Abstract
To explore the proper prognostic markers for the likelihood of metastasis in CRC patients. Seventy-seven fresh CRC samples were collected to evaluate the mRNA level of the selected marker using Real-time PCR. Moreover, 648 formalin-fixed paraffin-embedded CRC tissues were gathered to evaluate protein expression by immunohistochemistry (IHC) on tissue microarrays. The results of Real-Time PCR showed that low expression of Talin1 was significantly associated with advanced TNM stage (p = 0.034) as well as gender (p = 0.029) in mRNA levels. Similarly, IHC results indicated that a low level of cytoplasmic expression of Talin1 was significantly associated with advanced TNM stage (p = 0.028) as well as gender (p = 0.009) in CRC patients. Moreover, decreased expression of cytoplasmic Talin1 protein was found to be a significant predictor of worse disease-specific survival (DSS) (p = 0.011) in the univariate analysis. In addition, a significant difference was achieved (p = 0.039) in 5-year survival rates of DSS: 65% for low, 72% for moderate, and 88% for high Talin1 protein expression. Observations showed that lower expression of Talin1 at both the gene and protein level may drive the disparity of CRC patients’ outcomes via worse DSS and provide new insights into the development of progression indicators because of its correlation with increased tumor aggressiveness.
Subject terms: Cancer, Biomarkers, Oncology
Introduction
Colorectal cancer (CRC) is one of the frequent forms of solid tumor diagnosed in both sexes worldwide1,2. It is ranked as third in incidence but second in mortality in different countries, and most of deaths are caused by metastasis; hence, the prevention of subsequent metastasis is the main focus of clinical research3,4. Robust determinants of CRC prognosis include local involvement, lymphovascular invasion, positive surgical margin, regional lymph node metastasis, pre-operative elevation of CEA, and high tumor grade5,6. Moreover, traditional biomarkers, colono/sigmoidoscopy, imaging techniques (magnetic resonance imaging (MRI) and computed tomography (CT)) play specific roles as the gold standard in colorectal screening; however, they are not satisfactory enough7. Unfortunately, the early spread of tumor cells is usually not detectable, and access to more detailed information related to the identification of metastasis-specific molecular markers is urgently warranted.
Histopathological analysis as a prevalent standard test is applied to assess novel molecular biomarkers through simultaneous analysis of hundreds of tissue samples in the clinical setting8. The cancer patient's blood contains diverse tumor-derived materials, including circulating tumor cells (CTCs), and exosomes enable the minimally invasive monitoring of tumor evolution over time in the clinic9,10. It seems that achieving the molecular signature based on CTCs and exosomes, which is completely representative of the tumor, plays a significant role in precise theranostics and can be utilized in combination with clinicopathological staging. From a prognostic perspective, the identification of tumor metastasis-specific molecular markers which cover cancer heterogeneity and increase longitudinal surveillance of patients is essential for precision medicine11.
Although large‐scale studies have been carried out to identify novel biomarkers with great prognostic value, investigations of metastasis-related molecular markers should be increased to actualize the exact prediction of the time of metastasis. Advances in high throughput technology in omics data, especially transcriptomic approaches such as microarrays and RNA sequencing, would increase our knowledge of the molecular markers involved in CRC metastasis, resulting in the identification of potential therapeutic targets for CRC patients12,13. To the best of our knowledge, just a couple of studies have investigated the direct correlation between molecular metastasis cascades in CTCs and exosomes. Hence in the present study, databases were screened in order to analyze the common expression profile of these biomarkers as an essential step in understanding the mechanism of metastasis and in discovering the prognostic markers for CRC14. This study was performed using integrated analysis of bioinformatics data related to the molecular markers involved in CRC metastasis in both CTCs and exosomes. Using the GEO, Exocarta, and Vesiclepedia databases, similarities in the expressed genes among the above-mentioned biomarkers were identified. Both gene ontology (GO) analysis and KEGG pathway15 were examined to identify key candidate genes, and it was found that Talin1 could provide significant prognostic value in CRC metastasis.
The evaluation and validation of biological fluids markers as debatable matter should be assessed on tissue samples because of the lack of a consensus over isolation and characterization. Therefore, this step was taken in the current study to optimize the novel findings in the broad range of samples. The mRNA expression of Talin1 in fresh colorectal tumor samples and their adjacent normal tissues as the control were evaluated using Real-Time PCR. Then, to explore the protein localization and expression level of Talin1 in CRC samples as well as its correlation with clinicopathological features and survival outcome in CRC patients, the findings from the analysis of gene expression data were validated and confirmed on a large series of CRC tissue microarrays (TMAs) using the immunohistochemistry (IHC) technique.
Results
Bioinformatics approach
To find the valuable molecular marker between CTCs and tumor-derived exosomes that was affected in metastasis after online analysis of the total similarity of their expressed genes, the following 15 genes were found: TLN1, TMEM51, GNPNAT1, RPS4X, MARCKSL1, FARP1, RHOB, HSP90AB1, LFNG, AHNAK, RPSA, CNPY2, PRDX1, SLC25A5, and TGFBI, as shown in Fig. 1. A detailed analysis of these genes was applied in online web tools such as EnrichR as a collection of databases, and the results based on a p value < 0.05 as the cut-off criteria are summarized in Supplementary Table 2.
Figure 1.
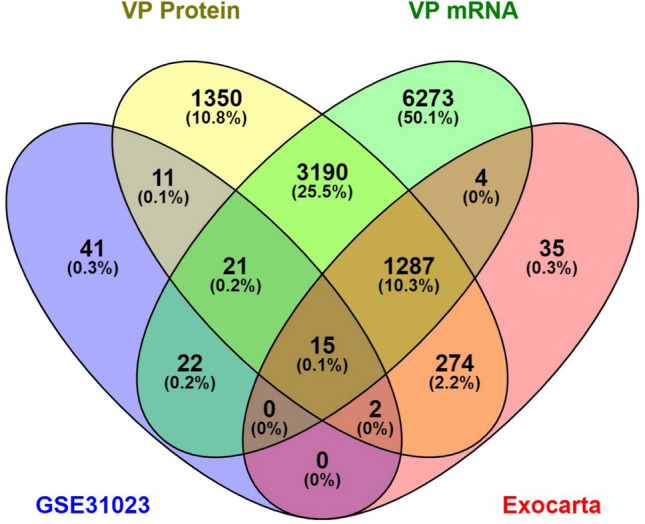
The common molecular markers between CTCs and exosome in databases. Fifteen hub genes were extracted from our mRNA and protein entrance mentioned databases.
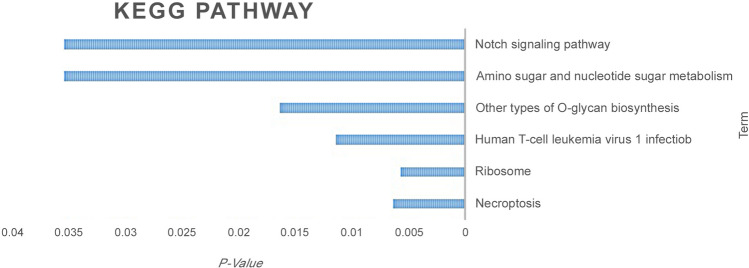
After data analysis of the included genes related to CTCs and exosomes in the literature, it was proven that the overexpression of various integrins can promote cancer cells to be more motile and more invasive. These adhesion molecules can be considered as a major potential breakthrough in oncologic treatment16,17, even as a regulators of cancer stemness, metastasis, and drug resistance18. Talin because of structural and functional significance with integrin cytoplasmic tails resulting in extracellular integrin compartments rearrangements and integrin activation19. Specifically, Talin1, which functionally contributes to the extravasation and metastasis of colon cancer CTCs20, was selected for the current evaluation. Talin1 protein expression was significantly localized in the cytoplasm that mediates integrin interactions with the extracellular matrix (ECM). From another side, for cell spreading and migration, the link between ECM- bound integrins and intracellular F actin is vital21. Talin1 mediates both inside-out and outside-in signaling, which leads to subsequent cascades22,23 and also has a direct effect on the regulation of insulin signaling, actin dynamic, and granule trafficking24. Talin serves as tension-sensing anchoring as well as a unique mechanotransductive point to link the cells to the ECM. It has been proven using cryoelectron microscopy (cryo-EM) full-length structure examination that the function of Talin could be regulated as a two-way mode of auto-inhibition, and the fast switching between its inactive conformations and active could regulate Focal adhesion kinase (FAK) turnover25. In our evaluation, focal adhesion among cellular components, cadherin, and RNA binding between molecular function were spotlighted. Also, cytoskeletal anchoring at plasma membrane, platelet aggregation, cell–cell adhesion, and junction throughout the biological process were considered significantly highlighted. Additionally, the KEGG pathway (as shown in Fig. 2) was common, and it was also related to cancer, like MAPK and integrin.
Figure 2.
KEGG pathway list related to the 15 excluded genes based on P-Value.
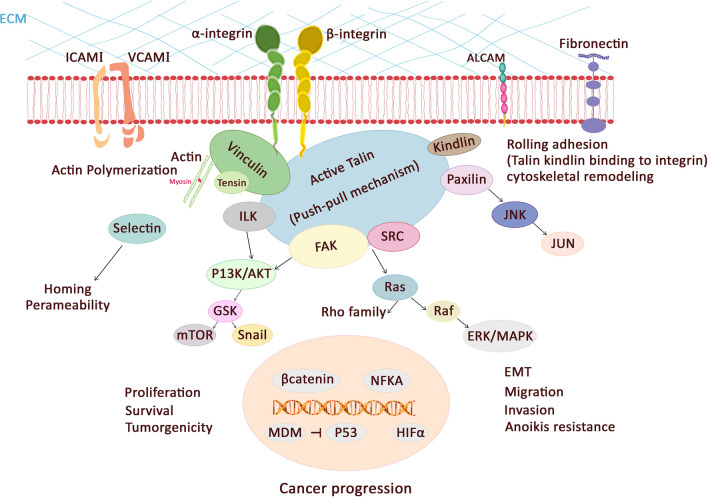
Overall, the major mediators of such cellular responses are integrins, which link to the actin cytoskeleton via Talin as a cytoskeletal protein26. Talin1 plays a key role in malignancies through implication in cell adhesion, migrations, proliferation, invasion, apoptosis, cytoskeleton remodeling, anchorage-independent growth, and tumorigenicity. In Fig. 3, the interaction of Talin and other molecules in malignancies are shown.
Figure 3.
