingerprint plot revealed the contribution of various individual intermolecular contacts. The O...H contacts are found to be the major interaction with the contribution of 25.4%. Further, the C–H...O interactions involved in the formation of supramolecular synthon on the 3D dnorm surface with the hydrogen bond distances (Figure 5). HSA also revealed the role of C–H...π and π...π interactions in stability of the crystal structure. The shape index, and curvedness generated in the range of -1.0 au to 1.0 au (Figure 4b), and -4.0 au to 0.4 au (Figure 4c), respectively to analyze the π-stacking interactions [53, 58, 59].
Figure 4.
(a) dnorm; (b) shape index; and (c) curvedness mapped with Hirshfeld surface.
Figure 5.
The C–H...O intermolecular H-bond interactions involved in the formation of supramolecular synthon.
3.5. Density functional theory calculations
The quantum computational calculations help to study the chemical and electronic behaviour of the compounds. The FMO energies and MEP surface analysis results in identification of the chemically active sites of the molecule. The DFT/B3LYP hybrid functional with 6311++G (d,p) basis set was used to optimize the geometrical coordinates of molecule, 3a (Figure 6) in gas phase. The computed structural parameters were compared with the experimental results. The optimized structure is well correlated with the experimental results, confirmed by the correlation coefficient values of bond lengths (0.9969), bond angles (0.9909), and torsion angles (0.9997) (Table S1–S3). Further, the frontier molecular orbital energies (EHOMO, ELUMO and Eg) and chemical reactive descriptors [60, 61, 62, 63, 64] calculated were summarized in Table 2.
Figure 6.
Optimized structure of the compound 3a.
Table 2.
Calculated FMO energies and chemical reactive descriptors of the compound, 3a.
| Parameters | Value [B3LYP/6–311++G (d,p)] (eV) |
|---|---|
| EHOMO | -7.0039 |
| ELUMO | -2.9396 |
| ΔEgap | 4.0643 |
| Ionization potential (I) | 7.0039 |
| Electron affinity (A) | 2.9396 |
| Electronegativity (χ) | 4.9718 |
| Chemical hardness (η) | 2.0321 |
| Global softness (σ) | 0.4921 |
| Electrophilicity (ω) | 6.0819 |
| Chemical potential (μ) | -4.9718 |
| Dipole moment (Debye) | 1.6356 |
Where, χ = (I + A)/2, η = (I-A)/2, σ = 1/η and ω = μ2/2η.
These global reactive parameters are very important to know the molecular stability and reactivity of the compound. The ionization potential (I) play a key role in reactivity of atoms and molecules. High ionization energy (6.8897 eV) specifies high stability and chemical inertness, electron affinity (2.9396 eV) value indicates that the molecule is less electrophilic. The highest value of EHOMO (-7.0039 eV) indicates a better inhibition efficiency and lower the ELUMO (-2.9396 eV), the easier is the acceptance of electrons. Global hardness (2.0321 eV) and softness (0.4921 eV) are very important properties indicating the low inhibition efficiency. The electronegativity (4.9718 eV) and chemical potential (-4.9718 eV) values signifies that the molecule is less reactive and has medium inhibition property. The low value of dipole moment (1.6356 Debye) specifies the low adsorption property of the molecule.
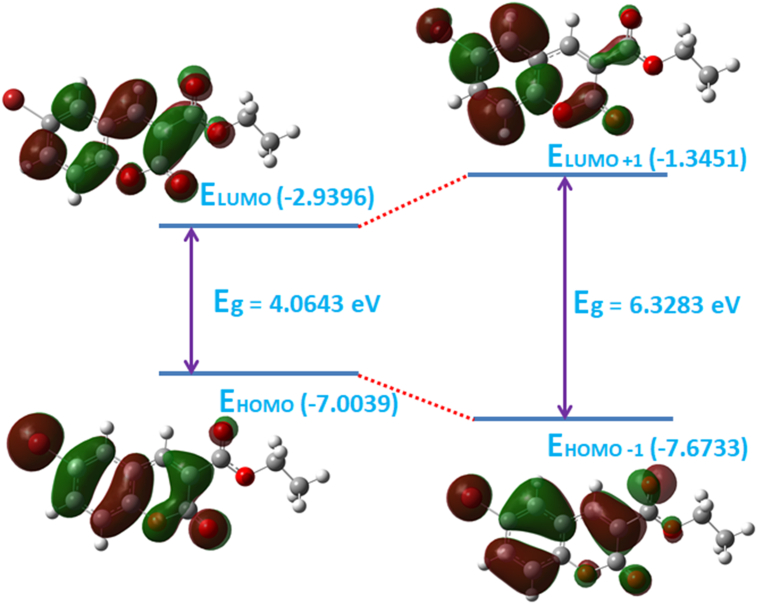
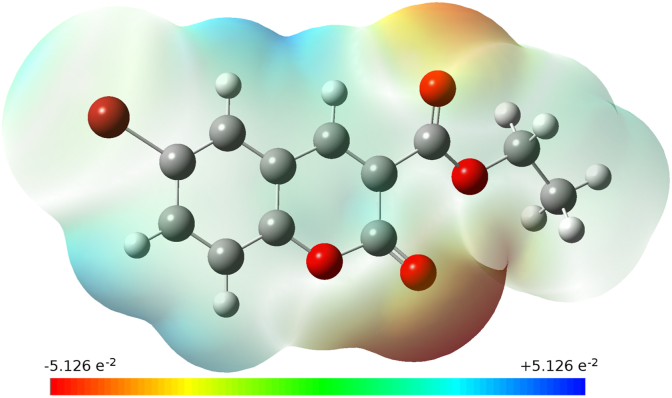
The energy gap has a significant role in case of organic molecules as it relates to the specific movement of the electrons from one energy state to another. The HOMO and LUMO behaves as an electron donor and acceptor, respectively. The polarizability and chemical reactivity can be evaluated with the band gap energy. The energy gap of molecule 3a show its chemically hard and less reactive nature. Figure 7 illustrates the HOMO-LUMO energies and their energy gap (ΔEgap = 4.0643 eV); H-1 and L+1 energies and their energy gap (6.3283 eV). The red and green color of the HOMO-LUMO signifies the positive and the negative phases of the wave functions. The MEP map (Figure 8) is drawn with the color range of -5.126 e−2 au (deepest red) to +5.126 e−2 au (deepest blue). The blue coloured region indicates the electrophilic reactivity with positive scale, green coloured region with negative scale indicates the nucleophilic reactivity, and red colour representing the sites of electrophilic attack. The red colour spread over the oxygen, and blue colour is concentrated around the hydrogens of carbon atoms.
