Abstract
Background
Multi-drug resistance (MDR) and extensive-drug resistance (XDR) associated with extended-spectrum beta-lactamases (ESBLs) and carbapenemases in Gram-negative bacteria are global public health concerns. Data on circulating antimicrobial resistance (AMR) genes in Gram-negative bacteria and their correlation with MDR and ESBL phenotypes from Nepal is scarce.
Methods
A retrospective study was performed investigating the distribution of ESBL and carbapenemase genes and their potential association with ESBL and MDR phenotypes in E. coli, Klebsiella spp., Enterobacter spp. and Acinetobacter spp. isolated in a major tertiary hospital in Kathmandu, Nepal, between 2012 and 2018.
Results
During this period, the hospital isolated 719 E. coli, 532 Klebsiella spp., 520 Enterobacter spp. and 382 Acinetobacter spp.; 1955/2153 (90.1%) of isolates were MDR and half (1080/2153) were ESBL producers. Upon PCR amplification, blaTEM (1281/1771; 72%), blaCTXM-1 (930/1771; 53%) and blaCTXM-8 (419/1771; 24%) were the most prevalent ESBL genes in the enteric bacilli. BlaOXA and blaOXA-51 were the most common blaOXA family genes in the enteric bacilli (918/1771; 25%) and Acinetobacter spp. (218/382; 57%) respectively. Sixteen percent (342/2153) of all isolates and 20% (357/1771) of enteric bacilli harboured blaNDM-1 and blaKPC carbapenemase genes respectively. Of enteric bacilli, Enterobacter spp. was the most frequently positive for blaKPC gene (201/337; 60%). The presence of each blaCTX-M and blaOXA were significantly associated with non-susceptibility to third generation cephalosporins (OR 14.7, p < 0.001 and OR 2.3, p < 0.05, respectively).The presence of each blaTEM, blaCTXM and blaOXA family genes were significantly associated with ESBL positivity (OR 2.96, p < 0.001; OR 14.2, p < 0.001 and OR 1.3, p < 0.05 respectively) and being MDR (OR 1.96, p < 0.001; OR 5.9, p < 0.001 and OR 2.3, p < 0.001 respectively).
Conclusions
This study documents an alarming level of AMR with high prevalence of MDR ESBL- and carbapenemase-positive ESKAPE microorganisms in our clinical setting. These data suggest a scenario where the clinical management of infected patients is increasingly difficult and requires the use of last-resort antimicrobials, which in turn is likely to intensify the magnitude of global AMR crisis.
Keyword: Multi-drug resistance, Antimicrobial resistance, Nepal, ESBL, Gram-negative bacilli, Carbapenemase, ESKAPE
Background
Gram-negative bacilli, particularly those in the bacterial family Enterobacteriaceae (e.g. Klebsiella spp. and Enterobacter spp.) and Acinetobacter spp. are common causes of serious community and hospital-acquired infections. These Gram-negative bacilli are also members of the ESKAPE group of pathogens [1].These are notoriously associated with antimicrobial resistance (AMR) and frequently carry genes that induce resistance to three or more classes of antimicrobials, making them multi-drug resistant. Such multi-drug resistant Gram-negative bacilli represent a significant global public health problem as they are more commonly associated with worse outcomes than susceptible isolates [2–5].The situation with multi-drug resistant Gram-negative bacilli is particularly alarming in South Asia, which is considered as a global epicentre of these microorganisms [6, 7].
Extended-spectrum beta-lactamases (ESBLs) are enzymes that can hydrolyse and therefore inactivate beta-lactam antimicrobials such as penicillins, cephalosporins and monobactams. ESBL activity is an important mechanism by which Gram-negative bacilli exhibit resistance against beta-lactam antimicrobials. Multiple ESBL variants have been detected and grouped into several structural and evolutionary families which include TEM, SHV, CTX-M, PER, VEB, GES, BES, TLA and OXA [8]. Of these, CTX-M and TEM are the most common ESBLs, while OXA enzymes such as OXA-48, 23, 24, 51, 58 are common carbapenemases that are associated with Gram-negative bacteria that cause nosocomial infections [8, 9].
The possession of ESBLs increases the risk of treatment failure with beta-lactam antimicrobials, may contribute to the spread of AMR in Gram-negative bacteria and may complicate infection control in hospital [9, 10]. Therefore, detection and reporting for the presence of ESBLs in bacterial pathogens are important for clinical care. This is also essential for developing optimal infection control measures in hospital [10]. Modified disc diffusion tests are routinely used to detect the presence of ESBLs and establish the nature of ESBL phenotypes [11]. Though being useful, these phenotypic detection methods are labour intensive and do not determine the classes of genes that are associated with ESBL activity. Molecular methods like PCR, DNA hybridization, or whole genome sequencing (WGS) may be used alongside phenotypic test methods to detect the presence of ESBL and carbapenemase associated genes and generate additional information regarding the epidemiological distribution of key resistance genes in sentinel locations [10].
South Asia is a key location for antimicrobial resistant pathogens. A high prevalence of MDR and ESBL positive Enterobacteriaceae family bacteria and Acinetobacter have been reported across the region [12–19]. However, little is known about various ESBL and carbapenemase genes associated with these phenotypes in Gram-negative bacilli isolated in hospitals of Nepal. Furthermore, there are little data originating from this region on the association of these genotypes with the observed phenotypes. Such data are important for assessing the contribution of combined phenotypic and genetic tests on clinical care and providing better understanding of the impact of resistance genes on the epidemiology of circulating pathogens.
Here, aiming to better understand the epidemiology of multi-drug resistant microorganisms and improve clinical care in our healthcare facility, we determined the AMR profile of all E. coli, Klebsiella spp., Enterobacter spp. and Acinetobacter spp. isolated from in- and out-patients at a major tertiary hospital in Kathmandu, Nepal, between June 2012 and December 2018. We additionally screened them for the phenotypic presence of ESBL activity by disc diffusion method and performed PCR amplification to detect any ESBL and carbapenemase associated AMR genes. Lastly, we investigated the relationship between the presence of blaCTX-M, blaTEM and blaOXA genes with an ESBL or MDR phenotype. Understanding the association between AMR genotype and phenotype is crucial to assess the value of genetic detection methods in clinical settings.
