Abstract
Liquid biopsy can provide a strong basis for precision medicine. We aimed to identify novel single-nucleotide variants (SNVs) in circulating tumor DNA (ctDNA) in patients with hepatocellular carcinoma (HCC). Deep sequencing of plasma-derived ctDNA from 59 patients with HCC was performed using a panel of 2924 SNVs in 69 genes. In 55.9% of the patients, at least one somatic mutation was detected. Among 25 SNVs in 12 genes, four frequently observed SNVs, MLH1 (13%), STK11 (13%), PTEN (9%), and CTNNB1 (4%), were validated using droplet digital polymerase chain reaction with ctDNA from 62 patients with HCC. Three candidate SNVs were detected in 35.5% of the patients, with a frequency of 19% for MLH1 chr3:37025749T>A, 11% for STK11 chr19:1223126C>G, and 8% for PTEN chr10:87864461C>G. The MLH1 and STK11 SNVs were also confirmed in HCC tissues. The presence of the MLH1 SNV, in combination with an increased ctDNA level, predicted poor overall survival among 107 patients. MLH1 chr3:37025749T>A SNV detection in ctDNA is feasible, and thus, ctDNA can be used to detect somatic mutations in HCC. Furthermore, the presence or absence of the MLH1 SNV in ctDNA, combined with the ctDNA level, can predict the prognosis of patients with HCC.
Subject terms: Gastroenterology, Molecular medicine, Oncology
Introduction
Liver cancer is the sixth most commonly diagnosed cancer and the fourth most common cause of cancer-related deaths globally1. Recently, systemic therapy for advanced hepatocellular carcinoma (HCC) has progressed drastically. In addition to sorafenib, several molecular-targeted agents and an immune checkpoint inhibitor have succeeded in clinical trials and have been used in the clinic2. However, the efficacy of these systemic therapies is still clinically insufficient3–7, likely due to factors such as the trial design, lack of patient stratification based on tumor mutations, and off-target activity of these agents.
In the current era of precision medicine, the detection of molecular drivers of tumorigenesis and DNA mutations in tumor samples has become a routine clinical practice to evaluate biomarkers that predict the response or resistance to targeted agents8. Liquid biopsy is based on convenient and minimally invasive collection of blood or urine samples, which can be acquired at multiple time points over the course of disease. Unlike most solid tumors, HCC can be confidently diagnosed using imaging techniques, which limits the availability of tissue biopsies for retrospective biomarker studies. Therefore, liquid biopsy can potentially provide a strong basis for individualized treatment of patients with HCC8.
Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA that is found in the bloodstream of patients with cancer and varies substantially in composition, from < 0.01 to > 60% of alleles in circulation9,10. The ctDNA of a patient contains the information regarding tumor-specific genetic or epigenetic alterations, including point mutations, copy-number variations, chromosomal rearrangements, and DNA methylation patterns of the tumor. Quantitative or qualitative analysis of ctDNA may reveal valuable information for early diagnosis, treatment, and tumor progression monitoring11.
The amount of ctDNA may represent a novel tool for screening, detecting, treatment monitoring, and predicting metastatic potential of HCC12–18. Studies have reported that tumor-specific mutations in TP5319, ITH20, HCK21, CTNNB122, and TERT22 are commonly detected in ctDNA from the peripheral blood of patients with HCC. However, there is no standard cutoff for the amount of ctDNA or a specific mutation in ctDNA that can be applied in clinical practice for predicting the HCC prognosis. Therefore, we aimed to screen for novel single nucleotide variants (SNVs) in HCC using targeted deep sequencing (TDS) of 69 candidate genes and validate the candidate SNVs using droplet digital polymerase chain reaction (ddPCR) for both blood and tissues. We also aimed to find the clinical correlation between candidate SNVs and the prognosis of patients with HCC.
Results
Patients’ demographics
The demographic and clinical parameters of the 107 patients with HCC who were included in the TDS or ddPCR analysis are shown in Supplementary Table S1. In total, 80 patients (74.8%) were older than 50 years, and 88 patients (82.2%) were males. The major etiology of HCC was hepatitis B virus infection, and 89 patients (83.2%) had good liver function (Child–Pugh class A). Serum levels of alpha-fetoprotein were higher than 200 ng/mL in approximately one-third of the patients (36.4%). The proportions of patients with stages 0, A, B, C, and D per the Barcelona Clinic Liver Cancer (BCLC) classification criteria were 5.6%, 42.1%, 15.9%, 32.7%, and 3.7%, respectively.
