The abilities of the new Vitek 2 AST-YS08 (YS08) and Sensititre YeastOne (SYO) systems to detect the resistances of Candida isolates to azoles and echinocandins were evaluated. In total, 292 isolates, including 28 Candida albicans (6 Erg11 and 2 Fks mutants), 57 Candida parapsilosis (26 Erg11 mutants), 24 Candida tropicalis (10 Erg11 and 1 Fks mutants), and 183 Candida glabrata (39 Pdr1 and 13 Fks mutants) isolates, were tested.
KEYWORDS: Candida, azole, echinocandin, Vitek 2 AST-YS08, Sensititre YeastOne
ABSTRACT
The abilities of the new Vitek 2 AST-YS08 (YS08) and Sensititre YeastOne (SYO) systems to detect the resistances of Candida isolates to azoles and echinocandins were evaluated. In total, 292 isolates, including 28 Candida albicans (6 Erg11 and 2 Fks mutants), 57 Candida parapsilosis (26 Erg11 mutants), 24 Candida tropicalis (10 Erg11 and 1 Fks mutants), and 183 Candida glabrata (39 Pdr1 and 13 Fks mutants) isolates, were tested. The categorical agreements (CAs) between the Clinical and Laboratory Standards Institute (CLSI) method and YS08 fluconazole MICs obtained using clinical breakpoints were 92.4% (C. albicans), 96.5% (C. parapsilosis), and 87.0% (C. tropicalis), and the CAs between the CLSI and SYO MICs were 92.3% (C. albicans), 77.2% (C. parapsilosis), 100% (C. tropicalis), and 98.9% (C. glabrata). For C. glabrata, the CAs with the CLSI micafungin MICs were 92.4% and 55.5% for the YS08 micafungin and caspofungin MICs, respectively; they were 100%, 95.6%, and 98.9% for the SYO micafungin, caspofungin, and anidulafungin MICs, respectively. YS08 does not provide fluconazole data for C. glabrata; the CA with the CLSI fluconazole MIC was 97.8% for the YS08 voriconazole MIC, using an epidemiological cutoff value (ECV) of 0.5 μg/ml. Increased CAs with the CLSI MIC were observed for the YS08 MIC using CLSI ECVs (for fluconazole and C. tropicalis, 100%; for micafungin and C. glabrata, 98.9%) and for the SYO MIC using method-specific ECVs (for fluconazole and C. parapsilosis, 91.2%; for caspofungin and C. glabrata, 98.9%). Therefore, the YS08 and SYO systems may have different abilities to detect mechanisms of azole and echinocandin resistance in four Candida species; the use of method-specific ECVs may improve the performance of both systems.
INTRODUCTION
Antifungal susceptibility testing (AFST) against Candida is useful for optimizing the treatment of Candida bloodstream infections (BSIs) (1, 2). In the clinical microbiology laboratory, two commercial broth microdilution (BMD)-based AFSTs, the Vitek 2 system (bioMérieux, Marcy l’Etoile, France) and the Sensititre YeastOne (SYO) system (Thermo Scientific, Cleveland, OH, USA), are widely used, particularly for Candida BSI isolates (3–6). The Vitek 2 system is a fully automated susceptibility testing system used for both bacteria and yeasts in many laboratories and has the advantages of rapidity, reduced costs, and ease of use (5–7). A new Vitek 2 AST-YS08 yeast susceptibility system (referred to as YS08) was recently updated to reflect the revised species-specific clinical breakpoints (CBPs) defined by the Clinical and Laboratory Standards Institute (CLSI) for common Candida species, but YS08 does not provide fluconazole MIC data for any of the Candida glabrata isolates (7, 8). On the other hand, SYO, the colorimetric broth microdilution method, has good concordance with the CLSI reference method for Candida susceptibility testing (4, 9) but requires manual setup and visual reading of the results (10, 11). To date, only one study has evaluated the new YS08 using revised CLSI CBPs (8). In this study, the essential agreement (EA) and categorical agreement (CA) between YS08 and SYO were suboptimal using 68 clinical Candida isolates skewed for resistant isolates. They suggest that validation data with resistant isolates need to be obtained (8).
Candida develops resistance to echinocandins through point mutations in the FKS1 (all Candida species) or FKS2 (C. glabrata only) gene, which are linked to resistance to all three echinocandins, including micafungin, caspofungin, and anidulafungin (12, 13). Several studies have evaluated the performance of echinocandin susceptibility testing by using a panel of Candida isolates with and without known FKS mutation-related echinocandin resistance mechanisms (14, 15), but there are limited data regarding the evaluation of AFSTs for the detection of azole-resistant Candida isolates using molecularly defined isolates (16). Multiple mechanisms of azole resistance, such as ERG11 mutations and overexpression of efflux pumps, have been reported for Candida albicans, Candida parapsilosis, and Candida tropicalis (17–19); however, the overwhelming singular mechanism of resistance identified in clinical isolates of C. glabrata is mutation of the transcription factor PDR1, which leads to increased expression of multidrug transporters that act as efflux pumps (20, 21).
The evaluation of AFST methods for Candida species is difficult, partly because the results can be influenced by the composition of resistant Candida isolates within a collection. When evaluation studies are performed using an abundance of antifungal-susceptible isolates in the collection, very major errors (VMEs) and substantial major discrepancies are uncommon, while errors are more common when studies are performed using an abundance of resistant isolates (22, 23). In addition, CA between two systems may vary depending on the interpretive criteria of the MICs used. Although most commercial AFST systems use CLSI CBPs for categorical interpretation of MICs (24, 25; bioMérieux), method-specific epidemiological cutoff values (ECVs) have recently been proposed as the interpretive criteria for SYO or Etest (26, 27). Given that the performance of AFST relies on categorical interpretive criteria of the MIC results, we performed a comparative evaluation of two commercial AFSTs (YS08 and SYO) and the reference CLSI BMD method for the detection of azole- and echinocandin-resistant isolates of four common Candida species, applying different MIC interpretive criteria, including CLSI CBPs, CLSI ECVs, or the method-specific ECVs of SYO (SYO ECVs). We tested a panel of well-characterized Candida strains from a South Korean collection composed of 97 azole- or echinocandin-resistant isolates with molecularly defined resistance mechanisms that reflect the antifungal resistance mechanisms most commonly identified in isolates of four common Candida species from South Korean hospitals (19, 28); we also tested 195 control isolates without defined resistance mechanisms. Furthermore, we evaluated the ability of voriconazole MIC determination using YS08 to correctly detect fluconazole-resistant (FR) C. glabrata isolates.
RESULTS
Fluconazole testing for C. albicans, C. parapsilosis, and C. tropicalis.
