Meiotic anaphase has been reported to be mediated by kinetochore-independent pushing forces in C. elegans. Danlasky et al. find that depletion of kinetochore proteins reduces pulling forces and prevents successful separation of homologous chromosomes during C. elegans meiosis.
Abstract
Anaphase chromosome movement is thought to be mediated by pulling forces generated by end-on attachment of microtubules to the outer face of kinetochores. However, it has been suggested that during C. elegans female meiosis, anaphase is mediated by a kinetochore-independent pushing mechanism with microtubules only attached to the inner face of segregating chromosomes. We found that the kinetochore proteins KNL-1 and KNL-3 are required for preanaphase chromosome stretching, suggesting a role in pulling forces. In the absence of KNL-1,3, pairs of homologous chromosomes did not separate and did not move toward a spindle pole. Instead, each homolog pair moved together with the same spindle pole during anaphase B spindle elongation. Two masses of chromatin thus ended up at opposite spindle poles, giving the appearance of successful anaphase.
Introduction
In eukaryotes, segregation of chromosomes during mitosis and meiosis is widely thought to require kinetochores, multiprotein structures that usually assemble at a single defined region of a chromosome called a centromere and that mediate binding to spindle microtubules. The outer kinetochore KMN network, composed of KNL-1, the MIS-12 complex, and the NDC-80 complex, is thought to directly mediate attachment of chromosomes to spindle microtubules (Cheeseman et al., 2006). End-on attachments of microtubule plus ends to NDC-80 complexes on the outer face of chromosome pairs is coupled with depolymerization of the plus ends to generate pulling forces that separate chromosomes from each other and move chromatids toward spindle poles. This results in a chromatid-to-pole movement called anaphase A (Inoué and Ritter, 1978; Vukušić et al., 2019). During anaphase B, the spindle lengthens and chromosomes move with the separating poles (Inoué and Ritter, 1978). Anaphase B is thought to be driven by outward pushing through a combination of anti-parallel sliding and microtubule polymerization between the separating chromosomes (Vukušić et al., 2019). Several observations have led to the idea that the outer faces of kinetochores are still attached to spindle microtubules during anaphase B. Bundles of microtubules called K-fibers extend from pole to kinetochore during anaphase B (Mastronarde et al., 1993; McDonald et al., 1992), and chromosome arms trail behind kinetochores during anaphase B (Inoué and Ritter, 1978). Cutting the spindle between a kinetochore and spindle pole sometimes caused the chromosome to transiently stop moving (Spurck et al., 1997). In addition, depletion of kinetochore proteins blocked anaphase B chromosome movement in the Caenorhabditis elegans embryonic mitotic spindle (Oegema et al., 2001).
Several observations in C. elegans oocyte meiotic spindles have suggested that different mechanisms are at work. (1) KNL-1, which is required for recruiting NDC-80 to the kinetochore, was reported to dissociate from chromosomes during anaphase and not be required for anaphase chromosome movement (Dumont et al., 2010). (2) Anaphase chromosome movement was not affected by laser ablation on the poleward side of chromosomes but was stopped by ablation between separating chromosomes, suggesting that microtubules push outward on the inner faces of separating chromosomes (Laband et al., 2017). (3) End-on microtubule attachments have not been observed on the poleward face of chromosomes. Only lateral microtubule attachments have been observed in metaphase and early anaphase, and end-on attachment to the inner face of chromosomes has been observed in late anaphase (Laband et al., 2017; McNally et al., 2006; Muscat et al., 2015; Redemann et al., 2018; Wignall and Villeneuve, 2009; Yu et al., 2019). (4) The kinetochore protein KNL-3 dissociated from chromosomes during anaphase, and KNL-3 depletion actually rescued anaphase in embryos depleted of the nuclear envelope and kinetochore protein MEL-28 (Hattersley et al., 2016). This latter result suggested that MEL-28–dependent removal of the KMN network might be important for anaphase.
In organisms with localized centromeres, the cohesin that holds sister centromeres together is protected from separase during anaphase I, ensuring that homologs separate during anaphase I but sisters remain together until anaphase II (Watanabe, 2005). Because C. elegans chromosomes do not have localized centromeres, they cannot use this strategy for ensuring homolog segregation before sister separation during meiosis. Instead, a single crossover generates a bivalent with two long arms and two short arms. Cohesion is protected on the long arms during anaphase I (de Carvalho et al., 2008), and conserved kinetochore proteins form large cup-shaped structures that envelop each long arm (Dumont et al., 2010; Howe et al., 2001; Monen et al., 2005). The aurora B kinase assembles along the short arms, forming a large “midbivalent ring” between the kinetochore cups (Dumont et al., 2010; Kaitna et al., 2002; Rogers et al., 2002; Wignall and Villeneuve, 2009). Aurora B kinase mediates separase cleavage of short-arm cohesin and homolog separation at anaphase I (Ferrandiz et al., 2018a; Rogers et al., 2002). At anaphase onset, there is no apparent kinetochore protein on the inner face of separating homologs, raising the question of how microtubules might be attached to the inner face of chromosomes as suggested by Laband et al. (2017). To address this question, we first analyzed the localization of KNL-1 and the MIS-12 subunit KNL-3 during anaphase in C. elegans female meiosis.
Results
Endogenously GFP-tagged KNL-1 and KNL-3 transition from cups to rings at the start of anaphase B
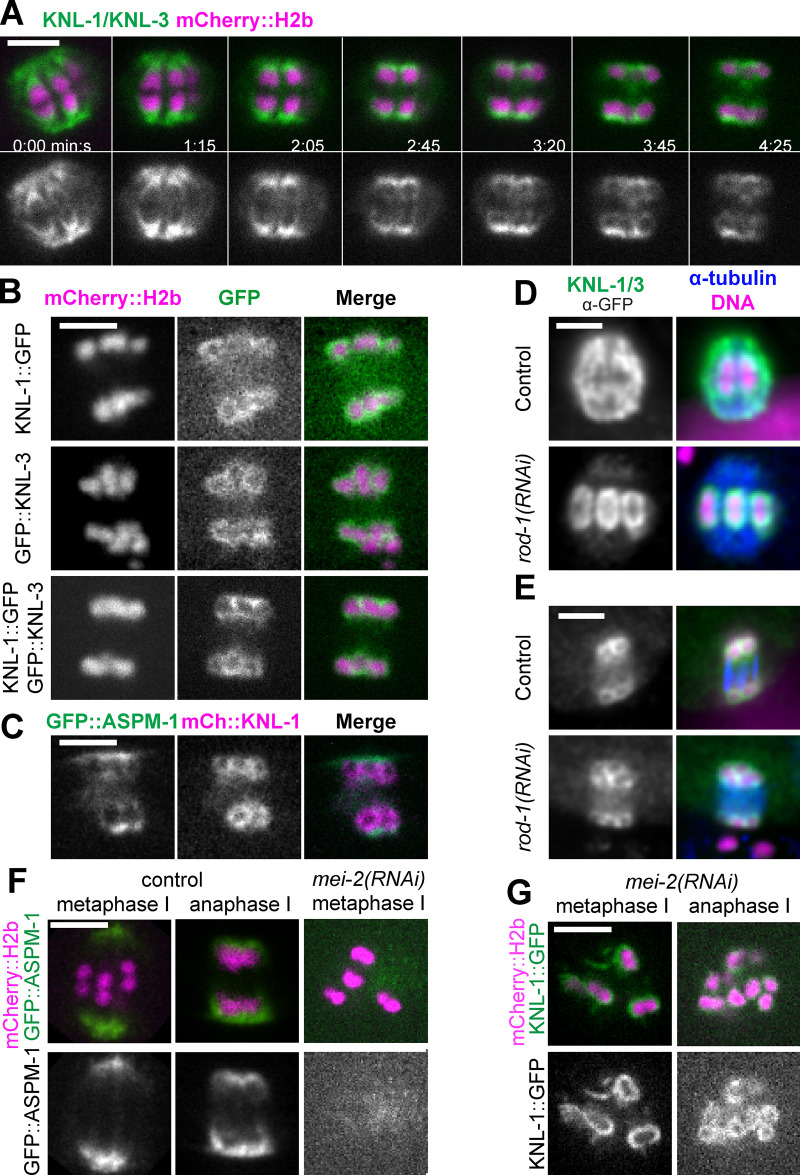
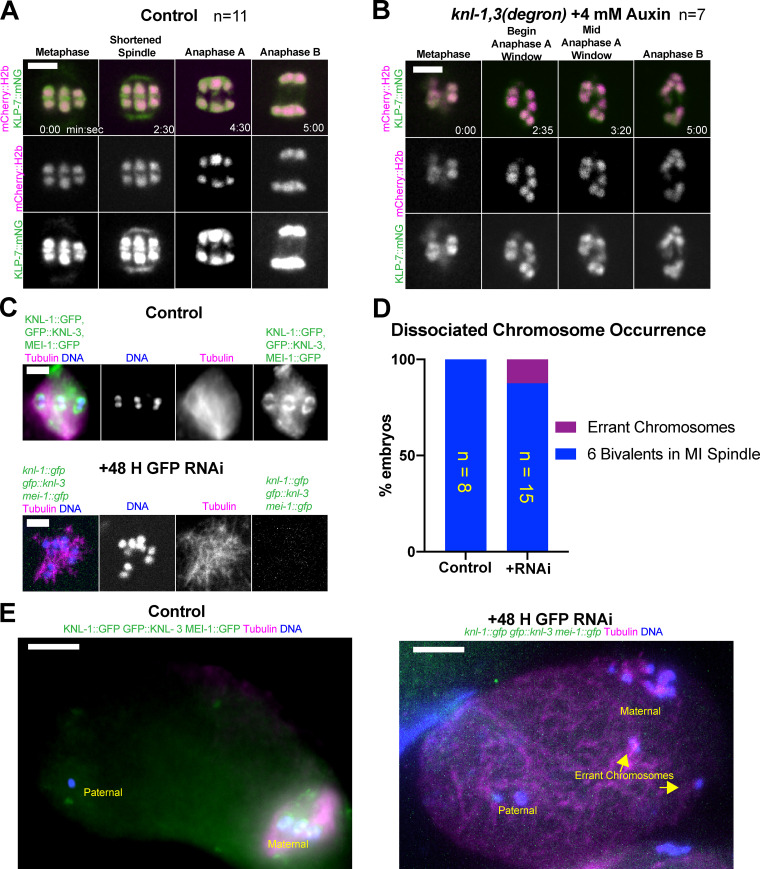
Live imaging of a C. elegans strain bearing in-frame GFP insertions at the endogenous knl-1 and knl-3 loci revealed cup-shaped structures enveloping the poleward face of mCherry::histone H2b–labeled chromosomes during metaphase and early anaphase (n = 10/10; Fig. 1 A, 0–2:05; Video 1). At the onset of anaphase B, defined as the switch from spindle shortening to spindle elongation, GFP-labeled KNL-1 and KNL-3 transitioned to enveloping all sides of each separating homolog so that they appeared as rings in optical sections (n = 10/10; Fig. 1 A, 2:45–4:25). This transition was previously observed by antibody staining of un-tagged KNL-1 (Monen et al., 2005). The fluorescence intensity of chromosome-associated KNL-1::GFP + GFP::KNL-3, measured as a unitless ratio to background fluorescence, decreased significantly between metaphase, 4.45 ± 0.68 min before initiation of spindle elongation, and the ring stage of anaphase B, 0.63 ± 0.11 min after initiation of spindle elongation (metaphase: mean 5.09 ± 0.71; n = 11; anaphase B: mean 1.96 ± 0.15; n = 11; P = 0.0003), whereas the fluorescence intensity of mCherry::H2b did not decrease significantly (metaphase: mean 2.80 ± 0.31; n = 11; anaphase B: mean 3.00 ± 0.36; n = 11; P = 0.70). This decrease in intensity might explain why KNL-1 and KNL-3 were previously described as dissociating from chromosomes during anaphase (Davis-Roca et al., 2017; Dumont et al., 2010; Hattersley et al., 2016). These rings also appeared at the onset of anaphase B in strains with only KNL-1::GFP or only GFP::KNL-3 (Fig. 1 B). The localization of kinetochore proteins on the inner face of separating homologs could provide attachment points for midzone microtubules during anaphase B. Consistent with this idea, KNL-1::mCherry rings were surrounded by GFP::ASPM-1, a microtubule minus-end binding protein (Fig. 1 C).
Figure 1.
Kinetochores transition from cups to rings at anaphase onset. (A–C) Single–focal plane time-lapse imaging of meiosis I embryos expressing the indicated fusions. (A) 0 min is mid spindle rotation. 1:15 is the beginning of homolog separation. (B) Individual localization of KNL-1::GFP and GFP::KNL-3 in late anaphase, during spindle elongation. (C) KNL-1::mCherry is surrounded by GFP::ASPM-1 at anaphase. (D and E) Fixed immunofluorescence images show that metaphase kinetochore cups (D) and anaphase kinetochore rings (E) remain on chromosomes after depletion of linear elements. (F and G) Time-lapse images reveal that KNL-1/3 still transition from cups to rings in the absence of ASPM-1–labeled spindle poles. Bars = 3 µm. mCh, mCherry.
Video 1.
Control meiosis I filmed in utero in a strain with endogenously GFP-tagged knl-1 and knl-3 genes and a transposon insertion expressing mKate::tubulin. Playback speed is six frames per second.
