Abstract
Most recently, the new coronavirus (SARS-CoV-2) has been recognized as a pandemic by the World Health Organization (WHO) while this virus shares substantial similarity with SARS-CoV. So far, no definitive vaccine or drug has been developed to cure Covid-19 disease, since many important aspects about Covid-19 such as pathogenesis and proliferation pathways are still unclear. It was proven that human ACE2 is the main receptor for the entry of Covid-19 into lower respiratory tract epithelial cells through interaction with SARS-CoV-2 S protein. Based on this observation, it is expected that the virus infection can be inhibited if protein-protein interaction is prevented. In this study, using structure-based virtual screening of FDA databases, several lead drugs were discovered based on the ACE2-binding pocket of SARS-CoV-2 S protein. Then, binding affinity, binding modes, critical interactions, and pharmaceutical properties of the lead drugs were evaluated. Among the previously approved drugs, Diammonium Glycyrrhizinate, Digitoxin, Ivermectin, Rapamycin, Rifaximin, and Amphotericin B represented the most desirable features, and can be possible candidates for Covid-19 therapies. Furthermore, molecular dynamics (MD) simulation was accomplished for three S protein/drug complexes with the highest binding affinity and best conformation and binding free energies were also computed with the Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) method. Results demonstrated the stable binding of these compounds to the S protein; however, in order to confirm the curative effect of these drugs, clinical trials must be done.
Keywords: ACE2, Covid-19, molecular docking, SARS-CoV-2, structure-based virtual screening
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
Coronaviruses are a genus of the Coronaviridae family; belong to the Coronavirinae subfamily, and the order of Nidovirales. They are categorized into four genera including Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Shanmugaraj et al., 2020; Siddell et al., 1983). They are enveloped viruses with a large plus-strand RNA genome which are typically present among several species of animals such as cows, bats, camels, cats, and avian. They may transmit from animals to humans, a process termed “spill over” (Mukhtar & Mukhtar, 2020; Shanmugaraj et al., 2020). More recently, a new Betacoronavirus has been found out provisionally named 2019 novel coronavirus (2019-nCoV) (Elfiky, 2020b; Zhu et al., 2020). This virus is now officially known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which causes the COVID-19 disease. This virus is probably originated from an animal repository and has recently triggered the epidemic in humans because of rapid transmission from human to human as well as its high mortality rate (Elfiky, 2020a; Mukhtar & Mukhtar, 2020; Wrapp et al., 2020). Therefore, inhibiting the SARS-CoV-2 virus has been a serious challenge for researchers and clinicians and they have become motivated to introduce and develop vaccines and therapeutic antibodies as well as drugs against this virus. Hence, the first genome sequencing of SARS-CoV-2 was published by Fan Wu et al. from china. They performed Phylogenetic analysis of the whole-genome sequence, containing 29,903 nucleotides, and reported that the virus has 89.1% nucleotide similarity to a group of SARS-like coronaviruses. Comparison of their conserved domains revealed that the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 was closely related to those of SARS-CoVs (73.8–74.9% amino acid identity), which makes it capable to use the human ACE2 (Angiotensin-Converting Enzyme 2) receptor for cell entry (Wu et al., 2020). Additionally, recent studies have shown that the ACE2 is also the receptor for SARS-CoV-2’s entry into lower respiratory tract epithelial cells, (Agostini et al., 2018; Chang et al., 2020; Morgenstern et al., 2005; Shang et al., 2020; Wrapp et al., 2020; Xu et al., 2020).
Designing novel drugs against a new virus through experimental techniques is very time-consuming; however, it is required to find an effective drug immediately to treat the infection and decrease death cases. Therefore, it seems to be logical to search for potential therapeutics among previously approved drugs.
Based on the above statement, in this study, potential agents were identified to inhibit the interaction of RBD domain of the SARS-CoV-2 with ACE2 receptor by using virtual screening approaches. Our results showed that among the studied drugs, Diammonium Glycyrrhizinate, Digitoxin, Ivermectin, Rapamycin, Rifaximin, and Amphotericin B might be effective therapeutics for the treatment of Covid-19 infection due to their better binding affinities and conformations. Lastly, MD simulation analysis and binding free energy calculations were accomplished for SARS-CoV-2-RBD/Diammonium Glycyrrhizinate, SARS-CoV-2-RBD/Digitoxin, and SARS-CoV-2-RBD/Ivermectin complexes, which had the highest binding affinity and the best conformations. Results of this study indicated that in silico approaches can be effectively used to develop a drug discovery pipeline using FDA approved drug databases, and it may lead to introduce novel potentials for the old drugs.
2. Materials and methods
Virtual screening approaches are extensively being applied in designing and development of new drugs. In this regard, one of the most common virtual screening techniques is structure-based virtual screening (SBVS) which only needs the three-dimensional structure of the interested protein and identifying its potential binding pockets to choose drugs, which interact strongly with these binding pockets, from large databases (Kalhor, Rahimi, et al., 2020; Kalhor, Sadeghi, et al., 2020; Shiri et al., 2018, 2019).
2.1. Receptor selection and preparation
In the SBVS method, identification and preparation of the target receptor is an essential step. Hence, the crystallographic structure of SARS-CoV-2 S protein (RBD domain) in complex with ACE2 (PDB entry: 6VW1) was applied for molecular docking studies (Shang et al., 2020). Also, other complexes of the SARS-CoV/ACE2 (PDB IDs: 6acj, 6acg, 6ack, 2ajf) ( F. Li et al., 2005; Song et al., 2018 ) were used for structural alignment analysis. Subsequently, the selected SARS-CoV-2-RBD was prepared as known receptor using AutoDock Tools 4.2. First, water molecules were removed, and then atoms were adjusted to the AutoDock atom types. In the next step, bond orders were assigned, and hydrogen atoms and Gasteiger-Marsili charges were added to the crystal structure. In order to conduct molecular docking, the receptor structure was prepared in the PDBQT format (Morris et al., 2009).
2.2. Binding pocket selection and grid box preparation for docking studies
In order to perform virtual screening, the binding pocket of SARS-CoV-2- RBD was selected based on the binding pocket inferred from the crystallographic structure of SARS-CoV-2/ACE2 complex. To determine the main residues involved in the binding regions, the crystal structure was studied using PDBsum web tool and LigPlot+ software (Laskowski, 2009; Laskowski & Swindells, 2011). Accordingly, Tyr449, Tyr453, Lus455, Phe456, Ala475, Gly476, Phe486, Asn487, Tyr489, GLn493, Gly496, Gln498, Tyr500, Asn501, Gly502 and Tyr505 residues were chosen as the reference amino acids for virtual screening. By using AutoDock Tools4.2, Grid box for docking studies was predicted into X = 26 Å, Y = 42 Å, Z = 26 Å grid points, and the grid spacing was 1 Å.
2.3. Ligand selection and preparation
In this work, two different FDA approved drug databases were downloaded from the zinc database15 (1681 compounds) and used for virtual screening, including FDA Approved drugs in per DrugBank(https://zinc.docking.org/substances/subsets/fda/) and FDA-approved-Drug-Library (1364 compounds). All compounds in this FDA-approved-Drug-library have well-characterized biological activity, clear targets, safety, and bioavailability properties which could significantly make drug development and optimization faster and easier. Also, since it covers various research areas, it is an comprehensive and ideal library for drug repurposing (https://www.targetmol.com/compound-library/FDA-approved-Drug-Library) In this study, by using AutoDock version 4.2, each drug of the databases was prepared as follows: non-polar hydrogen bonds were merged, Gasteiger-Marsili charges were added, atoms were adjusted to the AutoDock atom types, and the rotatable bonds were assigned, then were saved in the PDBQT format. Finally, drugs were converted to the SDF using Open Babel software (O'Boyle et al., 2011) to be prepared for docking with the receptor.
2.4. Virtual screening and molecular docking studies
In this step, approximately all of the FDA approved drugs from two databases were docked in the selected binding pocket using AutoDock Vina (Koes et al., 2013) . (http://smina.sf.net). Subsequently, based on the highest binding affinity, the result of docking was sorted and then were clustered based on the structural similarities and physicochemical features by ChemMine Web Tools (http://chemmine.ucr.edu/). Afterward, the selected drugs’ physiochemical properties were evaluated using OSIRIS Data Warrior (Sander et al., 2015). Ultimately, the potential drugs were chosen against the binding pocket of SARS-CoV-2-RBD.
