Delayed clearance of Plasmodium falciparum by artemisinin-based combination therapies (ACTs) has already been observed for African isolates. Here, we aimed to investigate the prevalence, among travelers returning from African countries, of polymorphisms in two genes correlated with delayed parasite clearance (encoding P. falciparum Kelch 13 [PfK13] and ubiquitin-specific protease 1 [pfubp1]) reported in eastern China and to provide baseline data for antimalarial drug resistance (ART) surveillance and evaluation.
KEYWORDS: Plasmodium falciparum, Kelch 13, pfubp1, antimalarial drug resistance, ACT, Africa
ABSTRACT
Delayed clearance of Plasmodium falciparum by artemisinin-based combination therapies (ACTs) has already been observed for African isolates. Here, we aimed to investigate the prevalence, among travelers returning from African countries, of polymorphisms in two genes correlated with delayed parasite clearance (encoding P. falciparum Kelch 13 [PfK13] and ubiquitin-specific protease 1 [pfubp1]) reported in eastern China and to provide baseline data for antimalarial drug resistance (ART) surveillance and evaluation. A total of 153 filter paper blood spots collected in 2017–2019 from patients with uncomplicated P. falciparum cases in Anhui and Shandong Provinces were included in this study. Among them, 3.3% (5/153) of the isolates carried PfK13 mutations, and 3 of them harbored the same synonymous mutation, C469C. A total of 13.1% (20/153) of the isolates were found to contain pfubp1 mutations, and all were nonsynonymous. The pfubp1 genotypes associated with ART that occurred in this study included E1528D (6.5% [10/153]) and D1525E (2.6% [4/153]). However, a high prevalence of the previously unreported mutation E1531D (5.9% [9/153]) was also detected. In addition, two types of deletions (encoding KID and KIE, respectively) and two types of insertions (encoding KYE and KYDKYD, respectively) were found in 16 isolates and 6 isolates, respectively. This study showed limited variation in PfK13 among travelers returning from African countries and suggested other potential molecular markers, such as pfubp1, for use in the surveillance of African isolates in ACT susceptibility studies. Further clinical trial research is under way to investigate these PfK13 and pfubp1 mutations, as well as other candidate molecular markers, and their roles in delaying parasite clearance.
INTRODUCTION
Imported malaria, especially imported Plasmodium falciparum malaria from Africa, has become a great threat to malaria elimination in China (1). In 2019, 1,013 cases of imported P. falciparum malaria from five African countries, including Nigeria, the Democratic Republic of the Congo, Côte d’Ivoire, Guinea, and Ghana, have been reported, accounting for 51.9% of the total P. falciparum cases reported in China (2). Malaria still remains a major challenge in sub-Saharan Africa, where 228 million cases and 405,000 malaria-related deaths were reported in 2018 (3). Artemisinin-based combination therapies (ACTs) were the first-line drugs recommended by the World Health Organization (WHO), and an estimated 214 million ACT treatments, most of which were used in African countries, were delivered in 2018. However, the emergence and development of Plasmodium falciparum resistance to ACTs, which initially occurred near the Thailand-Cambodia border, have become a grave concern (4, 5). To solve this problem, the WHO has launched efforts to contain antimalarial drug resistance (ART), including therapeutic efficacy studies (TESs) and integrated drug efficacy surveillance (iDES), particularly in the Greater Mekong Subregion (GMS) (6). In addition, molecular markers revealing genetic changes in these parasites were found to be highly associated with ACT resistance. Therefore, knowledge of these molecular markers at the country or regional level and monitoring of the genomes of the parasite population are crucial for the early detection of emerging resistance.
The gene encoding Plasmodium falciparum Kelch 13 (PfK13) was identified as a molecular marker for ACT-resistant isolates in 2014 (4). To date, the WHO has reported a total of nine single nucleotide polymorphism (SNP) sites (F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, and C580Y) that were validated in vivo and in vitro as associated with ACT resistance (7). These SNPs were found mainly in the GMS and were closely associated with delayed clearance following ACT treatment (8, 9).