Kinetochore proteins are found in at least three pools during oocyte meiosis. In addition to cups or rings around chromosomes, KNL-1 and 3 accumulated on spindle poles just before spindle rotation (Fig. 1 A; 0:00; 10/10 spindles), as previously reported for ZWL-1 (McNally et al., 2016), and were found in ROD-1–dependent linear elements on the spindle and embryo cortex during metaphase I (Fig. 1 D; 11/11 control embryos: cups and linear elements; 13/13 rod-1(RNAi) embryos: cups but no linear elements), as previously reported (Dumont et al., 2010). KNL-1 and 3 still localized in rings around late anaphase homologs after ROD-1 depletion (Fig. 1 E; 11/11 controls had rings, and 12/12 rod-1(RNAi) had rings), indicating they are not just caused by occlusion of linear elements that have accumulated at spindle poles. In control spindles, chromosomes are surrounded by the spindle pole protein ASPM-1 during anaphase and occlude GFP::ASPM-1 fluorescence (Fig. 1 F). Thus, the anaphase rings of KNL-1/3 could simply result from occlusion of spindle pole KNL-1,3 by chromosomes. However, in mei-2(RNAi) spindles, which have no discrete spindle poles (Fig. 1 F; Connolly et al., 2014; McNally and McNally, 2011), GFP-tagged KNL-1,3 still transitioned from cups to rings (Fig. 1 G; 3/3 time-lapse sequences). These results suggest that the KNL rings are not just an optical artifact of chromosomes occluding spindle pole KNL-1,3 and that the inner face of these rings could provide microtubule attachment points during anaphase B, as further suggested by time-lapse images of mKate2::tubulin and GFP-tagged KNL-1,3 (Fig. 2 A; 3:10–5:30) and by EM results (Laband et al., 2017; Redemann et al., 2018).
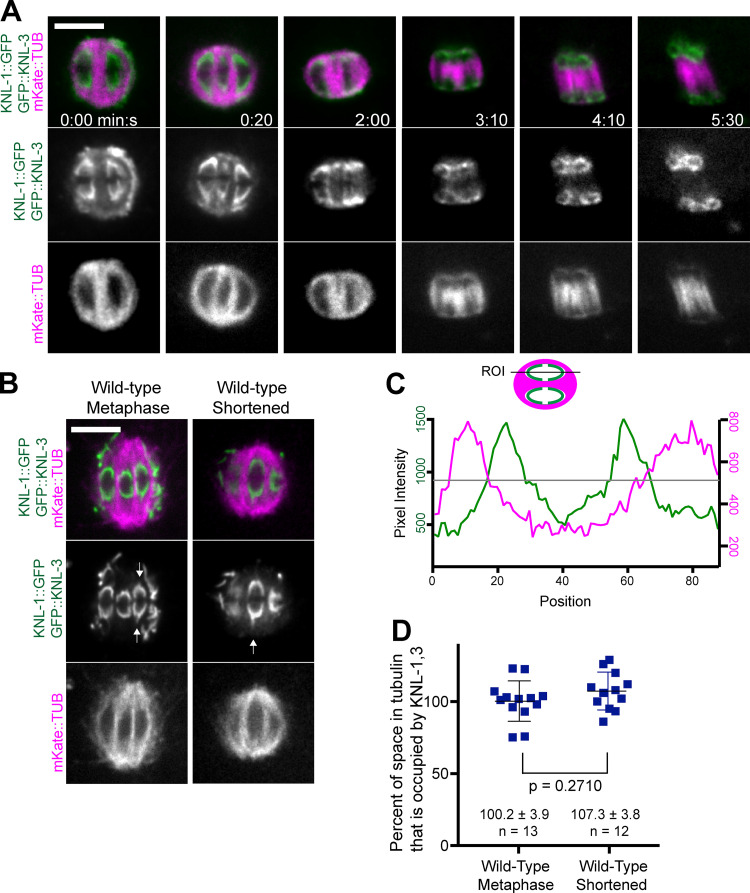
Figure 2.
Microtubule-free channels are filled with kinetochores. (A and B) Single–focal plane time-lapse sequences of embryos expressing KNL-1::GFP, GFP::KNL-3, and mKate::tubulin. Arrows indicate points of KNL-1,3 extending poleward. (C) Fluorescence intensity profile down the length of a shortened spindle just before anaphase onset. Gray line indicates the half-maximal intensity of mKate::tubulin. The inside edge of the microtubule-free channel was defined as the position of the inner red half-maximal intensity, and the outside edge of KNL was defined as the position of the outer green half-maximal intensity. (D) The length of KNL signal divided by the length of its corresponding microtubule-free channel. Values and error bars indicate mean ± SEM. Bars = 4 µm. ROI, region of interest; TUB, tubulin.
Microtubules are in close proximity to the poleward face of kinetochore cups
It has been suggested that C. elegans oocyte bivalents lack end-on microtubule attachments on their poleward face for two reasons. First, mCherry::H2b–labeled chromatin (McNally et al., 2006, 2016; Wignall and Villeneuve, 2009) and DAPI-stained chromatin (Davis-Roca et al., 2017; Muscat et al., 2015) reside in microtubule-free channels during metaphase and early anaphase. Second, microtubule ends were not observed within 250 nm of the poleward chromatin surface at metaphase or anaphase by EM (Redemann et al., 2018). The 250-nm distance was chosen because of EM observations of a ribosome-free layer around C. elegans chromosomes that has been interpreted as the kinetochore (Howe et al., 2001; O’Toole et al., 2003). Endogenously GFP-tagged KNL-1 and 3, however, extended much farther poleward than mCherry::H2b (Fig. 1 A; 0:00). Indeed, simultaneous live imaging of mKate::tubulin and GFP-tagged KNL-1,3 revealed that the KNL cups completely fill the microtubule-free channels (Fig. 2, B–D; and Video 1). In contrast, gaps are apparent between the poleward face of mCherry::H2b and spindle poles (Fig. 3 A). This observation raises the possibility that KNL-1,3 extend farther poleward than the ribosome-free zone. If microtubule plus ends are present in this farther-poleward volume, KNL-1–dependent NDC-80 (Dumont et al., 2010) might transiently engage with these plus ends as the spindle shortens. Alternatively, the microtubules on the poleward face of KNL-1,3 could generate pulling forces through lateral interactions with the KNL cups, as suggested by EM analysis (Redemann et al., 2018). The existence of pulling interactions is supported by the observation of points of GFP-tagged KNL-1,3 extending poleward from the tips of kinetochore cups (Fig. 2 B).
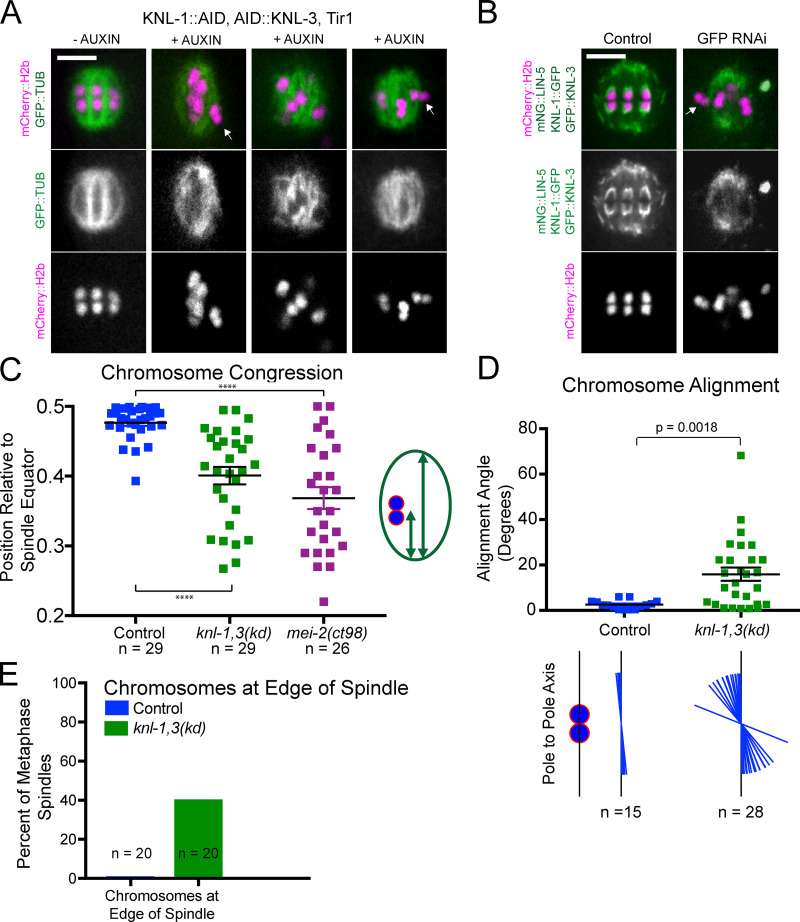
Figure 3.
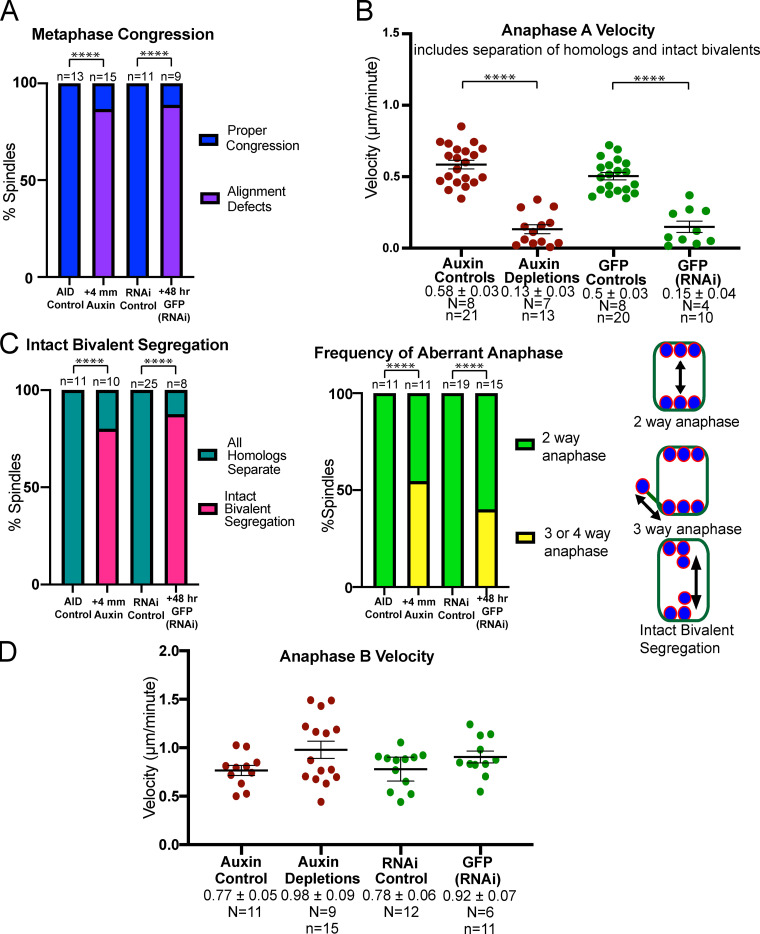
knl-1,3(kd) metaphase spindles have chromosome congression and alignment defects. (A and B) Single time points from time-lapse sequences of metaphase spindles depleted of KNL-1 and KNL-3 by auxin-induced degradation (A) or GFP(RNAi) (B). Arrows indicate chromosomes on periphery of spindle. The bright green body in the last image is that of a nearby sperm. (C) The relative position of each bivalent along the pole-to-pole axis of its spindle. A congressed bivalent has an expected ratio of 0.5. (D) The angle of individual chromosomes relative to the pole-to-pole axis was determined in five control and nine knl-1,3(kd) metaphase spindles. The alignment of each chromosome is illustrated below the graph. (E) The percentage of knl-1,3(kd) spindles with chromosomes on the outside edge of the spindle. Error bars indicate mean ± SEM. Bars = 4 µm. ****, P < 0.0001. mNG, mNeonGreen; TUB, tubulin.
KNL-1 and KNL-3 are required for metaphase chromosome congression and biorientation
Because Dumont et al. (2010) found that GFP::KNL-3 remained in kinetochore cups after knl-1(RNAi), we chose to double deplete KNL-1 and KNL-3 to more completely reduce kinetochore function during meiosis. Two previously described methods were used, auxin-induced degradation in strains with auxin-induced degrons (AIDs) appended to the endogenous knl-1 and knl-3 genes and GFP(RNAi) on strains with GFP appended to the endogenous knl-1 and knl-3 genes (Vargas et al., 2019). After 48-h RNAi, GFP fluorescence was not observed around chromosomes in living embryos (Fig. 3 B); however, staining of fixed embryos with a polyclonal anti-GFP antibody revealed faint puncta associated with meiotic chromosomes (Fig. S1, A and B). The anti-GFP staining intensity of these residual puncta, measured as a unitless ratio to background, was sixfold lower than that of the cup structures around control RNAi chromosomes (control RNAi: mean 12.6 ± 1.2; n = 44 embryos; GFP(RNAi): mean 1.9 ± 0.2; n = 16 embryos; P = 0.0001). Both auxin-induced degradation (Fig. 3 A) and GFP(RNAi) depletion of KNL-1 and KNL-3 (Fig. 3 B) resulted in failure to position bivalents at the metaphase plate. Because no significant differences were found between the two depletion methods (Fig. S2), phenotypic data were pooled and are referred to as knl-1,3(kd) for “knockdown.” In addition to failure of bivalents to position at a metaphase plate (Fig. 3 C), bivalents frequently failed to orient down the pole-to-pole axis of the spindle (Fig. 3 D). These two defects were previously reported for knl-1(RNAi) (Dumont et al., 2010). In addition, 40% of double-depleted metaphase spindles had at least one bivalent on the outside edge of the spindle (Fig. 3, A, B, and E). knl-1,3(kd) spindles frequently had two or more bivalents stacked end to end along the pole-to-pole axis of the spindle (Fig. 3 A) as previously described (Vargas et al., 2019). Finally, knl-1,3(kd) spindles did not have apparent microtubule-free channels oriented in the pole-to-pole axis (Fig. 3 A). This might indicate that intact kinetochore cups contribute to the organization of spindle microtubules. However, these congression defects were not due to a complete lack of spindle bipolarity as occurs in NDC-80–depleted mouse oocytes (Gui and Homer, 2013; Yoshida et al., 2020). Z-stacks of 16/18 knl-1,3(kd) live metaphase spindles labeled with GFP::ASPM-1 exhibited two spindle poles (Video 2). 2/18 z-stacks exhibited a smaller third pole. Instead, these congression defects might be due to a lack of poleward pulling forces on chromosomes.
