Abstract
Adjuvant chemotherapy(AC) plays a substantial role in the treatment of locally advanced gastric cancer (LAGC), but the response remains poor. We aims to improve its efficacy in LAGC. Therefore, we identified the expression of eight genes closely associated with platinum and fluorouracil metabolism (RRM1, RRM2, RRM2B, POLH, DUT, TYMS, TYMP, MKI67) in the discovery cohort (N=291). And we further validated the findings in TCGA (N=279) and GEO. Overall survival (OS) was used as an endpoint. Univariate and multivariate Cox models were applied. A multivariate Cox regression model was simulated to predict the OS. In the discovery cohort, the univariate Cox model indicated that AC was beneficial to high-RRM1, high-DUT, low-RRM2, low-RRM2B, low-POLH, low-KI67, low-TYMS or low-TYMP patients, the results were validated in the TCGA cohort. The multivariate Cox model showed consistent results. Cumulative analysis indicated that patients with low C-Score respond poorly to the AC, whereas the high and medium C-Score patients significantly benefit from AC. A risk model based on the above variables successfully predicted the OS in both cohorts (AUC=0.75 and 0.67, respectively). Further validation in a panel of gastric cancer cell (GC) lines (N=37) indicated that C-Score is significantly associated with IC50 value to fluorouracil. Mutation profiling showed that C-Score was associated with the number and types of mutations. In conclusion, we successfully simulated a predictive signature for the efficacy of AC in LAGC patients and further explored the potential mechanisms. Our findings could promote precision medicine and improve the prognosis of LAGC patients.
Keywords: Locally advanced gastric cancer, Adjuvant chemotherapy, Overall survival, Prediction, Mutation
Highlights
-
•
We successfully simulated a predictive signature for the efficacy of chemotherapy in LAGC patients and a GC cell line panel.
-
•
We further explored the potential mechanisms that it may be associated with the number and type of mutations.
-
•
Our findings could promote precision medicine and improve the prognosis of LAGC patients.
Introduction
Gastric cancer (GC) is the third most common cause of cancer mortality worldwide, kills over 780,000 patients in 2018 [1]. For early GC, which is confined to mucosa or submucosa (T1), endoscopic submucosal resection could improve 5-year survival rate to 84% [2,3]. However, though curative surgery and adjuvant therapies are employed, the prognosis of locally advanced GC (LAGC, stage II/III) remains poor [4]. Post-operative chemotherapy has been proved effective; still, many patients suffer relapse of the disease and eventually have poor outcomes [[5], [6], [7], [8]]. Therefore, how to improve the efficacy of chemotherapy in LAGC is an urgent problem to be solved.
Multiple strategies have been implemented to enhance the efficacy of chemotherapy, like neoadjuvant chemotherapy. The MAGIC trial demonstrates that patients of LAGC received pre-operative ECF (epirubicin, cisplatin, and fluorouracil) regimen shows improved 5-year OS rate (from 23% to 36%) compare to postoperative setting [9]. FFCD9703 trial improves 5-year OS rate from 24% to 38% by using cisplatin and fluorouracil pre-operatively [5,10]. ARTIST trial indicates that post-operative chemoradiation fails to reduce the recurrence rate after curatively resection of GC compared to chemotherapy alone [11]. Notably, ACTS-GC and CLASSIC trials highlight the efficacy of S-1 and capecitabine plus oxaliplatin regimens in improving the 5-year OS rate of patients with stage II/III GC [6,7]. However, over 30% of LAGC patients suffer progressive disease (PD) after chemotherapy, in which mechanisms remain unknown.
Platinum and fluorouracil are the most common reagents in chemotherapy of LAGC [12]. It is suggested ribonucleotide reductase (RR), thymidine synthase (TYMS), thymidine phosphorylase (TYMP), deoxyuridine triphosphatase (DUT) and dihydropyrimidine dehydrogenase (DPD) playing substantial roles in the metabolism of 5-fluorouracil [13]. The genetic variants, mRNA, and protein expression of TYMS, TYMP, RR, and DPD were identified as prognostic factors or predictors of chemotherapy response in GC patients [[14], [15], [16], [17], [18], [19]]. DNA repair-related genes like excision repair cross-complementing 1 (ERCC1), DNA polymerase η (POLH) were reported to be associated with platinum resistance [14,20]. A 72-gene signature of acquired resistance to cisplatin and 5-fluorouracil chemotherapy was identified in 123 metastatic GC patients, which could predict time to progress [21]. Another Singaporean study with 318 gastric cancer patients categorized the patients into three subtypes that may respond to different chemo-reagents [22]. Among these studies, RR, TYMS, DUT, TYMP, POLH were considered playing key roles in the resistance to cisplatin and 5-fluorouracil [23]. However, to date, no consensus has been reached in signature for chemotherapy in LAGC.
To identify an optimal signature for chemotherapy in LAGC, we retrospectively enrolled 291 post-operative patients with stage II/III GC, and we quantitated the signature of 8 biomarkers (RRM1, RRM2, RRM2B, TYMS, TYMP, DUT, POLH and Ki67) in the fluorouracil and platinum metabolism pathways on slides of patients by immunohistochemistry (IHC). Furthermore, we validated our model on TCGA (The Cancer Genome Atlas, N = 279) and a cell lines microarray (N = 37) from GEO (Gene Expression Omnibus). The findings were further analyzed in mutation profiles of TCGA and GEO datasets, which proposed the possible mechanism. Finally, a multivariable prediction model was addressed in both cohorts to predict the efficacy of fluorouracil and platinum-based chemotherapy for LAGC.
Materials and methods
Ethics statement
The protocol of this study was reviewed and approved by the institutional review board (IRB) of Zhejiang University Affiliated Sir Run Run Shaw Hospital (SRRSH) (Approval code: 2016-0628-3). Written informed consent was obtained from all the patients enrolled in this study.