Methods
Study design
This was a retrospective study of anonymised routine microbiology laboratory results originating from Patan Hospital in Lalitpur metropolitan city of the Kathmandu Valley, Nepal. All data regarding E. coli, Klebsiella spp., Enterobacter spp. and Acinetobacter spp. that were isolated from June 2012 to December 2018 were included in this study. These data were devoid of any personal identification information as this work was performed as a component of routine surveillance for infection control at Patan Hospital.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed at the time of bacterial isolation by modified Kirby-Bauer disc diffusion method, as previously described [20]. Zone size interpretations were performed following the appropriate Clinical and Laboratory Standards Institute (CLSI) guidelines [21]. The antimicrobials against which organisms were tested are listed in Additional file 1: Table S1; not all isolates were tested for all antimicrobials.
The bacteria producing resistant or intermediate response against tested antimicrobials were grouped as “non-susceptible” for the purposes of analysis. MDR was defined as an acquired non-susceptibility (without intrinsic resistance) to at least one agent of three or more antimicrobial classes. Intrinsic resistance was defined according to the CLSI guideline of 2014 [21]. The following intrinsic resistance were reported but ignored for the purpose of generating MDR profiles: (i) Acinetobacter spp. resistant against amoxicillin or penicillin, (ii) E. coli, Klebsiella, Enterobacter, or Acinetobacter spp. against vancomycin, teicoplanin, erythromycin or azithromycin. Phenotypic testing for ESBL positivity was conducted using the combination disc diffusion method with a beta-lactam antimicrobial disc alone and that in combination with a beta-lactamase inhibitor (clavulanic acid) [22]. The isolate was considered as ESBL positive if the zone of inhibition around the beta-lactamase inhibitor supplemented disc was ≥ 5 mm in comparison to the respective beta-lactam antimicrobial alone. The products of Mast diagnostics (Mast group Ltd., Liverpool, UK) namely D62C and D68C were used in this study.
Detection of resistance genes
Bacterial DNA was extracted by suspending bacterial colonies in Phosphate buffered saline (PBS) and subjecting them to 100 °C for five minutes; the suspensions were centrifuged and the supernatant was used as template for PCR amplifications. The PCR amplifications were performed in multiplex following previously described primers and conditions. The ESBL targets were blaCTXM-1, blaCTXM-2, blaCTXM-8, blaCTXM-9, blaCTXM-25, blaTEM and blaSHV [23]. The carbapenem resistance genes tested were blaOXA, blaKPC, blaOXA48 [24], blaNDM-1 [25], blaOXA1_4_30, blaOXA23, blaOXA24, blaOXA51, blaOXA58 [26], blaVIM and blaIMP [27].
For analysis purpose, the detection of any of blaOXA, blaOXA1_4_30, blaOXA23, blaOXA24, blaOXA48, blaOXA51 or blaOXA58 was designated as blaOXA positive. Similarly, the detection of any of blaCTXM-1, blaCTXM-2, blaCTXM-8, blaCTXM-9 or blaCTXM-25 was designated as blaCTXM positive. Fisher’s exact test was used to test for an association between the detection of a resistance gene and a specific resistance phenotype (ESBL positive, MDR, or resistance to third and fourth generation cephalosporins). All analyses were performed using the statistical software R version 3.5.3 and R Studio version 1.0.143. Venn diagrams were generated using the UpSetR R package [28].
Results
Bacterial isolates
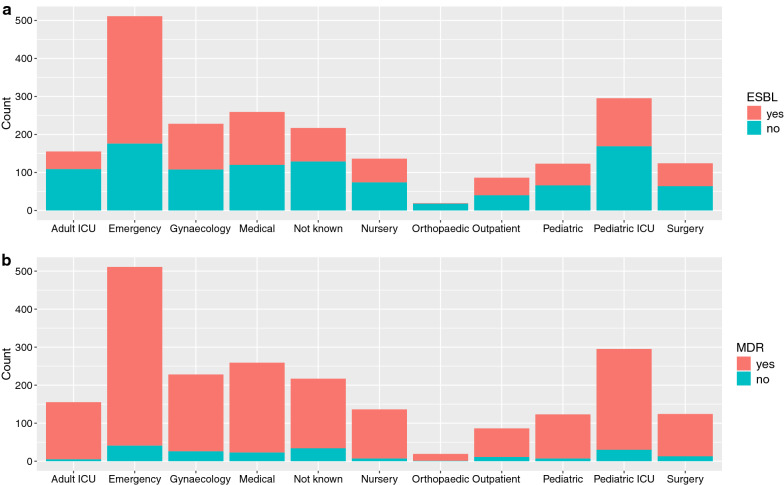
Between June 2012 and December 2018, the microbiology laboratory at Patan hospital isolated 719 E. coli, 532 Klebsiella spp., 520 Enterobacter spp. and 383 Acinetobacter spp., totalling 2153 isolates (Table 1). A majority (512/2153; 23.8%) of isolates originated from the samples that were taken from the patients visiting emergency department followed by paediatric intensive care unit (PICU, 295/2153; 13.7%), gynaecology ward (287/2153; 13.3%) and general medical ward (208/2153; 9.7%). The distribution of ESBL-positivity and MDR in microorganisms by hospital departments is shown in Fig. 1. Half (1080/2153; 50.2%) of all bacterial isolates were ESBL positive (Table 2), which was predictably more common (> 50%) in E. coli, Klebsiella spp. and Enterobacter spp. isolates than in Acinetobacter spp. isolates (< 1%). Approximately 90% of E. coli, Klebsiella spp. and Enterobacter spp. were multi-drug resistant; this proportion was relatively lower in Acinetobacter spp. (82%).
Table 1.
Summary of microorganisms
| n | % | |
|---|---|---|
| Microorganisms (Total = 2153) | ||
| E. coli | 719 | 33.4 |
| Klebsiella spp. | 532 | 24.7 |
| Enterobacter spp. | 520 | 24.2 |
| Acinetobacter spp. | 382 | 17.7 |
| Departments (Total = 2153) | ||
| Emergency | 512 | 23.8 |
| Paediatric ICU | 295 | 13.7 |
| Gynaecology | 287 | 13.3 |
| Medical | 208 | 9.7 |
| Adult ICU | 154 | 7.2 |
| Nursery | 136 | 6.3 |
| Surgery | 116 | 5.4 |
| Paediatric | 125 | 5.8 |
| Not known | 219 | 10.2 |
| Outpatient | 82 | 3.8 |
| Orthopaedic | 19 | 0.9 |
| ESBL positive (Total = 2153) | ||
| No | 1073 | 49.8 |
| Yes | 1080 | 50.2 |
Fig. 1.