Identification of novel SNVs in ctDNA from patients with HCC
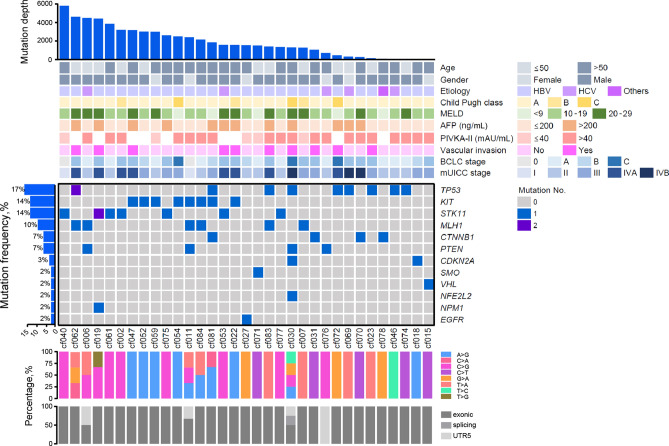
A total of 61 ctDNA sequencing datasets were generated from 59 patients with HCC and two controls (duplicate data using HapMap NA12878 cell line) and analyzed using a panel consisting of 2924 SNVs in 69 genes. Mutations were most frequently detected in TP53 (17%), followed by KIT and STK11 (14% each), MLH1 (10%), CTNNB1 and PTEN (7% each), CDKN2A (3%), and SMO, VHL, NFE2L2, NPM1, and EGFR (2% each). Finally, a total of 25 SNVs in 12 genes were identified in 33 patients (55.9%). The SNV landscape of these 33 patients with HCC is shown in Fig. 1. The associations between the presence of mutations in these 12 genes and clinical variables of the patients, including tumor characteristics, were evaluated, and CDKN2A, EGFR, MLH1, NFE2L2, PTEN, TP53, and VHL were found to be associated with several clinical parameters (Supplementary Table S2). Overall survival of the patients with MLH1 or NPM1 mutations was poor compared with that of the patients without these mutations (both log-rank P = 0.02; Supplementary Fig. S1).
Figure 1.
SNV landscape of 33 patients with HCC who tested positive for SNVs using ctDNA sequencing of 69 cancer genes. The patients are sorted in descending order according to the number of genes in which mutations were detected. Genes are sorted in descending order according to the number of patients who had a mutation in a particular gene. The depth of the mutations detected in a patient and the clinical and pathological characteristics of the patient are shown in the upper panel. Genes with SNVs are shown in the middle panel. Mutation frequencies of each gene are shown on the left. The mutation spectrum is shown at the bottom. ctDNA circulating tumor DNA, HCC hepatocellular carcinoma, SNV single-nucleotide variant.
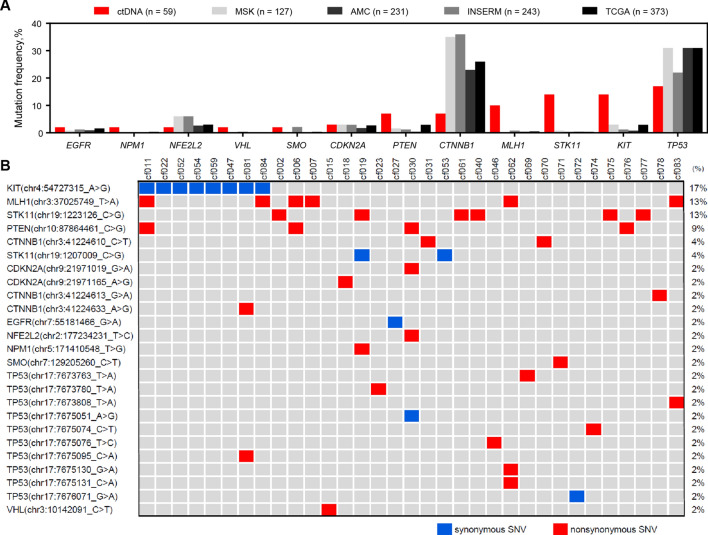
The mutation frequencies in these 12 genes were investigated in HCC tissues using published data for four next-generation sequencing cohorts, including The Cancer Genome Atlas, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets, French National Institute of Health and Medical Research, and Asan Medical Center ones (Fig. 2a)23–26. The most frequently mutated genes in the published data for the HCC tissue cohorts were CTNNB1 (30%) and TP53 (28.8%). On the contrary, KIT, STK11, MLH1, PTEN, VHL, NPM1, and EGFR variants were detected more frequently in ctDNA than in HCC tissues.
Figure 2.