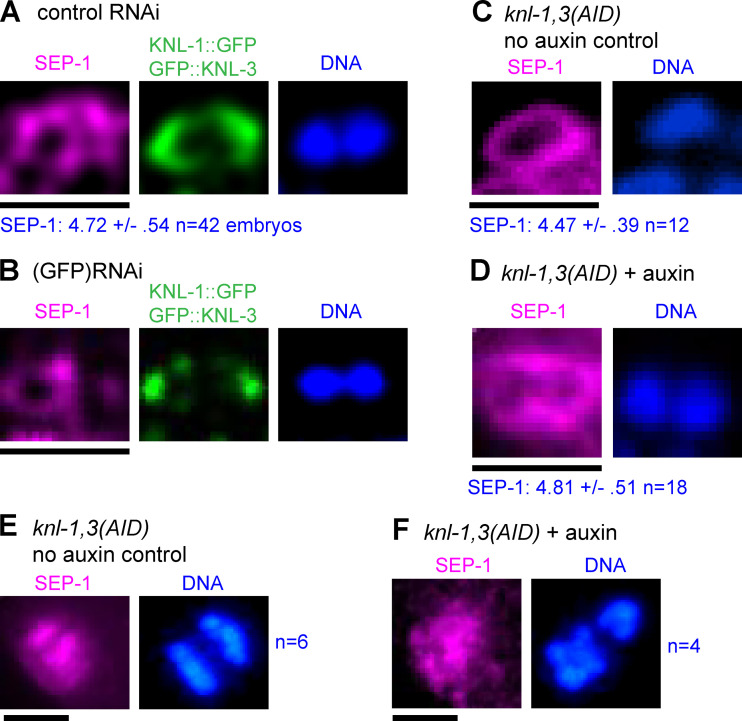
Figure S1.
Separase localizes normally after KNL-1,3 depletion. Meiotic embryos were fixed and stained with anti-separase (SEP-1), anti-GFP, and DAPI. (A–D) Single bivalents in metaphase I embryos of the indicated genotype. Values for SEP-1 are the mean pixel value of the brightest region around a bivalent ± SEM. Results of Student's t test: control RNAi versus GFP(RNAi), P = 0.037; and AID no auxin control versus AID + auxin, P = 0.71. (E and F) Early anaphase spindles with separase localized in between separating chromosomes. Bars = 2.6 µm.
Figure S2.
Comparison between auxin-inducible degron and GFP(RNAi) double depletions of endogenously tagged KNL-1 and KNL-3. (A) Frequency of congression defects observed in time-lapse sequences. Auxin versus GFP(RNAi), P = 1; Fisher’s exact test. (B) Rates of anaphase A chromosome separation during the time interval that anaphase A occurred in controls (−90 s from anaphase B onset). Auxin versus GFP(RNAi), P = 0.99; ANOVA. (C) Frequency of the anaphase defects depicted in the cartoon, determined from time-lapse sequences. Auxin versus GFP(RNAi) three- or four-way anaphase, P = 0.69; intact bivalent segregation, P = 1; Fisher’s exact test. (D) Anaphase B velocities determined from time-lapse sequences. Auxin versus GFP(RNAi), P = 0.87; ANOVA. Error bars and values are mean ± SEM. ****, P < 0.0001. N, number of embryos; n, number of chromosomes.
Video 2.
Demonstration of spindle bipolarity after KNL-1,3 double depletion. z-Stack of images taken at 1-µm steps through a KNL-1, KNL-3 double-depleted, 4 mm auxin, metaphase I spindle filmed in utero in a strain with auxin-induced degrons appended to the endogenous knl-1 and knl-3 genes and GFP appended to the endogenous aspm-1 gene and expressing TIR1, GFP::tubulin, and mCherry::histone H2b from transgenes.
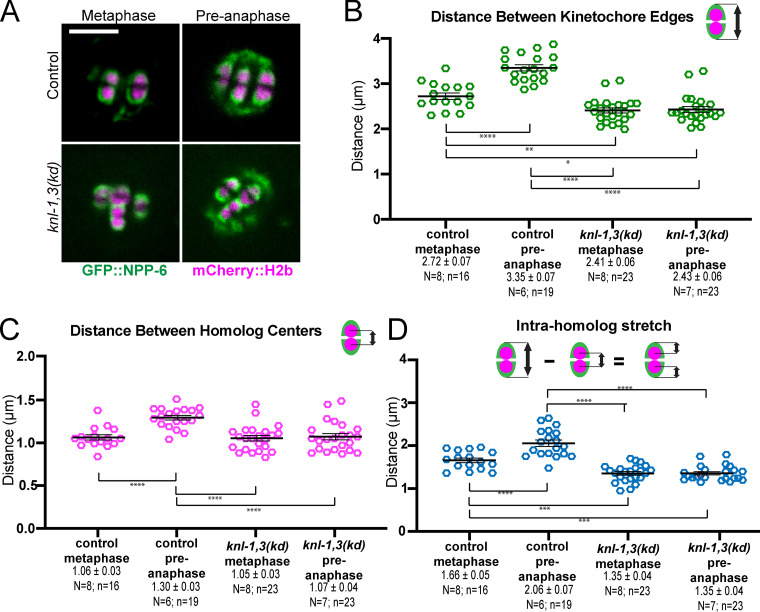
KNL-1 and KNL-3 are required for bivalent stretching
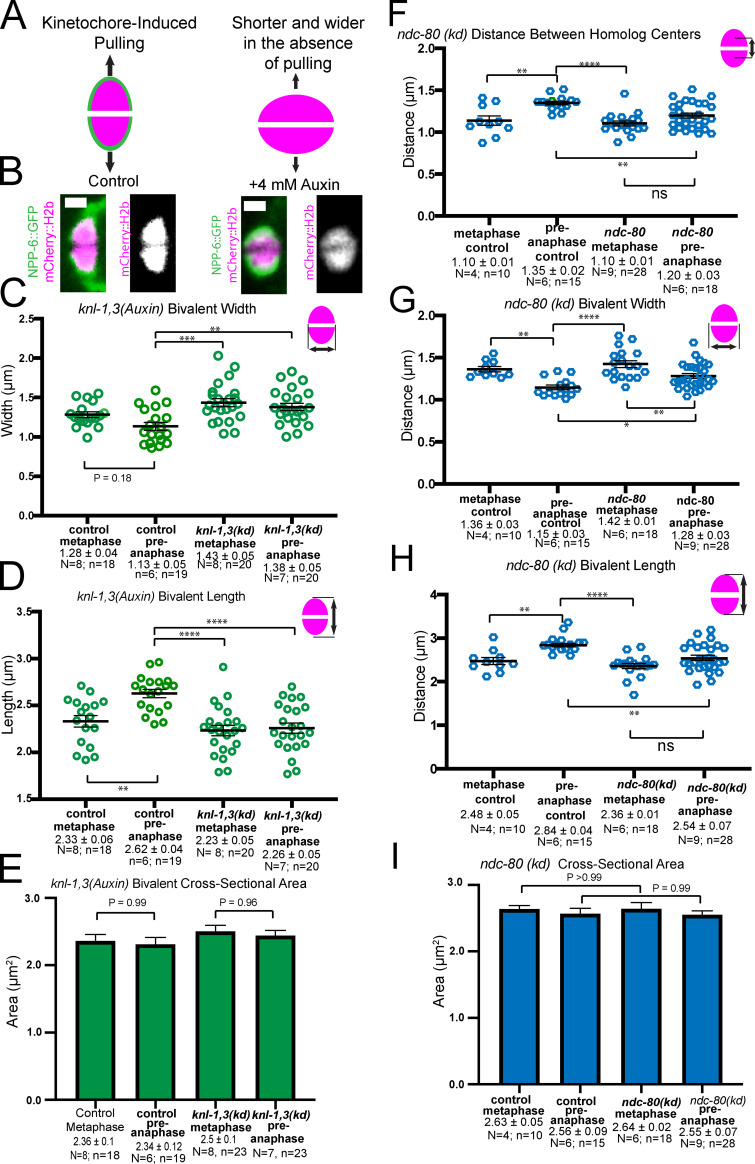
Poleward pulling forces on the two halves of a bivalent before cohesin cleavage should deform elastic chromatin, making bivalents longer and narrower. To specifically test for kinetochore-dependent chromosome stretching, we monitored bivalent length in control or knl-1,3(kd) embryos expressing mCherry::H2b and GFP::NPP-6, which labels kinetochore cups independently of KNL-1 and 3 (Hattersley et al., 2016; Fig. 4 A). We previously found that ZWL-1–labeled kinetochore cups stretch dramatically just before homolog separation (McNally et al., 2016). Because homolog separation is defective in knl-1,3(kd) embryos (see below), we measured the change in bivalent length between 5.7 ± 0.47 min and 2.2 ± 0.15 min before initiation of spindle elongation. Both inter-homolog (Fig. 4, B and C) and intra-homolog (Fig. 4 D) distances increased significantly between these time points (referred to as metaphase and preanaphase) in control embryos. In knl-1,3(kd) embryos, inter-homolog and intra-homolog distances did not increase between metaphase and preanaphase, and these distances were significantly smaller than in control embryos (Fig. 4, A–D; and Fig. S3, A–D). The increase in bivalent length between metaphase and preanaphase in control embryos might be due to ROD-1–dependent expansion of kinetochore volume (Pereira et al., 2018) or to stretching. However, the cross-sectional area of control bivalents did not change significantly at preanaphase when length increased (Fig. S3 E), and bivalent length still increased after rod-1(RNAi) (distance between homolog centers, metaphase: 1.15 ± 0.05 µm and preanaphase: 1.40 ± 0.05 µm; P = 0.0013). This result indicated that KNL-1 and 3 are required for stretching that would be indicative of preanaphase bipolar pulling forces on bivalents. Bivalent length (Fig. 4 B) and intra-homolog stretch (Fig. 4 D) were also significantly reduced in knl-1,3(kd) relative to controls at metaphase but to a lesser extent than at preanaphase. These results suggest that reduced pulling forces could contribute to the congression defect and that pulling forces increase just before homolog separation (2.2 ± 0.15 min before initiation of spindle elongation).
Figure 4.
Depletion of KNL-1 and KNL-3 results in loss of bivalent stretching. (A) Single frames from time-lapse sequences showing the stages when measurements were made. Metaphase: 5.7 min and preanaphase: 2.2 min before initiation of spindle elongation. (B) Distances between GFP::NPP-6–labeled kinetochore edges. (C) Distances between mCherry::H2b–labeled homolog centers. (D) Distances between GFP::NPP-6–labeled kinetochore edges and mCherry::H2b homolog centers, as a metric for intra-homolog stretching. Error bars and values are mean ± SEM. Bar = 4 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. N, number of embryos; n, number of bivalents.
Figure S3.
Bivalent stretching in KNL-1,3–depleted embryos and NDC-80–depleted embryos measured from mCherry::histone live images. (A) Diagram depicting increased length and decreased width expected from pulling. (B) Images of single bivalents taken from time-lapse sequences. (C) KNL-1,3–depleted bivalents are wider than controls due to decreased end-on pulling forces. (D) KNL-1,3–depleted bivalents are shorter than controls. (E) No significant difference in cross-sectional area between control and knl-1,3(kd) bivalents. (F) Distances between homolog centers in GFP::NDC-80 embryos treated with control or GFP(RNAi) indicate reduced stretching between metaphase and preanaphase. (G) Bivalent width decreases in NDC-80–depleted embryos, indicating residual stretching. (H) End-to-end bivalent length measurements indicate reduced stretching after NDC-80 depletion. (I) Cross-sectional area does not change in control or NDC-80 depletion. Bar = 1 µm. Error bars and values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ANOVA. N, number of embryos; n, number of bivalents.
KNL-1 and KNL-3 are required for anaphase A and homolog separation
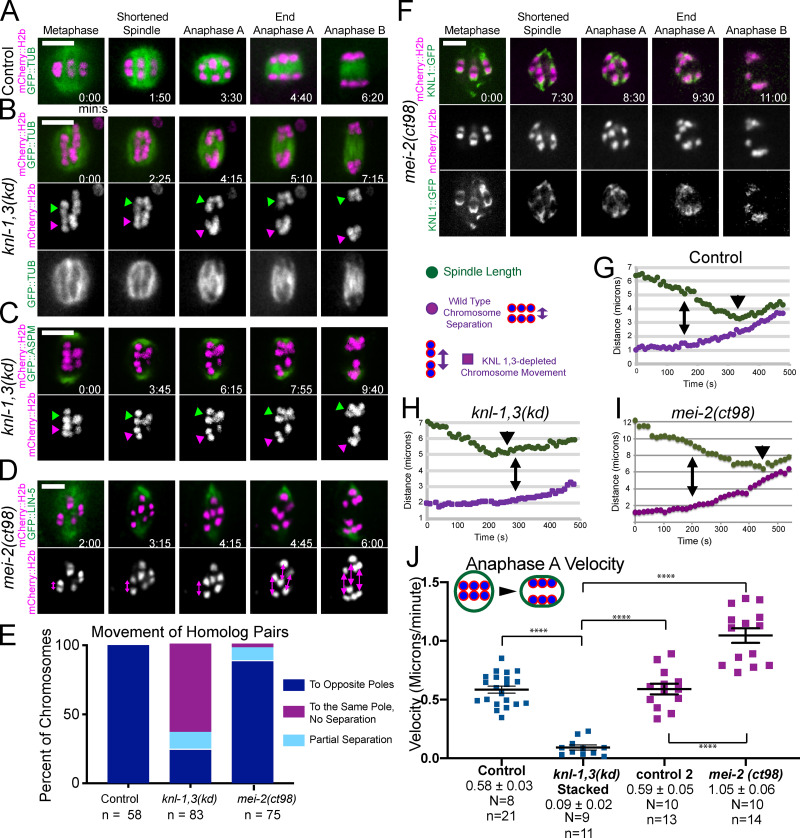
In control time-lapse sequences, all bivalents split into two homologs that moved to opposite spindle poles before the initiation of spindle elongation (Fig. 5, A and E; and Video 3). In knl-1,3(kd) embryos, 60% of bivalents did not split into two homologs before the initiation of spindle elongation and instead moved intact with one spindle pole during spindle elongation (Fig. 5, B, C, and E; Video 4; and Video 5). 19 bivalents in 14 embryos that segregated intact with one spindle pole eventually separated at heterogeneous times during spindle elongation (1.12 ± 0.91 min; range, 0.5–3.2 min after initiation of spindle elongation; Video 6). Only five bivalents remained intact through the end of the time-lapse sequence. As judged by size, no bivalents were observed in 11/11 metaphase II spindles, and both sister chromatids of 14/23 metaphase II univalents ended up at the same end of the anaphase II spindle. The delay in bivalents splitting into two homologs is not entirely an indirect consequence of the preceding congression defect since bivalents separated into two homologs in a mei-2(ct98) mutant that also had a severe congression defect (Fig. 3 C; and Fig. 5, D and E) but had intact kinetochore cups (Fig. 5 F). The congression defect could contribute, however, as most of the bivalents segregating intact were either stacked end to end at metaphase or were close to one pole at the onset of anaphase B. Separase localization on metaphase chromosomes and between anaphase chromosomes appeared normal in KNL-1,3–depleted spindles (Fig. S1), although a delay in relocalization might not be detected. AIR-2, which is required for cleavage of cohesin by separase (Ferrandiz et al., 2018b; Rogers et al., 2002), also localized normally between homologs at metaphase (Fig. S4, A and B). AIR-2 also dissociated from chromosomes and associated with midzone microtubules during anaphase in knl-1,3(kd) spindles as it did in controls (Fig. S4, A and B).
