Abstract
Autophagy is a dynamic circulatory system that occurs in all eukaryotic cells. Cytoplasmic material is transported to lysosomes for degradation and recovery through autophagy. This provides energy and macromolecular precursors for cell renewal and homeostasis. The Hippo-YAP pathway has significant biological properties in controlling organ size, tissue homeostasis, and regeneration. Recently, the Hippo-YAP axis has been extensively referred to as the pathophysiological processes regulating autophagy. Understanding the cellular and molecular basis of these processes is crucial for identifying disease pathogenesis and novel therapeutic targets. Here we review recent findings from Drosophila models to organisms. We particularly emphasize the regulation between Hippo core components and autophagy, which is involved in normal cellular regulation and the pathogenesis of human diseases, and its application to disease treatment.
Subject terms: Macroautophagy, Cancer
Facts
The Hippo pathway and autophagy have complex and reciprocal interactions. These bidirectional links coordinate the autophagic flux to the overall microenvironmental signal and regulate homeostasis and tumorigenesis.
Autophagy is involved in the biological effects of the Hippo pathway and vice versa. Hippo-mediated modalities profoundly influence autophagic flux and are extensively involved in the intracellular quality control, tissue homeostasis, regeneration, development, and differentiation.
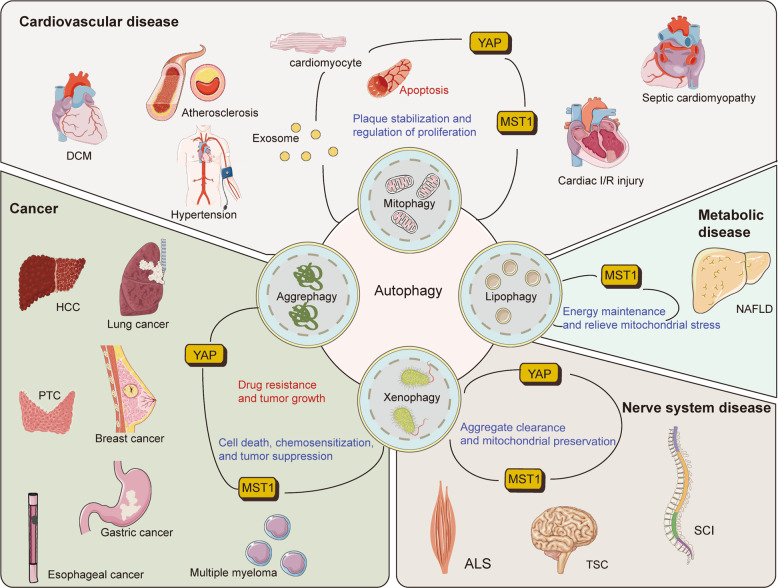
The association between the Hippo pathway and autophagy is relevant to the pathogenesis of a wide range of human diseases, from metabolic and neurodegenerative diseases, cardiovascular diseases to a variety of human solid tumors.
The emerging link between the Hippo pathway and autophagy means that targeting the Hippo-YAP-autophagy axis may provide new insights to prevent or promote autophagy in a variety of contexts, influencing metabolic reprogramming, cellular mechanical signals, mitochondrial quality control, and YAP/TAZ transcriptional activity.
Open questions
How are the Hippo pathway and autophagy interconnected, and what are the implications of these bidirectional links in homeostasis and tumorigenesis?
How prevalent is the Hippo-autophagy axis and what is its significance in disease development?
Will the emerging link between the Hippo pathway and autophagy make the role of targeted autophagy in cancer therapy clearer?
Introduction
Autophagy (also known as macroautophagy) is an accommodative process that occurs in different forms of cell stresses, including starvation, hypoxia, infection, high reactive oxygen content, and endoplasmic reticulum stress1. During this process, cells capture intracellular proteins and organelles, and transport them to lysosomes for degradation and export the products of autophagic degradation from lysosomes to the cytoplasm for recycling2,3. The first genetic screening of autophagy was conducted in Ohsumi’s laboratory, which analyzed this process in yeast and identified 15 autophagy-related proteins (ATGs)4. So far, over 30 ATGs have been identified5. The autophagy pathway is frequently divided into various individual stages: initiation, vesicle nucleation, elongation of the autophagy membrane, fusion with lysosomes, and degradation of intravesicular products (Fig. 1)6. Previously, autophagy has been considered a non-selective process. Recent studies have shown that autophagy can selectively eliminate harmful cytosols such as invading pathogens, dysfunctional organelles, and protein aggregates (called selective autophagy, including lipophagy, mitophagy, xenophagy, and aggrephagy), thereby contributing to the protection of cells in various environmental and metabolic stress3,7. Autophagy is strongly associated with neurodegeneration, cancer, metabolic diseases, immune and heart diseases, especially the role of autophagy in cancer7. Autophagy plays a dual role in cancer. Under tumorigenesis pressure, autophagy can clear oncogenic protein substrates and toxic unfolded proteins, inhibiting tissue damage and genomic instability8–10. Conversely, after tumor formation, increased autophagic flux often allows tumor cells to survive and grow9,11. This makes autophagy an interesting target for pharmacologists and clinicians.
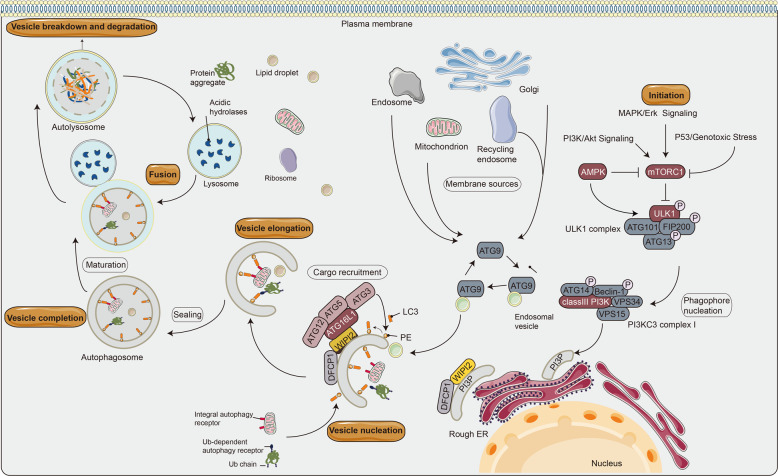
Fig. 1. Schematic diagram of the autophagy process in mammalian cells.
The mTOR complex 1 (mTORC1) contributes to the initiation of autophagy, integrates upstream signals such as PI3K/Akt pathway, AMPK, P53, and Bcl-2 protein family, which play different regulatory roles in autophagy135. The ULK1 complex induces vesicle nucleation and translocates to a characteristic endoplasmic reticulum (ER) structure called omegasome, where it phosphorylates PI3KC3 complex I to produce phosphatidylinositol-3-phosphate (PI3P) in omegasome. Specifically, Beclin1, a Bcl-2-homology (BH)-3 domain-only protein, is phosphorylated by ULK1 and acts as a scaffold for the PI3KC3 complex I, which facilitates localization of autophagy proteins to the phagophore. Atg9 is a transmembrane protein, which participates in the early stage of phagophore formation. PI3P recruits specific autophagy effectors, such as WIPIs (mammalian homolog of yeast Atg18) and zinc finger FYVE-type containing 1 (DFCP1). WIPIs directly binds to ATG16L1 under the regulation of the ubiquitin-like conjugation system to form the ATG12-ATG5-ATG16L1 complex and LC3 (mammalian homolog of yeast Atg8)-phosphatidylethanolamine (PE) binding. Ultimately, the isolation membrane is elongated and closed to form the autophagosome136. This binding reaction results in the conversion of LC3-I to LC3-II, a common autophagosome marker. When the autophagosome matures, it sheds the ATG proteins and fuses with the lysosome to produce autophagolysosome. Both the inner membrane of the autophagic vesicle and the luminal contents are degraded by lysosomal hydrolases (cathepsins B, D, and L). The resulting monomer molecules (such as amino acids and lipids) are recycled into the cytoplasm for reuse137. The pointed and blunt arrowheads indicate activation and inhibitory interactions, respectively. Ub, ubiquitin.