Genetic variations detected in ctDNA of patients with HCC. (a) Frequencies of variations in the ctDNA of 59 patients with HCC and frequencies of mutations in tissue genomic DNA in four HCC tissue cohorts. (b) ctDNA landscape of 25 variant sites in 12 genes from 33 patients with HCC. Samples are shown on the top, genes with variant sites on the left, and frequencies of these genes on the right. ctDNA circulating tumor DNA, HCC hepatocellular carcinoma.
Among the 25 SNVs that were detected 47 times in 33 patients with HCC (Fig. 2b), the most frequently detected were the KIT chr4:54727315A>G (rs55986963; 17%), MLH1 chr3:37025749T>A (rs63750447; 13%), STK11 chr19:1223126C>G (rs59912467; 13%), PTEN chr10:87864461C>G (rs11202592; 9%), CTNNB1 chr3:41224610C>T (rs121913400; 4%), and STK11 chr19:1207009C>G (rs79175212; 4%). Considering their high detection frequencies and nonsynonymous substitutions, MLH1 chr3:37025749T>A, STK11 chr19:1223126C>G, PTEN chr10:87864461C>G, and CTNNB1 chr3:41224610C>T were selected for further validation using ddPCR analysis.
Detection of SNVs in blood ctDNA of patients with HCC using ddPCR
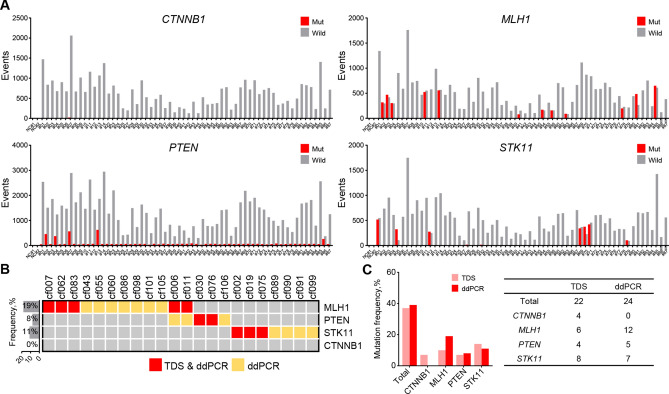
The four selected SNVs were analyzed using ddPCR in ctDNA of 62 patients with HCC. The MLH1, PTEN, and STK11 mutation-positive and -negative droplet clusters could be clearly distinguished. However, the CTNNB1 mutation was not detected, as no positive droplet clusters were visualized (Fig. 3a and Supplementary Fig. S2). The most frequently detected mutation was that in MLH1 (19%, 12/62 patients), followed by STK11 (11%, 7/62 patients) and PTEN (8%, 5/62 patients) (Fig. 3b). In the 62 patients, the MLH1 SNV detection rate of ddPCR was twice as high as that of TDS (Fig. 3c).
Figure 3.
Validation of four genes with SNVs detected in ctDNA in a validation set of 62 patients with HCC using ddPCR. (a) Total number of positive droplet events recorded via ddPCR of the four genes. Gray bars represent the number of wild-type-positive droplets, and red bars represent the number of mutant-positive droplets. (b) ctDNA landscape of the four genes with SNVs. (c) Mutation frequencies in the four genes detected in patients with HCC using TDS and ddPCR. ctDNA circulating tumor DNA, ddPCR droplet digital polymerase chain reaction, HCC hepatocellular carcinoma, SNV single-nucleotide variant, TDS targeted deep sequencing.
Prognostic potential of the MLH1, PTEN, STK11, and CTNNB1 SNVs in patients with HCC
The associations between clinical parameters and the MLH1, PTEN, STK11, and CTNNB1 SNVs were evaluated in the 107 patients whose SNVs were analyzed by TDS or ddPCR. Only the MLH1 SNV was significantly associated with the BCLC stage (P = 0.025; Supplementary Table S3), but no SNV was significantly associated with overall survival. However, subgroup analysis of the patients with advanced-stage HCC (BCLC stage C/D or modified Union for International Cancer Control stage IV) showed that the patients with the MLH1 SNV had a significantly lower survival than did those without this SNV (Supplementary Fig. S3). The CTNNB1, PTEN, and STK11 SNVs were not associated with the patient prognosis (Supplementary Fig. S4).