Figure 7.
Frontier molecular orbitals with energy gap of the compound, 3a.
Figure 8.
Molecular electrostatic potential map of the compound 3a.
3.5. α-Amylase inhibition activity
The results of the α-amylase inhibition study are summarized in Table 3.
Table 3.
Percentage inhibition of α-amylase enzyme of synthesized compounds 8 (a-e) at different concentrations.
| Concentration (μM) | 8a | 8b | 8c | 8d | 8e |
|---|---|---|---|---|---|
| 2 | 26.67 ± 0.45 | 36.33 ± 0.78 | 25.13 ± 0.80 | 37.80 ± 0.27 | 25.00 ± 0.58 |
| 4 | 30.93 ± 0.05 | 44.07 ± 0.32 | 30.80 ± 0.28 | 46.93 ± 0.40 | 32.53 ± 0.24 |
| 6 | 33.73 ± 0.12 | 49.53 ± 0.53 | 32.40 ± 0.82 | 46.87 ± 0.17 | 33.27 ± 0.27 |
| 8 | 45.87 ± 0.24 | 54.87 ± 0.25 | 59.80 ± 0.28 | 48.27 ± 0.18 | 54.93 ± 0.05 |
| 10 | 52.27 ± 0.75 | 53.07 ± 0.05 | 79.20 ± 0.75 | 58.27 ± 0.28 | 78.73 ± 0.72 |
| 12 | 75.53 ± 0.24 | 69.73 ± 0.52 | 86.05 ± 0.27 | 54.27 ± 0.77 | 79.67 ± 0.58 |
The synthesized compounds 8 (a-e) shows varying effect on glucose utilization at the tested concentrations. Amongst the series, the compounds 8c and 8e showed maximum inhibition of the enzyme with the values ranging from 5.43μM and 5.98μM respectively. Moderate inhibition was showed by the compounds 8a and 8b with values of 8.02 μM and 9.14μM, respectively. The compound 8d showed least α-amylase inhibition from 11.54μM. The compound 8c (Figure S7) shows maximum inhibition of the enzyme with the IC50 values of 5.43μM. This study investigated the ability of synthesized compounds 8 (a-e) to serve as effective anti-diabetic agents.
3.6. Statistical analysis
Five compounds 8 (a-e) are analyzed for their ability to inhibit α-amylase enzyme at four different concentration levels by Two–way analysis of variance (ANOVA) (Table S5), it is observed that the compounds show statistically significant results. In other words, mean values differences amongst the compounds are greater than the expected by chance with statistically significant difference (P = <0.001). Since the results are significant, we computed pair-wise comparison test by Duncan's multiple range tests to know the pair-wise effect for pair of compounds 8 (a-e) (Table S6) using SPSS (version 20) statistical software. It was observed that, all the pair of compounds shows statistically significant pair-wise effects, except 8e vs. 8c, 8d vs. 8b and 8b vs. 8a. Obviously, from the statistical analysis (ANOVA), the compounds 8c vs. 8a and 8c vs. 8b exhibit more statistical significant results at different concentrations as compared to other pair of compounds.
