Vga(A) protein variants confer different levels of resistance to lincosamides, streptogramin A, and pleuromutilins (LSAP) by displacing antibiotics from the ribosome. Here, we show that expression of vga(A) variants from Staphylococcus haemolyticus is regulated by cis-regulatory RNA in response to the LSAP antibiotics by the mechanism of ribosome-mediated attenuation. The specificity of induction depends on Vga(A)-mediated resistance rather than on the sequence of the riboregulator.
KEYWORDS: ABCF proteins, Staphylococcus haemolyticus, Vga(A), antibiotic resistance, clindamycin, lincosamides, pleuromutilins, regulation of gene expression, ribosome-mediated attenuation
ABSTRACT
Vga(A) protein variants confer different levels of resistance to lincosamides, streptogramin A, and pleuromutilins (LSAP) by displacing antibiotics from the ribosome. Here, we show that expression of vga(A) variants from Staphylococcus haemolyticus is regulated by cis-regulatory RNA in response to the LSAP antibiotics by the mechanism of ribosome-mediated attenuation. The specificity of induction depends on Vga(A)-mediated resistance rather than on the sequence of the riboregulator. Fine tuning between Vga(A) activity and its expression in response to the antibiotics may contribute to the selection of more potent Vga(A) variants because newly acquired mutation can be immediately phenotypically manifested.
INTRODUCTION
The expression of antibiotic resistance genes is commonly activated in response to ribosome-targeting antibiotics by ribosome-mediated attenuation (1). In the absence of antibiotic, the secondary structure of the 5′ untranslated RNA (5′ UTR) inhibits the expression of the resistance gene by either masking the ribosome binding site (RBS) or by generating a premature transcriptional terminator. Upon antibiotic binding, the ribosomes stall during translating at a short upstream regulatory open reading frame (uORF), which disrupts the formation of the inhibitory 5′ UTR structure, thus releasing the gene repression (1). The ability of an attenuator to sense the expression of a respective resistance gene and the antibiotic specificity of the protein together determine the overall resistance phenotype. Therefore, it is of great importance to understand the molecular mechanisms behind the specificity of antibiotic-driven expression of resistance determinants.
Antibiotic resistance proteins of the ATP binding cassette F (ARE-ABCF) family collectively confer resistance to antibiotics targeting the ribosomal peptidyl transferase center (PTC) by the ribosome protection mechanism (reviewed in references 2–5). At least seven phylogenetic lineages of ARE-ABCF are widely distributed, namely in the genomes of Firmicutes (AREs 1 to 3, 6, and 7) and Actinobacteria (AREs 4 and 5) species (6). Antibiotic resistance phenotypes of characterized ARE-ABCFs correspond to the spatial overlap in antibiotic binding sites (2, 4), namely, proteins Vga, Lsa, Sal, VmlR, and Lmo0919 confer resistance to lincosamides, streptogramin A, and pleuromutilins (LSAP), while Msr proteins confer resistance to macrolides, ketolides, and streptogramin B (MKSB), and proteins OptrA and PoxtA mediate resistance to phenicols and oxazolidinones (PhO).
Vga(A) is one of the best-studied ARE-ABCFs (7–11). Naturally occurring Vga(A) variants confer different levels of resistance to LSAP antibiotics in staphylococci (9, 12–18). Among human isolates, the incidence of vga(A) is usually low in Staphylococcus aureus (19–21), while it is more common in Staphylococcus haemolyticus, where it is often combined with msr(A), which encodes the macrolide-streptogramin B resistance protein of the same ARE1 subfamily (9, 22). A considerably higher incidence of vga(A) and related vga resistance genes (up to 33%) are present in S. aureus isolates of animal origin (18, 23–25), where they have spread in response to extensive usage of pleuromutilins in food-producing animals (26). In connection with the recent approval of lefamulin, the first systemic pleuromutilin in human medicine, vga resistance genes may also spread rapidly among S. aureus strains of human origin in the future.
According to the current model of the ARE-ABCF-mediated ribosome protection mechanism (3–5), Vga(A) binds the exit site (E site for uncharged tRNA) of the ribosome and displaces the antibiotic by a linker connecting the two nucleotide binding domains (NBD). The linker is also termed the antibiotic resistance domain (ARD) (27), since it is essential for resistance (11). The variable stretch of 15 amino acids in the ARD determines the antibiotic specificity of Vga(A) variants (9, 10). Although it has been suggested that vga(A) expression is regulated by ribosome-mediated attenuation of transcription (28), the mechanism was not experimentally proven. Moreover, how antibiotics differentially induce the expression of vga(A) variants with the different antibiotic specificity is an open question.