Talin1 plays significant roles in cancer related pathway.
Patients’ characteristics of fresh tissue samples
The current study included seventy-seven fresh tissue samples from CRC patients, 52 (67.4%) of which came from males and 25 (32.6%) from females (male/female ratio = 2.12). The median age of patients was 60 years (SD = 14.79, range 20–87); 37 (48%) patients were aged ≤ 60 years and 40 (52%) were aged > 60. Eight (10.4%) cases were diagnosed as stage I, 25 (32.6%) stage II, 27 (35%) and 17 (22%) stage III and IV respectively. Thirty-two (41.6%) cases were low-grade adenocarcinomas, and 14 (18.2%) and 31 (40.2%) were moderated and poorly differentiated adenocarcinomas, respectively.
Associations between Talin1 mRNA expression and clinicopathological parameters in fresh tissue samples
Talin1 is ubiquitously expressed. Websites like Uniprot27 and Genecards28 showed it as localized within the cytosol, extracellular, and in the cytoskeleton. The expression profiles of Talin1 were decreased in patients with CRC compared with the healthy group (p < 0.043) according to Real-Time PCR (Fig. 4).
Figure 4.
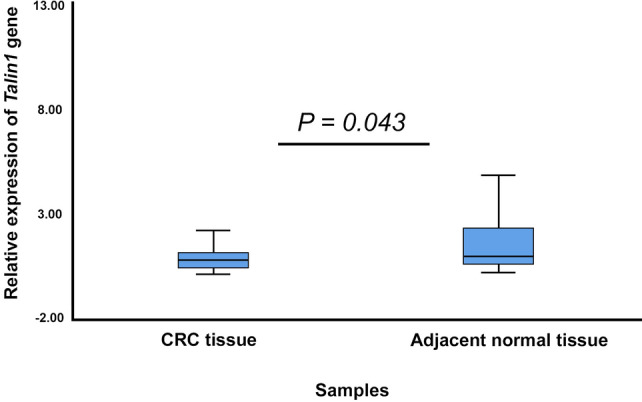
The expression levels of Talin1 using Real-Time PCR in fresh tissue samples of CRC.
Pearson’s χ2 test revealed that a statistically significant association among expression levels of Talin1 mRNA and TNM stage (p = 0.034) as well as gender (p = 0.029), indicating an association between the low level expression of Talin1 and an increased TNM stage (Table 1).
Table 1.
The association between mRNA Talin1 expression and clinicopathological parameters of fresh tissue colorectal cancer (CRC) samples.
| Patients and tumor characteristics | Total samples N (%) | mRNA Talin1 expression | P value | |
|---|---|---|---|---|
| Low N (%) | High N (%) | |||
| Median age, years (Range) | ||||
| ≤ Median age | 37 (48) | 24 (31.2) | 13 (16.9) | |
| > Median age | 40 (52) | 28 (36.3) | 12 (15.6) | 0.379 |
| Gender | ||||
| Male | 52 (67.4) | 39 (50.6) | 13 (16.9) | 0.027 |
| Female | 25 (32.6) | 24 (31.2) | 1 (1.3) | |
| (Male/Female) | 2.12 | |||
| Tumor differentiation | ||||
| Well | 32 (41.6) | 29 (37.7) | 3 (3.9) | 0.055 |
| Moderate | 14 (18.2) | 10 (13) | 4 (5.2) | |
| Poor | 31 (40.2) | 21 (27.2) | 10 (13) | |
| TNM stages | ||||
| I | 8 (10.4) | 7 (9) | 1 (1.3) | 0.034 |
| II | 25 (32.6) | 23 (29.9) | 2 (2.6) | |
| III | 27 (35) | 19 (24.7) | 8 (10.4) | |
| IV | 17 (22) | 15 (19.5) | 2 (2.6) | |
| Vascular invasion (VI) | ||||
| Present | 21 (27.2) | 16 (20.7) | 5 (6.5) | 0.396 |
| Absent | 56 (72.8) | 47 (61.1) | 9 (11.7) | |
| Lymph node invasion (LNI) | ||||
| Involved | 42 (54.5) | 32 (41.6) | 10 (13) | 0.162 |
| None | 35 (45.5) | 31 (40.2) | 4 (5.2) | |
| Neural invasion (NI) | ||||
| Involved | 25 (32.6) | 22 (28.6) | 3 (3.9) | 0.347 |
| None | 52 (67.4) | 41 (53.2) | 11 (14.3) | |
| Distant metastasis | ||||
| Present | 42 (54.5) | 32 (41.6) | 10 (13) | 0.162 |
| Absent | 35 (45.5) | 31 (40.2) | 4 (5.2) | |
| Tumor recurrence | ||||
| Yes | 7 (9) | 7 (9) | 0 (0.0) | 0.221 |
| No | 70 (91) | 56 (72.7) | 14 (18.3) | |
P value; Pearson’s χ2 test.
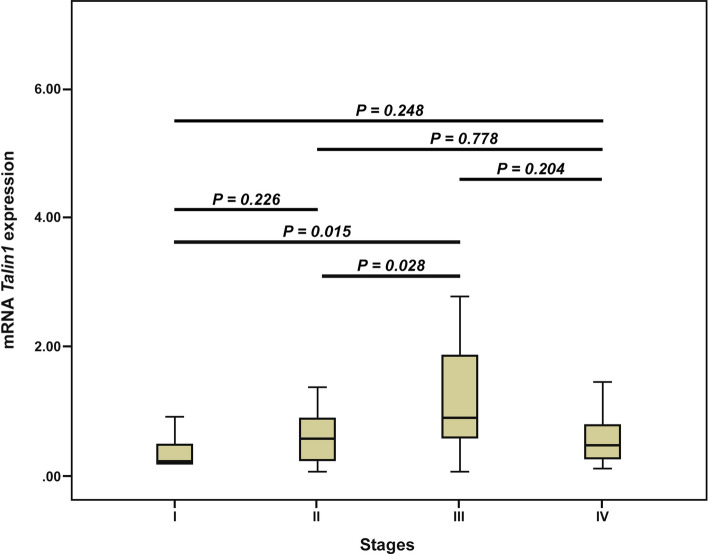
The nonparametric Kruskal–Wallis and Mann–Whitney U tests were utilized to measure the differences between the median expressions of Talin1 among the groups. The results of the Mann–Whitney U test also showed a statistically significant difference in the median level of Talin1 mRNA expression between stages I and III (p = 0.015) and stages II and III (p = 0.028). Further, we did not observe any statistically significant differences in the median level of Talin1 mRNA expression between other stages (Fig. 5). No association was found between Talin1 mRNA expressions or with other clinicopathological characteristics (Table 1).
Figure 5.
Box plot analysis of mRNA Talin1 expression levels in stage I to IV. Based on the standard definitions, each box-plot shows the median (bold line) and interquartile lines (box). The result of Mann–Whitney U test showed that there is an association for the median of Talin1 mRNA expression between stages I and III (p = 0.015) as well as stages II and III (p = 0.028). There were no statistically significant differences in the median level of Talin1 mRNA expression between other stages.
Prognostic value of Talin1 mRNA expression for clinical outcome in CRC patients
From the 77 CRC samples, 70 (91%) patients had no history of recurrence, while 7 (9%) patients were positive. In 42 (54.5%) metastasis occurred, while 35 (45.5%) patients were negative for it. Thirty (39.0%) patients had no history of metastasis, recurrence, or cancer-related death, while 47 (61.0%) patients for these parameters were positive. During the follow-up period, cancer-related death in 26 (33.8%) patients was reported. The mean duration of follow-up time was 20 months (SD = 11.3(, median was 25 months (8, 28), and the range was from 1 to 33 months. The mean DSS time for patients with high and low expression of Talin1 mRNA were 23.8 (SD = 1.6) and 27.2 (SD = 3.4) months, respectively.
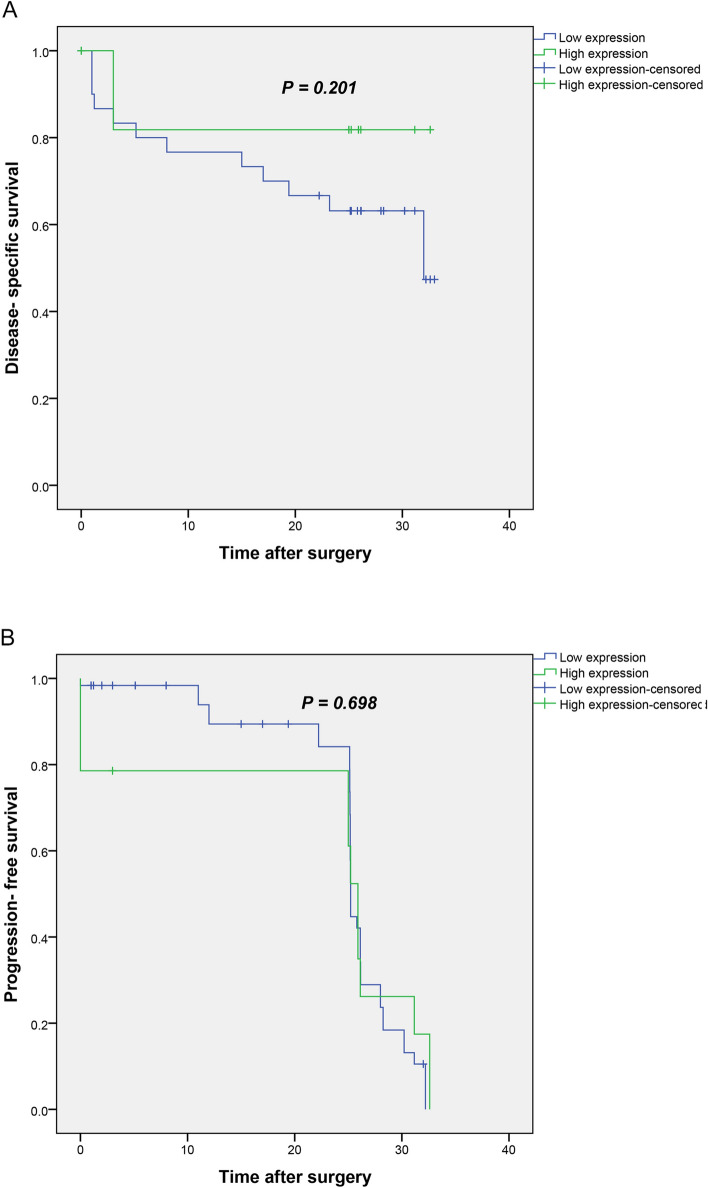
The results of the Kaplan–Meier curve demonstrated that patients with low Talin1 mRNA expression had shorter DSS compared to patients with high expression of Talin1 but without a statistically significant association (Log Rank test, p = 0.201) (Fig. 6A).