ESBL and MDR positivity status of isolates by ward. Count of ESBL-positive and -negative (a) and MDR and non-MDR isolates (b) stratified by hospital wards from where the samples were taken
Table 2.
ESBL positivity and MDR phenotype stratified by microorganisms
| Microorganisms (N = 2153) | ESBL-neg | ESBL-pos | Non-MDR | MDR | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | |
| E. coli (N = 719) | 294 | 40.9 | 425 | 59.1 | 76 | 10.6 | 643 | 89.4 |
| Klebsiella spp. (N = 532) | 210 | 39.5 | 322 | 60.5 | 44 | 8.3 | 488 | 91.7 |
| Enterobacter spp. (N = 520) | 189 | 36.3 | 331 | 63.7 | 10 | 1.9 | 510 | 98.1 |
| Acinetobacter spp. (N = 382) | 380 | 99.5 | 2 | 0.5 | 68 | 17.8 | 314 | 82.2 |
The distribution of AMR genes
The presence of screened AMR genes, stratified by the number of microorganisms tested is presented in Table 3. In E. coli isolates, 90.1% (648/719) were positive for blaTEM, 58.0% (417/719) were positive for blaCTXM-8 and 37.4% (269/719) were positive for blaOXA. In the Enterobacter spp., 56.2% (292/520) were positive for blaOXA, 59.6% (201/520) were positive for blaKPC, 80.3% (417/520) were positive for blaCTXM-1 and 57.9% (301/520) were positive for blaTEM. In Klebsiella spp., 77.4% (408/532) were positive for blaCTXM-1, 67.2% (357/532) were positive for blaOXA, 4.1% (22/532) were positive for blaKPC and 62.5% (332/532) of tested isolates were positive for blaTEM. In Acinetobacter spp., 58.6% (218/382) were positive for blaOXA51, 36.0% (134/382) were positive for blaOXA23 and 20.7% (79/382) were positive for blaNDM-1.
Table 3.
Summary of detection of AMR genes
| Organisms | Genes | CTX-M1 | CTX-M2 | CTX-M8 | CTX-M9 | CTX-M25 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | %a | n | %a | n | %a | n | %a | n | %a | ||
| E. coli | neg | 612 | 85.4 | 717 | 100 | 302 | 42.0 | 714 | 99.3 | 710 | 98.7 |
| pos | 105 | 14.6 | 0 | 0 | 417 | 58.0 | 5 | 0.7 | 9 | 1.3 | |
| n.t.b | 2 | 2 | 0 | 0 | 0 | ||||||
| Enterobacter | neg | 102 | 19.7 | 0 | n.a | 0 | n.a | 518 | 99.8 | 0 | n.a |
| pos | 417 | 80.3 | 0 | n.a | 0 | n.a | 1 | 0.2 | 0 | n.a | |
| n.t.b | 1 | 520 | 520 | 1 | 520 | ||||||
| Klebsiella | neg | 119 | 22.6 | 527 | 100 | 525 | 99.6 | 524 | 99.4 | 521 | 98.9 |
| pos | 408 | 77.4 | 0 | 0 | 2 | 0.4 | 3 | 0.6 | 6 | 1.1 | |
| n.t.b | 5 | 5 | 5 | 5 | 5 | ||||||
| Acinetobacter | neg | 368 | 98.9 | 372 | 100 | 372 | 100 | 372 | 100.0 | 372 | 100 |
| pos | 4 | 1.1 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0 | 0 | |
| n.t.b | 10 | 10 | 10 | 10 | 10 | ||||||
| Organisms | Genes | OXA | OXA1_4_30 | OXA23 | OXA24 | OXA48 | OXA51 | OXA58 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | %a | n | %a | n | %a | n | %a | n | %a | n | %a | n | %a | ||
| E. coli | neg | 450 | 62.6 | 0 | n.a | 0 | n.a | 0 | n.a | 715 | 99.4 | 0 | n.a | 0 | n.a |
| pos | 269 | 37.4 | 0 | n.a | 0 | n.a | 0 | n.a | 4 | 0.6 | 0 | n.a | 0 | n.a | |
| n.t.b | 0 | 719 | 719 | 719 | 0 | 719 | 719 | ||||||||
| Enterobacter | neg | 228 | 43.8 | 0 | n.a | 0 | n.a | 0 | n.a | 221 | 65.6 | 0 | n.a | 0 | n.a |
| pos | 292 | 56.2 | 0 | n.a | 0 | n.a | 0 | n.a | 116 | 34.4 | 0 | n.a | 0 | n.a | |
| n.t.b | 0 | 520 | 520 | 520 | 520 | 520 | |||||||||
| Klebsiella | neg | 174 | 32.8 | 0 | n.a | 0 | n.a | 0 | n.a | 475 | 89.5 | 0 | n.a | 0 | n.a |
| pos | 357 | 67.2 | 0 | n.a | 0 | n.a | 0 | n.a | 56 | 10.5 | 0 | n.a | 0 | n.a | |
| n.t.b | 1 | 532 | 532 | 532 | 1 | 532 | 532 | ||||||||
| Acinetobacter | neg | 0 | n.a | 342 | 91.9 | 238 | 64.0 | 363 | 97.6 | 0 | n.a | 154 | 41.4 | 357 | 96.0 |
| pos | 0 | n.a | 30 | 8.1 | 134 | 36.0 | 9 | 2.4 | 0 | n.a | 218 | 58.6 | 15 | 4.0 | |
| n.t.b | 382 | 10 | 10 | 10 | 382 | 10 | 10 | ||||||||
| Organisms | Genes | IMP | KPC | NDM1 | SHV | TEM | VIM | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | %a | n | %a | n | %a | n | %a | n | %a | n | %a | ||
| E. coli | neg | 577 | 99.7 | 719 | 100 | 681 | 94.7 | 719 | 100 | 71 | 9.9 | 578 | 99.8 |
| pos | 2 | 0.3 | 0 | 0.0 | 38 | 5.3 | 0 | 648 | 90.1 | 1 | 0.2 | ||
| n.t.b | 140 | 0 | 0 | 0 | 0 | 140 | |||||||
| Enterobacter | neg | 90 | 100 | 136 | 40.4 | 412 | 79.2 | 515 | 99.0 | 219 | 42.1 | 90 | 100 |