When 42 FR isolates (6 FR C. albicans isolates harboring the K143R mutation in Erg11p, 26 C. parapsilosis isolates harboring the Y132F mutation in Erg11p, and 10 FR C. tropicalis isolates with ERG11 overexpression or the Y132F mutation in Erg11p) and 64 fluconazole-susceptible (FS) isolates (20 C. albicans, 31 C. parapsilosis, and 13 C. tropicalis isolates without defined resistance mechanisms) were tested, the EAs between the CLSI M27 method and YS08 with fluconazole were 96.2%, 96.5%, and 100% for C. albicans, C. parapsilosis, and C. tropicalis, respectively; the EAs between CLSI M27 and SYO were 92.3%, 100%, and 95.7%, respectively (Table 1). When the categorical results for fluconazole with YS08 determined using two different interpretive criteria (CLSI CBPs or ECVs) were compared to those produced by the CLSI method, the agreements of YS08 using CLSI CBPs for C. albicans, C. parapsilosis, and C. tropicalis were 92.4%, 96.5%, and 87.0%, respectively; agreements determined using CLSI ECVs were 73.1%, 96.5%, and 100%, respectively. When the results for fluconazole produced by SYO were compared to those produced by the CLSI method, the CAs of the fluconazole MICs produced by SYO using CLSI CBPs for C. albicans, C. parapsilosis, and C. tropicalis were 92.3%, 77.2%, and 100%, respectively; the CAs obtained using SYO ECVs were 88.5%, 91.2%, and 100%, respectively.
TABLE 1.
CAs between fluconazole MICs obtained by the CLSI BMD method and those obtained by each of the two commercial BMD assays for 197 molecularly defined isolates of four common Candida species according to different MIC interpretive criteriaa
| Species (no. of isolates tested) | Antifungal agent | Test method | Interpretive criterionb | % of isolates with the following categorical MIC interpretationc
: |
CA (%) | No. (%) of isolates with discrepanciesd
|
EA (%)e | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R or non-WT | SDD | S or WT | VME | ME | Minor | ||||||
| C. albicans (26) | Fluconazole | CLSI M27 | CLSI CBP (8) | 23.1 | 0.0 | 76.9 | |||||
| YS08 | CLSI CBP (8) | 15.4 | 3.8 | 80.8 | 92.4 | 1 (3.8) | 0 (0.0) | 1 (3.8) | 96.2 | ||
| YS08 | CLSI ECV (1) | 42.3 | 57.7 | 73.1 | 1 (3.8) | 6 (23.1) | 0 (0.0) | ||||
| SYO | CLSI CBP (8) | 30.8 | 0.0 | 69.2 | 92.3 | 0 (0.0) | 2 (7.7) | 0 (0.0) | 92.3 | ||
| SYO | SYO ECV (2) | 34.6 | 65.4 | 88.5 | 0 (0.0) | 3 (11.5) | 0 (0.0) | ||||
| C. parapsilosis (57) | Fluconazole | CLSI M27 | CLSI CBP (8) | 45.6 | 0.0 | 54.4 | |||||
| YS08 | CLSI CBP (8) | 42.1 | 3.5 | 54.4 | 96.5 | 0 (0.0) | 0 (0.0) | 2 (3.5) | 96.5 | ||
| YS08 | CLSI ECV (2) | 49.1 | 50.9 | 96.5 | 0 (0.0) | 2 (3.5) | 0 (0.0) | ||||
| SYO | CLSI CBP (8) | 24.6 | 15.8 | 59.6 | 77.2 | 4 (7.0) | 0 (0.0) | 9 (15.8) | 100.0 | ||
| SYO | SYO ECV (4) | 40.4 | 59.6 | 91.2 | 4 (7.0) | 1 (1.8) | 0 (0.0) | ||||
| C. tropicalis (23) | Fluconazole | CLSI M27 | CLSI CBP (8) | 43.5 | 0.0 | 56.5 | |||||
| YS08 | CLSI CBP (8) | 30.5 | 13.0 | 56.5 | 87.0 | 0 (0.0) | 0 (0.0) | 3 (13.0) | 100.0 | ||
| YS08 | CLSI ECV (2) | 43.5 | 56.5 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| SYO | CLSI CBP (8) | 43.5 | 0.0 | 56.5 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 95.7 | ||
| SYO | SYO ECV (8) | 43.5 | 56.5 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| C. glabrata (91) | Fluconazole | CLSI M27 | CLSI CBP (64) | 42.9 | 57.1 | ||||||
| SYO | CLSI CBP (64) | 44.0 | 56.0 | 98.9 | 0 (0.0) | 0 (0.0) | 1 (1.1) | 95.6 | |||
| SYO | SYO ECV (128) | 40.7 | 59.3 | 97.8 | 2 (2.2) | 0 (0.0) | 0 (0.0) | ||||
| Voriconazole | YS08 | P-CLSI ECV (1) | 42.9 | 57.1 | 97.8 | 1 (1.1) | 1 (1.1) | 0 (0.0) | NA | ||
| YS08 | CLSI ECV (0.5) | 58.2 | 41.8 | 82.4 | 1 (1.1) | 15 (16.5) | 0 (0.0) | ||||
CLSI, Clinical and Laboratory Standards Institute; CLSI M27, CLSI M27-ED4 broth microdilution method (BMD); YS08, Vitek 2 AST-YS08 yeast susceptibility system; SYO, Sensititre YeastOne system; CLSI CBP, clinical breakpoint provided by CLSI; ECV, epidemiological cutoff value; SYO ECV, method-specific ECV of SYO; P-CLSI ECV, previous CLSI epidemiological cutoff value; R, resistant; non-WT, non-wild type; SDD, susceptible, dose dependent; S, susceptible; WT, wild type; CA, categorical agreement; VME, very major error; ME, major error; minor, minor error; EA, essential agreement; NA, not available.
The values in parentheses denote interpretive fluconazole MIC criteria (in micrograms per milliliter) from the CLSI CBPs (MICs greater than or equal to the MIC for resistance), CLSI ECVs (MICs greater than or equal to the non-WT MIC), or SYO ECVs (MICs greater than or equal to the non-WT MIC) against each Candida species (27, 30, 34). The results for voriconazole obtained with YS08 were alternatively evaluated for C. glabrata isolates because YS08 does not provide fluconazole results for C. glabrata. Although a CLSI clinical breakpoint is not yet proposed for voriconazole and C. glabrata, both the previous CLSI ECV (0.5 μg/ml) and the revised CLSI ECV (0.25 μg/ml) were used (34, 45).
Each of the categorical results were determined to indicate resistant, susceptible, dose dependent, and susceptible using speciesspecific CLSI CBPs of the CLSI M60, 1st edition, method (30) and as non-WT and WT using CLSI ECVs of the CLSI M59, 2nd edition, method (34) or the SYO ECVs (27).
Details regarding isolates with categorical errors obtained using CLSI CBPs were as follows. For C. albicans, YS08 misclassified two fluconazole-resistant (FR) isolates harboring a K143R mutation in Erg11 as fluconazole susceptible, dose dependent (F-SDD), and fluconazole susceptible (FS; MIC, ≤0.5 μg/ml), respectively. SYO misclassified two FS isolates (MICs, 1 and 2 μg/ml) as FR (MIC, >256 μg/ml). For C. parapsilosis, YS08 misclassified two FR isolates harboring Y132F in Erg11 as F-SDD, and SYO misclassified four and eight FR isolates as FS (MIC, 2 μg/ml) and F-SDD, respectively, and one FS isolate (MIC, 2 μg/ml) as F-SDD. For C. tropicalis, YS08 misclassified 3 FR isolates as F-SDD. For C. glabrata, SYO misclassified one F-SDD isolate (MIC, 8 μg/ml) as FR (MIC, 64 μg/ml).
Essential agreement (±2 log2 dilutions) between the MICs obtained by the CLSI M27 method and each of the Vitek 2 YS08 and YeastOne MICs.
Azole testing for C. glabrata.
