QPX7728 is a recently discovered ultra-broad-spectrum beta-lactamase inhibitor (BLI) with potent inhibition of key serine and metallo-beta-lactamases. QPX7728 enhances the potency of many beta-lactams, including carbapenems, in beta-lactamase-producing Gram-negative bacteria, including Acinetobacter spp. The potency of meropenem alone and in combination with QPX7728 (1 to 16 μg/ml) was tested against 275 clinical isolates of Acinetobacter baumannii (carbapenem-resistant A. baumannii [CRAB]) collected worldwide that were highly resistant to carbapenems (MIC50 and MIC90 for meropenem, 64 and >64 μg/ml).
KEYWORDS: QPX7728, OXA carbapenemases, metallo-beta-lactamases, CRAB
ABSTRACT
QPX7728 is a recently discovered ultra-broad-spectrum beta-lactamase inhibitor (BLI) with potent inhibition of key serine and metallo-beta-lactamases. QPX7728 enhances the potency of many beta-lactams, including carbapenems, in beta-lactamase-producing Gram-negative bacteria, including Acinetobacter spp. The potency of meropenem alone and in combination with QPX7728 (1 to 16 μg/ml) was tested against 275 clinical isolates of Acinetobacter baumannii (carbapenem-resistant A. baumannii [CRAB]) collected worldwide that were highly resistant to carbapenems (MIC50 and MIC90 for meropenem, 64 and >64 μg/ml). Addition of QPX7728 resulted in a marked concentration-dependent increase in meropenem potency, with the MIC90 of meropenem alone decreasing from >64 μg/ml to 8 and 4 μg/ml when tested with fixed concentrations of QPX7728 at 4 and 8 μg/ml, respectively. In order to identify the mechanisms that modulate the meropenem-QPX7728 MIC, the whole-genome sequences were determined for 135 isolates with a wide distribution of meropenem-QPX7728 MICs. This panel of strains included 116 strains producing OXA carbapenemases (71 OXA-23, 16 OXA-72, 16 OXA-24, 9 OXA-58, and 4 OXA-239), 5 strains producing NDM-1, one KPC-producing strain, and 13 strains that did not carry any known carbapenemases but were resistant to meropenem (MIC ≥ 4 μg/ml). Our analysis indicated that mutated PBP3 (with mutations localized in the vicinity of the substrate/inhibitor binding site) is the main factor that contributes to the reduction of meropenem-QPX7728 potency. Still, >90% of isolates that carried PBP3 mutations remained susceptible to ≤8 μg/ml of meropenem when tested with a fixed 4 to 8 μg/ml of QPX7728. In the absence of PBP3 mutations, the MICs of meropenem tested in combination with 4 to 8 μg/ml of QPX7728 did not exceed 8 μg/ml. In the presence of both PBP3 and efflux mutations, 84.6% of isolates were susceptible to ≤8 μg/ml of meropenem with 4 or 8 μg/ml of QPX7728. The combination of QPX7728 with meropenem against CRAB isolates with multiple resistance mechanisms has an attractive microbiological profile.
INTRODUCTION
Recent decades have seen a surge in the frequency and severity of infections caused by Acinetobacter, which was initially considered a low-virulence pathogen of minimal significance (1, 2). There are an estimated 45,000 cases of Acinetobacter infections per year in the United States and 1 million cases worldwide per year (3), with most infections occurring in intensive care units (ICUs) (3).
The significant challenges associated with the treatment of infections caused by Acinetobacter baumannii stem from its high levels of intrinsic (4) and acquired (5) antibiotic resistance, which are routinely manifested as an extensively drug-resistant (XDR) phenotype, or resistance to all available systemic antibiotics except those that are known to be less effective or more toxic than first-line agents (6). Until recently, carbapenems were the agents of choice to treat A. baumannii infections due to their well-documented efficacy against susceptible strains and excellent safety profiles. Unfortunately, the rate of resistance to carbapenems in A. baumannii is around 50% among ICU isolates in the United States (7) and appears to be even higher in Asia and Latin America (8). ICU-acquired infections with carbapenem-resistant Acinetobacter species, mainly A. baumannii, are associated with a high risk of death (3). In 2017, carbapenem-resistant A. baumannii was named a top-priority pathogen requiring research and development of new antibiotics by the World Health Organization (9), and its multidrug-resistant (MDR) strains were recently reclassified as an urgent resistance threat by the U.S. CDC (10).
The most frequently observed mechanism of carbapenem resistance in A. baumannii, which also provides the highest degree of resistance, is the production of carbapenemases that are able to hydrolyze carbapenems and other beta-lactam antibiotics (11). The predominant carbapenemases in acinetobacters belong to a diverse group of serine beta-lactamases from molecular class D (12), exemplified by OXA-23/OXA-24 (13, 14). Some clinical isolates may also produce metallo-beta-lactamases, such as New Delhi metallo-beta-lactamase (NDM) (15), that are structurally and mechanistically different from serine enzymes (16). The ubiquitous role that carbapenemase-hydrolyzing enzymes play in carbapenem resistance in A. baumannii provides an excellent rationale for the beta-lactamase inhibition approach. Various non-beta-lactamase-mediated mechanisms, such as inactivation or reduced expression of porin genes (4, 17), changes in expression (18), or mutations in the genes encoding penicillin binding proteins (19) and efflux pumps (4, 19), have also been shown to contribute to carbapenem resistance.
Restoration of beta-lactam antibiotic potency through fixed drug combination products with beta-lactamase inhibitors (BLIs) is a well-precedented clinical strategy (20, 21). Four beta-lactam–BLI combination products, ceftazidime-avibactam, ceftolozane-tazobactam, meropenem (MER)-vaborbactam, and imipenem-relebactam, were recently approved by the FDA (22). Three of these BLIs, avibactam, relebactam, and vaborbactam, have a broader spectrum of inhibition that includes Klebsiella pneumoniae carbapenemase (KPC), the major carbapenemase found in Enterobacterales; however, none of these BLIs have any activity against class D carbapenemases found in Acinetobacter or against class B metallo-beta-lactamases. QPX7728 is a recently discovered ultra-broad-spectrum beta-lactamase inhibitor from a class of cyclic boronates (23). It has two major improvements in spectrum compared to the currently marketed agents (the diazabicyclooctanes [DBO] avibactam and relebactam and the cyclic boronate vaborbactam): it is an efficient inhibitor of class D carbapenemases from A. baumannii, such as OXA-23, OXA-24/OXA-40, and OXA-58, and it inhibits various class B metallo-beta-lactamases from the B1 subclass, such as NDM, Verona integron-encoded metallo-β-lactamase (VIM), and imipenemase (IMP). Similar to these BLIs, it is a covalent low-off-rate inhibitor of serine enzymes. At the same time, inhibition of metallo-beta-lactamases by QPX7728 proceeds through a simple one-step complex formation typical of “fast on, fast off” inhibitors (24). QPX7728 is able to reverse resistance to numerous beta-lactam antibiotics in strains producing beta-lactamases (25, 26). This makes it an ideal product that could be coadministered with multiple different beta-lactam antibiotics, depending on the resistance mechanisms present in the specific pathogen, for individualized treatment of drug-resistant pathogens. We initiated a systematic evaluation of QPX7728 potency in combination with multiple partner antibiotics against panels of clinical isolates of target pathogens with varying resistance mechanisms. Against carbapenem-resistant Enterobacterales producing either serine or metallo-beta-lactamases with or without permeability defects, QPX7728, in combination with cephalosporins or carbapenems, including meropenem, was found to have excellent activity (27). The concentration of QPX7728 required to shift meropenem MICs for >90% of these carbapenem-resistant Enterobacteriales isolates to below the pharmacokinetic-pharmacodynamic (PK-PD) breakpoint for meropenem (≤8 μg/ml for a dose of 2 g every 8 h [q8h] as a 3-h infusion), the 90% target potentiation concentration (TPC90), was 8 μg/ml or less. As a continuation of this evaluation, we investigated QPX7728 potency in combination with meropenem against carbapenem-resistant A. baumannii.
