Abstract
Intrinsic resistance to ionizing radiation is the major impediment in the treatment and clinical management of esophageal squamous cell carcinoma (ESCC), leading to tumor relapse and poor prognosis. Although several biological and molecular mechanisms are responsible for resistance to radiotherapy in ESCC, the molecule(s) involved in predicting radiotherapy response and prognosis are still lacking, thus requiring a detailed understanding. Recent studies have demonstrated an imperative correlation amongst several long non-coding RNAs and their involvement in complex cellular networks like DNA damage and repair, cell cycle, apoptosis, proliferation, and epithelial-mesenchymal transition. Additionally, accumulating evidence has suggested abnormal expression of lncRNAs in malignant tumor cells before and after radiotherapy effects in tumor cells’ sensitivity. Thus, lncRNAs indeed represent unique molecules that can influence tumor cell susceptibility for various clinical interventions. On this note, herein, we have summarized the current status of lncRNAs in augmenting resistance/sensitivity in ESCC against radiotherapy. In addition, we have also discussed various strategies to increase the radiosensitivity in ESCC cells under clinical settings.
Keywords: radioresistance, radiotherapy, radiosensitivity, esophageal squamous cell carcinoma, long non-coding RNAs
1. Introduction
Globally, esophageal cancer (EC) is one of the most lethal cancer types and is the eighth most common cancer [1]. Esophageal squamous cell carcinoma (ESCC) accounts for almost 80% of all EC cases globally with an improper disposition and a higher incidence in the Asian ethnic population [2,3]. Plaintively, to the conventional notion, the current standard treatment strategies employed for metastatic ESCC patients are concurrent definitive chemo and radiotherapy. However, therapy resistance and tumor recurrence by radioresistant tumor cells are the major obstacles for ESCC treatment contributing to poor prognosis and dismal survival in ESSC patients (<20%) [4,5]. Furthermore, effective radiation-based treatment against radioresistant tumors involves higher radiation doses causing undesired side effects and damaging healthy tissue via “bystander effects” [6,7]. Therefore, identification of novel predictive and prognostic biomarkers is urgently needed to manage patients undergoing radiotherapy treatment.
Several studies have demonstrated the association of non-coding RNAs in regulating various cancer hallmarks, including the radioresistance processes in cells [8,9]. Our previous study suggested several microRNAs (miRNAs) dictating sensitivity and resistance against radiotherapy EC patients [10]. Furthermore, recent studies have reported the association of long non-coding RNAs (lncRNAs) with altered patients’ responses towards radiation therapy due to their active involvement in DNA damage response (DDR), apoptosis, and cell cycle arrest [11,12].
With this idea, we have collated scientific literature correlating several dysregulated lncRNAs corresponding to ESCC diagnostic when exposed to radiation therapy. Several long non-coding RNAs (Table 1) impede radiation therapy in ESCC by regulating the expression of several miRNAs modulating downstream targets involved in DDR, base excision repair pathway, ataxia telangiectasia mutated (ATM) pathway, and the respective DNA repair pathways. Additionally, another set of lncRNAs, such as FAM201A, sensitize the cancer cells towards radiation therapy in ESCC via targeting cyclin-dependent kinase subunit 1 (Cks1), cyclin D1, Mammalian target of rapamycin (mTOR) and activating Non-homologous end joining (NHEJ) pathway [13] thereby enhancing the radiosensitivity of cancer cells.
Table 1.
Characteristics of lncRNA in conferring radioresistance and radiosensitivity in ESCC.
| lncRNA | Genomic Location | Mean Fold Change in Expression Compared to Controls | Radiation Type (Dose) | Property | Validation Methods | Biological Significance | Genes/Proteins/Pathways Affected | Ref. |
|---|---|---|---|---|---|---|---|---|
| DNM3OS | 1q24.3 | KYSE-30 ~39.4-folds KYSE-50 ~35.8-folds KYSE-180 ~28.5-folds KYSE-140 ~35-folds KYSE-450 ~7-folds Tissues ~6.3-folds KYSE-150R~4.3-folds |
X-ray radiation (12 Gy, 12 days) |
Oncogenic | qRT-PCR, Western blot analyses, ChIP assay | Increased levels of growth factors, reduced tumor suppressor levels, reduce DNA damage response leading to sustained proliferative signals, reduced apoptotic rate and enhanced radioresistance | γH2AX ↓, cPARP ↓, p-ATM ↑, Rad50 ↑, p-Chk2 ↑, Ku80 ↑, MRE1 ↑, NBS1 ↑, DNA-PKcs ↑, and p53 ↑, PDGFα/β signaling ↑ | [12] |
| LINC00473 | 6q27 | TE-1 ~6.2-folds EC9706 ~6.8-folds ECA109 ~3.6-folds KYSE-450 ~1.6-folds Tissues- ~2.6-folds |
X-ray radiation (4 Gy) |
Oncogenic | qRT-PCR, Western blot analyses | Reduced DNA damage response, reduced tumor suppressor levels, stimulated cell cycle progression leading to enhanced proliferation, reduced apoptotic rates, and enhanced radioresistance | mir-374a-5p ↓ PARP ↑ SPIN1 ↑ miR-497-5p ↓ CdC25A ↑ |
[14,15] |
| LINC00657 | 20q11.23 | KY-SE ~1.4-folds Eca-109 ~1.5-folds TE-1 ~1.4-folds Tissues ~1.3-folds |
X-ray radiation (0, 4, 8 Gy) |
Oncogenic | qRT-PCR, Western blot analyses, BrdU assay | Reduced tumor suppressor levels, causing increased cell proliferation, metastasis, invasion, and enhanced radioresistance | miR-615-3p ↓ JunB ↑ TGFβ ↑ TGF signaling ↑ |
[16] |
| POU6F2-AS2 | 7p14.1 | KYSE-140 ~5-folds KYSE-510 ~6.1-folds KYSE-30 ~2.0-folds KYSE-70 ~1.0-fold |
Ionizing radiation (0, 4, 6 Gy) |
Oncogenic | qRT-PCR, microarray analyses, immunoblot, RIP assay, RNA pulldown assay, ChIP-seq assay | Reduced DNA damage response, reduced tumor suppressor levels, stimulated cell cycle progression leading to enhanced proliferation and enhanced radioresistance | Ybx1 ↑ | [17] |
| FAM201A | 9p13.1 | NA | X-ray radiation (0, 2, 4, 6, 8 and 10 Gy) |
Oncogenic | qRT-PCR, microarray analyses, Western blot analyses | Enhanced DNA damage response, reduced tumor suppressor levels leading to poorer radiosensitivity | miR-101 ↓ mTOR ↑ ATM ↑ |
[13] |
| MALAT1 | 11q13.1 | Eca109 ~3.1-folds TE-1 ~3.4-folds |
γ-radiation (5 Gy, 2.4 Gy/min) X-ray radiation (3.2 Gy/min) |
Oncogenic | qRT-PCR, Western blot analyses, RIP assay | Enhanced invasion and metastasis, reduced radiosensitivity | Cks1 ↑, YAP ↑ |
[18,19] |
| LOC285194 | 3q13.31 | KYSE-30 ~2.0-folds KYSE-510 ~1.7-folds KYSE-109 ~2-folds KYSE-70 ~1.1-folds KYSE-150 ~1.3-folds Tissues ~1.1-folds |
X-ray radiation(40 Gy, 20 fractions of 2 Gy, 5 fractions/week for 4 weeks) | Oncogenic | qRT-PCR | Enhanced cell proliferation, enhanced apoptosis and reduced radiosensitivity | NA | [20] |
| AFAP1-AS1 | 14p16.1 | KYSE-30 ~1.0-fold KYSE-70 ~15.0-folds KYSE-150 ~3.0-folds KYSE-450 ~5.0-folds KYSE-510 ~9.0-folds TE-10 ~3.0-folds Tissues ~0.8-folds |
X-ray radiation(60–70 Gy, 1.8–2 Gy/day for 5 days/week) | Oncogenic | qRT-PCR | NA | NA | [21] |
| LncTUG1 | 22q12.2 | EC9706 ~3.0-folds KYSE-30~2.9-folds KYSE-140 ~2.8-folds TE-13~2.8-folds |
X-ray radiation (2 Gy) |
Oncogenic | qRT-PCR, Western blot, RIP assay | Enhanced cell proliferation, invasion and reduced radiosensitivity | miR-144-3p ↓, MET ↑, p-Akt ↑ |
[22] |
↑ = Upregulation; ↓ = Downregulation.