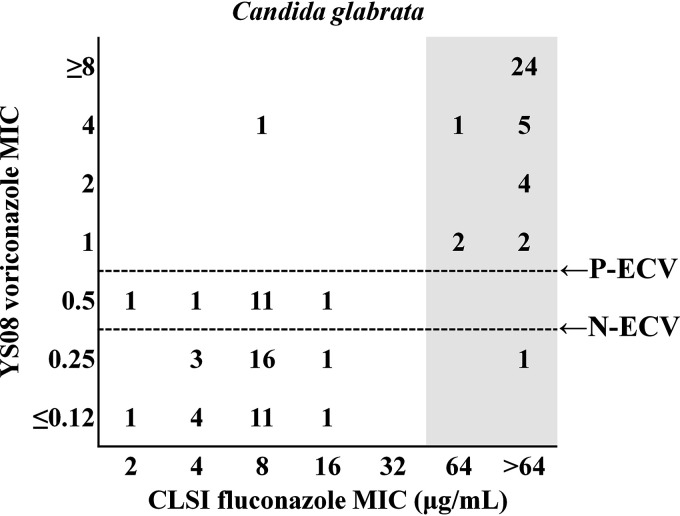
When the fluconazole MIC results for 91 isolates of C. glabrata (39 FR isolates harboring a Pdr1 mutation and 52 fluconazole-susceptible, dose dependent [F-SDD], isolates without Pdr1 mutations) obtained with SYO were compared to those produced by the CLSI method, the CAs for fluconazole produced by SYO using the CLSI CBPs and the SYO ECVs were 98.9% and 97.8%, respectively (Table 1). The EA for fluconazole between CLSI M27 and SYO was 95.6%. Because YS08 does not provide fluconazole data for C. glabrata, the results for voriconazole obtained with YS08 were used to predict the fluconazole susceptibility category for C. glabrata. Although a species-specific CLSI CBP has not yet been proposed for voriconazole and C. glabrata, categorical results for the YS08 voriconazole MICs based on the previous CLSI ECV of 0.5 μg/ml showed 97.8% agreement with the results of the CLSI method for fluconazole, whereas the categorical result of YS08 for voriconazole based on the new CLSI ECV (0.25 μg/ml) showed 82.4% agreement with the results of the CLSI method for fluconazole. Figure 1 shows that the voriconazole results obtained with YS08 with the previous CLSI ECV (P-ECV; 0.5 μg/ml) demonstrated a clearer separation between Pdr1 mutants and non-Pdr1 mutants than the results obtained with the new CLSI ECV (N-ECV; 0.25 μg/ml).
FIG 1.
A scattergram comparing the fluconazole MICs obtained by the Clinical and Laboratory Standards Institute (CLSI) method and voriconazole MICs obtained with the Vitek 2 AST-YS08 (YS08) system. Numbers represent the number of isolates of Candida glabrata at each MIC pair. The gray zone indicates fluconazole-resistant Pdr1 mutants of C. glabrata. The voriconazole MIC results obtained with YS08 with the previous CLSI ECV (P-ECV; 0.5 μg/ml) showed a clearer separation between Pdr1 mutants and non-Pdr1 mutants than those obtained with the new CLSI ECV (N-ECV; 0.25 μg/ml).
Echinocandin testing with YS08 and SYO.
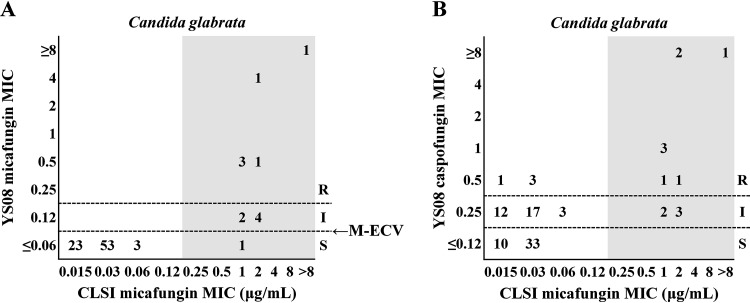
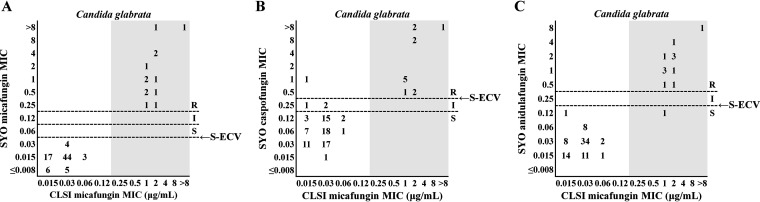
Table 2 summarizes the agreements between the micafungin susceptibility results obtained by the CLSI method and those obtained by each of the echinocandin tests with the YS08 or SYO method for 95 isolates of three common Candida species. Two micafungin-resistant (MR) C. albicans isolates and one MR C. tropicalis isolate harboring an Fks1 mutation were correctly classified as resistant or non-wild type (non-WT) to both micafungin and caspofungin with YS08 or SYO, irrespective of the interpretive criteria. Notably, one C. albicans isolate harboring S645P in Fks1 showed an anidulafungin MIC value of 0.25 μg/ml, which was misclassified as susceptible in the anidulafungin test of SYO using the CLSI CBP; however, it was correctly classified as non-WT to anidulafungin using the SYO ECVs. For 92 isolates of C. glabrata (13 MR isolates harboring Fks mutations and 79 micafungin-susceptible [MS] isolates without Fks mutations), the EA for micafungin between the CLSI M27 method and YS08 was 92.4%, while the EA between CLSI M27 and SYO was 97.8%. Figure 2 depicts two scattergrams comparing the micafungin MICs for C. glabrata isolates obtained by the CLSI method with the micafungin or caspofungin MICs obtained with YS08, using the CLSI CBPs or ECVs. Because YS08 does not show micafungin MIC results of ≤0.03 μg/ml, a modified ECV of 0.06 μg/ml instead of the CLSI ECV (0.03 μg/ml) was used for the micafungin and C. glabrata combination (Fig. 2A). For assessment of the caspofungin MICs obtained with YS08, only the CLSI CBP was used because of the lack of a CLSI ECV for caspofungin (Fig. 2B). The categorical results for the YS08 micafungin MIC obtained using the CLSI CBP showed that 7 out of 13 MR C. glabrata isolates with Fks mutation were misclassified as micafungin intermediate (six isolates) or MS (one isolate), whereas all 79 MS isolates were correctly classified as susceptible, resulting in a CA of 92.4% with the CLSI micafungin MIC. The categorical results for the YS08 micafungin MIC obtained using a modified ECV of 0.06 μg/ml showed that 12 of 13 MR isolates were correctly classified as non-WT, resulting in a CA of 98.9% with the CLSI micafungin MIC results. The categorical results for the YS08 caspofungin MIC obtained using the CLSI CBP did not differentiate between MR and MS isolates of C. glabrata, resulting in a CA of 55.5% with the CLSI micafungin MIC results. Figure 3 demonstrates that all three echinocandin (micafungin, caspofungin, and anidulafungin) tests with SYO showed excellent performance (>95%) to differentiate between MR Fks mutants of C. glabrata and MS non-Fks mutants, irrespective of the MIC interpretive criteria. The CA of SYO for micafungin was 100% using both the CLSI CBP and SYO ECV, while the CA of SYO for caspofungin was 95.6% using the CLSI CBP and 98.9% using the SYO ECV. In addition, the CAs of anidulafungin for C. glabrata obtained with SYO using the CLSI CBP and the SYO ECV were both 98.9%.