MLH1 and STK11 SNV detection in tissue genomic DNA (tDNA) using ddPCR
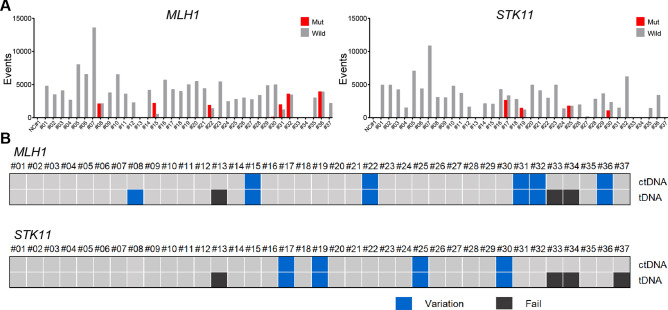
To determine whether the mutations detected in ctDNA were derived from HCC tissue, tDNA was extracted from available HCC tissues of 37 out of the 62 patients who were analyzed using ddPCR. MLH1 and STK11, the most frequently mutated genes detected in ctDNA using ddPCR, were selected for tDNA ddPCR analysis. The results are shown in Fig. 4a. In all cases, both MLH1 and STK11 mutations detected in ctDNA were also detected in tDNA. Only one patient, who had the MLH1 mutation in tDNA, did not show the same mutation in ctDNA (Fig. 4b).
Figure 4.
MLH1 and STK11 SNV detection in paired ctDNA and tumor DNA samples using ddPCR. (a) Total number of positive droplet events in ddPCR analysis of the two genes. Gray bars represent the number of wild-type-positive droplets, and red bars represent the number of mutant-positive droplets. (b) Comparison of SNV detection results for the two genes between paired ctDNA and tumor DNA samples. Samples with positive, negative, and failed SNV detection are shown in blue, gray, and black, respectively. ctDNA circulating tumor DNA, ddPCR droplet digital polymerase chain reaction, SNV single-nucleotide variant.
Association of the ctDNA level and presence of the MLH1 SNV with patient clinicopathological features and overall survival
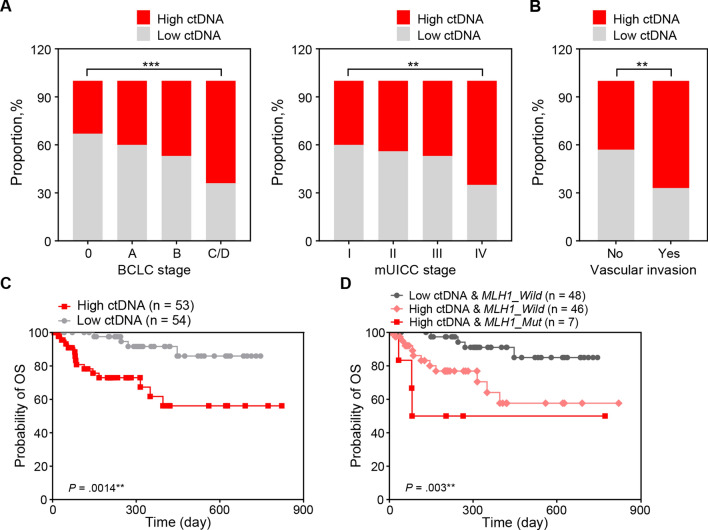
The amount of ctDNA was quantified in each of the 107 patients with HCC. Using the median concentration of recovered ctDNA (5.77 ng/mL), the patients were divided into high (≥ 5.77 ng/mL)- and low (< 5.77 ng/mL)-ctDNA groups. The proportion of patients with high ctDNA levels significantly increased with the HCC stage progression (P < 0.001 for BLCL stages; P < 0.01 for modified Union for International Cancer Control stages; Fig. 5a). Patients with vascular invasion also had increased ctDNA levels (P < 0.01; Fig. 5b). The overall survival of the patients in the high-ctDNA group was lower than that of the patients in the low-ctDNA group (log-rank P = 0.0014; Fig. 5c). Moreover, patients with the ctDNA MLH1 mutation in the high-ctDNA group demonstrated worse overall survival, whereas those with no ctDNA MLH1 mutation in the low-ctDNA group showed the most favorable overall survival (P = 0.003; Fig. 5d). On the other hand, when MLH1 SNV was combined with AFP level, the group showing low AFP (< 20 ng/mL) and no ctDNA MLH1 mutation had the best overall survival (P = 0.005; Supplementary Fig. S5).
Figure 5.
Association of the ctDNA level and MLH1 SNV with clinicopathological parameters and overall survival of patients with HCC. (a) Proportion of patients in the high-versus low-ctDNA group according to the BCLC stage. (b) Proportion of patients in the high-versus low-ctDNA group according to the mUICC stage. (c) Overall survival curves according to the ctDNA levels in the plasma of patients with HCC. (d) Overall survival curves according to the ctDNA levels and presence of the MLH1 SNV. BCLC Barcelona Clinic Liver Cancer, ctDNA circulating tumor DNA, HCC hepatocellular carcinoma, mUICC modified Union for International Cancer Control, SNV single-nucleotide variant.