Herein, we have discussed several lncRNAs associated with the radioresistance and radiosensitivity of ESCC. We have also highlighted the plausible molecular pathways in decoding the radioresistance/sensitivity of ESCC. Furthermore, no review articles or meta-analysis were reported to date in lncRNAs and ESCC radiotherapy.
2. Literature Search and Selection Methodology
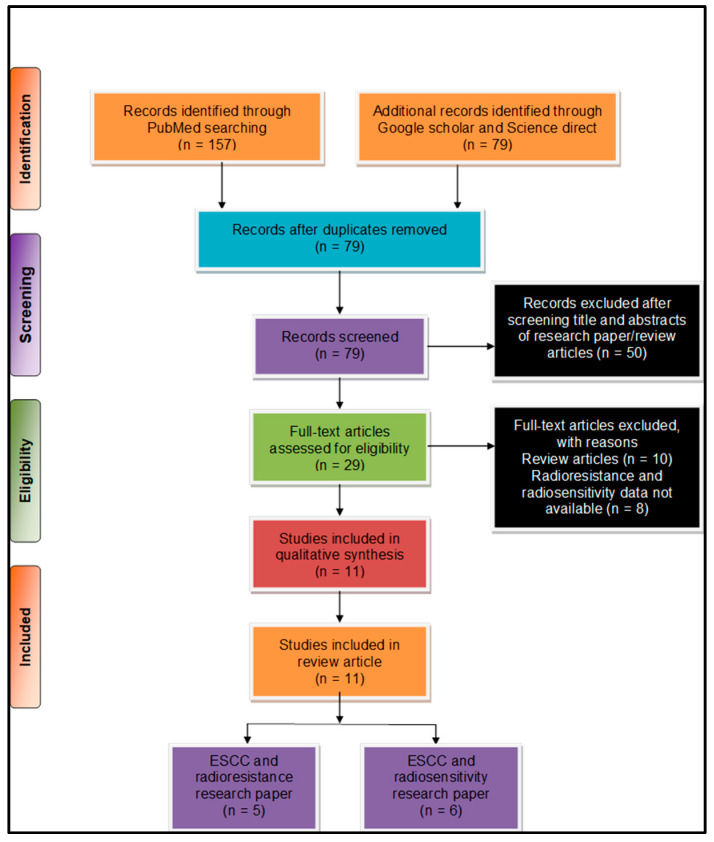
We used preferred reporting items for systematic reviews and meta-analysis (PRISMA) [23] for systematic analysis of the pieces of literature for this work. The literature search was performed by two individual authors (US and TSB) and further consulted with another researcher (AJ), whenever necessary. We considered the literature from January 2015–August 2020 to provide the updated information to the readers. We performed primary literature searches through various online databases, such as PubMed, ScienceDirect, and Google Scholar. We used a combination of several search keywords such as (“esophageal squamous cell carcinoma” OR “oesophageal squamous cell carcinoma” OR “ESCC”) AND (“radioresistance” OR Radio-resistance”) AND (“long non-coding RNA” OR “lncRNA” OR “Long noncoding RNA”); (“esophageal squamous cell carcinoma” OR “oesophageal squamous cell carcinoma OR “ESCC”) AND (“radio-resistance” OR “radio-resistance” OR “radio resistance”) AND (“lncRNA” OR “Long noncoding RNA” OR “Long non-coding RNA”) NOT (“miRNA” OR “micro RNA” OR “mi-RNA”). Furthermore, we were able to collate a total of (n = 236) pieces of literature from our primary literature search. A secondary search was subsequently performed post duplicate removal based on bibliographic data and a total of (n = 79) articles selected for final screening. After a careful scientific assessment, a total of (n = 11) articles were found suitable for further consideration eliminating book chapters, reviews, and conference abstracts (Figure 1).
Figure 1.
Preferred reporting items for systematic reviews and meta-analysis (PRISMA) flow chart describing the process of literature search and study selection related to esophageal squamous cell carcinoma (ESCC) and radioresistance/radiosensitivity. The total number of 11 relevant research articles were included in the review.
As per the PRISMA analysis, various long non-coding RNAs involved in ESCC radioresistance (Section 3) and radiosensitivity (Section 4) are discussed below.
3. Detailed Regulation of Radioresistance in ESCC through lncRNAs
3.1. Dynamin 3 Opposite Strands
Dynamin 3 Opposite Strand (DNM3OS) spans ~7.9 kb on chromosome 1q.24.3 and transcribes in an antisense orientation of the 14th intron of the DNM3 gene [24]. The DNM3OS was found to be expressed in a stage-dependent manner during several fetal development processes, such as the nasal process, pharyngeal arches, limb buds, and somites [24]. Furthermore, dysregulated DNM3OS is implicated in several cancers, including gastric cancer [25], ovarian cancer [25], and esophageal squamous cell carcinoma [12].
Zhang et al. [12] observed an upregulated expression (~6.30-fold) of DNM3OS in ESCC patients tissues (n = 16) compared to the matched healthy esophageal epithelial tissues. In addition, the upregulated expression levels of DNM3OS are associated with higher tumor grades. Additionally, the elevated expression of DNM3OS was also observed in several esophageal cancer cell lines viz. KYSE-30 (~39.40-fold), KYSE-150 (~35.80-fold), KYSE-180 (~28.50-fold), compared to the normal esophageal epithelial cell line Het-1A. To unveil the role of DNM3OS as a predictive biomarker for radiotherapy, the investigators irradiated ESCC cells (KYSE-150 and KYSE-150R) with 12 Gy radiations. They obtained a ~4.30-fold increase in the expression levels of DNM3OS in radioresistant KYSE-150R cells compared to parental cell KYSE-150 [12]. Furthermore, DNM3OS loss-of-function in radioresistant KYSE-150R cells led to an increase in sensitivity towards radiotherapy treatment. Thus, in vitro data suggest a positive correlation between DNM3OS expression and increased radioresistance in ESCC cells. Consistent with the above observations, slower tumor growth was observed in KYSE-30 xenografts Balb/c nude mice transfected with siDNM3OS cells compared to the control group mice irradiation with 12 Gy. Moreover, the tumor inhibition rate was higher for the DNM3OS knockout mice group, compared to the control group mice (76.59% vs. 47.03%) [12].
Further, an upregulated expression of DNM3OS in KYSE-30 (~39.20-fold) and KYSE-140 (~38.30-fold) observed in condition-media containing cancer-associated fibroblasts (CAFs) compared to the cells cultured in the standard medium [12]. It is well known that CAFs promote tumor growth in the tumor microenvironment by altering the extracellular matrix [12,26]. In addition, the dose enrichment ratio in KYSE-30 and KYSE-40 cultured in CAF containing condition-media was 3.3015 and 1.8368, respectively, compared to the control (standard medium), which confirmed the association of CAFs in modulating the expression of DNM3OS expression and thereby conferring radioresistance in ESCC [12]. Furthermore, to elucidate the underlying pathways responsible for DNM3OS deregulation, a molecule called “crenolanib” identified with kinase inhibitor signaling. Crenolanib is known to inhibit platelet-derived growth factor α/β (PDGFR α/β) signaling by targeting PDGF itself, thereby having the potential to inhibit DNM3OS. Moreover, a higher expression of PDGF β-polypeptide (PDGFβ), and platelet-derived growth factor receptor β-polypeptide (PDGFβR) observed in ESCC cell lines (KYSE-30 and KYSE-140) cultured in CAF-containing condition-media as compared to the controls [12]. Their association with DNM3OS expression regulation was further analyzed by transfection with PDGFRβ/PDGFβ siRNA in KYSE-30 and KYSE-140 cells before the treatment with CAF-containing condition-media. The above data suggest that PDGFRB/PDGF signaling pathway is involved in radioresistance in ESCC [12]. Furthermore, chromatin immunoprecipitation (ChIP) assay confirmed that the transcription factor Forkhead box protein O1 (FOXO1, a downstream molecule of PDGFRβ/PDGF pathway, regulates the transcription of DNM3OS by binding to DNM3OS promoter region [12].