2.5. Molecular dynamics simulation
In order to evaluate the conformational changes of the selected drugs in complex with SARS-CoV-2-RBD, Molecular Dynamics (MD) simulations were performed by Gromacs version 5.1.2 for a period of 100 ns. Firstly, topology file and force field parameters of the selected drugs were produced for Gromacs utilizing the PRODRG server (Schüttelkopf & Van Aalten, 2004). After that, the topology file of the receptor was generated using pdb2gmx and the united-atom GROMOS 96 43A1 force field (Abraham et al., 2015). In the next step, the receptor/ligand complexes were solvated in a rectangular box with SPC (simple point charge) water molecules with a 10 Å marginal radius (Van Tilbeurgh et al., 1999). In order to make the simulation system electrically neutral, we replaced solvent molecules with Cl − or Na + ions. Then, Particle Mesh Ewald (PME) summation was considered to obtain the long range electrostatic interactions (Darden et al., 1993). The Linear Constraint Solver (LINCS) algorithm was used for covalent bond constraints (Hess et al., 1997). The system was relaxed with several energy minimization steps. Subsequently, the system was balanced for 500 ps using NVT (constant Number of particles, Volume, and Temperature), following another 500 ps using NPT (constant Number of
particles, Pressure, and Temperature) ensembles at 300 K. Thermostat and barostat of the system were adjusted through the V-rescale22 and Parrinello–Rahman barostat algorithms, respectively (Bussi et al., 2007; Parrinello & Rahman, 1981). Finally, the balanced systems were simulated for a period of 100 ns with 2 fs time steps. The resulted trajectories of the MD simulations were used for further processing.
2.6. Binding energy analysis using MM-PBSA approach
The interaction free energies of each receptor-ligand complex were evaluated using Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) method. In this study, the last 1000 ps of the MD trajectories were used for the calculation of binding free energies, van der Waals energy, electrostatic energy, surface accessible surface area, and polar solvation energy, by using the equations as described by Kumari et al. (2014).
2.7. Visual presentations
Three dimensional structures of the protein and receptor/ligand complexes were visualized using PyMOL software, also the hydrophobic interactions and hydrogen bonds between protein and compounds were analyzed using discovery studio software (DeLano, 2002; Visualizer, 2017).
3. Results
3.1. Analysis of the structure of SARS-CoV-2-RBD/ACE2 complex and identification of binding pocket
It is well proved that the active form of SARS-CoV-2 spike (S) glycoprotein contains important structural domains; NTD (N-terminal domain), RBD (receptor binding domain), SD1/SD2 (subdomain1/2), and S2 domain. The RBD of SARS-CoV-2 S has efficient role in the binding to human ACE2 enzyme. The SARS-CoV-2 S is closely related to SARS-CoV S which also binds to ACE2 through RBD (Shang et al., 2020). According to the previous studies, the ACE2 binding site of SARS-RBD including; Tyr436, Tyr440, Tyr442, Lus443, Lus472, N473, Tyr 475, Gly482, Tyr484, Thr486, Thr487, Gly488, Ile489, Tyr491, and Gln492 residues and the SARS-RDB binding site of ACE2 including; Gln24, Thr27, Phe28, Asp30, Lys31, His34, Asp38, Tyr41, Gln42, Lus45, Lus79, Met82, Tyr83, Gln325, Gly326, Asn330, Lys353, A355 were key residues that were involved in SARS-RBD/ACE2 interactions (Song et al., 2018). Interestingly, the ACE2 binding site of SARS-CoV-2 -RBD including; Tyr449, Tyr453, Lus455, Phe456, Ala475, Gly476, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502 and Tyr505 residues and the SARS-CoV-2-RBD binding site of ACE2 including; Ser19, Gln24, Thr27, Phe28, Lys31, His34, Glu35, Glu37, Asp38, Tyr41, Gln42, Lus45, Lus79, Met82, Tyr83, Asn330, Lys353, Gly354, Asp355, Arg357, and Arg393 were recognized as the key residues which were involved in SARS-CoV-2-RBD/ACE2 interactions (Shang et al., 2020). Consistent with this findings, superimposition of SARS-CoV-2-RBD/ACE2 (PDB ID: 6vw1) and SARS-RBD/ACE2 (PDB ID: 6cag) complexes demonstrated that the secondary structure of residues involved in SARS-CoV-2-RBD binding site were highly similar to SARS-RBD binding site, so that both SARS-CoV-2 and SARS similarly interact with human ACE2 (Figure 1A). Taken together, the aforementioned residues of SARS-CoV-2 were selected as the binding pocket which was located in the RBD, so this selected binding pocket was applied for further virtual screening.
Figure 1.
Sequence and structure alignment of SARS-CoV and SARS-CoV-2-RBDs and their important residues in the interaction with ACE2: (A) Sequence alignment of SARS-CoV and SARS-CoV-2-RBDs. Red boxes indicate ACE2-interacting residues common in both SARS CoV and SARS-CoV-2, and green boxes indicate unique interacting residues in each. (B) Structural alignment of SARS-CoV-2 (gray) and SARS-CoV (blue) RBDs in complex with ACE2 (purple). Binding residues are colored in green, red and yellow in SARS-CoV-2, SARS-CoV, and ACE2, respectively. Close-up view of interacting residues (labeled and shown in stick representation) of ACE2 (C) and SARS-CoV-2 (D).
3.2. Structure-based virtual screening and identification of potential inhibitors of protein-protein interaction
In order to conduct structure-based virtual screening (SBVS), approved drugs libraries (FDA databases) was used to select potential drugs which may bind to the RBD of SARS-CoV-2. Molecular docking between the binding sites of SARS-CoV-2-RBD and selected libraries was performed by applying AutoDock Vina. Results were sorted based on the binding affinity. The binding affinity threshold was set as −10 kcal/mol. After analyzing the result, 20 drugs were found against the binding pocket with the binding affinity higher than −10 kcal/mol. To remove drugs with identical structure, they were grouped based on the structure similarity using ChemMine Web Tools. Eventually, a library with 12 hit drugs with highest binding affinity and best conformations were chosen as hit drugs and recruited for further analysis.
3.3. Analysis of the complexes of selected lead drugs with SARS-CoV-2-RBD
To select the best lead drugs, interactions between selected hit drugs with SARS-CoV-2-RBD, binding affinity and number of hydrogen bonds as well as interacting residues were studied. Subsequently, drugs with the highest binding affinity and hydrogen bonds were chosen from the hit drugs as the potential lead drugs (Table 1). As it can be deduced from the results Diammonium Glycyrrhizinate showed the highest binding affinity (-11.7545 kcal/mol), and, Digitoxin, Ivermectin, Rapamycin, Rifaximin, and Amphotericin B followed average ranged from −11.2534 to −10.5021 (kcal/mol), respectively.
Table 1.
Evaluation of the binding affinity (kcal/mol) and interacting residues in the SARS-CoV-2- RBD/drug complexes.
| Namedrug | binding affinity(kcal/mol) | HB-AAsa | NH-AAsb |
|---|---|---|---|
| Diammonium Glycyrrhizinate | −11.7545 | Tyr449,Gln493, Ser494, Gly496,Gln498, Tyr505 | Tyr453,Gly485,Phe486, Asn487,Tyr489,Tyr495, Asn501 |
| Digitoxin | −11.2534 | Lys403,Asp406,Gln409, Val417,Gly485 | Arg408,Ile418,Tyr453, Cys488,Glu484,Gly485, Phe486,Asn487,Cys488, Tyr489 |
| Ivermectin | −10.8697 | Tyr453,Leu492,Gln493, Ser494,Asn502 | Lys403,Tyr449,Lys452, Phe490,Tyr495,Gly502, Tyr505 |
| Rapamycin (Sirolimus) | −10.5657 | Lys403,Asp406,Arg408, Tyr453,Tyr489, Glu493, Ser494 | Glu409,Gly416,Val417, Ile418,Tyr421,Tyr449, Leu455,Phe456,Tyr495, Gly496 |
| Rifaximin | −10.5365 | Tyr449,Tyrs453,Gln493, Ser494 | Lys403,Leu455,Tyr495, Gly496,Tyr505 |
| Amphotericin B | −10.5021 | Tyr449,Lys452,Tyr453, Gln493,Ser494 | Leu455,Phe456,Glu484, Phe490 |
aHydrogen bonds forming Amino Acids, bNon-bonded contacts forming Amino Acids.
Moreover, the two-dimensional structure of the selected lead drugs as well as their predicted binding modes in complex with SARS-CoV-2-RBD were analyzed using pyMOL and discovery studio softwares, respectively.