The residual plot (Figure S8a) was used to determine evaluation of the goodness of a fitted model and homoscedasticity plot (Figure S8b) represents the variance around the regression line, and is the same for all predictor variable percentage of inhibition. The Figure S8c shows that the compound 8c exhibit more α-amylase inhibition activity compare to 8e. The column mean difference at 95% confidence intervals of ligands 8(a-e) is given in Figure S8d, wherein the % of inhibition plots the predicted residual (or weighted residual) assuming sampling from a Gaussian distribution.
3.7. Molecular docking and ADME predictions
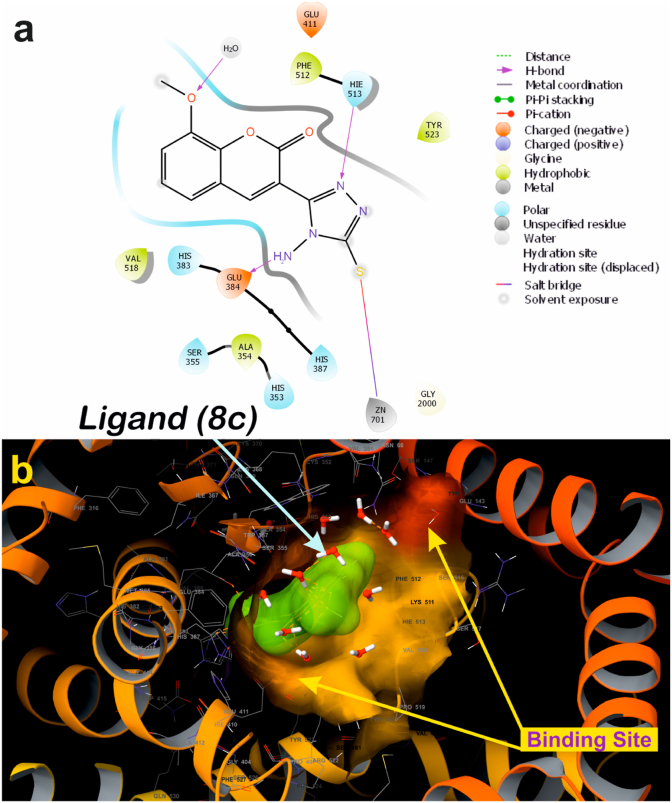
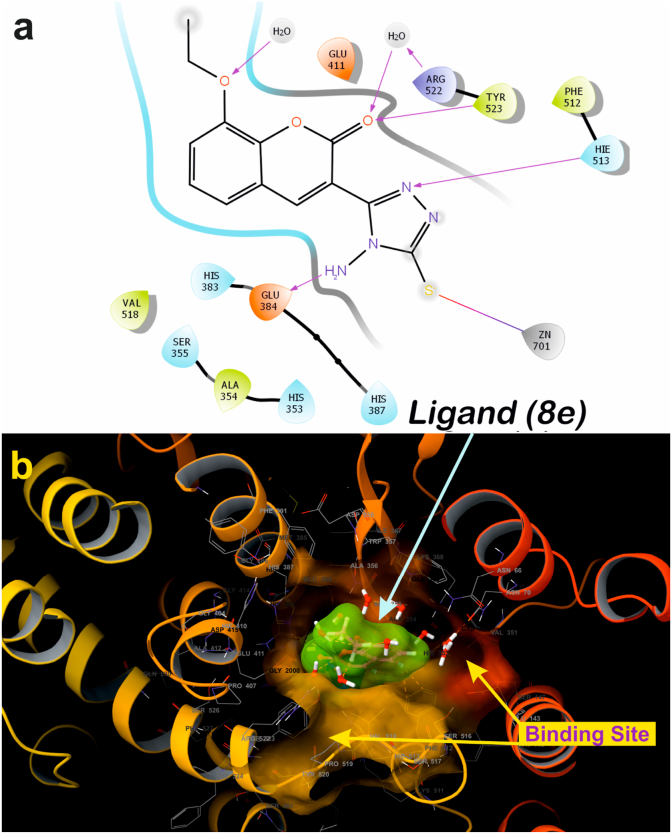
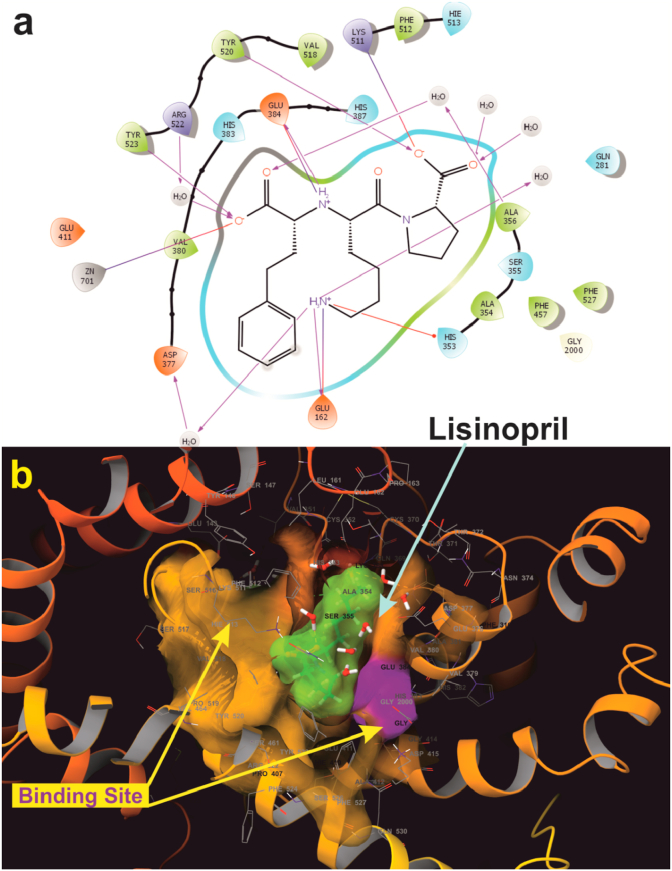
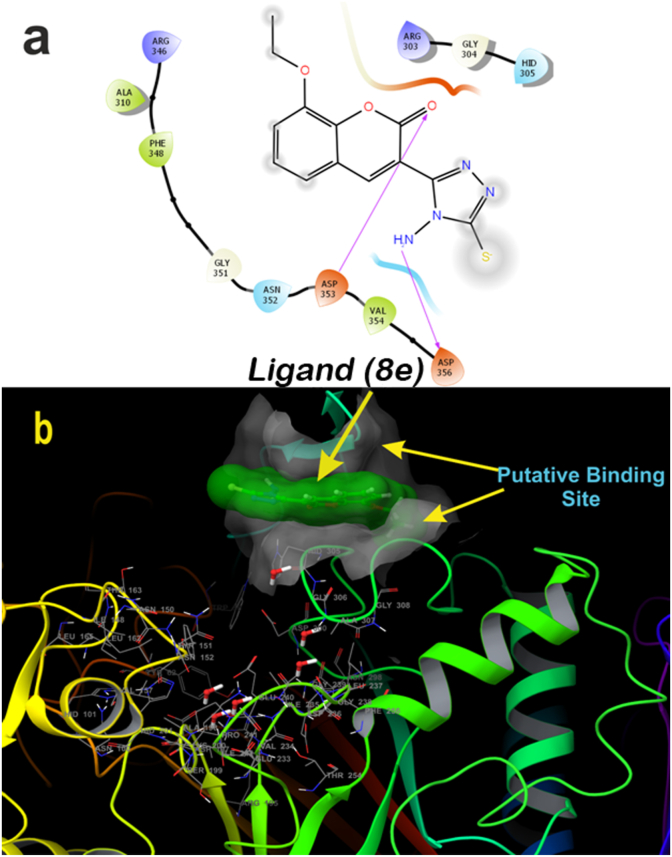
A molecular docking study gives an insight view of potent molecule interacting in a putative binding site. With this in background to understand the mechanism of action, and the binding site, the synthesized small molecules were executed on to molecular docking platform for virtual screening. Molecular docking results suggest that ligand 8c and 8e possess high affinity towards the active site of α-amylase and ACE [65]. Ligand 8c forms hydrogen bond with Glu384, His513 along with catalytically water molecule and also forms interact with Zinc metal via salt bridge (Figure 9a). ACE, a metalloproteinase containing Zinc at the activity site, is very essential for catalysis. Any interaction with Zinc suggests that the ligand might have competitive binding with the enzyme, which though it is very essential criteria for a good inhibitor in drug discovery [65]. Three-dimensional view of 8c suggests that the ligand is deeply embedded with docking score of -5.60 (kcal/mol) (Figure 9b). Whereas ligand 8e also bind with similar fashion as that of 8c at active site by forming hydrogen bond with amino acids His513, Glu384, Tyr523 and with catalytic water molecules surrounding at the active site, along with salt bridge with Zinc atom (Figure 10a), with the docking core of -5.87 (kcal/mol). Molecular docking results were validated by using a standard inhibitor Lisinopril, which forms hydrogen bond with His513, Glu384, Tyr523, Try520, Lys511, Arg523, Ala356, Glu162 and salt bridge with Zinc and Glu162 (Figure 11a). These interactions with amino acids are very important during catalysis [66, 67]. The three-dimensional view of the standard Lisinopril shows that the Lisinopril deeply binds at the active site (Figure 11b).