The Hippo-Yes-associated protein (YAP) pathway is an evolutionarily conserved pathway that controls organ size and tissue homeostasis12. The core kinase cassettes of the mammalian Hippo-YAP pathway consist of the mammalian sterile 20-like protein kinase 1 (STK3/MST2 and STK4/MST1) and an adapter protein, salvador family WW domain-containing protein 1 (SAV1)13,14, which may phosphorylate and activate the large tumor suppressor kinase 1/2 (LATS1/2)15. Adapter protein MOB kinase activator 1A (MOB1A) and MOB1B are also involved in the phosphorylation process16. YAP and PDZ-binding motif (TAZ, also known as WW domain-containing transcription regulator 1) are the major downstream transcription coactivators of the Hippo pathway. The phosphorylation of YAP/TAZ by the upstream kinase cascades MST1/2-LATS1/2 promotes the interaction of YAP and TAZ with cytoskeletal proteins, retains YAP and TAZ in the cytoplasm, and prevents their importation into the nucleus for transcriptional activation15,17. In contrast, when dephosphorylated, YAP can enter the nucleus and bind to the transcription factor TEA domain family member (TEADs) to control the expression of target genes15,18. YAP and TAZ rapidly shuttle between the nucleus and the cytoplasm by complex upstream components. LATS1/2-mediated phosphorylation limits the rate at which YAP and TAZ are imported into the nucleus. In addition, tethering of YAP and TAZ to the cytoskeletal proteins inhibit them as cellular mechanotransduction receptors19. The NDR (nuclear Dbf2-related) protein kinase family, including NDR1/STK38 (Serine/Threonine Kinase 38) and NDR2/STK38L (Serine/Threonine Kinase 38 Like), has identified additional kinases of Hippo signaling, similar to the LATS1/2 status in the Hippo signaling pathway (Fig. 2A)20,21. It is established that the Hippo-YAP pathway is regulated by cell–cell contact, cell polarity, cellular mechanotransduction, and G protein-coupled receptor ligands. However, recent studies have shown that autophagy has a series of crosstalk with the Hippo-YAP pathway. In physiological settings, the two conserved pathways, autophagy and Hippo-YAP signaling are essential in the protection of homeostasis. It has been shown that the deletion of autophagy-related genes interacting with the Hippo kinase cascades is associated with an accrued propensity of laboratory animals to spontaneously develop various disorders (Table 1). In this review, we summarize the regulation of autophagy by the Hippo-YAP pathway and discuss the multidisciplinary function of Hippo-YAP-autophagy in cells and various disorders.
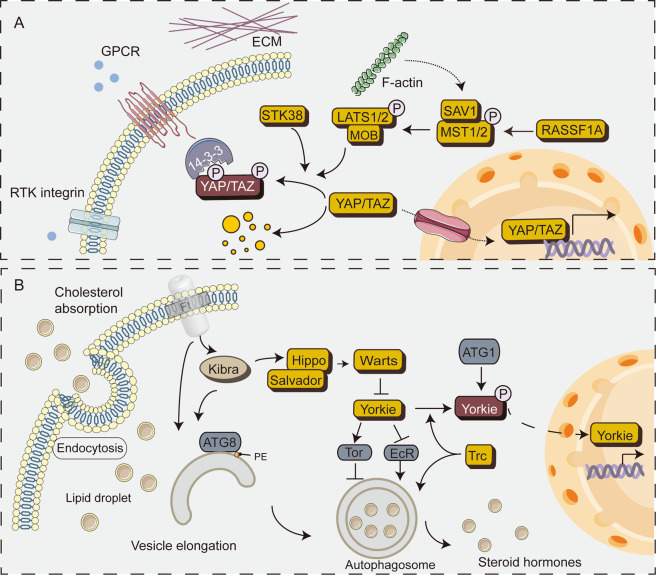
Fig. 2. Schematics diagram of the Hipoo pathway in mammals and the crosstalk between the Hippo pathway and autophagy in Drosophila.
A Schematics diagram of the Hippo pathway. In mammalian cells, phosphorylation of MST1/2 activates LATS1/2, which then phosphorylates YAP/TAZ at different Ser residues. Notably, STK38 can directly phosphorylate YAP. Phosphorylated YAP/TAZ is inhibited mainly through two mechanisms: (i) cytoplasmic retention through 14-3-3 binding and (ii) proteasome degradation. Inversely, inhibition of Hippo kinase leads to nuclear accumulation of YAP/TAZ, which bind to TEADs and other transcription factors. B Schematic diagram of the crosstalk between the Hippo pathway and autophagy in Drosophila. Typically, autophagy inhibits overgrowth of epithelial tissue. When the Hippo function is reduced, this mechanism is restricted. Atg1 phosphorylates Yorkie in a Hippo-Warts-independent manner, blocks the binding of Scalloped (TEADs in mammals) and decreases the activity of Yorkie. In addition, Warts (LATS1/2 in mammals) regulates autophagy via the EcR and Tor pathways. Trc (NDR1 in mammals) promotes the formation of autophagosome. Furthermore, Kibra (WWC1/2 in mammals) and Ft (FAT1-4 in mammals) are novel autophagy-regulated genes that promote ATG8-mediated elongation of the isolated membrane. The Ft mutant increases autophagy flux. The pointed and blunt arrowheads indicate activation and inhibitory interactions, respectively. Abbreviations: EcR, ecdysone receptor; Tor, target of rapamycin kinase; Trc, tricornered.
Table 1.
The core components of Hippo pathway affect various disorders via autophagy.
| Author, year | Disorders | Experimental models | Effector cell | Effects |
|---|---|---|---|---|
|
Zhang et al.45 Lin et al.138 You et al.139 |
DCM | Streptozotocin induce experimental diabetes in mice | CMEC/cardiomyocyte | MST1 knockdown upregulated autophagy and prevented apoptosis in cardiomyocytes and CMEC. |
| Shi et al.140 | DCM | Streptozotocin constructed diabetic model in endothelium-specific MST1 Tg mice | CMEC/cardiomyocyte | The MST1-enriched exosomes released from CMECs inhibit autophagy and glucose metabolism, thereby promote apoptosis in cardiomyocyte. |
| Yuan et al.141 | Atherosclerosis | ApoE−/− mice | HUVECs | Laminar flow protects the endothelium, inhibits Hippo-YAP signaling by promoting endothelial autophagy and SIRT1 expression, and blocks the formation of atherosclerotic plaques. |
| Wang et al.142 | Atherosclerosis | ApoE−/−: Mst1−/− and ApoE−/−: Mst1 Tg mice | Murine macrophage | In ApoE (−/−) mice, MST1 may stabilize atherosclerotic plaques by inhibiting macrophage autophagy and promoting macrophage apoptosis. |
| Shang et al.143 | Septic cardiomyopathy | Lipopolysaccharide (LPS)-induced septic cardiomyopathy MST1−/− mice | Cardiomyocyte | Septic cardiomyopathy is characterized with MST1 upregulation and deletion of MST1-activated mitophagy, thereby attenuated LPS-mediated mitochondrial damage. |
| Yu et al.144 | Cardiac I/R injury | Mst1−/− mice | Cardiomyocyte | MST1 deficiency activates protective mitophagy, thereby reducing cardiomyocyte mitochondrial apoptosis and regulating mitochondrial homeostasis. |
| Yao et al.145 | Hypertension | Infusion of Ang II induces hypertension in mice | HUVECs | In endothelial cells, mTORC1 regulates autophagy-dependent YAP degradation and controls blood pressure via COX-2/mPGES-1/PGE 2 cascade. |
| Lee et al.146 | ALS | ALS mouse model | Mouse motor neuron-like NSC34 cells | The activation of MST1 by SOD1 leads to autophagosome accumulation and blocking autophagy flux, which contribute to the demise of motor neurons both in vitro and in vivo. |
| Zhang et al.147 | SCI | MST1−/− and MST1 Tg SCI-induction mice | — | MST1 deficiency promotes posttraumatic spinal motor neuron survival via enhancement of autophagy flux. |
| Hsu et al.98 | Barth syndrome | — | MEFs | TAZ deficiency in MEFs caused defective mitophagosome biogenesis (the mitophagy in mitochondria quality control) and leads to impaired oxidative phosphorylation and oxidative stress. |
| Liang et al.70 | TSC | TSC mouse model | Mouse embryonic fibroblast | YAP is upregulated by mTOR in mouse and human perivascular epithelioid cell tumors (PEComas), and autophagy impairs YAP degradation in TSC-deficient cells, suggesting that the regulatory effects of YAP by mTOR and autophagy are therapeutic targets. |
| Xiao et al.148 | Doxorubicin-induced cardiotoxicity | DOX-induced cardiotoxicity model in mice | Rat cardiomyocytes | YAP/Parkin pathway presented DOX-induced cardiotoxicity in mouse heart by enhancing mitophagy. |
| Zhou et al.149 | NAFLD | MST1−/− and MST1 WT NAFLD mouse model | Mouse primary hepatocytes | MST1 deletion reversed Parkin-related mitophagy, suppressed hepatocyte mitochondrial stress, prevented diet-induced NAFLD. |
| Li et al.49 | HCC | Induction of HCC by intraperitoneal injection of diethylamine (DEN) in wild-type and RASSF1A-knockout mice. | Mouse primary hepatocytes | RASSF1A inhibits PI3K-AKT-mTOR pathway through MST1 to enhance autophagic flux, further inhibiting HCC and improving survival. |
| Li et al.92 | HCC | Induction of HCC by intraperitoneal injection of diethylamine (DEN) in wild-type and liver-specific LRPPRC-knockout mice. | Mouse primary hepatocytes | LRPPRC acts through YAP-P27 to control cell ploidy and P62 hence regulating autophagy maturation. |
| Lee et al.88 | HCC |
Liver-specific Atg7-knockout mice Atg7/YAP double-knockout mice |
The murine and human hepatocyte lines | Atg7 knockdown suppressed autophagy and YAP nuclear localization. YAP acts as an autophagic substrate in liver differentiation and carcinogenesis. |
| Liu et al.150 | PTC | Clinical thyroid papillary carcinoma tissue microarray analysis | PTC cell lines | In papillary thyroid cancer, YAP expression correlates with clinicopathological parameters. In vitro, YAP inhibits autophagy but enhances cell proliferation. |
| Li et al.151 | Breast cancer |
Human breast tissue microarray; MCF-7 cells were subcutaneously injected into BALB/c athymic nude mice |
Breast cell line and breast cancer cell line | HBXIP inhibits MST1 acetylation, leading to autophagy-dependent degradation of MST1, HBXIP-mediated reduction of tumor suppressor MST1 promotes the growth of breast cancer cells in vitro and in vivo. |
| Yan et al.69 | Gastric cancer | — | Normal gastric mucosal cell line and gastric cancer cell line | Knockdown of YAP causes mitochondrial apoptosis and cellular oxidative stress, which subsequently inhibits mitophagy, cancer cell survival, and migration. |
| Wang et al.89 | Lung cancer | Lung cancer and adjacent normal tissues | Lung cancer cell line | Aurora A upregulates YAP expression by blocking autophagy and Aurora A kinase expression is positively correlated with YAP. |
| Zhang et al.152 | Esophageal cancer | — | Esophageal cancer cell line | MST1 overexpression inhibits mitophagy activity, augments IL-24-induced esophageal cancer death via enhanced mitochondrial stress. |
| Fan et al.153 | Multiple myeloma |
PINK1-knockout mice and C57BL/6 WT controls Myeloma xenograft mouse model |
Multiple myeloma cell line | Activation of PINK1-dependent mitophagy inhibits migration, suppresses myeloma cell homing to calvarium, and decreases osteolytic bone lesions via the MOB1B-mediated Hippo-YAP/TAZ pathway. |
| Hu et al.154 | Pancreatic cancer | — | Normal ductal epithelial cell line and pancreatic cancer cell line | MST1 upregulation regulates pancreatic cancer cell apoptosis through mitofusin 2 (Mfn2)‑mediated mitophagy. |
| Wei et al.155 | Colorectal cancer | Colorectal cancer xenograft mouse model | Colorectal cancer cell line | FAT4 suppresses colorectal cancer by promoting autophagy and inhibiting the epithelial-to-mesenchymal transition (EMT). |
ALS amyotrophic lateral sclerosis, ApoE−/− apolipoprotein E-deficient, CMEC cardiac microvascular endothelial cell, DCM diabetic cardiomyopathy, HBXIP hepatitis B Virus X interacting protein, HCC hepatocellular carcinoma, HUVECs human umbilical vein endothelial cells, I/R ischemia-reperfusion, MEFs primary mouse embryonic fibroblasts, MST1−/− MST1 knockout, MST1 Tg MST1 transgenic, NAFLD non-alcoholic fatty liver disease, PINK1 PTEN-induced putative kinase 1, PTC papillary thyroid carcinoma, SCI spinal cord injury, SOD1 superoxide dismutase 1, TSC tuberous sclerosis complex, WT wild type.