RESULTS AND DISCUSSION
Increasing concentrations of QPX7728 enhance the in vitro potency of meropenem against carbapenem-resistant A. baumannii.
The tested panel of 275 strains of Acinetobacter (268 A. baumannii and 7 A. nosocomialis) was highly resistant to multiple antibiotics, with MIC50/MIC90 values for minocycline, tigecycline, sulbactam, polymyxin B, and meropenem similar to or 2- to 4-fold higher than those for the recently reported global collections (28, 29). The meropenem MIC50 and MIC90 were 64 μg/ml and >64 μg/ml, respectively, and only 1.5% and 3.6% of strains were inhibited at meropenem MICs of 4 μg/ml and 8 μg/ml (Table 1). MIC testing of meropenem in combination with increasing concentrations of QPX7728 (1 to 16 μg/ml) resulted in a concentration-related increase in the proportion of strains inhibited at each meropenem MIC; >75% and >90% of strains were inhibited at meropenem MICs of 4 μg/ml and 8 μg/ml in the presence of QPX7728 at ≥4 μg/ml (Table 1). QPX7728 alone exhibited some weak intrinsic antibacterial activity; 26.5% of strains were inhibited by QPX7728 at 16 μg/ml. A much smaller proportion of strains were inhibited by QPX7728 at 8 μg/ml (3.3%), and only two strains (0.7%) in the panel were inhibited by QPX7728 at 4 μg/ml (see Fig. S1S in the supplemental material). Hence, while QPX7728 at 16 μg/ml has some intrinsic antimicrobial activity that accounts for some of the observed MIC shifts with meropenem tested in combination with QPX7728, the observed shifts of meropenem MIC distributions when tested in combinations with QPX7728 at concentrations of ≤8 μg/ml are mainly attributable to beta-lactamase inhibition.
TABLE 1.
MIC distributions for meropenem, alone and combined with QPX7728 at various concentrations, and comparator antibiotics against 275 carbapenem-resistant strains of A. baumannii (n = 268) and A. nosocomialis (n = 7)
| Drug(s)a | % inhibited at MIC (μg/ml) of: |
MIC50 (μg/ml) | MIC90 (μg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||
| MER | ||||||||||||||
| Alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 3.6 | 14.9 | 46.9 | 78.2 | 100 | 64 | >64 |
| W/QPX 1 μg/ml | 0.4 | 0.4 | 0.7 | 1.5 | 2.2 | 7.3 | 18.5 | 38.2 | 56.4 | 72.7 | 88.7 | 100 | 16 | >64 |
| W/QPX 2 μg/ml | 0.7 | 1.1 | 1.5 | 6.2 | 15.6 | 32.0 | 57.5 | 74.2 | 89.8 | 96.4 | 98.2 | 100 | 4 | 32 |
| W/QPX 4 μg/ml | 1.1 | 1.8 | 4.7 | 16.4 | 33.1 | 60.4 | 80.0 | 94.5 | 99.3 | 100 | 100 | 100 | 2 | 8 |
| W/QPX 8 μg/ml | 5.8 | 12.4 | 24.7 | 34.2 | 56.4 | 75.6 | 91.3 | 98.5 | 100 | 100 | 100 | 100 | 1 | 4 |
| W/QPX 16 μg/ml | 33.1 | 39.3 | 50.2 | 65.5 | 77.1 | 86.9 | 97.5 | 100 | 100 | 100 | 100 | 100 | 0.25 | 4 |
| LVFX | 0.4 | 0.4 | 0.4 | 0.7 | 1.5 | 2.2 | 6.5 | 24.4 | 56.4 | 74.2 | 89.5 | 100 | 16 | >64 |
| MINO | 0.0 | 0.0 | 0.4 | 4.0 | 16.4 | 28.7 | 36.7 | 50.5 | 82.5 | 97.1 | 99.6 | 100 | 8 | 32 |
| TIG | 0.0 | 1.1 | 2.5 | 4.7 | 15.6 | 42.2 | 84.0 | 94.5 | 99.6 | 100 | 100 | 100 | 4 | 8 |
| SUL | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 4.4 | 11.6 | 26.2 | 61.1 | 88.4 | 100 | 32 | >64 |
| PmB | 0.0 | 0.0 | 1.8 | 30.5 | 74.9 | 88.0 | 91.3 | 93.8 | 96.0 | 97.5 | 99.3 | 100 | 1 | 4 |
MER, meropenem; QPX, QPX7728; LVFX, levofloxacin; MINO, minocycline; TIG, tigecycline; SUL, sulbactam; PmB, polymyxin B; W/, with.
We recently introduced a metric to enable comparison of the potency of a beta-lactamase inhibitor tested in combination with different partner beta-lactam antibiotics (27). This metric, the target potentiation concentration (TPC), describes the concentration of beta-lactamase inhibitor that is required to lower the MIC of the partner beta-lactam below a PK-PD or susceptibility breakpoint. The PK-PD breakpoint for meropenem at a dose of 2 g administered q8h by 3-h infusion is 8 μg/ml (30, 31); this concentration of meropenem was used to determine the TPC90 of QPX7728 in the 275 carbapenem-resistant A. baumannii (CRAB) isolates. The TPC90 of QPX7728 required to shift >90% of CRAB isolates to a meropenem MIC of ≤8 μg/ml was 4 μg/ml (Table 1). The MIC90s of meropenem tested in combination with 4 or 8 μg/ml of QPX7728 were 8 μg/ml and 4 μg/ml, respectively.
QPX7728 enhances the potency of meropenem against strains with diverse carbapenemases.
In order to better understand the impact of QPX7728, depending on the mechanism of carbapenem resistance, whole-genome sequence analysis was performed on 135 carbapenem-resistant isolates with a wide distribution of QPX7728-meropenem MICs, from ≤0.06 to 16 μg/ml (tested with a fixed QPX7728 concentration of either 4 or 8 μg/ml) (Fig. 1). Based on this analysis, the following acquired carbapenemases were identified: OXA-23 (n = 71), OXA-72 (n = 16), OXA-24 (n = 16; two strains also contained OXA-72), OXA-58 (n = 9), OXA-239 (n = 4), KPC (n = 1), and NDM-1 (n = 5); 13 isolates did not produce any known carbapenemases.
FIG 1.
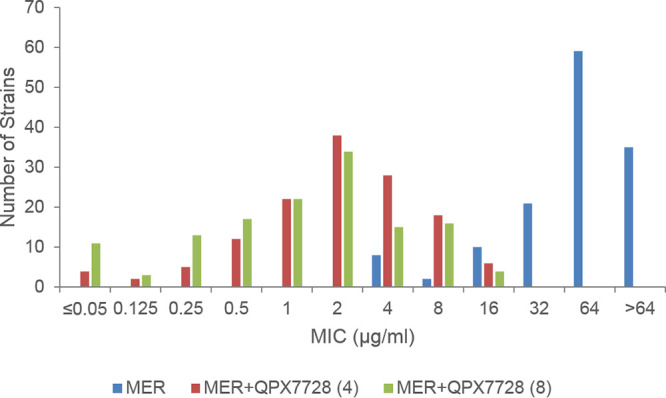
MIC distributions of meropenem alone or in combination with QPX7728 [(4), 4 μg/ml; (8), 8 μg/ml] against carbapenem-resistant A. baumannii with genome sequences available (n = 135). The 135 strains produced OXA-23 (n = 71), OXA-72 (n = 16), OXA-24 (n = 16; 2 strains also with OXA-72), OXA-58 (n = 9), OXA-239 (n = 4), KPC (n = 1), NDM-1 (n = 5), and no known carbapenemases (n = 13).