| pos | 0 | 0 | 201 | 59.6 | 108 | 20.8 | 5 | 1.0 | 301 | 57.9 | 0 | 0.0 | |
| n.t.b | 183 | 183 | 0 | 0 | 0 | 430 | |||||||
| Klebsiella | neg | 100 | 100 | 509 | 95.9 | 414 | 78.0 | 415 | 78.2 | 199 | 37.5 | 100 | 100 |
| pos | 0 | 0 | 22 | 4.1 | 117 | 22.0 | 116 | 21.8 | 332 | 62.5 | 0 | 0.0 | |
| n.t.b | 1 | 1 | 1 | 1 | 432 | ||||||||
| Acinetobacter | neg | 0 | n.a | 0 | n.a | 303 | 79.3 | 361 | 97.0 | 300 | 78.5 | 0 | n.a |
| pos | 0 | n.a | 0 | n.a | 79 | 20.7 | 11 | 3.0 | 82 | 21.5 | 0 | n.a | |
| n.t.b | 382 | 382 | 90 | 10 | 90 | 382 | |||||||
aPercentage of positive or negative in all isolates tested for a particular gene
bNot tested
The association of ESBL genes with ESBL phenotypes
For all subsequent analysis, isolates were classified as blaOXA or blaCTXM positive if any gene in the blaOXA or blaCTXM gene families were detected respectively. First, we assessed the association between phenotypic ESBL positivity and the presence of blaOXA, blaCTXM, and blaTEM (Table 4). The detection of each of these genes was significantly associated with an ESBL phenotype. The strength of association was variable between different classes with an odds ratio (OR) of 1.3 (95% CI 1.1–1.5, p < 0.05) for blaOXA, 2.96 (95% CI 2.5–3.6, p < 0.001) for blaTEM and 14.2 (95% CI 11.2–18.1, p < 0.001) for blaCTXM in Fisher’s test.
Table 4.
Association between ESBL positivity, MDR or non-susceptibility to cephalosporins and presence of resistance genes
| Genes | ESBL | non-ESBL | OR (95% CI)c | p-valued |
|---|---|---|---|---|
| blaOXAa | ||||
| OXA-neg | 406 | 458 | ||
| OXA-pos | 674 | 604 | 1.3 (1.1–1.5) | 0.009 |
| blaCTXMb | ||||
| CTXM-neg | 112 | 660 | ||
| CTXM-pos | 968 | 401 | 14.2 (11.2–18.1) | < 0.001 |
| blaTEM | ||||
| TEM-neg | 262 | 517 | ||
| TEM-pos | 818 | 545 | 3.0 (2.5–3.6) | < 0.001 |
| Genes | MDR | non-MDR | OR (95% CI)c | p-valued |
|---|---|---|---|---|
| blaOXAa | ||||
| OXA-neg | 751 | 113 | ||
| OXA-pos | 1198 | 80 | 2.3( 1.7–3.1) | < 0.001 |
| blaCTXMb | ||||
| CTXM-neg | 629 | 143 | ||
| CTXM-pos | 1319 | 50 | 6.0 ( 4.2–8.6) | < 0.001 |
| blaTEM | ||||
| TEM-neg | 680 | 99 | ||
| TEM-pos | 1269 | 94 | 2.0 ( 1.4–2.7) | < 0.001 |
| Genes | Cephal. ¾ non-susceptible | Cephal. ¾ susceptible | OR (95% CI)c | p-valued |
|---|---|---|---|---|
| blaOXAa | ||||
| OXA-neg | 825 | 35 | ||
| OXA-pos | 1244 | 26 | 2.0 ( 1.2–3.5) | 0.008 |
| blaCTXMb | ||||
| CTXM-neg | 710 | 54 | ||
| CTXM-pos | 1358 | 7 | 14.7 ( 6.6–38.6) | < 0.001 |
| blaTEM | ||||
| TEM-neg | 746 | 27 | ||
| TEM-pos | 1323 | 34 | 1.4 (0.8–2.4) | 0.22 |
Cephal. 3/4, 3rd or 4th generation cephalosporin
aAn isolate was classified as blaOXA positive if blaOXA1_4_30, blaOXA, blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-51 or blaOXA-58 was detected
bAn isolate was classified as blaCTXM positive if blaCTXM-1, blaCTXM-2, blaCTXM-8, blaCTXM-9 or blaCTXM-25 was detected
cOdds ratio (OR and 95% CI) of bacterial isolate to be ESBL positive, multi-drug resistant or resistant to 3rd or 4th generation cephalosporin in the presence of a respective gene (gene family)
dp-value from Fisher’s exact test
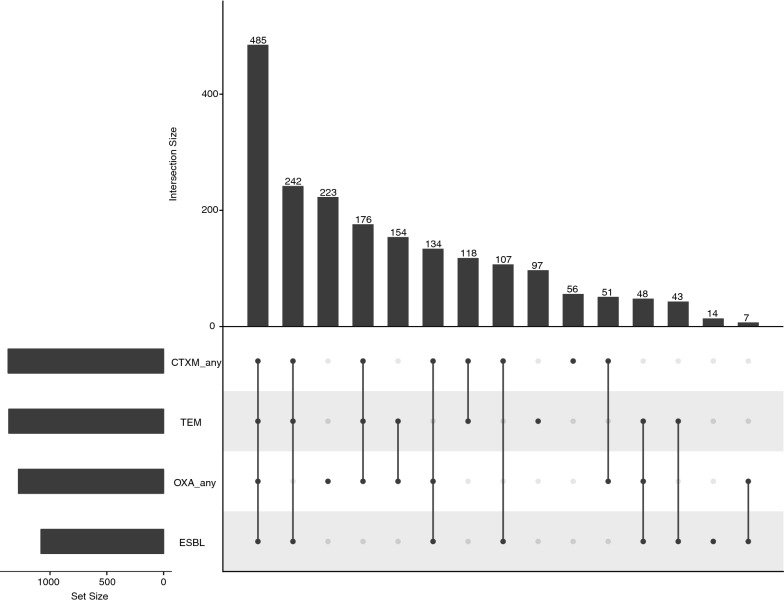
Next, the association between ESBL positivity and various combinations of blaOXA, blaCTXM and blaTEM resistance genes were investigated. The isolates that were tested for at least one member of blaOXA family, one member of blaCTXM family and blaTEM were analysed. A distribution of different combinations of blaOXA, blaCTXM and blaTEM resistance genes with ESBL positivity is shown in Fig. 2. The most frequent (485/1955; 24.8%) combination of genes that was associated with ESBL positivity was at least one member of blaOXA family gene, with at least a member of blaCTXM family gene and blaTEM gene. A smaller proportion of isolates (176/1955; 9.0%) was PCR positive for blaOXA, blaCTXM and blaTEM, but was phenotypically ESBL negative. The second most common (242/1955; 12.3%) gene combination associated with ESBL positivity was blaCTXM with blaTEM. The proportion of isolates that were phenotypically ESBL positive, but PCR amplification negative for blaOXA, blaCTXM and blaTEM was 0.7% (14/1955).