Here, we investigated the resistance phenotype and expression profile of vga(A) variants in clinical and commensal isolates of Staphylococcus haemolyticus (22, 29), and we further dissected how the expression of different vga(A) variants is regulated in response to different antibiotics.
RESULTS
Naturally occurring vga(A) variants are expressed differentially in response to clindamycin.
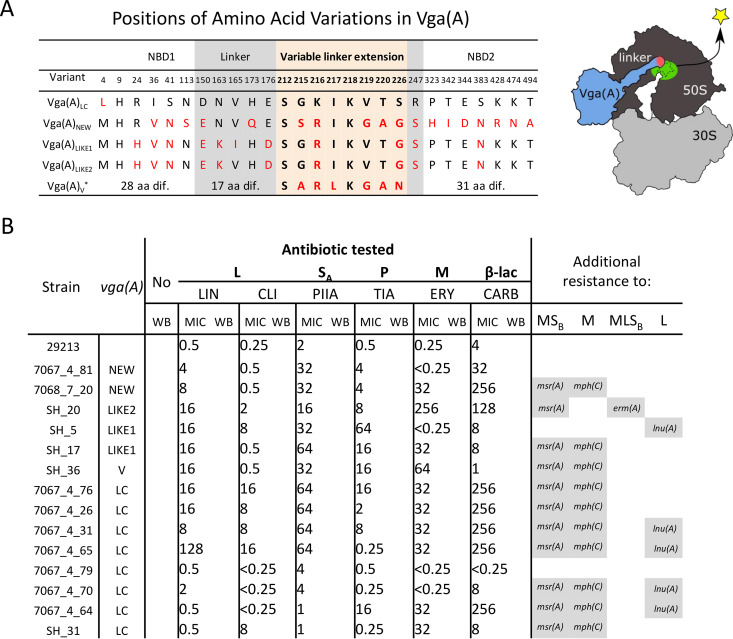
A search for vga(A) in 182 previously whole-genome sequenced S. haemolyticus strains (22, 30) revealed five different protein variants in 23 strains (Fig. 1A; see also Table S3 in the supplemental material). Vga(A)LC present in 17 strains was described previously (9); one Vga(A)V variant present in one strain differed from the previously described Vga(A)V (31) by a P(12)H substitution. Three protein variants, Vga(A)NEW (in two strains), Vga(A)LIKE1 (in two strains), and Vga(A)LIKE2 (in one strain), where the last two differ by the V(165)I substitution, are identified here for the first time (Fig. 1A). Despite each Vga(A) variant having a unique combination of seven amino acids in the 15-amino-acid-long region of the linker crucial for antibiotic specificity (Fig. 1A) (10), 18 out of 23 strains were resistant to lincomycin (lincosamide), pristinamycin IIA (streptogramin A), and tiamulin (pleuromutilin). The Vga(A) variants only differed in resistance to the lincosamide clindamycin (see Table S4 in the supplemental material). All but two clindamycin-resistant strains (SH5 and SH20) harbored vga(A)LC, which is consistent with the previously reported specificity of this variant (9). In the two clindamycin-resistant strains (SH20 and SH5) without vga(A)LC, the clindamycin resistance could be explained by the presence of erm(A) and lnu(A), encoding a 23S rRNA methyltransferase and a lincosamide nucleotidyltransferase, respectively. Thus, we conclude that the vga(A)LC variant confers resistance to clindamycin in the analyzed S. haemolyticus isolates, as all strains apart from SH5 and SH20 did not harbor any additional resistance genes that could explain the observed clindamycin resistance.
FIG 1.
Vga(A) variants differ in their expression profiles in response to antibiotics in S. haemolyticus. (A) Differences in amino acid sequences of Vga(A) variants among Staphylococcus haemolyticus strains. Only positions of amino acid variations are shown. Amino acid variations from Vga(A)LC are shown in red; variations within a 15-amino-acid-long linker stretch that was proven to affect resistance specificity are highlighted by the pink background. Model of Vga(A) binding to the ribosome showing the position of the variable region at the Vga(A) linker. (B) MICs of lincomycin (LIN), clindamycin (CLI), erythromycin (ERY), pristinamycin IIA (PIIA), tiamulin (TIA), and carbenicillin (CARB) for S. haemolyticus strains with vga(A) variants, and Western blots (WBs) showing Vga(A) production in response to the subinhibitory concentration of respective antibiotic. Representative WBs of at least two independent experiments each are shown. All WBs are shown in Fig. S1 in the supplemental material. Full WBs of seven representative strains together with loading WB control are shown in Fig. S2 in the supplemental material, and the growth of these strains in the presence of antibiotics is shown in Fig. S3. Additional macrolide (M), lincosamide (L), streptogramin B (SB) resistance determinants identified in the genomes of tested strains are shown on the right.