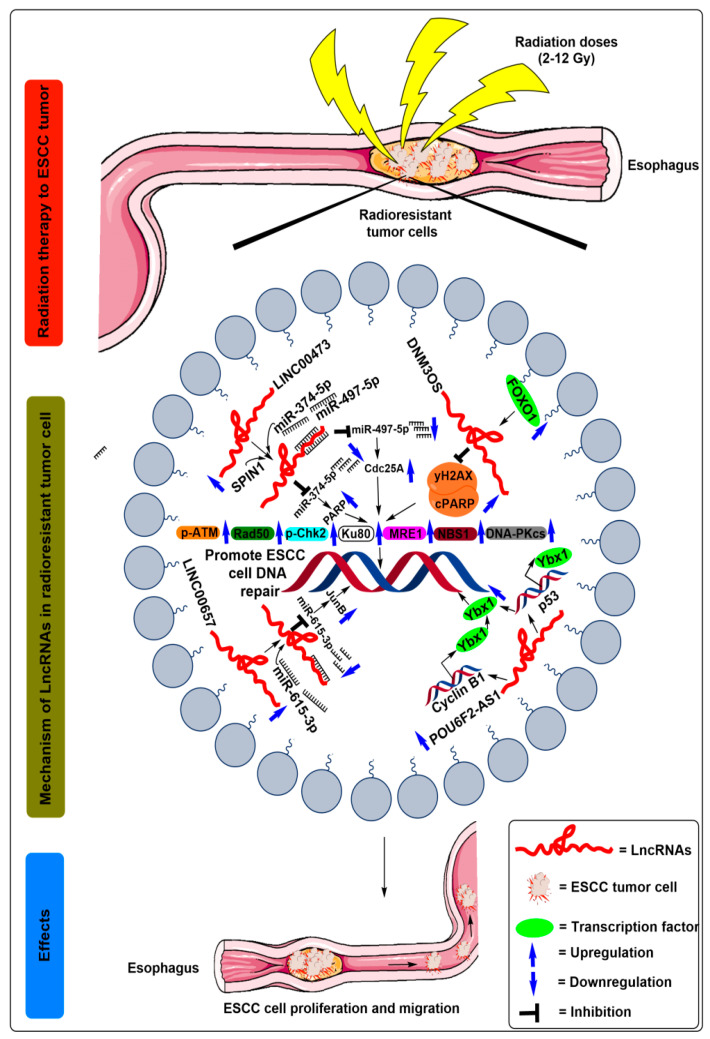
Since the alteration in DNA repair enzymes regulates cell fate in response to radiotherapy treatment, the same authors investigated the involvement of DNM3OS in DNA damage response after radiation treatment to ESCC cells. Mechanistically, the levels of double-strand break proteins, such as H2A histone family member X (γH2AX) protein and cleaved poly ADP ribose polymerase (PARP), raised in irradiated siDNM3OS transfected ESCC cells compared to control cells [12]. Furthermore, the levels of DNA repair enzymes, such as pATM, Rad50, phosphorylated checkpoint kinase 2 (pChk2), Ku80, meiotic recombination 11 homolog 1 (MRE1), Nijmegen breakage syndrome 1 (NBS1), DNA protein kinase (DNA-PKcs), decreases in irradiated siDNM3OS transfected ESCC cells [12] (Figure 2 and Table 1).
Figure 2.
Schematic illustration of the molecular mechanisms of lncRNAs in the regulation of radioresistance in ESCC treatment. After radiation exposure to the ESCC cells, increased expression of FOXO1 transcribes the lncRNA DNM3OS by binding to its promoter region. Moreover, the increased expression of DNM3OS suppressed the levels of double-strand break proteins, such as H2A histone family member X (γH2AX) protein and cleaved poly ADP ribose polymerase (PARP) followed by increased levels of DNA repair enzymes, such as pATM, Rad50, phosphorylated checkpoint kinase 2 (pChk2), Ku80, meiotic recombination 11 homolog 1 (MRE1), Nijmegen breakage syndrome 1 (NBS1), DNA protein kinase (DNA-PKcs), and ultimately promote ESCC cell DNA repair. After radiation exposure to the ESCC cells, SPIN1, miR-374-5p, and miR-497-5p demonstrated competitive binding to upregulated lncRNA LINC00473. Moreover, miR-374-5p and miR-497-5p was suppressed after binding to the LINC00473, which is followed by the upregulated expression of PARP and Cdc25A, respectively, which are ultimately unable to break the double-strand DNA of ESCC cells. After radiation exposure to the ESCC cells, miR-615-5p competes for its binding to the upregulated lncRNA LINC00657. Moreover, the expression of miR-615-5p was suppressed after binding to the LINC00657, which is followed by the upregulated expression of JunB, which ultimately promotes the double-strand DNA repair of ESCC cells. After radiation exposure to the ESCC cells, POU6F2-AS1 expression was increased, which further recruits Ybx1 to the promoters of cyclin B1 (CCNB1) and p53 gene and the DNA damage sites and thus increases the Ybx1 protein levels, which ultimately promotes ESCC cell DNA repair.
The above findings suggest that the DNM3OS in association with CAFs induced radioresistance via targeting PDGFβ and FOXO1 genes in ESCC, which indicate the importance of DNM3OS in ESCC as a target molecule.
3.2. Long Intergenic Non-Protein Coding RNA 473
Long intergenic non-protein coding RNA 473 (LINC00473) is located on the long arm of chromosome 6 at 6q27 [27]. Several studies have reported the association of dysregulated LINC00473 in head and neck squamous cell carcinoma (HNSCC) [28] and ESCC [14], enhancing tumor progression inhibiting radiation response. Unfortunately, limited information is available on the association of LINC00473 with radioresistance in ESCC.
In this regard, Chen et al. [14] observed an upregulated expression of LINC00473 (~2.60-fold) in ESCC tumor tissues (n = 96) compared to the adjacent healthy tissues. Similar results were validated in the ESCC cell lines (TE-1, EC9706, Eca109, and KYSE-450), where TE-1 (~6.20-fold) and EC9706 (~6.80-fold) ESCC cell line demonstrated a higher expression level of LINC00473 compared to normal esophageal epithelial cell line Het-1A (Table 1). The role of LINC00473 was confirmed by irradiating the ESCC cell lines with an increasing dose of X-ray radiation (0, 2, 4, and 8 Gy) [14]. The increased expression of LINC00473 positively correlated with clinicopathological characteristics, such as tumor node metastasis (TNM) stage and poor prognosis of ESCC [14]. Moreover, the shRNA-mediated knockdown of LINC00473 paired with an accumulated dose of irradiation caused a lower survival fraction in the ESCC cell lines (TE-1 and EC9706) as assessed using the colony survival assay [14] (Table 1). Furthermore, another group suggested LINC00473 upregulation in ESCC cells KYSE-30 (~1.20-fold) and TE-5 (~2.2-fold) after 8-doses of X-ray irradiation of 6 Gy each was given every 3 h in a total period of one day compared to the control cells [15]. Furthermore, increased cell proliferation and colony formation potential were observed in ESCC cell lines KYSE-30 and TE-5 when treated with augmented radiation (0, 2, 4, 6, and 8 Gy) doses; this validates the pivotal association of LINC00473 in radioresistance [15]. Mechanistically, the authors observed reduced expression levels of poly (ADP-ribose) polymerase (PARP) protein in ESCC cells irradiated with 4 Gy radiations compared to the control group [14]. The PARP protein plays an important role in repairing DNA damage caused by chemo/radiotherapeutic drugs in cancer [29]. Subsequently, several investigators recognized miR-374a-5p and miR-497-5p as critical downstream targets of LINC00473, downregulating the expression profile against miR-374a-5p and miR-497-5p via competitive sponging [14,15]. Validating the role of LINC00473 in association with miR-374a-5p and miR-497-5p in ESCC radioresistance, these researchers observed enhanced cell viability, colony survival, PARP, and CdC25A protein expression levels in miR-374a-5p and miR-497-5p knockdown in ESCC cell [14,15]. These results demonstrated the oncogenic character of LINC00473, augmenting the radioresistance in ESCC cells.