TABLE 2.
CAs of micafungin results obtained by the reference CLSI BMD method and echinocandin (micafungin, caspofungin, or anidulafungin) results obtained by two commercial BMD assays for 95 isolates of three common Candida species according to different MIC interpretive criteriaa
| Species (no. of isolates tested) | Antifungal agent | Test method | Interpretive criterionb | % of isolates with the following categorical MIC interpretationc
: |
CA (%) | No. (%) of isolates with discrepancies |
EA (%)e | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R or non-WT | I | S or WT | VME | ME | Minor | ||||||
| C. albicans (2) | Micafungin | CLSI M27 | CLSI CBP (1) | 100.0 | |||||||
| YS08 | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100.0 | ||||
| YS08 | M-CLSI ECV (0.12) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| SYO | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100.0 | ||||
| SYO | SYO ECV (0.12) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| Caspofungin | YS08 | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| SYO | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| SYO | SYO ECV (0.5) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| Anidulafungin | SYO | CLSI CBP (1) | 50.0 | 50.0 | 50.0 | 1 (50.0)d | 0 (0.0) | 0 (0.0) | |||
| SYO | SYO ECV (0.25) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| C. tropicalis (1) | Micafungin | CLSI M27 | CLSI CBP (1) | 100.0 | |||||||
| YS08 | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100.0 | ||||
| YS08 | CLSI ECV (0.12) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| SYO | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100.0 | ||||
| SYO | SYO ECV (0.12) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| Caspofungin | YS08 | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| SYO | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| SYO | SYO ECV (0.5) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| Anidulafungin | SYO | CLSI CBP (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| SYO | SYO ECV (1) | 100.0 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| C. glabrata (92) | Micafungin | CLSI M27 | CLSI CBP (0.25) | 14.1 | 85.9 | ||||||
| YS08 | CLSI CBP (0.25) | 6.5 | 6.5 | 87.0 | 92.4 | 1 (1.1) | 0 (0.0) | 6 (6.5) | 92.4 | ||
| YS08 | M-CLSI ECV (0.12) | 13.0 | 87.0 | 98.9 | 1 (1.1) | 0 (0.0) | 0 (0.0) | ||||
| SYO | CLSI CBP (0.25) | 14.1 | 85.9 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 97.8 | |||
| SYO | SYO ECV (0.06) | 14.1 | 85.9 | 100.0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| Caspofungin | YS08 | CLSI CBP (0.5) | 13.0 | 40.2 | 46.8 | 55.5 | 0 (0.0) | 4 (4.3) | 37 (40.2) | ||
| SYO | CLSI CBP (0.5) | 15.2 | 3.3 | 81.5 | 95.6 | 0 (0.0) | 1 (1.1) | 3 (3.3) | |||
| SYO | SYO ECV (0.5) | 15.2 | 84.8 | 98.9 | 0 (0.0) | 1 (1.1) | 0 (0.0) | ||||
| Anidulafungin | SYO | CLSI CBP (0.5) | 13.0 | 87.0 | 98.9 | 1 (1.1) | 0 (0.0) | 0 (0.0) | |||
| SYO | SYO ECV (0.25) | 13.0 | 87.0 | 98.9 | 1 (1.1) | 0 (0.0) | 0 (0.0) | ||||
CLSI, Clinical and Laboratory Standards Institute; CLSI M27, CLSI M27-ED4 broth microdilution method; YS08, Vitek 2 AST-YS08 yeast susceptibility system; SYO, Sensititre YeastOne system; CLSI CBP, clinical breakpoint provided by the CLSI; ECV, epidemiological cutoff value; M-CLSI ECV, modified CLSI ECV; SYO ECV, SYO-specific ECV; R, resistant; non-WT, non-wild type; I, intermediate; S, susceptible; WT, wild type; CA, categorical agreement; VME, very major error; ME, major error; minor, minor error; EA, essential agreement.
The values in parentheses denote interpretive echinocandin MIC criteria (in micrograms per milliliter) from the CLSI CBPs (MICs greater than or equal to the MIC for resistance), CLSI ECVs (MICs greater than or equal to the non-WT MIC), or SYO ECVs (MICs greater than or equal to the non-WT MIC) against each Candida species (26, 30, 34). Because YS08 does not give the micafungin MIC results of ≥0.03 μg/ml, a modified ECV of 0.06 μg/ml instead of the CLSI ECV (0.03 μg/ml) was used for micafungin for YS08.
Each of the categorical results were determined to indicate resistant, intermediate, and susceptible using the species-specific CLSI CBPs of the CLSI M60, 1st edition, method (30) and as non-WT and WT using the CLSI ECVs of the CLSI M59, 2nd edition, method (34) or SYO ECVs (26).
One C. albicans harboring Fks1 S645P was misclassified as susceptible in the SYO anidulafungin test using the CLSI CBP.
Essential agreement (±2 log2 dilutions) between the MICs obtained by the CLSI M27 method and each of the Vitek 2 YS08 and YeastOne MICs.
FIG 2.
Two scattergrams comparing the micafungin MICs obtained by the CLSI method and the micafungin (A) or caspofungin (B) MICs obtained with the Vitek 2 AST-YS08 (YS08) system. Numbers represent the number of isolates of Candida glabrata at each MIC pair. The gray zone indicates micafungin-resistant Fks mutants of C. glabrata. The resistant (R), intermediate (I), and susceptible (S) designations represent categorical MIC interpretations determined by the use of CLSI clinical breakpoints (CLSI CBPs). The micafungin MIC results obtained with YS08 with the modified CLSI ECV (M-ECV; 0.06 μg/ml) showed a clearer separation between Fks mutants and nonmutants than those obtained with YS08 with the CLSI CBP (A), whereas the caspofungin MIC results obtained with YS08 did not discriminate between Fks mutants and nonmutants (B).
FIG 3.
Three scattergrams of the micafungin MICs obtained by the CLSI method versus the micafungin (A), caspofungin (B), and anidulafungin (C) MICs obtained by the Sensititre YeastOne (SYO) system. Numbers represent the number of isolates of Candida glabrata at each MIC pair. The gray zone indicates micafungin-resistant Fks mutant isolates of C. glabrata. The resistant (R), intermediate (I), and susceptible (S) designations represent the categorical MIC interpretations determined by the use of CLSI clinical breakpoints (CLSI CBPs). The SYO results obtained for all three echinocandins, including micafungin, caspofungin, and anidulafungin, correctly discriminated Fks mutants and nonmutants of C. glabrata (>95%), irrespective of the interpretive criteria used, i.e., CLSI CBPs or SYO-specific ECVs (S-ECV).
DISCUSSION
By testing 292 isolates of four common Candida species with or without molecular mechanisms of acquired azole and echinocandin resistance, the present study revealed the different abilities of the YS08 and SYO systems to detect various mechanisms of azole and echinocandin resistance. YS08 showed performance characteristics superior to those of SYO for the detection of FR C. parapsilosis isolates harboring the Y132F mutation in Erg11p, but caspofungin testing with YS08 seemed to be unreliable for C. glabrata. SYO showed excellent performance for all three echinocandins, irrespective of the interpretive criteria used, CLSI CBPs or SYO ECVs. This is the first study to show that YS08 voriconazole testing may be used as a surrogate marker for fluconazole testing of C. glabrata and that the use of method-specific ECVs may improve the performance of commercial AFST systems.