Among P. falciparum isolates from Africa, A578S was the most common mutation site and showed no relationship to clinical or in vitro ACT resistance (10, 11). In eastern China, the ART-related PfK13 mutation R561H was identified as the main mutation site in Zhejiang Province among migrant workers from Rwanda (12). In 2017, Jiangsu Province reported a patient who had returned from Equatorial Guinea and was harboring the M579I site, which was confirmed to be linked to ACT resistance, with a 2.29% in vitro survival rate by ring-stage survival assay (13), suggesting that careful surveillance of African parasite populations is still warranted.
Another molecular marker, encoding P. falciparum ubiquitin-specific protease (pfubp1), was adopted as a major molecular marker for monitoring ACT resistance in Africa by recent surveillance studies (14, 15). It is known that ubp1 encodes a deubiquitinating (DUB) enzyme that is responsible for the cleavage of ubiquitin from any protein or peptide to which it is joined (16). A homologue of pfubp1 was first discovered in the rodent malaria parasite Plasmodium chabaudi (pcubp1 encodes ubiquitin carboxy-terminal hydrolase 1) and found to be associated with ACT resistance (17). Two studies in Kenya and Ghana showed that the codon changes D1525E (from aspartic acid to glutamic acid) and E1528D (from glutamic acid to aspartic acid) are closely associated with delayed parasite clearance (18, 19). More studies are therefore required to validate pfubp1 and its role in ACT resistance.
The increase in imported P. falciparum cases from Africa, which has become the main infection source in China (20), is a major risk factor for the spread of ACT resistance. Here, we aimed to investigate the genetic diversity of the PfK13 propeller and pfubp1 alleles in Anhui and Shandong Provinces in eastern China and to examine the PfK13 and pfubp1 mutation status of the pathogen population so as to monitor and evaluate the potential emergence of ACT resistance.
RESULTS
Epidemiological and clinical study.
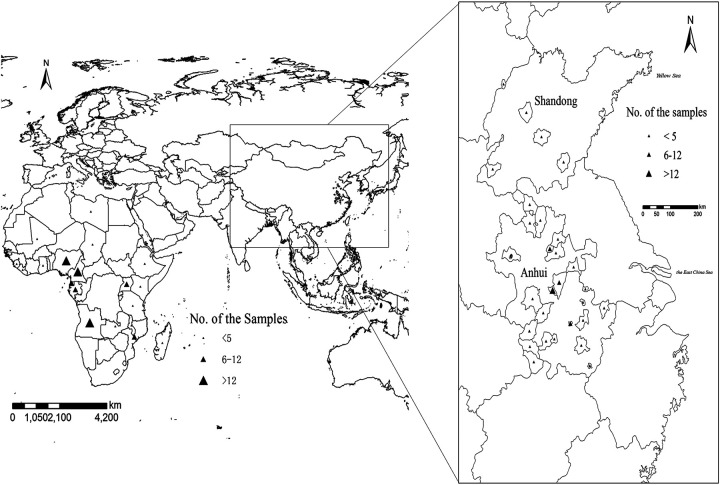
A total of 153 P. falciparum malaria cases were involved in this study, and all were imported from African countries (Fig. 1). The average patient age was 42 years, and 147 patients (147/153 [96.1%]) were male. The numbers of cases reported in 2017, 2018, and 2019 were 41, 44, and 68, respectively. The P. falciparum cases were mainly imported from the Democratic Republic of the Congo (n = 28), Nigeria (n = 27), and Angola (n = 16). None of the patients who stayed in hospital (n = 86) were positive on day 3 after ACT administration as detected by microscopy.
FIG 1.
Study sample collection sites in source countries in Africa and in counties in Shandong and Anhui. All countries and counties are labeled according to the number of samples obtained using ArcGIS 10.1.
Polymorphisms in the PfK13 propeller.
To investigate PfK13 propeller polymorphisms, all 153 P. falciparum samples were successfully sequenced and analyzed (Table 1). A total of five isolates with PfK13 propeller mutation sites were observed. Among them, three isolates harbored the same synonymous mutation (C469C) and had come from Nigeria, Ghana, and Liberia. Another isolate, from the Democratic Republic of the Congo, reported in 2019, harbored the synonymous mutation site T537T. The remaining isolate, from Uganda, was found to carry V589A (Table 1).