Figure 6.
(A) Kaplan–Meier curves for disease-specific survival (DSS) and progression-free survival (PFS) based on mRNA Talin1 expression levels in CRC patients. In CRC patients, lower level of Talin1 mRNA expression was associated with shorter DSS compared to the tumors with high expression of Talin1 mRNA expression (P = 0.201). (B) Kaplan–Meier curve showed that low level of Talin1 expression is not significantly related to PFS (P = 0.698).
Furthermore, the mean PFS time for patients with low and high expression rates of Talin1 mRNA were 24.9 (SD = 0.89) and 21.7 (SD = 3.2) months, respectively. The Kaplan–Meier curve showed that low level Talin1 expression is not significantly related to PFS (p = 0.698) (Fig. 6B).
Characteristics of patients’ FFPE tissue samples
Because the application of tissue microarray (TMA) technology has covered the evaluation of clinical values of huge number of candidate markers, 760 cases were included in this research, and for final scoring, 648 CRCs tissues were left. The FFPE population samples consisted of 303 (46.8%) male and 262 (40.4%) female patients, with a male/female ratio of 1.15. The median age of patients was calculated as 60 years (SD = 14.79, range 23–92). To clarify, 298 (46.0%) were ≤ 60 and 267 (41.2%) were > 60. Tumor size ranged from 1 to 20 cm, and tumors were classified into two groups. Three hundred and eighty-two (59.0%) of cases had tumors that were smaller in size than the mean (size ≤ 4.5) and 181 (27.9%) had tumors larger than the mean (size > 4.5). In this study, 90 (13.9%) patients had stage I, 254 (39.2%) stage II, 198 (30.6%) stage III, and 22 (3.4%) stage IV tumors. Moreover, 212 (32.7%) cases were grade I, 316 (48.8%) were grade II, and 34 (5.2%) were grade III; VI was found in 87 (13.4%) cases, and LNI and NI were observed in 212 (32.7%) cases. Tumor recurrence was seen in 96 (14.8%) patients, and 90 (13.9%) patients showed distant metastasis during the follow-up period.
Expression of Talin1 protein in CRC and adjacent normal tissues samples
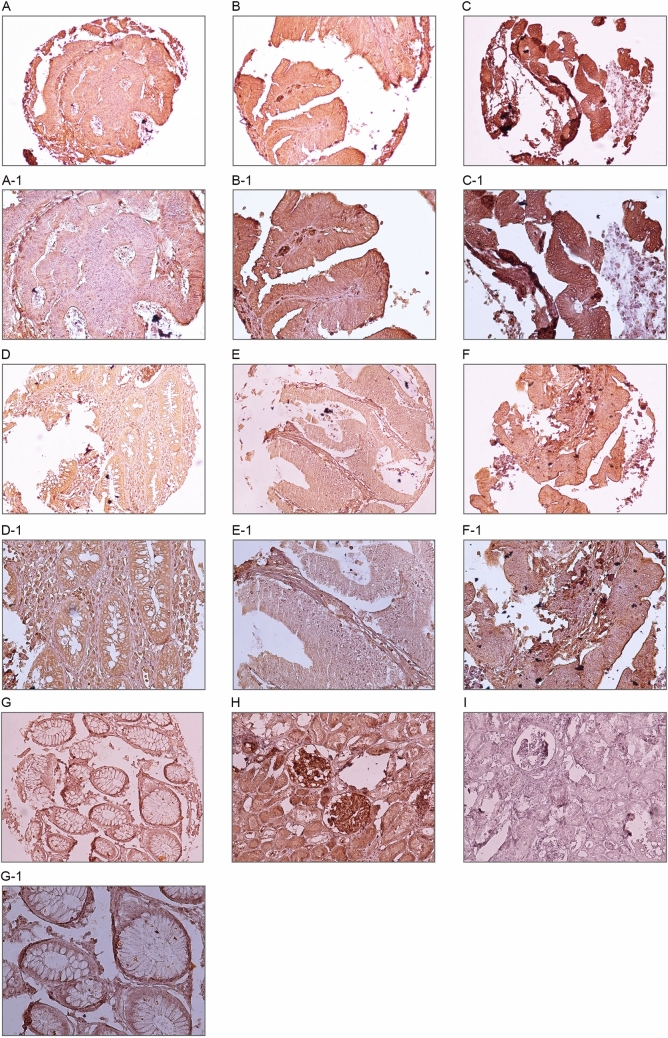
The expression level of Talin1 protein was evaluated using IHC on TMA sections by three scoring methods. H-score consist of staining intensity and percentage of positive tumor cells. In the CRC samples, Talin1 was expressed at varying intensities in the cell cytoplasm and ECM. Cytoplasmic and ECM expressions of Talin1 were detected in 643 (99.2%) and 628 (96.9%) CRC cases, respectively. The mean expression of Talin1 was 149 in cytoplasm and 138 in ECM. Talin1 expression based on the H-score demonstrated that low, moderate, and high levels of cytoplasmic expression of Talin1 were observed in 205 (31.6%), 349 (53.9%), and 94 (14.5%) CRC samples, respectively. Moreover, low, moderate, and high ECM expression of Talin1 was found in 235 (36.3%), 379 (58.5%), and 34 (5.2%) CRC patients, respectively (Table 2). The expression of Talin 1 was observed in the cytoplasm and ECM in all cases of adjacent normal tissue (Table 2). The mean expression of Talin1 was 151 in cytoplasm and 161 in ECM; therefore, Talin1 expression was greater in the adjacent normal tissue samples than in CRC samples, particularly in ECM expression. Moreover, human kidney tissue, showed strong staining as a positive control (Fig. 7).
Table 2.
Cytoplasmic and extra cellular matrix (ECM) Talin1 expression (Intensity of staining, percentage of positive tumor cells, and H-score in FFPE colorectal cancer (CRC) and adjacent normal tissues.
| Scoring system | Cytoplasmic expression N (%) | ECM expression N (%) | ||
|---|---|---|---|---|
| CRC samples | Adjacent normal tissues | CRC samples | Adjacent normal tissues | |
| Intensity of staining | ||||
| Negative (0) | 5 (0.8) | 24 (35.8) | 20 (3.1) | 10 (14.9) |
| Weak (+ 1) | 187 (28.9) | 33 (49.3) | 189 (29.2) | 50 (74.6) |
| Moderate (+ 2) | 361 (55.7) | 3 (4.5) | 404 (62.3) | 0 (0.0) |
| Strong (+ 3) | 95 (14.7) | 0 (0.0) | 35 (5.4) | 0 (0.0) |
| Not identified | 0 (0.0) | 7 (10.4) | 0 (0.0) | 7 (10.4) |
| Percentage of positive tumor cells | ||||
| < 25% | 18 (2.8) | 0 (0.0) | 30 (4.6) | 0 (0.0) |
| 25–50% | 25 (3.9) | 0 (0.0) | 51 (7.9) | 1 (1.5) |
| 51- 75% | 156 (24.1) | 6 (9.0) | 162 (25.0) | 11 (16.4) |
| > 75% | 449 (69.3) | 54 (80.6) | 405 (62.5) | 48 (71.6) |
| Not identified | 0 (0.0) | 7 (10.4) | 0 (0.0) | 7 (10.4) |
| H-score (3 groups) | ||||
| 0–100 | 205 (31.6) | 25 (37.3) | 235 (36.3) | 11 (16.4) |
| 101–200 | 349 (53.9) | 32 (47.8) | 379 (58.5) | 49 (73.1) |
| 201–300 | 94 (14.5) | 3 (4.5) | 34 (5.2) | 0 (0.0) |
| Not identified | 0 (0.0) | 7 (10.4) | 0 (0.0) | 7 (10.4) |
| Total | 648 (100) | 67 (100) | 648 (100) | 67 (100) |
H-score indicates Histological score.
Figure 7.
Immunohistochemical (IHC) analysis of Talin1 expression in different colorectal cancer (CRC) samples. CRC samples expressed Talin1 at various levels. Cytoplasmic expression of Talin1 at various levels: weak (A), moderate (B), and strong (C) in magnification of × 200; Weak (A-1), moderate (B-1), and strong (C-1) in magnification × 400. ECM expression of Talin1 in CRC at various levels is as follows: weak (D), moderate (E), and strong (F) in magnification × 200. Weak (D-1), moderate (E-1), and strong (F-1) in magnification × 400. IHC staining of kidney tissue was presented as positive (H) and negative (I) controls, adjacent normal tissue sample (G, G-1) in magnification × 200; and × 400.
Associations between Expression of Talin1 protein and clinicopathological characteristics in CRC patients
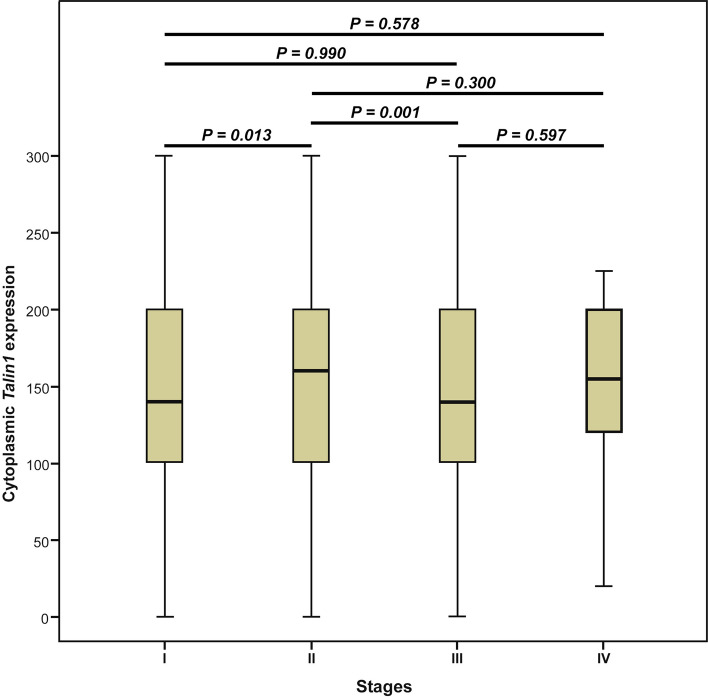
The results of Pearson’s χ2 test showed that there is an association between expression levels of Talin1 and TNM stage (p = 0.028) as well as gender (p = 0.009) (Table 3). The non-parametric Kruskal–Wallis test indicated a difference between the median expression level of Talin1 protein and various TNM stages (I–IV) (p = 0.004). The results of the Mann–Whitney U test revealed a statistically significant difference in the median level of Talin1 protein expression between stages I and II (p = 0.013) and stages II and III (p = 0.001). We did not find any statistically significant differences in the median level of Talin1 protein expression between other stages of CRC cases (Fig. 8).