Expression analysis of vga(A) in response to the subinhibitory concentration of antibiotics in 14 strains revealed that in most cases, vga(A) was expressed only after exposure to LSAP antibiotics, and that the vga(A) expression profile of all but two tested strains (SH31 and SH20) correlated with the resistance phenotype (Fig. 1B; see also Fig. S2 and S4 in the supplemental material). Clindamycin, in particular, induced the expression of vga(A)LC in strains that were clindamycin resistant, while in four vga (A)LC-positive but clindamycin-susceptible strains, the expression of vga(A)LC was not detected upon clindamycin induction (Fig. 1B). Strains in which the resistance phenotype did not correlate with the vga (A) expression profile (Fig. 1B) include strain SH31, in which vga(A)LC expression was not detected upon induction of clindamycin despite phenotypic clindamycin resistance, and strain SH20, in which antibiotic-induced expression of vga(A)LIKE2 seems to be suppressed due to the presence of ermA. As ErmA renders ribosomes resistant to macrolide-lincosamide-streptogramin B antibiotics (see Fig. S1B in the supplemental material), our observation indicates that the ribosome-mediated attenuation mechanism regulates vga(A) expression.
vga(A) expression is regulated by the mechanism of ribosome-mediated attenuation.
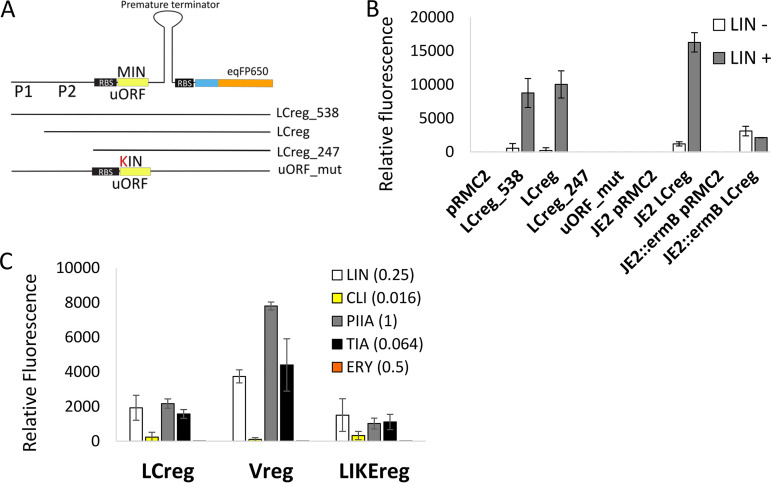
Next, we addressed the mechanism of regulation of vga(A) expression. It was experimentally proven that two homologs of vga(A), lmo0919 and vml(R) from Listeria monocytogenes and Bacillus subtilis, respectively, are regulated by ribosome-mediated attenuation of transcription in response to lincosamides (28, 32). Simultaneously, it was shown that despite the nucleotide sequence of the upstream regulatory region of vga(A) and lmo0919 homologs diverging significantly, the position and size of a 3-amino-acid uORF are conserved across large evolutionary distances; therefore, the same mechanism for their regulation has been suggested (28). To validate the attenuation mechanism of vga(A) regulation, regions 538, 375, and 247 nucleotides upstream of vga(A)LC, together with the first 19 codons of vga(A)LC, were translationally fused with the red fluorescent protein eqFP650 (33), leading to plasmids LCreg_538, LCreg, and LCreg_247, respectively. Also, a mutation of the uORF (MIN) start codon (ATG to AAG) in the LCreg_538 construct leading to plasmid uORF_mut was generated (Fig. 2A). Relative fluorescence in response to the subinhibitory concentration of lincomycin (0.25 mg/liter) of S. aureus carrying reporter constructs showed that expression of eqFP650 was induced only in strains with LCreg_538 and LCreg plasmids (Fig. 2B). The signal was not detected in LCreg_247, which does not contain the predicted promoter or the uORF_mut-lacking start codon. These results indicate that vga(A) is transcribed together with its leader sequence and that an intact uORF is required for induction. In addition, induction of eqFP650 was also abolished when the LCreg fusion construct was coexpressed with the constitutively expressed ErmB 23S rRNA methyltransferase (34), which prevents binding of lincomycin to ribosomes and thus the formation of stalled ribosome complex (Fig. 2B). Together, our results imply that vga(A) expression is regulated by a ribosome-mediated attenuation mechanism in response to ribosome-bound lincomycin.