TABLE 1.
Prevalence of P. falciparum PfK13 polymorphisms in travelers returning from Africa in Shandong and Anhui Provinces, 2017–2019
| Study site | No. of wild-type genes |
PfK13 mutation |
Yr | ||
|---|---|---|---|---|---|
| Site | No. of isolates | Prevalence (%) | |||
| Anhui | 41 | 0 | 0 | 0 | 2017 |
| 42 | C469C | 2 | 4.8 | 2018 | |
| 41 | C469C | 1 | 2.4 | 2019 | |
| Shandong | 24 | T573T V589A | 2 | 8.3 | 2019 |
Polymorphisms in pfubp1.
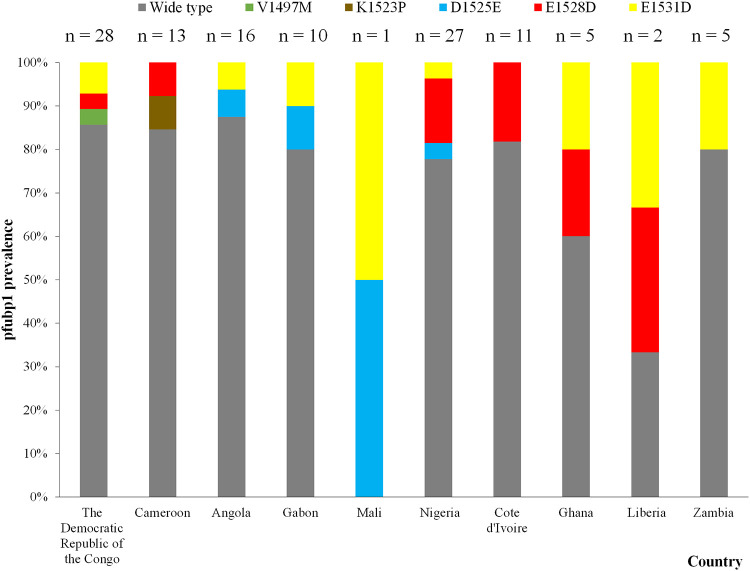
For pfubp1 investigation and evaluation, a 300-bp region was amplified and sequenced. All 153 samples were successfully sequenced. A total of 20 isolates (13.1% [20/153]) were found to harbor seven different SNPs. Among them, E1528D was the major polymorphism (n = 10) (6.5% [10/153]), followed by E1531D (n = 9) (5.9% [9/153]) and D1525E (n = 4) (2.6% [4/153]) (Table 2). The E1528D and E1531D sites were found mainly in patients returning from Nigeria (n = 4) and the Democratic Republic of the Congo (n = 2), respectively (Fig. 2). Fourteen of the isolates had single mutation sites, while the remaining six were found to harbor mixed mutant alleles: three isolates were found to harbor both D1525E and E1531D, two isolates were found to harbor both E1528D and E1531D, and one isolate was found with both V1497M and E1531D.
TABLE 2.
Prevalence of pfubp1 polymorphisms in travelers returning from Africa in Shandong and Anhui Provinces, 2017–2019
| Study site | No. of wild-type genes |
pfubp1 mutation |
Yr | ||
|---|---|---|---|---|---|
| Site | No. of isolates | Prevalence (%) | |||
| Anhui | 40 | D1525E | 1 | 2.4 | 2017 |
| E1528D | 1 | 2.4 | |||
| 38 | D1525E | 2 | 4.5 | 2018 | |
| E1528D | 3 | 6.7 | |||
| E1531D | 5 | 11.1 | |||
| 32 | V1497M | 1 | 2.4 | 2019 | |
| D1525E | 1 | 2.4 | |||
| E1528D | 6 | 14.3 | |||
| E1531D | 4 | 9.5 | |||
| Shandong | 24 | K1523P | 1 | 3.8 | 2019 |
| E1531G | 1 | 3.8 | |||
FIG 2.
Prevalences of pfubp1 in travelers returning from source countries. The numbers of samples for the source countries, including wild-type and mutated sequences, are indicated.
Insertions and deletions in pfubp1 isolates.