Discussion
The present study identified novel HCC-associated SNVs using a panel of 2924 SNVs in 69 genes to probe ctDNA from 59 patients with HCC. We identified 25 SNVs in 12 genes and validated four (MLH1, STK11, PTEN, and CTNNB1) SNV candidates using ddPCR on plasma from other 62 patients with HCC. The MLH1 SNV (chr3:37025749T>A) was detected in ctDNA and confirmed in tDNA of patients with HCC. Of the four candidate SNVs, only the MLH1 SNV correlated with the HCC stage and could predict overall survival in patients with advanced HCC. Patients with high levels of ctDNA (≥ 5.77 ng/mL) in the plasma and ctDNA positivity for the MLH1 SNV exhibited the worst survival rate. To our knowledge, this is the first study to detect the MLH1 chr3:37025749T>A SNV in the ctDNA of patients with HCC and to show its prognostic value for overall survival.
MLH1 plays a key role in DNA mismatch repair via recognition and repair of mismatched bases during DNA replication. Additionally, MLH1 recruits other mismatch repair proteins to the mismatch site to correct DNA replication errors27,28. Defects in repair genes may lead to genomic instability and cancer development, suggesting that polymorphisms in specific repair genes may contribute to individual differences in the risk of developing cancer29–31. It has been reported that MLH1 polymorphisms correlate with the pathogenesis of colorectal cancer, lung cancer, breast cancer, prostate cancer, head and neck squamous cell carcinoma, and oral squamous cell carcinoma29–36. However, there are only a few studies on the relationship between MLH1 polymorphisms and HCC37,38. Two previous case–control studies found an increased risk of HCC and reported a poor prognosis in patients with the MLH1 rs1800734 single-nucleotide polymorphism37,38. However, these studies did not include the MLH1 chr3:37025749T>A (rs63750447) SNV nor did they analyze ctDNA of patients with HCC. Therefore, the present study is the first to clarify the relationship between the MLH1 chr3:37025749T>A SNV, particularly in ctDNA, and HCC.
The MLH1 chr3:37025749T>A SNV, which is located in an exon, induces an amino acid substitution of valine to aspartate. The global minor allele frequency of the MLH1 chr3:37025749T>A SNV is very low (at 0.519%)39. We found the incidence of the MLH1 chr3:37025749T>A SNV to be approximately 13–19% using TDS and ddPCR, which suggests a surprisingly high frequency of this MLH1 SNV 37025749T>A in patients with HCC. Considering that the reported minor allele frequency of the same polymorphism is 3–8% in Chinese and Japanese patients with other cancers, such as colorectal and lung cancer40–42, the incidence of the MLH1 chr3:37025749T>A SNV in Korean patients with HCC is quite high. Therefore, the MLH1 chr3:37025749T>A SNV may be more specific to HCC, especially in Korean patients.
Previous studies have reported that the detection of the MLH1 chr3:37025749T>A polymorphism in the blood is associated with an increased risk of gastric, colorectal, and endometrial cancer in Asian populations43. The possible underlying mechanism for the influence of the MLH1 chr3:37025749T>A SNV on cancer development may be associated with the mismatch repair function of the MLH1 protein. The amino acid affected by the MLH1 chr3:37025749T>A SNV is located between the NH2-terminal ATP-binding domain and the COOH-terminal PMS2 interaction domain of the MLH1 protein. The MLH1 chr3:37025749T>A SNV alters the charge of the protein and is likely to disrupt its structure and stability. Previous studies have demonstrated decreased in vitro mismatch repair and β-galactosidase activities of the MLH1 protein encoded by the MLH1 chr3:37025749T>A variant gene44,45. Taken together, the MLH1 chr3:37025749T>A mutation is a partial loss-of-function variant that may increase the risk of cancer in the carriers.
The current study has a few limitations. First, we did not include patients without HCC, such as patients with liver cirrhosis. The HapMap NA12878 cell line was used as a control for the analysis of the TDS data. Second, only one MLH1 SNV (chr3:37025749T>A) was included in the panel comprising 2924 variants in 69 genes. Therefore, we did not study the correlation between other MLH1 SNVs and HCC. Third, hepatitis B virus was the main etiology of HCC in the present study. Therefore, the study finding may not be generalizable to population with different etiology and external validation is crucial.
Despite outstanding advances in the precision medicine of various cancers, individualized cancer therapy using companion diagnostics is still challenging in HCC. This drawback may be due to genetic heterogeneity of HCC itself or a difficulty in obtaining liver tissue from patients because this cancer is uniquely diagnosed based on imaging modalities. Liquid biopsy can be considered the most promising tool to overcome these limitations in HCC precision therapy. The current study demonstrated the possibility of using ctDNA to detect HCC mutations and predict the prognosis of patients with HCC.