Mechanism of autophagy
Autophagy initiation begins with the activation of the Unc-51-like autophagy activating kinase 1 (ULK1, also known as ATG1) complex, including ULK1, ULK2, RB1 inducible coiled-coil 1 (FIP200), and ATG13. This leads to the recruitment of ATGs to the specific subcellular location called the phagophore assembly site, which activates class III phosphatidylinositol-3-kinase (PI3KC3) complex I, including VPS34, Beclin1 (mammalian homolog of yeast Atg6), p150 (mammalian homolog of yeast VPS15), and Atg14 or ultraviolet radiation resistance-associated gene protein (also known as P63), and nucleation of an annular structure of the isolation membrane, called phagophore22,23. The ATG5–ATG12 complex conjugates with ATG16 to expand the autophagosome membrane, causing the phagosome to expand into a sphere. The enzymolysis of Atg4 to LC3 (Atg8 family protein) produces cytoplasmic LC3-I, which conjugates to lipid phosphatidylethanolamine to form LC3-II and then recruits to the autophagosome membrane. Eventually, the autophagosome fuses with the lysosome and the contents are degraded, thereby enabling cellular metabolic pathways and the renewal of specific organelles (Fig. 1)1,23–26. Furthermore, p62 (also known as SQSTM1) is an autophagic modifier of the LC3 family that acts as a bridge between LC3 and ubiquitinated substrates27,28. p62-bound polyubiquitinated proteins are integrated into the completed autophagosome and degraded in autolysosomes29,30. Whereas increased p62 levels are associated with autophagy inhibition, decreased p62 levels are associated with autophagy activation, revealing that the steady-state levels of this protein could reflect the state of autophagy29–31. Thus, p62 combined with LC3 completely monitors the autophagic flux32.
Crosstalk between the Hippo pathway and autophagy in Drosophila
Hippo signaling is essential for proper growth control in Drosophila and the loss of hippo (MST1/2 in mammals) causes tissue overgrowth33. Interestingly, autophagy induction actively suppresses hippo-induced tissue overgrowth. Meanwhile, Atg1 overexpression inhibits Yorkie (YAP in mammals), further suppressing epithelial overgrowth and cell proliferation34. Mechanistically, Atg1/ULK1 phosphorylates Yorkie at two serine residues, S74 and S97, thereby blocking transcriptional activation and inhibition of Yorkie activity. Atg1-mediated phosphorylation is an additional inhibitory input independent of the Hippo-Warts pathway35.
Steroid hormones are critical signaling molecules for growth regulation36. Warts (LATS1/2 in mammals) regulate steroid hormone production through an autophagy-dependent pathway (also called lipophagy). Precisely, Warts control the production of Drosophila steroid ecdysone through their effector microRNA bantam, which responds to nutrients, thus mobilizing the transport of the steroid precursor cholesterol37. Notably, YAP (mammalian Yki homolog) regulates steroidogenesis in tumor cells38, indicating that the regulation of steroidogenesis by the Wts-Yki pathway may be an evolutionarily conserved mechanism.
Trc (NDR1 in mammals) acts as a conserved regulator of autophagy and is required for early autophagosome formation in fly larvae39. Kibra, upstream components of the Drosophila Hippo pathway, act as autophagy regulatory factors required for proper autophagy function40. Drosophila protocadherin Fat (Ft) is a cell adhesion molecule in the Hippo pathway that regulates growth and planar cell polarity41. Ft mutations cause neurodegenerative changes through autophagy defects; autophagosomes accumulate in the Ft mutant photoreceptors, which are filled with partially degraded material and damaged mitochondria41. In conclusion, the core components of the Drosophila Hippo pathway are involved in the regulation of autophagy at multiple levels and several crosstalks exist in these two conserved pathways (Fig. 2B).
Regulation of autophagy by the Hippo pathway core kinase cassettes in mammals
MST1/2 protein kinases
Posttranslational modifications caused by the Hippo pathway kinases have become a powerful means to regulate autophagy in mammals. STK3/MST2 and STK4/MST1 are critical components of the Hippo pathway, which play a pivotal role in organ size control and tumor suppression. Recent studies have shown that STK3/STK4 can also be involved in the regulation of autophagy, which dynamically interacts with the Atg8 family of autophagy proteins in vitro. Specifically, they both phosphorylate LC3 at threonine 50. STK3/STK4-mediated phosphorylation is critical for the fusion of autophagosomes with lysosomes and the ability of cells to clear intracellular cargo (such as bacteria)42,43. STK3/STK4 deletion leads to protein aggregate accumulation of autophagic substrates p62 and LC3-II42,44,45. STK4/MST1 phosphorylates Beclin1 in its BH3 domain at Thr108, thereby inhibiting the Beclin1–Vps34 complex, which directly inhibits autophagy. Phosphorylation cascade can enhance Beclin1-Bcl-2 interaction and induce apoptosis44–46. RASSF1A, a Hippo pathway scaffold protein, binds to MST1, promotes the activation of MST1 and causes apoptosis (induced by the death receptor signaling pathway)47,48. The loss of RASSF1A can also lead to the blockage of the autophagic flux49. The regulation of autophagy by MST1/2 is involved in several human diseases (see Table 1).
NDR protein kinases
NDR1/2 (STK38/STK38L) is regulated through alterations in the subcellular localization and phosphorylation status, which influence cell cycle, apoptosis, and autophagy in mammalian cells21. Furthermore, STK38/STK38L acts as a major stress response and plays an essential role in autophagy. Precisely, STK38 regulates itself and XPO1 (exportin-1) nuclear export by phosphorylating XPO1 on serine 1055, thereby supporting autophagy regulator Beclin1 and Hippo effector YAP shuttle into the cytoplasm50. STK38 is also a new binding of Beclin1, which promotes autophagosome formation in mammalian cells. Conversely, STK38-depleted cells reduced PI3KC3 complex I (Beclin1-ATG14-Vps34) and PI3P formation, resulting in reduced autophagosome formation39. Moreover, STK38 regulates the chaperone-assisted selective autophagy (CASA), which initiates the CASA complex (including Hsc70, HspB8, synaptopodin-2 (SYNPO2), and the co-chaperone BAG3) and mediates the degradation of misfolded, damaged, and aggregation-prone proteins51. STK38 further inhibits BAG3-mediated autophagy in a kinase activity-independent manner, which relies on the remodeling of BAG3 chaperone complexes and disrupts the interaction of HspB8 and SYNPO252. The underlying mechanism by which Hippo pathway core kinase cassettes regulate autophagy is shown in Fig. 3.