Study design
The study design was depicted in Fig. S1. Briefly, we enrolled 472 GC patients who were treated at the department of surgical oncology in SRRSH between 1995 and 2011. The inclusion criteria were as followed: 1. Stage II and III Gastric adenocarcinoma with pathology diagnosis, 2. Inform consent or waiver of consent provided by the patient. We excluded patients with the following criteria: 1. Metastatic or Stage I disease; 2. None-adenocarcinoma or multiple cancer; 3. No tissue sample obtained; 4. Failure to provide consent inform; 5. Loss of follow-up after surgery. A total of 291 out of 472 GC patients were finally enrolled in the study. All patients were Han Chinese. Among these patients, 173 of 291 patients received adjuvant chemotherapy (Table 1). The chemotherapy regimens included folinic acid, 5-fluorouracil and oxaliplatin (FOLFOX6; 93 Cases); epirubicin, oxaliplatin, and xeloda (EOX; 13 cases); epirubicin, oxaliplatin and 5-fluorouracil (EOF; 49 cases), multiple chemo-regimen (12 cases), mitomycin and 5-fluorouracil (4 cases) and oral S-1 regimen (2 cases). All patients were periodically followed up. The TNM stage was determined according to the NCCN guidelines for GC (Version 3, 2016) [12]. The human tissue samples were obtained from surgery and stored at room temperature after formalin-fixed and paraffin-embedded (FFPE). Correlation result displayed storage time did not affect gene expression in statistical significance (Table S1).
Table 1.
Host characteristics of SRRSH and TCGA cohorts.
| Variables/biomarkers | SRRSH | TCGA |
|---|---|---|
| All casesa | 291 | 279 |
| Age | ||
| Mean (SD) | 59.7 (11.6) | 65.8 (10.6) |
| Range | 29–80 | 29–80 |
| ≤60 | 133 | 81 |
| >60 | 158 | 193 |
| Sex | ||
| Male | 207 | 179 |
| Female | 84 | 100 |
| TNM stage | ||
| II | 111 | 117 |
| III | 180 | 162 |
| Tumor gradeb | ||
| Low | 35 | 6 |
| Medium | 71 | 81 |
| High | 185 | 192 |
| Histology | ||
| Papillary | 4 | NA |
| Mucinous | 24 | NA |
| Signet ring cell | 30 | NA |
| Poor differentiation | 231 | NA |
| Tumor location | ||
| Proximal | 61 | NA |
| Body | 63 | NA |
| Distal | 156 | NA |
| Linitis plastica | 11 | NA |
| Tumor size | ||
| <5cm | 108 | NA |
| ≥5cm | 174 | NA |
| Chemotherapy | ||
| Yes | 173 | 146 |
| No | 118 | 133 |
Among all biomarkers, there are part of patients' expression information missing: 49 patients in RRM1,48 patients in RRM2, 46 patients in RRM2B, 49 patients in POLH, 47 patients in DUTPASE, 48 patients in KI-67, 46 patients in TYMS and 39 patients in TYMP in SRRSH cohort; In TCGA dataset, age information in 5 cases were missing.
33 cases in the SRRSH cohort and 4 cases in the TCGA cohort are undifferentiated, which were included in high tumor grade.
We validated our results in TCGA [24] and GC cell lines dataset (GSE22183) [22] from GEO. All clinical, pathological information of 279 LAGC cases in 391 GC patients were enrolled in our study (Table 1). In the GSE22183 dataset, genome wide mRNA expression profiles of 37 GC lines were achieved and analyzed. Among them, 28 gastric cancer cell lines were tested for the 50% growth inhibition (IC50) to fluorouracil (detailed information was listed in Table S2).
Immunohistochemistry
The immunohistochemistry staining (IHC) was applied to determine the protein levels of biomarkers on FFPE samples. To optimize the reaction conditions, we reassembled all FFPE tissue samples into a Multiple Tissue Array as we previously described [15]. The accuracy of IHC was validated by quantitative RT-PCR (qRT-PCR) on two parallel samples. Briefly, sections were deparaffinized by xylene and rehydrated, the endogenous peroxidase activity was blocked with 3% H2O2 (hydrogen peroxide) in methanol for 15 min at room temperature. Then the sections were placed in citrate buffer (pH 6.0) and heated in a microwave oven for 10 min for antigen retrieval. The sections were later incubated with normal goat serum and then applied with a primary antibody. Further, the slides were incubated with horseradish peroxidase-labeled polymer conjugated diaminobenzidine and then counterstained with hematoxylin (DAKO).
An automated imaging system was employed to obtain digital images of the stained sections for subsequent quantitative analyses. Two investigators scored the expression of biomarkers for each sample in a double-blind manner. Score criteria of RRM1, RRM2, and RRM2B were similar to our previous studies [15,25]. Both immunoreactivities in nucleus and cytoplasm were evaluated using a weighted histoscore method [26]. All the images were scored based on the following categories: subcellular localization, staining intensity, and/or percentage of stained cells. The biomarkers' expressions were classified as negative (−), weakly positive (+), positive (++), or strongly positive (+++) (Fig. S2). An expression score of − or + was designated as low expression, and a score of ++ or +++ was defined as high expression.
Antibodies
The primary antibodies of RRM1 (1:50), RRM2 (1:10), and RRM2B (1:200) were generated, selected, and tested in our previous experiments [25,27]. Thymidine synthase antibody (1:5, Catalog#: MBS190051), Ki67 (1:100, Clone: B56), dUTPase (DUT, 1:500, Clone: 1C9), thymidine phosphorylase (1:5, Catalog#: ab180783) and DNA polymerase η (1:200, PA5-29063) were purchased from MyBioSource (San Diego, CA), BD Bioscience (San Jose, CA), Novus (Littleton, CO), Abcam (Cambridge, MA) and Thermo Fisher (Waltham, MA), respectively.
Statistics analysis
All demographic data, clinicopathological information, and IHC results were coded and resembled as a database. Double data entry and logic checks were used for error reduction. SAS 9.4 (SAS Institute, USA), JMP 8.0 Software (SAS Institute, USA) and GraphPad Prism 5.0 (GraphPad Software, USA) were used for the statistical analysis and plotting. Contingency tables and Fisher's exact test were used for the categorical variables to evaluate the association between markers and clinical variables. The OS was calculated from the date of diagnosis to the date of death. Patients who were alive at the last follow-up were censored. The association between biomarker and survival was analyzed using the Kaplan–Meier method and log-rank test. Multivariate analyses were carried out using the Cox proportional hazards model adjust with age, gender, and the interaction effect between chemotherapy and covariates of interest in both the SRRSH cohort and TCGA cohort. All tests were two-sided, and P < 0.05 was considered statistically significant.