Figure 5.
KNL-1 and KNL-3 are required for anaphase A chromosome segregation. (A–D) Representative time-lapse sequences of control (A), knl-1,3(kd) (B and C), and mei-2(ct98) (D) embryos. Arrowheads in B and C indicate intact bivalents segregating. Double arrows in D indicate homolog separation. (E) The fate of individual homolog pairs. (F) KNL-1 localizes properly to cups and rings in mei-2(ct98). (G–I) Representative plots of spindle length and distance between homolog centers over time. For knl-1,3(kd) embryos, distance between separating intact bivalents is shown. Time 0 is initiation of spindle shortening. Double arrows indicate the initiation of homolog separation in G and I and initiation of bivalent separation in H. Arrowheads indicate initiation of spindle elongation. (J) Homolog separation velocities during the 90 s before initiation of spindle elongation. For knl-1,3(kd) embryos, velocities shown are between midbivalents of intact bivalents that later moved apart during anaphase B. Error bars and values are mean ± SEM. Bars = 4 µm. ****, P < 0.0001. N, number of embryos; n, number of chromosomes; TUB, tubulin.
Video 3.
Control, no auxin, meiosis I filmed in utero in a strain with auxin-induced degrons appended to the endogenous knl-1 and knl-3 genes and expressing TIR1, GFP::tubulin, and mCherry::histone H2b from transgenes. Playback speed is six frames per second.
Video 4.
KNL-1, KNL-3 double depletion, 4 mm auxin, meiosis I filmed in utero in a strain with auxin-induced degrons appended to the endogenous knl-1 and knl-3 genes and expressing TIR1, GFP::tubulin, and mCherry::histone H2b from transgenes. Playback speed is six frames per second.
Video 5.
KNL-1, KNL-3 double depletion, 4 mm auxin, meiosis I filmed in utero in a strain with auxin-induced degrons appended to the endogenous knl-1 and knl-3 genes, endogenously GFP-tagged aspm-1 gene, and expressing TIR1 and mCherry::histone H2b from transgenes. Playback speed is six frames per second.
Video 6.
Delayed separation of homologs after KNL-1, KNL-3 double depletion. Meiosis I filmed in utero in a 4-mM auxin–treated strain with auxin-induced degrons appended to the endogenous knl-1 and knl-3 genes and expressing TIR1, GFP::tubulin, and mCherry::histone H2b from transgenes. Left arrow: bivalent splits and the two homologs segregate to opposite poles. Upper right arrow: bivalent splits after segregating intact to one pole. Lower right arrow: bivalent segregates intact to one pole and remains intact at the end of the time-lapse sequence. Playback speed is six frames per second.
Figure S4.
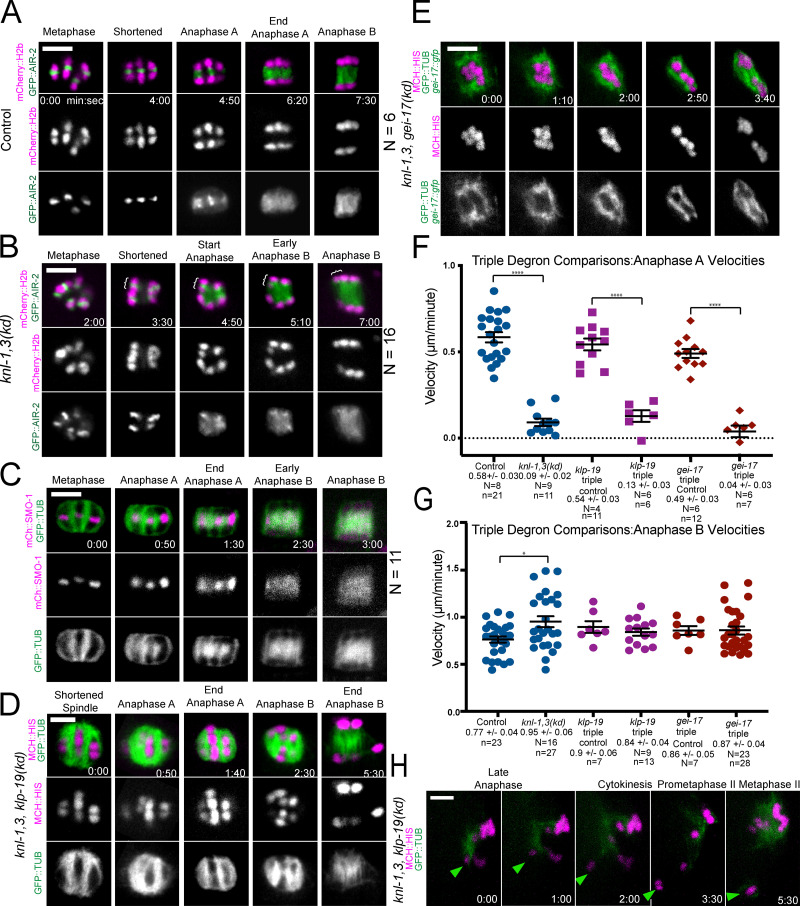
AIR-2 localizes normally in knl-1,3(kd) bivalents. AID triple depletions of KNL-1,3 and chromokinesin KLP-19 or SUMO E3 ligase GEI-17. (A–C) Representative time-lapse sequences of GFP-tagged AIR-2 (A and B) or mCherry-tagged SUMO (SMO-1; C) in control (A and C) or knl-3(kd) (B) embryos. Brackets in B indicate an intact bivalent segregating. (D) Time-lapse images of a spindle following overnight auxin-induced degradation of KNL-1,3, and KLP-19. (E) Time-lapse images of a spindle following overnight auxin-induced degradation of KNL-1,3, and GEI-17 displaying an abnormal disorganized structure. (F and G) No significant difference is observed in anaphase A or B velocities for KNL-1,3 double depletion or either triple depletion. (H) In 2/14 live time-lapse sequences, an intact bivalent (arrowhead) escaped the spindle in KNL-1,3 and KLP-19–depleted spindles during late anaphase I. It did not reincorporate into the MII spindle. Error bars and values are mean ± SEM. *, P < 0.05; ****, P < 0.0001. Bars = 3 µm. N, number of embryos; n, number of chromosomes. mCh MCH, mCherry; TUB, tubulin.
In control spindles, anaphase occurred in three phases. (1) Spindles first shortened in the pole-to-pole axis. (2) Chromosomes then separated a short distance as the spindle continued to shorten (anaphase A). (3) Spindles abruptly transitioned to lengthening (anaphase B), during which chromosomes moved at the same speed as the separating spindle poles (Fig. 5, A and G; McNally et al., 2016). In knl-1,3(kd) spindles, spindle shortening occurred at wild-type velocities (control: −1.03 ± 0.06 µm/min; n = 22; knl-1,3(kd): −0.98 ± 0.06 µm/min; n = 18), but chromosomes only began to move apart when spindle elongation started (Fig. 5, B, C, and H). This failure of anaphase A was not due only to the preceding congression defect, as anaphase A occurred in mei-2(ct98) spindles, which also have a congression defect (Fig. 3 C; and Fig. 5, D and I).
To more carefully document anaphase A velocities, we measured the velocity of homolog separation during the 90 s before the initiation of spindle elongation because anaphase A occurs during this period in control spindles (Fig. 5 G). Chromosome movement was greatly reduced during this period in knl-1,3(kd) spindles. These measurements included both distances between homologs that later segregated to opposite poles (Fig. S2 B) and distances between stacked bivalents that later separated from each other intact (Fig. 5 J). In contrast, anaphase A velocities were significantly faster in mei-2(ct98) spindles than in control spindles (Fig. 5 J). These results suggest that KNL-1 and 3 are required for pulling forces that drive anaphase A while the spindle is still shortening.
Anaphase B occurs in KNL-1,3–depleted spindles but is highly error prone
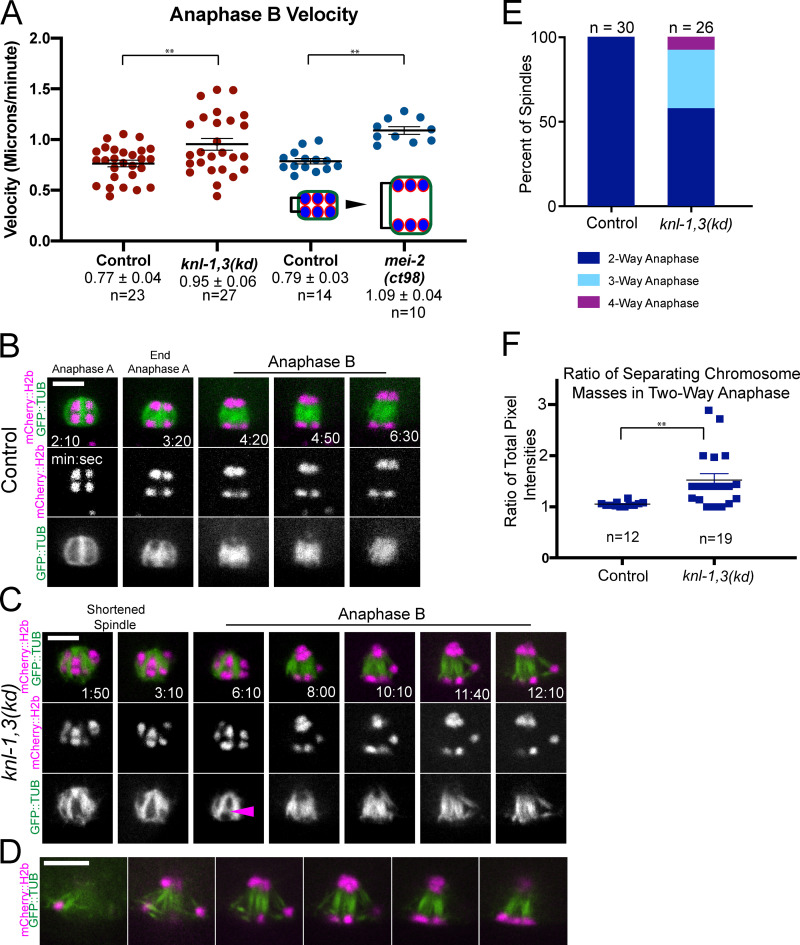
The majority of chromosomes in knl-1,3(kd) spindles moved at the same velocity as the separating spindle poles, and this anaphase B velocity was significantly faster than controls (Fig. 6 A and Fig. S2 D). GFP::tubulin fluorescence appeared between separating chromosomes in both control (Fig. 6 B) and knl-1,3(kd) spindles (Fig. 6 C, arrowhead), consistent with fibers pushing chromosomes apart. However, anaphase B was highly error prone in knl-1,3(kd) spindles. One chromosome frequently moved in a direction nearly perpendicular to the two major separating masses of chromosomes, with bundles of microtubules lengthening between each major mass and the errant chromosome (Fig. 6 C, 10:10–12:10; Fig. 6, D and E; and Fig. S2 C). The observations of intact bivalents moving apart (Fig. 5, C–E) and microtubule bundles lengthening in three-way anaphases (Fig. 6 C) suggested that microtubule bundles push any two chromosome masses apart in knl-1,3(kd) spindles. The result of this promiscuous pushing between intact bivalents was that the total mass of chromatin in the two separated masses of chromosomes, measured as the 3D sum of mCherry::H2b pixel values, was unequal in knl-1,3(kd) spindles at the end of anaphase B (Fig. 6 F).
Figure 6.
Aberrant anaphase B occurs in knl-1,3(kd) embryos. (A) Velocities of chromosome separation during spindle elongation. (B and C) Representative time-lapse sequences of anaphase B. Arrowhead in C indicates tubulin appearing between separating chromosomes. 8:00–12:10 min:sec in C shows an example of “three-way anaphase” in which a chromosome moved perpendicular to the spindle axis while attached to the spindle by two microtubule bundles. Arrowhead indicates microtubules between separating chromosomes. (D) z-Stack of a single time point in a knl-1,3(kd) embryo shows a “four-way anaphase.” (E) Frequencies of aberrant anaphases. (F) Fluorescence intensity of separated chromosome masses at the end of anaphase B. Error bars and values are mean ± SEM. **, P < 0.01. Bars = 4 µm. TUB, tubulin.