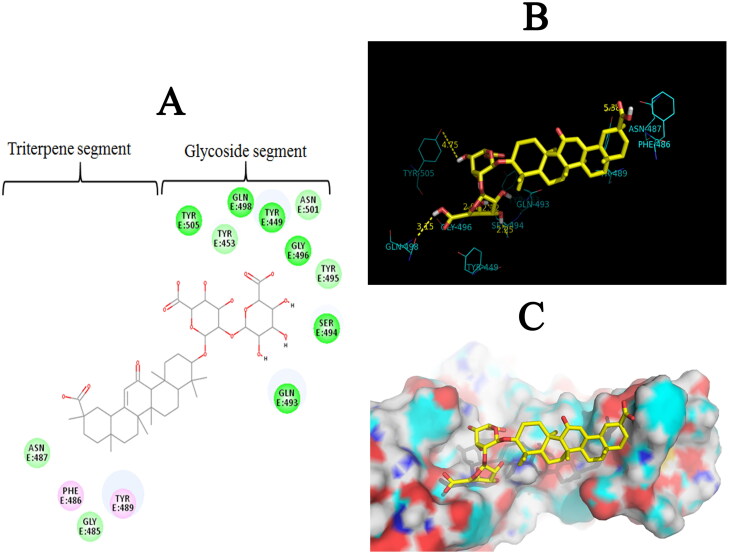
The Diammonium Glycyrrhizinate small molecule consists of two parts: (1) a Glycoside segment, and (2) a triterpene segment (Figure 2A). Analysis of the docking results revealed that seven hydrogen bonds were formed between hydroxyl (OH) and oxygen (—O— and C = O) of Glycoside segment of Diammonium Glycyrrhizinate and Gln493 (angle O—H—N = 160.83°, distance = 2.17 Å), Ser494 (angle O—H—O = 154.98°, distance = 2.25 Å), Ser494 (angle O—H—O = 142.66°, distance = 2.12 Å), Gly496 (angle HO—H—N = 104.99°, distance = 2.92 Å), Tyr449 (angle O—H—O = 91.46°, distance = 2.86 Å), Gln498 (angle O—H—O = 156.37°, distance = 3.15 Å), Tyr505 (angle O—H—O = 89.91°, distance = 4.75 Å) residues. Also, one hydrogen bond was formed between triterpene segment of Diammonium Glycyrrhizinate and Tyr489 (angle O—H—O = 78.84°, distance = 5.38 Å) residue in the binding pocket of SARS-CoV-2-RBD (Figure 2B). In addition, the triterpene segment of the docked compound was embedded in a hydrophobic pocket formed by Tyr489, Asn487 and Phe486 residues (Figure 2C).
Figure 2.
Two-dimensional structure of Diammonium Glycyrrhizinate and three-dimensional illustration of itsinteraction with the ACE2 binding pocket of SARS-CoV-2-RBD. (A) Representation of 2 D structure of Diammonium Glycyrrhizinate small molecule in complex. The green, light green, and pink spheres represent residues involved in hydrogen bond interactions, the hydrophobic interactions, and Pi-Alkyl interactions, respectively. (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Diammonium Glycyrrhizinate (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
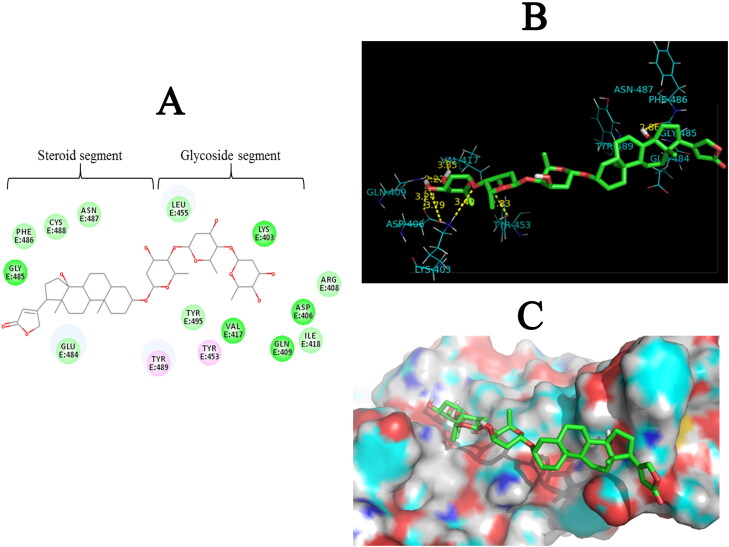
The structure of Digitoxin small molecule also consists of two parts: (1) a Glycoside segment and (2) a Steroid segment (Figure 3A). Detailed study of docking results of Digitoxin revealed the presence of six hydrogen bonds formed between hydroxyl (OH) and oxygen of Glycoside segment from Digitoxin and Tyr453 (angle O—H—O = 83.70°, distance = 2.83 Å), Lys403 (angle O—H—N = 152.02°, distance = 3.40 Å), Lys403 (angle HO—H—N = 102.20°, distance = 3.79 Å), Asp406 (angle O—H—O = 77.42°, distance = 3.24 Å), Gln409 (angle HO—H—N = 153.49°, distance = 2.22 Å), Val417 (angle O—H—N = 99.65°, distance = 3.35 Å) residues. Additionally, one hydrogen bond was formed between hydroxyl of Steroid segment from Digitoxin and Gly485 (angle O—H—O = 83.41°, distance = 2.86 Å) residue in the binding pocket of SARS-CoV-2-RBD (Figure 3B). In addition, Steroid segment of the docked compound was embedded in a hydrophobic pocket formed by Tyr489, Glu484, Gly485 and Phe486 residues (Figure 3C).
Figure 3.
Two-dimensional structure of Digitoxin and three-dimensional illustration of its interaction with the ACE2 binding pocket of SARS-CoV-2-RBD (A) Representation of 2 D structure of Digitoxin small molecule in complex. The green, light green, and pink spheres represent residues involved inhydrogen bond interactions, the hydrophobic interactions, and Pi-Alkyl interactions, respectively (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Digitoxin (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
The structure of Ivermectin small molecule consists of two parts: (1) a Glycoside segment, and (2) a Macrolide segment (Figure 4A). The analysis of Ivermectin demonstrated that five hydrogen bonds were formed between hydroxyl (OH) and oxygen of Ivermectin and Tyr453 (angle HO—H—O = 141.06°, distance = 2.43 Å), Leu492 (angle O—H—O = 82.36°, distance = 3.87 Å), Gln493 (angle HO—H—N = 118.86°, distance = 3.19 Å), Ser494 (angle O—H—O = 72.42°, distance = 4.26 Å) and Asn501 (angle O—H—N = 107.00°, distance = 4.49 Å) residues in the binding pocket of SARS-CoV-2-RBD (Figure 4B). In addition, the docked compound was embedded in a hydrophobic pocket formed by Gln493, Asn501, Tyr505 and Tyr449 residues (Figure 4C).
Figure 4.
Two-dimensional structure of Ivermectin and three-dimensional illustration of its interaction with the ACE2 binding pocket of SARS-CoV-2-RBD. (A) Representation of 2 D structure of Rifaximin small molecule in complex. The green, light green and pink spheres represent residues involved in hydrogen bond interactions, the hydrophobic interactions, and Pi-Alkyl interactions, respectively (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Ivermectin (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
The structure of Rapamycin small molecule consists of two parts: (1) a Cyclohexanol segment and (2) a Polyene macrolide segment (Figure 5A). Based on the docking results, eight hydrogen bonds were formed between hydroxyl (OH) and oxygen (—O— and C = O) from Rapamycin and Arg408 (angle O—H—N = 153.87°, distance = 3.92 Å), Asp406 (angle O—H—O = 49.79°, distance = 4.19 Å), Lys403 (angle O—H—N = 136.74°, distance = 1.98 Å), Lys403 (angle HO—H—N = 84.97°, distance = 2.82 Å), Tyr489 (angle O—H—O = 137.13°, distance = 5.09 Å), Gln493 (angle O—H—N = 151.31°, distance = 3.57 Å), Ser494 (angle O—H—O = 64.76°, distance = 3.09 Å), Tyr453 (angle HO—H—O = 81.97°, distance = 2.98 Å residues, and one hydrogen bond was formed between N of piperidine from Rapamycin and Tyr505 (angle N—H—O = 111.26°, distance = 5.60 Å) residue in the binding site of SARS-CoV-2-RBD (Figure 5B). In addition, the docked compound was embedded in a hydrophobic pocket formed by Tyr453, Gly416, Val417, Leu455, Phe456 and Gln409 residues (Figure 5C).
Figure 5.