Computational modeling and verification
The Cox proportional hazard regression was performed to develop a model to assess and predict the effect of chemotherapy on patients with different expressions of biomarkers adjusting for age, gender, and tumor grade. The proportional hazards assumption was tested, and no violation has been detected among variables of interest. Since the effect of chemotherapy may potentially be influenced by the expression of biomarkers, as well as age, gender and tumor grade, we included interaction terms between chemotherapy (yes/no) and each of the covariates above. The full model based on SRRSH cohort indicated that age, RRM1, POLH, TYMS and the interaction terms between chemo and POLH, TYMS and TYMP remained to be statistically significantly associated with survival after adjusting for other covariates and interaction effects. Backward stepwise elimination was applied to select variables and interaction terms to be included in the final mode based on Akaike information criterion (AIC). Harrell's C was calculated to assess model discrimination. To further simplify the model structure, we tried to remove interaction terms between chemo and age, gender and tumor grade, but the prediction accuracy (Harrell's C) compromised significantly; therefore, we kept those terms within the model. To test the predictability of the predictors identified in the SRRSH cohort we included the same predictors in the TCGA cohort model and assessed the Harrell's C. However, due to differences on several important characteristics (e.g. age, grade, follow-up time, median survival and etc.) between the SRRSH and TCGA cohort, the Harrell's C based on TCGA cohort is slightly lower than that based on the SRRSH cohort.
Results
Expression of 5-fluorouracil and platinum related markers in LAGC patients who received curative surgery in the SRRSH cohort and TCGA cohort
We constructed a Multiple Tissue Array (MTA), including all patients enrolled, as we previously reported [15]. RRM1, RRM2, RRM2B, DUT, TYMS, TYMP, POLH, and Ki67 were stained on the slides of MTAs. All demographic data and contingencies with biomarkers in the SRRSH cohort were listed in Table S3. Among all the GC cases, positive expression of RRM1, RRM2, RRM2B, POLH, DUT, Ki67, TYMS, TYMP were 110 (45.4%), 135 (55.6%), 137 (55.1%), 154 (63.6%), 140 (57.4%), 155 (63.8%), 141 (57.5%), 59 (23.4%), respectively. All the biomarkers were higher in elderly and male patients except TYMP, compared to younger and female patients. RRM1, DUT, and TYMS were preferentially higher in stage III, low tumor grade and proximal GC patients, whereas RRM2, TYMS and TYMP were associated with poor differentiation.
The baseline data and contingencies of biomarkers of the TCGA cohort were listed in Table S4. All the mRNA expression of selected biomarkers was dichotomized with median levels. In 279 LAGC cases, positive expression of RRM1, RRM2, RRM2B, POLH, DUT, MKI67, TYMS, TYMP were 136 (49.0%), 140 (50.0%), 126 (45.0%), 150 (54.0%), 146 (52.0%), 135 (48.0%), 136 (49.0%), 155 (56.0%), respectively. POLH, DUT was associated with high tumor grade and younger age.
5-Fluorouracil and platinum related biomarkers predict the efficacy of chemotherapy in LAGC
Univariate Cox proportional analysis indicated that adjuvant chemotherapy was beneficial to high RRM1 (HR, 0.52, 95% CI 0.33–0.84), high DUT (HR, 0.65, 95% CI 0.43–1.00), low RRM2 (HR, 0.51, 95% CI 0.30–0.84), low RRM2B (HR, 0.57, 95% CI 0.35–0.97), low POLH (HR, 0.34, 95% CI 0.19–0.60), low Ki67 (HR, 0.52, 95% CI 0.29–0.92), low TYMS (HR, 0.55, 95% CI 0.31–1.01) or low TYMP (HR, 0.60, 95% CI 0.40–0.89) LAGC patients (Table 2, Fig. 1). To avoid the confounding effect, we further conducted a multivariate Cox proportional analysis. In low RRM2B, low RRM2, low POLH, low Ki67 or low RRM1 patients, chemotherapy remained to protect factors in LAGC (P < 0.05), whereas other biomarkers showed a consistent trend with the univariate model.
Table 2.
Univariable and multi-variable Cox proportional hazard analysis in SRRSH cohort.
| Gene signatures | Treatment | Univariable model |
Multi-variable model |
||
|---|---|---|---|---|---|
| HR (95% CI) | Pa | HRb (95% CI) | Pa | ||
| RRM1 | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.74 (0.45–1.23) | 0.25 | 0.72 (0.42–1.23) | 0.23 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.52 (0.33–0.84) | 0.007 | 0.66 (0.40–1.10) | 0.11 | |
| RRM2B | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.57 (0.35–0.97) | 0.04 | 0.55 (0.31–0.99) | 0.045 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.70 (0.45–1.09) | 0.12 | 0.85 (0.72–1.92) | 0.51 | |
| RRM2 | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.51 (0.30–0.84) | 0.008 | 0.54 (0.32–0.89) | 0.02 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.78 (0.49–1.24) | 0.3 | 0.97 (0.57–1.66) | 0.92 | |
| POLH | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.34 (0.19–0.60) | 0.0003 | 0.37 (0.20–0.68) | 0.001 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.92 (0.60–1.42) | 0.71 | 1.05 (0.66–1.67) | 0.84 | |
| dUTPase (DUT) | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.63 (0.35–1.11) | 0.11 | 0.63 (0.35–1.31) | 0.13 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.65 (0.43–1.00) | 0.05 | 0.82 (0.51–1.31) | 0.41 | |
| Ki67 | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.52 (0.29–0.92) | 0.02 | 0.49 (0.25–0.95) | 0.034 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.74 (0.48–1.13) | 0.17 | 0.85 (0.55–1.33) | 0.48 | |
| TYMS | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.55 (0.31–1.01) | 0.05 | 0.60 (0.37–1.14) | 0.12 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.78 (0.52–1.18) | 0.25 | 0.90 (0.57–1.42) | 0.65 | |
| TYMP | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.60 (0.40–0.89) | 0.01 | 0.67 (0.44–1.03) | 0.07 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.84 (0.45–1.57) | 0.6 | 0.97 (0.47–1.97) | 0.92 | |
| Chemo-signature | |||||
| Low | Surgery alone | Reference | Reference | ||
| Surgery + AC | 1.37 (0.65–2.97) | 0.41 | 2.11 (0.84–5.56) | 0.11 | |
| Medium | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.61 (0.37–1.00) | 0.05 | 0.64 (0.38–1.10) | 0.1 | |
| High | Surgery alone | Reference | Reference | ||
| Surgery + AC | 0.39 (0.21–0.72) | 0.003 | 0.44 (0.23–0.83) | 0.01 | |
AC: adjuvant chemotherapy.
