As of March 9, 2020, more than 100,000 cases of coronavirus disease-2019 (COVID-19) were reported in more than 100 countries with thousands deaths globally. It is now known that Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is a new type of coronavirus causing COVID-19 infection (1). The most common clinical feature of SARS-CoV-2 infection is fever (2). Moreover, acute respiratory distress syndrome (ARDS) is the most frequent cause of admission to intensive care unit in COVID-19 patients (1). Lactate dehydrogenase (LDH), a key enzyme in the glycolytic pathway and a cytoplasmic enzyme found in most organs, has been linked to inflammation response and cell damage. Currently, the role of serum LDH levels in ARDS patients infected by SARS-CoV-2 is unclear.
Between January 30 and Feb 22, 2020, 77 fever patients diagnosed with SARS-CoV-2 infection were admitted to the hospital of Changsha Public Health Center. In all patients, fever was defined assessed as follows: reported a fever history during the time from the onset symptom to admission, fever was defined as a rise in body temperature and presence of axillary temperature ≥37.0 °C. Exclusion criteria included onset symptoms without fever, and patients with cancer. Clinical information of COVID-19 patients such as age, gender, days from onset of symptoms, medical history, physical examination, clinical presentation, laboratory tests, and imaging studies during admission were collected. Laboratory findings including erythrocyte sedimentation rate, C-reactive protein, procalcitonin, liver and renal function, blood chemistry, coagulation test, complete blood count, LDH and creatine kinase were collected. ARDS was diagnosed as a decrease in the PaO2/FiO2 index below 300 mmHg according to the Berlin definition.
Data were statistical analyzed with Student’s t-test, Mann-Whitney U-test, Fisher exact test and Chi-square analysis. Variables that were significant on univariate analysis were included in multivariate logistic regression analysis. Receiver-operator characteristic (ROC) analysis for ARDS was applied to determine the cut-off point and area under the curve (AUC). Survival curves without ARDS were established using Kaplan-Meier method and the log-rank test.
The 77 fever patients were categorized as non-ARDS group (n=63, 81.81%) and ARDS group (n=14, 18.19%). The baseline characteristics are shown in Table 1. Univariate and multivariable regression analyses identified that serum LDH level was a predictor of ARDS in SARS-CoV-2-infected patients with fever (Table 2, OR: 1.02; 95% CI, 1.00–1.04). The AUC of ROC curve showing the ability of LDH levels to predict development of ARDS was 0.809, and the best threshold of ≥273 U/L on admission revealed a sensitivity of 57.1% and a specificity of 93.7% (Figure 1). Survival curves for development of ARDS are showed in Figure 2. Analysis of the curves indicate that patients with LDH ≥273 U/L are more likely to develop ARDS (P<0.001, log-rank test).
Table 1. Characteristics of SARS-CoV-2-infected patients with fever.
| Characteristics | Total (n=77) | Non-ARDS (n=63) | ARDS (n=14) | P value |
|---|---|---|---|---|
| Age, years, mean (SD) | 47.99±15.75 | 46.37±15.65 | 55.29±14.57 | 0.055 |
| Gender, n (%) | 0.073 | |||
| Female | 33 (42.86%) | 30 (47.62%) | 3 (21.43%) | |
| Male | 44 (57.14%) | 33 (52.38%) | 11 (78.57%) | |
| Hubei exposure | 52 (67.53%) | 42 (66.67%) | 10 (71.43%) | 0.731 |
| Systolic pressure, mmHg, mean (SD) | 124.47±11.87 | 124.95±11.88 | 122.29±12.03 | 0.451 |
| Diastolic pressure, mmHg, mean (SD) | 78.30±8.96 | 78.70±9.17 | 76.50±8.03 | 0.410 |
| Heart rate, beats per min, mean (SD) | 91.01±13.87 | 88.73±13.44 | 101.29±11.16 | 0.002 |
| Respiratory rate, IQR | 20 [20, 21] | 20 [20, 20] | 21.5 [20, 23] | <0.001 |
| Days from illness onset to first hospital admission (days), IQR | 5 [3, 7] | 5 [3, 7.5] | 4.5 [3, 6.75] | 0.779 |
| Signs and symptoms, n (%) | ||||
| Fatigue | 32 (41.56%) | 27 (42.86%) | 5 (35.71%) | 0.624 |