QPX7728 had a marked effect on the potency of meropenem against this subset of 135 isolates (Fig. 1). Overall, the geometric mean MICs for carbapenemase producers were reduced 14- to 119-fold and 12- to 268-fold with QPX7728 at 4 and 8 μg/ml, respectively (Table 2). For isolates producing OXA-23 (the most frequent beta-lactamase produced in this panel; 52.6%), the geometric mean MIC was reduced 20- to 30-fold and the MIC50 and MIC90 were reduced 16- to 32-fold and 8-fold by 4 and 8 μg/ml of QPX7728, respectively; MIC90 values were 8 μg/ml at both concentrations of QPX7728 versus 64 μg/ml for meropenem alone. The meropenem MIC was reduced to the 8-μg/ml PK-PD breakpoint for >90% (94.4%) of the isolates with QPX7728 at 4 μg/ml, making 4 μg/ml the TPC90 for this group of isolates (at 2 μg/ml of QPX7728, 66.2% of the strains were inhibited by 8 μg/ml of meropenem; see Table S1 in the supplemental material for the complete MIC distribution). For all other groups of class D carbapenemases, QPX7728 at 4 μg/ml shifted the meropenem MICs of all the isolates to the PK-PD breakpoint or below, with MIC90 values of 2 to 4 μg/ml. The TPC90 was 4 μg/ml for other OXA-producing isolates in the panel (see Table S2 in the supplemental material). The meropenem MIC of the single KPC-producing strain of A. baumannii in the panel was reduced from 64 to ≤0.06 μg/ml by QPX7728 at 4 μg/ml. The meropenem MIC values of 100% of non-carbapenemase-producing strains were reduced below 8 μg/ml with QPX7728 at 4 μg/ml. For the 5 NDM-producing strains tested with QPX7728 at 4 or 8 μg/ml, 60% and 100% of the strains became susceptible at the meropenem PK-PD breakpoint (8 μg/ml); thus, the QPX7728 TPC90 is 8 μg/ml. The 2-fold-lower QPX7728 TPC90 for OXA-producing strains compared to NDM-producing strains is consistent with its higher potency for inhibition of the OXA carbapenemases compared to NDM-1: dissociation constant (Kd), 0.58 to 3.2 nm versus 32 nm, respectively (24).
TABLE 2.
In vitro potencies of meropenem alone and combined with QPX7728 at 4 μg/ml and 8 μg/ml against 135 carbapenem-resistant Acinetobacter strains according to carbapenemase present
| CRB | n | Drug | No. of strains at a meropenem MIC (μg/ml) of: |
GMa
MIC |
MIC50 (μg/ml) | MIC90 (μg/ml) | %Sb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||||||
| All | 135 | MER | 8 | 2 | 10 | 21 | 59 | 35 | 51.1 | 64 | 65 | 7.4 | ||||||
| QPX 4 μg/ml | 4 | 2 | 5 | 12 | 22 | 38 | 28 | 18 | 6 | 1.9 | 2 | 8 | 95.6 | |||||
| QPX 8 μg/ml | 11 | 3 | 13 | 17 | 22 | 34 | 15 | 16 | 4 | 1.1 | 2 | 8 | 97.0 | |||||
| OXA-23 | 71 | MER | 3 | 14 | 50 | 4 | 54.7 | 64 | 64 | 0.0 | ||||||||
| QPX 4 μg/ml | 3 | 6 | 8 | 18 | 17 | 15 | 4 | 2.7 | 4 | 8 | 94.4 | |||||||
| QPX 8 μg/ml | 4 | 6 | 6 | 11 | 16 | 10 | 14 | 4 | 1.8 | 2 | 8 | 94.4 | ||||||
| OXA-72 | 16 | MER | 4 | 12 | 107.6 | 65 | 65 | 0.0 | ||||||||||
| QPX 4 μg/ml | 1 | 1 | 3 | 4 | 3 | 2 | 2 | 1.1 | 1 | 8 | 100 | |||||||
| QPX 8 μg/ml | 1 | 1 | 2 | 6 | 1 | 4 | 1 | 0.6 | 0.5 | 2 | 100 | |||||||
| OXA-24 | 16 | MER | 2 | 14 | 117.4 | 65 | 65 | 0.0 | ||||||||||
| QPX 4 μg/ml | 1 | 2 | 2 | 3 | 6 | 2 | 1.0 | 1 | 4 | 100 | ||||||||
| QPX 8 μg/ml | 2 | 2 | 3 | 1 | 5 | 3 | 0 | 0.4 | 0.5 | 2 | 100 | |||||||
| OXA-58 | 9 | MER | 3 | 4 | 2 | 11.8 | 16 | NDc | 33.3 | |||||||||
| QPX 4 μg/ml | 1 | 1 | 1 | 2 | 3 | 1 | 0.8 | 1 | ND | 100 | ||||||||
| QPX 8 μg/ml | 1 | 1 | 1 | 1 | 3 | 2 | 0.9 | 2 | ND | 100 | ||||||||
| OXA-239 | 4 | MER | 3 | 1 | 38.1 | 32 | ND | 0.0 | ||||||||||
| QPX 4 μg/ml | 2 | 2 | 1.4 | 1 | ND | 100 | ||||||||||||
| QPX 8 μg/ml | 1 | 1 | 2 | 0.6 | 0.5 | ND | 100 | |||||||||||
| KPC | 1 | MER | 1 | >64 | ND | ND | 0.0 | |||||||||||
| QPX 4 μg/ml | 1 | ≤0.06 | ND | ND | 100 | |||||||||||||
| QPX 8 μg/ml | 1 | ≤0.06 | ND | ND | 100 | |||||||||||||
| NDM | 5 | MER | 5 | 128.0 | 65 | ND | 0.0 | |||||||||||
| QPX 4 μg/ml | 1 | 2 | 2 | 6.1 | 4 | ND | 60 | |||||||||||
| QPX 8 μg/ml | 2 | 1 | 2 | 0.7 | 2 | ND | 100 | |||||||||||
| None | 13 | MER | 5 | 2 | 3 | 2 | 1 | 0 | 10.4 | 8 | 32 | 53.8 | ||||||
| QPX 4 μg/ml | 3 | 5 | 5 | 2.2 | 2 | 4 | 100 | |||||||||||
| QPX 8 μg/ml | 2 | 2 | 7 | 2 | 1.6 | 2 | 4 | 100 | ||||||||||
GM, geometric mean. For calculations, a MIC of >64 μg/ml was assumed to be 128 μg/ml.
%S (susceptible) at meropenem PK-PD breakpoint of 8 μg/ml.
ND, not done.
Known non-beta-lactamase-mediated mechanisms of resistance to meropenem are associated with an increased meropenem-QPX7728 MIC in A. baumannii.
As described above, the QPX7728 TPC90s for all groups of OXA-producing strains were the same, 4 μg/ml, which corresponds well with the narrow range of inhibition potencies determined against purified enzymes; Kd, 0.58 to 3.2 nm (24). At the same time, MIC50/MIC90 values for OXA-23-producing isolates were 2- to 4-fold higher than those for isolates producing OXA-72, OXA-24/OXA-40, and OXA-58. Since the MICs for beta-lactam–BLI combinations can be increased due to the presence of non-beta-lactamase-mediated mechanisms of resistance to the partner beta-lactam antibiotic, we analyzed the sequences of genes known to be associated with increased meropenem MICs in A. baumannii (4, 17, 19, 32–34).