Indicate p-value <0.05.
Adjusted by age and sex.
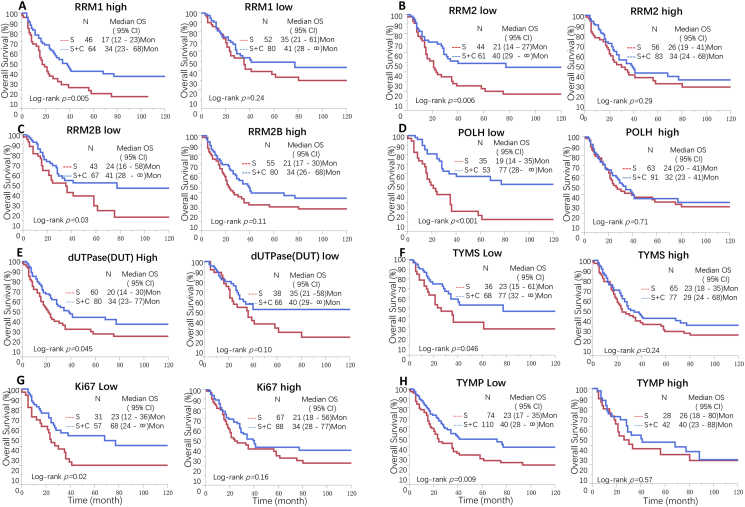
Fig. 1.
C-Score biomarkers predict the efficacy of chemotherapy in LAGC in both SRRSH and TCGA cohorts.
C-Score biomarkers were evaluated in both SRRSH and TCGA cohorts, Kaplan-Meier analysis indicated that high RRM1 (A), low RRM2 (B), low RRM2B (C), low POLH (D), high DUT (E), low TYMS (F), low KI67 (G) or low TYMP (H) were associated with better response to adjuvant chemotherapy compare to low RRM1, high RRM2, high RRM2B, high POLH, low DUT, high TYMS, high KI67 or high TYMP, respectively.
The result was validated in the TCGA cohort. Univariate Cox model showed high RRM1 (HR, 0.47, 95% CI 0.24–0.90), low RRM2 (HR, 0.42, 95% CI 0.23–0.76), low RRM2B (HR,0.38, 95% CI 0.20–0.69), low POLH (HR,0.34, 95% CI 0.19–0.60), low MKI67 (HR, 0.40, 95% CI 0.21–0.72), low TYMS (HR, 0.36, 95% CI 0.19–0.66) or low TYMP (HR, 0.35, 95% CI 0.16–0.69) LAGC patients had better survival after chemotherapy. High DUT was marginally associated with chemotherapy efficacy (P = 0.05), whereas low DUT demonstrated high response to chemotherapy (P = 2.0E-04). The multivariate Cox model also showed consistent results with the univariate model (Table S5).
C-Score is an ideal predictor for efficacy of adjuvant therapy in LAGC
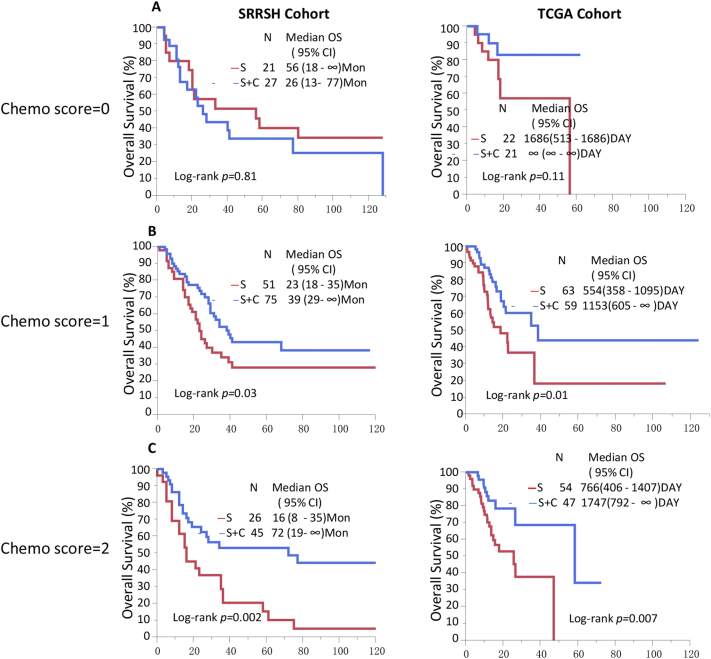
Cumulative analyses were conducted based on the risk scores (C-Score) generated from the identified candidate biomarkers that were significantly associated with chemotherapy efficacy. The C-Score was calculated as the sum of dichotomized biomarkers (0/1), the value of RRM1 and DUT were labeled as positive for their expressions associating with high efficacy, whereas the rest biomarkers were labeled as a minus for low efficacy. The formula was as following: C-Score = RRM1 + DUT − RRM2 − RRM2B − POLH − Ki67 − TYMS − TYMP. The sum was further categorized as three groups, score −4 and −5 were group low, score −2 and −3 were group medium, score −1, 0, 1 were group high. The univariate COX model indicated that high group response to chemotherapy better than group medium (HR, 0.39, 95% CI 0.21–0.72 vs. HR, 0.61, 95% CI 0.37–1.00), but the group low had an inadequate response to chemotherapy (HR, 1.37, 95% CI 0.65–2.97). The multivariate Cox model indicated similar results (Table 2). Kaplan-Meier analysis showed that patients who received chemotherapy had significantly better OS than patients who received surgery only in group high and group medium (log-rank P = 0.003, and 0.05, respectively, Fig. 2). The validation in the TCGA cohort also showed similar results that patients in group high and medium response to chemotherapy better than patients who only received surgery (HR, 0.36, 95% CI 0.15–0.75, HR, 0.47, 95% CI 0.25–0.84, respectively). The patients in group low demonstrated no difference in death risk and OS in surgery plus chemotherapy and surgery alone GC patients.