Two-dimensional structure of Rapamycin and three-dimensional illustration of its interaction with the ACE2 binding pocket of SARS-CoV-2-RBD. (A) Representation of 2 D structure of Rapamycin small molecule in complex. The green, light green and pink spheres represent residues involved in hydrogen bond interactions, the hydrophobic interactions, and Pi-Alkyl interactions, respectively (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Rapamycin (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
The structure of Rifaximin small molecule consists of two parts: (1) Macrolide segment, and (2) a Naphto Imidazopyridine segment (Figure 6A). Four hydrogen bonds were formed between hydroxyl (OH) and oxygen (—O— and C = O) of Rifaximin and Tyr453 (angle O—H—O = 123.49°, distance = 3.02 Å), Gln493 (angle HO—H—N = 119.47°, distance = 2.71 Å), Gln493 (angle O—H—N = 140.94°, distance = 4.16 Å), Gln493 (angle O—H—O = 68.92°, distance = 4.56 Å) residues, and two hydrogen bonds were formed between N of amine and pyridine from Rifaximin and Ser494 (angle N—H—O = 161.38°, distance = 4.27 Å) and Tyr449 (angle N—H—OH =129.54°, distance = 4.35 Å) residues in the binding pocket of SARS-CoV-2-RBD (Figure 6B). Moreover, the docked compound was embedded in a hydrophobic pocket formed by Gln493, Tyr505, Tyr453 and Tyr449 residues (Figure 6C).
Figure 6.
Two-dimensional structure of Rifaximin and three-dimensional illustration of its interaction with the ACE2 binding pocket of SARS-CoV-2-RBD. (A) Representation of 2 D structure of Rifaximin small molecule in complexs. The green, light green and pink spheres represent residues involved in carbon hydrogen bond interactions, the hydrophobic interactions, and Pi-Alkyl interactions, respectively (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Rifaximin (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
Like all the previously mentioned drugs, the structure of Amphotericin B small molecule consists of two parts: (1) a Glycoside segment, and (2) a Polyene macrolide segment (Figure 7A). Analysis of Amphotericin B/SARS-CoV-2-RBD complex showed that two hydrogen bonds were formed between amine group of Glycoside segment from Amphotericin B and Lys452 (angle N—H—N = 116.47°, distance = 4.46 Å) and Leu492 (angle N—H—O = 94.10°, distance = 4.87 Å) residues, and three hydrogen bonds were formed between hydroxyl (OH) group of Glycoside segment of Amphotericin B and Leu492 (angle O—H—O = 114.46°, distance = 3.19 Å), Gln493 (angle O—H—N = 104.23°, distance = 3.71 Å), Ser494 (angle HO—H—N = 110.36°, distance = 2.47 Å) residues. Also, five hydrogen bonds were formed between hydroxyl (OH) group of Polyene macrolide segment from Amphotericin B and Ser494 (angle O—H—O = 59.47°, distance = 3.40 Å), Ser494 (angle HO—H—O = 137.85°, distance = 2.37 Å), Ser494 (angle O—H—O = 116.63°, distance = 2.40 Å),Tyr449 (angle O—H—O = 135.84°, distance = 4.66 Å), Gln493 (angle HO—H—N = 153.01°, distance = 3.13 Å), Tyr453 (angle O—H—O = 153.22°, distance = 4.59 Å) residue in the binding pocket of SARS-CoV-2-RBD (Figure 7B). In addition, the docked compound was embedded in a hydrophobic pocket formed by Leu492, Leu455, Phe456 and Tyr449 residues (Figure 7C).
Figure 7.
Two-dimensional structure of Amphotericin B and three-dimensional illustration of its interaction with the ACE2 binding pocket of SARS-CoV-2-RBD. (A) Representation of 2 D structure of Amphotericin B small molecule in complex. The green, light green and pink spheres represent residues involved in carbon hydrogen bond interactions, the hydrophobic interactions, Pi-Alkyl interactions, respectively (B) Hydrogen bonds in protein-ligand complex are shown as yellow dotted lines. (C) 3 D illustration of Amphotericin B (stick representation) in the binding pocket of SARS-CoV-2-RBD (surface representation) (PDB ID: 6vw1).
3.4. Evaluation of the physiochemical properties of the lead drugs
In this study, several important physiochemical parameters of the selected lead drugs were evaluated using OSIRIS Data Warrior; including molecular weight (MW), LogP, LogS, number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), polar surface area (PSA), and number of rotatable bonds (Rb). The results showed that the majority of lead drugs possessed following physicochemical properties; MW< 1000, −9 <LogS < −5, 0.3 <cLogP < 6.5, HBD< 12, HBA< 18, PSA< 320 A°, and RB< 8. Detailed calculation of these parameters is tabulated in Table 2. It is well documented that 90% of orally active compounds follow Lipinski’s rule of five (RO5) .Currently, the RO5 is one of the most important factors in choosing hits and leads. The RO5 asserts following criteria for an orally active compound: MW < 500 Da, logp < 5, HBA < 10, and HBD < 5 (Lipinski et al., 2012). As it can be deduced from Table 2, these lead drugs do not fallow the RO5 but have been proved to be potentially successful drugs. Digitoxin and Rifaximin had lower MW than other drugs. Also, the number of HBA, HBD and Rb in Digitoxin were lower than other drugs. Ivermectin and Rapamycin were identical in both HBA, and HBD. Interestingly, the predicted results of drug likeness properties of the lead drugs were considerably high, except Diammonium Glycyrrhizinate (-4.291) and Amphotericin B (-0.137); however, Rifaximin was predicted to be mutagenic and tumorigenic.
Table 2.
Evaluation of physicochemical properties and toxicity risk parameters of selected lead drugs using OSIRIS Data Warrior.
|
Name drug |
Physicochemical properties |
Toxicity
risks parameters |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MWa | CLPb | CLSc | HBAd | HBDe | PSAf | RBg | DLh | MUTi | TUMj | |
| Diammonium Glycyrrhizinate | 857.000 | 0.982 | −5.144 | 16 | 8 | 267.04 | 7 | −4.291 | NO | NO |
| Digitoxin | 764.946 | 2.747 | −6.086 | 13 | 5 | 182.83 | 7 | 3.325 | NO | NO |
| Ivermectin | 1736.18 | 5.410 | −6.61 | 14 | 3 | 170.06 | 8 | 5.231 | NO | NO |
| Rapamycin (Sirolimus) | 914.182 | 6.510 | −6.713 | 14 | 3 | 195.43 | 6 | 2.378 | NO | NO |
| Rifaximin | 785.888 | 5.843 | −9.904 | 14 | 5 | 198.38 | 3 | 6.195 | High | high |
| Amphotericin B | 924.087 | 0.323 | −5.077 | 18 | 12 | 319.61 | 3 | −0.137 | NO | NO |
a Molecular weight (g/mol),b cLogP(lipophilicity),c cLogS(solubility), d H-bond acceptors,eH-bond donors , f Polar Surface Area(A2), g Rotatable bonds, h Drug likeness, iMutagenic, j Tumorigenic.