In conclusion, MLH1 SNV detection in ctDNA is feasible, and thus, ctDNA can be used to confidently detect somatic mutations in HCC tissue. Furthermore, ctDNA positivity or negativity for the MLH1 chr3:37025749T>A mutation can provide a prognostic value, with or without measurement of the amount of ctDNA in the plasma of patients with HCC.
Methods
Patients and sample collection
A total of 146 consecutive treatment-naïve patients with HCC were enrolled at Ajou University Hospital between May 2016 and April 2019 were enrolled. Patients were diagnosed with HCC if their tumor had a minimum diameter of > 1 cm, and typical features of HCC (arterial phase hyperenhancement, washout in the portal venous or delayed phase, threshold growth, and capsule appearance) were observed using multiphase computed tomography and/or magnetic resonance imaging. If these criteria were met but the diagnosis of HCC was only considered probable, a liver biopsy was performed for final diagnosis46. Patients were excluded if they had other concurrent malignancies. A volume of 10 mL of blood was collected from each patient. Sufficient ctDNA (≥ 7 ng/mL) for further analysis using TDS or ddPCR was only obtained from 107 patients. Snap-frozen HCC tissues from 35 patients who underwent hepatectomy and two patients who were diagnosed using liver biopsy were used for validation analysis with the ddPCR technique (Supplementary Fig. S6). All experiments were conducted in accordance with the ethical guidelines of the 1975 Declaration of Helsinki. Written informed consent was obtained from all patients, and the study was approved by the Institutional Review of Board of Ajou University of School of Medicine (approval No: AJIRB-BMR-OBS-16-344).
Extraction of ctDNA and tDNA
ctDNA was extracted from plasma using the QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The purity of ctDNA was determined using the Agilent high-sensitivity DNA kit and a Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA, USA). tDNA was extracted from snap-frozen tissues using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions and was eluted in a volume of 200 μL. The purity of the extracted genomic DNA was assessed by electrophoresis on a 1% agarose gel, and the DNA concentration was quantified using a Qubit 2.0 fluorometer and the Agilent high-sensitivity DNA kit.
Library construction and sequencing
A customized panel was designed that targeted 2924 SNVs in 69 genes (Supplementary Table S4). A sequencing library was constructed from ctDNA using the Celemics library preparation kit (Celemics, Seoul, Republic of Korea). The capture of hybridization targets was performed using Celemics customized capture probes and the Celemics target capture kit. PCR amplification of captured libraries was performed using the KAPA HiFi HotStart PCR kit (Kapa Biosystems, Boston, MA, USA). Captured samples were pooled and sequenced using the Illumina NextSeq 500 platform (Illumina, San Diego, CA, USA) with 2 × 150-bp paired-end runs.
ctDNA sequencing and variant calling
Sequencing of ctDNA was performed on the Illumina NextSeq 500 platform with 151-bp paired-end reads, with a coverage greater than 2000 reads in the panel regions per sample. The raw image data were transformed to and stored in a FASTQ format. Low-quality sequence reads with a PHRED score of less than 30 and adapter sequence reads were trimmed using the Trim Galore! tool and then mapped to a human reference genome (hg38) using Burrows–Wheeler Aligner with default parameters. Local realignment of indels and the normalization of base quality scores were performed using the Genome Analysis Toolkit. Sequence variations were detected using VarScan2 and annotated using the ANNOVAR software. Somatic variants were filtered as follows: allele frequencies of the variants in the normal population were obtained from the 1000 Genomes Project and Exome Aggregation Consortium dataset, and variants with allele frequencies greater than 5% in either of the normal populations were filtered out. Variants in the control cell line were also filtered out (Supplementary Fig. S7).
Public omics data analysis
For comparative analysis of ctDNA target variations, we used the data for four large liver cancer tissue cohorts (from The Cancer Genome Atlas, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets, French National Institute of Health and Medical Research, and Asan Medical Center), with the whole-exome sequencing data listed on cBioPortal (https://www.cbioportal.org/)23.
ddPCR
ddPCR was performed using a previously reported method47. Each 20-μL reaction contained 10 μL of a ddPCR supermix (no dUTP), 6 μL of a primer–probe premix (0.4 μL each of 10 μmol/L upstream and downstream primers, 0.2 μL of 10 μmol/L probe, and 5 μL of deionized water), and 4 μL of a nucleic acid extract. Each sample reaction was added to the middle of a DG8 cartridge. Next, 70 μL of oil was added to the bottom row of each lane to avoid bubble formation, and the wells were covered. The reaction system and droplet-forming cartridge were placed in a droplet generator and subjected to microdroplet treatment. Droplets were produced in the top row of the wells, and the suction volume was adjusted to 40 μL. The samples were then gently moved to 96-well plates, and a preheated PX1 heat sealing device was used with a sealing film (with the red line up) at 180 °C for 5 s. The PCR conditions were as follows: predenaturation at 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 55 °C for 1 min, with a temperature change rate of 2 °C/s. The 96-well plate containing PCR-amplified products was then positioned on a QX200 microdrop reader and analyzed using the QuantaSoft software (Bio-Rad).