Table 3.
The association between cytoplasmic Talin1 expression and clinicopathological parameters of colorectal cancer (CRC) samples (Intensity of staining and H-score).
| Patients and tumor characteristics | Total samples N (%) | Intensity of staining N (%) | P value | H-score N (%) | P value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 (Negative) | 1 + (Weak) | 2 + (Moderate) | 3 + (Strong) | H-score 0–100 | H-score 101–200 | H-score 201–300 | ||||
| Median age, years (Range) | 60 (23–92) | |||||||||
| ≤ Median age | 298 (46.0) | 2 (50.0) | 87 (50.9) | 161 (51.8) | 48 (60.8) | 0.492 | 95 (51.6) | 156 (51.5) | 47(60.3) | 0.359 |
| > Median age | 267 (41.2) | 2 (50.0) | 84 (49.1) | 150 (48.2) | 31 (39.2) | 89 (48.4) | 147 (48.5) | 31 (39.7) | ||
| Not identified | 83 (12.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Gender | ||||||||||
| Male | 303 (46.8) | 0 (0.0) | 93 (54.4) | 157 (50.5) | 53 (67.1) | 0.009 | 96 (52.2) | 154 (50.8) | 53 (67.9) | 0.023 |
| Female | 262 (40.4) | 4 (100.0) | 78 (45.6) | 154 (49.5) | 26 (32.9) | 88 (47.8) | 149 (49.2) | 25 (32.1) | ||
| Not identified (Male/Female) | 83 (12.8) 1.15 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Mean tumor size (cm) | 4.5 (1–20) | |||||||||
| ≤ Mean | 382 (59.0) | 2 (50.0) | 115 (67.6) | 212 (68.2) | 53 (67.9) | 0.896 | 123 (67.2) | 207 (68.3) | 52 (67.5) | 0.967 |
| > Mean | 181 (27.9) | 2 (50.0) | 55 (32.4) | 99 (31.8) | 25 (32.1) | 60 (32.8) | 96 (31.7) | 25 (32.5) | ||
| Not identified | 85 (13.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Tumor differentiation | ||||||||||
| Well | 212 (32.7) | 1 (25.0) | 66 (38.8) | 113 (36.5) | 32 (41.0) | 0.832 | 68 (37.2) | 112 (37.1) | 32 (41.6) | |
| Moderate | 316 (48.8) | 3 (75.0) | 91 (53.5) | 179 (57.7) | 43 (55.1) | 102 (55.7) | 172 (57.0) | 42 (54.5) | 0.852 | |
| Poor | 34 (5.2) | 0 (0.0) | 13 (7.6) | 18 (5.8) | 3 (3.8) | 13 (7.1) | 18 (6.0) | 3 (3.9) | ||
| Not identified | 86 (13.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| TNM stages | ||||||||||
| I | 90 (13.9) | 1 (25.0) | 32 (18.7) | 46 (14.8) | 11 (14.1) | 0.254 | 35 (19.0) | 44 (14.5) | 11 (14.3) | |
| II | 254 (39.2) | 1 (25.0) | 66 (38.6) | 143 (46.0) | 44 (56.4) | 68 (37.0) | 142 (46.9) | 44 (57.1) | 0.028 | |
| III | 198 (30.6) | 2 (50.0) | 68 (39.8) | 106 (34.1) | 22 (28.2) | 76 (41.3) | 101 (33.3) | 21 (27.3) | ||
| IV | 22 (3.4) | 0 (0.0) | 5 (2.9) | 22 (28.2) | 1 (1.3) | 5 (2.7) | 16 (5.3) | 1 (1.3) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Vascular invasion (VI) | ||||||||||
| Present | 87 (13.4) | 1 (25.0) | 24 (14.0) | 49 (15.8) | 13 (16.7) | 0.884 | 28 (15.2) | 47 (15.5) | 12 (15.6) | 0.995 |
| Absent | 477 (73.6) | 3 (75.0) | 147 (86.0) | 262 (84.2) | 65 (83.3) | 156 (84.8) | 256 (84.5) | 65 (84.4) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Lymph node invasion (LNI) | ||||||||||
| Involved | 212 (32.7) | 2 (50.0) | 71 (41.5) | 114 (36.7) | 25 (32.1) | 0.471 | 79 (42.9) | 109 (36.0) | 24 (31.2) | 0.14 |
| None | 352 (54.3) | 2 (50.0) | 100 (58.5) | 197 (63.3) | 53 (67.9) | 105 (57.1) | 194 (64.0) | 53 (68.8) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Neural invasion (NI) | ||||||||||
| Involved | 212 (32.7) | 1 (25.0) | 33 (19.3) | 71 (22.8) | 10 (12.8) | 0.254 | 36 (19.6) | 70 (23.1) | 9 (11.7) | 0.08 |
| None | 352 (54.3) | 3 (75.0) | 138 (80.7) | 240 (77.2) | 68 (87.2) | 148 (80.4) | 233 (76.9) | 68 (88.3) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Distant metastasis | ||||||||||
| Present | 90 (13.9) | 2 (40.0) | 30 (28.0) | 48 (26.2) | 10 (18.5) | 0.509 | 35 (29.2) | 46 (26.1) | 9 (17.0) | 0.238 |
| Absent | 259 (40.0) | 3 (60.0) | 77 (72.0) | 135 (73.8) | 44 (81.5) | 85 (70.8) | 130 (73.9) | 44 (83.0) | ||
| Not identified | 299 (46.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Tumor recurrence | ||||||||||
| Yes | 96 (14.8) | 2 (40.0) | 28 (26.2) | 55 (30.1) | 11 (20.0) | 0.45 | 33 (27.5) | 53 (30.1) | 10 (18.5) | 0.248 |
| No | 254 (39.2) | 3 (60.0) | 79 (73.8) | 128 (69.9) | 44 (80.0) | 87 (72.5) | 123 (69.9) | 44 (81.5) | ||
| Not identified | 298 (46.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
H-score indicates Histological score.
Values in bold are statistically significant.
P value; Pearson’s χ2 test.
Figure 8.
Box plot analysis of Talin1 expression levels in stage I to IV. Based on the standard definitions, each box-plot shows the median (bold line) and interquartile lines (box). The result of Mann–Whitney U test showed that there is a statistically significant association for the median of cytoplasmic Talin1 expression between stages I and II (P = 0.013) as well as stages II and III (P = 0.001). There were no statistically significant differences in the median level of Talin1 protein expression between other stages.
Further, the results of Spearman’s correlation exhibited a significant adverse correlation between Talin1 protein expression and advanced TNM stages (p = 0.038). No association was found between cytoplasmic Talin1 protein expression and the other clinicopathological characteristics (Table 3).
The results of Pearson’s χ2 and Spearman’s correlation tests showed that there is no association or correlation between ECM Talin1 expression and the clinicopathological parameters of CRCs patients (Table 4).
Table 4.
The association between extra cellular matrix (ECM) Talin1 expression and clinicopathological parameters of colorectal cancer (CRC) samples (Intensity of staining and H-score).
| Patients and tumor characteristics | Total samples N (%) | Intensity of staining N (%) | P value | H-score N (%) | P value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 (Negative) | 1 + (Weak) | 2 + (Moderate) | 3 + (Strong) | H-score 0–100 | H-score 101–200 | H-score 201–300 | ||||
| Median age, years (Range) | 60 (23–92) | |||||||||
| ≤ Median age | 298 (46.0) | 14 (73.7) | 82 (49.1) | 187 (54.0) | 15 (45.5) | 0.160 | 109 (52.9) | 174 (53.2) | 15 (46.9) | 0.789 |
| > Median age | 267 (41.2) | 5 (26.3) | 85 (50.9) | 159 (46.0) | 18 (54.5) | 97 (47.1) | 153 (46.8) | 17 (53.1) | ||
| Not identified | 83 (12.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Gender | ||||||||||
| Male | 303 (46.8) | 6 (31.6) | 82 (49.1) | 195 (56.4) | 20 (60.6) | 0.79 | 99 (48.1) | 185 (56.6) | 19 (59.4) | 0.126 |
| Female | 262 (40.4) | 23 (68.4) | 85 (50.9) | 151 (43.6) | 13 (39.4) | 107 (51.9) | 142 (43.4) | 13 (40.6) | ||
| Not identified (Male/Female) | 83 (12.8) 1.15 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Mean tumor size (cm) | 4.5 (1–20) | |||||||||
| ≤ Mean | 382 (59.0) | 11 (57.9) | 118 (70.7) | 231 (67.2) | 22 (66.7) | 0.667 | 141 (68.4) | 220 (67.7) | 21 (65.6) | 0.947 |
| > Mean | 181 (27.9) | 8 (42.1) | 49 (29.3) | 113 (32.8) | 11 (33.3) | 65 (31.6) | 105 (32.3) | 11 (34.4) | ||
| Not identified | 85 (13.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Tumor differentiation | ||||||||||
| Well | 212 (32.7) | 6 (31.6) | 64 (38.3) | 128 (37.3) | 14 (42.4) | 0.952 | 81 (39.3) | 117 (36.1) | 14 (43.8) | |
| Moderate | 316 (48.8) | 12 (63.2) | 91 (54.5) | 195 (56.9) | 18 (54.5) | 112 (54.4) | 187 (57.7) | 17 (53.1) | 0.833 | |
| Poor | 34 (5.2) | 1 (5.3) | 12 (7.2) | 20 (5.8) | 1 (3.0) | 13 (6.3) | 20 (6.2) | 1 (3.1) | ||
| Not identified | 86 (13.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| TNM stages | ||||||||||
| I | 90 (13.9) | 0 (0.0) | 35 (21.0) | 49 (14.2) | 6 (18.2) | 0.199 | 38 (18.4) | 46 (14.1) | 6 (18.8) | |
| II | 254 (39.2) | 10 (52.6) | 69 (41.3) | 159 (46.1) | 16 (48.5) | 88 (42.7) | 151 (46.3) | 15 (46.9) | 0.252 | |
| III | 198 (30.6) | 7 (36.8) | 54 (32.3) | 127 (36.8) | 10 (30.3) | 67 (32.5) | 121 (37.1) | 10 (31.3) | ||
| IV | 22 (3.4) | 2 (10.5) | 9 (5.4) | 10 (2.9) | 1 (3.0) | 13 (6.3) | 8 (2.5) | 1 (3.1) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Vascular invasion (VI) | ||||||||||
| Present | 87 (13.4) | 3 (15.8) | 25 (15.0) | 56 (16.2) | 3 (9.1) | 0.749 | 32 (15.5) | 52 (16.0) | 3 (9.4) | 0.616 |
| Absent | 477 (73.6) | 16 (84.2) | 142 (85.0) | 289 (83.8) | 30 (90.9) | 174 (84.5) | 274 (84.0) | 29 (90.6) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Lymph node invasion (LNI) | ||||||||||
| Involved | 212 (32.7) | 11 (57.9) | 60 (35.9) | 130 (37.7) | 11 (33.3) | 0.285 | 79 (38.3) | 122 (37.4) | 11 (34.4) | 0.907 |
| None | 352 (54.3) | 8 (42.1) | 107 (64.1) | 215 (62.3) | 22 (66.7) | 127 (61.7) | 204 (62.6) | 21 (65.6) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Neural invasion (NI) | ||||||||||
| Involved | 212 (32.7) | 5 (26.3) | 32 (19.2) | 76 (22.0) | 2 (6.1) | 0.15 | 42 (20.4) | 71 (21.8) | 2 (6.3) | 0.115 |
| None | 352 (54.3) | 14 (73.7) | 135 (80.8) | 269 (78.0) | 31 (93.9) | 164 (79.6) | 255 (78.2) | 30 (93.8) | ||
| Not identified | 84 (13.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Distant metastasis | ||||||||||
| Present | 90 (13.9) | 5 (35.7) | 29 (26.9) | 54 (26.0) | 2 (10.5) | 0.376 | 37 (26.8) | 51 (26.6) | 2 (10.5) | 0.294 |
| Absent | 259 (40.0) | 9 (64.3) | 79 (73.1) | 154 (74.0) | 17 (89.5) | 101 (73.2) | 141 (73.4) | 17 (89.5) | ||
| Not identified | 299 (46.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Tumor recurrence | ||||||||||
| Yes | 96 (14.8) | 5 (35.7) | 31 (28.7) | 53 (25.5) | 7 (35.0) | 0.672 | 39 (28.3) | 50 (26.0) | 7 (35.0) | 0.667 |
| No | 254 (39.2) | 9 (64.3) | 77 (71.3) | 155 (74.5) | 13 (65.0) | 99 (71.7) | 142 (74.0) | 13 (65.0) | ||
| Not identified | 298 (46.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
H-score indicates Histological score.