3.5. Molecular dynamic simulation analysis
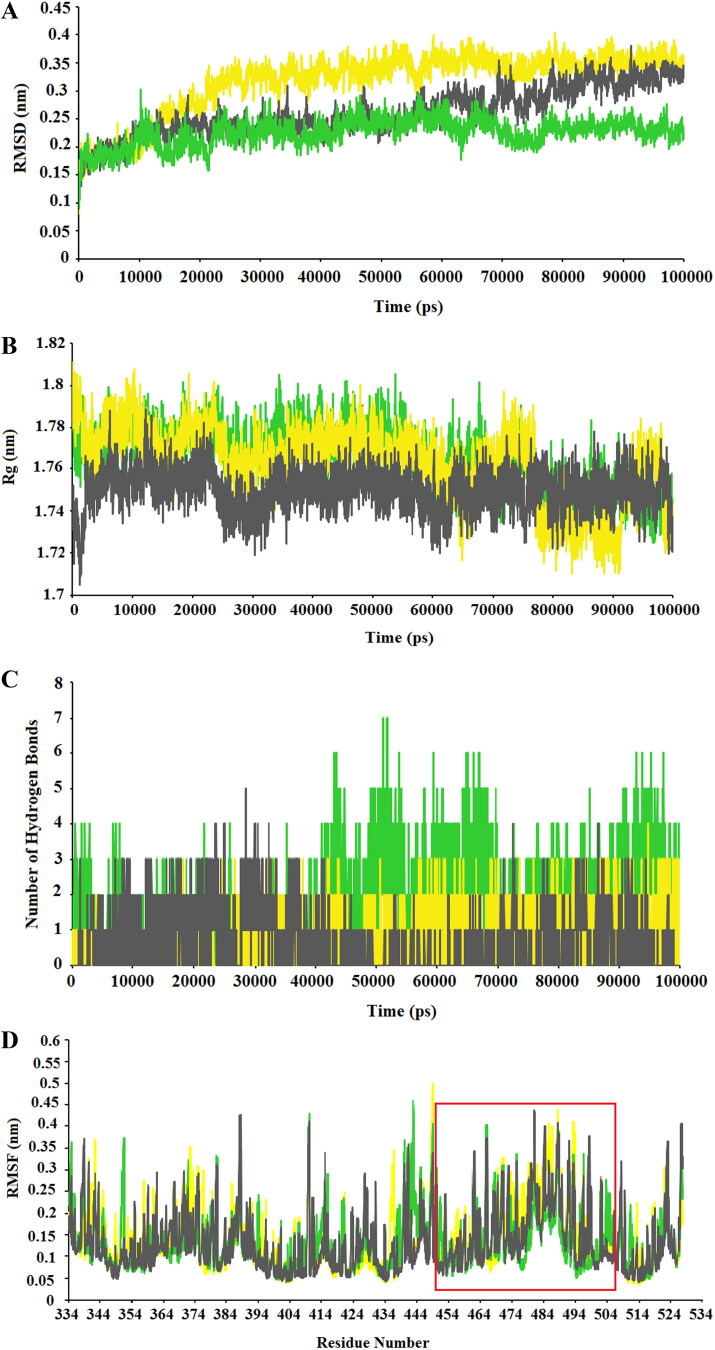
In the present study, to assess conformational behavior, stability, and flexibility of the docked complexes, MD simulations were accomplished for three complexes with the highest binding affinity and best conformations: SARS-CoV-2-RBD/Diammonium Glycyrrhizinate, SARS-CoV-2-RBD/Digitoxin, and SARS-CoV-2-RBD/Ivermectin complexes. RMSD (Root Mean Square Deviation) value was computed considering the proteins backbone with respect to the initial conformations. RMSD plots for three SARS-CoV-2-RBD/drug complexes during 100 ns of simulations are showed in Figure 8A. It can be seen that the RMSD value of SARS-CoV-2-RBD/Diammonium Glycyrrhizinate complex was in the range of 0.08 to 0.4 nm, and reached its steady state at∼ 0.35 nm from 60000 ps to 100000 ps. the RMSD value for SARS-CoV-2-RBD/Digitoxin complex was fluctuated from 0.09 to 0.38 nm and reached its steady state at 0.24 nm between 10000 and 50000 ps, then slightly increased and persisted at ∼0.32 nm from 70000 ps till the end of the simulation, while the RMSD value for SARS-CoV-2-RBD/Ivermectin complex was in range of about 0.09 to 0.30 nm at the first 10000 ps, however it decreased and remained stable at 0.24 nm from 10000 to 100000 ps, except between 70000 and 80000 ps timescale. The pattern of changes in RMSD was almost similar for three complexes; nonetheless, SARS-CoV-2-RBD/Ivermectin complex displayed the least fluctuation of RMSD plot during MD simulations.
Figure 8.
Analysis of MD simulations results: (A) RMSD plots of the SARS-CoV-2-RBD/drug complexes during100 ns of simulations. (B) Rg plots of the SARS-CoV-2-RBD/drug complexes during MD simulations. (C) The number of H-bonds between the SARS-CoV-2-RBD and drugs. (D) RMSF of backbone Cα atoms of the complexes versus residue number in the sequence, the binding pocket is shown in the red box. In all plots, SARS-CoV-2-RBD/Diammonium Glycyrrhizinate, SARS-CoV-2-RBD/Digitoxin, and SARS-CoV-2-RBD/Ivermectin complexes are indicated as yellow, green, and gray, respectively.
Rg (Radius of gyration) value was calculated as an indicator of stability and also protein structure compactness. As depicted in Figure 8B, SARS-CoV-2-RBD/Ivermectin complex had the least Rg value, but it exhibited the highest fluctuation at the first 25000 ps. The result showed that the Rg plot of SARS-CoV-2-RBD/Diammonium Glycyrrhizinate fluctuated more than Ivermectin and Digitoxin complexes.
Numbers of hydrogen bonds (H-bonds) were calculated to investigate the stability of H-bonds between SARS-CoV-2-RBD/drug complexes during MD simulations (Figure 8C). As it can be deduced from the results, SARS-CoV-2-RBD/Diammonium Glycyrrhizinate possessed 0-4 H-bonds, SARS-CoV-2-RBD/Digitoxin had 0-7 H-bonds, and SARS-CoV-2-RBD/Ivermectin complexes presented 0-5 hydrogen bonds. However, in all of the complexes, two H-bonds were observed in average which were strongly stable (especially, in the SARS-CoV-2-RBD/Diammonium Glycyrrhizinate complex) throughout MD simulations.
RMSF (Root Mean Square Fluctuation) value was computed from the trajectories of each complex to evaluate the flexibility of individual residues (Figure 8D). The RMSF plot of complex structures uncovered that all residues fluctuated between 0.05 and 0.5 nm in Diammonium Glycyrrhizinate, 0.05 nm and 0.45 nm in Digitoxin, and 0.05 nm and 0.47 nm in Ivermectin. The RMSF plot of complexes showed that all residues located in the binding pocket of SARS-CoV-2-RBD had fluctuations between 0.08 nm and 0.43 nm, which indicates that the selected drugs kept close contact with their binding pocket throughout MD simulations.
3.6. Binding free energy calculations
The MM/PBSA method is applied to evaluate the interactions between SARS-CoV-2-RBD and Diammonium Glycyrrhizinate, Digitoxin, and Ivermectin drugs. Free binding energy of the all SARS-CoV-2-RBD/drug complexes was calculated for the last 1000 ps trajectories and the results of energy components of the complexes are given in Table 3. Diammonium Glycyrrhizinate, Digitoxin, and Ivermectin presented binding energy of −182.900, −133.789 and −89.360 kJ/mol, respectively, suggesting Diammonium Glycyrrhizinate having the highest affinity for SARS-CoV-2-RBD. Subsequently, van der Waals (ΔEvdw), electrostatic (ΔEele), polar solvation energy (ΔGpol), and nonpolar interactions (ΔGnonpol) were computed to evaluate the most significant interaction term which impacts the calculated binding energy.
Table 3.
Evaluation of the binding free energies (kJ/mol)) and energy components (kJ/mol)) in the SARS-CoV-2-RBD/drug complexes.
|
Name drug |
ΔEvdwa | ΔEeleb | ΔGpolc | ΔGnonpold | ΔGbinde |
|---|---|---|---|---|---|
| Diammonium Glycyrrhizinate |
−201.495 | −11.734 | 44.418 | −14.418 | −182.900 |
| Digitoxin | −192.755 | −1.784 | 76.812 | −16.042 | −133.789 |
| Ivermectin | −131.07 | −3.956 | 53.812 | −8.146 | −89.360 |
aEvdw, van der Waals contribution; bΔEele, electrostatic contribution; cΔGpol, polar solvation energy; dΔGnonpol, nonpolar solvation energy; eΔGbind, binding energy; ΔGbind = ΔEvdw + ΔEele + ΔGpol + ΔGnonpol.
As it can be deduced from Table 3, ΔEvdw and ΔGnonpol played remarkable roles in the formation of these complexes; however, van der Waals interactions showed more impact on the binding of SARS-CoV-2-RBD to the selected drugs. Therefore, it suggests the importance of the non-covalent interactions in these complexes.