Statistical analysis
The IBM SPSS software version 22.0 (SPSS, Inc., Chicago, IL, USA) and the GraphPad Prism 7.01 software (GraphPad Software, San Diego, CA, USA) were used for statistical analyses. All tests were two-sided and considered statistically significant if P was < 0.05. Continuous variables were compared using independent sample t-tests. Analysis of variance was used to compare continuous variables between more than three groups. Categorical data were compared using a Pearson χ2 test or a Fisher exact test. Survival analysis was performed using the Kaplan–Meier method with a log-rank test.
Supplementary information
Acknowledgements
The biospecimens for this study were provided by the Ajou University Human Bio-Resource Bank, a member of the Korea Biobank Network, which is supported by the Ministry of Health and Welfare. This research was supported by Grants (HI18C0531 and HI15C1551) of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea. This research was also supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF-2018M3A9E8023861, NRF-2017R1D1A1B03033996, and NRF-2019R1C1C1007366), funded by the Korean government (MSIT).
Author contributions
H.G.W. and J.Y.C. designed the conception of the study; J.C. and J.W.E. developed the methodology and analyzed the data; H.R.A. and C.W.S acquired the data; S.S.K., J.W.E, and J.C. wrote the manuscript; H.J.C. and G.O.B constructed the databases; and S.W.C. supervised the study. All authors reviewed the manuscript.
Data availability
The data used or analyzed in this article is available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Soon Sun Kim and Jung Woo Eun.
Supplementary information
is available for this paper at 10.1038/s41598-020-74494-y.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver EASL Clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AL, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/s1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/s0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 6.Kudo M, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 7.El-Khoueiry AB, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J. Exp. Clin. Cancer Res. 2018;37:213. doi: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell JA, Sharma R. The clinical role of 'liquid biopsy' in hepatocellular carcinoma. Hepat. Oncol. 2016;3:45–55. doi: 10.2217/hep.15.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren N, et al. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2006;132:399–407. doi: 10.1007/s00432-005-0049-5. [DOI] [PubMed] [Google Scholar]
- 13.Iizuka N, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res. 2006;26:4713–4719. [PubMed] [Google Scholar]
- 14.Yang YJ, et al. Quantification of plasma hTERT DNA in hepatocellular carcinoma patients by quantitative fluorescent polymerase chain reaction. Clin. Invest. Med. 2011;34:E238. doi: 10.25011/cim.v34i4.15366. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z, et al. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol. Oncol. Res. 2012;18:271–276. doi: 10.1007/s12253-011-9438-z. [DOI] [PubMed] [Google Scholar]
- 16.Piciocchi M, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol. Int. 2013;7:1050–1057. doi: 10.1007/s12072-013-9481-9. [DOI] [PubMed] [Google Scholar]
- 17.Huang A, et al. Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J. Cancer. 2016;7:1798–1803. doi: 10.7150/jca.15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan L, et al. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int. J. Infect. Dis. 2018;67:92–97. doi: 10.1016/j.ijid.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Yu L, et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis B virus-related hepatocellular carcinoma after hepatectomy: a cohort study for 10 years' follow up. Hepatol. Res. 2016;46:765–774. doi: 10.1111/hepr.12611. [DOI] [PubMed] [Google Scholar]
- 20.Huang A, et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J. Hepatol. 2017;67:293–301. doi: 10.1016/j.jhep.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Cai ZX, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int. J. Cancer. 2017;141:977–985. doi: 10.1002/ijc.30798. [DOI] [PubMed] [Google Scholar]