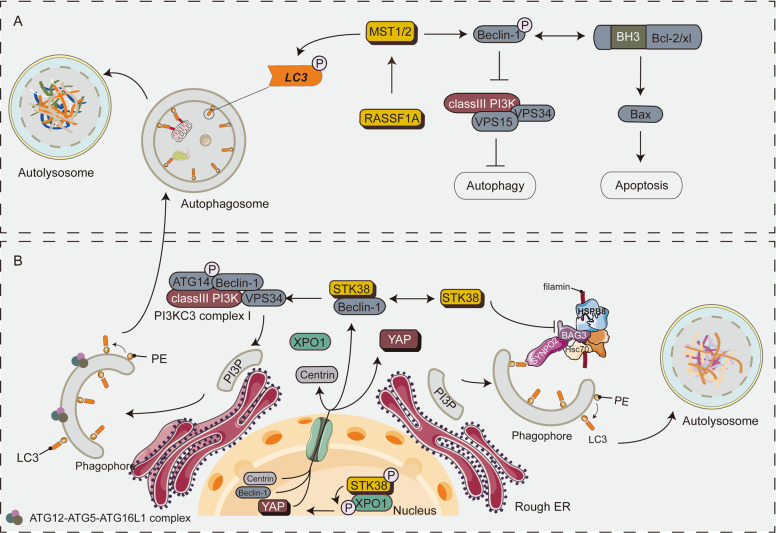
Fig. 3. A schematic diagram showing the core components of the Hippo pathway regulating autophagy in mammals.
A STK3/STK4 kinases are essential for autophagy. Specifically, STK3/STK4 directly phosphorylates LC3 at threonine 50 (Thr50) in mammalian cells, promotes the fusion of autophagosomes with lysosomes and the degradation of cargo in autolysosomes. MST1/STK4 phosphorylates the BH3 domain of Beclin1 at Thr108 and inhibits Vps34 kinase activity, thereby preventing the formation of autophagosome. RASSF1A promotes the initiation and maturation of autophagy by regulating MST1. In addition, MST1 mediates the interaction between Beclin1 and Bcl-2 thereby inducing apoptosis. B STK38 phosphorylation of XPO1 on S1055 is vital for the nuclear export of crucial intracellular signal sensors such as Beclin1, YAP1, and Centrin1. Cytoplasmic STK38 interacts with Beclin1 and promotes the formation of the Beclin1-ATG14-Vps34 complex, leading to the formation of PI3P. CASA is activated in mechanically stressed cells and tissues under the regulation ofSTK38. STK38 disrupts the interaction of BAG3 with HSPB8 and SYNPO2. Moreover, CASA activation is independent of the STK38 targets BECN1.
Crosstalk between transcriptional coactivators YAP/TAZ and autophagy
Autophagy acts as a downstream regulator of YAP/TAZ. Although YAP/TAZ controls autophagic flux by regulating the degradation of autophagosomes, YAP/TAZ is also essential for the maturation of autophagosomes into autolysosomes53,54. The use of autophagy inhibitors or endogenous knockdown of autophagy-related genes (e.g., ATG7/10 or ATG16L1) can inhibit YAP-mediated cell proliferation. Similarly, double YAP/TAZ knockdown and verteporfin (the inhibitors of YAP/TAZ55) treatment significantly impaired autophagy53,54. These demonstrated that the Hippo pathway maintains autophagy. In YAP/TAZ-activated cells, especially the aggressive solid tumor cells, the autophagic flux may be increased, thereby enhancing proliferation, invasion and metastasis of these cells. The underlying mechanism by which YAP/TAZ regulates autophagy is shown in Fig. 4.
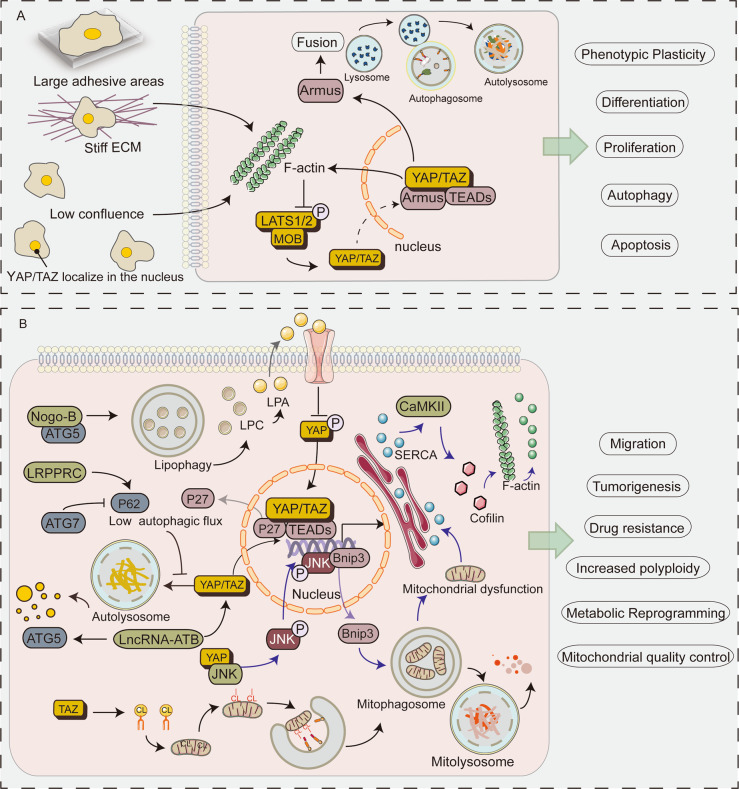
Fig. 4. Schematic diagram showing the role of YAP and TAZ in autophagy.
A When cells are at low density and on a stiff extracellular matrix (ECM), F-actin level is elevated leading to activation and nuclear import of YAP/TAZ, and upregulation of YAP/TAZ targets (such as myosin II and Armus). Activation of YAP/TAZ promotes F-actin accumulation. Cell mechanics control autophagic flux by regulating the transcriptional activity of YAP/TAZ. The YAP/TAZ-autophagy axis regulates a series of biological processes, such as proliferation, apoptosis, differentiation and phenotypic plasticity. B Loss of Atg7 or LRPPRC decreases autophagic flux. As an autophagic substrate, YAP cannot be degraded by autophagy, which increases nuclear localization of YAP. Activated YAP triggers accumulation of p27, which in turn leads to cellular polyploidy. lncRNA-ATB influence autophagy by participating in the transcriptional regulation of ATG5. In addition, lncRNA-ATB promotes autophagy by regulating YAP activation. Nogo-B interacts with ATG5 to promote lipophagy leading to LPC-dependent inhibition of YAP phosphorylation and enhances the oncogenic activity of YAP. YAP promotes metastasis via the mitophagy-SERCA-CaMKII pathways and cofilin/F-actin/lamellipodium axis. YAP binds to JNK in the cytoplasm, inducing JNK phosphorylation and nuclear localization, enhancing Bnip3 transcriptional activity. The Bnip3-induced mitophagy leads to mitochondrial dysfunction and ATP deficiency. Insufficient ATP inactivates SERCA and triggers [Ca2+]i overload; [Ca2+]i which phosphorylates CaMKII and inactivates cofilin, ultimately leading to F-actin degradation and abrogation of lamellipodium-based migration. Cardiolipin (CL) is a phospholipid found in the inner mitochondrial membrane. TAZ is required for catalyzation of CL. When mitochondria are damaged, cardiolipin is externalized and LC3 contains CL-binding sites to initiate mitophagy, thereby maintaining mitochondrial quality control. CaMKII, Ca/calmodulin-dependent protein kinases ΙΙ; CL, cardiolipin; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase.
Contact inhibition
Contact inhibition is a fundamental characteristic of normal cells. However, the loss of contact inhibition is an important feature of cancer cells56. The mechanical signals exerted by the physical state of cells in tissues and contact inhibition have recently been linked to the Hippo-YAP axis18,57. The decreased proliferation and cell survival resulting from contact inhibition is partly autophagy-dependent54. The Hippo pathway serves as the functionality of YAP and TAZ by regulating their subcellular localization and protein levels58. At high cell density, YAP/TAZ is redistributed into the cytoplasm and becomes inactive18,59, failing to regulate the expression of myosin II gene. This results in a drastic reduction in the formation of the F-actin stress fibers, ultimately impairing autophagosome formation54. Conversely, at low cell density, YAP/TAZ localizes in the nucleus and becomes active, resulting in increased F-actin formation, thus promoting autophagosome formation. In conclusion, YAP/TAZ regulates autophagy. The inhibition of YAP/TAZ function, caused by high cell density, reduces basal autophagy54. These signal crosstalks regulate the autophagy-dependent clearance of aggregation-prone proteins, survival under metabolic stress, as well as cell proliferation and differentiation54,60.
Mechanotransduction
Cells sense microenvironmental factors through a mechanotransduction system. They translate these stimuli into biochemical signals that control cell growth, differentiation, and cancer progression. Notably, YAP/TAZ is the medium for the mechanical cues indicated by the microenvironment18. YAP/TAZ is essential in the degradation of autophagosomes in both the steady-state and induced autophagy contexts. Autophagy is a downstream event of the YAP/TAZ mechanotransduction: mechanical signals act as the upstream inputs to control YAP/TAZ-mediated transcriptional activation of the TBC1D family members (such as Armus). This promotes the fusion of autophagosomal vesicles with lysosomes and regulates autophagy efficiency. Similar to the YAP/TAZ knockdown effect, low mechanical signal input slows down the autophagic flux53. Overall, mechanical signals can control autophagic flux through the regulation of YAP/TAZ transcriptional activity.