| Cough | 48 (62.34%) | 38 (60.32%) | 10 (71.43%) | 0.438 |
| Anorexia | 2 (2.60%) | 2 (3.17%) | 0 (0.00%) | 0.499 |
| Myalgia | 8 (10.39%) | 6 (9.52%) | 2 (14.29%) | 0.597 |
| Dyspnea | 11 (14.29%) | 5 (7.94%) | 6 (42.86%) | <0.001 |
| Expectoration | 20 (25.97%) | 18 (28.57%) | 2 (14.29%) | 0.270 |
| Sore throat | 5 (6.49%) | 5 (7.94%) | 0 (0.00%) | 0.276 |
| Dizziness | 3 (3.90%) | 3 (4.76%) | 0 (0.00%) | 0.405 |
| Headache | 7 (9.09%) | 5 (7.94%) | 2 (14.29%) | 0.455 |
| Any comorbidity, n (%) | ||||
| Hypertension | 14 (18.18%) | 9 (14.29%) | 5 (35.71%) | 0.060 |
| Cardiovascular disease | 3 (3.90%) | 2 (3.17%) | 1 (7.14%) | 0.488 |
| Diabetes | 4 (5.19%) | 3 (4.76%) | 1 (7.14%) | 0.717 |
| Laboratory tests | ||||
| White blood cell count, ×109/L, mean (SD) | 4.52±1.52 | 4.62±1.41 | 4.07±1.91 | 0.076 |
| Neutrophil count, ×109/L, mean (SD) | 3.05±1.30 | 3.04±1.20 | 3.14±1.72 | 0.787 |
| Lymphocyte count, ×109/L, mean (SD) | 1.52±3.63 | 1.69±4.00 | 0.75±0.34 | 0.001 |
| Hemoglobin, g/L, mean (SD) | 130.16±21.49 | 128.98±22.67 | 135.43±14.62 | 0.313 |
| Platelet count, ×109/L, mean (SD) | 173.06±113.71 | 177.16±123.21 | 154.64±52.43 | 0.506 |
| C-reactive protein, mg/L, mean (SD) | 19.8 (8.6, 39) | 16.2 (7.6, 38.1) | 33 (21.9, 74.8) | 0.011 |
| Procalcitonin, ng/mL, IQR | 0.05 (0.05, 0.05) | 0.05 (0.05, 0.05) | 0.05 (0.05, 0.05) | 0.465 |
| Erythrocyte sedimentation rate, mm/h, IQR | 48 [27, 72] | 46 [25, 77] | 51.5 [45.5, 66] | 0.837 |
| Alanine aminotransferase, U/L, mean (SD) | 24.90±12.84 | 24.44±13.01 | 26.98±12.29 | 0.507 |
| Aspartate aminotransferase, U/L, mean (SD) | 30.09±14.03 | 27.87±11.63 | 40.08±19.32 | 0.003 |
| Albumin, g/L, IQR | 37 [34.7, 39.1] | 38 [35, 40] | 34 [31.9, 35.6] | 0.002 |
| Total bilirubin, mmol/L, mean (SD) | 12.64±5.21 | 12.61±5.29 | 12.79±5.07 | 0.909 |
| Direct bilirubin, mmol/L, mean (SD) | 4.64±2.39 | 4.52±2.30 | 5.19±2.78 | 0.343 |
| Lactate dehydrogenase, U/L, mean (SD) | 204.30±75.70 | 187.88±61.97 | 278.20±89.41 | <0.001 |
| Creatinine, μmol/L, mean (SD) | 56.17±24.66 | 56.78±26.57 | 53.42±13.36 | 0.648 |
| Blood urea nitrogen, mmol/L, mean (SD) | 4.43±1.87 | 4.26±1.89 | 5.18±1.60 | 0.097 |
| Uric acid, μmol/L, mean (SD) | 258.68±99.46 | 261.41±104.09 | 246.41±77.22 | 0.613 |
| Prothrombin time, s, mean (SD) | 11.95±0.98 | 11.83±1.00 | 12.50±0.67 | 0.020 |
| Activated partial thromboplastin time, s, mean (SD) | 32.94±3.57 | 33.06±3.66 | 32.39±3.20 | 0.526 |
| D-dimer, ug/mL, IQR | 0.2 (0.1-0.4) | 0.2(0.1, 0.4) | 0.4 (0.1, 0.5) | 0.065 |
| Chest radiography, n (%) | ||||
| Unilateral pneumonia | 6 (7.79%) | 6 (9.52%) | 0 (0.00%) | 0.229 |
| Bilateral pneumonia | 64 (83.12%) | 50 (79.37%) | 14 (100.00%) | 0.062 |
SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; ARDS, acute respiratory distress syndrome; IQR, interquartile range; SD, standard deviation.
Table 2. Univariate and Multivariate analyses for the association between SARS-CoV-2-infected patients with ARDS and without ARDS.
| Characteristics | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | ||
| Lactate dehydrogenase, U/L | 1.01 | (1.01, 1.02) | 0.001 | 1.02 | (1.00, 1.04) | 0.020 | |
| Aspartate aminotransferase, U/L | 1.05 | (1.01, 1.10) | 0.008 | 1.00 | (0.93, 1.07) | 0.964 | |
| Albumin, g/L | 0.81 | (0.69, 0.95) | 0.009 | 1.00 | (0.70, 1.35) | 0.862 | |
| Prothrombin time, s | 2.07 | (1.09, 3.93) | 0.026 | 1.42 | (0.46, 4.35) | 0.617 | |
| Lymphocyte count, ×109/L | 0.05 | (0.01, 0.40) | 0.005 | 0.19 | (0.01, 3.38) | 0.261 | |
| C-reactive protein, mg/L | 1.03 | (1.00, 1.05) | 0.016 | 0.99 | (0.94, 1.03) | 0.487 | |
| Heart rate, beats per min | 1.08 | (1.02, 1.14) | 0.004 | 1.10 | (1.00, 1.21) | 0.059 | |
| Respiratory rate, breaths per min | 1.76 | (1.16, 2.67) | 0.008 | 2.00 | (0.97, 4.13) | 0.062 | |
| Dyspnea | 8.70 | (2.15, 35.22) | 0.002 | 2.15 | (0.11, 42.83) | 0.617 | |
SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; OR, odds ratio; CI, confidence interval.