Values in bold are statistically significant.
P value; Pearson’s χ2 test.
Prognostic value of Talin1 protein expression for clinical outcome in CRC
Out of 648 CRC samples included in the present study, survival analysis was performed on 350 patients for whom follow-up data was accessible. From these 350 patients, 239 (68.3%) patients had no history of metastasis, recurrence, or cancer-related death, while 111 (31.7%) patients for these parameters were positive. In 91 (26.0%) patients metastasis and in 96 (27.4%) ones recurrence occurred. In 97 (27.7%), patients cancer-related death was documented during the mean duration of 45.0 months (SD = 27.6) as follow-up time with a median of 41.0 months (27, 66); the range was from 1 to 117 months.
Survival outcomes based on expression of Talin1 in CRC
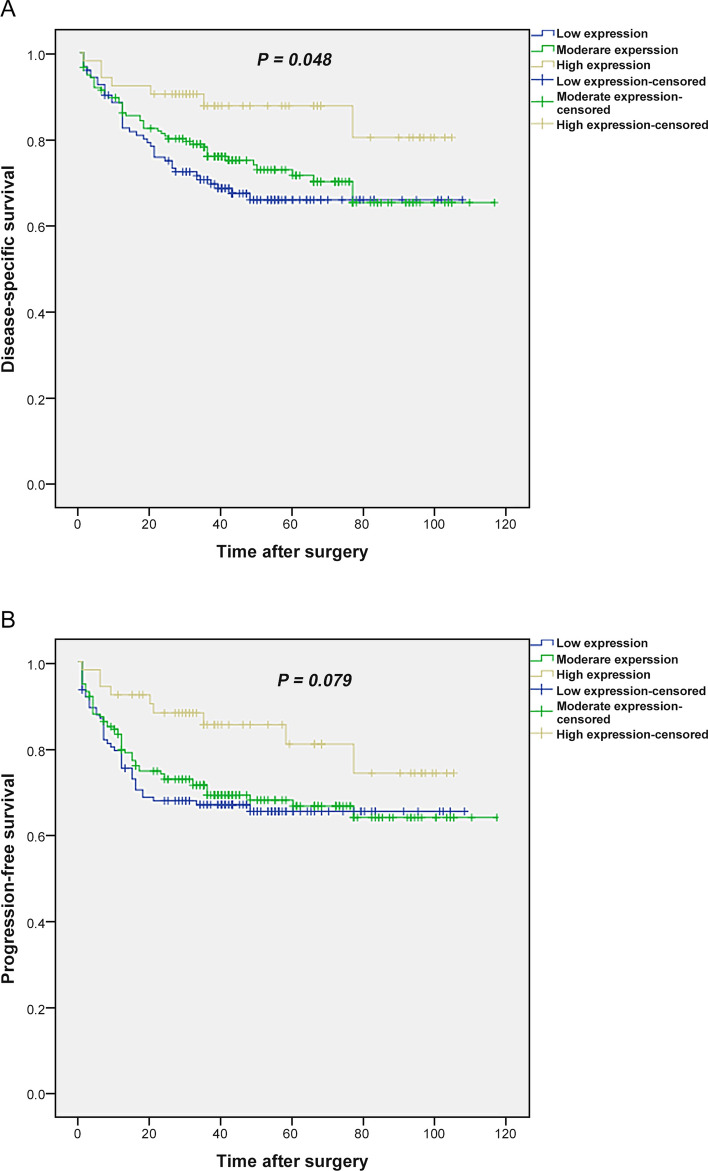
Kaplan–Meier curve results indicated significant differences between the patients with three groups of cytoplasmic expression rates of Talin1 and DSS (Log Rank test, p = 0.048) (Fig. 9A). The mean DSS times for patients with high, moderate, and low cytoplasmic expression of Talin1 were 91 (SD = 4.6), 86 (SD = 3.7), and 47 (SD = 4.0) months, respectively. Furthermore, the 5-year survival rates for DSS in patients whose samples expressed three groups of cytoplasmic Talin1 expression was 88%, 72%, and 65%, respectively (p = 0.039). Besides, there was no significant differences between PFS and patients with cytoplasmic Talin1 expression levels (p = 0.079) (Fig. 9B). The mean PFS times for patients with three groups of cytoplasmic expression of Talin1 were 87 (SD = 5.1), 81 (SD = 3.9), and 74 (SD = 4.3) months, respectively.
Figure 9.
Kaplan–Meier curves for disease-specific survival (DSS) and progression-free survival (PFS) based on cytoplasmic Talin1 protein expression level in CRC patients. (A) In CRC patients, lower level of Talin1 protein expression was associated with shorter DSS compared to the tumors with high expression of this protein (P = 0.048). (B) Kaplan–Meier survival analysis showed that low levels of Talin1 protein expression are not significantly related to PFS (P = 0.079).
Univariate and multivariate analyses were performed to investigate whether Talin1 expression was an independent prognostic factor of DSS or PFS and to assess the clinical significance of various parameters that might influence survival outcomes in CRC patients29. Among all of the clinicopathological parameters, only cytoplasmic Talin1 expression (p = 0.050), LNI (p = 0.010), NI (p = 0.008), distant metastasis (p < 0.001), tumor recurrence (p < 0.001), and age (p = 0.009) affecting DSS in univariate analysis. In addition, in multivariate analysis, only age (p = 0.015) was significantly related to DSS in these cases (Table 5).
Table 5.
Univariate and multivariate Cox regression analyses of potential prognostic factor for disease-specific survival (DSS) in patients with colorectal cancer.
| Covariate | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| H-score | 0.050 | 0.174 | ||
| High versus Low | 0.814 (0.532–1.245) | 0.342 | 0.909 (0.582–1.418) | 0.673 |
| High versus Moderate | 0.378 (0.169–0.847) | 0.018 | 0.437 (0.183–1.042) | 0.062 |
| Age | 1.780 (1.155–2.743) | 0.009 | 1.717 (1.113–2.649) | 0.015 |
| Lymph node invasion (LNI) | 0.571 (0.372–0.877) | 0.010 | – | – |
| Neural invasion (NI) | 0.523 (0.324–0.845) | 0.008 | – | – |
| Distant metastasis | 0.104 (0.066–0.162) | < 0.001 | – | – |
| Tumor recurrence | 0.131 (0.084–0.203) | < 0.001 | – | – |
H-score indicates Histological score.
Values in bold are statistically significant.
The variables with P value less than 0.05 were included in multivariable analyses.
HR hazard ratio, CI confidence interval.
Kaplan–Meier analysis showed that there were no differences between DSS or PFS and the patients with three groups of ECM expression of Talin1 (Log Rank test; p = 0.815, p = 0.595), respectively. Moreover, the results of univariate and multivariate analyses indicated that the listed clinicopathologic variables were not affecting the DSS or PFS of CRC patients.
Discussion
Because cross-talking between cancer cells and exosomes plays a vital role in the CRC dynamic network, especially in metastasis30, the identification and characterization of both molecular markers as powerful tools in non-invasive cancer monitoring are significant steps to improving our understanding of molecular and cellular mechanisms of cancer metastasis31. Furthermore, the identification of new prognostic markers for the likelihood of metastasis that can stratify patients into various classification is clearly warranted. Therefore, considering the heterogeneity and complexity of CRC, this study was conducted to predict prognoses of CRC patient based on the evaluation of their gene expression network and to the clinical significance of their common molecular marker by bioinformatics analysis in multiple fashions. We take advantage of the limited available data related to CTC and exosome collection assays throughout online web tools for tracking CRC metastasis. “Wet-lab” experiments using patient specimens for identifying cancer biomarkers through bioinformatics analysis were considered vital. The best application of that can be develop a practically manageable gene based on applying p values and enrichment pathway analysis32.