There were marked differences in the frequencies of mutations in PBP3 between OXA-23 and other OXA-producing isolates. The substitutions A515V and A515T in PBP3 occurred in 43.7% of OXA-23-producing strains (31 of 71 strains) compared to none in the “other OXA” group (see Table S3 in the supplemental material). Multilocus sequence typing (MLST) analysis indicated that 29 out of 31 isolates and 29 out of 40 isolates with and without A515 substitutions, respectively, were sequence type 2 (ST2). Meropenem-QPX7728 (8 μg/ml) MIC frequency distributions of A515V/A515T-positive (n = 31) and A515V/A515T-negative (n = 40) PBP3 sets demonstrated good separation (Fig. 2). Of 40 A515V/A515T PBP3-negative strains, 27 (65.8%) had meropenem-QPX7728 (8 μg/ml) MIC values of ≤1 μg/ml and only one strain had a MIC value of 4 μg/ml. Importantly, 100% of A515V/A515T PBP3-negative strains were inhibited at MIC values of ≤8 μg/ml. Of 31 A515V/A515T PBP3-positive strains, 27 (87%) had meropenem-QPX7728 (8 μg/ml) MIC values of ≥4 μg/ml, and none had MIC values of ≤1 μg/ml.
FIG 2.
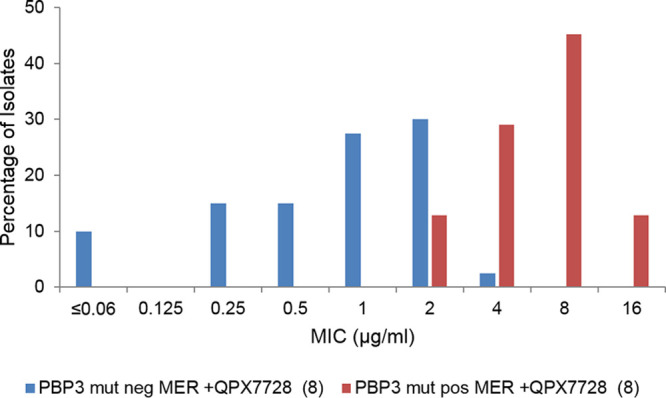
Distributions of meropenem-QPX7728 (8 μg/ml) MICs against OXA-23-producing strains (n = 71) based on the presence of an A515V or A515T mutation in PBP3. PBP3 mut pos, presence of an A515V or A515T mutation (n = 31); PBP3 mut neg, absence of these mutations.
Based on this analysis, it appears that the 2- to 4-fold difference in the meropenem-QPX7728 MIC90 observed against OXA-23-producing strains versus strains producing other OXAs (Table 2) is due to the high proportion of strains with A515V/A515T mutations in PBP3 affecting meropenem potency. Of note, the isolates in the panel are clinical isolates collected over multiple years, from 2010 to 2017, and are from 14 different countries located in Europe, the Middle East, and Southeast Asia. This mutation was recently identified as a mechanism of meropenem resistance in A. baumannii isolates recovered from a patient undergoing meropenem treatment (19).
Several other amino acid substitutions in PBP3 were identified in the set of 135 strains, but many of them were found in strains with both meropenem-QPX7728 (8 μg/ml) MICs of ≥1 μg/ml and ≤0.5 μg/ml (see Table S3). Five substitutions (found in 12 non-OXA-23-producing isolates) similar to the previously described A515V and A515T substitutions were present only in the strains with meropenem-QPX7728 (8 μg/ml) MICs of ≥1 μg/ml. All these substitutions were mapped onto the three-dimensional (3D) crystal structure of A. baumannii PBP3 (Fig. 3). Most of the mutations, A515V/T, A583V, T526S, T506P, and T512S, occur at buried positions either within the 506-to-517 loop adjacent to the active site or in its immediate vicinity. These mutations are expected to perturb the loop conformation, resulting in unfavorable interactions with the bulky C-2 substituent of the bound meropenem. The fifth substitution that is associated with an increased meropenem-QPX7728 MIC, H370Y, is harder to interpret. Based on one published report (19), H370Y was not associated with an increase in the imipenem MIC; however, the mutation was found in four isolates with meropenem-QPX7728 MIC values of 2 to 4 μg/ml (see Table S3). Direct PBP binding assays will be required to confirm and clarify the roles of these mutations.
FIG 3.
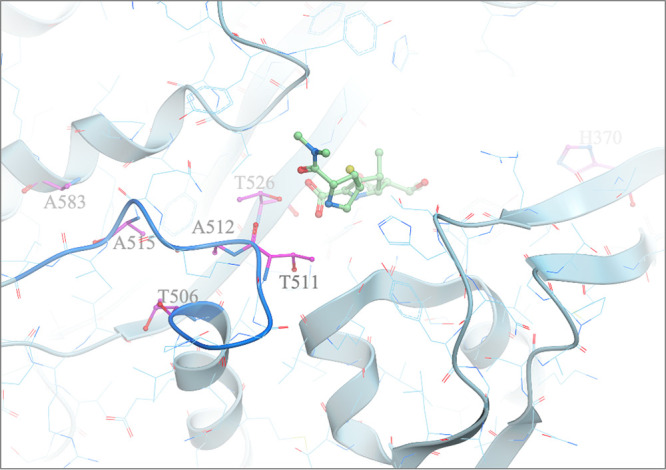
Mapping of potential meropenem resistance mutations in PBP3 from A. baumannii. Potential meropenem resistance mutations (shown as magenta carbon atoms) in PBP3 from A. baumannii were mapped onto its 3D crystal structure (Protein Data Bank [PDB] code 3UE3 [PMID 22050378]). Most of the mutations occur at buried positions 506 to 517 within the loop (blue ribbon segment), either adjacent to the active site or in its immediate vicinity. These mutations are expected to perturb the loop conformation, resulting in unfavorable interactions with the bulky C-2 substituent of the bound meropenem (light-green carbon ball and stick). Meropenem was modeled by superposition of P. aeruginosa PBP3/meropenem complex crystal structure (PDB code 3PBR [PMID 21135211]) onto PBP3 from A. baumannii.
The impacts of PBP3 mutations on meropenem-QPX7728 MIC distributions were assessed further in 130 non-NDM-containing strains of A. baumannii (Table 3 and Fig. 4). Similar to the MIC frequency distributions of A515V/A515T-positive and A515V/A515T-negative PBP3 sets of OXA-23-containing isolates, the MIC frequency distributions of isolates producing all types of OXA carbapenemases and non-carbapenemase-producing isolates with significant PBP3 mutations were associated with higher meropenem-QPX7728 MICs (Fig. 4A). In the absence of mutations in PBP3, meropenem-QPX7728 (8 μg/ml) MICs were ≤2 μg/ml in 97.6% of the isolates, whereas 66% of PBP3 mutation-positive strains had MICs of ≥4 μg/ml. (A chi square test confirmed significant association between PBP3 status and the meropenem-QPX7728 MIC [P < 0.001]). PBP3 mutations resulted in 4- to 8-fold increases in MIC50/MIC90 values. Of note, despite the decreased potency impact of PBP3 mutations, QPX7728 at both 4 μg/ml and 8 μg/ml reduced meropenem MICs to or below 8 μg/ml in >90% of both PBP3 mutation-negative and -positive strains (Table 3).
TABLE 3.