Furthermore, Cheng et al. [14] verified potential downstream targets against miR-374a-5p, where the SPIN1 gene competed with LINC00473 for miR-374a-5p binding (Figure 2 and Table 1). Additionally, SPIN1 overexpression promoted proliferation and colony formation in vitro, and induced tumor formation and reduced apoptosis in nude mice through PI3K/AKT signaling pathways in ovarian cancer [30,31]. Likewise, depletion of SPIN1 oncogene suppressed the proliferation and survival fraction of ESCC cells and reduced the PARP protein levels, while its overexpression reversed the effects [14].
Overall, the above findings suggest that LINC00473 weakened radiation therapy’s effect in ESCC patients via the cumulative activity of miR-374a-5p/miR-497-5p/SPIN1/PARP1; therefore, LINC00473 could act as a novel prognostic biomarker and a potential therapeutic target for ESCC.
3.3. Non-Coding RNA Activated by DNA Damage
NORAD is a non-coding RNA activated by DNA damage (earlier known as LINC00657) associated intergenic lncRNA located on the long arm of chromosome 20 at 20q11.23. It is evolutionarily conserved among species and ubiquitously expressed in human tissues [32]. Further, its inactivation leads to chromosomal instability in diploid human cell lines. It is ascertained that this lncRNA acts as a multivalent binding platform for the Pumilio-Fem3-binding factor (PUF) family of protein, which plays an essential role in cell cycle control and neuronal activity [32].
Sun et al. [16] by quantitative real-time polymerase chain reaction (qRT-PCR) observed that LINC00657 was upregulated in ESCC tissues by ~1.30-fold and in ESCC cell lines Eca109 (~1.50-fold), and KY-SE (~1.40-fold) after treatment with graded X-ray irradiations of 0, 4, and 8 Gy (Table 1) [16]. Further, it was observed that depletion of LINC00657 in ESCC cells rendered them more sensitive to the radiotherapy treatments, as indicated by the reduced bromodeoxyuridine/5-bromo-2’-deoxyuridine (BrdU+) cell fraction and attenuated proliferation and migration potential of ESCC cells [16] (Figure 2 and Table 1). Additionally, when the ESCC cells were irradiated with a dose of 8 Gy, LINC00657 sponged the miR-615-3p activity and reduced its level in the ESCC cells [16]. Notably, miR-615-5p are involved in lung cancer [33], hepatocellular carcinoma [34], and pancreatic ductal adenocarcinoma [35]. Additionally, by utilizing bioinformatics analysis, the authors identified JunB as the downstream target genes of miR-615-3p. Furthermore an inverse relationship amongst miR-615-3p and JunB protein expression profiles observed using Western blot assays, which suggests that LINC00657 could increase the expression of JunB through sponging miR-615-3p and promotes the radioresistance of ESCC [16] (Figure 2, Table 1). Moreover, Kaplan–Meier analyses of a cohort of 183 ESCC patients showed shorter overall survival in the JunB-high expression group patients when compared with JunB-low expression group patients [16].
From the above results, we conclude that LINC00657 increases the proliferation and migration potential of ESCC cells through targeting miR-615-3p and JunB [16], and enhances the radioresistance in ESCC cells.
3.4. POU6F2 Antisense RNA 2
POU6F2 antisense RNA 2 (POU6F2–AS2) encoded by the p14.1 locus of chromosome 7 and explicitly implicated to enhance ESCC progression [17]. Recently, Liu et al. [17] reported that POU6F2-AS2 is involved in ionizing radiation-induced DNA damage response in ESCC. Moreover, an upregulated expression of POU6F2-AS2 in irradiated ESCC cell lines KYSE-140~5-fold) KYSE-510 (~6.1-fold), KYSE-30 (~2-fold), and KYSE-70 (~1-fold), compared to the control cells [17] (Table 1). Further, depletion of POU6F2-AS2 levels increased the ESCC cell sensitivity towards ionizing radiation. Subsequent experimental evidence, such as RNA pull-down, mass spectrometry, Western blot, and immunoprecipitation assays, suggested that Y-box binding protein (Ybx1) acted as the downstream target of POU6F2-AS2 [17]. Ybx1 is a chromatin-binding protein that regulates RNA/DNA binding events depending upon its localization and implicated in DNA damage repair. Moreover, knockdown of POU6F2–AS2 inhibited the recruitment of Ybx1 to the promoters of cyclin B1 (CCNB1) and p53 gene as well as to the DNA damage sites, thereby, confirming a close relationship between POU6F2-AS2 and DNA repair pathways [17] (Figure 2, Table 1). In addition, ChIP-seq indicated a reduction in the signal corresponding to the binding of Ybx1 with chromatin in response to reduced POU6F2-AS2 expression.
Therefore, these results demonstrated that POU6F2-AS2 is essential for Ybx1′s chromatin localization, which subsequently aids in the growth of ESCC cells after DNA damage, hence collectively enhancing their radioresistance.
4. Detailed Regulation of Radiosensitivity in ESCC through lncRNA4
4.1. Family with Sequence Similarity 201 Member A
Family with sequence similarity 201 member (FAM201A) is a 2.9 kb long-non protein-coding gene, with five splice variants present on the p13.1 region of chromosome 9 [13]. Despite lacking the protein-coding capacity, it plays an essential role in the pathogenesis of various diseases. For instance, several of its single nucleotide polymorphisms (SNPs) associated with both obsessive-compulsive disorders of Tourette’s syndrome [36]. Additionally, its role was observed in osteonecrosis of the femoral head (ONFH) [37] in colorectal cancer [38].
Recently, Chen et al. [13] found significant upregulation of FAM201A in ESCC radioresistant patients (n = 15) compared to radiosensitive (n = 20) patients. Further analyses suggested that ESCC patients belonging to the FAM-high expression group (n = 13, ∆Ct = 8.437) exhibited an inadequate short term response to radiotherapy and had a lower survival time, contrary to the FAM-low expression group (n = 22, ∆Ct = 6.155), whereas, neither of the groups correlated with either T or N stage of tumor stage [13]. Moreover, the authors observed reduced survival and increased apoptotic rates after graded doses of radiation (0, 2, 4, 6, 8, and 10 Gy) in Eca109 radiosensitive cells compared to the Eca109R radioresistant cells [13] (Table 1). Furthermore, siRNA-mediated knockdown of FAM201A exhibited decreased proliferation in ESCC cell lines Eca109 and Eca109R. However, when Eca109R cells transfected with FAM201A mimics, the authors did not observe any increased proliferation because the expression level of FAM201A was already at a higher level in Eca109R cells [10]. Moreover the knockdown of FAM201A in a xenograft tumor mouse model significantly blocked tumor growth with decreased tumor volume and weight, indicating that FAM201A could induce radiosensitivity both in vitro and in vivo in the model system [13].