The multiple nucleotide sequence alignment results showed that nine isolates were genotyped with deletions of AAATACGAC (encoding KID) at amino acid residues 1520 to 1522, and seven isolates were identified with deletions of AAATACGAA (encoding KIE) at amino acid residues 1526 to 1528 (Fig. S1). In addition, we found that four isolates had insertions of AAATATGAA (encoding KYE) at amino acid residues 1526 to 1528, and two isolates had insertions of AAATATGACAAATATGAC (encoding KYDKYD) at amino acid residues 1519 to 1524 (Fig. S1).
DISCUSSION
The resistance of P. falciparum to ACTs is one of the major challenges facing malaria elimination worldwide (21). The previous work of our group focused on the molecular markers found in migrant populations along the China-Myanmar and China-Vietnam borders, because these regions were at high risk due to the ART that emerged and spread from the Thailand-Myanmar border in the GMS (11, 22, 23). In this study, we aimed to determine genetically whether new candidate SNPs in K13 or pfubp1 existed among patients returning from African countries, in order to understand the population genetics of K13 and pfubp1 polymorphisms and to explore the relationship between molecular markers and clinical drug effectiveness.
Our findings provide evidence that a limited number of PfK13 polymorphisms were present among isolates from Africa, in agreement with many other reports (24–26). Interestingly, synonymous mutations accounted for the majority of the K13 mutant alleles observed. We found four isolates harboring two synonymous mutations, C469C and T537T, which have no correlation with ART. However, the widely reported mutation A578S was not observed in this study. The treatment outcomes of these patients showed that all the isolates were sensitive to ACT, and no positive parasite results were observed on day 3 after ACT administration.
Since a limited set of PfK13 molecular markers occurred in the African isolates, we introduced another marker, pfubp1, to explore its role in delayed parasite clearance among African isolates. This assay detected the known delayed parasite clearance genotypes D1525E and E1528D, which were observed in 2.6% (n = 4) and 6.5% (n = 10) of the isolates, respectively. D1528E was also found in isolates from Ghana, Kenya, and Tanzania (14, 15, 27), and the prevalence of E1528D was lower than those among the Ghanaian (7.4% [6/81]), Kenyan (17.1% [6/35]), and Tanzanian (10.5% [2/19]) isolates. Notably, among the African isolates, especially those from Nigeria (which reported four isolates with pfubp1 D1528E), the ART of the parasite populations increases in the presence of long-term drug use. In addition, we observed a novel mutation, E1531D, at rates as high as 5.9% in the African isolates. Therefore, new mutations such as E1531D may be selected, and their prevalence may increase with the continuing use of ACTs to treat African isolates.
Combining the SNP results of PfK13 and pfubp1, we found that one isolate from Ghana simultaneously harbored the C469C K13 allele and the E1528D and E1531D alleles of pfbup1, while another two isolates harbored the T537T and V589A K13 alleles and the KID and KIE pfubp1 deletions. This surprising result may provide a basis for further investigation into possible correlations among these mutations using additional K13 and pfubp1 genotypes from different geographical isolates.
Interestingly, we found two types of deletions and two types of insertions in the multiple sequence alignment between the mutated isolates and wild-type isolates. The KYE insertion at bp 4576 to 4584 was also observed in isolates from Ghana, Kenya, and Burkina Faso and in a patient returning from Liberia to the United Kingdom (14, 15, 28). The KYDKYD insertion at bp 4555 to 4572 had not been observed previously, nor had either of the deletions (KID and KIE). The most common deletions and insertions in pfubp1 in this study were lysine, asparagine, and glutamic acid residues. These resulted from deletions/insertions of the amino acids KID, KIE, KYE, and KYDKYD at positions ranging from 1519 to 1528. These deletions/insertions led to frameshifts that were caused by a deletion or insertion of either a thymidine or a guanosine nucleotide. Although the treatment outcomes of the patients harboring these isolates indicated that they were still sensitive to ACTs, more studies of the roles of these deletions and insertions in the relationship between phenotype and clinical outcome are needed to validate these results.
Conclusion.