Figure 1.

Receiver operating characteristic curves. The AUC value of LDH in predicting ARDS. Blue shading shows the bootstrap estimated 95% CI with AUC. LDH, lactate dehydrogenase; ARDS, acute respiratory distress syndrome.
Figure 2.
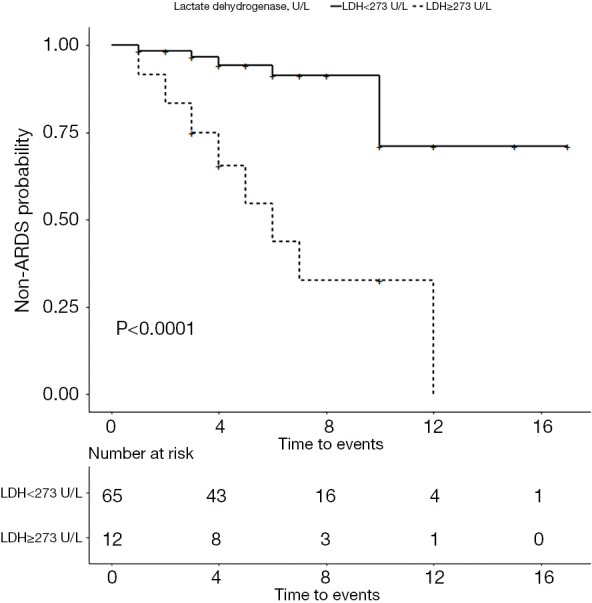
Kaplan-Meier curve showing development of ARDS in SARS-CoV-2-infected patients with fever stratified by LDH levels < and ≥273 U/L on admission. LDH, lactate dehydrogenase; ARDS, acute respiratory distress syndrome.
LDH is an important enzyme in anaerobic metabolism in almost all living organisms (3). Several studies suggested that serum LDH was elevated in severe COVID-19 patients (4,5). Consistently, we show that patients infected by SARS-CoV-2 with high levels of LDH on admission are more likely to develop ARDS. Inflammation and cell damage play an important role in the pathological processes of pulmonary tissues (6). Higher LDH levels have been found in COVID-19 patients than in patients with SARS-CoV-2 negative confirmed pneumonia (7). Yuan et al. found that COVID-19 mRNA clearance ratio was highly associated with LDH levels (8). Research has shown that SARS-CoV-2 as a positive-sense RNA virus may activate inflammasomes, leading to cellular pyroptosis and aggressive symptoms (9). This may partly explain the association of LDH with ARDS in COVID-19 patients. However, we found that the best threshold for predicting ARDS was 273 U/L. LDH level was independently associated with ARDS, and could strongly predict the incidence of ARDS. To our knowledge, this is the first study reporting the association of LDH with ARDS in COVID-19 patients with fever on admission. Our findings will help physicians to evaluate the condition of the illness at an earlier stage.
However, we note the following limitations to our study. First, this is a single-center retrospective study. Secondly, the serum level of LDH was only tested on admission. Thus, it should be tested at different times for better evaluation of its predicting value. Third, since all patients were from Changsha, our results may not apply to other regions since the clinical features of COVID-19 may vary in other regions.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank all the patients involved in the study.
Funding: This work was supported by the Special Emergency Treatment for Novel Coronavirus Pneumonia, Science and Technology department of Hunan province, China (grant number: 2020SK3004).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the Second Xiangya Hospital, Central South University (Changsha, China, No. LYF2020044) and informed consent were waived due to its retrospective nature.
Provenance and Peer Review: This article was a free submission to the journal. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2411). The authors have no conflicts of interest to declare.
References
- 1.Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 2020;9:313-9. 10.1080/22221751.2020.1725399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su Y, Ju MJ, Ma JF, et al. Lactate dehydrogenase as a prognostic marker of renal transplant recipients with severe community-acquired pneumonia: a 10-year retrospective study. Ann Transl Med 2019;7:660. 10.21037/atm.2019.10.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Zheng L, Liu L, et al. Liver impairment in COVID-19 patients: A retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int 2020. [Epub ahead of print]. [DOI] [PubMed]
- 5.Mo P, Xing Y, Xiao Y, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis 2020. [Epub ahead of print].
- 6.Drent M, Cobben NA, Henderson RF, et al. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur Respir J 1996;9:1736-42. 10.1183/09031936.96.09081736 [DOI] [PubMed] [Google Scholar]
- 7.Zhao D, Yao F, Wang L, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis 2020;71:756-61. 10.1093/cid/ciaa247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Zou R, Zeng L, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm Res 2020;69:599-606. 10.1007/s00011-020-01342-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and Pyroptosis as Therapeutic Targets for COVID-19. J Immunol 2020;205:307-12. 10.4049/jimmunol.2000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as