Figure 9.
Two-dimensional putative binding pose of ligand 8c with ACE (PDB ID: 1O86) (a); Three-dimensional geometric pose wherein 8c deeply embedded at the active site of ACE (b).
Figure 10.
Two-dimensional putative binding pose of ligand 8e with ACE (PDB ID: 1O86) (a); Three-dimensional geometric pose wherein 8e deeply embedded at the active site of ACE (b).
Figure 11.
Two-dimensional binding pose of standard inhibitor Lisinopril with ACE (PDB ID: 1O86) (a). Three-dimensional geometric pose wherein Lisinopril deeply embedded at the active site of ACE (b).
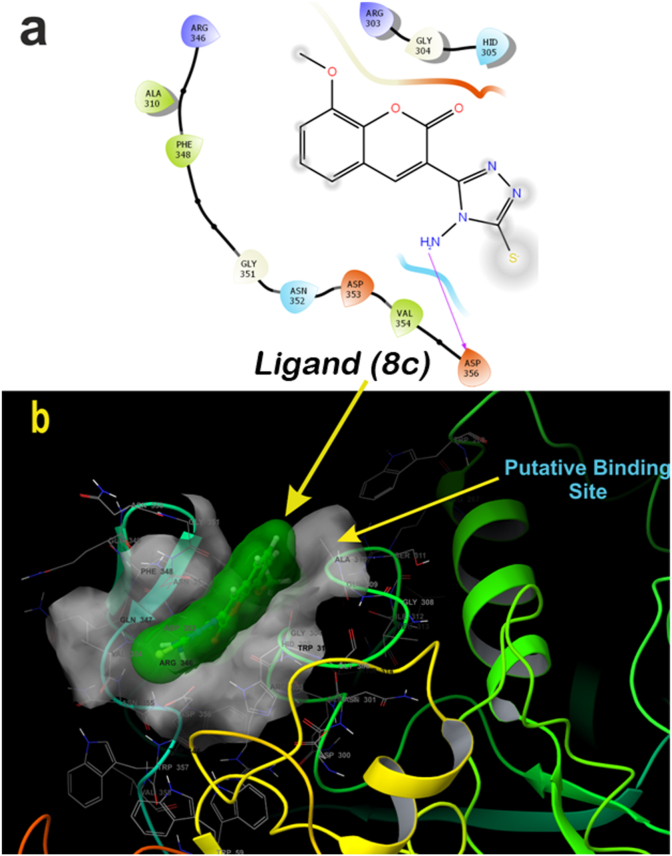
Ligand 8c and 8e also binds as similar fashion as that of standard inhibitor binds with ACE. Whereas in case of α-amylase, ligand 8c and 8e also binds efficiently with docking score of -3.42 and -3.34 (kcal/mol) respectively. Ligand 8c binds with hydrogen bond with Asp356 (Figure 12a) and three-dimensional view depicts it bind a bit far of 5Å from chloride ion (Figure 12b). Ligand 8e also follows the same pattern of binding with Asp353 and Asp356 amino acids via hydrogen bond (Figures 13a and 13b). A maximum violation of 4 or above is considered as the ligand is not able to pass through ADME studies. Whereas in case of 8(a-e) possess all the different criteria ranges, which a 95% of drug warranties to have, with these cut-off ranges it is suggested that 8(a-e) possess good ADME values within the range as mentioned in (Table 4). Hence these results suggest that the ligand 8c and 8e is potent ligands among the series of 8(a-e) based on molecular docking (Table 5) and ADME studies up to 95% of drugs with zero violation criteria of Lipinski's rule as mentioned (Table 4).
Figure 12.
Two-dimensional binding pose of standard inhibitor ligand 8c with α-amylase (PDB ID: 4W93) (a). Three-dimensional geometric pose wherein ligand 8c deeply embedded at putative binding site of α-amylase (b).
Figure 13.
Two-dimensional binding pose of standard inhibitor ligand 8e with α-amylase (PDB ID: 4W93) (a). Three-dimensional geometric pose wherein ligand 8e deeply embedded at putative binding site of α-amylase (b).
Table 4.
Computer aided ADME screening of the synthesized compounds, 8 (a-e).
| Ligand | a∗ | b∗ (Å3) | c∗ (Å3) | d∗ (Å3) | e∗ (Å3) | f∗ (Å3) | g∗ (Å3) | h∗ (Å3) | i∗ (nm/sec) | j∗ (Å3) | k∗ (nm/sec) | l∗ (Kp in m/hr) | m∗ (Å3) | n∗ (%) | o∗ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8a | 26.70 | 9.78 | 18.66 | 13.27 | 0.73 | -2.32 | -3.32 | -5.33 | 71.69 | -0.23 | 229.40 | -5.78 | -0.40 | 64.40 | 0 |
| 8b | 26.35 | 9.67 | 18.39 | 13.27 | 0.66 | -2.23 | -2.50 | -5.29 | 71.68 | -0.24 | 213.35 | -5.78 | -0.42 | 63.99 | 0 |
| 8c | 26.89 | 9.41 | 19.47 | 13.74 | 0.31 | -1.81 | -2.18 | -5.28 | 73.45 | -0.47 | 88.81 | -5.68 | -0.49 | 62.13 | 0 |
| 8d | 26.81 | 9.40 | 19.77 | 13.72 | 0.30 | -1.77 | -2.18 | -5.23 | 73.42 | -0.46 | 88.76 | -5.69 | -0.49 | 62.09 | 0 |
| 8e | 29.18 | 10.02 | 19.90 | 13.63 | 0.69 | -2.33 | -2.43 | -5.76 | 73.67 | -0.63 | 89.09 | -5.59 | -0.41 | 64.41 | 0 |
| Lisinopril | 40.97 | 14.93 | 26.56 | 19.58 | -1.19 | -1.70 | -1.20 | -2.31 | 0.16 | -1.96 | 0.11 | -8.09 | -0.92 | 5.66 | 0 |
| A# | 13 to 70 | 4 to 18 | 8 to 35 | 4 to 45 | -2 to 6.5 | -6.5 to 0.5 | < -5 | <25 poor to >500 good | -3 to 1.2 | <25 poor to >500 good | -- | -1.5 to 1.5 | <25% poor | Max is 4 |
A# - 95% drug possess these criteria ranges: Note: a∗-QPpolrz; b∗-QPlogPC16; c∗-QPlogPoct; d∗-QPlogPw; e∗-QPlogPo/w; f∗-QPlogS; g∗-CIQPlogS; h∗-QPlogHERG; i∗-QPPCaco; j∗-QPlogBB; k∗-QPPMDCK; l∗-QPlogKp; m∗-QPlogKhsa; n∗-Percent Human Oral Absorption; o∗-Rule of Five.