The present results showing molecular markers including PfK13 and pfubp1 in African isolates have implications for the development of ART with continuing use of ACTs. The findings of this study indicated the existence of limited PfK13 polymorphisms among African isolates and suggested the adoption of other potential molecular markers of ART, such as pfubp1, to carry out molecular surveillance for clinical analysis of P. falciparum ART. Further research is under way to investigate and elucidate both PfK13 and pfubp1, as well as other candidate molecular markers, and their roles in delayed parasite clearance in clinical trials.
MATERIALS AND METHODS
Study sites and samples.
This study was conducted in Anhui and Shandong Provinces in eastern China, which were predominant centers of imported P. falciparum malaria. Anhui Province covers 105 counties with 70.6 million people and experienced a malaria resurgence in 2005–2008 that was due mainly to the accumulation of residual foci of P. vivax (29). Shandong Province has a long coastline measuring 3,024.4 km. It contains 137 counties and has a population of 97.9 million. Economic trade overseas is frequent. The number of imported P. falciparum cases, especially those from Africa, has increased significantly in these two provinces, and 240 P. falciparum cases were reported in 2018, accounting for 13.6% of all P. falciparum cases nationwide (2).
A total of 153 P. falciparum-infected blood samples of travelers returning from Africa from 2017 to 2019 were collected and examined at enrollment, including 127 cases from Anhui and 26 cases from Shandong (Fig. 1). Approximately 100 μl of finger prick blood was spotted onto a piece of 3MM Whatman filter paper (GE Healthcare, Boston, MA, USA) and air dried. Each of the samples was labeled with a study number and stored at −4°C until extraction.
Individual epidemiological information was also collected from a Web-based reporting system (China Information System for Disease Control and Prevention) and analyzed in this study.
Treatment and follow-up.
Patients were treated with dihydroartemisinin and piperaquine according to national antimalarial regulations, with a total adult dose of 2.5 mg dihydroartemisinin/kg of body weight and 20 mg/kg piperaquine for 3 days (30). The thick and thin blood smears of the patients who stayed in the hospital for treatment were collected on day 3. Giemsa-stained blood slides were prepared for the identification of Plasmodium to species level. Slides were examined and read by an expert microscopist certified as level 1 by the WHO. Slides were considered negative when no asexual parasites were found after 1,000 white blood cells (WBCs) had been counted.
Genetic polymorphisms.
The Plasmodium falciparum genomic DNA from the approximately 10-μl blood samples collected was extracted with a QIAamp DNA blood kit (Qiagen, Valencia, CA) as described previously (4). Polymorphisms in the PfK13 gene (PF3D7_1343700) were determined by nested PCR amplification of an 849-bp fragment (from amino acids 427 to 709) as described previously (14). Polymorphisms in pfubp1 (PF3D7_0104300) were identified in the 300-bp region (from amino acids 1463 to 1563) by using a slightly revised PCR method (14). A DNA sample extracted from the 3D7 parasite strain was used as a positive control. The primers used for the pfubp1 nested PCR were as follows: nest 1 forward primer, CGCCCGTACTATGAAGAAGATC; nest 1 reverse primer, GGCTTTTACCTGAACTGTTCAGG; nest 2 forward primer, CGTAAACAGAATATTCAGGATTGG; nest 2 reverse primer, CTAGCCCTTTATTATCATTATCGT.
Amplification of the pfubp1 polymorphisms was performed with 40 cycles of a 3-step PCR procedure with 30 s annealing at 52°C (nest 1) or 55°C (nest 2) and elongation at 72°C for 90 s. The PCR products were collected and sent for Sanger sequencing (Shanghai Bunan Biological Co., Ltd., Shanghai, China).
Data analysis.
Sequences were analyzed with the BLAST program (http://blast.ncbi.nlm.nih.gov/). Multiple nucleotide sequence alignments and analysis were carried out using the MAFFT Web-based tool with Clustal Omega Sequence Alignment Editor (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences with poor quality after three sequencing attempts were not included in the analysis. A map showing the study sites and the numbers of isolates was created by ArcGIS 10.1 (Environmental Systems Research Institute, Inc.). Version 4.0.2 of the R statistical software (R Foundation for Statistical Computing, Vienna, Austria) was used to conduct statistical analyses, and the chi-square test was employed to test the different constituent ratios of pfubp1 and PfK13 polymorphisms between Anhui and Shandong in 2017–2019. GraphPad Prism 8.3.0 (GraphPad Software Inc., San Diego, CA, USA) was used to plot the distribution of pfubp1 and PfK13 mutations.