Distributions of meropenem-QPX7728 against characterized strains of Acinetobacter according to the presence of PBP3 and efflux mutationsa
| Strain (drug[s]) | % inhibited at a MIC (μg/ml) of: |
MIC50 (μg/ml) | MIC90 (μg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||
| All (n = 130) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.2 | 7.7 | 15.4 | 31.5 | 76.9 | 100.0 | 64 | >64 |
| W/QPX 4 μg/ml | 3.1 | 4.6 | 8.5 | 17.7 | 34.6 | 63.1 | 83.1 | 96.9 | 100.0 | 100.0 | 100.0 | 100.0 | 2 | 8 |
| W/QPX 8 μg/ml | 5.4 | 9.2 | 19.2 | 32.3 | 49.2 | 74.6 | 86.2 | 96.9 | 100.0 | 100.0 | 100.0 | 100.0 | 2 | 8 |
| PBP3 mutation positive (n = 47) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.6 | 12.8 | 21.3 | 40.4 | 95.7 | 100.0 | 64 | 64 |
| W/QPX 4 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 | 25.5 | 57.4 | 91.5 | 100.0 | 100.0 | 100.0 | 100.0 | 4 | 8 |
| W/QPX 8 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 6.4 | 34.0 | 61.7 | 91.5 | 100.0 | 100.0 | 100.0 | 100.0 | 4 | 8 |
| PBP3 mutation negative (n = 83) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 4.8 | 12.0 | 26.5 | 66.3 | 100.0 | 64 | >64 |
| W/QPX 4 μg/ml | 4.8 | 7.2 | 13.3 | 27.7 | 51.8 | 84.3 | 97.6 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 1 | 4 |
| W/QPX 8 μg/ml | 8.4 | 14.5 | 30.1 | 50.6 | 73.5 | 97.6 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 0.5 | 2 |
| AdeN NF (n = 64) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 | 9.4 | 15.6 | 28.1 | 79.7 | 100.0 | 64 | >64 |
| W/QPX 4 μg/ml | 1.6 | 1.6 | 3.1 | 9.4 | 20.3 | 51.6 | 75.0 | 93.8 | 100.0 | 100.0 | 100.0 | 100.0 | 2 | 8 |
| W/QPX 8 μg/ml | 3.1 | 6.3 | 9.4 | 20.3 | 35.9 | 65.6 | 78.1 | 93.8 | 100.0 | 100.0 | 100.0 | 100.0 | 2 | 8 |
| AdeN F (n = 41) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.3 | 7.3 | 9.8 | 24.4 | 73.2 | 100.0 | 64 | >64 |
| W/QPX 4 μg/ml | 4.9 | 9.8 | 17.1 | 34.1 | 58.5 | 70.7 | 87.8 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 1 | 8 |
| W/QPX 8 μg/ml | 9.8 | 17.1 | 36.6 | 58.5 | 68.3 | 80.5 | 92.7 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 0.5 | 4 |
| PBP3 mutation positive AdeN NF (n = 26) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.7 | 11.5 | 15.4 | 26.9 | 92.3 | 100.0 | 64 | 64 |
| W/QPX 4 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 3.8 | 11.5 | 46.2 | 84.6 | 100.0 | 100.0 | 100.0 | 100.0 | 8 | 16 |
| W/QPX 8 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 7.7 | 19.2 | 46.2 | 84.6 | 100.0 | 100.0 | 100.0 | 100.0 | 8 | 16 |
| PBP3 mutation positive AdeN F (n = 11) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 9.1 | 9.1 | 27.3 | 100.0 | 100.0 | 64 | 64 |
| W/QPX 4 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 54.5 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 4 | 8 |
| W/QPX 8 μg/ml | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 27.3 | 72.7 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 4 | 8 |
| PBP3 mutation negative AdeN F (n = 30) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.7 | 6.7 | 10.0 | 23.3 | 63.3 | 100.0 | 64 | >64 |
| W/QPX 4 μg/ml | 6.7 | 13.3 | 23.3 | 46.7 | 80.0 | 93.3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 1 | 2 |
| W/QPX 8 μg/ml | 13.3 | 23.3 | 50.0 | 80.0 | 93.3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 0.25 | 1 |
| PBP3 mutation negative AdeN NF (n = 37) | ||||||||||||||
| MER alone | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | 5.4 | 13.5 | 27.0 | 70.3 | 100.0 | 64 | 65 |
| W/QPX 4 μg/ml | 2.7 | 2.7 | 5.4 | 16.2 | 32.4 | 78.4 | 94.6 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 2 | 4 |
| W/QPX 8 μg/ml | 5.4 | 10.8 | 16.2 | 35.1 | 56.8 | 97.3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 1 | 2 |
Five NDM-containing strains were excluded from the analysis. For the purpose of the analysis, PBP3 mutations were considered to potentially affect the meropenem-QPX7728 MIC (i.e., significant mutations) if they were present only in the strains with meropenem-QPX7728 MICs of ≥1 μg/ml; 83 strains contained either unaltered PBP3 (compared to the genome of ATCC 19606 [PBP3 negative]) or PBP3 with nonsignificant mutations (present in the strains with MICs of <1 μg/ml), 47 strains contained PBP3 with significant mutations, including H307Y (PBP3 positive) (see Table S2). AdeN was considered nonfunctional if nonsense or frameshift mutations or insertions were found in the adeN gene (n = 64). Strains containing AdeN with amino acid substitutions due to missense mutations were excluded from the analysis.
FIG 4.
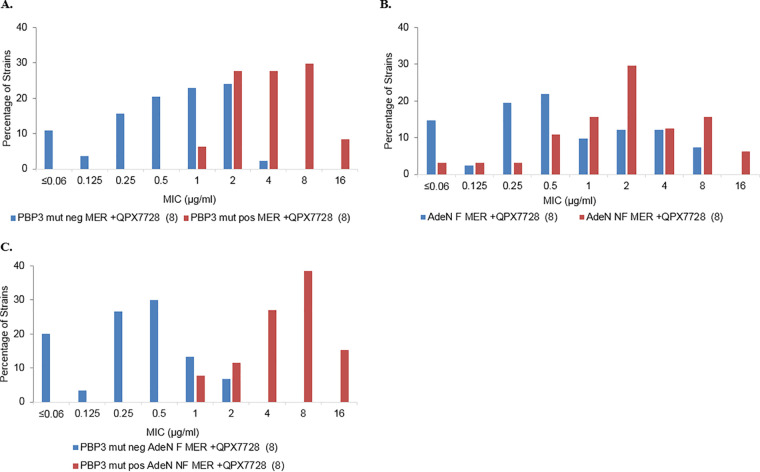
Distributions of meropenem-QPX7728 (8 μg/ml) MICs against carbapenem-resistant A. baumannii (n = 182 [9 NDM-containing strains were excluded from the analysis]) based on the presence of a significant PBP3 mutation and/or AdeN status. (A) MIC distribution based on the presence of PBP3 mutations. For the purpose of the analysis, PBP3 mutations were considered to potentially affect meropenem-QPX7728 MICs (i.e., significant mutations) if they were present only in the strains with meropenem-QPX7728 MICs of ≥1 μg/ml. A total of 107 strains contained either unaltered PBP3 (compared to the genome of ATCC 19606) or PBP3 with nonsignificant mutations (present in the strains with MICs of <1 μg/ml) (PBP3 mut neg), and 75 strains contained PBP3 with significant mutations plus H307Y (PBP3 mut pos) (see Table S2). (B) MIC distribution based on the presence of AdeN mutations. AdeN was considered nonfunctional if nonsense or frameshift mutations or insertions were found in the adeN gene (n = 83). Functional AdeN was present in 99 isolates. Strains containing AdeN with amino acid substitutions due to missense mutations were excluded from the analysis. (C) MIC distribution based on the presence of PBP3 and AdeN mutations. Thirty-five strains contained both PBP3 with significant mutations (PBP3 mut pos) and a nonfunctional AdeN (AdeN NF); 40 strains did not contain significant PBP3 mutations (PBP3 mut neg) and had functional AdeN (AdeN F).
Overexpression of the AdeIJK efflux pump due to mutations in the negative regulator AdeN has been shown to be associated with the increase in the meropenem MIC (34). We evaluated MIC frequency distributions for strains with functional (n = 41) or nonfunctional (n = 64) AdeN. AdeN was considered nonfunctional if the adeN gene contained frameshifts, nonsense mutations, or insertions. Strains with missense mutations leading to single amino acid substitutions were excluded from the analysis. There appeared to be a higher proportion of strains with elevated meropenem-QPX7728 MICs with nonfunctional AdeN. This was also evident when we compared the impacts of AdeN mutations on the meropenem-QPX7728 MIC based on the presence of significant PBP3 mutations (Table 3). However, no clear separation of MIC distributions was apparent based on the functional status of AdeN (Fig. 4B).