Cancer stem cell
Autophagy is critical in quality control, remodeling, and metabolic functions of adult and cancer stem cells (CSCs)61. Similarly, YAP/TAZ regulates the biological properties of the stem cells during normal organ development and tumorigenesis62. The YAP/TAZ-autophagy connection maintains the transformed properties of the tumor cells. It also influences the acquisition of CSC status in benign cells53. Increased levels of YAP/TAZ result in the transformation of terminally differentiated epithelial cells (such as primary mammary gland cells) into stem/progenitor cells63. These reprogramming events require YAP/TAZ-dependent regulation of autophagy. YAP expression greatly increases autophagosome clearance in induced differentiated cells, whereas inactivated autophagy-related genes impair the YAP-mediated reprogramming steps53. These data suggest that YAP/TAZ requires an effective autophagic flux to maintain CSC-inducing phenotypic plasticity.
Apoptosis
YAP overexpressed in multiple human solid tumors and inhibited apoptosis64,65. YAP located in the nucleus interacts with p73 and promotes apoptosis in response to DNA damage, suggesting a dual role of YAP on apoptosis66. The role of YAP in inhibiting cell apoptosis is at least partially autophagy-dependent. In ovarian and breast cancer cells, YAP knockdown increased cisplatin-induced apoptosis by decreasing autophagy67,68. Besides, YAP maintains mitophagy, the selective degradation of mitochondria by autophagy, which can block the caspase-9 apoptotic pathway, contributing to the gastric cancer cell survival and migration69. In tuberous sclerosis complex (TSC) 1-TSC2-deficient cells, the autophagic system impairs the degradation of YAP, leading to YAP accumulation. This subsequently causes abnormal proliferation, inducing apoptosis70. The interaction between autophagy and YAP is important in the control and modulation of apoptosis and apoptotic thresholds.
Metastasis
The migration of cancer cells into the circulatory or lymphatic system to form metastases is an extremely complex process in which the Hippo-YAP-autophagy axis is extensively involved71–73. Recent functional studies suggest that YAP mediates cancer metastasis via the modulation of actin dynamics74,75 and the control of transcriptional activity76,77, and along with the long non-coding RNA (lncRNA)-dependent manner78,79. Once the cancer cells spread to the systemic circulation and colonize distant organs, autophagic flux is induced to respond to the stressful microenvironments, including hypoxia, nutritional deficiencies and the extracellular matrix detachment80,81. F-actin polymerization drives the cellular membrane extension in lamellipodia, leading to cytoskeletal rearrangement, thus promoting migration74,82. YAP deficiency promoted the phosphorylation of JNK (c-Jun N-terminal kinases), which activated Bnip3 transcriptional activity and contributed to the Bnip3-required mitophagy. Higher Bnip3 caused mitochondrial dysfunction and ATP shortage, degraded F-actin via SERCA/[Ca2+] i/CaMKII/cofilin axis, and attenuated lamellipodium-based migration75. In the triple-negative breast cancer (TNBC) cells, autophagy promoted YAP nuclear localization, promoting TNBC cell migration and invasion83.
Hepatocarcinogenesis
YAP and TAZ are widely activated in human malignancies, which plays a vital role in tumorigenesis and the growth of most solid tumors84. For example, overexpression of YAP causes prominent hepatomegaly and induces tumor stem cell attributes, and hepatocarcinogenesis85,86. Autophagy maintains hepatic organ size and differentiation and, when autophagy is impaired, YAP is a driver of tissue remodeling and tumorigenesis87. In vivo, the liver-specific Atg7-deletion (Atg7 knockout (KO)) mice showed an 8.5-fold increase in the relative liver weight compared to the control mice at three months. Dysplastic nodules appeared at 8 months, whereas hepatocellular carcinoma (HCC) developed at 12 months. Meanwhile, the Atg7/YAP double KO mice attenuated hepatomegaly and hepatocarcinogenesis with significantly lower tumor size and number than the Atg7-KO mice88.
As expected, knockdown of Atg7 or Atg5 reduced autophagic flux. Interestingly, shAtg7 or shAtg5 induced nuclear translocation of YAP leading to the activation of TEAD4. Furthermore, YAP colocalized with autophagosomes, so that the cytoplasmic degradation of YAP was at least partially autophagy-dependent88,89. As YAP is an essential downstream mediator of tissue remodeling, progenitor cell activation, tumorigenesis, and drug resistance in the autophagy-deficient liver, the concomitant loss of YAP attenuates these abnormalities88,90. LRPPRC is a mitochondrion-associated protein91. The loss of LRPPRC expression promotes hepatocarcinogenesis92. Specifically, the deletion of LRPPRC leads to liver-specific YAP nuclear accumulation and induces accumulation of the cyclin-dependent kinase inhibitor p27, which in turn leads to cell polyploidy92,93. Concurrently, the deletion of mitochondrion-associated protein LRPPRC reduces p62 protein levels and impairs autophagic maturation. In vitro, LRPPRC knockdown synergistically enhances the diethylamine-induced genomic instability and hepatocarcinogenesis92. The lncRNAs are associated with clinicopathological parameters of HCC and can be used as biomarkers for HCC diagnosis94. Precisely, lncRNA-ATB activates the YAP-dependent autophagy and increases the expression of ATG5. The high expression of lncRNA-ATB is associated with poor prognosis and pathological characteristics of HCC95. Nogo-B, an endoplasmic reticulum residential protein, is highly expressed and promotes tumorigenesis in HCC. Mechanistically, Nogo-B interacts with ATG5 to encourage droplet lipid degradation and induces lipophagy-mediated oxidized low-density lipoprotein metabolism and subsequent lysophosphatidic acid-stimulated YAP oncogenic activity96.
Mitochondrial quality control
Mitophagy removes damaged mitochondria through autophagy, which is essential for mitochondrial quality control, metabolic homeostasis, and energy supply97. Notably, TAZ is required for mitophagy but not autophagosome biogenesis98. TAZ is a phospholipid transacylase that catalyzes the remodeling of cardiolipin, a mitochondrial endosomal phospholipid. The redistribution of cardiolipin controls the initiation of mitophagy98,99. Mechanistically, TAZ knockdown and inducible TAZ depletion prevent LC3 vesicles from recognizing mitophagosomes, thereby inhibiting mitophagy initiation. This leads to impaired oxidative phosphorylation and oxidative stress. Thus, TAZ is required for the initiation of mitophagy. It is involved in mitochondrial quality control. Mutations of the TAZ gene can cause Barth syndrome98,100.
Hippo-YAP-autophagy axis in clinical applications
Autophagy is an attractive therapeutic target in numerous diseases. As autophagy has a wide correlation with normal homeostasis, targeting it is particularly challenging. In contrast, the role of the Hippo pathway in cancer is widely described. It plays a vital role in tissue renewal and repair. Therefore, targeting the Hippo-YAP-autophagy axis might provide several promising targets. The emerging link between the Hippo pathway and autophagy is now largely implicated in pathophysiological processes, such as cancer, metabolic and neurodegenerative diseases, and cardiovascular diseases (Fig. 5)101–103. Here we introduce some small molecules or drugs that target the Hippo core components autophagy regulatory network (Table 2).
Fig. 5. The core components of hippo pathway affect various disorders via autophagy.
Hippo-YAP axis regulates autophagy and affects the development of disease progression. Inhibition of disease progression (beneficial process) is shown in blue, whereas promotion of disease progression (harmful process) is shown in red. In many diseases, autophagy clears dysfunctional mitochondria and protein aggregates. As two conserved signaling pathways, the Hippo pathway and autophagy intersect in the regulation of cell death and proliferation, tumorigenesis, and survival and growth of tumor cells. ALS, amyotrophic lateral sclerosis; DCM, diabetic cardiomyopathy; HCC, hepatocellular carcinoma; I/R, ischemia-reperfusion; NAFLD, non-alcoholic fatty liver disease; PTC, papillary thyroid carcinoma; SCI, spinal cord injury; TSC, tuberous sclerosis complex.
Table 2.
Small molecules or drugs that target the Hippo core components autophagy regulatory network.
| Organ | Diseases | Small molecules or drugs | Effects | Reference | |
|---|---|---|---|---|---|
 |
Brain | SAH | Melatonin | Melatonin play a neuroprotective role by regulating the homeostasis between apoptosis and autophagy through the oxygen species (ROS)-MST1 pathway | Shi et al.134 |
| Glioblastoma | Silibinin | Silibinin induced glioblastoma cell apoptosis and autophagy via inhibition of mTOR and YAP. | Bai et al.110 | ||
 |
Heart | DCM | Melatonin | Melatonin protects against DCM by increasing autophagy and reducing apoptosis through MST1/Sirt3 signaling | Zhang et al.131 |
| Cardiotoxicity | Adriamycin | HMGB1 is functionally related to YAP and participates in adriamycin-induced cardiotoxicity by upregulating autophagy. | Luo et al.129 | ||
 |
Liver | HCC | Sorafenib | Sorafenib promotes autophagy and is the standard treatment for advanced HCC, LATS1 restricts lethal autophagy in sorafenib-induced HCC cells. | Tang et al.119 |
| Hepatic fibrosis | Dihydrotanshinone I | Dihydromorphone I exerts anti-fibrotic effects by blocking the YAP-TEAD2 complex and stimulating autophagy | Ge et al.123 | ||
 |
Pancreas | Pancreatic cancer | Neratinib | Neratinib degrades MST4 via autophagy and is essential for the inactivation of YAP/TAZ. | Dent et al.118 |
 |
Colon | Colon cancer | Curcumin | Curcumin induces autophagy via inhibition of YAP. | Zhu et al.109 |
| Colon cancer | Shikonin | Shikonin effectively suppress colon cancer cell viability and migration, and induces autophagy via inhibiting the activity of YAP. | Zhu et al.112 |
DCM diabetic cardiomyopathy, HCC hepatocellular carcinoma, SAH subarachnoid hemorrhage.