KNL-1,3 surround late-lagging univalents as they are stretched
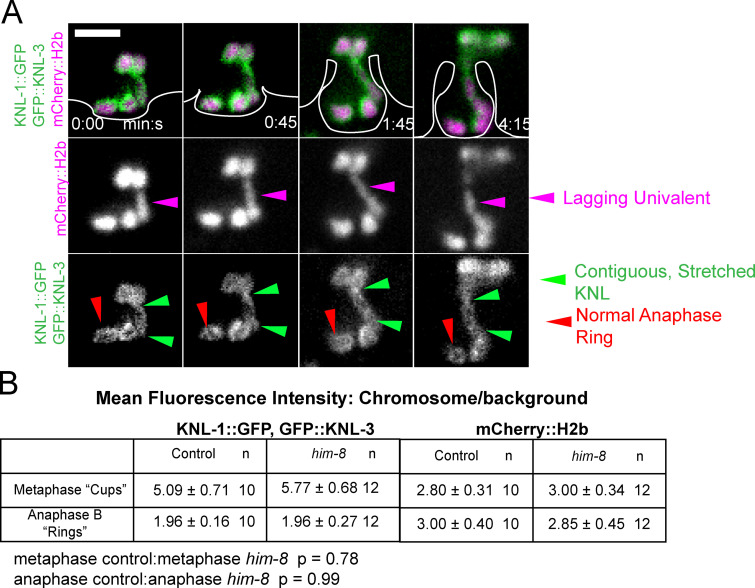
Univalent X chromosomes, resulting from crossover failure in a him-8 mutant, do not lose cohesion, frequently lag during late anaphase, and appear to be stretched as they are captured by the ingressing polar body membrane (Cortes et al., 2015; Fig. 7 A). The explanation for stretching has been unclear because chromosome stretching is associated with bipolar pulling forces and because laser cutting experiments suggested that pushing forces predominate in late anaphase (Laband et al., 2017). In 9/9 him-8 anaphase I spindles with a late-lagging univalent, GFP-tagged KNL-1,3 enveloped the univalent as it stretched (Fig. 7 A and Video 7). Because KNL-1,3 are required for metaphase chromosome stretching (Fig. 4), this result suggested that KNL-1,3 could still be exerting pulling forces during late anaphase. The naturally univalent X chromosome in spermatocytes is also stretched during anaphase and is associated with both end-on and lateral microtubule attachments to kinetochores (Fabig et al., 2020). Davis-Roca et al. (2017) reported that a variety of conditions causing meiotic errors result in a delay in the dissociation of KNL-3 from anaphase chromosomes. However, we observed no difference between control and him-8 embryos in the decrease of KNL-1::GFP + GFP::KNL-3 between metaphase and anaphase (Fig. 7 B).
Figure 7.
Univalent X chromosomes in a him-8 mutant are coated in KNL-1,3 as they are stretched during late anaphase. (A) Representative time-lapse sequence of late anaphase in a him-8 mutant. Polar body ingression, which is only visible with enhanced contrast, has been drawn for clarity. (B) The decrease in fluorescence intensity of KNL-1::GFP, GFP::KNL-3 between metaphase and anaphase is not significantly different between control and him-8. Values are mean ± SEM. Bar = 3 µm.
Video 7.
Lagging univalent X chromosome segregation during meiosis I filmed in utero in a him-8 strain with endogenously GFP-tagged knl-1 and knl-3 genes and a transposon insertion expressing mCherry::histone H2b. Playback speed is six frames per second. mNG, mNeonGreen.
NDC-80 is required for KNL-1,3–dependent anaphase A homolog separation but not anaphase A chromosome-to-pole movement
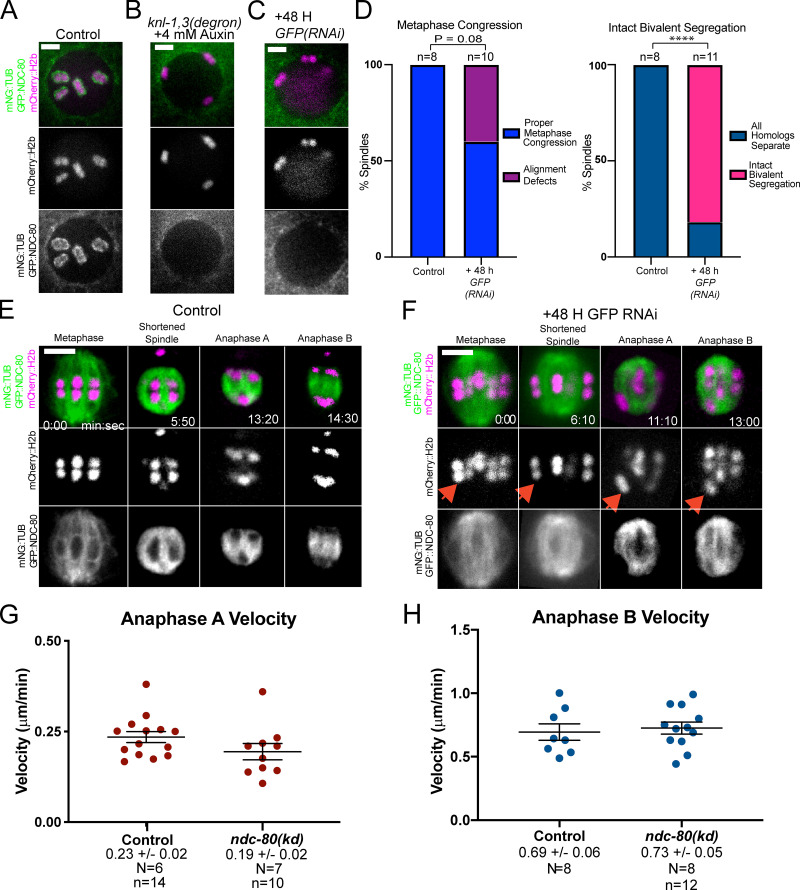
NDC-80 and RZZ-dependent dynein are the most obvious candidates for KNL-1,3–dependent poleward force generators at kinetochores because of their in vitro activities and because both NDC-80 and the RZZ component ZWL-1 require KNL-1 for association with kinetochore cups (Dumont et al., 2010). To test whether the phenotypes of knl-1,3(kd) spindles could be caused only by loss of NDC-80, we constructed a strain with a GFP tag at the endogenous ndc-80 gene that also expressed mNeonGreen-tagged tubulin and mCherry::H2b and that also contained the KNL-1 and KNL-3 degrons. Auxin treatment indicated that association of endogenously GFP-tagged NDC-80 is dependent on KNL-1,3 (Fig. 8, A and B; n = 6/6 −1 oocytes). We then subjected the ndc-80(GFP) worms to GFP(RNAi). This treatment resulted in a complete loss of NDC-80 from kinetochore cups (Fig. 8 C; n = 14/15 −1 oocytes). Metaphase congression defects were observed in a significantly lower fraction of NDC-80–depleted embryos than in knl-1,3(kd) embryos (Fig. 8 D vs. Fig. 3 C; 21/24 spindles in knl-1,3(kd) versus 4/10 ndc-80 spindles; Fisher’s exact test; P = 0.008). Bivalent stretching in NDC-80–depleted embryos was intermediate between that of control and knl-1,3(kd) embryos (Fig. S3, F–I). Similar to knl-1,3(kd) embryos (Fig. 5 E), intact bivalents were observed moving apart from each other at high frequency in NDC-80–depleted embryos (Fig. 8, D–F; Video 8; and Video 9). In contrast with intact bivalents in knl-1,3(kd) embryos, which exhibited no anaphase A–like chromosome movement (Fig. 5, H and J), intact bivalents in NDC-80–depleted spindles moved toward one pole at wild-type velocities (Fig. 8 F, arrows; and Fig. 8 G). Anaphase B velocity was the same between control, NDC-80–depleted (Fig. 8 H), and knl-1,3(kd) embryos (Fig. 6). The weaker phenotypes of NDC-80 depletion suggested that either KNL-1,3 have some NDC-80–independent function or that residual amounts of NDC-80, which were not detectable by GFP fluorescence (Fig. 8 C), remain after GFP(RNAi) but not after knl-1,3(kd).
Figure 8.
NDC-80 depletion results in intact bivalents moving poleward at wild-type anaphase A velocity. (A–C) Maximum intensity projections of −1 oocytes. GFP::NDC-80 was not detected on bivalents in knl-1,3(kd) (B) or GFP(RNAi) oocytes (C). (D) Frequency of congression and anaphase defects in control and NDC-80–depleted embryos from time-lapse sequences. ****, P < 0.0001. (E and F) Representative time-lapse sequences of control (E) and NDC-80–depleted (F) embryos. Arrows indicate an intact bivalent moving poleward. (G) Chromatid-to-pole velocities in control and NDC-80–depleted embryos. Note these control velocities are one half the homolog separation velocities reported in Fig. 5. (H) Anaphase B velocities of control and NDC-80–depleted embryos. Error bars and values are mean ± SEM. Bars = 3 µm. N, number of embryos; n, number of chromosomes. mNG, mNeonGreen; TUB, tubulin.
Video 8.
RNAi control meiosis I filmed in utero in a strain with endogenously GFP-tagged ndc-80 gene and a transposon insertion expressing mNeonGreen::tubulin and mCherry::histone H2b. Playback speed is six frames per second. mNG, mNeonGreen.
Video 9.
NDC-80 depletion, via GFP(RNAi), meiosis I filmed in utero in a strain with endogenously GFP-tagged ndc-80 gene and a transposon insertion expressing mNeonGreen::tubulin and mCherry::histone H2b. Playback speed is six frames per second.
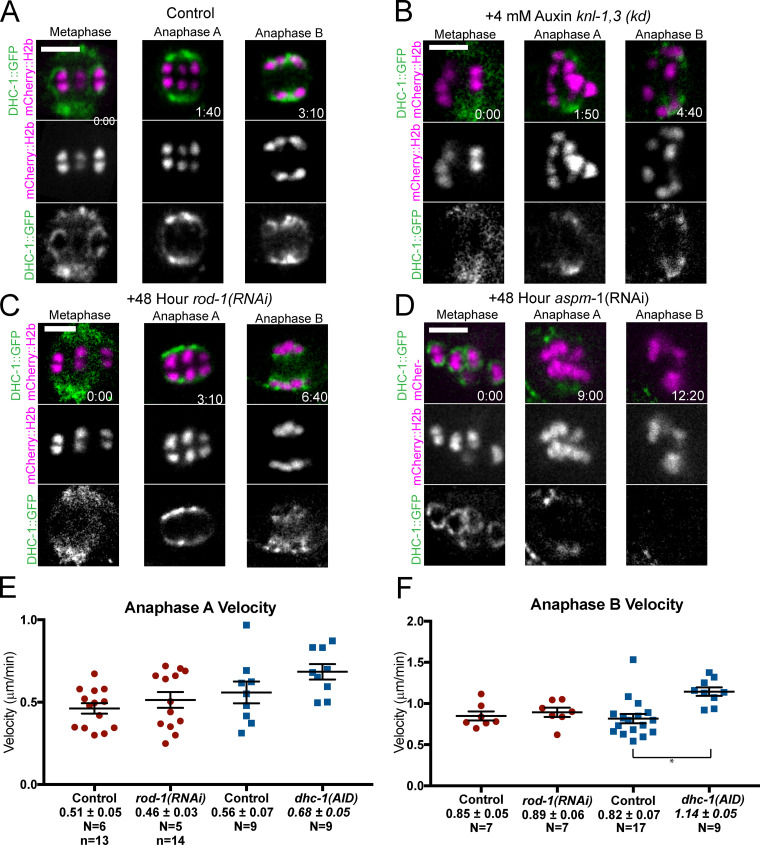
Cytoplasmic dynein is not associated with anaphase chromosomes, is not required for anaphase A, and restrains anaphase B
Using endogenously GFP-tagged dynein heavy chain, we first found that dynein was associated with metaphase I kinetochore cups in time-lapse sequences of 8/10 control embryos (Fig. 9 A). Previous studies of C. elegans mitosis (Gassmann et al., 2008) or meiosis (Muscat et al., 2015) found kinetochore dynein only in experimentally induced monopolar spindles. Our robust detection in control embryos allowed us to demonstrate that association of dynein with metaphase kinetochore cups was dependent on KNL-1,3 (Fig. 9, A and B) and on ROD-1 (Fig. 9 C; mean background subtracted pixel values at bivalents: control, 135 ± 24; n = 10; knl-1,3(kd), 32 ± 11; n = 10; rod-1(RNAi), 16 ± 8; n = 8). During anaphase A, kinetochore dynein merges with spindle pole dynein (Fig. 9 A), and spindle pole dynein is ASPM-1 dependent (van der Voet et al., 2009). To test whether DHC-1 is associated with chromosomes during anaphase B, we imaged endogenously tagged DHC-1 in aspm-1(RNAi) spindles. In aspm-1(RNAi) embryos, DHC-1 localized in kinetochore cups during metaphase (8/10 embryos) but completely dissociated from chromosomes before anaphase A (Fig. 9 D; n = 5/5 embryos, mean background subtracted pixel values at aspm-1(RNAi) chromosomes: metaphase, 152 ± 34; n = 10; anaphase, 30 ± 11; n = 5). These results indicated that KNL-1,3 and ROD-1 target dynein to kinetochore cups during metaphase but that dynein near chromosomes during anaphase A is not directly associated with chromosomes and is instead associated with ASPM-1 and microtubules that surround the chromosomes.
Figure 9.