4. Discussion
Recently, a new and highly pathogenic coronavirus (SARS-CoV-2) has caused a pandemic in the world known as Covid-19 disease (Kandeel & Al-Nazawi, 2020). Although, different drugs based on clinical presentation have been used against Covid-19 infection, no drug or vaccine has yet been approved to treat this novel human coronavirus because of its unknown pathogenesis. Hence, the outbreak of Covid-19 has become a global challenge. Rational drug discovery can identify quick, testable and novel drugs out of the approved small molecule (Jin et al., 2020). It is approved that the binding of SARS-CoV-2 S protein to human ACE2 leads to facilitate its infection like SARS-CoV (Rismanbaf, 2020). In this regard, superimposition of SARS-CoV-2-RBD and SARS-CoV-RBD complexes with ACE2 disclosed that the residues of the ACE2 involved in the interaction with RBD were identical in both SARS-CoV-2 and SARS-CoV (Gln24, Thr27, Phe28, Lys31, His34, Asp38, Tyr41, Gln42, Lus45, Lus79, Met82, Tyr83, Asn330, Lys353, and Asp355). Similarly, the main interacting residues of the RBD domain of SARS-CoV-2 (Tyr449, Tyr453, Asn487, Tyr489, Gly496, Thr500, Gly502 and Tyr505 residues) were also identical to SARS-CoV (Figure 1A). In addition, MSA studies showed that these binding pockets are highly conserved in both SARS-CoV-2 and SARS-CoV (data not shown). Therefore, it can be suggested that the binding pocket of SARS-CoV-2 to ACE2 might be a potential target for drug discovery to disrupt the viral S protein/ACE2 interface. In this regard, we implemented SBVS approach and molecular docking studies to identify more effective drug for treating Covid-19 infection. The molecular docking was accomplished between flexible residues of the selected binding pocket of SARS-CoV-2-RBD (Tyr449, Tyr453, Lus455, Phe456, Ala475, Gly476, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502 and Tyr505 residues) and selected FDA drug libraries using AutoDock Vina tools; docking results were sorted based on the binding affinity. The results of SBVS approach proved that this strategy was efficient in detecting lead drugs with highest binding affinity and best conformations. These lead drugs from our screening are: Diammonium Glycyrrhizinate (pubchem CID: 656656), Digitoxin (pubchem CID: 441207), Ivermectin (pubchem CID: 6321424), Rapamycin (pubchem CID: 5284616), Rifaximin (pubchem CID: 6436173), and Amphotericin B (pubchem CID: 5280965) (Table 1). The detailed analysis of 2 D structure of the selected lead drugs demonstrated that two classes of drugs were obtained in silico; (1) Glycosides (Diammonium Glycyrrhizinate and Digitoxin) and (2) macrolides (Ivermectin, Rapamycin, Rifaximin, Amphotericin B). Glycosides are molecules attached to sugar which binds to functional groups (aglycone) through a glycosidic bond. Glycosides are grouped based on the chemical nature of the aglycane. Aglycone segment in the Diammonium Glycyrrhizinate structure is a triterpenoid, while in the Digitoxin structure is a steroidal nucleus, which involved in the glycosidic linkage via oxygen atom (O-glycosides). Based on the glycoside’s aglycone type, they not only demonstrate various physicochemical properties, but also have different therapeutic uses (Thakur & Raj, 2017; Tinku Gupta, 2018).
Also, results showed that Ivermectin, Rapamycin, Rifaximin, and Amphotericin B have macrocyclic ring (Figures 4A, 5A, 6A, 7A), the most successful classes of macrocyclic drugs in clinical practice. The macrolides drugs are natural products mostly extracted from Streptomyces spp. They consist of a large macrocyclic lactone ring, which may be attached to one or more deoxy sugars. Some macrolides have antiparasite, antibiotic, and antifungal activity (Yu & Sun, 2013). Taken together, it seems that drugs with Glycoside and macrolide segments have a great potential to inhibit the interaction of SARS-CoV-2 with ACE2 and can be applied as favorable pharmacophore models for further studies which aim to discover novel therapeutic drugs against Covid-19 infection.
Analysis of the docking results showed that Diammonium Glycyrrhizinate had the highest binding affinity. Diammonium glycyrrhizinate, a traditional Chinese medicine (TCM), is extracted and purified from licorices (Glycyrrhiza glabra). It is known for its anti-inflammatory effects, resistance to biologic oxidation and membranous protection. Glycyrrhiza glabra is able to reduce inflammatory injury via suppression of NF-κB, TNF-α (Feng et al., 2007). Several studies reported that this compound have potential anti-viral effect against a wide range of viruses including HSV-1 (Utsunomiya et al., 1995), CMV (Numazaki et al., 1994), VZV (Baba & Shigeta, 1987), HBV (Sato et al., 1996), HCV (Arase et al., 1997), HIV (Ito et al., 1988), influenza virus (Utsunomiya et al., 1997), Epstein-Barr virus (Lin, 2003), and interestingly, SARS-CoV (Cinatl et al., 2003). In the 2003, J Cinat et al. published that glycyrrhizin is one of the most potent inhibitors of SARS-CoV replication in Vero cells. Glycyrrhizin not only inhibits virus replication, but also hinders adsorption and penetration of the virus—early steps of the replicative cycle. Furthermore, glycyrrhizin can induce nitrous oxide synthase and production of nitrous oxide in macrophages. Intriguingly, it is well documented that nitrous oxide inhibits replication of viruses (Cinatl et al., 2003).
The next highlighted lead drug, Digitoxin, is a cardiac glycoside that inhibits Na-K pump ATPase activity. Currently, it has been reported that this compound have anti-viral action against both DNA and RNA viruses such as Cytomegalovirus, Herpes simplex, Influenza, and Coronavirus (Amarelle & Lecuona, 2018; Athanasios et al., 2020; Mohammadi et al., 2001). In the study, Pollard etal reported that Digitoxin might be effective therapeutics for the treatment of both influenza and COVID-19 through reduced expression of cytokines such as TNFa, GRO/KC, MIP2, MCP1, TGFb, and IFNg (Pollard et al., 2020). Also, there are various theoretical studies in the literature which have aimed to identify potential inhibitors against SARS-CoV-2 virus using structure-based virtual screening of different databases. Interestingly, several studies have recommended Digitoxin as a favorable drug, For instance, de Oliveira etal performed virtual screening calculations of SWEETLEAD library, and they found Digitoxin as one of the top-scoring compounds (de Oliveira et al., 2020). In same way, the Sekhar used molecular docking based virtual screening of SuperDRUG2 database. In his docking calculations, Digitoxin was found as a candidate drug for inhibiting SARS-CoV-2 virus (Sekhar, 2020). According to molecular docking studies and literature-review, Digitoxin can be considered as a potential drug against Covid-19.
The next selected compound, Ivermectin, is an antiparasit which previously shown to have broad-spectrum antiviral activity against both DNA and RNA viruses in vitro (Götz et al., 2016; Lundberg et al., 2013; Tay et al., 2013; Wagstaff et al., 2012) . Recently, it has been reported that this compound can inhibit Covid-19, and with a single addition to Vero-hSLAM cells 2 h post infection with SARS-Cov-2 was able to cause ∼5000-fold reduction in viral RNA at 48 h (Caly et al., 2020). According to the results, the three of the highlighted leads are related to well-known antibiotics; Rapamycin, Rifaximin, and Amphotericin B. Rapamycin which is a macrolide compound produced by Streptomyces hygroscopicu that is used to treat a rare lung disease called lymphangioleiomyomatosis. This compound has immunosuppressant functions in humans and plays an important role in preventing the rejection of kidney transplants, also it inhibits activation of T cells and B cells by reducing their sensitivity to IL-2 through mTOR inhibition (J. Li et al., 2014). It has been reported that the Rapamycin can be a potent inhibitor for HCV RNA replication as well as primary human hepatocytes (Stöhr et al., 2016). Likewise, Yadi Zhou et al. reported that this compound can be a candidate drug against Covid-19, in virtual screening (Zhou et al., 2020). Another antibiotic of the selected lead drugs is Rifaximin, a gastrointestinal-selective antibiotic, with a broad spectrum of antimicrobial activity. This compound is currently approved for treating traveler's diarrhea, irritable bowel syndrome, and hepatic encephalopathy (Shayto et al., 2016). Also, Amphotericin B is another antibiotic produced by Streptomyces nodosus which is a macrocyclic antifungal agent belongs to a chemical class of polyene antifungal drugs. In fact, this compound is an antifungal agent with a broad spectrum of activity against many fungal species (Alobid et al., 2001). There is an interesting report that claims using Amphotericin B could significantly improve the antiviral properties of Rifamycin derivatives in murine cell lines; more probably through increasing the cell membrane permeability (Hackett et al., 1972).The detailed analysis of interactions between SARS-Cov-2-RBD binding pocket residues and the selected lead drugs is presented in Figures 2–7. In all cases lead drugs are anchored by the hydrogen bonds with at least one of these critical binding residues, Tyr449, Tyr 453, Gln493, and Ser494, However, Diammonium Glycyrrhizinate and Rapamycin showed higher hydrogen bonds than the other selected lead drugs. Although, the number of hydrogen bonds was considerable, hydrophobic interactions were the most frequent interactions in complexes. Moreover, our results indicated that all selected lead drugs could efficiently bind to SARS-CoV-2-RBD. On the whole, these leads can selectively interact with the RBD binding residues of SARS-CoV-2 S protein and can result in the prevention of Covid-19 infection through blocking ACE2-binding pocket of SARS-CoV-2 S protein.
It has been observed that about 90% of orally active compounds follow RO5. In this study important physicochemical properties of the selected lead drugs were calculated by OSIRIS Data Warrior software (Table 2). The result demonstrated that these drugs do not fallow the RO5, but they have been applied for treating some disease, so they are proved to be safe for human. likewise, Silvio Roggo has reported some natural products which do not fallow the RO5 but are proved to be potentially successful drugs, so he has suggested that natural products can be exempted from RO5 (Roggo, 2007).