Fig. 2.
Association between ESBL positivity and combinations of resistance genes blaOXA, blaCTXM and blaTEM. Black circles represent ESBL positive and/or the detection of resistance genes of blaOXA family, blaCTXM family or blaTEM. A grey circle indicates that a characteristic was not detected. The vertical bars indicate the number of isolates in the group summarized directly below. The horizontal bars indicate the total number of isolates positive for resistance genes of blaOXA family, blaCTXM family, blaTEM or for ESBL positivity. An isolate was classified as blaOXA positive if any gene of blaOXA family was detected. The same is true for blaCTXM genes. Only the isolates that were tested for ESBL positivity, at least one member of blaOXA family, at least one member of blaCTXM family and blaTEM were included
The association of ESBL genes with non-susceptibility to third and fourth generation cephalosporins
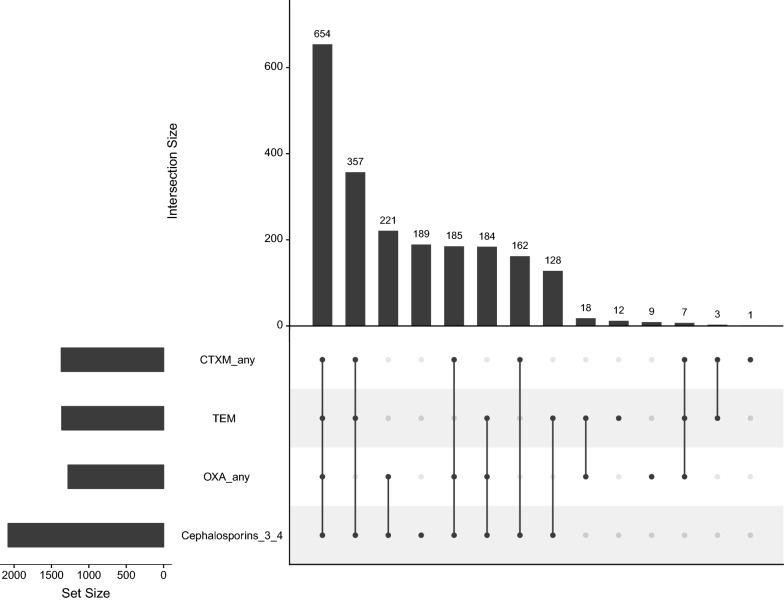
We also tested for the association between non-susceptibility to third and fourth generation cephalosporins (Cefotaxime, Cefepime, Ceftriaxone and Cefixime) and the presence of blaOXA, blaCTXM, or blaTEM (Table 4). The blaCTXM and blaOXA gene families were significantly associated with non-susceptibility to third and fourth generation cephalosporins with odds ratios of 14.7 (95% CI 6.6–38.6, p < 0.001) and 2.03 (95% CI 1.2–3.5, p < 0.05) respectively. The most frequent combination (654/2121; 30.8%) associated with non-susceptibility against third and fourth generation cephalosporins was the presence of all three blaOXA, blaCTXM, and blaTEM genes, followed by the presence of blaCTXM and blaTEM only (357/2121; 16.8%). Only 8.9% (189/2121) of isolates that were phenotypically non-susceptible to third and fourth generation cephalosporins were PCR amplification negative for blaOXA, blaCTXM, and blaTEM (Fig. 3).
Fig. 3.
Association between non-susceptibility to 3rd and 4th generation cephalosporins and combinations of resistance genes blaOXA, blaCTXM and blaTEM. Black circles represent non-susceptibility to cephalosporins (3rd/4th gen.) and/or the detection of resistance genes of blaOXA family, blaCTXM family or blaTEM. A grey circle indicates that a characteristic was not detected. The vertical bars indicate the number of isolates in the group summarized directly below. The horizontal bars indicate the total number of isolates positive for resistance genes of blaOXA family, blaCTXM family, for blaTEM or for non-susceptibility to 3rd and 4th generation cephalosporins. An isolate was classified as blaOXA positive if any gene of blaOXA family was detected. The same is true for blaCTXM genes. Only isolates that had a known 3rd and 4th generation cephalosporin resistance profile and were tested for at least one member of blaOXA family, at least one member of blaCTXM family and blaTEM were included
The association of ESBL genes with MDR
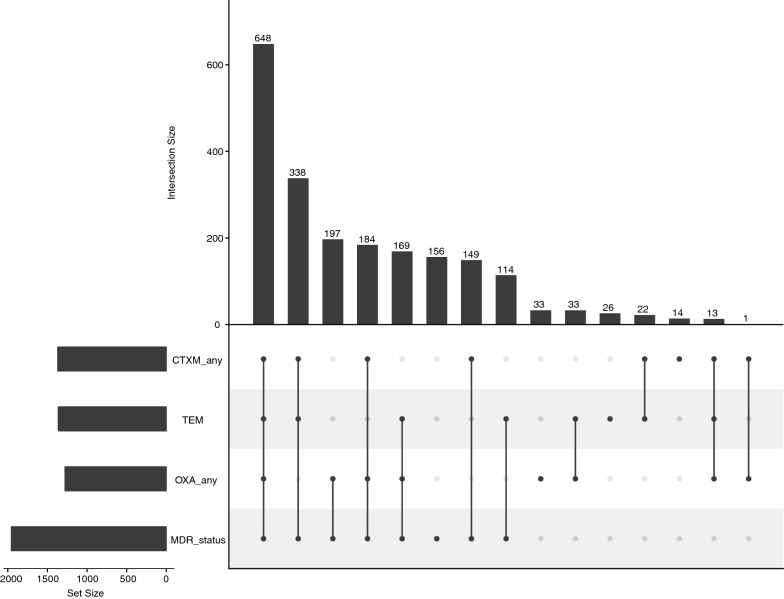
We similarly investigated for any potential association between the presence of blaOXA, blaCTXM or blaTEM genes with MDR (Table 4). A multi-drug resistant phenotype was significantly associated with the presence of blaCTXM (OR 5.99, 95% CI 4.2–8.6, p < 0.001), blaOXA (OR 2.3, 95% CI 1.7–3.1, p < 0.001) and blaTEM (OR 1.96, 95% CI 1.4–2.7, p < 0.001). Additionally, the most common gene combinations associated with MDR were blaOXA and blaCTXM with blaTEM genes (648/2097; 30.9%), followed by blaCTXM with blaTEM (338/2097; 16.1%). A small fraction of isolates (156/2097; 7.4%) were PCR amplification negative for blaOXA, blaCTXM and blaTEM, but were multi-drug resistant phenotypes (Fig. 4).