Dynein is not present on anaphase chromosomes, and auxin-induced degradation of dynein does not slow anaphase. (A–D) Representative time-lapse images of endogenously GFP-tagged DHC-1. (A) In control embryos, DHC-1 is in kinetochore cups at metaphase and spindle poles during anaphase A and overlaps with poles and chromosomes during anaphase B. (B and C) Neither knl-1,3(kd) bivalents (B) nor rod-1(RNAi) bivalents (C) exhibit DHC-1 on kinetochore cups at metaphase. (D) aspm-1(RNAi) embryos retain DHC-1 on kinetochore cups at metaphase but have no DHC-1 on poles or anaphase chromosomes. (E and F) Auxin-induced degradation of DHC-1 does not slow anaphase A or B. *, P < 0.05. Error bars and values are mean ± SEM. Bars = 3 µm. N, number of embryos; n, number of chromosomes.
To address the role of dynein in anaphase chromosome movement, we monitored anaphase A velocities after 1 h of auxin-induced degradation of DHC-1 (Zhang et al., 2015) and after rod-1(RNAi). DHC-1::degron + auxin spindles failed to rotate and were long with pointed poles as previously reported for partial RNAi depletion (Crowder et al., 2015; Ellefson and McNally, 2009), but anaphase A velocities were not significantly slower than controls in DHC-1::AID + auxin or in rod-1(RNAi) spindles (Fig. 9 E). Anaphase B velocities were significantly faster in DHC-1::AID + auxin but not in rod-1(RNAi) (Fig. 9 F). These results are consistent with previously reported wild-type or faster than wild-type anaphase velocities in dhc-1(RNAi) (McNally et al., 2016), dhc-1(ts), dnc-6(null), zwl-1(RNAi), and spdl-1(RNAi) (Laband et al., 2017). These results indicate that chromosomal dynein does not play a role in anaphase A or B but that nonchromosomal dynein slows spindle elongation during anaphase B. We were unable to test for redundancy between NDC-80 and dynein because endogenously tagged NDC-80::GFP was not completely depleted in worms treated with both GFP(RNAi) and rod-1(RNAi). However, because DHC-1 is not associated with chromosomes during anaphase, it is not likely to be an anaphase force generator at chromosomes.
Robust mechanisms mediate attachment of chromosomes to microtubules
Kinetochores are widely thought to mediate attachment of chromosomes to microtubules, but chromosomes were never observed to completely dissociate from spindle microtubules in knl-1,3(kd) spindles. This attachment might be mediated by a small residual number of molecules of KMN network proteins or by KNL-1,3–independent microtubule-binding proteins. Microtubule-binding proteins known to associate with chromosomes in the absence of KNL-1 during metaphase include the midbivalent ring components KLP-19, HCP-1,2, and CLS-2 (Dumont et al., 2010). The SUMO ligase, GEI-17, and SUMO are required for recruitment of KLP-19 and BUB-1 to the midbivalent ring (Pelisch et al., 2019, 2017). Midbivalent rings elongate in microtubule-free channels between separating homologs during early anaphase, and then some components transfer to microtubules in late anaphase (Fig. S4, A and C; Davis-Roca et al., 2018; Dumont et al., 2010; Laband et al., 2017; Muscat et al., 2015; Pelisch et al., 2019). Meiosis in auxin-treated knl-1, knl-3, klp-19, and knl-1, knl-3, gei-17 triple-degron strains proceeded essentially identically to knl-1,3(kd) meiosis (Fig. S4, D–G; and Video 10). No chromosomes completely detached from the spindle in 21 GEI-17 triple-depleted meioses and in only 2/14 KLP-19 triple-depleted meioses, a chromosome that moved perpendicular to the long axis of the spindle completely detached during late anaphase I and was not incorporated into the metaphase II spindle (Fig. S4 H).
Video 10.
KNL-1, KNL-3, GEI-17 triple depletion, 4 mm auxin, meiosis I filmed in utero in a strain with auxin-induced degrons appended to the endogenous knl-1, knl-3, and gei-17 genes and expressing TIR1, GFP::tubulin, and mCherry::histone H2b from transgenes. Playback speed is six frames per second.
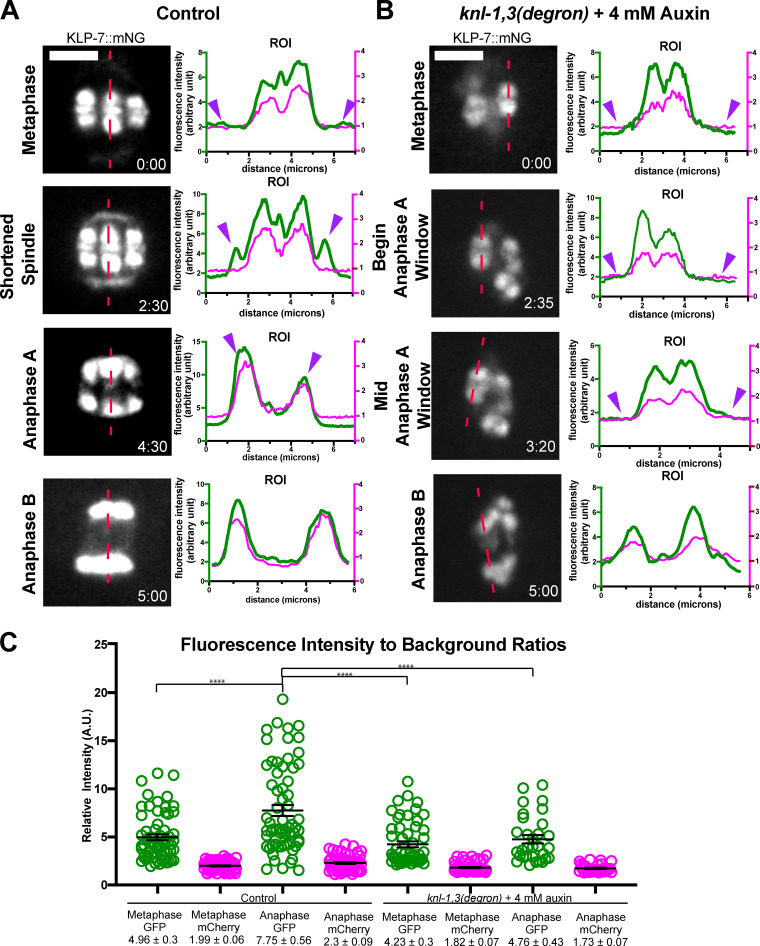
The kinesin 13, KLP-7, is also a microtubule-binding protein that is associated with chromosomes during meiosis and thus might provide redundant attachments between chromosomes and microtubules. KLP-7 endogenously tagged at the C-terminus with mNeonGreen (Heppert et al., 2018) and, observed live, was most concentrated on meiotic chromosomes in a bi-lobed pattern similar to histone H2b–labeled chromatin throughout metaphase and anaphase, with lower amounts in the midbivalent ring during metaphase (Fig. 10 A, 0 and 2:30; Fig. S5 A; 11/11 control embryos). This pattern was similar to that previously observed (Connolly et al., 2015; Gigant et al., 2017; Han et al., 2015). KLP-7::mNeonGreen was lost from midbivalent rings in 6/7 knl-1,3(kd) embryos but remained associated with chromatin throughout metaphase and anaphase in 7/7 knl-1,3(kd) embryos (Fig. 10 B; and Fig. S5, A and B). In agreement with previous studies (Connolly et al., 2015; Gigant et al., 2017; Han et al., 2015), KLP-7 accumulated on spindle poles during spindle shortening in control embryos (Fig. 10 A, 2:30). In contrast, KLP-7 did not accumulate on spindle poles in 6/7 knl-1,3(kd) embryos (Fig. 10 B). In control but not knl-1,3(kd) embryos, the mean fluorescence intensity of KLP-7::mNeonGreen increased between metaphase and anaphase (Fig. 10 C). These results indicate that KNL-1,3 recruited KLP-7 to spindle poles but not to chromatin and that the increase in apparent chromosomal intensity during anaphase in control embryos was due to merging of pole and chromosomal pools. These results also suggest that KLP-7 could mediate chromosome–microtubule attachments in the absence of KNL-1,3 during error-prone anaphase B. We were unable to directly test this because knl-1,3(degron) klp-7(RNAi) oocytes would not ovulate.
Figure 10.
The kinesin-13 KLP-7 is retained on bivalent chromatin in KNL-1,3–depleted embryos. (A and B) Representative time-lapse sequences of endogenously tagged KLP-7::mNeonGreen in control and knl-1,3(kd) embryos. Fluorescence intensity plots along the dotted red lines are shown for KLP-7::mNeonGreen in green and mCherry::H2b in red. Purple arrowheads indicate spindle poles. (C) KLP-7::mNeonGreen fluorescence associated with chromosomes. Error bars and values are mean ± SEM. Bars = 3 µm. ****, P < 0.0001. mNG, mNeonGreen; ROI, region of interest.
Figure S5.
The kinesin-13 KLP-7 is retained on bivalents following KNL-1,3 depletion. (A and B) Representative time-lapse sequences. (A) In control spindles, endogenously tagged KLP-7::mNG localizes to meiotic spindle poles, the main chromatin masses, and in midbivalent rings. (B) Following KNL-1,3 depletion, KLP-7::mNG is retained on the main chromatin masses but is no longer present at spindle poles or midbivalent rings. (C–E) Results of fixed immunofluorescence. (C and D) Following triple GFP(RNAi) depletion of endogenously GFP-tagged KNL-1,3 and MEI-1, all bivalents were associated with apolar meiotic spindles in the majority of embryos. (E) Example of a rare occurrence of errant maternal chromosomes that appear to have left the meiotic spindle. Bars (A–C) = 3 µm. Bars (E) = 7 µm. MI, meiosis one; mNG, mNeonGreen.
The C. elegans katanin, MEI-1/MEI-2, binds microtubules (Joly et al., 2016; McNally et al., 2014; McNally and McNally, 2011) and colocalizes with DAPI or mCherry::H2b inside the kinetochore cups (McNally et al., 2006; Srayko et al., 2000). mei-1(null) mutants assemble apolar meiotic spindles (Connolly et al., 2014; McNally and McNally, 2011). mei-1(RNAi) also results in an increased velocity of cytoplasmic streaming during meiosis (Kimura et al., 2017), which should scatter any chromosomes not attached to the apolar spindle. Anti-GFP staining of fixed embryos from a strain with endogenously GFP-tagged knl-1, knl-3, and mei-1 genes after GFP(RNAi) did not reveal any GFP remaining on chromosomes. In 14/16 of these embryos, all maternal chromosomes were associated with the apolar spindle (Fig. S5, C and D). In only two triple-depleted embryos (Fig. S5 E), chromosomes were observed far from the apolar spindle. Attachment of chromosomes to microtubules in knl-1,3(kd) thus may be mediated by KLP-7, by very low amounts of MEI-1/2 or KLP-19, or by unknown factors.
Discussion
The results presented here extend the findings of Dumont et al. (2010) by showing that KNL-1,3 are required for homolog separation and anaphase A chromosome-to-pole movement. Our results are consistent with their study in that anaphase B velocities are not affected by depletion of the KMN network and this kinetochore-independent anaphase is highly error prone. Dumont et al. (2010) reported that the number of chromosomes on the metaphase II plate of knl-1(RNAi) embryos was altered to 5.6 from the wild-type number of 6. This number was reported as a mean, and since error-prone anaphase I segregation can result in loss or gain of chromosomes, the only clue to the severity of this phenotype was the reported standard error of 1.1.
The finding that depletion of KNL-1,3 or NDC-80 diminishes the bivalent stretching that occurs between metaphase and preanaphase strongly supports the existence of poleward pulling forces that may be established during spindle shortening. Purified NDC-80 complexes generate force at the plus end of depolymerizing microtubules (Powers et al., 2009), but only the sides of microtubules have been observed in close proximity to the ribosome-free layer of C. elegans chromosomes during late metaphase by EM (Redemann et al., 2018). Possible explanations for this discrepancy include the following: (1) Plus-end attachments may have been missed in EM analysis because the kinetochore extends much farther poleward than the ribosome-free zone, as suggested in Fig. 2. (2) Plus-end attachments may occur only transiently just before homolog separation, a stage not yet captured by EM. (3) The lateral contacts observed by EM are converted to a transient plus-end attachment when each microtubule depolymerizes from its plus end. By analogy, the plus-end contacts with the embryo cortex that mediate spindle positioning have an average lifetime of 1.4 s (Kozlowski et al., 2007). Photobleaching experiments indicate that meiotic spindle microtubules turn over extremely rapidly (Yu et al., 2019). (4) NDC-80 and KNL-1 (Espeut et al., 2012) might bind the side of a microtubule, while the minus end of that microtubule is transported toward the minus end of another microtubule by ROD-1–independent dynein (Elting et al., 2014; Sikirzhytski et al., 2014; Tan et al., 2018). (5) Pulling on the lateral surface of microtubules is generated either by an unknown KMN-dependent protein or by an unknown mechanism of the known KMN components. Whatever the mechanism, pulling is likely achieved by arrays of very short overlapping microtubules rather than microtubules running contiguously from chromosome to pole (Laband et al., 2017; Redemann et al., 2018; Srayko et al., 2006).
The observation that both homologs that later separate during anaphase B and bivalents that remain intact during anaphase B undergo attenuated anaphase A movement toward spindle poles in knl-1,3(kd) suggests a lack of poleward pulling forces that persist during normal anaphase A. In addition, the presence of GFP::KNL rings around him-8 univalent X chromosomes that are stretched during late anaphase also supports the persistence of pulling forces into late anaphase B. The homolog separation failure after depletion of KNL-1,3 or NDC-80 thus could indicate that homologs must be pulled apart even after cohesin cleavage. Pulling might help resolve remaining topological entanglements (Hong et al., 2018a, 2018b), or molecular crowding might slow the separation of chromosome-sized objects by Brownian motion (Delarue et al., 2018) in the absence of pulling. Alternatively, KMN might act as a positive regulator of separase through an unknown mechanism. In Drosophila melanogaster oocyte meiosis, the KNL-1 homolog SPC105R is a negative regulator of separase (Wang et al., 2019). The end-to-end stacking of bivalents at metaphase of knl-1,3(kd) spindles may also inhibit homolog separation. Anaphase A homolog separation may be important to generate a sufficient distance to allow the microtubule polymerization between homologs that drives anaphase B (Dumont et al., 2010; Laband et al., 2017).