Fig. 2.
C-Score is an ideal predictor for adjuvant therapy efficacy in LAGC.
A. In LAGC patients with low C-Score, adjuvant chemotherapy demonstrated poor efficacy in prolong OS (log-rank p = 0.81 and 0.11 in SRRSH and TCGA cohort, respectively). B. In LAGC patients with medium chemo-score, adjuvant chemotherapy showed significant improved OS compare to surgery only patients (log-rank p = 0.03 and 0.01 in SRRSH and TCGA cohort, respectively). C. In LAGC patients with high C-Score, adjuvant chemotherapy showed significant improved OS compare to surgery only patients (log-rank p = 0.002 and 0.007 in SRRSH and TCGA cohort, respectively).
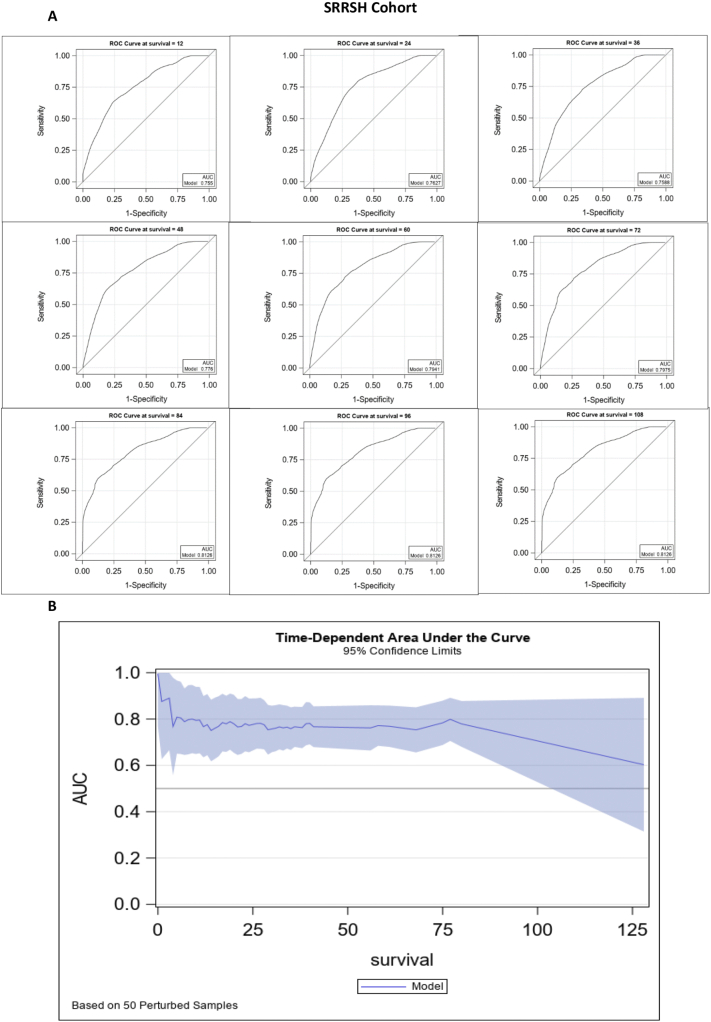
To further explore the value of C-Score, we developed a predictive model by including interaction terms between chemotherapy and each of the other covariates. The concordance test indicated that including the interaction effect has significantly improved the model accuracy. After performing model selection logic, the final model included age, gender, chemotherapy, tumor grade, RRM1, POLH, TYMD, TYMP, and interaction terms between chemo and each of the other covariates. The model accuracy based on the SRRSH cohort (Harrel's C = 0.7212) was slightly higher compared to that of the TCGA cohort (Harrel's C 0.6823). Also, due to the limited number of observations with survival time longer than three years in the TCGA cohort, here we only reported the model parameters estimated based on the SRRSH cohort. The ROC curves and AUC at specific time points have been presented in Fig. 3A. The AUC fluctuated between 0.75 and 0.81 across different survival tie points. Due to the limited number of observations, with survival time longer than 84 months, the ROC value remained the same after the 84th month measurement. The time-dependent AUC and corresponding 95% confidence limits were presented in Fig. 3B, which indicated that the model prediction accuracy was comparatively stable before the 84th month. Therefore, we suggest applying this model for estimation of no-longer than 5 to 6-year survival outcomes.
Fig. 3.
Prediction model based on C-Score could successfully predict efficacy of adjuvant chemotherapy in LAGC.
A. The ROC curves and AUC of the model, AUC values fluctuated between 0.75 and 0.81 across different survival tie points, with the highest AUC in 84, 96 and 108 months (0.8126). B. The time-dependent AUC and corresponding 95% confidence limits were presented.
Based on this model, we developed a calculator to predict the hazard ratio of receiving chemotherapy against not receiving chemotherapy for individual patients with different characteristics and biomarker profiles. A sample calculator is available in the supplementary materials in the excel spreadsheet format. The information needed to calculate the HR (chemo/non-chemo) includes gender (0-male/1-female), age in year, grade [[1], [2], [3], [4]], RRM1, POLH, TYMS, and TYMP (0-no/1-yes) (Attached file1). One limitation of this calculator is that the prediction for low-grade (grade I) patients may not be applicable. Since chemotherapy has a very significant protective effect for most low-grade patients, which could have potentially masked the modification effect from biomarker profile differences, our predictive model cannot detect the difference due to the biomarker profile variance among grade I patients.