Table 5.
The Docking scores of the compounds, 8 (a-e) against α-Amylase and ACE. Docking scores and Glide energies (kcal/mol).
| Protein |
α-Amylase (PDB ID: 4W93) |
Angiotensin-converting enzyme (PDB ID: 1O86) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | Docking Score | Glide Evdw | Glide Energy | Glide Emodel | XP Hbond | Docking Score | Glide Evdw | Glide Energy | Glide Emodel | XP Hbond |
| 8a | -2.96 | -24.58 | -25.61 | -29.02 | -0.61 | -2.78 | -19.17 | -23.99 | -20.13 | -1.14 |
| 8b | -3.09 | -24.96 | -26.76 | -31.14 | -0.90 | -2.88 | -17.98 | -22.91 | -17.84 | -1.18 |
| 8c | -3.42 | -23.42 | -33.01 | -41.38 | -0.77 | -5.60 | -6.13 | -19.01 | -26.81 | -2.87 |
| 8d | -2.47 | -23.91 | -22.77 | -23.43 | 0.00 | -5.33 | -15.14 | -30.52 | -9.84 | -1.98 |
| 8e | -3.34 | -27.60 | -28.71 | -31.72 | -0.85 | -5.87 | -6.05 | -19.67 | -30.01 | -3.07 |
| Lisinopril | -- | -- | -- | -- | -- | -16.70 | -39.88 | -75.39 | -125.99 | -3.95 |
4. Conclusion
In summary, a series of new coumarin-triazole hybrids 8 (a-e) synthesized and characterized, and the crystal and molecular structure of the intermediate 3a was confirmed by crystallographic and computational studies. The compound 3a is stabilized through the formation of supramolecular ring motif [R22 (14), R22 (10) and R21 (6)]. The HSA shows that, the intermolecular O…H (25.4%) interactions are the main contributor amongst the other interactions. The DFT studies revealed the good correlation between the calculated and experimental structural parameters. The energy gap (4.0463 eV) value indicating the molecule 3a is chemically hard and is less reactive. Further, the α-amylase inhibition assay results of compounds 8 (a-e) show promising inhibitory potentials. Amongst them, compounds 8c and 8e found remarkable with the IC50 values of 5.43μM and 5.98μM, and therefore these might act as leads as diabetic agents, which was supported by the comparative statistical analysis and ADME calculation results.
Declarations
Author contribution statement
Vagish Channa Basappa: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Vivek Hamse Kameshwar, Karthik Kumara: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Dileep Kumar Achutha, Lokanath Neratur Krishnappagowda: Analyzed and interpreted the data.
Ajay Kumar Kariyappa: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors are grateful to the Department of Physics, and IOE Instrumentation Facility, Vijnana Bhavana, University of Mysore, for recording X-ray diffraction and spectral analysis.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Odeyemi S., Dewar J. In vitro antidiabetic activity affecting glucose uptake in hepg2 cells following their exposure to extracts of Lauridia tetragona (L.f.) R.H. Archer. Processes. 2020;8:33. [Google Scholar]
- 2.Holman N., Young B., Gadsby R. Current prevalence of Type 1 and Type 2 diabetes in adults and children in the UK. Diab. Med. 2015;32:1119–1120. doi: 10.1111/dme.12791. [DOI] [PubMed] [Google Scholar]
- 3.Petrie J.R., Guzik T.J., Touyz R.M. Diabetes, hypertension, and cardiovascular disease: clinical insights and vascular mechanisms. Can. J. Cardiol. 2018;34:575–584. doi: 10.1016/j.cjca.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo H.J., Youn S.W. Zn(II)-Catalyzed one-pot synthesis of coumarins from ynamides and salicylaldehydes. Org. Lett. 2019;21:3422–3426. doi: 10.1021/acs.orglett.9b01181. [DOI] [PubMed] [Google Scholar]
- 5.Tasior M., Poronik Y.M., Vakuliuk O., Sadowski B., Karczewski M., Gryko D.T. V-Shaped bis-coumarins: synthesis and optical properties. J. Org. Chem. 2014;79:8723–8732. doi: 10.1021/jo501565r. [DOI] [PubMed] [Google Scholar]
- 6.Bouasla S., Amaro-Gahete J., Esquivel D., Lopez M.I., Jimenez-Sanchidrian C., Teguiche M., Romero-Salguero F.J. Coumarin derivatives solvent-free synthesis under microwave irradiation over heterogeneous solid catalysts. Molecules. 2017;22 doi: 10.3390/molecules22122072. 2072-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrowska K. Coumarin-piperazine derivatives as biologically active compounds. Saudi Pharmaceut. J. 2020;28:220–232. doi: 10.1016/j.jsps.2019.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renuka N., Ajay Kumar K. Synthesis and biological evaluation of fused pyrans bearing coumarin moiety as potent antimicrobial agents. Philip. J. Sci. 2015;144:91–96. [Google Scholar]
- 9.Peng X.-M., Damu G.L.V., He Zhou C. Current developments of coumarin compounds in medicinal chemistry. Curr. Pharmaceut. Des. 2013;19:3884–3930. doi: 10.2174/1381612811319210013. [DOI] [PubMed] [Google Scholar]
- 10.Nagamallu R., Gurunanjappa P., Kariyappa A.K. Synthesis of coumarin appended 1,3-oxazines as potent antimicrobial and antioxidant agents. Pharm. Chem. J. 2017;51:582–589. [Google Scholar]
- 11.Hwu J.R., Lin S.-Y., Tsay S.-C., De Clercq E., Leyssen P., Neyts J. Coumarin−Purine ribofuranoside conjugates as new agents against Hepatitis C Virus. J. Med. Chem. 2011;54:2114–2126. doi: 10.1021/jm101337v. [DOI] [PubMed] [Google Scholar]