We found excellent (>90%) EAs and CAs for the fluconazole MICs between the CLSI method and the YS08 or SYO method for three species, including C. albicans, C. parapsilosis, and C. tropicalis; however, the CA of YS08 was 87.0% for C. tropicalis and the CA of SYO was 77.2% for C. parapsilosis. The only minor errors (13%) of YS08 with fluconazole were observed with C. tropicalis due to the misclassification of 3 of 10 FR isolates as F-SDD, suggesting that the F-SDD results from the fluconazole test of YS08 should be interpreted cautiously for C. tropicalis. A Y132F amino acid substitution in Erg11p is reportedly the most frequent fluconazole resistance mechanism in clinical isolates of C. parapsilosis in South Korea; this substitution has also been reported in Brazil, the United States, and Kuwait (28). In a recent Italian study, SYO was able to correctly detect FR C. parapsilosis isolates harboring the Y132F ERG11 gene (Y132F isolates), in which the fluconazole MICs for most (44 of 49 isolates) Y132F isolates were ≥64 μg/ml using the SYO method; all but 1 of the isolates were also resistant to voriconazole using the CLSI method (29). Notably, the fluconazole MICs of most Y132F isolates in the present study were 8 to 16 μg/ml using the CLSI (24 of 26 isolates) or YS08 (24 of 26 isolates) method, and only 7.7% of them were resistant to voriconazole and 80.8% were susceptible, dose dependent (SDD), to voriconazole by the CLSI method (data not shown); therefore, SYO does not efficiently detect Y132F isolates with lower fluconazole MICs. These results indicate that YS08 appears to be a superior method for the detection of FR C. parapsilosis isolates harboring a Y132F mutation in Erg11p from the South Korean collection.
According to the revised CLSI CBP, C. glabrata isolates are no longer considered susceptible to fluconazole but are classified only as either F-SDD or FR (30). In the present study, the fluconazole test with SYO was able to differentiate between 39 FR isolates and 52 F-SDD isolates of C. glabrata, resulting in a CA of 98.9% with the CLSI method. However, the new YS08 does not provide fluconazole results for C. glabrata. A previous study showed that the CA for fluconazole between the Vitek 2 AF03 yeast susceptibility test and the CLSI BMD method was greater than 90% for all Candida species except C. glabrata (87.8%) (23). A recent study also showed that a modified YS08 to assess the adequacy of fluconazole MICs for C. glabrata by setting the tested isolates to C. albicans within the Vitek 2 software produced VMEs (3 of 26 isolates) compared to the results obtained with the previous version of the Vitek 2 AST-YS07 card (7). This suggests the need for an alternative approach to detect FR C. glabrata isolates with YS08.
Acquired resistance to both fluconazole and voriconazole in C. glabrata is mostly governed by a single mechanism through a gain-of-function mutation in the transcription factor for Pdr1p, regulating the drug-efflux transporter genes (20). Accordingly, cross-resistance between fluconazole and voriconazole in C. glabrata has already been reported (31, 32); in one study, 98.8% of FR isolates were not susceptible to voriconazole (MIC > 0.5 μg/ml) (32). Given that the Infectious Diseases Society of America guidelines for the management of candidiasis recommend routinely performing AFST for determination of C. glabrata susceptibility to azoles and echinocandins (1), we alternatively investigated the voriconazole MIC determination of YS08 to correctly discriminate FR C. glabrata isolates harboring a Pdr1 mutation from F-SDD isolates without Pdr1 mutations. Because a CLSI CBP for the voriconazole-C. glabrata combination has not been proposed, we used both the previous and the new CLSI ECVs (0.5 and 0.25 μg/ml, respectively) to differentiate wild-type (WT) from non-WT strains of C. glabrata. The voriconazole result obtained with YS08 with the previous CLSI ECV (0.5 μg/ml) showed a clearer separation between Pdr1 mutants and non-Pdr1 mutants, resulting in a CA of 97.8% with the results for fluconazole by the CLSI method, while the CA of the YS08 voriconazole test using the new ECV (0.25 μg/ml) was only 82.4%. These findings indicate that the use of the YS08 voriconazole test with an ECV of 0.5 μg/ml could provide a reliable discrimination between Pdr1 mutants and non-Pdr1 mutants of C. glabrata for clinical laboratories.
Because the significant interlaboratory variability of the in vitro caspofungin test by the reference CLSI BMD method contributes to false resistance, the echinocandin results obtainded with the two systems were compared to the micafungin results obtained using CLSI methods (30, 33). When the micafungin and caspofungin MIC results obtained with YS08 were compared to the micafungin MIC results for C. glabrata produced by the CLSI method, the CA of YS08 for micafungin was 92.4% using the CLSI CBP, whereas it was 55.5% for the YS08 caspofungin test. Our findings are consistent with the results of a recent study in which the YS08 caspofungin test for C. glabrata had low CAs (5/15 isolates) with the SYO caspofungin test, although the results for the isolates that they evaluated were not molecularly confirmed (8). In the YS08 instructions, the Vitek 2 system also recommends the use of an alternative method before reporting YS08 caspofungin test results for C. glabrata (bioMérieux). All of these findings confirm that YS08 micafungin test results are more reliable than YS08 caspofungin test results.
The CBPs classify isolates as susceptible or resistant, while the ECVs are useful for distinguishing between WT isolates without resistance mechanisms and non-WT isolates harboring resistance mechanisms (34). Both CBPs and ECVs can be used to determine the CA between any two methods. When the micafungin, caspofungin, and anidulafungin MIC results for all 95 isolates (16 Fks mutants and 79 nonmutants) of three Candida species produced by SYO were compared to the micafungin MIC results produced by the CLSI method, excellent CAs (>95%) were found for all three echinocandins, irrespective of the interpretive criteria used, i.e., CBPs or SYO ECVs. Notably, the CA of the SYO caspofungin test results for C. glabrata were 95.6% using the CLSI CBP and 98.9% using the SYO ECV. In addition, the CAs of the SYO fluconazole test results for C. glabrata were 77.2% using the CLSI CBP and 91.2% using the SYO ECV, suggesting that the use of method-specific ECVs may improve the performance of SYO.
YS08 uses CLSI CBPs for interpreting the results for azoles or echinocandins. Using CLSI CBPs, the fluconazole test for C. tropicalis with YS08 showed a 13.0% minor error rate without VMEs or major errors (ME); the micafungin test for C. glabrata with YS08 showed a 6.5% minor error rate with 1.1% VMEs. The reasons for the high rates of minor errors for YS08 are not fully clear, but the discrepancies may reflect the tendency of the YS08 MIC results to be slightly lower than the CLSI or SYO MIC results. We found that adjusting the fluconazole CLSI ECV of 1 μg/ml for C. tropicalis improved the performance of YS08, yielding a CA of 100% and reducing the minor error rate from 13.0% to 0%. In addition, we applied a modified ECV of 0.06 μg/ml to the YS08 micafungin test for C. glabrata, which reduced the minor error rate from 6.5% to 0%, while it increased the CA from 92.4% to 98.9%. Therefore, we emphasize that erroneous results may be observed if the results obtained by one method are interpreted by use of the ECVs/CBPs determined for another method. Considering that echinocandin resistance is more frequently found among C. glabrata isolates worldwide (35) than among other Candida species, we suggest that broadening the micafungin MIC detection range to 0.03 μg/ml and applying the CLSI ECV (0.03 μg/ml) for micafungin and C. glabrata might further improve the performance of YS08 for the detection of echinocandin resistance. In addition, YS08 will be most useful if either method- or species-dependent ECVs are available for the Candida isolate and antifungal agent evaluated, such as SYO (26, 27).