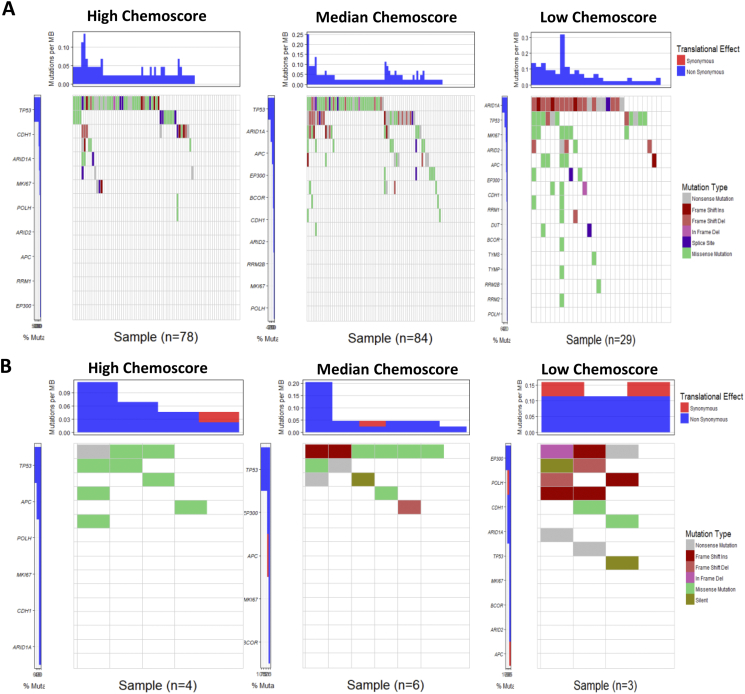
The C-Score is associated with mutation type and burden in LAGC
To identify the possible mechanisms underlying the predictive efficacy of C-Score, we analyzed the mutation profiles of the LAGC from the TCGA cohort. The mutation profiles in group low demonstrated a significantly higher rate of functional mutations than group median and high group (Fig. 4). Notably, the mutation type in low group cases was mainly frame shift ins/del, non-sense, and splice site, which could distinctly alter the protein translating, whereas the primary mutation type in group median and high was missense mutation. The highest frequency of the mutated gene in group low was ARID1A, and in group median and high was TP53.
Fig. 4.
The C-Score is associated with mutation type and burden in LAGC.
A. The waterfall plots of mutation profiles of LAGCs from TCGA were depicted, the patients with low C-Score demonstrated a significantly higher rate of functional mutations than group median and high group. B. The waterfall plots of mutation profiles of 28 gastric cancer cell lines. Cell lines with low chemo-signature (group low) showed high frequency of frameshift mutations in and high frequency of missense mutation than group median and group high cell lines.
To further validate our findings, we analyzed the association between C-Score biomarkers' expression and IC50 value to 5-fluorouracil in GC cell lines from a published study [22]. The result demonstrated that the IC50 value was significantly correlated with chemo-signature in GC cell lines (P = 0.044, Table 3). Moreover, the mutation profile showed a similar distribution pattern to the data from the TCGA cohort, which also showed that high frequency of frameshift mutations in group low and high frequency of missense mutation in group median and group high. The highest mutated gene was EP300 in group low and TP53 in group median and high (Fig. 4).
Table 3.
Correlation analysis between C-Score and IC50 in 28 GC cell lines.
| IC50/C-Score | 0 | 1 | 2 | |
|---|---|---|---|---|
| Resistant | 2 (100%) | 10 (58.8%) | 2 (22.2%) | |
| Sensitive | 0 (0%) | 7 (41.2%) | 7 (77.8%) | P = 0.044⁎ |
Chi-square test.
Discussion
In this study, we identified an optimal signature for LAGC in the SRRSH cohort and further validated the results in the TCGA cohort. Eight biomarkers closely correlated with oxaliplatin and 5-fluorouracil metabolisms were detected in 291 LAGC patients in the SRRSH cohort and further validated in 279 LAGC patients in the TCGA cohort. Univariate COX model indicated that adjuvant chemotherapy was beneficial in high RRM1, high DUT, low RRM2, low RRM2B, low POLH, low Ki67, low TYMS or low TYMP LAGC patients in both SRRSH and TCGA cohorts. In the multivariable COX model, all the biomarkers demonstrated similar results except DUT in the TCGA cohort, which only showed a consistent trend. C-Score based on these biomarkers indicated that patients with high C-Score responded to chemotherapy better than the patients with medium and low C-Score in both cohorts. A Cox regression model including age, gender, chemotherapy (yes/no), tumor grade and four of the eight biomarkers was employed to predict the OS of LAGC in both cohorts, which showed that the AUC of SRRSH cohort was 0.7212, and the AUC of TCGA cohort was 0.6823. A calculator based on our findings was developed to predict the benefit of chemotherapy. We found that the C-Score of GC cell lines were significantly associated with an IC50 value of 5-fluorouracil, the GC cell lines with low C-Score had significant higher functional mutation rate compare to median and high C-Score, which suggested potential mechanisms. Further, a Calculator was established based on C-Score and other clinic-pathological factors, which could successfully predict OS of LAGC. Thus, we propose that C-Score could be a predictive factor in assisting stratification and treatment decisions of LAGC patients.
The prediction model for chemotherapy efficacy remains controversial in LAGC. A recently published study suggested that the immunoScore signature could predict response to adjuvant chemotherapy in stage II and III GC patients in subgroup analysis, but the result was not validated [28]. Another two studies reported that serum biomarkers based model in GC patients from single-center could classify the patients who are suitable for chemoradiation or chemotherapy [29,30]. However, these models were conducted in a small sample size and were not independently validated. The most extensive study in the prediction model derived from the CLASSIC study, which includes 746 patients in the discovery set and 943 in the validation set. However, it is only a survival predicting model with clinical variables [31]. In the present study, we demonstrated a prediction model based on metabolic genes of chemo-reagents and clinic-pathological variables with high accuracy in the prediction of chemotherapy efficacy in two independent cohorts of LAGC patients. The discriminative capability of our model is comparable to other existing models (0.7–0.8), though the Harrell's C in the TCGA cohort is relatively lower (0.67). It could be useful in the decision-making chemotherapy of LAGC.