Prior observations that spindle cutting on the poleward face of C. elegans female meiotic spindles did not alter anaphase velocity (Laband et al., 2017) and that cutting between separating chromosomes immediately stopped chromosome movement (Laband et al., 2017; Yu et al., 2019) are consistent with a long history of spindle cutting experiments in mitotic and meiotic spindles of a variety of species (Spurck et al., 1997, 1990). Elting et al. (2014) and Sikirzhytski et al. (2014) found that poleward chromosome movement after cutting on the poleward side was caused by dynein-mediated transport of the minus end of a kinetochore fiber on other spindle microtubules. Thus, cutting on the poleward face of a chromosome does not necessarily block pulling forces on that chromosome. Cutting between homologs in meiosis I or between sister chromatids in meiosis II or mitosis is not possible until after proper chromosome separation has started. Thus, midzone cutting experiments cannot show that poleward pulling is dispensable for proper chromosome separation. In some studies (Nicklas, 1989), neither cutting in front of nor behind an anaphase chromosome was able to stop anaphase motion.
The idea that anti-parallel microtubule bundles push on the inner faces of separating chromosomes must be resolved with the observation that chromosome arms lag behind kinetochores during anaphase B in species with localized centromeres (Inoué and Ritter, 1978) and that chromosomes move in unison without tumbling or dissociating from the spindle even in the presence of cytoplasmic streaming, as occurs in the C. elegans meiotic embryo (McNally et al., 2010; Yang et al., 2003). Our finding that KNL-1, KNL-3, and NDC-80, representatives of the three components of the KMN network, all transition into rings that cover the inner face of anaphase B chromosomes suggests a possible attachment point for pushing fibers.
Materials and methods
C. elegans strains were generated by standard genetic crosses, and genotypes were confirmed by PCR. Genotypes of all strains are listed in Table S1.
Live in utero imaging
L4 larvae were incubated at 20°C overnight on MYOB plates seeded with OP50, except for auxin treatments in which L4 larvae were incubated at 16°C overnight, and FGP7 L4 larvae were treated at 25°C overnight. Worms were anesthetized by picking 20 adult hermaphrodites with a platinum wire into a solution of 0.1% tricaine, 0.01% tetramisole in PBS in a watch glass for 30 min as described in Kirby et al. (1990) and McCarter et al. (1999). Worms were then transferred in a small volume (@ 20 μl) to the center of a thin agarose pad (2% in water) on a slide. Additional PBS was pipetted around the edges of the agarose pad, and a 22-×-30-mm cover glass was placed on top. The slide was then inverted and placed on the stage of an inverted microscope. Meiotic embryos adjacent to the spermatheca were then identified by bright-field microscopy before initiating time-lapse fluorescence. For all live imaging, the stage and immersion oil temperature was 22°C–24°C. For all live imaging data other than Video 2 and Fig. 8, A–C, single–focal plane time-lapse images were acquired with a Solamere spinning disk confocal microscope equipped with an Olympus IX-70 stand, Yokogawa CSU10, Hamamatsu ORCA FLASH 4.0 CMOS (complementary metal oxide semiconductor) detector, Olympus 100×/1.35 objective, 100-mW Coherent Obis lasers set at 30% power, and MicroManager software control. Pixel size was 65 nm. Exposures were 100 ms. Time interval between image pairs was 10 s with the following exceptions. Fig. 1, A–C; Fig. 3, A and B; Fig. 5, A–C; and Fig. 6, B and C were acquired at 5-s intervals. Fig. 5, D and F was captured at 15-s intervals. Focus was adjusted manually during time-lapse imaging. Only images in which both spindle poles and/or both bivalent halves were in focus were used for quantitative analysis. Both poles or both half-bivalents were considered to be in focus when both exhibited equal brightness and sharpness.
For scoring spindle bipolarity by imaging GFP::ASPM-1 (Video 2 and data in text), nine images were acquired at 1-µm z-steps for the first time point. Subsequently, single-plane images were acquired at 5-s intervals.
For documenting NDC-80 depletion from bivalents in Fig. 8, A–C, z-stacks were acquired at 1-µm steps and 100-ms exposures through the entire nucleus of the −1 oocyte, the most mature oocyte before nuclear envelope breakdown. −1 oocytes were used because mNeonGreen::tubulin, which is required for documenting anaphase A versus B, is excluded from the nucleus, allowing documentation of GFP::NDC-80 depletion. Only two or three focal planes were subjected to maximum intensity projection for the figure because of cytoplasmic mNeonGreen::tubulin fluorescence in the focal planes above and below.
Timing
Control spindles maintain a steady-state length of 8 µm for 7 min before initiating APC-dependent spindle shortening, followed by spindle rotation, followed by homolog separation as the spindle continues to shorten (anaphase A), and followed by a transition to spindle elongation (anaphase B; McNally et al., 2016; Yang et al., 2005). Because knl-1,3(kd) affects spindle dimensions, spindle rotation (Vargas et al., 2019), chromosome congression, and anaphase movement, we used time relative to the initiation of spindle elongation to compare parameters between control and knl-1,3(kd).
Auxin
C. elegans strains with knl-1 and knl-3 endogenously tagged with auxin-inducible degrons and aTIR1 transgene were treated with 4 mm auxin overnight on seeded plates. dhc-1(AID) (Zhang et al., 2015) strains were treated with 4 mm auxin for 1 h on seeded plates.
RNAi
L4 larvae were placed on IPTG-induced lawns of HT115 bacteria bearing L4440-based plasmids. After 48 h, worms were fixed or imaged live. RNAi clones for aspm-1, rod-1, and mei-2 were from the genomic RNAi feeding library (Medical Research Council Gene Services, Source BioScience, Nottingham, UK; Kamath et al., 2003). C. elegans optimized GFP was cloned into L4440 for the GFP(RNAi) experiments.
Fixed immunofluorescence
C. elegans meiotic embryos were extruded from hermaphrodites in 0.8× egg buffer by gently compressing worms between coverslip and slide, flash frozen in liquid N2, permeabilized by removing the coverslip, and then fixed in ice-cold methanol before staining with antibodies and DAPI. The primary antibodies used in this work were mouse monoclonal anti-tubulin (DM1α; Sigma-Aldrich; 1:200), rabbit anti–SEP-1 (Bembenek et al., 2007; 1:350), and rabbit anti-GFP (NB600-308SS; Novus Biologicals; 1:600). The secondary antibodies used were Alexa Fluor 488 anti-mouse (A-11001; Thermo Fisher Scientific; 1:200) and Alexa Fluor 594 anti-rabbit (A11037; Thermo Fisher Scientific; 1:200). z-stacks were captured at 1-µm steps for each meiotic embryo. Images in Fig. 1, D and E; Fig. S1, C–F; and Fig. S5, C and E were captured with an inverted microscope (Olympus IX-81) equipped with a 60× PlanApo 1.42 objective, a disk-scanning unit (Olympus), and an ORCA Flash 4.0 CMOS detector and controlled with MicroManager software. Exposure time for anti-GFP, DAPI, and anti-tubulin was 250 ms. Exposure time for anti–SEP-1 was 500 ms. Pixel size is 130 nm. The images shown are maximum intensity projections of five focal planes. Images in Fig. S1, A and B were captured with the Solamere system described under live imaging. Pixel size is 65 nm. Images in Fig. S1, A and B were deconvolved with Huygens Essentials, and the images shown are maximum intensity projections of four focal planes.
Fluorescence intensity measurements
Fluorescence intensity line profiles in Fig. 2 and Fig. 10 are from single–focal plane images. Quantification of fluorescence intensity change in KNL-1::GFP/GFP::KNL-3 when transitioning from cups to rings was done on single focal plane images using the segmentation function in Ivision software (BioVision Technologies). Mean pixel intensities of kinetochores as metaphase cups and anaphase rings were divided by mean background fluorescence intensity to generate a ratio for comparison between embryos. mCherry::H2b values were also measured at these time points as a control for photobleaching. All other quantifications were done through ImageJ software. Fluorescence intensity of KLP-7::mNeongreen was measured by drawing ellipses around individual homologs. Mean pixel values were then divided by background mean pixel values to generate a ratio for comparison. mCherry::H2b values were also measured at these time points as a control for photobleaching. A region of interest line was drawn through spindles progressing through meiosis in Fig. 10, A and B to show a change in KLP-7 localization over time relative to mCherry::H2b as a control.
Bivalent stretching measurements
For Fig. 4, Fig. S3, and Fig. S5, spindles were filmed in utero at 10-s intervals. Metaphase values were measured 5.7 ± 0.47 min before initiation of spindle elongation. “Preanaphase” values were measured 2.2 ± 0.15 min before initiation of spindle elongation because this corresponds to the time just before homolog separation in controls. In the first method, a fluorescence intensity profile was generated along a 1-pixel-wide line drawn from a point halfway between one spindle pole and the proximal half-bivalent to a point halfway between the opposite half-bivalent and the opposite spindle pole. The peaks of mCherry fluorescence were used as the centers of mCherry histone half-bivalents. The outer edges of GFP::NPP-6 or mCherry histone were determined as the points of half-maximal GFP or mCherry fluorescence intensity. In the second method, a rectangular region of interest was drawn around each half-bivalent, and the auto contrast function in FIJI was used to set the dimmest pixel to black and the brightest pixel to white. Homolog centers were determined by zooming, such that the cross-shaped cursor was the same size as the homolog, and manually centering the cursor over the homolog. The outer edges of GFP::NPP-6 were then determined by visual inspection. Comparison of these two methods on 10 bivalents by one-way ANOVA yielded P values of 0.88 for NPP-6 outer edges and 0.98 for homolog centers. To ensure that homologs had not yet separated at the time of each preanaphase measurement, the ratio between peak mCherry pixel values and the pixel values of the trough between homologs was made for each bivalent. No significant difference in this ratio was found between metaphase and preanaphase or between control and knl-1,3(kd). Intrahomolog stretch in Figs. 4, D and E, was determined by subtracting the distance between mCherry::H2b homolog centers from the distance between the outside edges of NPP-6::GFP–labeled outer edges of bivalents. Cross-sectional areas of chromosomes in Fig. S3 D and Fig. S5 D were calculated by treating bivalents as ellipses, using the equation 1/2(width) 1/2(length) π.
Anaphase velocity assays
For anaphase chromosome separation velocities in control embryos for Fig. 5, Fig. 6, Fig. 8, Fig. 9, Fig. S2, and Fig. S4, the change in distance between homolog centers was divided by time elapsed in minutes. For intact bivalent movement measurements of auxin-depleted knl-1,3(kd) embryos in Fig. 5 J, the change in distance was measured between the midbivalent regions of each intact bivalent. Anaphase B velocities in Fig. 6 A and Fig. S2 C included only chromosomes separating parallel with the main spindle axis. Chromosomes segregating oblique to the main spindle axis in three- or four-way anaphases were not included in these figures; however, the velocities of these oblique anaphase B movements were not significantly different from the displayed values. For Fig. 8 anaphase A measurements, one half of the pole-to-pole spindle shortening velocity was subtracted from the rate of decrease in chromosome-to-pole distance. For wild-type–appearing anaphase B, only the number of embryos, N, is shown, since at this stage chromosomes grouped together and were treated as one moving mass.
Statistics
P values were calculated in GraphPad Prism using Student’s t test for comparing means of only two groups (e.g., Fig. 2 D), one-way ANOVA for comparing means of three or more groups, and Fisher’s exact test when comparing frequencies of contingent data.
Online supplemental material
Fig. S1 shows anti-GFP staining relevant to Fig. 3 and anti–SEP-1 staining relevant to Fig. 5. Fig. S2 compares depletion methods relevant to Fig. 3. Fig. S3 shows bivalent stretching data relevant to Fig. 4 and Fig. 8. Fig. S4 shows AIR-2 localization relevant to Fig. 5 and triple-depletion results. Fig. S5 shows KLP-7 localization relevant to Fig. 10 and triple-depletion results. Table S1 presents the C. elegans strain list. Video 1 is relevant to Fig. 1 and Fig. 2. Video 2 is relevant to Fig. 3. Video 3, Video 4, Video 5, and Video 6 are relevant to Fig. 5. Video 7 is relevant to Fig. 7. Video 8 and Video 9 are relevant to Fig. 8. Video 10 is relevant to Fig. S4 and triple-depletion results.
Supplementary Material
presents the C. elegans strain list.
Acknowledgments
We thank Jordan Ward (University of California, Santa Cruz, Santa Cruz, CA), Abby Dernburg (University of California, Berkeley, Berkeley, CA), Bruce Bowerman (University of Oregon, Eugene, OR), Fede Pelisch (University of Dundee, Dundee, UK), Sander van den Heuvel (Utrecht University, Utrecht, Netherlands), Akatsuki Kimura (National Institute of Genetics, Mishima, Japan), Bob Goldstein (University of North Carolina, Chapel Hill, NC), and Arshad Desai (Univerisity of California, San Diego, San Diego, CA) for strains. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, Office of Research Infrastructure Programs (P40 OD010440). We thank Lesilee Rose for critical reading of the manuscript.
This work was supported by National Institute of General Medical Sciences grants 1R01GM079421 and 1R35GM124889 and by the U.S. Department of Agriculture/National Institute of Food and Agriculture Hatch project (1009162 to F.J. McNally).
The authors declare no competing financial interests.
Author contributions: B.M. Danlasky, M.T. Panzica, K.P. McNally, E. Vargas, C. Bailey, W. Li, T. Gong, E.S. Fishman, and X. Jiang performed experiments, analyzed data, and prepared figures. F.J. McNally wrote the manuscript with contributions from all authors.