- 12.Xia J., Huang X., Cai M. Heterogeneous Copper(I)-catalyzed cascade addition–oxidative cyclization of nitriles with 2-aminopyridines or amidines: efficient and practical synthesis of 1,2,4-triazoles. Synthesis. 2019;51:2014–2022. [Google Scholar]
- 13.Yang N., Yuan G. A multicomponent electrosynthesis of 1,5-disubstituted and 1-aryl 1,2,4-triazoles. J. Org. Chem. 2018;83:11963–11969. doi: 10.1021/acs.joc.8b01808. [DOI] [PubMed] [Google Scholar]
- 14.Liu J.-Q., Shen X., Wang Y., Wang X.-S., Bi X. [3+ 2] cycloaddition of isocyanides with aryl diazonium salts: catalyst-dependent regioselective synthesis of 1,3- and 1,5-disubstituted 1, 2, 4-triazoles. Org. Lett. 2018;20:6930–6933. doi: 10.1021/acs.orglett.8b03069. [DOI] [PubMed] [Google Scholar]
- 15.Jatangi N., Tumula N., Palakodety R.K., Nakka M. I2-Mediated oxidative C–N and N–S bond formation in water: a metal-free synthesis of 4, 5-disubstituted/n-fused 3-amino-1, 2, 4-triazoles and 3-substituted 5-amino-1, 2, 4-thiadiazoles. J. Org. Chem. 2018;83:5715–5723. doi: 10.1021/acs.joc.8b00753. [DOI] [PubMed] [Google Scholar]
- 16.Azzouni S., Abdelli A., Gaucher A., Arfaouni Y., Efrit M.L., Prim D. From imidates to vinyl-1,2,4-triazoles: synthesis, mechanistic aspects and first issues of their reactivity. Tetrahedron. 2018;74:6972–6978. [Google Scholar]
- 17.Bechara W.S., Khazhieva I.S., Rodriguez E., Charette A.B. One-pot synthesis of 3,4,5-trisubstituted 1,2,4-triazoles via the addition of hydrazides to activated secondary amides. Org. Lett. 2015;17:1184–1187. doi: 10.1021/acs.orglett.5b00128. [DOI] [PubMed] [Google Scholar]
- 18.Beyzaei H., Ghanbari Kudeyani M., Samareh Delarami H., Aryan R. Synthesis, antimicrobial and antioxidant evaluation, and molecular docking study of 4,5-disubstituted 1,2,4-triazole-3-thiones. J. Mol. Struct. 2020;1215:128273. [Google Scholar]
- 19.Thakkar S.S., Thakor P., Doshi H., Ray A. 1,2,4-Triazole and 1,3,4-oxadiazole analogues: synthesis, MO studies, in silico molecular docking studies, antimalarial as DHFR inhibitor and antimicrobial activities. Bioorg. Med. Chem. 2017;25:4064–4075. doi: 10.1016/j.bmc.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 20.Agiso H.A., Esatu H., Hairat S., Zaki M. TBHP/TBAI–Mediated simple and efficient synthesis of 3,5-disubstituted and 1,3,5-trisubstituted 1H-1,2,4-triazoles via oxidative decarbonylation of aromatic aldehydes and testing for antibacterial activities. Tet. Lett. 2020;61:151989. [Google Scholar]
- 21.Vagish C.B., Sudeep P., Jayadevappa H.P., Ajay Kumar K. 1,2,4-Triazoles: Synthetic and medicinal perspectives. Int. J. Curr. Res. 2020;12(8):12950–12960. [Google Scholar]
- 22.Aboeldahab A.M.B., Beshi E.M.A., Shoman M.E., Rabea S.M., Aly O.M. Spirohydantoins and 1,2,4-triazole-3-carboxamide derivatives as inhibitors of histone deacetylase: design, synthesis, and biological evaluation. Eur. J. Med. Chem. 2018;146:79–92. doi: 10.1016/j.ejmech.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Dehestani L., Ahangar N., Hasemi S.M., Irannejad H., Masihi P.H., Shakiba A., Emami S. Design, synthesis, in vivo and in silico evaluation of phenacyl triazole hydrazones as new anticonvulsant agents. Bioorg. Chem. 2018;78:119–129. doi: 10.1016/j.bioorg.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Aouad M.R., Al-Mohammad H.M., Al-blewi F.F., Ihmaid S., Elbadawy H.M., Althagfan S.S., Rezki N. Introducing of acyclonucleoside analogues tethered 1,2,4-triazole as anticancer agents with dual epidermal growth factor receptor kinase and microtubule inhibitors. Bioorg. Chem. 2020;94:103446. doi: 10.1016/j.bioorg.2019.103446. [DOI] [PubMed] [Google Scholar]
- 25.Fadaly W.A.A., Elshaier Y.A.M.M., Hassanein E.H.M., Abdellatif K.R.A. New 1,2,4-triazole/pyrazole hybrids linked to oxime moiety as nitric oxide donor celecoxib analogs: synthesis, cyclooxygenase inhibition anti-inflammatory, ulcerogenicity, anti-proliferative activities, apoptosis, molecular modeling and nitric oxide release studies. Bioorg. Chem. 2020;98:103752. doi: 10.1016/j.bioorg.2020.103752. [DOI] [PubMed] [Google Scholar]
- 26.Renuka N., Vivek H.K., Pavithra G., Ajay Kumar K. Synthesis of coumarin appended pyrazolyl-1,3,4-oxadiazoles and pyrazolyl-1,3,4-thiadiazoles: evaluation of their in vitro antimicrobial and antioxidant activities and molecular docking studies. Russ. J. Bioorg. Chem. 2017;43:197–210. [Google Scholar]
- 27.Avco A., Tasci H., Kandemir U., Can O.D., Gokhan Kelekci N., Tozkoparan B. Synthesis, characterization, and in vivo pharmacological evaluation of novel mannich bases derived from 1,2,4-triazole containing a naproxen moiety. Bioorg. Chem. 2020;100:103892. doi: 10.1016/j.bioorg.2020.103892. [DOI] [PubMed] [Google Scholar]
- 28.Bakri Y.E., Marmouzi I., Jemli M.E., Anouar E.H., Karthikeyan S., Harmaoui A., Faouzi M.E.A., Mague J.T., Essassi E.M. Synthesis, biological activity and molecular modeling of a new series of condensed 1,2,4-triazoles. Bioorg. Chem. 2019;92:103193. doi: 10.1016/j.bioorg.2019.103193. [DOI] [PubMed] [Google Scholar]