- 22.Huang A, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J. Cancer. 2016;7:1907–1914. doi: 10.7150/jca.15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding JJ, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin. Cancer Res. 2019;25:2116–2126. doi: 10.1158/1078-0432.Ccr-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn SM, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 27.Cohen SA, et al. Frequent PIK3CA mutations in colorectal and endometrial tumors with 2 or more somatic mutations in mismatch repair genes. Gastroenterology. 2016;151:440–447. doi: 10.1053/j.gastro.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riazy M, et al. Mismatch repair status may predict response to adjuvant chemotherapy in resectable pancreatic ductal adenocarcinoma. Mod. Pathol. 2015;28:1383–1389. doi: 10.1038/modpathol.2015.89. [DOI] [PubMed] [Google Scholar]
- 29.Ma G, et al. Functional annotation of colorectal cancer susceptibility loci identifies MLH1 rs1800734 associated with MSI patients. Gut. 2016;65:1227–1228. doi: 10.1136/gutjnl-2016-311543. [DOI] [PubMed] [Google Scholar]
- 30.Nogueira GA, et al. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int. J. Cancer. 2015;137:810–818. doi: 10.1002/ijc.29435. [DOI] [PubMed] [Google Scholar]
- 31.van der Klift HM, et al. Comprehensive mutation analysis of PMS2 in a large cohort of probands suspected of lynch syndrome or constitutional mismatch repair deficiency syndrome. Hum. Mutat. 2016;37:1162–1179. doi: 10.1002/humu.23052. [DOI] [PubMed] [Google Scholar]
- 32.Harkness EF, et al. Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: a cohort study. J. Med. Genet. 2015;52:553–556. doi: 10.1136/jmedgenet-2015-103216. [DOI] [PubMed] [Google Scholar]
- 33.Lin LH, et al. The hMLH1 -93G>A promoter polymorphism is associates with outcomes in oral squamous cell carcinoma patients. Ann. Surg. Oncol. 2014;21:4270–4277. doi: 10.1245/s10434-014-3897-x. [DOI] [PubMed] [Google Scholar]
- 34.Lo YL, et al. Polymorphisms of MLH1 and MSH2 genes and the risk of lung cancer among never smokers. Lung Cancer. 2011;72:280–286. doi: 10.1016/j.lungcan.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Niu L, Li S, Liang H, Li H. The hMLH1 -93G>A polymorphism and risk of ovarian cancer in the Chinese population. PLoS ONE. 2015;10:e0135822. doi: 10.1371/journal.pone.0135822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomlinson IP, Houlston RS, Montgomery GW, Sieber OM, Dunlop MG. Investigation of the effects of DNA repair gene polymorphisms on the risk of colorectal cancer. Mutagenesis. 2012;27:219–223. doi: 10.1093/mutage/ger070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CC, et al. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int. J. Epidemiol. 2005;34:1310–1318. doi: 10.1093/ije/dyi191. [DOI] [PubMed] [Google Scholar]
- 38.Zhu X, et al. Single nucleotide polymorphisms in MLH1 predict poor prognosis of hepatocellular carcinoma in a Chinese population. Oncotarget. 2017;8:80039–80049. doi: 10.18632/oncotarget.16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Center for Biotechnology Information. ClinVar; [VCV000041632.1], https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000041632.1.
- 40.Wang Y, et al. A novel missense mutation in the DNA mismatch repair gene hMLH1 present among East Asians but not among Europeans. Hum. Hered. 1998;48:87–91. doi: 10.1159/000022786. [DOI] [PubMed] [Google Scholar]
- 41.Chiu CH, et al. MLH1 V384D polymorphism associates with poor response to EGFR tyrosine kinase inhibitors in patients with EGFR L858R-positive lung adenocarcinoma. Oncotarget. 2015;6:8407–8417. doi: 10.18632/oncotarget.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohsawa T, et al. Colorectal cancer susceptibility associated with the hMLH1 V384D variant. Mol. Med. Rep. 2009;2:887–891. doi: 10.3892/mmr_00000187. [DOI] [PubMed] [Google Scholar]
- 43.Li S, et al. Pooling-analysis on hMLH1 polymorphisms and cancer risk: evidence based on 31,484 cancer cases and 45,494 cancer-free controls. Oncotarget. 2017;8:93063–93078. doi: 10.18632/oncotarget.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M, et al. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–4604. doi: 10.1158/0008-5472.Can-06-3509. [DOI] [PubMed] [Google Scholar]
- 45.Fan Y, et al. Analysis of hMLH1 missense mutations in East Asian patients with suspected hereditary nonpolyposis colorectal cancer. Clin. Cancer Res. 2007;13:7515–7521. doi: 10.1158/1078-0432.Ccr-07-1028. [DOI] [PubMed] [Google Scholar]
- 46.Marrero JA, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 47.Hui Y, et al. Micro-droplet digital polymerase chain reaction and real-time quantitative polymerase chain reaction technologies provide highly sensitive and accurate detection of Zika Virus. Virol. Sin. 2018;33:270–277. doi: 10.1007/s12250-018-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used or analyzed in this article is available from the corresponding author upon reasonable request.