In this study, MD simulation of Diammonium Glycyrrhizinate and Digitoxin as well as Ivermectin drugs in complex with SARS-CoV2-RBD was performed. As it has been mentioned previously, Digitoxin and Ivermectin drugs displayed significantly favorable results, in silico and in vitro. Here, for the first time, we identified Diammonium Glycyrrhizinate as potential natural drug to inhibit the interaction of SARS-CoV-2-RBD with ACE2, based on the SBVS approach. Therefore, these three drugs, which had the best features, were selected for conducting MD simulations. RMSD values of Ivermectin, Digitoxin, and Diammonium Glycyrrhizinate remained less than 0.3 nm, 0.36 nm and 0.4 nm, throughout the simulation periods, respectively (Figure 8A). These results indicated the convenient stability of the complexes. Also, Rg values of complexes were diminished during MD simulations especially during the last 30000 ps (Figure 8.B). The pattern of changes in RMSF plots was almost similar for three complexes (Figure 8D). Comparing results obtained from MD simulations revealed that these drugs could form stable complexes with SARS-CoV-2-RBD. Analysis of the energy components revealed that Ivermectin had the least binding free energy (-89.360 kJ/mol) compared to the Diammonium Glycyrrhizinate (-182.900 kJ/mol) and Digitoxin (-133.789 kJ/mol) complexes. Therefore, it seems that Diammonium Glycyrrhizinate and Digoxin displayed significantly favorable interactions with SARS-CoV-2-RBD.Considering the need for dealing with the worldwide spread of Covid-19 infection, the lead drugs identified here would be reasonable initial compounds for experimental surveys in limiting Covid-19’s virus-/host interactions and may be noteworthy for the researchers interested in identifying novel small-molecule therapeutics.
5. Conclusion
In this study, based on structure-based virtual screening of clinically approved drugs and the crystallographic structure of SARS-CoV-2-RBD, several lead drugs were identified. The results obtained from docking pose analysis and molecular interactions between SARS-CoV-2-RBD and the lead drugs demonstrated that Diammonium Glycyrrhizinate, Digitoxin, Ivermectin, Rapamycin, Rifaximin, and Amphotericin B drugs had better binding affinities and conformation than other drugs, subsequently, among the selected lead drugs, MD simulation and binding free energy calculation were accomplished for three complexes with highest binding affinity: Diammonium Glycyrrhizinate and Digitoxin and Ivermectin. All three drugs were formed stable complexes with SARS-CoV-2-RBD during simulation periods. Additionally, binding free energy analysis showed that Diammonium Glycyrrhizinate had the lowest free energy of binding in SARS-CoV-2-RBD complex. The results of this study can be utilized for selecting candidate drugs for in vitro and in vivo surveys, and also can provide information for next studies.
Glossary
Abbreviations
- ACE2
Angiotensin Converting Enzyme 2
- COVID-19
coronavirus disease 2019
- FDA
Food and Drug Administration
- HBD
Hydrogen bond donor
- HBA
Hydrogen bond acceptor
- LBSV
Ligand-based virtual screening
- MD
Molecular dynamics
- MM/PBSA
Molecular Mechanics/Poisson–Boltzmann Surface Area
- MW
Molecular weight
- NPT
constant Number of particles, Pressure, and Temperature
- NTD
N-terminal domain
- NVT
constant Number of particles, Volume, and Temperature
- PDB
Protein data bank
- PDBQT
Protein data bank, partial charge and atom type
- PSA
Polar surface area
- RB
Rotatable bonds
- RBD
Receptor-Binding Domain
- RMSD
Root Mean Square Deviation
- RMSF
Root Mean Square Fluctuations
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SBVS
Structure-based virtual screening
- ΔEele
Electrostatic contribution
- ΔEvdw
Van der Waals contributions
- ΔGnonpol
Non-polar contributions
- ΔGpol
Polar solvation contribution
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., & Lindahl E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, , 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., Baric R. S., & Denison M. R. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio, (2), e00221 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alobid I., Bernal M., Calvo C., Vilaseca I., Berenguer J., & Alós L. (2001). Treatment of rhinocerebral mucormycosis by combination of endoscopic sinus debridement and amphotericin B. American Journal of Rhinology, (5), 327–331. 10.1177/194589240101500508 [DOI] [PubMed] [Google Scholar]
- Amarelle L., & Lecuona E. (2018). The antiviral effects of Na, K-ATPase inhibition: A minireview. International Journal of Molecular Sciences, (8), 2154 10.3390/ijms19082154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase Y., Ikeda K., Murashima N., Chayama K., Tsubota A., Koida I., Suzuki Y., Saitoh S., Kobayashi M., & Kumada H. (1997). The long term efficacy of glycyrrhizin in chronic hepatitis C patients. Cancer, (8), 1494–1500. [DOI] [PubMed] [Google Scholar]
- Athanasios T., Psychos C., & Domenikos S. (2020). Communication: Novel drug combination Doxycycline-Melatonin-Digoxin (D-M-D) for possible Covid-19 treatment. Journal of Modern Medicinal Chemistry, (1), 1–3. 10.12970/2308-8044.2020.08.01 [DOI] [Google Scholar]
- Baba M., & Shigeta S. (1987). Antiviral activity of glycyrrhizin against varicella-zoster virus in vitro. Antiviral Research, (2), 99–107. 10.1016/0166-3542(87)90025-8 [DOI] [PubMed] [Google Scholar]
- Bussi G., Donadio D., & Parrinello M. (2007). Canonical sampling through velocity rescaling. The Journal of Chemical Physics, (1), 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Caly L., Druce J. D., Catton M. G., Jans D. A., & Wagstaff K. M. (2020). The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, , 104787 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y., Tung, Y., Lee, K., Chen, T., Hsiao, Y. , Chang, H. , Hsieh, T., Su, C., Wang, S., Yu, J., Shih, S., Lin, Y., Lin, Y., Tu, Y.E., Hsu, C., Juan, H., Tung, C., Chen, C. Potential Therapeutic Agents for COVID-19 Based on the Analysis of Protease and RNA Polymerase Docking. Preprints 2020, 2020020242 (doi: 10.20944/preprints202002.0242.v2). [DOI]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., & Doerr H. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. The Lancet, (9374), 2045–2046. 10.1016/S0140-6736(03)13615-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., & Pedersen L. (1993). Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of Chemical Physics, (12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- de Oliveira O. V., Rocha G. B., Paluch A. S., & Costa L. T. (2020). Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1772885. ISSN: 0739-1102 (Print) 1538-0254 (Online) [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, , 82–92. [Google Scholar]
- Elfiky A. A. (2020. a). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, , 117477 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, , 117592 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Wang H., Yao C., Zhang J., & Tian Z. (2007). Diammonium glycyrrhizinate, a component of traditional Chinese medicine Gan-Cao, prevents murine T-cell-mediated fulminant hepatitis in IL-10-and IL-6-dependent manners. International Immunopharmacology, (10), 1292–1298. 10.1016/j.intimp.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Götz, V., Magar, L., Dornfeld, D., Giese, S., Pohlmann, A., Höper, D., Kong, B. W., Jans, D. A., Beer, M., Haller, O., & Schwemmle, M. (2016). Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Scientific Reports, 6(1 ), 23138–15. 10.1038/srep23138 [DOI] [PMC free article] [PubMed]
- Hackett A. J., Sylvester S., Joss U. R., & Calvin M. (1972). Synergistic effect of rifamycin derivatives and amphotericin B on viral transformation of a murine cell line. Proceedings of the National Academy of Sciences, (12), 3653–3654. 10.1073/pnas.69.12.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H. J., & Fraaije J. G. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, (12), 1463–1472. [DOI] [Google Scholar]
- Ito M., Sato A., Hirabayashi K., Tanabe F., Shigeta S., Baba M., De Clercq E., Nakashima H., & Yamamoto N. (1988). Mechanism of inhibitory effect of glycyrrhizin on replication of human immunodeficiency virus (HIV). Antiviral Research, (6), 289–298. 10.1016/0166-3542(88)90047-2 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You,T., Liu, X.,.. . . Yang, H. ( 2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. doi: 10.1038/s41586-020-2223-y [DOI] [PubMed]
- Kalhor H., Rahimi H., Akbari Eidgahi M. R., & Teimoori-Toolabi L. (2020). Novel small molecules against two binding sites of Wnt2 protein as potential drug candidates for colorectal cancer: A structure based virtual screening approach. Iranian Journal of Pharmaceutical Research, (2), 160–174. 10.22037/IJPR.2019.15297.13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor H., Sadeghi S., Marashiyan M., Kalhor R., Aghaei Gharehbolagh S., Akbari Eidgahi M. R., & Rahimi H. (2020). Identification of new DNA gyrase inhibitors based on bioactive compounds from streptomyces: Structure-based virtual screening and molecular dynamics simulations approaches. Journal of Biomolecular Structure & Dynamics, (3), 791–806. 10.1080/07391102.2019.1588784 [DOI] [PubMed] [Google Scholar]