- 29.Yeye E.O., Kanwal, Khan K.M., Chigurupati S., Wadood A., Rehman A.U., Perveen S., Maharajan M.K., Shamim S., Hameed S., Aboaba S.A., Taha M. Syntheses, in vitro α-amylase and α-glucosidase dual inhibitory activities of 4-amino-1,2,4-triazole derivatives their molecular docking and kinetic studies. Bioorg. Med. Chem. 2020;28:115467. doi: 10.1016/j.bmc.2020.115467. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick G. Phase annealing in SHELX-90: direct methods for larger structures. Acta Crystallogr. 1990;A46:467–473. [Google Scholar]
- 31.Sheldrick G.M. SHELXT - integrated space-group and crystal-structure determination. Acta crysta. 2015;A71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spek A. PLATON, an integrated tool for the analysis of the results of a single crystal structure determination. Acta Crystallogr. 1990;A46:c34. [Google Scholar]
- 33.Macrae C.F., Bruno I.J., Chisholm J.A., Edgington P.R., McCabe P., Pidcock E., Rodriguez-Monge L., Taylor R., van de Streek J., Wood P.A. Mercury CSD 2.0– new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41:466–470. [Google Scholar]
- 34.Turner M.J., McKinnon J.J., Wolff S.K., Grimwood D.J., Spackman P.R., Jayatilaka D., Spackman M.A. University of Western Australia; 2018. CrystalExplorer (Version 17.5) [Google Scholar]
- 35.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. 1988;B37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 36.Becke A.D. A new mixing of Hartree–Fock and local density-functional theories. J. Chem. Phys. 1993;98:1372–1377. [Google Scholar]
- 37.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., Caricato M., Marenich A.V., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., Izmaylov A.F., Sonnenberg J.L., Williams, Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M.J., Heyd J.J., Brothers E.N., Kudin K.N., Staroverov V.N., Keith T.A., Kobayashi R., Normand J., Raghavachari K., Rendell A.P., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Morokuma K., Farkas O., Foresman J.B., Fox D.J. Rev. C.01; Wallingford, CT: 2016. Gaussian 16. [Google Scholar]
- 38.Frisch A., Hratchian H.P., Dennington R.D., Keith T.A., Millam J., Nielsen B., Holder A.J., Hiscocks J. Gaussview 6.0.8. In: Wallingford C., editor. USA. 2009. Wallingford, CT. [Google Scholar]
- 39.Sudha P., Zinjarde S.S., Bhargava S.Y., Kumar A.R. Potent α-amylase inhibitory activity of Indian Ayurvedic medicinal plants, BMC Complement. Altern. Med. 2011;11:5. doi: 10.1186/1472-6882-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natesh R., Schwager S.L.U., Sturrock E.D., Acharya K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature. 2003;421(6922):551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 41.Williams L.K., Zhang X., Caner S., Tysoe C., Nguyen N.T., Wicki J., Williams D.E., Coleman J., McNeill J.H., Yuen V., Andersen R.J., Withers S.G., Brayer G.D. The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif. Nat. Chem. Biol. 2015;11:691–696. doi: 10.1038/nchembio.1865. [DOI] [PubMed] [Google Scholar]
- 42.Kameshwar V.H., Kumar J.R., Priya B.S., Swamy S.N. Synthesis, characterization and bioactivity studies of novel 1,3,4-oxadiazole small molecule that targets basic phospholipase A2 from Vipera russelli. Mol. Cell. Biochem. 2016;426:161–175. doi: 10.1007/s11010-016-2888-6. [DOI] [PubMed] [Google Scholar]
- 43.Chimenti F., Secci D., Bolasco A., Chimenti P., Granese A., Befani O., Turini P., Alcaro S., Ortuso F. Inhibition of monoamine oxidases by coumarin-3-acyl derivatives: biological activity and computational study. Bioorg. Med. Chem. Lett. 2004;14:3697–3703. doi: 10.1016/j.bmcl.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Horning E.C., Horning M.G. Coumarins from 2-hydroxy-3-methoxybenzaldehyde. J. Am. Chem. Soc. 1947;69:968–969. doi: 10.1021/ja01196a506. [DOI] [PubMed] [Google Scholar]
- 45.Han S., Zhang F.-F., Qian H.-Y., Chen L.-L., Pu J.-B., Xie X., Chen J.-Z. Design, syntheses, structure–activity relationships and docking studies of coumarin derivatives as novel selective ligands for the CB2 receptor. Eur. J. Med. Chem. 2015;93:16–32. doi: 10.1016/j.ejmech.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 46.Patil S.A., Unki S.N., Kulkarni A.D., Naik V.H., Badami P.S. Synthesis, characterization, in vitro antimicrobial and DNA cleavage studies of Co(II), Ni(II) and Cu(II) complexes with ONOO donor coumarin Schiff bases. J. Mol. Struct. 2011;985:330–338. [Google Scholar]
- 47.Ghalehshahi H.G., Balalaie S., Sohbati H.R., Azizian H., Alavijeh M.S. Synthesis, CYP 450 evaluation, and docking simulation of novel 4-aminopyridine and coumarin derivatives. Arch. der Pharm. 2019;352 doi: 10.1002/ardp.201800247. [DOI] [PubMed] [Google Scholar]
- 48.Raslan M.A., Khalil M.A. Heterocyclic synthesis containing bridgehead nitrogen atom: synthesis of 3-[(2H)-2-oxobenzo[b]pyran-3-yl]-s-triazolo[3,4-b]-1,3,4-thiadiazine and thiazole derivatives. Heteroat. Chem. 2003;14:114–120. [Google Scholar]