Fig. 4.
Association between MDR and combinations of resistance genes blaOXA, blaCTXM and blaTEM. Black circles represent MDR and/or the detection of resistance genes of blaOXA family, blaCTXM family or blaTEM. A grey circle indicates that a characteristic was not detected. The vertical bars indicate the number of isolates in the group summarized directly below. The horizontal bars indicate the total number of isolates positive for resistance genes of blaOXA family, blaCTXM family, for blaTEM or for MDR phenotype. An isolate was classified as blaOXA positive if any gene of blaOXA family was detected. The same is true for blaCTXM genes. Only isolates that had an AMR profile and were tested for at least one member of blaOXA family, at least one member of blaCTXM family and blaTEM were included
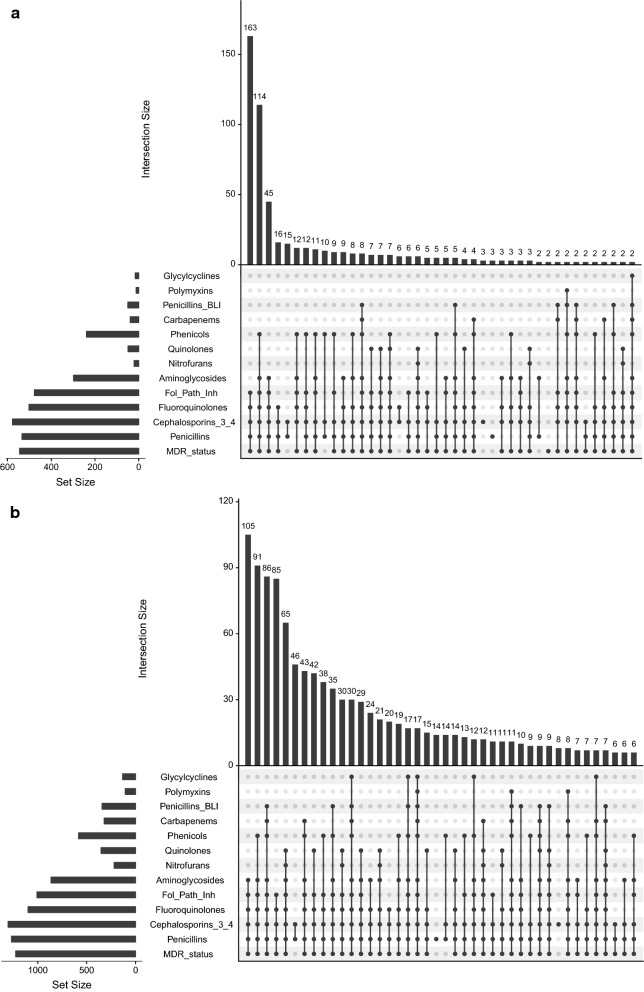
As MDR was defined as an acquired non-susceptibility to at least one agent in three different classes of antimicrobials, a multi-drug resistant phenotype can originate from a multitude of individual antimicrobial resistant phenotypes. Therefore, we compared the pattern of resistance for the isolates originating from outpatients (emergency ward and outpatient department, Fig. 5a) and that from inpatients (all other wards, Fig. 5b). For the microorganisms originating from the outpatients, the three most frequent AMR combinations accounted for over half (322/535; 60.2%) of all outpatient isolates. For the microorganisms originating from the inpatients, the three most frequent AMR combinations accounted only for 26% of all inpatient isolates (282/1057) and the diversity of resistant phenotypes was much broader than that from outpatient isolates.
Fig. 5.
AMR patterns for outpatients and inpatients. AMR patterns are presented for outpatients (emergency and outpatients departments) in (a) and for inpatients (all other hospital wards) in (b). Only the 42 most frequent combinations are shown. Black circles represent resistance to the antimicrobials groups and/or multidrug resistance (MDR). The vertical bars represent the number of isolates characterized by the AMR pattern described under the bar. The horizontal bars indicate the total number of isolates resistant to the different groups, or MDR
Discussion
In this study, very high prevalence of ESBL positivity and MDR was found in Enterobacteriaceae family bacteria (E. coli, Klebsiella spp. and Enterobacter spp.). Similar MDR prevalence has been reported in other studies from Nepal, but the ESBL positive prevalence we observed (over 50%) is higher than others have reported. For example, one study from Nepal found that in community-acquired infections caused by E. coli, 24% were ESBL positive and 78% had MDR [12]. Another hospital-based study from Nepal showed that in urine samples, 27% of E. coli were ESBL positive and 97% of Enterobacteriaceae bacteria (mostly E. coli and Klebsiella spp.) were multi-drug resistant [13]. For Acinetobacter spp., we report over 81% MDR, similar to what others have reported in ICUs in Nepal, but the ESBL prevalence we found (< 1% in Acinetobacter spp.) is much lower than what has been reported (around 12–13%) [15, 16, 29]. In the context of South Asia, 79% of Acinetobacter spp., 71% of Klebsiella spp. and 54% of E. coli isolated from young infants were estimated to be multi-drug resistant strains [19]. ESBL positivity was confirmed in 42% of K. pneumoniae and 33% of E. coli isolated from several hospitals in India [30], 40% in Enterobacteriaceae isolates from Pakistan [18] and 16% in enteric and non-enteric Gram-negative bacilli from Bangladesh [31].