Traditional biomarkers (CEA, CA19-9 and FOBT), as well as colon/sigmoidoscopy, play an unsatisfactory specificity role in CRC screening7. Since the limitation of these various methods are considerable33 shifting to CTCs and TDEs as a liquid biopsy approach34,35 attracted much attention and help to discover markers which are important in diagnostic, prognostic and cancer staging10,36,37, estimation of relapse risk, therapeutic targets identification38,39, and proper monitoring of patients8,40. Regardless of the metastatic site, CTC enumeration are enough as a proper cancer-monitoring index whenever CEA and other marker levels are not measurable41,42.
Although several mechanisms are involved in cancer progression, recent evidence points to EMT and CSCs as having significant roles in these processes43. EMT plays an important role in the invasion and metastasis of CRC, which is mediated by FAK. However, whether FAK participates in EMT, through the EGF/EGFR signaling pathway, remains unknown44. Talin1 can activate integrin's as core regulators45 and play an important role as a focal adhesion proteins as well as be mediators of adhesion to the ECM46. Therefore, it must be considered extensively to determine its role clearly. FAK-Talin binding is required for adhesion turnover and cell motility and in our view has a significant point in common with molecules between circulating tumor cells (CTCs) and tumor-derived exosomes (TDEs).
In malignant tumors, EMT is crucial for acquisition of a mesenchymal phenotype with invasive and metastatic properties leading to tumor progression. The important markers related to this process include Twist 47–49, E-cadherin, vimentin, N-cadherin, Snail, Slug, integrin's, cytokeratin's, fibronectin, b-catenin, ZEB1, ZEB2, and TGFb50,51. The correlation analysis was performed between two markers (Talin1 and Twist1) and the results showed that there is no significant correlation between cytoplasmic expression of them in these series of CRC patients (p-value = 0.104).
Additionally, Most of the fifteen genes that were obtained from our bioinformatics analysis, have been previously implicated in cancer development, but Talin1 was selected due to its most important biological roles in migrant cells. Assembly and disassembly of FAK including Talin are essential for cell migration; meanwhile, molecular mechanisms details remained poorly understood and must be elucidated52. Up and down regulation of FAK protein expression might have a profound effect on signal transduction; for example, FAK expression levels were increased in primary CRC compared with normal mucosa and decreased in liver metastases53. In addition, our findings point towards Talin1 targeting as a promise for the future, and we supposed that EMT and stemness have been extensively investigated in the field of CTC-related research54. With regard to metastasis, the identification of molecular markers that robustly define a subset of CTCs which are enriched in CSCs are vital55. Thus, an improved understanding of the EMT/CTC/CSC connections may uncover the novel therapeutic targets and underline tumorigenicity mechanisms56.
Several studies have previously investigated levels of Talin1 expression in CRC in various areas such as by western blot analysis, HPLC-Chip/MS analysis, and enzyme-linked immunosorbent assay (ELISA). Nevertheless, no study was found that addressed the expression of Talin1 either in mRNA or protein levels in CRC tissue. Hence, due to the existence of information gaps regarding the expression of Talin1 in both mRNA and protein levels, the present study was designed to examine that in a well-characterized series of 77 fresh and 648 FFEP CRC tissue samples. This is also the first report of localization of Talin1 expression in cytoplasm and ECM in a large number of tumor samples with evaluation of its impact on CRC prognosis. The current results showed that low level mRNA expression of Talin1 is associated with increases in TNM stages. Furthermore, the results of TMA revealed an adverse association between the cytoplasmic expression levels of Talin1 and increasing TNM stage and gender, while no association between the ECM expression of Talin1 and the clinicopathological parameters was found. It was determined that the localization of Talin expression depends completely upon its function related to integrins. Moreover, a statistically significant difference was observed in the median level of Talin1 mRNA and protein expression between different stages. To clarify, the comparison of low stages (I–II) and high stages (III) showed that Talin1 mRNA and protein cytoplasmic expression levels are associated with CRC aggressiveness. It is noteworthy that the continuous refinement of the TNM system, T (tumor), N (node), and M (metastasis) staging define the prognosis and patient outcome and play a pivotal role in decision-making for treatment57,58. Accumulating data underlines the importance of structuring the management of CRC patient's survival by stages at diagnosis and prognostic stratification59,60. In addition, LNI, NI, distant metastasis, tumor recurrence, and age were prognostic variables in univariate analysis that depicted the associations between these parameters and more aggressive tumor behavior. In cytoplasmic Talin1 expression, only age was a prognosis factor affecting the DSS in multivariate analysis. It was previously shown that age and comorbidities worsened the survival rate of CRC patients61. In this study, the results exhibited that low level cytoplasmic expression of Talin1 compared with ECM expression is related to the degree of malignancy and progression in CRC cases. Talin1 cytoplasmic expression can be considered more significant in comparison with ECM ones62,63.
Previous articles have shown that there are many challengeable reports regarding Talin1 expression, analysis, and localization in various tissues. The first study related to Talin expression showed that it increased64, but eight years later the reduced expression of Talin in liver metastases compared with matched primary human CRC was reported65. Barbazan et al. introduced one molecular pattern in CRC for CTC detection66,67. Their research was continued by Insua et al., who included Talin1 as a gene involved in CTC extravasation during metastatic dissemination in their gene expression panel, which was composed of GAPDH, VIL1, CLU, TIMP1, TLN1, LOXL3, and ZEB2. This panel was checked as a prognostic and predictive tool in metastatic CRC blood samples. Patients with higher gene panel expression compared with low ones had reduced PFS and OS rates (p = 0.003 and p = 0.001), respectively65. According to the current results, this prognostic value seems to be related to Talin1 expression accompanied by the other markers, which were important in the panel53. One recent study revealed that Talin1 expression was upregulated in CRC, and the proliferation, migration, and invasive ability of the CRC cell line was significantly reduced by its knockdown compared with the control. In the current study, however, it was downregulated in advanced tumor progression68. Analysis of Talin1 using ELISA showed no significant correlation between its preoperative levels in the serum of patients and age and gender; however, a significant correlation between Talin1 levels and tumor grade, TNM stage, and lymph node metastasis was found69.
The results of the Kaplan–Meier curve on protein determined that tumors with low-level expression of the Talin1 protein tend to have a worse prognosis for DSS. Moreover, CRC patients who expressed a lower level of cytoplasmic Talin1 had a worse 5-year survival rate for DSS compared with those with high expression. Thus, low expression level of Talin1 can be considered a worse prognostic factor of DSS in CRC patients. Talin1 levels could not be used as an independent prognostic factor of DSS. More prolonged follow-up time may increase the prognostic value of Talin1. In mRNA expression, the current finding showed that patients with low Talin1 mRNA expression had shorter DSS compared to patients with high expression of Talin1, but the difference was not statistically significant. This might be due to the limited sample size and shorter follow-up period in comparison with FFEP samples, because all fresh tissue samples were collected on 2017.
The current study was similar to other studies conducted on Talin1 in some malignancies in its importance in cancer development, invasion, migration, and metastasis, and even introduces it as a cancer stem cell (CSCs) marker, because in the current study, Talin1 was a common marker between CTCs and exosomes68,70–72. Therefore, it must be mentioned that Talin1 can be an extremely helpful factor for a worse prognosis of DSS in CRC patients. Moreover, the use of IHC can verify the Real-Time PCR results and establish Talin1 status in routine clinical practice.
Conclusion
The current study is the first to exhibit that low level Talin1 mRNA expression is associated with more aggressive tumor behavior in CRC patients. Furthermore, we found that decreased cytoplasmic expression of Talin1 rather than its ECM expression has a clinical significance in CRC cases and is associated with increased invasiveness and poor prognosis risk for DSS in univariate analysis. To sum up, investigation of the expression pattern of Talin1 in the cytoplasm of tumor biopsies as a predictor and prognostic indicator of cancer progression in CRC patients’ tumor biopsies is useful. Extending the follow-up time might increase the prognostic value of Talin1 as an independent prognostic factor, but that must be confirmed in further studies.
Methods
Data set collection
To find the common molecular marker between CTCs and exosomes, first, GSE31023 gene expression and CTC profiling by array were obtained from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo) based on the GPL13497 (Agilent-026652 Whole Human Genome Microarray 4 × 44 K v2) platforms, containing the CTC in CRC patients (n = 6) and healthy subjects (n = 3). The GEO2R online analysis tool to detect the DEGs was applied, using a p value < 0.05 and |logFC|≥ 2 as cut-off criteria.
Secondly, the exosome markers were sought in two known exosome-related websites, https://www.exocarta.org and https://microvesicles.org, which were sequentially searched. A search for CRC cells in Vesiclepedia revealed 37 research sets containing 22,375 proteins and 10,813 mRNA. After duplicates were removed, a total of 6150 proteins and 10,813 mRNA were determined. Another 1617 related CRC exosome proteins were extracted from Exocarta, and all of them are summarized in Supplementary Table 1.
Molecular markers are valuable tools in the development of cancer screening and monitoring strategies. GO and GSEA are the common useful methods employed in this study to suggest new markers among the CTCs and exosomes in CRC samples. GO is useful for annotating genes and identifying characteristic biological attributes for high-throughput data analysis73. The GO project describe the attributes of gene products in the molecular functions, biology processes, and cellular components74. Gene set enrichment analysis (GSEA) determines whether a prior defined set of genes shows statistical significance75,76. Thus, the initially extracted markers in this article were subjected to STRING (https://string-db.org/) and EnrichR (amp.pharm.mssm.edu/Enrichr/) to obtain a better comprehensive of the significantly related pathway, which is described in Supplementary Table 2.
Patients’ characteristics and tissue collection
In the current study, 77 CRC fresh tumor tissues and their adjacent normal tissue samples from 2018 to 2019 were collected from university-based hospitals (Firoozgar and Bahman) in Tehran, Iran. Patients who had undergone surgery without receiving any relevant chemotherapy or radiotherapy were included in this study. The specimens were embedded in RNA later solution, transferred to the laboratory, and frozen in liquid nitrogen. Patient information including gender, age, TNM stage, tumor differentiation, and distant metastasis was also recorded. To evaluate the protein expression of Talin1, 760 FFPE tissues of CRC taken from 2010 to 2019 were collected from university-based hospitals (Firoozgar, Bahman, Rasool, and Hasheminejad) in Tehran, Iran. Hematoxylin and eosin (H&E) stained slides and archived medical records were retrieved to obtain patients’ clinicopathological characteristics, including age, gender, tumor size (maximum tumor diameter), tumor differentiation, TNM stage, venous invasion (VI), lymph node invasion (LNI), neural invasion (NI), distant metastasis, and tumor recurrence. In addition, 60 adjacent normal tissue samples were used to compare expression levels of the Talin1 markers in a range of tissue samples. From the date of surgery to the date of death was measured as Disease-specific survival (DSS). The interval between the primary surgery and the last follow-up visit was considered as Progression-free survival (PFS) if the patient showed no evidence of disease, metastasis, recurrence, CRC or disease-related death.