The sensitivity to chemotherapy was suggested associating with the genomic and epigenomic landscape of GC. According to TCGA, GC was characterized into four molecular subtypes: Epstein–Barr virus (EBV)-positive, micro-satellite instable (MSI), genomically stable (GS), and chrom instability (CIN). Among them, ARID1A was the most frequently identified mutated gene in EBV subtype, whereas TP53 and APC were the genes enriched in CIN subtype, PI3KCA, and CDH1 were identified in MSI subtype and GS subtype [24], however, no specific drug sensitivity was tested in this study. Another Korean study indicated that GC with mesenchymal phenotype was resistant to standard chemotherapy [32]. In our study, besides RRM1, DUT, TYMS, RRM2, TYMP, POLH, RRM2B and Ki67, which were accessed in our two cohorts, the difference in chemo-sensitivity may be also associated with the mutation numbers and types in LAGC patients. The most commonly mutated gene in the high and medium chemo-signature group was TP53, and ARID1A mutations were mostly identified in low C-Score LAGC, which might be ascribed to CIN and EBV subtypes, respectively. Regarding the epithelial and mesenchymal phenotypes, C-Score was not associated with tumor grade in both cohorts (P > 0.05, data not shown), but we did find that high tumor grade was higher in low C-Score LAGC compare to the median and high C-Score patients. Further, a high functional mutation rate identified in the low C-Score group indicated a high frequency of neo-antigens, which suggested that immunotherapy could be an ideal synergistic therapy with chemotherapy [33]. Therefore, we proposed that C-Score was associated with molecular subtypes and immunotherapy of LAGC, though more research is warranted to decipher the underlying mechanisms.
Our study has the distinct advantages of relatively large sample size, two-phase design, validation in GC cell lines, and multi-variate predictive model. We acknowledge several limitations. First, we validated the mRNA level in the TCGA cohort, which was not consistent with the SRRSH cohort, which evaluated the protein level (IHC) of the biomarkers. However, the principle of protein transcription and previous studies indicated the linear correlation between mRNA level and protein level [34], though epigenetic factors could not be ruled out [35]. More independent cohorts using IHC detection for protein were required to validate our findings. Second, the heterogeneity between the two cohorts, like race, age, and gender, could not be alleviated, albeit the tumor stage, tumor grade and chemotherapy information were parallel. Third, the chemo-regimens of included LAGC were not unified, which may affect the results. However, all the regimens were based on oxaliplatin and 5-fluorouracil, which means the fundamental anti-tumor mechanisms were identical. Lastly, we did not include other reported genes associating with fluorouracil and platinum in this study, like ERCC1, which has been reported as biomarker of cisplatinum resistance in multiple malignancies [18,36].
Conclusion
In this study, we identified and validated an eight gene signature associating with the responsiveness of chemotherapy in two independent cohorts of LAGC. Based on the signature and other clinical variables, we generated a risk model that could predict the survival of LAGC patients. Moreover, we also validated the results in a panel of GC cell lines. Mutation profiling based on the C-Score indicated that poor response to the chemotherapy in the low C-Score group might be associated with the number and type of mutations. The C-Score identified in this study could not only contribute to the stratification of the patients but also could help reduce the resistance to chemotherapy in LAGC.
Abbreviations
Consent for publication
All authors agreed on the publication of this manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 81972453, No. 81972597, No. 81602471 and No. 81672729), Natural Science Foundation of Zhejiang (Q16H160010, LY18H160005), Zhejiang Medical Scientific Research Foundation (2020RC022).
CRediT authorship contribution statement
Wang Qinchuan: Conceptualization, Investigation, Writing - original draft preparation, Writing - review & editing; Liu Xiyong: Conceptualization, Methodology, Writing - review & editing; Chen Chen: Formal analysis, Software; Chen Jida: Methodology; Xu Beisi: Formal analysis; Chen Lini: Investigation; Zhou Jichun: Investigation; Huang Yasheng: Writing - review & editing; Chen Wenjun: Writing - review & editing; Teng Rongyue: Writing - review & editing; Zhao Wenhe: Writing - review & editing; Jin Lidan: Writing - review & editing; Shen Jun: Writing - review & editing; Shen Jianguo: Writing - review & editing; Wang Linbo: Resources, Supervision; Yun Yen: Conceptualization, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We thank Mrs. Mariko Lee from the Microscope core Lab, City of Hope, for technical assistance with acquisition of immunohistochemistry microscopy images; and Mrs. Yafan Wang from the Translational Medicine core Lab, City of Hope, for patients' information collection in the City of Hope set, and Ms. Yanyan Chai from the Oncology Surgery Dept, Sir Run Run Shaw Hospital, for patients' information collection and follow-up in Zhejiang University set.
Data availability statement
All data are available upon request after publication pending approval from the authors and subject to SRRSH rules and regulations regarding human subject research and material transfer agreement.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100901.
Contributor Information
Yun Yen, Email: yyen@tmu.edu.tw.
Linbo Wang, Email: linbowang@zju.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Isomoto H., Shikuwa S., Yamaguchi N., Fukuda E., Ikeda K., Nishiyama H. Endoscopic submucosal dissection for early gastric cancer: a large-scale feasibility study. Gut. 2009;58(3):331–336. doi: 10.1136/gut.2008.165381. [DOI] [PubMed] [Google Scholar]
- 3.Nakajima T., Nashimoto A., Kitamura M., Kito T., Iwanaga T., Okabayashi K. Adjuvant mitomycin and fluorouracil followed by oral uracil plus tegafur in serosa-negative gastric cancer: a randomised trial. Gastric Cancer Surgical Study Group. Lancet. 1999;354(9175):273–277. doi: 10.1016/s0140-6736(99)01048-x. [DOI] [PubMed] [Google Scholar]
- 4.Neri B., de Leonardis V., Romano S., Andreoli F., Pernice L.M., Bruno L. Adjuvant chemotherapy after gastric resection in node-positive cancer patients: a multicentre randomised study. Br. J. Cancer. 1996;73(4):549–552. doi: 10.1038/bjc.1996.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Ugo D., Rausei S., Biondi A., Persiani R. Preoperative treatment and surgery in gastric cancer: friends or foes? Lancet Oncol. 2009;10(2):191–195. doi: 10.1016/S1470-2045(09)70021-X. [DOI] [PubMed] [Google Scholar]