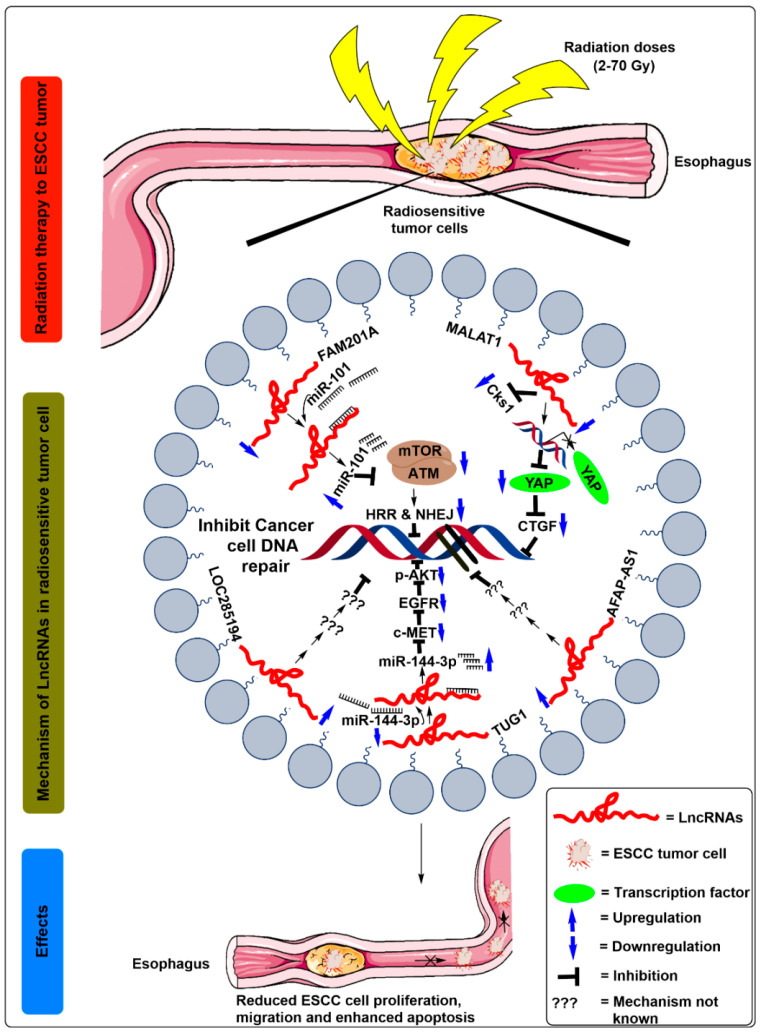
It was further observed that FAM201A confers radiosensitivity in ESCC by sponging its putative downstream target miR-101, and by regulating the expression of mTOR and ATM gene. A previous study by Yan et al. [39] showed that miR-101 functioned as a tumor suppressor in ESCC and regulated the radiosensitivity by inhibiting the mTOR and ATM expression. However, Chen et al. [13] by luciferase reporter assays, observed that FAM201A mutants suppressed the expression of miR-101 in Eca109 cells, suggesting a negative correlation between FAM201A and miR-101. Additionally, FAM201A overexpression using FAM201A mimics significantly reduced the miR-101 expression levels and enhanced the mTOR and ATM protein levels in Eca109/Eca109R cells [13] (Figure 3, Table 1). These results suggest that the increased ATM and mTOR levels due to FAM201A upregulation decrease the apoptotic rate, possibly by enhancing DNA repair by homologous recombination repair pathway (HRR) and non-homologous end joining (NHEJ) pathways.
Figure 3.
Schematic illustration of the molecular mechanisms of lncRNAs in the regulation of radiosensitivity in ESCC treatment. After radiation exposure to the ESCC cells, miR-101 binds to downregulate lncRNA FAM201A. Moreover, the expression of miR-101 was increased after binding to the FAM201A, which is followed by downregulated expression of mTOR and ATM, which decreases the homologous recombination repair (HRR) and non-homologous end joining (NHEJ) pathway and thus promotes the breakdown of double-strand DNA of ESCC cells. After radiation exposure to the ESCC cells, MALAT1 expression was decreased, which further inhibits Cks1 levels at both mRNA and protein levels. In addition, it also decreased YAP’s translational activity, reducing the expression levels of connective tissue growth factor (CTGF), thus enhancing the breakdown of the double-strand DNA in ESCC cells. After radiation exposure to the ESCC cells, downregulated expression of lncRNAs LOC285194 and AFAP-AS1 inhibits DNA repairing of ESCC cells. After radiation exposure to the ESCC cells, miR-144-3p competes with binding to the downregulated lncRNA TUG1. Moreover, the expression of miR-144-3p was increased after binding to the TUG1, which is followed by downregulated expression of c-MET, EGFR, and p-Akt protein, which ultimately promotes the breakdown of double strand DNA of ESCC cells and confers radiosensitivity.
Altogether, FAM201A regulates ESCC radiosensitivity via targeting miR-101/ATM/HRR axes and miR-101/mTOR/NHEJ axes, which suggests the potential of FAM201A as a diagnostic/prognostic biomarker and an excellent therapeutic target for ESCC.
4.2. Metastasis-Associated Lung Adenocarcinoma Transcript 1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is alternatively known as nuclear enriched abundant transcript 2, (NEAT2) is a ubiquitously expressed, 8.7 kb lncRNA that regulates alternative splicing and transcription functions in cells [40]. MALAT1 has been implicated in various malignancies and confers radiosensitivity, including gastric cancer [41], cervical cancer [42], and esophageal squamous cell carcinoma [18].
Li et al. [18] by qRT-PCR observed a downregulated expression of MALAT1 in ESCC cells (EC9706 and KYSE-150) treated with ionizing radiation of 5 Gy for 8 h. Additionally, the overexpression of MALAT1 in irradiated EC9706 and KYSE-150 cells reduced the apoptotic rate leading to enhanced cell viability, suggesting the role of MALAT1 in reducing the radiosensitivity of ESCC cells [18]. The authors further constructed the ESCC tumor xenograft model using a subcutaneous injection of KYSE-150 cells into the BALB/c nude mice and irradiated for five days with a total dose of 10 Gy γ-radiation. As a result, reduced tumor volume observed in the xenograft model compared to the control group [18]. Studies by Yao et al. [19] also revealed a similar downregulated expression of MALAT1 in ESCC cell lines Eca-109, and TE-1 irradiated with a treatment dose of 3.2 Gy/min. In addition, these cells showed reduced spheroid formation capacity, stemness marker expression (SOX2 and Nanog), aldehyde dehydrogenase 1 (ALDH1) activity, and migration potential ESCC.
In order to better understand the mechanisms of the MALAT1 in regulating radiosensitivity in ESCC cells, the authors identified cyclin-dependent kinase subunit 1 (Cks1) and Yes-associated protein (YAP) oncogenic transcriptional factor as the potential targets of MALAT1 (Figure 3, Table 1). The authors further demonstrated that the depletion of MALAT1 in ESCC cells inhibited the Cks1 levels at both mRNA and protein levels. In addition, it also decreased the translational activity of YAP and reduced the expression levels of connective tissue growth factor thus, collectively, enhancing apoptosis and reducing the stemness, cell proliferation, and migration potential of the ESCC cells when subjected to radiation therapy [18,19,43] (Figure 3, Table 1).
The above data confirmed that MALAT1 modulates radiosensitivity and stemness of cancer stem cells by regulating Cks1 and YAP expression levels in ESCC.
4.3. Tumor Suppressor Candidate 7 or Limbic System Associated Membrane Protein
Tumor suppressor candidate 7 (TUSC7) or limbic system associated membrane protein (LOC285194), a 2 kb long lncRNA, consists of four exons and is located on 3q13.31 [44].
Tong et al. [20] by qRT-PCR, examined the expression of LOC285194 in 142 ESCC tissues and observed that the expression of LOC285194 was significantly reduced (~1.10-fold) (Table 1) compared to adjacent healthy tissues. The investigators further analyzed the expression levels of LOC285194 by qRT-PCR in ESCC cell lines. They observed downregulated levels of LOC285194 in KYSE-30 (~2.0-fold), KYSE-510 (~1.7-fold), KYSE-109 (~2.0-fold) cells lines compared to a standard esophageal epithelial cell line, Het-1A [20] (Table 1). Moreover, the LOC285194 low expressing group showed a positive correlation with increased tumor size, higher histologic grade, i.e., G2 and G3/G4, advanced TNM stage, enlarged lymph node metastasis and distant metastasis as compared to the group with a high expression of LOC285194 [20].