MATERIALS AND METHODS
Fungal isolates.
A total of 292 clinical isolates of four common Candida species from a South Korean collection were tested. Table 3 described the 292 isolates evaluated in this study. The South Korean collection of Candida isolates included 284 BSI isolates and 8 isolates from other clinical specimens (4 from urine, 1 from ascitic fluid, 1 from sputum, 1 from skin, and 1 from ear pus) submitted to Chonnam National University Hospital, Gwangju, South Korea, from several South Korean hospitals during a nationwide surveillance conducted in South Korea (2003 to 2018) (19, 28, 36, 37). Candida isolates were identified to the species level using biochemical tests, matrix-assisted laser desorption ionization–time of flight mass spectrometry, and/or molecular methods (37, 38). For all Candida isolates, antifungal MICs were determined using the CLSI M27, 4th edition (M27-ED4), broth microdilution method (39). Sequence analyses of ERG11 and three transcription factor genes (TAC1, MRR1, and UPC2) were performed for all 26 C. albicans and 57 C. parapsilosis isolates, irrespective of their fluconazole susceptibility (28, 40); the expression levels of CDR1, MDR1, and ERG11 were quantified, and the ERG11 and UPC2 genes were sequenced for all 23 isolates of C. tropicalis (19). The PDR1 genes were sequenced for all 39 FR isolates and 52 F-SDD isolates of C. glabrata (41). The FKS1 (for Candida species) and FKS2 (for C. glabrata) genes were sequenced for both the 16 MR isolates of three Candida species and the 79 MS C. glabrata isolates (42, 43). All isolates were stored at −70°C in Trypticase soy broth supplemented with 15% glycerol until they were tested. These isolates were passaged twice on potato dextrose agar at 35°C prior to testing.
TABLE 3.
Resistance mechanisms and categorical results for fluconazole and micafungin by the CLSI M27 method for all 292 isolates of four common Candida species evaluated in this studya
| Candida species | Result by CLSI M27b
|
Defined mechanism(s) of resistance | No. of isolates |
|||
|---|---|---|---|---|---|---|
| Drug | Category | Blood | Other | Total | ||
| C. albicans | Fluconazole | R | Erg11, K143R mutation | 5 | 1 | 6 |
| S | No mutations (Erg11, Tac1, Mrr1, and Upc2) defined | 16 | 4 | 20 | ||
| Micafungin | R | Fks1, S645P mutation | 2 | 2 | ||
| C. parapsilosis | Fluconazole | R | Erg11, Y132F mutation | 26 | 26 | |
| S | No mutations (Erg11, Tac1, Mrr1, and Upc2) defined | 31 | 31 | |||
| C. tropicalis | Fluconazole | R | ERG11 overexpression | 6 | 3 | 9 |
| R | Erg11, Y132F mutation | 1 | 1 | |||
| S | No mutations (Erg11 and Upc2) defined | 13 | 13 | |||
| Micafungin | R | Fks1, S654S/P | 1 | 1 | ||
| C. glabrata | Fluconazole | R | Pdr1 mutations N132S, G1099S (1),c S236N (1), P258L (1), P258S (2), V260A (1), L280S (1), S316I (1), P327L (1), S337F (1), E340G, D919Y (1), E340K (1), G346S (2), L347F (2), L366P, E555D (1), A731I (1), R376Q (1), S391L (1), Y556C (1), H576Y (2), Y584D (1), E714D (1), T752I (1), N768D (2), R772K (1), K776E (1), G788W (1), L825P (1), F832L, L833V (1), Y932C (1), G943S (1), S947L (1), D954E (1), T1080N (1), and G1088E (1) | 39 | 39 | |
| SDD | No Pdr1 mutation | 52 | 52 | |||
| Micafungin | R | Fks mutations Fks1, S629P (1),c Fks1, P633T and Fks2, D666N (1), Fks2, S663P (8), Fks2, R665G (1), Fks2, I661_S662insFL (1), Fks2, and P667T (1) | 13 | 13 | ||
| S | No Fks1 or Fks2 mutations | 79 | 79 | |||
| Total | Fluconazole | R/SDD | Presence of defined azole resistance mechanisms | 77 | 4 | 81 |
| SDD/S | No azole mutation defined | 112 | 4 | 116 | ||
| Micafungin | R | Fks1 or Fks2 mutations | 16 | 16 | ||
| S | No Fks1 or Fks2 mutations | 79 | 79 | |||
CLSI, Clinical and Laboratory Standards Institute; R, resistant; S, susceptible; SDD, susceptible, dose dependent.
Antifungal MICs were obtained by the CLSI M27-ED4 broth microdilution method (39) and categorized using the CLSI M60, 1st edition, method (30).
Numbers in parentheses indicate the number of strains with that resistance mechanism. The new Pdr1 sequences of FR C. glabrata isolates have been deposited in GenBank with accession numbers MT077187, MT077189, MT077190, MT077193, MT077196, MT077197, MT077200, MT077202, MT077205 to MT077207, MT090722, MT090725, MT090726, MT090728 to MT090735, MT090737, and MT090738 (24 isolates); the Fks sequence has been given GenBank accession number MT090721 (1 isolate).
Antifungal susceptibility testing.
The antifungal MICs for each isolate were determined by three methods: by the CLSI M27-ED4 BMD method and with the Vitek 2 and Sensititre systems. For all Candida isolates, the MICs of fluconazole and micafungin were redetermined by the CLSI M27-ED4 broth microdilution method after 24 h of incubation; they were defined as the lowest concentrations that produced an approximately 50% growth reduction compared to the growth of a drug-free control (39). The Vitek 2 system AST-YS08 and the Sensititre YeastOne YO10 system were used following the manufacturers’ instructions (25; bioMérieux). The ranges of antifungal drug concentrations used to determine the MICs for fluconazole, voriconazole, micafungin, and caspofungin were 0.5 to 64 μg/ml, 0.12 to 8 μg/ml, 0.06 to 8 μg/ml, and 0.12 to 8 μg/ml, respectively, for YS08, and those for fluconazole, micafungin, caspofungin, and anidulafungin were 0.12 to 256 μg/ml, 0.008 to 8 μg/ml, 0.008 to 8 μg/ml, and 0.015 to 8 μg/ml, respectively, for SYO. Antifungal MICs were automatically determined with the Vitek 2 system, while the MIC endpoints in the SYO were visually determined after 24 h of incubation as changes in color (purple or blue) from that for the drug-free control growth (red). Two reference strains (C. parapsilosis ATCC 22019 and Candida krusei ATCC 6258) were tested in each antifungal susceptibility test as quality control isolates. The reproducibility of YS08 was determined by testing five isolates (C. albicans ATCC 64550, C. parapsilosis ATCC 22019, C. tropicalis ATCC 750 and ATCC 201380, and C. glabrata ATCC 90030) in triplicate on three different days (44); the results showed 100% reproducibility for all azole and echinocandin agents.