Cancer
Autophagy is involved in several tumor progression stages, including tumorigenesis, progression, and malignant status maintenance73. In the early stages of tumorigenesis, autophagy maintains genome stability by removing oncogenic protein substrates, toxic unfolded proteins, and damaged organelles. This prevents chronic tissue damage, cell damage, and inflammation. Moreover, autophagy inhibits the accumulation of carcinogenic p62 protein aggregates, thereby promoting tumor suppression104–106. At an advanced tumor stage, the autophagic flux increases to cope with various environmental pressures, including hypoxia, nutritional deficiencies, DNA damage, metabolic stress, and chemotherapy. This maintains the survival and growth of tumor cells, and promotes tumor invasion and metastasis9,73,107. However, when blocking or promoting autophagy at different stages of tumor development, the biological effects on the tumor cell behavior may vary.
As previously mentioned, YAP acts as an autophagic substrate. The expression of YAP protein and YAP target genes is regulated by the autophagic flux88,89. Thus, some small molecules that induce autophagy can reduce the oncogenic activity of YAP/TAZ. Curcumin, a natural polyphenolic compound108, induces autophagy in colon cancer cells, further inhibiting cell proliferation and YAP expression109. Silibinin, a flavonolignan from the seeds of milk thistle, induced glioblastoma cell apoptosis and autophagy via inhibition of mammalian target of rapamycin and YAP110. Shikonin is the main bioactive ingredient extracted from the root of Lithospermum erythrorhizon111, which exerts anti-colon cancer effects similar to silibinin and inhibits YAP activity by inducing autophagy112.
As of May 2020, a search for “autophagy and cancer” on ClinicalTrials.gov revealed 72 studies focusing on the inhibition and evaluation of autophagy to improve the clinical prognosis for cancer patients. Targeted drugs either as single agents or in combinations can exert antitumor effects by enhancing both apoptotic and toxic autophagic processes113. For instance, neratinib (ERBB1/2/4 inhibitor) enhanced [pazopanib (the kinase inhibitor) + entinostat (histone deacetylase inhibitor)] lethality against sarcoma and other tumor cell types in vitro and in vivo. Specifically, the triplet combination increases the phosphorylation of YAP/TAZ and promotes the conversion of LC3 and expression of Beclin1 and ATG13, which together enhance autophagosome formation114,115. The mammalian STK 26/MST4 stimulates ATG4B activity and increases autophagic flux by phosphorylating ATG4B116. The MST4–MOB4 complex can disrupt the assembly of the MST1–MOB1 complex by alternative pairing, thereby increasing YAP activity117. Neratinib degrades MST4 via autophagy, enhancing LATS1/2 phosphorylation, and is also required for YAP/TAZ inactivation118.
Typically, although LATS1 plays a tumor suppressor role in the Hippo pathway, it also exerts a pro-survival function in the HCC cells119. Sorafenib (Srf), a multi-kinase inhibitor that promotes autophagy, is the standard treatment for advanced HCC120,121. The blockade of LATS1 expression resulted in increased Srf-induced apoptosis and decreased cell viability in vitro, as well as reduced tumor growth in vivo. LATS1 promotes K27-ubiquitination of Beclin1 on lysines K32 and K263, which inhibits autophagy induction and autophagic flux in HCC cells after Srf treatment. In Srf-nonrespondent patient, LATS1 expression is significantly increased, suggesting that LATS1 is a clinically relevant biomarker for Srf sensitivity119. The revelation of LATS1 functionally independent of the kinase activity in autophagy regulation requires consideration for targeted LATS1 kinases therapy.
Non-cancerous diseases
Autophagy plays a pivotal role in protein quality control, especially in maintaining metabolic homeostasis122. Dihydrotanshinone I, a natural monomeric compound isolated from Salvia miltiorrhiza Bunge, can improve liver function and reduce liver fibrosis. The underlying mechanism is associated with the cytoplasmic retention of YAP, thereby causing downregulation of fibrogenic gene expression, which stimulates autophagic flux and accelerates the degradation of the liver collagen123.
Autophagy offers promising targets for the prevention and treatment of cardiovascular diseases122. It has been reported that HMGB1, a chromosomal protein, acts as an autophagy sensor and induces autophagy after prolonged cellular stress124,125. The expression of HMGB1 is highly correlated with YAP activity, which is involved in tumorigenesis and acquisition of the tumor stem cell characteristics126,127. Adriamycin, an anthracycline chemotherapy drug, can also cause cardiotoxicity128. Specifically, adriamycin upregulates HMGB1 expression and induces cardiomyocyte autophagy followed by cardiac damage, whereas YAP reverses adriamycin-induced cardiac damage by downregulating HMGB1129. Melatonin regulates autophagy and has both chronobiotic and cytoprotective properties130. Melatonin significantly alleviates left ventricle remodeling and cardiac dysfunction in dilated cardiomyopathy by inducing autophagy and alleviating mitochondrial dysfunction, which is partially dependent on MST1/Sirt3 signaling131,132.
Autophagy is essential for maintaining proteostasis and a healthy mitochondrial pool, especially in maintaining the homeostasis of non-dividing nerve cells133. Melatonin plays a cytoprotective role in a variety of neurodegenerative diseases130. In subarachnoid hemorrhage-induced rats, melatonin can regulate the homeostasis between apoptosis and autophagy by inhibiting the ROS-MST1 pathway134.
Conclusions and perspectives
The understanding of the regulatory network between the Hippo-YAP pathway and autophagy has gradually been enriched in recent years. In this review, we show that these two highly conserved signaling pathways are widely involved in pathophysiological processes such as apoptosis, cell proliferation, cell differentiation, and metabolism, and can influence the pathogenesis of human diseases. This multidisciplinary view improves our understanding of why these two signaling pathways have been preserved throughout evolution. However, variations in the activity between autophagy and the Hippo-YAP pathway in different tissue types, tumor microenvironments, and disease states are some of the fundamental puzzles yet to be resolved. In addition, the paradoxical effect of autophagy in cancer makes autophagy-targeted therapy in cancer controversial. However, the Hippo pathway dysregulation occurs in a wide range of human cancers. This is essential in the development of novel and more specific drugs. For example, a combination of Hippo pathway-targeted drugs with autophagy inhibitors and inducers may be potential therapies for various human diseases. A better understanding and targeting of the Hippo-YAP-autophagy axis is an auspicious direction.