Additionally, to investigate the effect of radiation doses on the ESCC patients, 55 patients were irradiated with a total radiation dose of 40 Gy which was administered in 20 fractions each of 2 Gy, for four weeks with five fractions per week, during the first cycle of chemotherapy [20]. As expected, the low LOC285194 expression group was more resistant towards radiotherapy as inferred from a 15% pathCR rate contrary to the 57% pathCR rate in the high LOC285194 expression group [20]. The above results indicate that LOC285194 could be used as a biomarker for screening and treating ESCC patients before esophagectomy. Furthermore, to study the prognostic significance of LOC285194 expression, Kaplan–Meier analysis was performed, which revealed that low expression of LOC285194 was associated with wretched disease-free survival (DFS) and overall survival (OS) [20]. Meanwhile, multivariate analysis revealed that low expression of LOC285194 and distant metastases were independent factors that affected DFS and OS [20]. However, the molecular mechanisms that influence the radiotherapy sensitivity caused by LOC285194 dysregulation are not understood well. Therefore, these results suggest that the downregulated expression of LOC285194 signifies a higher risk of disease recurrence and treatment failure.
4.4. Actin Filament Associated Protein 1
Actin filament associated protein 1 (AFAP1-AS1), a 6810 bp long lncRNA, is transcribed from the actin filament associated protein 1 (AFAP1) gene in the antisense orientation and is located in the 14p16.1 region of chromosome 14. AFAP1 gene codes the protein responsible for the cross-linking of actin filaments [45].
Zhou et al. [21], screened 18 lncRNAs, implicated in esophageal cancer in association with chemoradiotherapy resistance using qRT-PCR. AFAP1-AS1, along with two other lncRNAs, was deregulated more than 2-fold in the paired cell lines KYSE-30 and its resistant counterpart KYSE-30R (exhibited cross-resistance to 5-FU by ~3-folds and paclitaxel by ~5-fold). A similar result was observed in the parental TE-10 and cisplatin-resistant TE-10R cell lines. Furthermore, 204 patients underwent a total radiation dose of 60–70 Gy, i.e., 1.8–2.0 Gy/day for five days/week. A total of 162 out of the 204 patients were selected for further investigation, amongst which 81 cases belonged to a high AFAP1-AS1 expression group, and the other 81 cases belonged to the low AFAP1-AS1 expression group. In another cohort of 48 patients, the expression of AFAP1-AS1 upregulation of ~0.81-fold was observed compared to adjacent healthy tissues [21]. This upregulation further correlated with lymph node metastasis, distant metastasis, advanced clinical stage, and resistance to definitive chemoradiotherapy (dCRT). However, no clinicopathological feature was observed to be associated with dCRT response. A similar upregulation of about ~2.2–15-fold was found in the cell lines KYSE-30, KYSE-70, KYSE-150, KYSE-450, KYSE-510, and KYSE-10 as compared to the healthy esophageal mucosa cell Het-1A [21] (Table 1).
Additionally, statistical analysis revealed that AFAP1-AS1 could efficiently distinguish between ESCC samples and healthy esophageal mucosa (79.4% specificity and 73.3% sensitivity) and could also detect early ESCC (stage I + II, n = 79) with a specificity and sensitivity of 92.3% and 44.6% respectively [21]. However, results corresponding to the Kaplan–Meier survival analysis showed that higher levels of AFAP1-AS1 were negatively associated with progression-free survival (PFS) and OS before dCRT [21]. Consistently, multivariate analysis revealed the upregulation of AFAP1-AS1 as an influential independent prognosis factor of inferior PFS but the most significant unfavorable prognostic factor of OS [21].
These composite results demonstrate that AFAP1-AS1 could be a predictive marker of clinical outcomes in ESCC patients treated with dCRT.
4.5. Taurine-Upregulated Gene 1
lncRNA taurine-upregulated gene 1 (TUG1) is a 6.7 kb-long transcript with six reading frames and mapped to the chromosome region 22q12.2. It was first identified as an upregulated transcript in the developing retinal cells [46].
Wang et al. [22] demonstrated that TUG1 significantly upregulated in radiosensitive ESCC tissues (~1.5-fold) and cell lines, such as EC9706 and KYSE-30 (-3.0-fold and 2.3-fold, respectively) to their resistant counterparts (Table 1). The depletion of TUG1 by siRNA mediated knockdown resulted in suppressed proliferation, migration, invasion, and increased apoptosis of EC9706 and KYSE-30 cell lines. Additionally, it was observed that exposure of TUG1-depleted cells with 2 Gy irradiation inhibited their proliferation, colony-forming ability, and induced apoptosis. These observations collectively concluded that TUG1 is associated with ESCC radiosensitivity and radiotherapy [22]. Furthermore, the authors constructed an in vivo model by establishing KYSE-30 xenografts in Balb/c nude mice to confirm their previous findings. Interestingly, reduced tumor volume, tumor growth, and tumor weight observed in response to TUG1 knockdown in the KYSE-30 xenografts compared to the controls after irradiation of 2 Gy [22].
Mechanistically, hsa-miR-144-3p and hsa-miR-145-5p were found as the potential target of TUG1 [22]. The authors further indicated by Luciferase reporter assays that miR-144-3p was indeed a downstream target of TUG1, where these two transcripts were inversely correlated as established by Pearson’s correlation coefficient. miR-144-3p acts as a tumor suppressor in ESCC, and depletion of miR-144-3p restored the effects of TUG1 suppression on radiotherapy, consequently validating its role as a regulator of radiation [22] (Figure 3, Table 1). Additionally, met proto-oncogene (MET) harbored a binding site for miR-144-3p at its 3’-UTR region. [43]. Further, the expression levels of MET protein and its mRNA levels were decreased in the cell lines transfected with miR-144-3p mimics. However, miR-144-3p inhibitors restored the MET expression levels that were suppressed in response to si-TUG1, suggesting the existence of lncTUG1/miR-144-3p/MET axis [22].
c-MET is a proto-oncogene that codes for the protein c-MET receptor tyrosine kinase (c-MET RTK). It expresses in the epithelial tissues of many organs during embryogenesis adulthood [47]. In addition, MET induced upregulation of epidermal growth factor (EGFR) and increased Akt phosphorylation, which further reflects a resistance effect on cancer radiotherapy [22] (Figure 3, Table 1). Consistent with these findings, knockdown of MET decreased EGFR and phosphorylated-Akt (p-Akt) protein levels, possibly inhibiting cell proliferation [43]. However, reduced tumor levels of Ki67 and lowest levels of MET and p-Akt observed in the 2 Gy radiation group depleted with si-TUG1 [22].
These composite results indicate that lncTUG1 regulates the radiosensitivity of ESCC via the TUG1/miR-144-3p/MET axis and can utilize it as a potential biomarker for radiotherapy.
5. Conclusion and Future Perspectives
We summarize the current scientific insights into the association of lncRNAs with a response to radiotherapy and a predictive biomarker for radiation treatment in ESCC patients. Further, we observed that lncRNAs might also act as potential therapeutic targets because of their differential expression patterns in ESCC cancerous tissue/cell lines before and after radiation therapy. The lncRNAs DNM3OS, LINC00473, LINC00657, POU6F2–AS2 found to be involved in radioresistance, while FAM201A, MALAT1, LOC285194, AFAP1-AS1, TUG1 lncRNAs linked to radiosensitivity in ESCC. As the upregulation of DNM3OS and POU6F2–AS2 expression enhances radioresistance in ESCC cells. However, knockdown of lncRNA DNM3OS and POU6F2–AS2 increased the extent of radiosensitivity in ESCC cells, as demonstrated by higher expression of yH2AX and reduced levels of DNA repair proteins followed by impaired double-strand break (DSB) repair [17,48]. The reviewed literature data indicate the potential efficacy of DNM3OS and POU6F2–AS2 as targets to improve the outcome of radiotherapy. We have also highlighted the cumulative association of miRNAs and lncRNAs in many aspects of cellular response to radiation therapy. For example, oncogenic LINC00473 favors radiation therapy resistance combined with mir-374a-5p, followed by alteration in DNA damage repair protein [14]. Furthermore, collated functions of LINC00657 and miR-615-3p in ESCC cells hamper the TGF signaling pathways and impair the cellular response to radiation [16]. On the other hand, oncogenic FAM201A and TUG1 favor radiosensitivity by inhibiting miR-101 and miR-144-3p, respectively. These findings suggest lncRNA and miRNA’s functional relevance, which together possess great potential in radiation therapy. Moreover, the preclinical studies and increased success rates of nucleic acid therapeutics provide an opportunity to target lncRNAs-miRNA for cancer treatment [16].