Data analyses.
EA was determined with respect to the MIC results of YS08 and SYO, with EA being a result within 2 dilutions (two wells) of the result obtained by CLSI BMD. The MIC results obtained byYS08 were interpreted according to two criteria, including CLSI CBPs and CLSI ECVs (Tables 1 and 2) (30, 34, 45). The MIC results obtained with SYO were interpreted according to two criteria, including CLSI CBPs and SYO ECVs (26, 27, 30). CA was defined as the percentage of isolates classified by the two commercial tests into the same susceptible (or WT) or resistant (or non-WT) category into which they were classified by the CLSI reference method. Errors were considered VMEs when an isolate classified as “resistant” by the CLSI method using the CLSI CBP was categorized as “susceptible” or “WT” by a test, MEs when an isolate classified as “susceptible” by the CLSI method using the CLSI CBP was classified as “resistant” or “non-WT” by a test, and minor errors for other discordances.
Ethics statement.
This study was approved by the Institutional Review Board of Chonnam National University Hospital (approval number CNUH-2018-120).
Data availability.
The new Pdr1 sequences of FR C. glabrata isolates have been deposited in GenBank with accession numbers MT077187, MT077189, MT077190, MT077193, MT077196, MT077197, MT077200, MT077202, MT077205 to MT077207, MT090722, MT090725, MT090726, MT090728 to MT090735, MT090737, and MT090738 (24 isolates), while the Fks sequence has been given GenBank accession number MT090721 (1 isolate) (Table 3).
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (grant NRF-2019R1A2C1004644). bioMérieux supplied Vitek 2 AST-YS08 cards free of charge.
We declare no conflicts of interest.
REFERENCES
- 1.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Executive summary: clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:409–417. doi: 10.1093/cid/civ1194. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan TY, Hsu LY, Alejandria MM, Chaiwarith R, Chinniah T, Chayakulkeeree M, Choudhury S, Chen YH, Shin JH, Kiratisin P, Mendoza M, Prabhu K, Supparatpinyo K, Tan AL, Phan XT, Tran TT, Nguyen GB, Doan MP, Huynh VA, Nguyen SM, Tran TB, Van Pham H. 2016. Antifungal susceptibility of invasive Candida bloodstream isolates from the Asia-Pacific region. Med Mycol 54:471–477. doi: 10.1093/mmy/myv114. [DOI] [PubMed] [Google Scholar]
- 4.Posteraro B, Spanu T, Fiori B, De Maio F, De Carolis E, Giaquinto A, Prete V, De Angelis G, Torelli R, D'Inzeo T, Vella A, De Luca A, Tumbarello M, Ricciardi W, Sanguinetti M. 2015. Antifungal susceptibility profiles of bloodstream yeast isolates by Sensititre YeastOne over nine years at a large Italian teaching hospital. Antimicrob Agents Chemother 59:3944–3955. doi: 10.1128/AAC.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. 2007. Multicenter comparison of the Vitek 2 yeast susceptibility test with the CLSI broth microdilution reference method for testing fluconazole against Candida spp. J Clin Microbiol 45:796–802. doi: 10.1128/JCM.01986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson JF, Pfaller MA, Diekema DJ, Rinaldi MG, Riebe KM, Ledeboer NA. 2011. Multicenter comparison of the Vitek 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing caspofungin, micafungin, and posaconazole against Candida spp. J Clin Microbiol 49:1765–1771. doi: 10.1128/JCM.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korem M, Geffen Y, Amit S. 2020. Don't mess with the machine—evaluation of fluconazole susceptibility testing for Candida glabrata using the new VITEK2 AST-YS08 card following species modification. Diagn Microbiol Infect Dis 96:114896. doi: 10.1016/j.diagmicrobio.2019.114896. [DOI] [PubMed] [Google Scholar]
- 8.Wong KY, Gardam D, Boan P. 2019. Comparison of Vitek 2 YS08 with Sensititre YeastOne for Candida susceptibility testing. Pathology 51:668–669. doi: 10.1016/j.pathol.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Espinel-Ingroff A, Pfaller M, Messer SA, Knapp CC, Holliday N, Killian SB. 2004. Multicenter comparison of the Sensititre YeastOne colorimetric antifungal panel with the NCCLS M27-A2 reference method for testing new antifungal agents against clinical isolates of Candida spp. J Clin Microbiol 42:718–721. doi: 10.1128/jcm.42.2.718-721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaller MA, Espinel-Ingroff A, Jones RN. 2004. Clinical evaluation of the Sensititre YeastOne colorimetric antifungal plate for antifungal susceptibility testing of the new triazoles voriconazole, posaconazole, and ravuconazole. J Clin Microbiol 42:4577–4580. doi: 10.1128/JCM.42.10.4577-4580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaller MA, Chaturvedi V, Diekema DJ, Ghannoum MA, Holliday NM, Killian SB, Knapp CC, Messer SA, Miskov A, Ramani R. 2008. Clinical evaluation of the Sensititre YeastOne colorimetric antifungal panel for antifungal susceptibility testing of the echinocandins anidulafungin, caspofungin, and micafungin. J Clin Microbiol 46:2155–2159. doi: 10.1128/JCM.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother 53:3690–3699. doi: 10.1128/AAC.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arendrup MC, Park S, Brown S, Pfaller M, Perlin DS. 2011. Evaluation of CLSI M44-A2 disk diffusion and associated breakpoint testing of caspofungin and micafungin using a well-characterized panel of wild-type and fks hot spot mutant Candida isolates. Antimicrob Agents Chemother 55:1891–1895. doi: 10.1128/AAC.01373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astvad KM, Perlin DS, Johansen HK, Jensen RH, Arendrup MC. 2013. Evaluation of caspofungin susceptibility testing by the new Vitek 2 AST-YS06 yeast card using a unique collection of FKS wild-type and hot spot mutant isolates, including the five most common Candida species. Antimicrob Agents Chemother 57:177–182. doi: 10.1128/AAC.01382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posteraro B, Martucci R, La Sorda M, Fiori B, Sanglard D, De Carolis E, Florio AR, Fadda G, Sanguinetti M. 2009. Reliability of the Vitek 2 yeast susceptibility test for detection of in vitro resistance to fluconazole and voriconazole in clinical isolates of Candida albicans and Candida glabrata. J Clin Microbiol 47:1927–1930. doi: 10.1128/JCM.02070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. doi: 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souza AC, Fuchs BB, Pinhati HM, Siqueira RA, Hagen F, Meis JF, Mylonakis E, Colombo AL. 2015. Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob Agents Chemother 59:6581–6587. doi: 10.1128/AAC.01177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi MJ, Won EJ, Shin JH, Kim SH, Lee WG, Kim MN, Lee K, Shin MG, Suh SP, Ryang DW, Im YJ. 2016. Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob Agents Chemother 60:3653–3661. doi: 10.1128/AAC.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. doi: 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healey KR, Perlin DS. 2018. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J Fungi (Basel) 4:105. doi: 10.3390/jof4030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkow EL, Lockhart SR, Ostrosky-Zeichner L. 2020. Antifungal susceptibility testing: current approaches. Clin Microbiol Rev 33:e00069-19. doi: 10.1128/CMR.00069-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaller MA, Diekema DJ, Procop GW, Wiederhold NP. 2014. Multicenter evaluation of the new Vitek 2 yeast susceptibility test using new CLSI clinical breakpoints for fluconazole. J Clin Microbiol 52:2126–2130. doi: 10.1128/JCM.00658-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanguinetti M, Posteraro B. 2018. Susceptibility testing of fungi to antifungal drugs. J Fungi (Basel) 4:110. doi: 10.3390/jof4030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.TREK Diagnostic Systems. 2012. Sensititre YeastOne: YeastOne susceptibility, v1.8. TREK Diagnostic Systems, Cleveland, OH. [Google Scholar]