To limit the spread of antimicrobial resistant microorganisms, to guide clinical care and improve patients’ outcome, it is essential to have precise knowledge on AMR profile of the infecting bacteria prior to starting antimicrobial therapy to select an appropriate drug. Antimicrobial sensitivity testing and detection of ESBL positivity can be performed by classical phenotypical methods such as disc diffusion, which is labour intensive and time consuming. Automated systems exist, but their installation, running and maintenance costs are often a barrier in low resource settings. Alternatively, simple genetic methods such as PCR can detect AMR genes rapidly and with high sensitivity [2, 10, 32]. A better understanding of the association between phenotype and genotype is a prerequisite to evaluate the usefulness of genetic detection methods in clinical settings. In this study, we assessed the correlation between the detection of blaCTXM, blaTEM and blaOXA genes by PCR and ESBL positivity or MDR as detected phenotypically by disc diffusion.
For E. coli, Enterobacter spp. and Klebsiella spp., the resistance genes most frequently detected were blaOXA, blaTEM, blaCTXM-1 and blaCTXM-8.This is consistent with a study performed in 2012–2013 at Kathmandu Medical College and Teaching Hospital, in which blaCTXM and blaTEM were frequently detected in ESBL positive E. coli [14]. A study from Bangladesh reported blaCTXM-1 (51%) and blaSHV (27%) to be the most common ESBL genes in Klebsiella pneumoniae [17]. In Pakistan and India, as in our study, blaOXA, blaTEM and blaCTXM, in addition to blaSHV were detected to be common AMR genes in Enterobacteriaceae isolates [18, 30]. For Acinetobacter spp., blaOXA-51, blaOXA-23 and blaNDM-1were the most frequently detected AMR genes, as found by others in Nepal, India and Bangladesh [33–35].
The detection of any gene of the blaCTXM family was strongly associated with ESBL positivity (OR 14.2, 95% CI11.2–18.1), MDR (OR 5.99, 95% CI 4.2–8.6) and resistance to 3rd and 4th generation cephalosporins (OR 14.7, 95% CI 6.6–38.6). The detection of blaTEM was strongly associated with ESBL positivity (OR 2.96, 95% CI 2.5–3.6) and MDR (OR 1.96, 95% CI 1.4–2.7). An association between blaTEM and resistance to 3rd and 4th generation cephalosporins was suggested by an OR of 1.4, but it was not statistically significant (95% CI 0.8–2.4), perhaps due to low number of isolates susceptible to 3rd and 4th generation cephalosporins. Finally, the detection of any gene of the blaOXA family was associated with multi-drug resistant phenotype (OR 2.3, 95% CI 1.7–3.1). This suggests that the detection of blaTEM, or any gene of the blaCTXM or blaOXA family is an important index for multi-drug resistant phenotype, and as expected, the detection of a blaCTXM family gene or blaTEM indicates a higher odds of ESBL positive phenotype. Collectively, this suggests that the detection of key AMR genes by molecular methods is an important index for ESBL positivity and MDR in bacterial isolates.
In this study, we also described different AMR patterns found in the isolates originating from emergency and outpatient wards (outpatients) compared to all other wards of the hospital (inpatients), which may reflect different usage of antimicrobials in community compared to the hospital. It can be assumed that isolates originating from outpatients reflect what is circulating in communities. For outpatients, the three most frequent AMR combinations accounted for more than half of all isolates. Perhaps people contracting community-acquired infections predominantly self-medicate, which results in exposure to the limited array of antimicrobials that are freely available in shops and pharmacies. Only infections that are (or have become) resistant to those drugs prompt a visit to the hospital. As a result, infections seen in outpatients have similar resistance profile. In contrast, a higher diversity of AMR combinations found in inpatients may reflect a wider array of antimicrobials available and used at hospitals.
Conclusions
MDR along with possession of ESBL and carbapenemase associated resistance genes among Gram-negative bacilli pose a serious problem in therapeutic management of patients. The compromised infection control and inadequate antimicrobial usage policies coupled with high burden of Gram-negative bacilli possessing transferable antimicrobial resistance genes in resource limited settings set out an ideal scenario for the emergence and dissemination of multi-drug resistant pathogens. In this extensive retrospective study, a high burden of multi-drug resistant clinical Gram-negative bacilli possessing diverse ESBL and carbapenemase resistance bla genes were evidenced. Further, our study signifies that there is a high probability of Gram-negative bacilli to be multi-drug resistant and ESBL positive in case of detection of any of blaTEM, blaCTXM or blaOXA family genes. Additionally, the detection of any of the blaCTXM family genes highly implies that the Gram-negative bacilli are non-susceptible to the extended-spectrum beta-lactam antimicrobials.
Supplementary information
Additional file 1: Table S1. Classes of antimicrobials and their members.
Acknowledgements
We wish to acknowledge all staffs in the clinical microbiology laboratory of Patan Hospital.
Abbreviations
- AMR
Antimicrobial resistance
- CI
Confidence interval
- CLSI
Clinical and laboratory standards institute
- DNA
Deoxy-ribonucleic acid
- ESBL
Extended-spectrum beta-lactamase
- MDR
Multi-drug resistance
- OR
Odds ratio
- PCR
Polymerase chain reaction
- PICU
Pediatric intensive care unit
- WGS
Whole genome sequencing
- XDR
Extensive-drug resistance
Authors’ contributions
AK conceptualized the research and has a major contribution in writing and editing the manuscript. BB performed manuscript editing. GT performed manuscript editing. KGP performed laboratory work and manuscript editing. NM performed statistical analysis of data. RMZ performed statistical analysis of data and preparation of graphs. SB reviewed manuscript and has a major contribution in manuscript editing. SD performed laboratory work and manuscript editing. SM conducted the research, performed laboratory work, data management, data analysis, results interpretation, major manuscript writing, editing and formatting. SMD performed manuscript editing. All authors read and approved the final manuscript.
Funding
This project was funded by the Wellcome Trust and the Royal Society (100087/14/Z). AK was funded as a leadership fellow through the Oak Foundation (OCAY-15–547).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
An ethical approval for conducting this study was taken from Nepal Health Research Council (NHRC) (Reference number 1017). Data used in this retrospective study were the anonymised routine microbiology laboratory results originating from Patan Hospital. These data were devoid of any patients’ identifying information. Further, the data used in this study were a part of routine surveillance for infection prevention and control at Patan Hospital. Thus, individual written informed consent was not required.