Acknowledgements
Dr. Tianmin Xu was supported by funding from the Department of Science and Technology of Jilin Province (grant number 20180201032YY and 20200602008ZP); the Department of Finance of Jilin Province (grant number 2019SCZT017).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Blandino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 5.Ktistakis NT, Tooze SA. Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 2016;26:624–635. doi: 10.1016/j.tcb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 7.Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 8.Kocaturk NM, et al. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 2019;134:116–137. doi: 10.1016/j.ejps.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 9.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H-Y, White E. Role of autophagy in cancer prevention. Cancer Prev. Res. 2011;4:973–983. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 13.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 14.Kango-Singh M, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 15.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 16.Xiong S, et al. Regulation of protein interactions by Mps one binder (MOB1) phosphorylation. Mol. Cell. Proteomics. 2017;16:1111–1125. doi: 10.1074/mcp.M117.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moroishi, T. et al. The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell167, 10.1016/j.cell.2016.11.005 (2016). [DOI] [PMC free article] [PubMed]
- 18.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 19.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharif AAD, Hergovich A. The NDR/LATS protein kinases in immunology and cancer biology. Semin. Cancer Biol. 2018;48:104–114. doi: 10.1016/j.semcancer.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Hergovich, A. The roles of NDR protein kinases in Hippo signalling. Genes7, 10.3390/genes7050021 (2016). [DOI] [PMC free article] [PubMed]
- 22.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 23.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 24.Abada A, Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep. 2014;15:839–852. doi: 10.15252/embr.201439076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy12, 10.1080/15548627.2015.1100356 (2016). [DOI] [PMC free article] [PubMed]
- 27.Jiang P, Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett BJ, et al. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011;7:572–583. doi: 10.4161/auto.7.6.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J-H, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019;20:211–226. doi: 10.1038/s41580-018-0086-y. [DOI] [PubMed] [Google Scholar]
- 34.Pérez E, Das G, Bergmann A, Baehrecke EH. Autophagy regulates tissue overgrowth in a context-dependent manner. Oncogene. 2015;34:3369–3376. doi: 10.1038/onc.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyra, L. K., Nandi, N., Tracy, C. & Krämer, H. Yorkie growth-promoting activity is limited by Atg1-mediated phosphorylation. Dev. Cell52, 10.1016/j.devcel.2020.01.011 (2020). [DOI] [PMC free article] [PubMed]
- 36.Danielsen ET, et al. A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Developmental cell. 2016;37:558–570. doi: 10.1016/j.devcel.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Texada, M. J. et al. Autophagy-mediated cholesterol trafficking controls steroid production. Dev. Cell48, 10.1016/j.devcel.2019.01.007 (2019). [DOI] [PubMed]
- 38.Farrow JM, Yang JC, Evans CP. Autophagy as a modulator and target in prostate cancer. Nat. Rev. Urol. 2014;11:508–516. doi: 10.1038/nrurol.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joffre C, et al. The pro-apoptotic STK38 kinase is a new Beclin1 partner positively regulating autophagy. Curr. Biol. 2015;25:2479–2492. doi: 10.1016/j.cub.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin A, Neufeld TP, Choe J. Kibra and aPKC regulate starvation-induced autophagy in Drosophila. Biochem. Biophys. Res. Commun. 2015;468:1–7. doi: 10.1016/j.bbrc.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Napoletano F, et al. Polyglutamine Atrophin provokes neurodegeneration in Drosophila by repressing fat. EMBO J. 2011;30:945–958. doi: 10.1038/emboj.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson DS, et al. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol. Cell. 2015;57:55–68. doi: 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrestha BK, et al. NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. J. Biol. Chem. 2020;295:1240–1260. doi: 10.1074/jbc.RA119.010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maejima Y, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, et al. MST1 coordinately regulates autophagy and apoptosis in diabetic cardiomyopathy in mice. Diabetologia. 2016;59:2435–2447. doi: 10.1007/s00125-016-4070-9. [DOI] [PubMed] [Google Scholar]
- 46.Lee EF, et al. Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1. Autophagy. 2019;15:785–795. doi: 10.1080/15548627.2018.1564557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pefani D-E, et al. TGF-β targets the Hippo pathway scaffold RASSF1A to facilitate YAP/SMAD2 nuclear translocation. Mol. Cell. 2016;63:156–166. doi: 10.1016/j.molcel.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Oh HJ, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 49.Li W, et al. Suppressor of hepatocellular carcinoma RASSF1A activates autophagy initiation and maturation. Cell Death Differ. 2019;26:1379–1395. doi: 10.1038/s41418-018-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin AP, et al. STK38 kinase acts as XPO1 gatekeeper regulating the nuclear export of autophagy proteins and other cargoes. EMBO Rep. 2019;20:e48150. doi: 10.15252/embr.201948150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulbricht A, et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 52.Klimek C, et al. The Hippo network kinase STK38 contributes to protein homeostasis by inhibiting BAG3-mediated autophagy. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1556–1566. doi: 10.1016/j.bbamcr.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Totaro A, et al. Cell phenotypic plasticity requires autophagic flux driven by YAP/TAZ mechanotransduction. Proc. Natl Acad. Sci. USA. 2019;116:17848–17857. doi: 10.1073/pnas.1908228116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pavel M, et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat. Commun. 2018;9:2961. doi: 10.1038/s41467-018-05388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, et al. Verteporfin inhibits YAP function through up-regulating 14-3-3sigma sequestering YAP in the cytoplasm. Am. J. Cancer Res. 2016;6:27–37. [PMC free article] [PubMed] [Google Scholar]
- 56.Holley RW. Control of growth of mammalian cells in cell culture. Nature. 1975;258:487–490. doi: 10.1038/258487a0. [DOI] [PubMed] [Google Scholar]
- 57.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 60.Gumbiner BM, Kim N-G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 2014;127:709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boya, P., Codogno, P. & Rodriguez-Muela, N. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development145, 10.1242/dev.146506 (2018). [DOI] [PubMed]
- 62.Park JH, Shin JE, Park HW. The role of Hippo pathway in cancer stem cell biology. Mol. Cell. 2018;41:83–92. doi: 10.14348/molcells.2018.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panciera T, et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 65.Furth N, Aylon Y, Oren M. p53 shades of Hippo. Cell Death Differ. 2018;25:81–92. doi: 10.1038/cdd.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lapi E, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol. Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 67.Xiao L, et al. YAP induces cisplatin resistance through activation of autophagy in human ovarian carcinoma cells. OncoTargets Ther. 2016;9:1105–1114. doi: 10.2147/OTT.S112358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang Y, et al. Cisplatin-induced autophagy protects breast cancer cells from apoptosis by regulating yes-associated protein. Oncol. Rep. 2017;38:3668–3676. doi: 10.3892/or.2017.6035. [DOI] [PubMed] [Google Scholar]
- 69.Yan H, et al. Yap regulates gastric cancer survival and migration via SIRT1/Mfn2/mitophagy. Oncol. Rep. 2018;39:1671–1681. doi: 10.3892/or.2018.6252. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Liang N, et al. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J. Exp. Med. 2014;211:2249–2263. doi: 10.1084/jem.20140341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee C-K, et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 2019;363:644–649. doi: 10.1126/science.aav0173. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Z. et al. OTUB2 promotes cancer metastasis via Hippo-independent activation of YAP and TAZ. Mol. Cell73, 10.1016/j.molcel.2018.10.030 (2019). [DOI] [PubMed]
- 73.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiao Y, et al. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep. 2017;19:1495–1502. doi: 10.1016/j.celrep.2017.04.075. [DOI] [PubMed] [Google Scholar]
- 75.Shi C, et al. Yap promotes hepatocellular carcinoma metastasis and mobilization via governing cofilin/F-actin/lamellipodium axis by regulation of JNK/Bnip3/SERCA/CaMKII pathways. Redox Biol. 2018;14:59–71. doi: 10.1016/j.redox.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang T, et al. YAP promotes breast cancer metastasis by repressing growth differentiation factor-15. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:1744–1753. doi: 10.1016/j.bbadis.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 77.Kim T, et al. MRTF potentiates TEAD-YAP transcriptional activity causing metastasis. EMBO J. 2017;36:520–535. doi: 10.15252/embj.201695137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017;19:106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr. Opin. Cell Biol. 2010;22:241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Avivar-Valderas A, et al. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK. Oncogene. 2013;32:4932–4940. doi: 10.1038/onc.2012.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gross SR. Actin binding proteins: their ups and downs in metastatic life. Cell Adh. Migr. 2013;7:199–213. doi: 10.4161/cam.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen W, Bai Y, Patel C, Geng F. Autophagy promotes triple negative breast cancer metastasis via YAP nuclear localization. Biochem. Biophys. Res. Commun. 2019;520:263–268. doi: 10.1016/j.bbrc.2019.09.133. [DOI] [PubMed] [Google Scholar]
- 84.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl Acad. Sci. USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patel SH, Camargo FD, Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee YA, et al. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat. Commun. 2018;9:4962. doi: 10.1038/s41467-018-07338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang P, et al. Activation of Aurora A kinase increases YAP stability via blockage of autophagy. Cell Death Dis. 2019;10:432. doi: 10.1038/s41419-019-1664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y, et al. YAP promotes multi-drug resistance and inhibits autophagy-related cell death in hepatocellular carcinoma via the RAC1-ROS-mTOR pathway. Cancer Cell Int. 2019;19:179. doi: 10.1186/s12935-019-0898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics. 2002;79:124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li W, et al. LRPPRC sustains Yap-P27-mediated cell ploidy and P62-HDAC6-mediated autophagy maturation and suppresses genome instability and hepatocellular carcinomas. Oncogene. 2020;39:3879–3892. doi: 10.1038/s41388-020-1257-9. [DOI] [PubMed] [Google Scholar]