- Kandeel M., & Al-Nazawi M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, , 117627 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes D. R., Baumgartner M. P., & Camacho C. J. (2013). Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. Journal of Chemical Information and Modeling, (8), 1893–1904. 10.1021/ci300604z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Consortium O. S. D. D., & Lynn A. (2014). g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, (7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Laskowski R. A. (2009). PDBsum new things. Nucleic Acids Research, (suppl_1), D355–D359. 10.1093/nar/gkn860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A., & Swindells M. B. (2011). LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. ACS Publications; 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Li J., Kim S. G., & Blenis J. (2014). Rapamycin: One drug, many effects. Cell Metabolism, (3), 373–379. 10.1016/j.cmet.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., & Harrison S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.), (5742), 1864–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Lin J.-C. (2003). Mechanism of action of glycyrrhizic acid in inhibition of Epstein-Barr virus replication in vitro. Antiviral Research, (1), 41–47. (03)00030-5 10.1016/S0166-3542(03)00030-5 [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., & Feeney P. J. (2012). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, , 4–17. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Lundberg L., Pinkham C., Baer A., Amaya M., Narayanan A., Wagstaff K. M., Jans D. A., & Kehn-Hall K. (2013). Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Research, (3), 662–672. 10.1016/j.antiviral.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Mohammadi K., Kometiani P., Xie Z., & Askari A. (2001). Role of protein kinase C in the signal pathways that link Na+/K+-ATPase to ERK1/2 . Journal of Biological Chemistry, (45), 42050–42056. 10.1074/jbc.M107892200 [DOI] [PubMed] [Google Scholar]
- Morgenstern B., Michaelis M., Baer P. C., Doerr H. W., & Cinatl J. (2005). Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochemical and Biophysical Research Communications, (4), 905–908. 10.1016/j.bbrc.2004.11.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., & Olson A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, (16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar F., & Mukhtar N. (2020). Coronavirus (COVID-19): Let’s prevent not panic. Journal of Ayub Medical College Abbottabad, (1), 141–144. [PubMed] [Google Scholar]
- Numazaki K., Nagata N., Sato T., & Chiba S. (1994). Effect of glycyrrhizin, cyclosporin A, and tumor necrosis factor α on infection of U‐937 and MRC‐5 cells by human cytomegalovirus. Journal of Leukocyte Biology, (1), 24–28. 10.1002/jlb.55.1.24 [DOI] [PubMed] [Google Scholar]
- O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., & Hutchison G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, (1), 33 10.1021/ci200227u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M., & Rahman A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, (12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pollard H. B., Pollard B. S., & Pollard J. R. (2020). Classical drug digitoxin inhibits influenza cytokine storm, with implications for COVID-19 therapy. bioRxiv. 10.1101/2020.04.09.034983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanbaf A. (2020). Potential treatments for COVID-19; a narrative literature review. Archives of Academic Emergency Medicine, (1), e29. [PMC free article] [PubMed] [Google Scholar]
- Roggo S. (2007). Natural products in drug discovery. CHIMIA International Journal for Chemistry, (6), 312–312. 10.2533/chimia.2007.312 [DOI] [Google Scholar]
- Sander T., Freyss J., von Korff M., & Rufener C. (2015). DataWarrior: An open-source program for chemistry aware data visualization and analysis. Journal of Chemical Information and Modeling, (2), 460–473. 10.1021/ci500588j [DOI] [PubMed] [Google Scholar]
- Sato H., Goto W., Yamamura J., Kurokawa M., Kageyama S., Takahara T., Watanabe A., & Shiraki K. (1996). Therapeutic basis of glycyrrhizin on chronic hepatitis B. Antiviral Research, (2-3), 171–177. 10.1016/0166-3542(96)00942-4 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf A. W., & Van Aalten D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallographica Section D Biological Crystallography, (8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Sekhar T. (2020). Virtual screening based prediction of potential drugs for COVID-19. Preprints.org. 10.20944/preprints202002.0418.v2 [DOI]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., & Li F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, (7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugaraj B., Malla A., & Phoolcharoen W. (2020). Emergence of novel coronavirus 2019-nCoV: Need for rapid vaccine and biologics development. Pathogens, (2), 148 10.3390/pathogens9020148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayto R. H., Mrad R. A., & Sharara A. I. (2016). Use of rifaximin in gastrointestinal and liver diseases. World Journal of Gastroenterology, (29), 6638 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiri F., Pirhadi S., & Ghasemi J. B. (2019). Dynamic structure based pharmacophore modeling of the Acetylcholinesterase reveals several potential inhibitors. Journal of Biomolecular Structure & Dynamics, (7), 1800–1813. 10.1080/07391102.2018.1468281 [DOI] [PubMed] [Google Scholar]
- Shiri F., Pirhadi S., & Rahmani A. (2018). Identification of new potential HIV-1 reverse transcriptase inhibitors by QSAR modeling and structure-based virtual screening. Journal of Receptor and Signal Transduction Research, (1), 37–47. 10.1080/10799893.2017.1414844 [DOI] [PubMed] [Google Scholar]
- Siddell S. G., Anderson R., Cavanagh D., Fujiwara K., Klenk H. D., Macnaughton M. R., Pensaert M., Stohlman S. A., Sturman L., & van der Zeijst B. A. (1983). Coronaviridae. Intervirology, (4), 181–189. 10.1159/000149390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., & Xiang Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLOS Pathogens, (8), e1007236 p 10.1371/journal.ppat.1007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr, S., Costa, R., Sandmann, L., Westhaus, S., Pfaender, S., Anggakusuma, Dazert, E., Meuleman, P., Vondran, F. W. R., Manns, M., Steinmann, E., Hahn, T., & Ciesek, S. ( 2016). Host cell mTORC1 is required for HCV RNA replication. Gut, (12), 2017–2028. 10.1136/gutjnl-2014-308971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M. Y. F., Fraser J. E., Chan W. K. K., Moreland N. J., Rathore A. P., Wang C., Vasudevan S. G., & Jans D. A. (2013). Nuclear ocalization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENVserotypes by the inhibitor Ivermectin. Antiviral Research, (3), 301–306. 10.1016/j.antiviral.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Thakur A., & Raj P. (2017). Pharmacological perspective of Glycyrrhiza glabra Linn: A mini-review. Journal of Analytical & Pharmaceutical Research, (5), 00156 10.15406/japlr.2017.05.00156 [DOI] [Google Scholar]
- Tinku Gupta M. M. (2018). Ammonium glycyrrhizinate: a compherensive review of its traditionaluse, phytochemistry, pharmacology & safety. Asian Journal of Pharmaceutical Education and Research, (1), 58–65. [Google Scholar]
- Utsunomiya T., Kobayashi M., Herndon D. N., Pollard R. B., & Suzuki F. (1995). Glycyrrhizin(20β-carboxy-11-oxo-30-norolean-12-en-3β-yl-2-O-β-d-glucopyranuronosyl-α-d-glucopyranosiduronic acid) improves the resistance of thermally injured mice to opportunistic infection of herpes simplex virus type 1. Immunology Letters, (1), 59–66. 10.1016/0165-2478(94)00183-R [DOI] [PubMed] [Google Scholar]
- Utsunomiya T., Kobayashi M., Pollard R. B., & Suzuki F. (1997). Glycyrrhizin, an active component of licorice roots, reduces morbidity and mortality of mice infected with lethal doses of influenza virus. Antimicrobial Agents and Chemotherapy, (3), 551–556. 10.1128/AAC.41.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tilbeurgh H., Bezzine S., Cambillau C., Verger R., & Carriere F. (1999). Colipase: Structure and interaction with pancreatic lipase. Biochimica et Biophysica Acta (Bba) - Molecular and Cell Biology of Lipids, (2–3), 173–184. 10.1016/S1388-1981(99)00149-3 [DOI] [PubMed] [Google Scholar]
- Visualizer B. D. S. (2017). Version 4.5 (software). http://mayavi.sourceforge.net
- Wagstaff K. M., Sivakumaran H., Heaton S. M., Harrich D., & Jans D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. The Biochemical Journal, (3), 851–856. 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), (6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., & Zhang Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, (7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., & Hao P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, (3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., & Sun D. (2013). Macrocyclic drugs and synthetic methodologies toward macrocycles. Molecules (Basel, Switzerland), (6), 6230–6268. 10.3390/molecules18066230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., & Cheng F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, (1), 14–18. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, (8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]