Radiotherapy is a well-established noninvasive treatment modality in which many patients do respond and get treated. However, several factors, such as familial mutation, epigenetic changes, and other comorbid pathological conditions, increase the resistance in a large cohort of patients, which is alarming and needs more in-depth analysis for improving interventions. Given this, our study addresses this and highlights lncRNAs as one molecular signature attributing to radiotherapy’s increased resistance. We have also proposed that modifying the lncRNA signature can influence the sensitivity of refractory patients. Furthermore, our study emphasized the role of lncRNAs in facilitating radiation therapy and suggested that LncRNA based therapeutics hold tremendous potential in improving cancer-directed therapies. lncRNA based intervention is an emerging area and expected to be a critical factor in deciding treatment modality for refractory or highly aggressive tumors ranging from gastric to lung and many more.
With the advancement of RNA-based therapeutics delivery, new combined radio-RNA therapies can emerge [49,50]. However, there would be challenges for the effective delivery of RNAi to cells; thus, there would be obstacles in the clinical trials-based study. Overall, multicentric studies in a large number of patients’ samples are warranted to utilize the lncRNAs based therapeutics in ESCC cells.
Abbreviations
| AFAP1-AS1 | Actin Filament Associated Protein 1 |
| ALDH1 | Aldehyde dehydrogenase 1 |
| AP-1 | Activator protein 1 |
| ATM | Ataxia telangiectasia mutated |
| BrdU | Bromodeoxyuridine/5-bromo-2’-deoxyuridine |
| CAFs | Cancer-associated fibroblasts |
| CCNB1 | Cyclin B1 |
| Cdc25A | Cell division cycle 25 A |
| ChIP | Chromatin immunoprecipitation |
| Cks1 | Cyclin-dependent kinase subunit 1 |
| c-MET RTK | c-MET receptor tyrosine kinase |
| CTGF | Connective tissue growth factor |
| dCRT | Definitive chemoradiotherapy |
| DDR | DNA damage response |
| DNA-PKcs | DNA protein kinase |
| DNM3OS | Dynamin 3 Opposite Strand |
| DOAJ | Directory of open access journals |
| EC | Esophageal cancer |
| ESCC | Esophageal squamous cell carcinoma |
| FAM201A | Family with sequence similarity 201 member A |
| FOXO1 | Forkhead box protein O1 |
| HNSCC | Head and neck squamous cell carcinoma |
| LD | Linear dichroism |
| LINC00473 | Long Intergenic Non-Protein Coding RNA 473 |
| lncRNAs | Long non-coding RNAs |
| MALAT1 | Metastasis-Associated Lung Adenocarcinoma Transcript 1 |
| MDPI | Multidisciplinary Digital Publishing Institute |
| miRNAs | MicroRNAs |
| MRE1 | Meiotic recombination 11 homolog 1 |
| mTOR | Mammalian target of rapamycin |
| NBS1 | Nijmegen breakage syndrome 1 |
| NHEJ | Non-homologous end joining |
| NORAD | Non-coding RNA activated by DNA damage |
| p-Akt | Phosphorylated-Akt |
| PARP | Protein and cleaved poly ADP ribose polymerase |
| pChk2 | Phosphorylated checkpoint kinase 2 |
| PDGFR α/β | Platelet-derived growth factor α/β |
| PDGFβ | PDGF β-polypeptide |
| PDGFβR | Platelet-derived growth factor receptor βpolypeptide |
| PFS | Progression-free survival |
| POU6F2—AS2 | POU6F2 Antisense RNA 2 |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SNPs | Single nucleotide polymorphisms |
| SPIN1 | Spindlin 1 |
| TNM | Tumor node metastasis |
| TUG1 | Taurine-Upregulated Gene 1 |
| TUSC7 | Tumor Suppressor Candidate 7 |
| YAP | Yes-associated protein |
| γH2AX | yH2A histone family member X |
Author Contributions
U.S., V.A., T.S.B., and A.J. conceived the idea. U.S., V.A., and T.S.B. wrote the first draft of the manuscript. U.S. prepared the figures and table. K.S., M.K.R., S.K.S., H.P., and A.J. revised the manuscript. A.B. edited the manuscript and suggested improvement for scientific quality, organization and style. All authors have read and agreed to the published version of the manuscript.
Funding
Uttam Sharma would like to acknowledge Department of Science and Technology (DST), India for the DST-INSPIRE fellowship grant (IF180680).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Liang H., Fan J.H., Qiao Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohashi S., Miyamoto S., Kikuchi O., Goto T., Amanuma Y., Muto M. Recent Advances from Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 3.Yang X., Zhu H., Qin Q., Yang Y., Yang Y., Cheng H., Sun X. Genetic variants and risk of esophageal squamous cell carcinoma: A GWAS-based pathway analysis. Gene. 2015;556:149–152. doi: 10.1016/j.gene.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su M., Xiao Y., Ma J., Cao D., Zhou Y., Wang H., Liao Q., Wang W. Long non-coding RNAs in esophageal cancer: Molecular mechanisms, functions, and potential applications. J. Hematol. Oncol. 2018;11:118. doi: 10.1186/s13045-018-0663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H., Song H., Chen G., Yang X., Liu J., Ge Y., Lu J., Qin Q., Zhang C., Xu L., et al. eEF2K promotes progression and radioresistance of esophageal squamous cell carcinoma. Radiother. Oncol. 2017;124:439–447. doi: 10.1016/j.radonc.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Podralska M., Ciesielska S., Kluiver J., van den Berg A., Dzikiewicz-Krawczyk A., Slezak-Prochazka I. Non-Coding RNAs in Cancer Radiosensitivity: MicroRNAs and lncRNAs as Regulators of Radiation-Induced Signaling Pathways. Cancers. 2020;12:1662. doi: 10.3390/cancers12061662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickman A.W., Jaramillo R.J., Lechner J.F., Johnson N.F. Alpha-particle-induced p53 protein expression in a rat lung epithelial cell strain. Cancer Res. 1994;54:5797–5800. [PubMed] [Google Scholar]
- 8.Chi H.C., Tsai C.Y., Tsai M.M., Yeh C.T., Lin K.H. Roles of Long Noncoding RNAs in Recurrence and Metastasis of Radiotherapy-Resistant Cancer Stem Cells. Int. J. Mol. Sci. 2017;18:1903. doi: 10.3390/ijms18091903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra A., Jain M., Prakash H., Vasquez K.M., Jain A. The regulatory roles of long non-coding RNAs in the development of chemoresistance in breast cancer. Oncotarget. 2017;8:110671–110684. doi: 10.18632/oncotarget.22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra A., Sharma U., Puhan S., Bandari N.C., Kharb A., Arifa P.P., Thakur L., Prakash H., Vasquez K.M., Jain A. Stabilization of miRNAs in esophageal cancer contributes to radioresistance and limits efficacy of therapy. Biochimie. 2019;156:148–157. doi: 10.1016/j.biochi.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L., Wang Q., Liu R., Chen Z., Zhang X., Zhou P., Wang Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46:717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Hua Y., Jiang Z., Yue J., Shi M., Zhen X., Zhang X., Yang L., Zhou R., Wu S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019;25:1989–2000. doi: 10.1158/1078-0432.CCR-18-0773. [DOI] [PubMed] [Google Scholar]
- 13.Chen M., Liu P., Chen Y., Chen Z., Shen M., Liu X., Li X., Li A., Lin Y., Yang R., et al. Long Noncoding RNA FAM201A Mediates the Radiosensitivity of Esophageal Squamous Cell Cancer by Regulating ATM and mTOR Expression via miR-101. Front. Genet. 2018;9:611. doi: 10.3389/fgene.2018.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W., Zhang Y., Wang H., Pan T., Zhang Y., Li C. LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy. J. Cell. Biochem. 2019;120:14562–14572. doi: 10.1002/jcb.28717. [DOI] [PubMed] [Google Scholar]