RNA isolation, reverse transcription, and real-time PCR
Briefly, tissue specimens were lysed and homogenized in the presence of a highly denaturing guanidine-thiocyanate-containing buffer that inactivates RNases. Trizol reagent (Sigma, USA) added to provide appropriate binding conditions were grounded. Then, the total RNA was isolated based on the RNAeasy Mini Kit (Qiagen Cat No. /ID: 74,104) protocol. One μg of RNA was reverse transcripted with PrimeScript 1st Strand complementary DNA Synthesis kit (Takara). Real-time PCR assay was performed on the Qiagen Rotor Gen Q system using the SYBR green Premix Dimer Eraser kit (TaKaRa cat number: RR820Q). The cycling conditions were an initial 30 s denaturation at 95 °C and 40 cycles (5 s at 95 °C, 30 s at 60 °C, and 45 s at 72 °C). The GAPDH gene was set as internal control; Talin1 expression level was detected in forty pairs of CRC and adjacent normal tissue samples. The primer sequences were as follows: GAPDH: 5′-AACTTTGGCATTGTGGAAGG-3′ F and 5′-CACATTGGGGGTAGGAACAC-3′ R. Talin1: 5′-.TTGGAGATGCCAGCAAGCGACT-3′ F and 5′-CCAGTTCTGTGGCTGCCTGATT-3′. The expression levels of Talin1 mRNA were normalized against GAPDH levels based on the 2−ΔΔCt approach77.
Construction of tissue microarrays (TMAs)
Colorectal tissue TMAs were prepared as described previously78. Briefly, our pathologist (Z.S.) examined the H&E slides to select the representative tumor area in each block. Then, using a precision arraying instrument (Tissue Arrayer Minicore; ALPHELYS, Plaisir, France), selected spots were punched into recipient blocks, and TMA blocks were constructed in three copies. In each TMA block, adjacent normal tissue samples were also included to compare the expression pattern of Talin1 with tumor tissue specimens79,80. Due to technical problems and an insufficient amount of tissue for scoring, some cases were lost during the staining and excluded from the study. Tumor heterogeneity is a major concern during the TMA procedure, and thus, at least two or three cores were evaluated from each sample to achieve better results81,82.
Immunohistochemistry (IHC) staining
The protein expression of Talin1 was evaluated using our IHC laboratory protocol y29,83. Sequential TMA sections at 60 °C for 30 min were dewaxed, in xylenes rehydrated, and then under ethanol treatment graded. 3% H2O2 (endogenous peroxidase) were applied as blocker for 20 min at room temperature. Antigen was retrieved by autoclaving tissue sections for 10 min in sodium citrate buffer (pH 6.0). Then, all of the slides were blocked (blocker protein, Dako, Denmark) for 20 min. After serial dilution, 1:1000 for anti-Talin1 antibody was prepared as an optimal dilution for subsequent use. The tissue sections were incubated overnight at 4 °C with the following antibody dilutions: anti Talin1 antibody (ab71333; Abcam, UK). The next day, after three washes in Tris-buffered saline (TBS), sections were incubated with anti-rabbit/anti-mouse envision IgG–HRPO (EnVision, Dako) as the secondary antibody for 1 h. 3,3′-diaminobenzidine (DAB) (Dako) substrate as a chromogen for treating the TMA slides were used in 3 min at room temperature. Sections were counterstained with hematoxylin (Dako), in alcohol dehydrated, with xylenes cleared and mounted. TBS was replaced for negative controls, and only the secondary antibody was used. Meanwhile, human kidney tissue was used as a positive control for Talin1 staining.
Evaluation of immunostaining
Our pathologist (Z.S.) using a semi-quantitative scoring examined the immunostained tissue arrays system in a coded manner. Consensus was achieved by the second investigator (Z.M.) in difficult cases for confirming the scoring.
Scoring system
In the CRC samples, the Talin1 marker was expressed at various intensities in the cell cytoplasm and ECM, each of which was analyzed separately. The intensities of Talin1 staining were scored on a 4-point scale, ranging from negative to strong (zero = negative, one = weak, two = moderate, and three = strong). The percentages of positive tumor cells varied between zero and 100. Histochemical score (H-score) as an overall score was achieved by multiplying the intensity of staining by the percentage of positive cells; then, a final score of 0 to 300 was given to each core. In the current study, the mean H-scores were classified into three groups: 1. low expression (0–100), 2. moderate expression (101–200), and 3. high expression (201–300).
Statistical analysis
SPSS software version 22.0 (IBM Corp, USA) was utilized to analyze the data. Categorical data was reported by N (percentage) and quantitative data as mean (SD) and median (Q1, Q3). Pearson’s χ2 and Spearman’s correlation tests were used to analyze the significance of associations and correlations between Talin1 expression and clinicopathological parameters. Kruskal–Wallis and Mann–Whitney U tests were applied for pairwise comparisons between groups. DSS and PFS curves were drawn using the Kaplan–Meier method, and the log-rank test was used to compare the estimated curves between groups with 95% confidence intervals (CI). To clarify that which variables affected DSS or PFS, the Cox proportional hazards regression model was applied. In all parts, a p-value of < 0.05 was considered statistically significant. As noted, in the first step, all quantified data was replicated an average of three times.
Ethical approval
The Research Ethics Committee of Iran University of Medical Sciences issued IR.IUMS.REC 1395.9221513203 for this study. All procedures, including obtaining informed consent from each human participant before surgery, were in accordance with the above-mentioned ethical standards.
Supplementary information
Acknowledgements
This study is part of a PhD thesis (Number: 1395.9221513203). Therefore, we are so thankful from the Oncopathology Research Center of Iran University of Medical Sciences.
Author contributions
Z.M. and M.E. designed and supervised the work; S.V. wrote the manuscript, collected the fresh and paraffin embedded tissues, collected the patient data and survival outcomes, and performed all experiments. L.S. analyzed and interpreted the data, helped to prepare the figures and Tables as well as contributed to write the some section of the manuscript. Z.H.S. marked the most representative areas in different parts of the tumor for preparing the TMAs blocks, scored TMAs slides after immunohistochemically staining and helped to prepare the Fig. 6; E.G. and M.N. contributed to gather the paraffin embedded tissues and prepared the SPSS survival data, F.F. contributed to gather the fresh and paraffin embedded tissues. M.A. was the colorectal surgery specialists who helped to collect the samples and access to patient's data. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marzieh Ebrahimi, Email: mebrahimi@royaninstitute.org.
Zahra Madjd, Email: majdjabari.z@iums.ac.ir.
Supplementary information
is available for this paper at 10.1038/s41598-020-74810-6.
References:
- 1.Meltzer S, et al. Circulating exosomal miR-141-3p and miR-375 in metastatic progression of rectal cancer. Transl. Oncol. 2019;12:1038–1044. doi: 10.1016/j.tranon.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Bray FG, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Rafiemanesh H, et al. Colorectal cancer in Iran: epidemiology and morphology trends. EXCLI J. 2016;15:738–744. doi: 10.17179/excli2016-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.5Compton, C. C. et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Archives of pathology & laboratory medicine124, 979–994, doi:10.1043/0003-9985(2000)124<0979:PFICC>2.0.CO;2 (2000). [DOI] [PubMed]
- 6.Kulendran M, Stebbing JF, Marks CG, Rockall TA. Predictive and prognostic factors in colorectal cancer: a personalized approach. Cancers. 2011;3:1622–1638. doi: 10.3390/cancers3021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamantis A, Magiorkinis E, Koutselini H. Fine-needle aspiration (FNA) biopsy: historical aspects. Folia Histochem Cytobiol. 2009;47:191–197. doi: 10.2478/v10042-009-0027-x. [DOI] [PubMed] [Google Scholar]
- 8.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 9.Shou X, et al. Six-gene Assay as a new biomarker in the blood of patients with colorectal cancer: establishment and clinical validation. Mol. Oncol. 2019;13:781–791. doi: 10.1002/1878-0261.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vafaei S, et al. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review. Cancer Manag Res. 2019;11:8669–8698. doi: 10.2147/CMAR.S219699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J, et al. Associations between the cyclooxygenase-2 expression in circulating tumor cells and the clinicopathological features of patients with colorectal cancer. J. Cell. Biochem. 2019;120:4935–4941. doi: 10.1002/jcb.27768. [DOI] [PubMed] [Google Scholar]
- 12.Ptitsyn AA, Weil MM, Thamm DH. Systems biology approach to identification of biomarkers for metastatic progression in cancer. BMC Bioinform. 2008;9(Suppl 9):S8. doi: 10.1186/1471-2105-9-S9-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiyama H, Noda H, Konishi F, Rikiyama T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J. Gastroenterol. 2014;20:8928–8938. doi: 10.3748/wjg.v20.i27.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.14Norcic, G. Liquid Biopsy in Colorectal Cancer-Current Status and Potential Clinical Applications. Micromachines (Basel)9, doi:10.3390/mi9060300 (2018). [DOI] [PMC free article] [PubMed]
- 15.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokeland G, Schumacher U. The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol. Cancer. 2019;18:12. doi: 10.1186/s12943-018-0937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desiniotis A, Kyprianou N. Significance of talin in cancer progression and metastasis. Int. Rev. Cell Mol. Biol. 2011;289:117–147. doi: 10.1016/B978-0-12-386039-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbazan J, et al. Liver metastasis is facilitated by the adherence of circulating tumor cells to vascular fibronectin deposits. Can. Res. 2017;77:3431–3441. doi: 10.1158/0008-5472.CAN-16-1917. [DOI] [PubMed] [Google Scholar]
- 21.Bachir AI, Horwitz AR, Nelson WJ, Bianchini JM. Actin-based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Biol. Cold Spring Harbor. Perspect. 2017 doi: 10.1101/cshperspect.a023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tadokoro S, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 23.Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai EP, et al. In vivo role of focal adhesion kinase in regulating pancreatic beta-cell mass and function through insulin signaling, actin dynamics, and granule trafficking. Diabetes. 2012;61:1708–1718. doi: 10.2337/db11-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dedden D, et al. The architecture of Talin1 reveals an autoinhibition mechanism. Cell. 2019;179:120–131. doi: 10.1016/j.cell.2019.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das M, Ithychanda S, Qin J, Plow EF. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochem. Biophys. Acta. 1838;579–588:2014. doi: 10.1016/j.bbamem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(GDPR), E. s. n. G. D. P. R. UniProtKB - Q9Y490 (TLN1_HUMAN), https://www.uniprot.org/uniprot/Q9Y490 (2020).