We next compared MIC frequency distributions for strains that had significant PBP3 mutations and nonfunctional (NF) AdeN (PBP3 mutation positive, AdeN NF; n = 26) to strains without significant PBP3 mutations and with functional (F) AdeN efflux (PBP3 mutation negative, AdeN F; n = 30). These sets of strains demonstrated the best separation of MIC frequency distributions (Fig. 3C). None of the PBP3 mutation-negative, AdeN F isolates had a MIC of >2 μg/ml, and 80% of such isolates had MIC values of ≤0.5 μg/ml. Conversely, 81% of the PBP3 mutation-positive, AdeN NF strains had MIC values of ≥4 μg/ml, and none had a MIC value of ≤0.5 μg/ml. Determination of meropenem MICs with 8 μg/ml QPX7728 showed a 16- to 32-fold increase in MIC50/MIC90 values caused by the presence of both PBP3 and efflux mutations; however, 84.6% of these isolates were inhibited at 8 μg/ml of meropenem with QPX7728 at 4 and 8 μg/ml, respectively (Table 3).
PBP3 mutation-positive, AdeN NF isolates either carried OXA-23 (21 strains) or did not carry any carbapenemases (6 strains). Relative copy numbers of OXA-23 were estimated for 21 PBP3 mutation-positive, AdeN NF strains (35); based on this analysis, 9 and 12 strains had relative copy numbers of ≥2 (due to gene duplication or plasmid location) and <2, respectively. All PBP3 mutation-positive, AdeN NF strains with an OXA-23 relative copy number of ≥2 had meropenem-QPX7728 MICs of ≥8 μg/ml, and 4 out of 9 strains had meropenem-QPX7728 MICs of 16 μg/ml. Approximately 42% (5 out 12) of PBP3-positive, AdeN NF strains with OXA-23 relative copy numbers of <2 had meropenem-QPX7728 MICs of ≥8 μg/ml, while none had an MIC of ≥16 μg/ml (Table 4). These data, taken together, indicate that a combination of PBP3 mutations, AdeN mutations (presumably leading to overexpression of the AdeIJK efflux pump), and an increase in the copy number of a carbapenemase gene are associated with an increased meropenem-QPX7728 MIC (8 μg/ml) in clinical isolates of carbapenem-resistant A. baumannii.
TABLE 4.
In vitro potencies of meropenem alone and combined with QPX7728 at 8 μg/ml against strains that carry A515V or A515T PBP3 mutations and nonfunctional AdeNa
| Strain | Country of origin | Ch-OXA type | ADC type | Acquired beta-lactamase | MLST ST | CarO status | PBP3 mutation | AdeN | RCNV of OXA-23b | MIC (μg/ml) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| MER | MER + 7728 (8 μg/ml) | ||||||||||
| AB1391 | China | OXA-66 | ADC-30 | OXA-23, TEM-1D | ST2 | No change | A515T | ISAba1 at nt 308 | 0.7 | 4 | 2 |
| AB1451 | Thailand | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | ISAba1 at nt 500 | 0.9 | 8 | 4 |
| AB1384 | China | OXA-66 | ADC-30 | OXA-23, TEM-1D | ST2 | No change | A515T | ISAba1 at nt 308 | 1.1 | 8 | 4 |
| AB1225 | Italy | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | ISAba1 at nt 337 | 1.1 | 8 | 8 |
| AB1302 | Belgium | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | ISAba10 interruption at nt 229 | 1.1 | 4 | 4 |
| AB1382 | China | OXA-66 | ADC-30 | OXA-23, TEM-1D | ST2 | No change | A515T | ISAba1 at nt 308 | 1.2 | 4 | 4 |
| AB1389 | China | OXA-66 | ADC-30 | OXA-23, TEM-1D | ST2 | No change | A515T | ISAba1 at nt 308 | 1.3 | 4 | 4 |
| AB1441 | Thailand | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS from aa 84 | 1.3 | 8 | 8 |
| AB1201 | Israel | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST604 | No change | A515V | ISAba1 at nt 373 | 1.4 | 8 | 8 |
| AB1199 | Sweden | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS from aa 82 | 1.5 | 16 | 8 |
| AB1181 | Israel | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST604 | K156stop | A515V | ISAba1 at nt 373 | 1.6 | 8 | 8 |
| AB1383 | China | OXA-66 | ADC-30 | OXA-23, TEM-1D | ST2 | No change | A515T | ISAba1 at nt 308 | 1.7 | 4 | 4 |
| AB1190 | Singapore | OXA-66 | ADC-73 A243T | OXA-23 | ST2 | No change | A515V | ISAba1 at nt376 | 2.0 | 4 | 16 |
| AB1226 | Italy | OXA-66 | ADC-73 | OXA-23 | ST2 | No change | A515V | ISAba1 at nt 373 | 2.0 | 4 | 8 |
| AB1227 | Italy | OXA-66 | ADC-73 | OXA-23 | ST2 | No change | A515V | ISAba1 at nt 337 | 2.3 | 8 | 8 |
| AB1320 | Greece | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS from aa 14 | 2.4 | 16 | 8 |
| AB1406 | Thailand | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS at aa 33 | 2.4 | 16 | 16 |
| AB1250 | Belgium | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS from aa 84 | 2.4 | 8 | 16 |
| AB1263 | Germany | OXA-66 | ADC-73 | OXA-23 | ST2 | No change | H370Y, A515V | ISAba1 at nt 301 | 2.7 | 16 | 16 |
| AB1275 | Italy | OXA-66 | ADC-73 | OXA-23 | ST2 | No change | A515V | ISAba1 at nt 337 | 2.8 | 8 | 8 |
| AB1257 | Germany | OXA-66 | ADC-73 | OXA-23, TEM-1D | ST2 | No change | A515V | FS from aa 84 | 3.0 | 8 | 8 |
All the strains had an ISAba1 insertion sequence upstream of the intrinsic ADC, which is known to result in increased expression (44). No changes were identified in Omp33-36 or OmpA. AB1406 had a frameshift mutation in PBP1a (from amino acid 45). Abbreviations: Ch-OXA, chromosomal OXA; ADC, Acinetobacter-derived cephalosporinase; nt, nucleotide; aa, amino acid; FS, frameshift.
The RCNV of the OXA-23 gene was determined using DNAStar with RPKM normalization (35).
The porin CarO is essential for the uptake of l-ornithine and has been shown to be associated with resistance to carbapenems (17). Four isolates in our set of strains carried loss-of-function mutations in CarO. Three isolates carried either PBP3 (H370Y) or various AdeN mutations and had a meropenem-QPX7728 MIC of 2 μg/ml (Table 5). One isolate carried PBP3 (A515V) and had nonfunctional AdeN; it had the highest meropenem-QPX7728 MIC of 8 μg/ml (Table 5). Although this represents a small number of isolates, the meropenem-QPX7728 MIC may similarly increase due to interactions of various independent intrinsic mechanisms.
TABLE 5.
In vitro potencies of meropenem alone and combined with QPX7728 at 8 μg/ml against strains that carry H370Y PBP3 mutations and/or nonfunctional CarOa
| Strain | MLST ST | Beta-lactamase | CarO | PBP3 mutation | AdeN | MIC (μg/ml) |
|
|---|---|---|---|---|---|---|---|
| MER alone | MER + QPX7728 (8 μg/ml) | ||||||
| AB1012 | ST2 | None | ISAba1 interruption at nt 240 | H370Y | R183Hb | 32 | 2 |
| AB1392 | ST2 | OXA-23, PER-1 | ISAba15 at nt 515 | No mutation | ISAba1 at nt 308 | 32 | 2 |
| AB1483 | ST2 | OXA-23 | ISAba125 at nt 240 | H370Y A346V | T58Nb | 32 | 2 |
| AB1181 | ST604 | OXA-23 | K156 stop | A515V | ISAba1 at nt 373 | 64 | 8 |
Changes that are known to affect protein function are in boldface.