Ethical considerations.
This study was reviewed and approved by the ethical committee of the National Institute of Parasitic Diseases, Chinese Centre for Disease Control and Prevention (NIPD, China CDC; no. 2019008).
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the key techniques in collaborative prevention and control of major infectious diseases in the Belt and Road Initiative (grant 2018ZX10101002-004), the National Natural Science Foundation of China (grant 81602904), and the State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (grant MMLKF14-03).
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Feng J, Xia Z, Vong S, Yang W, Zhou S, Xiao N. 2014. Preparedness for malaria resurgence in China: case study on imported cases in 2000–2012. Adv Parasitol 86:231–265. doi: 10.1016/B978-0-12-800869-0.00009-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Feng J, Zhang S, Xia Z, Zhou S. 2019. Epidemiological characteristics of malaria and the progress towards its elimination in China in 2018. Chin J Parasitol Parasit Dis 37:241–247. doi: 10.12140/j.issn.1000-7423.2019.03.001. [DOI] [Google Scholar]
- 3.World Health Organization. 2019. World malaria report 2019. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Tracking Resistance to Artemisinin Collaboration (TRAC), et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 2018. Accelerating malaria elimination in the Greater Mekong Subregion. World Health Organization, Geneva, Switzerland: https://www.who.int/malaria/areas/greater_mekong/overview/en/. [Google Scholar]
- 7.World Health Organization. 2018. Artemisinin resistance and artemisinin-based combination therapy efficacy: status report. World Health Organization, Geneva, Switzerland: https://apps.who.int/iris/handle/10665/274362?search-result=true&query=Artemisinin+and+artemisinin-based+combination+therapy+resistance&scope=&rpp=10&sort_by=score&order=desc. [Google Scholar]
- 8.Boulle M, Witkowski B, Duru V, Sriprawat K, Nair SK, McDew-White M, Anderson TJ, Phyo AP, Menard D, Nosten F. 2016. Artemisinin-resistant Plasmodium falciparum K13 mutant alleles, Thailand-Myanmar border. Emerg Infect Dis 22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJ, Nair S, McDew-White M, Flegg JA, Grist EP, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NP, Nosten F, White NJ, Woodrow CJ. 2015. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis 15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Thakur GD, Dieye A, Djalle D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino FE, Fandeur T, Ferreira-da-Cruz MF, Fola AA, Fuehrer HP, Hassan AM, Herrera S, Hongvanthong B, Houze S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati JB, Menard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niare K, Noedl H, KARMA Consortium, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng J, Kong X, Xu D, Yan H, Zhou H, Tu H, Lin K. 2019. Investigation and evaluation of genetic diversity of Plasmodium falciparum Kelch 13 polymorphisms imported from Southeast Asia and Africa in southern China. Front Public Health 7:95. doi: 10.3389/fpubh.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Ruan W, Zhou S, Huang F, Lu Q, Feng X, Yan H. 2020. Molecular surveillance of Pfcrt and k13 propeller polymorphisms of imported Plasmodium falciparum cases to Zhejiang Province, China between 2016 and 2018. Malar J 19:59. doi: 10.1186/s12936-020-3140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu F, Culleton R, Zhang M, Ramaprasad A, von Seidlein L, Zhou H, Zhu G, Tang J, Liu Y, Wang W, Cao Y, Xu S, Gu Y, Li J, Zhang C, Gao Q, Menard D, Pain A, Yang H, Zhang Q, Cao J. 2017. Emergence of indigenous artemisinin-resistant Plasmodium falciparum in Africa. N Engl J Med 376:991–993. doi: 10.1056/NEJMc1612765. [DOI] [PubMed] [Google Scholar]