- 15.Liu W.H., Qiao H.Y., Xu J., Wang W.Q., Wu Y.L., Wu X. LINC00473 contributes to the radioresistance of esophageal squamous cell carcinoma by regulating microRNA4975p and cell division cycle 25A. Int. J. Mol. Med. 2020;46:571–582. doi: 10.3892/ijmm.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y., Wang J., Pan S., Yang T., Sun X., Wang Y., Shi X., Zhao X., Guo J., Zhang X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed. Pharmacother. 2018;108:316–324. doi: 10.1016/j.biopha.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Sun X., Zhu H., Qin Q., Yang X., Sun X. Long noncoding RNA POU6F2-AS2 is associated with oesophageal squamous cell carcinoma. J. Biochem. 2016;160:195–204. doi: 10.1093/jb/mvw025. [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Zhou Y., Tu B., Bu Y., Liu A., Kong J. Long noncoding RNA MALAT1 affects the efficacy of radiotherapy for esophageal squamous cell carcinoma by regulating Cks1 expression. J. Oral Pathol. Med. 2017;46:583–590. doi: 10.1111/jop.12538. [DOI] [PubMed] [Google Scholar]
- 19.Yao Q., Yang J., Liu T., Zhang J., Zheng Y. Long noncoding RNA MALAT1 promotes the stemness of esophageal squamous cell carcinoma by enhancing YAP transcriptional activity. FEBS Open Bio. 2019;9:1392–1402. doi: 10.1002/2211-5463.12676. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Tong Y.S., Zhou X.L., Wang X.W., Wu Q.Q., Yang T.X., Lv J., Yang J.S., Zhu B., Cao X.F. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J. Transl. Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X.L., Wang W.W., Zhu W.G., Yu C.H., Tao G.Z., Wu Q.Q., Song Y.Q., Pan P., Tong Y.S. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol. Carcinog. 2016;55:2095–2105. doi: 10.1002/mc.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P., Yang Z., Ye T., Shao F., Li J., Sun N., He J. lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J. Exp. Clin. Cancer Res. 2020;39:7. doi: 10.1186/s13046-019-1519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gotzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009;62:e1–e34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe T., Sato T., Amano T., Kawamura Y., Kawamura N., Kawaguchi H., Yamashita N., Kurihara H., Nakaoka T. DNM3OS, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 25.Wang S., Ni B., Zhang Z., Wang C., Wo L., Zhou C., Zhao Q., Zhao E. Long non-coding RNA DNM3OS promotes tumor progression and EMT in gastric cancer by associating with Snail. Biochem. Biophys. Res. Commun. 2019;511:57–62. doi: 10.1016/j.bbrc.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Tao L., Huang G., Song H., Chen Y., Chen L. Cancer associated fibroblasts: An essential role in the tumor microenvironment. Oncol. Lett. 2017;14:2611–2620. doi: 10.3892/ol.2017.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C., Yang Y., Yu J., Meng F., Zhang T., Gao Y. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am. J. Cancer Res. 2017;7:2157–2168. [PMC free article] [PubMed] [Google Scholar]
- 28.Han P.B., Ji X.J., Zhang M., Gao L.Y. Upregulation of lncRNA LINC00473 promotes radioresistance of HNSCC cells through activating Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7305–7313. doi: 10.26355/eurrev_201811_16267. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Weaver D.T. The ups and downs of DNA repair biomarkers for PARP inhibitor therapies. Am. J. Cancer Res. 2011;1:301–327. [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y., Yue W., Zhang P., Li L., Xie X., Yuan H., Chen L., Liu D., Yan F., Pei X. Spindlin1, a novel nuclear protein with a role in the transformation of NIH3T3 cells. Biochem. Biophys. Res. Commun. 2005;335:343–350. doi: 10.1016/j.bbrc.2005.07.087. [DOI] [PubMed] [Google Scholar]
- 31.Bothig B., Schulze P., Schreier E., Diedrich S., Michel S. Atypical human rotaviruses in the G.D.R. Acta Virol. 1989;33:320–326. [PubMed] [Google Scholar]
- 32.Lee S., Kopp F., Chang T.C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y., Mendell J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong Y., Huo X., Sun R., Liu Z., Huang M., Yang S. lncRNA Gm15290 promotes cell proliferation and invasion in lung cancer through directly interacting with and suppressing the tumor suppressor miR-615-5p. Biosci. Rep. 2018;38 doi: 10.1042/BSR20181150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X., Deng L., Tang D., Ying G., Yao X., Liu F., Liang G. miR-615-5p prevents proliferation and migration through negatively regulating serine hydromethyltransferase 2 (SHMT2) in hepatocellular carcinoma. Tumour Biol. 2016;37:6813–6821. doi: 10.1007/s13277-015-4506-8. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y., Zhang T., Wang C., Jin X., Jia C., Yu S., Chen J. MiRNA-615-5p functions as a tumor suppressor in pancreatic ductal adenocarcinoma by targeting AKT2. PLoS ONE. 2015;10:e0119783. doi: 10.1371/journal.pone.0119783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D., Mathews C.A., Scharf J.M., Neale B.M., Davis L.K., Gamazon E.R., Derks E.M., Evans P., Edlund C.K., Crane J., et al. Cross-disorder genome-wide analyses suggest a complex genetic relationship between Tourette’s syndrome and OCD. Am J. Psychiatry. 2015;172:82–93. doi: 10.1176/appi.ajp.2014.13101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang G., Zhao G., Xia J., Wei Y., Chen F., Chen J., Shi J. FGF2 and FAM201A affect the development of osteonecrosis of the femoral head after femoral neck fracture. Gene. 2018;652:39–47. doi: 10.1016/j.gene.2018.01.090. [DOI] [PubMed] [Google Scholar]
- 38.Matsumura K., Kawasaki Y., Miyamoto M., Kamoshida Y., Nakamura J., Negishi L., Suda S., Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36:1191–1199. doi: 10.1038/onc.2016.282. [DOI] [PubMed] [Google Scholar]
- 39.Yan D., Ng W.L., Zhang X., Wang P., Zhang Z., Mo Y.Y., Mao H., Hao C., Olson J.J., Curran W.J., et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y., Pan J., Geng Q., Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. FEBS Open Bio. 2019;9:1212–1222. doi: 10.1002/2211-5463.12649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Lu H., He Y., Lin L., Qi Z., Ma L., Li L., Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37:1683–1691. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 43.Dong X., Meng L., Liu P., Ji R., Su X., Xin Y., Jiang X. YAP/TAZ: A promising target for squamous cell carcinoma treatment. Cancer Manag. Res. 2019;11:6245–6252. doi: 10.2147/CMAR.S197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H., Chen A., Shen J., Zhang X., Hou M., Li J., Chen J., Zou H., Zhang Y., Deng Q., et al. Long non-coding RNA LOC285194 functions as a tumor suppressor by targeting p53 in non-small cell lung cancer. Oncol. Rep. 2019;41:15–26. doi: 10.3892/or.2018.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu S., Yang D., Ye Y., Liu P., Chen Z., Lei T., Pu J., Liu L., Wang Z. Long noncoding RNA actin filament-associated protein 1 antisense RNA 1 promotes malignant phenotype through binding with lysine-specific demethylase 1 and repressing HMG box-containing protein 1 in non-small-cell lung cancer. Cancer Sci. 2019;110:2211–2225. doi: 10.1111/cas.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 47.Organ S.L., Tsao M.S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011;3(Suppl. 1):S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H., Gao S., De Geyter C. A natural antisense transcript, BOKAS, regulates the pro-apoptotic activity of human Bok. Int. J. Oncol. 2009;34:1135–1138. doi: 10.3892/ijo_00000241. [DOI] [PubMed] [Google Scholar]
- 49.Amodio N., Raimondi L., Juli G., Stamato M.A., Caracciolo D., Tagliaferri P., Tassone P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018;11:63. doi: 10.1186/s13045-018-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]