- 26.Espinel-Ingroff A, Alvarez-Fernandez M, Canton E, Carver PL, Chen SC, Eschenauer G, Getsinger DL, Gonzalez GM, Govender NP, Grancini A, Hanson KE, Kidd SE, Klinker K, Kubin CJ, Kus JV, Lockhart SR, Meletiadis J, Morris AJ, Pelaez T, Quindos G, Rodriguez-Iglesias M, Sanchez-Reus F, Shoham S, Wengenack NL, Borrell Sole N, Echeverria J, Esperalba J, Gomez GPE, Garcia Garcia I, Linares MJ, Marco F, Merino P, Peman J, Perez Del Molino L, Rosello Mayans E, Rubio Calvo C, Ruiz Perez de Pipaon M, Yague G, Garcia-Effron G, Guinea J, Perlin DS, Sanguinetti M, Shields R, Turnidge J. 2015. Multicenter study of epidemiological cutoff values and detection of resistance in Candida spp. to anidulafungin, caspofungin, and micafungin using the Sensititre YeastOne colorimetric method. Antimicrob Agents Chemother 59:6725–6732. doi: 10.1128/AAC.01250-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinel-Ingroff A, Turnidge J, Alastruey-Izquierdo A, Botterel F, Canton E, Castro C, Chen YC, Chen Y, Chryssanthou E, Dannaoui E, Garcia-Effron G, Gonzalez GM, Govender NP, Guinea J, Kidd S, Lackner M, Lass-Florl C, Linares-Sicilia MJ, Lopez-Soria L, Magobo R, Pelaez T, Quindos G, Rodriguez-Iglesia MA, Ruiz MA, Sanchez-Reus F, Sanguinetti M, Shields R, Szweda P, Tortorano A, Wengenack NL, Bramati S, Cavanna C, DeLuca C, Gelmi M, Grancini A, Lombardi G, Meletiadis J, Negri CE, Passera M, Peman J, Prigitano A, Sala E, Tejada M. 2018. Method-dependent epidemiological cutoff values for detection of triazole resistance in Candida and Aspergillus species for the Sensititre YeastOne colorimetric broth and Etest agar diffusion methods. Antimicrob Agents Chemother 63:e01651-18. doi: 10.1128/AAC.01651-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi YJ, Kim YJ, Yong D, Byun JH, Kim TS, Chang YS, Choi MJ, Byeon SA, Won EJ, Kim SH, Shin MG, Shin JH. 2018. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis 24:1768–1770. doi: 10.3201/eid2409.180625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martini C, Torelli R, de Groot T, De Carolis E, Morandotti GA, De Angelis G, Posteraro B, Meis JF, Sanguinetti M. 2020. Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a large Italian hospital. Front Cell Infect Microbiol 10:232. doi: 10.3389/fcimb.2020.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2017. Performance standards for antifungal susceptibility testing of yeasts, 1st ed CLSI supplement M60 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Panackal AA, Gribskov JL, Staab JF, Kirby KA, Rinaldi M, Marr KA. 2006. Clinical significance of azole antifungal drug cross-resistance in Candida glabrata. J Clin Microbiol 44:1740–1743. doi: 10.1128/JCM.44.5.1740-1743.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. 2012. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J Clin Microbiol 50:1199–1203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 2018. Epidemiological cutoff values for antifungal susceptibility testing, 2nd ed CLSI supplement M59 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect Dis 6:S79–S94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Won EJ, Shin JH, Choi MJ, Lee WG, Park YJ, Uh Y, Kim SY, Lee MK, Kim SH, Shin MG, Suh SP, Ryang DW. 2015. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One 10:e0118770. doi: 10.1371/journal.pone.0118770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HS, Shin JH, Choi MJ, Won EJ, Kee SJ, Kim SH, Shin MG, Suh SP. 2017. Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann Lab Med 37:223–230. doi: 10.3343/alm.2017.37.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byun SA, Won EJ, Kim MN, Lee WG, Lee K, Lee HS, Uh Y, Healey KR, Perlin DS, Choi MJ, Kim SH, Shin JH. 2018. Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes. Front Microbiol 9:1523. doi: 10.3389/fmicb.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th ed CLSI standard M27 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Morio F, Pagniez F, Besse M, Gay-Andrieu F, Miegeville M, Le Pape P. 2013. Deciphering azole resistance mechanisms with a focus on transcription factor-encoding genes TAC1, MRR1 and UPC2 in a set of fluconazole-resistant clinical isolates of Candida albicans. Int J Antimicrob Agents 42:410–415. doi: 10.1016/j.ijantimicag.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Arastehfar A, Daneshnia F, Zomorodian K, Najafzadeh MJ, Khodavaisy S, Zarrinfar H, Hagen F, Zare Shahrabadi Z, Lackner M, Mirhendi H, Salehi M, Roudbary M, Pan W, Kostrzewa M, Boekhout T. 2019. Low level of antifungal resistance in Iranian isolates of Candida glabrata recovered from blood samples in a multicenter study from 2015 to 2018 and potential prognostic values of genotyping and sequencing of PDR1. Antimicrob Agents Chemother 63:e02503-18. doi: 10.1128/AAC.02503-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park MS, Park JE, Song DJ, Huh HJ, Park S, Kang CI, Shin JH, Lee NY. 2017. First case of echinocandin-resistant Candida albicans in Korea. Ann Lab Med 37:556–558. doi: 10.3343/alm.2017.37.6.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho EJ, Shin JH, Kim SH, Kim HK, Park JS, Sung H, Kim MN, Im HJ. 2015. Emergence of multiple resistance profiles involving azoles, echinocandins and amphotericin B in Candida glabrata isolates from a neutropenia patient with prolonged fungaemia. J Antimicrob Chemother 70:1268–1270. doi: 10.1093/jac/dku518. [DOI] [PubMed] [Google Scholar]
- 44.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, Hardy D, Zimmer B, Butler-Wu S, Dien Bard J, Brasso B, Shawar R, Dingle T, Humphries R, Sei K, Koeth L, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. 2018. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 56:e01934-17. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaller MA, Diekema DJ. 2012. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol 50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The new Pdr1 sequences of FR C. glabrata isolates have been deposited in GenBank with accession numbers MT077187, MT077189, MT077190, MT077193, MT077196, MT077197, MT077200, MT077202, MT077205 to MT077207, MT090722, MT090725, MT090726, MT090728 to MT090735, MT090737, and MT090738 (24 isolates), while the Fks sequence has been given GenBank accession number MT090721 (1 isolate) (Table 3).