- 49.Bhat M.A., Siddiqui N., Khan S.A., Mohamed M.I. Synthesis of triazolothiazolidinone derivatives of coumarin with antimicrobial activity. Acta. Pol. Pharm. 2009;66:625–632. [PubMed] [Google Scholar]
- 50.Aliya I., Zaib S., Jabeen F., Iqbal J., Saeed A. Unraveling the alkaline phosphatase inhibition, anticancer, and antileishmanial potential of coumarin–triazolothiadiazine hybrids: design, synthesis, and molecular docking analysis. Arch. der Pharm. 2016;349:553–565. doi: 10.1002/ardp.201500392. [DOI] [PubMed] [Google Scholar]
- 51.Santos-Contreras R.J., Martínez-Martínez F.J., García-Báez E.V., Padilla-Martínez I.I., Peraza A.L., Höpfl H. Carbonyl–carbonyl, carbonyl–π and carbonyl–halogen dipolar interactions as the directing motifs of the supramolecular structure of ethyl 6-chloro-2-oxo-2H-chromene-3-carboxylate and ethyl 6-bromo-2-oxo-2H-chromene-3-carboxylate. Acta Crystallogr. 2007;C63:o239–o242. doi: 10.1107/S0108270107008712. [DOI] [PubMed] [Google Scholar]
- 52.Allen F.H. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr. 2002;B58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 53.Channabasappa V., Kumara K., Neratur L.K., Kariyappa A.K. Synthesis, crystal structure studies and Hirshfeld surface analysis of 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one. Chem. Data Coll. 2018;15(16):134–142. [Google Scholar]
- 54.Seth S.K. Structural elucidation and contribution of intermolecular interactions in O-hydroxy acyl aromatics: insights from X-ray and Hirshfeld surface analysis. J. Mol. Struct. 2014;1064:70–75. [Google Scholar]
- 55.Spackman M.A., McKinnon J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm. 2002;4(66):378–392. [Google Scholar]
- 56.McKinnon J.J., Jayatilaka D., Spackman M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Comm. 2007;37:3814–3816. doi: 10.1039/b704980c. [DOI] [PubMed] [Google Scholar]
- 57.Kumara K., Jyothi M., Naveen S., Khanum S.A., Lokanath N.K. Synthesis, characterization, crystal structure and Hirshfeld surface analysis of 1-(4-ethoxyphenyl)-3-(4-methylphenyl) prop-2en-1-one. Chem. Data Coll. 2017;9:152–163. [Google Scholar]
- 58.Seth S.K. Tuning the formation of MOFs by pH influence: X-ray structural variations and Hirshfeld surface analyses of 2-amino-5-nitropyridine with cadmium chloride. CrystEngComm. 2013;15:1772–1781. [Google Scholar]
- 59.Luo Y.H., Wu G.G., Mao S.L., Sun B.W. Complexation of different metals with a novel N-donor bridging receptor and Hirshfeld surfaces analysis. Inorg. Chim. Acta. 2013;397:1–9. [Google Scholar]
- 60.Bernstein J., Davis R.E., Shimoni L., Chang N.-L. Patterns in hydrogen bonding: functionality and graph set analysis in crystals. Angew Chem. Int. Ed. Engl. 1995;34:1555–1573. [Google Scholar]
- 61.Jyothi K.L., Kumara K., Hema M.K., Mahesha, Gautam R., Guru Row T.N., Lokanath N.K. Structural elucidation, theoretical insights and thermal properties of three novel multicomponent molecular forms of gallic acid with hydroxypyridines. J. Mol. Struct. 2020;1207:127828. [Google Scholar]
- 62.Kumara K., Harish K.P., Shivalingegowda N., Tandon H.C., Mohana K.N., Lokanath N.K. Crystal structure studies, Hirshfeld surface analysis and DFT calculations of novel 1-[5-(4-methoxy-phenyl)-[1,3,4]oxadiazol-2-yl]-piperazine derivatives. Chem. Data Coll. 2017;11–12:40–58. [Google Scholar]
- 63.Kumara K., Kumar A.D., Kumar K.A., Lokanath N.K. Synthesis, spectral and X-ray crystal structure of 3-(3-methoxyphenyl)-5-(3-methylthiophen-2-yl)-4,5-dihydro-1H-pyrazole-1-carboxamide: Hirshfeld surface, DFT calculations and thermo-optical studies. Chem. Data Coll. 2018;13–14:40–59. [Google Scholar]
- 64.Kumara K., Kumar A.D., Naveen S., Kumar K.A., Lokanath N.K. Synthesis, spectral characterization and X-ray crystal structure studies of 3-(benzo[d][1,3]dioxol-5-yl)-5-(3-methylthiophen-2-yl)-4,5-dihydro-1H-pyrazole-1-carboxamide: Hirshfeld surface, DFT and thermal analysis. J. Mol. Struct. 2018;1161:285–298. [Google Scholar]
- 65.Xavier S., Periandy S., Ramalingam S. NBO, conformational, NLO, HOMO–LUMO, NMR and electronic spectral study on 1-phenyl-1-propanol by quantum computational methods, Spectrochim. Acta Part A: Mol. Biomol. Spect. 2015;137:306–320. doi: 10.1016/j.saa.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 66.Savitha M.N., Siddesha J.M., Suvilesh K.N., Yariswamy M., Vivek H.K., D’Souza C.J.M., Umashankar M., Vishwanath B.S. Active-site directed peptide l-Phe-d-His-l-Leu inhibits angiotensin converting enzyme activity and dexamethasone-induced hypertension in rats. Pept. 2019;112:34–42. doi: 10.1016/j.peptides.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 67.Fang L., Geng M., Liu C., Wang J., Min W., Liu J. Structural and molecular basis of angiotensin-converting enzyme by computational modeling: insights into the mechanisms of different inhibitors. PloS One. 2019;14 doi: 10.1371/journal.pone.0215609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.