References
- Bembenek, J.N., Richie C.T., Squirrell J.M., Campbell J.M., Eliceiri K.W., Poteryaev D., Spang A., Golden A., and White J.G.. 2007. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 134:3837–3848. 10.1242/dev.011361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Connolly, A.A., Osterberg V., Christensen S., Price M., Lu C., Chicas-Cruz K., Lockery S., Mains P.E., and Bowerman B.. 2014. Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol. Biol. Cell. 25:1298–1311. 10.1091/mbc.e13-11-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, A.A., Sugioka K., Chuang C.H., Lowry J.B., and Bowerman B.. 2015. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J. Cell Biol. 210:917–932. 10.1083/jcb.201412010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes, D.B., McNally K.L., Mains P.E., and McNally F.J.. 2015. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. eLife. 4:e06056. 10.7554/eLife.06056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder, M.E., Flynn J.R., McNally K.P., Cortes D.B., Price K.L., Kuehnert P.A., Panzica M.T., Andaya A., Leary J.A., and McNally F.J.. 2015. Dynactin-dependent cortical dynein and spherical spindle shape correlate temporally with meiotic spindle rotation in Caenorhabditis elegans. Mol. Biol. Cell. 26:3030–3046. 10.1091/mbc.E15-05-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Roca, A.C., Muscat C.C., and Wignall S.M.. 2017. Caenorhabditis elegans oocytes detect meiotic errors in the absence of canonical end-on kinetochore attachments. J. Cell Biol. 216:1243–1253. 10.1083/jcb.201608042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Roca, A.C., Divekar N.S., Ng R.K., and Wignall S.M.. 2018. Dynamic SUMO remodeling drives a series of critical events during the meiotic divisions in Caenorhabditis elegans. PLoS Genet. 14:e1007626. 10.1371/journal.pgen.1007626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho, C.E., Zaaijer S., Smolikov S., Gu Y., Schumacher J.M., and Colaiácovo M.P.. 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22:2869–2885. 10.1101/gad.1691208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue, M., Brittingham G.P., Pfeffer S., Surovtsev I.V., Pinglay S., Kennedy K.J., Schaffer M., Gutierrez J.I., Sang D., Poterewicz G., et al. 2018. mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell. 174:338–349.E20. 10.1016/j.cell.2018.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont, J., Oegema K., and Desai A.. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901. 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson, M.L., and McNally F.J.. 2009. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol. Biol. Cell. 20:2722–2730. 10.1091/mbc.e08-12-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting, M.W., Hueschen C.L., Udy D.B., and Dumont S.. 2014. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 206:245–256. 10.1083/jcb.201401091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut, J., Cheerambathur D.K., Krenning L., Oegema K., and Desai A.. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482. 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabig, G., Kiewisz R., Lindow N., Powers J.A., Cota V., Quintanilla L.J., Brugués J., Prohaska S., Chu D.S., and Müller-Reichert T.. 2020. Male meiotic spindle features that efficiently segregate paired and lagging chromosomes. eLife. 9:e50988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz, N., Barroso C., Telecan O., Shao N., Kim H.M., Testori S., Faull P., Cutillas P., Snijders A.P., Colaiacovo M.P., et al. 2018a. Author Correction: Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 9:3558. 10.1038/s41467-018-05848-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz, N., Barroso C., Telecan O., Shao N., Kim H.M., Testori S., Faull P., Cutillas P., Snijders A.P., Colaiacovo M.P., et al. 2018b. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 9:834. 10.1038/s41467-018-03229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant, E., Stefanutti M., Laband K., Gluszek-Kustusz A., Edwards F., Lacroix B., Maton G., Canman J.C., Welburn J.P.I., and Dumont J.. 2017. Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7 prevents chromosome segregation and cytokinesis defects in oocytes. Development. 144:1674–1686. 10.1242/dev.147504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, L., and Homer H.. 2013. Hec1-dependent cyclin B2 stabilization regulates the G2-M transition and early prometaphase in mouse oocytes. Dev. Cell. 25:43–54. 10.1016/j.devcel.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Adames K., Sykes E.M., and Srayko M.. 2015. The KLP-7 Residue S546 Is a Putative Aurora Kinase Site Required for Microtubule Regulation at the Centrosome in C. elegans. PLoS One. 10:e0132593. 10.1371/journal.pone.0132593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley, N., Cheerambathur D., Moyle M., Stefanutti M., Richardson A., Lee K.Y., Dumont J., Oegema K., and Desai A.. 2016. A Nucleoporin Docks Protein Phosphatase 1 to Direct Meiotic Chromosome Segregation and Nuclear Assembly. Dev. Cell. 38:463–477. 10.1016/j.devcel.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppert, J.K., Pani A.M., Roberts A.M., Dickinson D.J., and Goldstein B.. 2018. A CRISPR Tagging-Based Screen Reveals Localized Players in Wnt-Directed Asymmetric Cell Division. Genetics. 208:1147–1164. 10.1534/genetics.117.300487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Sonneville R., Wang B., Scheidt V., Meier B., Woglar A., Demetriou S., Labib K., Jantsch V., and Gartner A.. 2018a. LEM-3 is a midbody-tethered DNA nuclease that resolves chromatin bridges during late mitosis. Nat. Commun. 9:728. 10.1038/s41467-018-03135-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Velkova M., Silva N., Jagut M., Scheidt V., Labib K., Jantsch V., and Gartner A.. 2018b. The conserved LEM-3/Ankle1 nuclease is involved in the combinatorial regulation of meiotic recombination repair and chromosome segregation in Caenorhabditis elegans. PLoS Genet. 14:e1007453. 10.1371/journal.pgen.1007453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, M., McDonald K.L., Albertson D.G., and Meyer B.J.. 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153:1227–1238. 10.1083/jcb.153.6.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué, S., and Ritter H. Jr. 1978. Mitosis in Barbulanympha. II. Dynamics of a two-stage anaphase, nuclear morphogenesis, and cytokinesis. J. Cell Biol. 77:655–684. 10.1083/jcb.77.3.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, N., Martino L., Gigant E., Dumont J., and Pintard L.. 2016. Microtubule-severing activity of the AAA+ ATPase Katanin is essential for female meiotic spindle assembly. Development. 143:3604–3614. 10.1242/dev.140830 [DOI] [PubMed] [Google Scholar]
- Kaitna, S., Pasierbek P., Jantsch M., Loidl J., and Glotzer M.. 2002. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr. Biol. 12:798–812. 10.1016/S0960-9822(02)00820-5 [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kimura, K., Mamane A., Sasaki T., Sato K., Takagi J., Niwayama R., Hufnagel L., Shimamoto Y., Joanny J.F., Uchida S., et al. 2017. Endoplasmic-reticulum-mediated microtubule alignment governs cytoplasmic streaming. Nat. Cell Biol. 19:399–406. 10.1038/ncb3490 [DOI] [PubMed] [Google Scholar]
- Kirby, C., Kusch M., and Kemphues K.. 1990. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev. Biol. 142:203–215. 10.1016/0012-1606(90)90164-E [DOI] [PubMed] [Google Scholar]
- Kozlowski, C., Srayko M., and Nedelec F.. 2007. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 129:499–510. 10.1016/j.cell.2007.03.027 [DOI] [PubMed] [Google Scholar]
- Laband, K., Le Borgne R., Edwards F., Stefanutti M., Canman J.C., Verbavatz J.M., and Dumont J.. 2017. Chromosome segregation occurs by microtubule pushing in oocytes. Nat. Commun. 8:1499. 10.1038/s41467-017-01539-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N., McDonald K.L., Ding R., and McIntosh J.R.. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489. 10.1083/jcb.123.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter, J., Bartlett B., Dang T., and Schedl T.. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205:111–128. 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- McDonald, K.L., O’Toole E.T., Mastronarde D.N., and McIntosh J.R.. 1992. Kinetochore microtubules in PTK cells. J. Cell Biol. 118:369–383. 10.1083/jcb.118.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.P., and McNally F.J.. 2011. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol. Biol. Cell. 22:1550–1560. 10.1091/mbc.e10-12-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K., Audhya A., Oegema K., and McNally F.J.. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.L., Martin J.L., Ellefson M., and McNally F.J.. 2010. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Dev. Biol. 339:126–140. 10.1016/j.ydbio.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K., Berg E., Cortes D.B., Hernandez V., Mains P.E., and McNally F.J.. 2014. Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol. Biol. Cell. 25:1037–1049. 10.1091/mbc.e13-12-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.P., Panzica M.T., Kim T., Cortes D.B., and McNally F.J.. 2016. A novel chromosome segregation mechanism during female meiosis. Mol. Biol. Cell. 27:2576–2589. 10.1091/mbc.e16-05-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen, J., Maddox P.S., Hyndman F., Oegema K., and Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. 10.1038/ncb1331 [DOI] [PubMed] [Google Scholar]
- Muscat, C.C., Torre-Santiago K.M., Tran M.V., Powers J.A., and Wignall S.M.. 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 4:e06462. 10.7554/eLife.06462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R.B. 1989. The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J. Cell Biol. 109:2245–2255. 10.1083/jcb.109.5.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, E.T., McDonald K.L., Mäntler J., McIntosh J.R., Hyman A.A., and Müller-Reichert T.. 2003. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163:451–456. 10.1083/jcb.200304035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch, F., Tammsalu T., Wang B., Jaffray E.G., Gartner A., and Hay R.T.. 2017. A SUMO-Dependent Protein Network Regulates Chromosome Congression during Oocyte Meiosis. Mol. Cell. 65:66–77. 10.1016/j.molcel.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch, F., Bel Borja L., Jaffray E.G., and Hay R.T.. 2019. Sumoylation regulates protein dynamics during meiotic chromosome segregation in C. elegans oocytes. J. Cell Sci. 132:jcs232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, C., Reis R.M., Gama J.B., Celestino R., Cheerambathur D.K., Carvalho A.X., and Gassmann R.. 2018. Self-Assembly of the RZZ Complex into Filaments Drives Kinetochore Expansion in the Absence of Microtubule Attachment. Curr. Biol. 28:3408–3421.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., and Asbury C.L.. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875. 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann, S., Lantzsch I., Lindow N., Prohaska S., Srayko M., and Muller-Reichert T.. 2018. A Switch in Microtubule Orientation during C. elegans Meiosis. Curr. Biol. 28:2991–2997.e2. [DOI] [PubMed]
- Rogers, E., Bishop J.D., Waddle J.A., Schumacher J.M., and Lin R.. 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157:219–229. 10.1083/jcb.200110045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikirzhytski, V., Magidson V., Steinman J.B., He J., Le Berre M., Tikhonenko I., Ault J.G., McEwen B.F., Chen J.K., Sui H., et al. 2014. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol. 206:231–243. 10.1083/jcb.201401090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurck, T.P., Stonington O.G., Snyder J.A., Pickett-Heaps J.D., Bajer A., and Mole-Bajer J.. 1990. UV microbeam irradiations of the mitotic spindle. II. Spindle fiber dynamics and force production. J. Cell Biol. 111:1505–1518. 10.1083/jcb.111.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurck, T., Forer A., and Pickett-Heaps J.. 1997. Ultraviolet microbeam irradiations of epithelial and spermatocyte spindles suggest that forces act on the kinetochore fibre and are not generated by its disassembly. Cell Motil. Cytoskeleton. 36:136–148. [DOI] [PubMed] [Google Scholar]
- Srayko, M., Buster D.W., Bazirgan O.A., McNally F.J., and Mains P.E.. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., O’toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Tan, R., Foster P.J., Needleman D.J., and McKenney R.J.. 2018. Cooperative Accumulation of Dynein-Dynactin at Microtubule Minus-Ends Drives Microtubule Network Reorganization. Dev. Cell. 44:233–247.E4. 10.1016/j.devcel.2017.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet, M., Berends C.W., Perreault A., Nguyen-Ngoc T., Gonczy P., Vidal M., Boxem M., and van den Heuvel S.. 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat. Cell Biol. 11:269–277. 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- Vargas, E., McNally K.P., Cortes D.B., Panzica M.T., Danlasky B.M., Li Q., Maddox A.S., and McNally F.J.. 2019. Spherical spindle shape promotes perpendicular cortical orientation by preventing isometric cortical pulling on both spindle poles during C. elegans female meiosis. Development. 146:dev178863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukušić, K., Buda R., and Tolic I.M.. 2019. Force-generating mechanisms of anaphase in human cells. J. Cell Sci. 132:jcs231985. [DOI] [PubMed] [Google Scholar]
- Wang, L.I., Das A., and McKim K.S.. 2019. Sister centromere fusion during meiosis I depends on maintaining cohesins and destabilizing microtubule attachments. PLoS Genet. 15:e1008072. 10.1371/journal.pgen.1008072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y. 2005. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 21:405–412. 10.1016/j.tig.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Wignall, S.M., and Villeneuve A.M.. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11:839–844. 10.1038/ncb1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Y., McNally K., and McNally F.J.. 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev. Biol. 260:245–259. 10.1016/S0012-1606(03)00216-1 [DOI] [PubMed] [Google Scholar]
- Yang, H.Y., Mains P.E., and McNally F.J.. 2005. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J. Cell Biol. 169:447–457. 10.1083/jcb.200411132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Nishiyama S., Lister L., Hashimoto S., Mishina T., Courtois A., Kyogoku H., Abe T., Shiraishi A., Choudhary M., et al. 2020. Prc1-rich kinetochores are required for error-free acentrosomal spindle bipolarization during meiosis I in mouse oocytes. Nat. Commun. 11:2652. 10.1038/s41467-020-16488-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.H., Redemann S., Wu H.Y., Kiewisz R., Yoo T.Y., Conway W., Farhadifar R., Muller-Reichert T., and Needleman D.. 2019. Central-spindle microtubules are strongly coupled to chromosomes during both anaphase A and anaphase B. Mol. Biol. Cell. 30:2503–2514. 10.1091/mbc.E19-01-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Ward J.D., Cheng Z., and Dernburg A.F.. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142:4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents the C. elegans strain list.