The mutation was not investigated for its impact on AdeN activity.
Based on our initial, relatively limited analysis, it appears that the absence of significant PBP3 mutations is associated with a meropenem-QPX7728 (8 μg/ml) MIC of ≤8 μg/ml, and when both PBP3 mutations and loss-of-function mutations in AdeN are absent, meropenem-QPX7728 (8 μg/ml) MIC values are ≤4 μg/ml. In our panel, we did not have any isolates with mutations in AdeJ, the RND (resistance-nodulation-division) component of the AdeIJK efflux pump, which has been shown to be associated with increased meropenem MICs (19), nor did we have any mutations in the porin Omp33-36 that was shown to play a role in carbapenem resistance (36). Studies involving more clinical isolates will enable evaluation of the impacts of these mutations on the meropenem-QPX7728 MIC, in particular, in combination with PBP3 and AdeN mutations. The overreaching goal of these studies is to be able to predict susceptibility or resistance to the meropenem-QPX7728 MIC based on molecular analysis.
Summary.
QPX7728 is an ultra-broad-spectrum beta-lactamase inhibitor that increases the potency of various beta-lactam antibiotics against Gram-negative bacteria producing diverse beta-lactamases (23, 25–27). In this study, we demonstrated that QPX7728 significantly enhances the potency of meropenem against a panel of highly carbapenem-resistant isolates of A. baumannii, where >85% of strains had MICs of meropenem alone of ≥32 μg/ml. QPX7728 (tested at 4 μg/ml or 8 μg/ml) significantly enhanced the potency of meropenem against all groups of OXA carbapenemases (OXA-23, OXA-24, OXA-72, and OXA-58), against several strains producing NDM-1 metallo-beta-lactamase, and against strains that did not produce any known carbapenemases, with the meropenem MIC90 decreasing from >64 μg/ml to 8 μg/ml.
A major objective of this study was to determine the concentration of QPX7728 that is required to shift meropenem MICs for >90% of isolates to below the PK-PD breakpoint for meropenem (≤8 μg/ml when administered at 2 g every 8 h as a 3-h infusion). This QPX7728 concentration, the TPC90, was 8 μg/ml or less for the majority of the subsets of carbapenem-resistant isolates of A. baumannii. The TPC90 should be taken into consideration as an important metric for translation of BLI potency into exposures that are expected to be associated with BLI activity in vivo.
While carbapenemase production is the major mechanism of carbapenem resistance, various non-beta-lactamase-mediated mechanisms, such as mutations in penicillin binding proteins or those affecting drug permeability and efflux, have been demonstrated to reduce the potency of meropenem in clinical isolates of A. baumannii (2, 4, 17–19, 34). Using a panel of strains with a broad distribution of meropenem-QPX7728 MICs, we demonstrated that the same mechanisms appear to affect the potency of meropenem-QPX7728, with mutations in PBP3 located in the vicinity of the substrate binding site playing the greatest role. Despite the presence of these previously described target-based mutations, meropenem with QPX7728 had excellent potency against strains with PBP3 mutations; although the MIC90 of meropenem plus QPX7728 was increased 4- to 8-fold compared to isolates without PBP mutations, the meropenem-QPX7728 MICs remained below the PK-PD breakpoint (8 μg/ml). The MIC90 for the subset of strains that had PBP3 mutations and AdeN mutations (and hence, presumably, an overexpressed AdeIJK efflux pump) was increased to 16 μg/ml, with 84.6% of the strains inhibited by meropenem-QPX7728 at 8 μg/ml of each. In the absence of PBP3 mutations, meropenem-QPX7728 MICs did not exceed 8 μg/ml (for NDM-1-negative strains), and when both PBP3 and efflux mutations were absent, the meropenem-QPX7728 MIC did not exceed 4 μg/ml. These findings, if corroborated in future studies, might be important for development of molecular tools to predict meropenem-QPX7728 susceptibility.
MATERIALS AND METHODS
Bacterial strains.
A worldwide collection of 275 nonduplicate clinical isolates of carbapenem-resistant A. baumannii (n = 268) and A. nosocomialis (n = 7) that were acquired from 1998 to 2018 (245 isolates were collected in 2011 to 2018) were from the Qpex Biopharma, Inc. (San Diego, CA), collection of strains. The majority of strains originated from various global survey programs and were acquired from JMI (North Liberty, IA) and IHMA (Schaumburg, IL). The collection was geographically diverse, representing 42 countries located on 6 continents, and grouped into 5 geographic regions: Asia-Pacific (43 isolates; 15.6% overall), Europe (131; 47.6%), Middle East-Africa (21 isolates; 7.6%), Latin America (37; 13.5%), and North America (43; 15.6%). Whole-genome sequencing performed on 135 isolates indicated that the collection was also genetically diverse, representing at least 22 different STs. ST2 (86/135; 63.7%) dominated among the sequenced isolates, followed by ST25 (9/135; 6.7%) and ST1 and ST156 (4/135; 3.0%).
Whole-genome sequencing and analysis.
Acinetobacter isolates were grown in Luria-Bertani Miller broth (LB) until late log phase (optical density at 600 nm [OD600], ∼0.8). Genomic DNA was extracted using a PureLink genomic-DNA minikit (Invitrogen; catalog no. K182001-2). DNA libraries were prepared and sequenced at the Institute of Genomic Medicine, University of California, San Diego, CA, using KAPA library preparation kits (Roche Sequencing and Life Science, Indianapolis, IN) with acoustic DNA shearing (sequencing was done on Illumina NovaSeq, MiSeq, or HiSeq platforms). All reads were deposited in the Sequence Read Archive (SRA) under accession number PRJNA627433. De novo assembly of sequence data was performed using SPAdes 3.13 (St. Petersburg University, St. Petersburg, Russia) (37). Assembled contigs were annotated using PROKKA 1.12 (Victorian Bioinformatics Consortium, Monash University, Melbourne, Australia) (38). The assembled contigs were used to identify resistance genes using AMRFinderPlus 3.2.1 (NCBI) (39). MLST was performed using MLST 2.11 (https://github.com/tseemann/mlst) (Victorian Bioinformatics Consortium, Monash University, Melbourne, Australia; Pasteur scheme). The amino acid sequences of genes of interest were aligned using MAFFT 7.407 (40) to identify substitutions, and raw FASTQ sequence data were subsequently aligned with a custom database using DNAStar 16.0.0 (DNAStar Inc., Madison WI) to identify miscalled bases and structural sequence changes near the genes. Insertion sequence (IS) elements identified near these genes were typed using ISfinder (Le Laboratoire de Microbiologie et Génétique Moléculaires, Toulouse, France) (41). ATCC 19606 was used as a wild-type reference. The relative copy number variation (RCNV) of beta-lactamase genes was also determined using DNAStar with reads per kilobase per million mapped reads (RPKM) normalization (35).
Antimicrobial susceptibility testing.
Bacterial isolates were subjected to broth microdilution susceptibility testing, performed according to Clinical and Laboratory Standards Institute (CLSI) methods (42) using panels prepared in house. A checkerboard assay conforming to the Moody procedures in the Clinical Microbiology Procedures Handbook (43) was used to evaluate the effects of varying concentrations of QPX7728 on the meropenem MIC. Meropenem MICs with QPX7728 at 4 μg/ml and 8 μg/ml were determined numerous times, and modal MIC values were used for the subsequent analyses. Meropenem was purchased from Carbosynth (Compton, United Kingdom); all other antibiotics were from Sigma-Aldrich. All beta-lactamase inhibitors were synthesized at Qpex Biopharma, San Diego, CA.