- 6.Bang Y.J., Kim Y.W., Yang H.K., Chung H.C., Park Y.K., Lee K.H. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379(9813):315–321. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 7.Sakuramoto S., Sasako M., Yamaguchi T., Kinoshita T., Fujii M., Nashimoto A. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N. Engl. J. Med. 2007;357(18):1810–1820. doi: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 8.Sasako M., Sakuramoto S., Katai H., Kinoshita T., Furukawa H., Yamaguchi T. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J. Clin. Oncol. 2011;29(33):4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham D., Allum W.H., Stenning S.P., Thompson J.N., Van de Velde C.J., Nicolson M. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 2006;355(1):11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 10.Ychou M., Boige V., Pignon J.P., Conroy T., Bouche O., Lebreton G. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J. Clin. Oncol. 2011;29(13):1715–1721. doi: 10.1200/JCO.2010.33.0597. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Lim D.H., Kim S., Park S.H., Park J.O., Park Y.S. Phase III trial comparing capecitabine plus cisplatin versus capecitabine plus cisplatin with concurrent capecitabine radiotherapy in completely resected gastric cancer with D2 lymph node dissection: the ARTIST trial. J. Clin. Oncol. 2012;30(3):268–273. doi: 10.1200/JCO.2011.39.1953. [DOI] [PubMed] [Google Scholar]
- 12.Ajani J.A., D'Amico T.A., Almhanna K., Bentrem D.J., Chao J., Das P. Gastric Cancer, Version 3.2016, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2016;14(10):1286–1312. doi: 10.6004/jnccn.2016.0137. [DOI] [PubMed] [Google Scholar]
- 13.Park D.J., Lenz H.J. Determinants of chemosensitivity in gastric cancer. Curr. Opin. Pharmacol. 2006;6(4):337–344. doi: 10.1016/j.coph.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Kwon H.C., Roh M.S., Oh S.Y., Kim S.H., Kim M.C., Kim J.S. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann. Oncol. 2007;18(3):504–509. doi: 10.1093/annonc/mdl430. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q., Liu X., Zhou J., Huang Y., Zhang S., Shen J. Ribonucleotide reductase large subunit M1 predicts poor survival due to modulation of proliferative and invasive ability of gastric cancer. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y., Cui J., Xi H., Cai A., Shen W., Li J. Association of thymidylate synthase expression and clinical outcomes of gastric cancer patients treated with fluoropyrimidine-based chemotherapy: a meta-analysis. Onco Targets Ther. 2016;9:1339–1350. doi: 10.2147/OTT.S98540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L., Liu S., Lei Y., Wang K., Xu M., Chen Y. Systemic immune-inflammation index, thymidine phosphorylase and survival of localized gastric cancer patients after curative resection. Oncotarget. 2016;7:44185–44193. doi: 10.18632/oncotarget.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzger R., Leichman C.G., Danenberg K.D., Danenberg P.V., Lenz H.J., Hayashi K. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J. Clin. Oncol. 1998;16(1):309–316. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- 19.Ichikawa W., Takahashi T., Suto K., Shirota Y., Nihei Z., Shimizu M. Simple combinations of 5-FU pathway genes predict the outcome of metastatic gastric cancer patients treated by S-1. Int. J. Cancer. 2006;119(8):1927–1933. doi: 10.1002/ijc.22080. [DOI] [PubMed] [Google Scholar]
- 20.Tomicic M.T., Aasland D., Naumann S.C., Meise R., Barckhausen C., Kaina B. Translesion polymerase eta is upregulated by cancer therapeutics and confers anticancer drug resistance. Cancer Res. 2014;74(19):5585–5596. doi: 10.1158/0008-5472.CAN-14-0953. [DOI] [PubMed] [Google Scholar]
- 21.Kim H.K., Choi I.J., Kim C.G., Kim H.S., Oshima A., Michalowski A. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei Z., Tan I.B., Das K., Deng N., Zouridis H., Pattison S. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145(3):554–565. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Longley D.B., Harkin D.P., Johnston P.G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X., Zhang H., Lai L., Wang X., Loera S., Xue L. Ribonucleotide reductase small subunit M2 serves as a prognostic biomarker and predicts poor survival of colorectal cancers. Clin. Sci. (Lond.) 2013;124(9):567–578. doi: 10.1042/CS20120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teo K., Gemmell L., Mukherjee R., Traynor P., Edwards J. Bad expression influences time to androgen escape in prostate cancer. BJU Int. 2007;100(3):691–696. doi: 10.1111/j.1464-410X.2007.07001.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou B., Phan V., Liu X., Juhasz A., Chu P.G., Yen Y. Production of a monoclonal antibody against the hRRM2 subunit of ribonucleotide reductase and immunohistochemistry study of human cancer tissues. Hybridoma (Larchmt.) 2006;25(5):264–270. doi: 10.1089/hyb.2006.25.264. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y., Zhang Q., Hu Y., Li T., Yu J., Zhao L. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann. Surg. 2018;267(3):504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 29.Qin R., Yang Y., Chen H., Qin W., Han J., Gu Y. Prediction of neoadjuvant chemotherapeutic efficacy in patients with locally advanced gastric cancer by serum IgG glycomics profiling. Clin. Proteomics. 2020;17:4. doi: 10.1186/s12014-020-9267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn H.S., Sohn T.S., Kim M.J., Cho B.K., Kim S.M., Kim S.T. SEPROGADIC - serum protein-based gastric cancer prediction model for prognosis and selection of proper adjuvant therapy. Sci. Rep. 2018;8(1):16892. doi: 10.1038/s41598-018-34858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Y., Li T., Liang X., Hu Y., Huang L., Liao Z. Association of adjuvant chemotherapy with survival in patients with stage II or III gastric cancer. JAMA Surg. 2017;152(7) doi: 10.1001/jamasurg.2017.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh S.C., Sohn B.H., Cheong J.H., Kim S.B., Lee J.E., Park K.C. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat. Commun. 2018;9(1):1777. doi: 10.1038/s41467-018-04179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y.T., Hsu M., Lee P., Shin S.J., Mhawech-Fauceglia P., Odunsi K. Cancer/testis antigen CT45: analysis of mRNA and protein expression in human cancer. Int. J. Cancer. 2009;124(12):2893–2898. doi: 10.1002/ijc.24296. [DOI] [PubMed] [Google Scholar]
- 35.Harvey Z.H., Chen Y., Jarosz D.F. Protein-based inheritance: epigenetics beyond the chromosome. Mol. Cell. 2018;69(2):195–202. doi: 10.1016/j.molcel.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friboulet L., Olaussen K.A., Pignon J.P., Shepherd F.A., Tsao M.S., Graziano S. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N. Engl. J. Med. 2013;368(12):1101–1110. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data are available upon request after publication pending approval from the authors and subject to SRRSH rules and regulations regarding human subject research and material transfer agreement.