- 93.Zhang, S. et al. Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell31, 10.1016/j.ccell.2017.04.004 (2017). [DOI] [PMC free article] [PubMed]
- 94.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 95.Wang C-Z, Yan G-X, Dong D-S, Xin H, Liu Z-Y. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J. Gastroenterol. 2019;25:5310–5322. doi: 10.3748/wjg.v25.i35.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tian Y, et al. ER-residential Nogo-B accelerates NAFLD-associated HCC mediated by metabolic reprogramming of oxLDL lipophagy. Nat. Commun. 2019;10:3391. doi: 10.1038/s41467-019-11274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 98.Hsu P, et al. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11:643–652. doi: 10.1080/15548627.2015.1023984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chu CT, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barth PG, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 1983;62:327–355. doi: 10.1016/0022-510X(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 101.Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 102.Chen H-T, et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer. 2019;18:101. doi: 10.1186/s12943-019-1030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu S, et al. Modulation of autophagy in human diseases strategies to foster strengths and circumvent weaknesses. Med. Res. Rev. 2019;39:1953–1999. doi: 10.1002/med.21571. [DOI] [PubMed] [Google Scholar]
- 104.Barnard RA, et al. Autophagy inhibition delays early but not late-stage metastatic disease. J. Pharm. Exp. Ther. 2016;358:282–293. doi: 10.1124/jpet.116.233908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mulcahy Levy JM, Thorburn A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020;27:843–857. doi: 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 2019;9:1167–1181. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singh SS, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 108.Giordano, A. & Tommonaro, G. Curcumin and cancer. Nutrients11, 10.3390/nu11102376 (2019). [DOI] [PMC free article] [PubMed]
- 109.Zhu J, et al. Curcumin induces autophagy via inhibition of Yes-associated protein (YAP) in human colon cancer cells. Med. Sci. Monit. 2018;24:7035–7042. doi: 10.12659/MSM.910650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bai Z-L, Tay V, Guo S-Z, Ren J, Shu M-G. Silibinin induced human glioblastoma cell apoptosis concomitant with autophagy through simultaneous inhibition of mTOR and YAP. Biomed. Res. Int. 2018;2018:6165192. doi: 10.1155/2018/6165192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo C, et al. Pharmacological properties and derivatives of shikonin-A review in recent years. Pharm. Res. 2019;149:104463. doi: 10.1016/j.phrs.2019.104463. [DOI] [PubMed] [Google Scholar]
- 112.Zhu J, Zhao L, Luo B, Sheng W. Shikonin regulates invasion and autophagy of cultured colon cancer cells by inhibiting yes-associated protein. Oncol. Lett. 2019;18:6117–6125. doi: 10.3892/ol.2019.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Booth, L. A., Roberts, J. L. & Dent, P. The role of cell signaling in the crosstalk between autophagy and apoptosis in the regulation of tumor cell survival in response to sorafenib and neratinib. Semin. Cancer Biol. 10.1016/j.semcancer.2019.10.013 (2019). [DOI] [PMC free article] [PubMed]
- 114.Booth L, Roberts JL, Poklepovic A, Dent P. The lethality of [pazopanib + HDAC inhibitors] is enhanced by Neratinib. Front. Oncol. 2019;9:650. doi: 10.3389/fonc.2019.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dent P, et al. Neratinib inhibits Hippo/YAP signaling, reduces mutant K-RAS expression, and kills pancreatic and blood cancer cells. Oncogene. 2019;38:5890–5904. doi: 10.1038/s41388-019-0849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang, T. et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell32, 10.1016/j.ccell.2017.11.005 (2017). [DOI] [PMC free article] [PubMed]
- 117.Chen M, et al. The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J. Biol. Chem. 2018;293:14455–14469. doi: 10.1074/jbc.RA118.003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dent, P. et al. Neratinib degrades MST4 via autophagy that reduces membrane stiffness and is essential for the inactivation of PI3K, ERK1/2, and YAP/TAZ signaling. J. Cell Physiol. 10.1002/jcp.29443 (2020). [DOI] [PMC free article] [PubMed]
- 119.Tang F, et al. LATS1 but not LATS2 represses autophagy by a kinase-independent scaffold function. Nat. Commun. 2019;10:5755. doi: 10.1038/s41467-019-13591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kudo M, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 121.Tai WT, et al. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. doi: 10.1038/cddis.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang, Y., Whaley-Connell, A. T., Sowers, J. R. & Ren, J. Autophagy as an emerging target in cardiorenal metabolic disease: From pathophysiology to management. Pharmacol. Ther.191, 10.1016/j.pharmthera.2018.06.004 (2018). [DOI] [PMC free article] [PubMed]
- 123.Ge M, et al. The anti-hepatic fibrosis effects of dihydrotanshinone I are mediated by disrupting the yes-associated protein and transcriptional enhancer factor D2 complex and stimulating autophagy. Br. J. Pharm. 2017;174:1147–1160. doi: 10.1111/bph.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang D, et al. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y-G, et al. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy. 2019;15:1935–1953. doi: 10.1080/15548627.2019.1596485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang L, et al. Dedifferentiation process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1α signaling enhances pancreatic cancer stemness. Cell Death Dis. 2019;10:724. doi: 10.1038/s41419-019-1956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen R, et al. High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology. 2018;67:1823–1841. doi: 10.1002/hep.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hardaway BW. Adriamycin-associated cardiomyopathy: where are we now? updates in pathophysiology, dose recommendations, prognosis, and outcomes. Curr. Opin. Cardiol. 2019;34:289–295. doi: 10.1097/HCO.0000000000000617. [DOI] [PubMed] [Google Scholar]
- 129.Luo P, et al. HMGB1 contributes to adriamycin-induced cardiotoxicity via up-regulating autophagy. Toxicol. Lett. 2018;292:115–122. doi: 10.1016/j.toxlet.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 130.Cardinali DP. Melatonin: clinical perspectives in neurodegeneration. Front. Endocrinol. 2019;10:480. doi: 10.3389/fendo.2019.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang, M. et al. Melatonin protects against diabetic cardiomyopathy through Mst1/Sirt3 signaling. J. Pineal. Res.63, 10.1111/jpi.12418 (2017). [DOI] [PubMed]
- 132.Wang S, et al. Melatonin activates Parkin translocation and rescues the impaired mitophagy activity of diabetic cardiomyopathy through Mst1 inhibition. J. Cell Mol. Med. 2018;22:5132–5144. doi: 10.1111/jcmm.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 134.Shi L, et al. Melatonin regulates apoptosis and autophagy via ROS-MST1 pathway in subarachnoid hemorrhage. Front. Mol. Neurosci. 2018;11:93. doi: 10.3389/fnmol.2018.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Polson HEJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 137.Man SM, Kanneganti T-D. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy. 2016;12:2504–2505. doi: 10.1080/15548627.2016.1239679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin J, et al. Mst1 inhibits CMECs autophagy and participates in the development of diabetic coronary microvascular dysfunction. Sci. Rep. 2016;6:34199. doi: 10.1038/srep34199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.You P, et al. Lin28a protects against diabetic cardiomyopathy through Mst1 inhibition. J. Cell Physiol. 2020;235:4455–4465. doi: 10.1002/jcp.29321. [DOI] [PubMed] [Google Scholar]
- 140.Hu J, et al. Exosomal Mst1 transfer from cardiac microvascular endothelial cells to cardiomyocytes deteriorates diabetic cardiomyopathy. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:3639–3649. doi: 10.1016/j.bbadis.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 141.Yuan P, et al. Laminar flow inhibits the Hippo/YAP pathway via autophagy and SIRT1-mediated deacetylation against atherosclerosis. Cell Death Dis. 2020;11:141. doi: 10.1038/s41419-020-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang T, et al. Mst1 participates in the atherosclerosis progression through macrophage autophagy inhibition and macrophage apoptosis enhancement. J. Mol. Cell Cardiol. 2016;98:108–116. doi: 10.1016/j.yjmcc.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 143.Shang X, et al. Mst1 deletion reduces septic cardiomyopathy via activating Parkin-related mitophagy. J. Cell Physiol. 2020;235:317–327. doi: 10.1002/jcp.28971. [DOI] [PubMed] [Google Scholar]
- 144.Yu W, Xu M, Zhang T, Zhang Q, Zou C. Mst1 promotes cardiac ischemia-reperfusion injury by inhibiting the ERK-CREB pathway and repressing FUNDC1-mediated mitophagy. J. Physiol. Sci. 2019;69:113–127. doi: 10.1007/s12576-018-0627-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yao L, et al. Regulation of YAP by mammalian target of rapamycin complex 1 in endothelial cells controls blood pressure through COX-2/mPGES-1/PGE cascade. Hypertension. 2019;74:936–946. doi: 10.1161/HYPERTENSIONAHA.119.12834. [DOI] [PubMed] [Google Scholar]
- 146.Lee JK, et al. MST1 functions as a key modulator of neurodegeneration in a mouse model of ALS. Proc. Natl Acad. Sci. USA. 2013;110:12066–12071. doi: 10.1073/pnas.1300894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang M, Tao W, Yuan Z, Liu Y. Mst-1 deficiency promotes post-traumatic spinal motor neuron survival via enhancement of autophagy flux. J. Neurochem. 2017;143:244–256. doi: 10.1111/jnc.14154. [DOI] [PubMed] [Google Scholar]
- 148.Xiao, D. et al. Enhanced mitophagy mediated by the YAP/Parkin pathway protects against DOX-induced cardiotoxicity. Toxicol. Lett. 330, 10.1016/j.toxlet.2020.05.015 (2020). [DOI] [PubMed]
- 149.Zhou T, Chang L, Luo Y, Zhou Y, Zhang J. Mst1 inhibition attenuates non-alcoholic fatty liver disease via reversing Parkin-related mitophagy. Redox Biol. 2019;21:101120. doi: 10.1016/j.redox.2019.101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu Z, et al. A potential role for the Hippo pathway protein, YAP, in controlling proliferation, cell cycle progression, and autophagy in BCPAP and KI thyroid papillary carcinoma cells. Am. J. Transl. Res. 2017;9:3212–3223. [PMC free article] [PubMed] [Google Scholar]
- 151.Li L, et al. Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene. 2016;35:4048–4057. doi: 10.1038/onc.2015.476. [DOI] [PubMed] [Google Scholar]
- 152.Zhang J, et al. Overexpression of macrophage stimulating 1 enhances the anti-tumor effects of IL-24 in esophageal cancer via inhibiting ERK-Mfn2 signaling-dependent mitophagy. Biomed. Pharmacother. 2019;114:108844. doi: 10.1016/j.biopha.2019.108844. [DOI] [PubMed] [Google Scholar]
- 153.Fan S, et al. PINK1-dependent mitophagy regulates the migration and homing of multiple myeloma cells via the MOB1B-mediated Hippo-YAP/TAZ pathway. Adv. Sci. 2020;7:1900860. doi: 10.1002/advs.201900860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hu Y, et al. Mammalian STE20‑like kinase 1 regulates pancreatic cancer cell survival and migration through Mfn2‑mediated mitophagy. Mol. Med. Rep. 2020;22:398–404. doi: 10.3892/mmr.2020.11098. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 155.Wei R, et al. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J. Exp. Clin. Cancer Res. 2019;38:112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]