- 28.28GenesCard, TLN1 Gene, https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLN1 (2020).
- 29.Saeednejad Zanjani L, et al. Cytoplasmic expression of B7–H3 and membranous EpCAM expression are associated with higher grade and survival outcomes in patients with clear cell renal cell carcinoma. Ann. Diagn. Pathol. 2020;46:151483. doi: 10.1016/j.anndiagpath.2020.151483. [DOI] [PubMed] [Google Scholar]
- 30.Fu Q, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018 doi: 10.1038/s41388-018-0391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vafaei S, et al. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review. Cancer Manag. Res. 2019;11:8669–8698. doi: 10.2147/CMAR.S219699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung Y, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. Offic. J. Am. Assoc. Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 33.Mousavi S, et al. Tumor-derived exosomes: Potential biomarkers and therapeutic target in the treatment of colorectal cancer. J. Cell. Physiol. 2019;234:12422–12432. doi: 10.1002/jcp.28080. [DOI] [PubMed] [Google Scholar]
- 34.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2017;14:235. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 35.Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, Sun H. Colorectal cancer heterogeneity and targeted therapy: clinical implications, challenges and solutions for treatment resistance. Semin. Cell Dev. Biol. 2017;64:107–115. doi: 10.1016/j.semcdb.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA, Rosell R. Real-time liquid biopsies become a reality in cancer treatment. Ann. Transl. Med. 2015;3:36–36. doi: 10.3978/j.issn.2305-5839.2015.01.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brock, G., Castellanos-Rizaldos, E., Hu, L., Coticchia, C. & Skog, J. Liquid biopsy for cancer screening, patient stratification and monitoring. Translational Cancer Research; Vol 4, No 3 (June 2015): Translational Cancer Research (Application of Genomic Technologies in Cancer Research) (2015).
- 38.Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J. Gastroenterol. 2017;23:5086. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez A, et al. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev. Anticancer Ther. 2018;18:19–38. doi: 10.1080/14737140.2018.1403320. [DOI] [PubMed] [Google Scholar]
- 40.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 41.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8:55632–55645. doi: 10.18632/oncotarget.17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vafaei S, Roudi R, Madjd Z, Aref AR, Ebrahimi M. Potential theranostics of circulating tumor cells and tumor-derived exosomes application in colorectal cancer. Cancer Cell Int. 2020;20:288. doi: 10.1186/s12935-020-01389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang K, et al. Correlation between FAK and EGF-induced EMT in colorectal cancer cells. J. Oncol. 2020;2020:5428920. doi: 10.1155/2020/5428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan RB, Goult BT. Adhesions assemble!-autoinhibition as a major regulatory mechanism of integrin-mediated adhesion. Frontiers Mol. Biosci. 2019;6:144. doi: 10.3389/fmolb.2019.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klapholz B, Brown NH. Talin - the master of integrin adhesions. J. Cell Sci. 2017;130:2435–2446. doi: 10.1242/jcs.190991. [DOI] [PubMed] [Google Scholar]
- 47.Oh BY, et al. Twist1-induced epithelial-mesenchymal transition according to microsatellite instability status in colon cancer cells. Oncotarget. 2016;7:57066–57076. doi: 10.18632/oncotarget.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen X, et al. The long noncoding RNA TUG1 is required for TGF-beta/TWIST1/EMT-mediated metastasis in colorectal cancer cells. Cell Death Dis. 2020;11:65. doi: 10.1038/s41419-020-2254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdel Raouf SM, Ibrahim TR, Abdelaziz LA, Farid MI, Mohamed SY. Prognostic Value of TWIST1 and EZH2 Expression in Colon Cancer. J. Gastrointest. Cancer. 2019 doi: 10.1007/s12029-019-00344-4. [DOI] [PubMed] [Google Scholar]
- 50.Busch EL, McGraw KA, Sandler RS. The potential for markers of epithelial-mesenchymal transition to improve colorectal cancer outcomes: a systematic review. Cancer Epidemiol. Prevent. Biomark. 2014;23:1164–1175. doi: 10.1158/1055-9965.EPI-14-0017. [DOI] [PubMed] [Google Scholar]
- 51.Ieda T, et al. Visualization of epithelial-mesenchymal transition in an inflammatory microenvironment-colorectal cancer network. Sci. Rep. 2019;9:16378. doi: 10.1038/s41598-019-52816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang HJ, Chen JZ, Zhang WL, Ding YQ. Focal adhesion plaque associated cytoskeletons are involved in the invasion and metastasis of human colorectal carcinoma. Cancer Invest. 2010;28:127–134. doi: 10.3109/07357900903147184. [DOI] [PubMed] [Google Scholar]
- 53.Ayaki M, et al. Reduced expression of focal adhesion kinase in liver metastases compared with matched primary human colorectal adenocarcinomas. Clin. Cancer Res. Offic. J. Am. Assoc. Cancer Res. 2001;7:3106–3112. [PubMed] [Google Scholar]
- 54.Papadaki MA, et al. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol. Cancer Ther. 2019;18:437–447. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 55.Gkountela S, Aceto N. Stem-like features of cancer cells on their way to metastasis. Biology direct. 2016;11:33. doi: 10.1186/s13062-016-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agnoletto C, et al. Heterogeneity in circulating tumor cells: the relevance of the stem-cell subset. Cancers (Basel) 2019 doi: 10.3390/cancers11040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puppa G, Sonzogni A, Colombari R, Pelosi G. TNM staging system of colorectal carcinoma: a critical appraisal of challenging issues. Arch. Pathol. Lab. Med. 2010;134:837–852. doi: 10.1043/1543-2165-134.6.837. [DOI] [PubMed] [Google Scholar]
- 58.Wang S, et al. Reconsidering the prognostic significance of tumour deposit count in the TNM staging system for colorectal cancer. Sci. Rep. 2020;10:89. doi: 10.1038/s41598-019-57041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joachim C, et al. Overall survival of colorectal cancer by stage at diagnosis: data from the martinique cancer registry. Medicine. 2019;98:e16941. doi: 10.1097/MD.0000000000016941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider NI, Langner C. Prognostic stratification of colorectal cancer patients: current perspectives. Cancer Manag. Res. 2014;6:291–300. doi: 10.2147/CMAR.S38827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamano T, et al. Influence of age and comorbidity on prognosis and application of adjuvant chemotherapy in elderly Japanese patients with colorectal cancer: a retrospective multicentre study. Eur. J. Cancer. 2017;81:90–101. doi: 10.1016/j.ejca.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 62.Li T, et al. Loss of vinculin and membrane-bound beta-catenin promotes metastasis and predicts poor prognosis in colorectal cancer. Mol. Cancer. 2014;13:263. doi: 10.1186/1476-4598-13-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atherton P, et al. Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 2015;6:10038. doi: 10.1038/ncomms10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porter RM, et al. Monoclonal antibodies to cytoskeletal proteins: an immunohistochemical investigation of human colon cancer. J. Pathol. 1993;170:435–440. doi: 10.1002/path.1711700406. [DOI] [PubMed] [Google Scholar]
- 65.Insua YV, et al. Predicting outcome and therapy response in mCRC patients using an indirect method for CTCs detection by a multigene expression panel: a multicentric prospective validation study. Int. J. Mol. Sci. 2017 doi: 10.3390/ijms18061265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barbazan J, et al. A logistic model for the detection of circulating tumour cells in human metastatic colorectal cancer. J. Cell Mol. Med. 2012;16:2342–2349. doi: 10.1111/j.1582-4934.2012.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barbazan J, et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PLoS ONE. 2012;7:e40476. doi: 10.1371/journal.pone.0040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji L, Jiang F, Cui X, Qin C. Talin1 knockdown prohibits the proliferation and migration of colorectal cancer cells via the EMT signaling pathway. Oncol. Lett. 2019;18:5408–5416. doi: 10.3892/ol.2019.10902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Bostanci O, et al. A novel screening test for colon cancer: Talin-1. Eur. Rev. Med. Pharmacol. Sci. 2014;18:2533–2537. [PubMed] [Google Scholar]
- 70.Kang W, et al. Talin1 targeting potentiates anti-angiogenic therapy by attenuating invasion and stem-like features of glioblastoma multiforme. Oncotarget. 2015;6:27239–27251. doi: 10.18632/oncotarget.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu N, et al. Upregulation of Talin-1 expression associates with advanced pathological features and predicts lymph node metastases and biochemical recurrence of prostate cancer. Medicine. 2016;95:e4326. doi: 10.1097/MD.0000000000004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang KP, Zhang JL, Ren YH, Qian YB. Talin-1 correlates with reduced invasion and migration in human hepatocellular carcinoma cells. APJCP. 2014;15:2655–2661. doi: 10.7314/apjcp.2014.15.6.2655. [DOI] [PubMed] [Google Scholar]
- 73.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris MA, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suarez-Farinas M, Lowes MA, Zaba LC, Krueger JG. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA) PLoS ONE. 2010;5:e10247. doi: 10.1371/journal.pone.0010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 78.Kalantari E, et al. Increased expression of ALDH1A1 in prostate cancer is correlated with tumor aggressiveness: a tissue microarray study of iranian patients. Appl. Immunohistochem. Mol. Morphol. 2017;25:592–598. doi: 10.1097/PAI.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 79.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J. Pathol. 2001;195:72–79. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 80.Zlobec I, Suter G, Perren A, Lugli A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. JoVE. 2014 doi: 10.3791/51893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fernebro E, Dictor M, Bendahl PO, Ferno M, Nilbert M. Evaluation of the tissue microarray technique for immunohistochemical analysis in rectal cancer. Arch. Pathol. Lab. Med. 2002;126:702–705. doi: 10.1043/0003-9985(2002)126<0702:EOTTMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 82.Hoos A, et al. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am. J. Pathol. 2001;158:1245–1251. doi: 10.1016/S0002-9440(10)64075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rasti A, et al. Co-expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell carcinomas. Sci. Rep. 2018;8:11739. doi: 10.1038/s41598-018-30168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.