Data availability. All reads were deposited in the Sequence Read Archive (SRA) under accession number PRJNA627433.
Supplementary Material
ACKNOWLEDGMENTS
This publication includes data generated at the UC San Diego IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (S10 OD026929). The project has been funded in whole or in part with federal funds from the Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA), under OTA number HHSO100201600026C.
We all meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship of the manuscript; we take responsibility for the integrity of the work as a whole and have given final approval of the version to be published.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Spellberg B, Bonomo RA. 2014. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit Care Med 42:1289–1291. doi: 10.1097/CCM.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent JL, Sakr Y, Singer M, Martin-Loeches I, Machado FR, Marshall JC, Finfer S, Pelosi P, Brazzi L, Aditianingsih D, Timsit JF, Du B, Wittebole X, Maca J, Kannan S, Gorordo-Delsol LA, De Waele JJ, Mehta Y, Bonten MJM, Khanna AK, Kollef M, Human M, Angus DC, for the EPIC III Investigators. 2020. Prevalence and outcomes of infection among patients in intensive care units in 2017. JAMA 323:1478. doi: 10.1001/jama.2020.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vila J, Martí S, Sánchez-Céspedes J. 2007. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother 59:1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell GA, Hamidian M, Hall RM. 2016. IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere 1:e00117-16. doi: 10.1128/mSphere.00117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infectious Diseases Society of America. 2012. White paper: recommendations on the conduct of superiority and organism-specific clinical trials of antibacterial agents for the treatment of infections caused by drug-resistant bacterial pathogens. Clin Infect Dis 55:1031–1046. doi: 10.1093/cid/cis688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zilberberg MD, Kollef MH, Shorr AF. 2016. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Hosp Med 11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 8.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 9.WHO. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 10.CDC. 2019. Antibiotic resistance threats in the United States, 2019. CDC, Atlanta, GA. [Google Scholar]
- 11.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Docquier JD, Mangani S. 2016. Structure-function relationships of class D carbapenemases. Curr Drug Targets 17:1061–1071. doi: 10.2174/1389450116666150825115824. [DOI] [PubMed] [Google Scholar]
- 13.Nigro SJ, Hall RM. 2016. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother 71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 14.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18:E362–E365. doi: 10.1111/j.1469-0691.2012.03928.x. [DOI] [PubMed] [Google Scholar]
- 16.Palzkill T. 2013. Metallo-beta-lactamase structure and function. Ann N Y Acad Sci 1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mussi MA, Relling VM, Limansky AS, Viale AM. 2007. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett 581:5573–5578. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Cuenca F, Martinez-Martinez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother 51:565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 19.Hawkey J, Ascher DB, Judd LM, Wick RR, Kostoulias X, Cleland H, Spelman DW, Padiglione A, Peleg AY, Holt KE. 2018. Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb Genom 4:e000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush K, Bradford PA. 2016. Beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush K, Bradford PA. 2019. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat Rev Microbiol 17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 22.Papp-Wallace KM. 2019. The latest advances in beta-lactam/beta-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opin Pharmacother 20:2169–2184. doi: 10.1080/14656566.2019.1660772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecker SJ, Reddy KR, Lomovskaya O, Griffith DC, Rubio-Aparicio D, Nelson K, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Parkinson J, Totrov M, Boyer SH, Glinka TW, Pemberton OA, Chen Y, Dudley MN. 2020. Discovery of cyclic boronic acid QPX7728, an ultra-broad-spectrum inhibitor of serine and metallo beta-lactamases. J Med Chem 63:7491–7507. doi: 10.1021/acs.jmedchem.9b01976. [DOI] [PubMed] [Google Scholar]
- 24.Tsivkovski R, Totrov M, Lomovskaya O. 2020. Biochemical characterization of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases. Antimicrob Agents Chemother 64:e00130-20. doi: 10.1128/AAC.00130-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomovskaya O, Nelson K, Rubio-Aparicio D, Tsivkovski R, Sun D, Dudley MN. 2020. Impact of intrinsic resistance mechanisms on potency of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob Agents Chemother 64:e00552-20. doi: 10.1128/AAC.00552-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomovskaya O, Tsivkovski R, Nelson K, Rubio-Aparicio D, Sun D, Totrov M, Dudley MN. 2020. Spectrum of beta-lactamase inhibition by the cyclic boronate QPX7728, an ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases: enhancement of activity of multiple antibiotics against isogenic strains expressing single beta-lactamases. Antimicrob Agents Chemother 64:e00212-20. doi: 10.1128/AAC.00212-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson K, Rubio-Aparicio D, Sun D, Dudley M, Lomovskaya O. 2020. In vitro activity of the ultra-broad-spectrum beta-lactamase inhibitor QPX7728 against carbapenem-resistant Enterobacterales (CRE) with varying intrinsic and acquired resistance mechanisms. Antimicrob Agents Chemother 64:e00757-20. doi: 10.1128/AAC.00757-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flamm RK, Shortridge D, Castanheira M, Sader HS, Pfaller MA. 2019. In vitro activity of minocycline against U.S. isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus species complex, Stenotrophomonas maltophilia, and Burkholderia cepacia complex: results from the SENTRY Antimicrobial Surveillance Program, 2014 to 2018. Antimicrob Agents Chemother 63:e01154-19. doi: 10.1128/AAC.01154-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLeod SM, Moussa SH, Hackel MA, Miller AA. 2020. In vitro activity of sulbactam-durlobactam against Acinetobacter baumannii-calcoaceticus complex isolates collected globally in 2016 and 2017. Antimicrob Agents Chemother 64:e02534-19. doi: 10.1128/AAC.02534-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuti JL, Dandekar PK, Nightingale CH, Nicolau DP. 2003. Use of Monte Carlo simulation to design an optimized pharmacodynamic dosing strategy for meropenem. J Clin Pharmacol 43:1116–1123. doi: 10.1177/0091270003257225. [DOI] [PubMed] [Google Scholar]
- 31.Lee LS, Kinzig-Schippers M, Nafziger AN, Ma L, Sorgel F, Jones RN, Drusano GL, Bertino JS. Jr.. 2010. Comparison of 30-min and 3-h infusion regimens for imipenem/cilastatin and for meropenem evaluated by Monte Carlo simulation. Diagn Microbiol Infect Dis 68:251–258. doi: 10.1016/j.diagmicrobio.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Cayo R, Rodriguez MC, Espinal P, Fernandez-Cuenca F, Ocampo-Sosa AA, Pascual A, Ayala JA, Vila J, Martinez-Martinez L. 2011. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 55:5907–5913. doi: 10.1128/AAC.00459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando DM, Xu W, Loewen PC, Zhanel GG, Kumar A. 2014. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob Agents Chemother 58:6424–6431. doi: 10.1128/AAC.03074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krumm N, Sudmant PH, Ko A, O'Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE, NHLBI Exome Sequencing Project. 2012. Copy number variation detection and genotyping from exome sequence data. Genome Res 22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novović K, Mihajlović S, Dinić M, Malešević M, Miljković M, Kojić M, Jovčić B. 2018. Acinetobacter spp. porin Omp33-36: classification and transcriptional response to carbapenems and host cells. PLoS One 13:e0201608. doi: 10.1371/journal.pone.0201608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu C-H, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder Tool and Resistance Gene Database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, M100, 30th ed CLSI, Wayne, PA. [Google Scholar]
- 43.Moody J. 2007. Synergism testing: broth microdilution checkerboard Clinical microbiology procedures handbook, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 44.Hamidian M, Hall RM. 2013. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 68:2682–2683. doi: 10.1093/jac/dkt233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.