- 14.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, van Schalkwyk DA, Sawa P, Omar SA, Clark TG, Bousema T, Sutherland CJ. 2014. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis 210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams T, Ennuson NAA, Quashie NB, Futagbi G, Matrevi S, Hagan OCK, Abuaku B, Koram KA, Duah NO. 2018. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates. Parasit Vectors 11:175. doi: 10.1186/s13071-018-2762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KD. 2005. The discovery of ubiquitin-dependent proteolysis. Proc Natl Acad Sci U S A 102:15280–15282. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. 2007. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol 65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrmann S, Straimer J, Mwai L, Abdi A, Rippert A, Okombo J, Muriithi S, Sasi P, Kortok MM, Lowe B, Campino S, Assefa S, Auburn S, Manske M, Maslen G, Peshu N, Kwiatkowski DP, Marsh K, Nzila A, Clark TG. 2013. Genome-wide screen identifies new candidate genes associated with artemisinin susceptibility in Plasmodium falciparum in Kenya. Sci Rep 3:3318. doi: 10.1038/srep03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rovira-Vallbona E, Bottieau E, Guetens P, Verschueren J, Rebolledo J, Nulens E, Van der Hilst J, Clerinx J, Van Esbroeck M, Rosanas-Urgell A. 2019. Imported malaria and artemisinin-based combination therapy failure in travellers returning to Belgium: a retrospective study. Travel Med Infect Dis doi: 10.1016/j.tmaid.2019.101505. [DOI] [PubMed] [Google Scholar]
- 20.Feng J, Zhang L, Huang F, Yin JH, Tu H, Xia ZG, Zhou SS, Xiao N, Zhou XN. 2018. Ready for malaria elimination: zero indigenous case reported in the People's Republic of China. Malar J 17:315. doi: 10.1186/s12936-018-2444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J, Zhou D, Lin Y, Xiao H, Yan H, Xia Z. 2015. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with Plasmodium falciparum and Plasmodium vivax isolates from the China-Myanmar border. Antimicrob Agents Chemother 59:2554–2559. doi: 10.1128/AAC.04843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Campino S, Diez Benavente E, Warhurst DC, Beshir KB, Lubis I, Gomes AR, Feng J, Jiazhi W, Sun X, Huang F, Tang LH, Sutherland CJ, Clark TG. 2019. Artemisinin resistance-associated markers in Plasmodium falciparum parasites from the China-Myanmar border: predicted structural stability of K13 propeller variants detected in a low-prevalence area. PLoS One 14:e0213686. doi: 10.1371/journal.pone.0213686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Li J, Yan H, Feng X, Xia Z. 2015. Evaluation of antimalarial resistance marker polymorphism in returned migrant workers in China. Antimicrob Agents Chemother 59:326–330. doi: 10.1128/AAC.04144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou R, Yang C, Li S, Zhao Y, Liu Y, Qian D, Wang H, Lu D, Zhang H, Huang F. 2019. Molecular surveillance of drug resistance of Plasmodium falciparum isolates imported from Angola in Henan Province, China. Antimicrob Agents Chemother 63:e00552-19. doi: 10.1128/AAC.00552-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, Xu X, Jiang J, Yu C, Tian C, Li W. 2018. Surveillance of antimalarial resistance molecular markers in imported Plasmodium falciparum malaria cases in Anhui, China, 2012–2016. Am J Trop Med Hyg 98:1132–1136. doi: 10.4269/ajtmh.17-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemu G, Nizar E, Göte S, Erasmus K, Angel C. 2014. Large differences in variability for genes associated with antimalarial drug resistance between samples from Tanzania and Ethiopia. Malar J 13(Suppl 1):P87. [Google Scholar]
- 28.Sutherland CJ, Lansdell P, Sanders M, Muwanguzi J, van Schalkwyk DA, Kaur H, Nolder D, Tucker J, Bennett HM, Otto TD, Berriman M, Patel TA, Lynn R, Gkrania-Klotsas E, Chiodini PL. 2017. pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob Agents Chemother 61:e02382-16. doi: 10.1128/AAC.02382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, Xiao H, Xia Z, Zhang L, Xiao N. 2015. Analysis of malaria epidemiological characteristics in the People's Republic of China, 2004–2013. Am J Trop Med Hyg 93:293–299. doi: 10.4269/ajtmh.14-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Health Commission. 2016. Technical regulations for application of antimalarials. National Health Commission, Beijing, China. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.