Consent for publication
All data used in this study were the anonymised microbiological data devoid of patients’ identification and personal information. Thus obtaining consent for publication is not applicable for this study. All authors consented for the publication of this research.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12941-020-00390-y.
References
- 1.Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197(8):1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 2.Exner M, Bhattacharya S, Christiansen B, Gebel J, Goroncy-Bermes P, Hartemann P, et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:5. doi: 10.3205/dgkh000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organisation Global report on surveillance: antimicrobial resistance. World Heal Organ. 2014;1:7. [Google Scholar]
- 4.Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27(2):128. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi Y, Bonomo RA, Hooper DC, Kaye KS, Johnson JR, Clancy CJ, et al. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the antibacterial resistance leadership group. Clin Infect Dis. 2017;64(1):S30–5. doi: 10.1093/cid/ciw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laxminarayan R, Chaudhury RR. Antibiotic resistance in india: drivers and opportunities for action. PLOS Med. 2016;13(3):e1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatia R, Narain JP. The growing challenge of antimicrobial resistance in the South-East Asia Region–are we losing the battle? Indian J Med Res. 2010;132(5):482–486. doi: 10.4103/0971-5916.73313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathers AJ, Peirano G, Pitout JDD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitout JDD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 11.Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansari S, Hari P, Gautam R, Shrestha S, Neopane P, Rimal B, Mandal F, Ansari SR, Chapagain ML, Ansari SRS, Nepal HP, Gautam R, Shrestha S, Neopane P, et al. Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care center of Nepal. Antimicrob Resist Infect Control. 2015;4:15. doi: 10.1186/s13756-015-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav KK, Adhikari N, Khadka R, Pant AD, Shah B. Multidrug resistant Enterobacteriaceae and extended-spectrum β-lactamase producing Escherichia coli: a cross-sectional study in National Kidney Center. Nepal Antimicrob Resist Infect Control. 2015;4:42. doi: 10.1186/s13756-015-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokhrel R, Thapa B, Kafle R, Shah P, Tribuddharat C. Co-existence of beta-lactamases in clinical isolates of Escherichia coli from Kathmandu. Nepal BMC Res Notes. 2014;7(1):694. doi: 10.1186/1756-0500-7-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanal S, Joshi DR, Bhatta DR, Devkota U, Pokhrel BM. β-Lactamase-producing multidrug-resistant bacterial pathogens from tracheal aspirates of intensive care unit patients at national institute of neurological and allied sciences. Nepal ISRN Microbiol. 2013;2013:1–5. doi: 10.1155/2013/847569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhandari P, Thapa G, Pokhrel BM, Bhatta DR, Devkota U. Nosocomial isolates and their drug resistant pattern in icu patients at national institute of neurological and allied sciences. Nepal Int J Microbiol. 2015;2015:1–6. doi: 10.1155/2015/572163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan ER, Aung MS, Paul SK, Ahmed S, Haque N, Ahamed F, et al. Prevalence and molecular epidemiology of clinical isolates of escherichia coli and klebsiella pneumoniae harboring extended-spectrum beta-lactamase and carbapenemase genes in Bangladesh. Microb Drug Resist. 2018;24(10):1568–1579. doi: 10.1089/mdr.2018.0063. [DOI] [PubMed] [Google Scholar]
- 18.Abrar S, Hussain S, Khan RA, Ul Ain N, Haider H, Riaz S. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrob Resist Infect Control. 2018;20(7):26. doi: 10.1186/s13756-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaurasia S, Sivanandan S, Agarwal R, Ellis S, Sharland M, Sankar MJ. Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ. 2019;364:5314. doi: 10.1136/bmj.k5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer A, Kirby W, Sherris J, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute . Standards for antimicrobial susceptibility testing; twenty-fourth informational supplement, CLSI document M100–S24. New York: Clinical and Laboratory Standards Institute; 2014. pp. 1–226. [Google Scholar]
- 22.Thomson KS, Sanders CC. Detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992;36(9):1877–1882. doi: 10.1128/AAC.36.9.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57(1):154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011;49(2):718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Shirani K, Ataei B, Roshandel F. Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes (VIM and IMP) in Pseudomonas aeruginosa strains producing MBL enzyme, isolated from patients with secondary immunodeficiency. Adv Biomed Res. 2016;5(1):124. doi: 10.4103/2277-9175.186986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20(12):1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra S, Rijal B, Pokhrel B. Emerging threat of multidrug resistant bugs—Acinetobacter calcoaceticus baumannii complex and Methicillin resistant Staphylococcus aureus. BMC Res Notes. 2013;6(1):98. doi: 10.1186/1756-0500-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautam V, Thakur A, Sharma M, Singh A, Bansal S, Sharma A, et al. Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res. 2019;149(2):208–215. doi: 10.4103/ijmr.IJMR_172_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jobayer M, Afroz Z, Nahar S, Begum A, Begum S, Shamsuzzaman S. Antimicrobial susceptibility pattern of extended-spectrum beta-lactamases producing organisms isolated in a Tertiary Care Hospital, Bangladesh. Int J Appl Basic Med Res. 2017;7(3):189–192. doi: 10.4103/ijabmr.IJABMR_28_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licker M, Anghel A, Moldovan R, Hogea E, Muntean D, Horhat F, et al. Genotype-phenotype correlation in multiresistant Escherichia coli and Klebsiella pneumoniae strains isolated in Western Romania. Eur Rev Med Pharmacol Sci. 2015;19(10):1888–1894. [PubMed] [Google Scholar]
- 33.Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of bla OXA-23 and bla NDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control. 2017;6(1):21. doi: 10.1186/s13756-017-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayakumar S, Mathur P, Kapil A, Das BK, Ray P, Gautam V, et al. Molecular characterization & epidemiology of carbapenem-resistant Acinetobacter baumannii collected across India. Indian J Med Res. 2019;149(2):240–246. doi: 10.4103/ijmr.IJMR_2085_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatun MN, Farzana R, Lopes BS, Shamsuzzaman SM. Molecular characterization and resistance profile of nosocomial Acinetobacter baumannii intensive care unit of tertiary care hospital in Bangladesh. Bangladesh Med Res Counc Bull. 2015;41(2):101–107. doi: 10.3329/bmrcb.v41i2.29991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Classes of antimicrobials and their members.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.