FIG 2.
Expression of vga(A) is regulated by a ribosome-mediated attenuation mechanism. (A) Testing the activity of transcriptional attenuator encoded upstream of vga(A)LC using fluorescent reporter fusion. Graphical overview of reporter constructs encompassing 538, 375, and 247 nucleotides upstream of the vga(A)LC, including predicted promoters P1 and P2 and the first 19 codons fused to eqFP650 fluorescent protein and LCreg_538 construct in which the start codon ATG of the predicted upstream regulatory open reading frame (uORF), was mutated to AAG (uORF_mut). (B) Relative fluorescence intensity of eqFP650 reporter constructs LCreg_538, LCreg, LCreg_247, and uORF_mut expressed in S. aureus RN4220 in the absence or the presence of lincomycin (LIN, 0.25 mg/liter) and LCreg reporter construct expressed in S. aureus JE2 without or with constitutively expressed ermB, inserted in the genome. ErmB prevents the binding of lincomycin to the ribosome. (C) Relative fluorescence intensity of S. aureus RN4220 expressing eqFP650 fluorescent protein under the control of vga(A)LC (LCreg), vga(A)V (Vreg), or vga(A)LIKE (LIKEreg) 5′ UTRs in response to lincomycin (LIN), clindamycin (CLI), pristinamycin IIA (PIIA), tiamulin (TIA), and erythromycin (ERY) at an antibiotic concentration which corresponded to the maximum level of induction. (B, C) Averages and standard deviations of three independent measurements are shown. The alignment of 5′ UTRs and secondary structure predictions are shown in Fig. S5 in the supplemental material. The full range of antibiotic concentrations and corresponding fluorescence levels are shown in Fig. S6a. The growth of S. aureus RN4220 in the presence of antibiotics is shown in Fig. S6b.
The sequence of the 5′ UTR does not determine the antibiotic specificity of vga(A) induction.
Sequence and secondary structure analysis of 5′ UTRs showed that a putative regulatory region of all vga(A) variants is homologous with predicted terminator and antiterminator structures and identical short uORFs coding for the 3-amino-acid peptide MIN; however, each variant has its unique 5′ UTR sequence differing in 4 to 36 nucleotides (see Fig. S5 in the supplemental material). To identify whether this polymorphism could be responsible for a different expression of vga(A) variants in response to antibiotics in S. haemolyticus, we compared the expression of eqFP650 fused with 375, 374, and 458 upstream regions of vga(A)LC, vga(A)LIKE1, and vga(A)V, respectively (plasmids LCreg, LIKEreg, and Vreg expressed in RN4220), after exposure to a range of lincomycin, clindamycin, pristinamycin IIA, tiamulin, and erythromycin concentrations. All tested vga(A) 5′-UTR reporters reached the maximal eqFP650 fluorescence in the presence of lincomycin, clindamycin, pristinamycin IIA, and tiamulin at an antibiotic concentration close to MIC50 for susceptible strains (Fig. 2C; see also Table S6 and Fig. S6 in the supplemental material). In contrast, erythromycin did not induce expression of eqFP650 (Fig. 2C and Fig. S6). However, the total level of fluorescence among antibiotics differed. Clindamycin was the weakest inducer of eqFP650 expression, independent of 5′ UTRs of vga(A)LC or other 5′-UTR variants, leaving unexplained the observation that vga(A)LC, but not other vga(A) variants, was induced by exposure to clindamycin in S. haemolyticus.
The specificity of vga(A) expression is shaped according to the antibiotic specificity of Vga(A) variants.
Since the polymorphism of 5′ UTRs did not explain differences in vga(A) expression in response to clindamycin, we hypothesized that the Vga(A) protein itself might determine the specificity of its expression. To test this hypothesis, we investigated the expression of the LCreg reporter in the presence of constitutively produced Vga(A) mutants. The mutants differed in only five amino acids in the middle of the inter-NBD linker (amino acid positions 212, 218, 219, 220, and 226; see Table S5 in the supplemental material) but exhibited different abilities to confer resistance to lincosamides (10). The experiment was done as described previously; however, a wider range of antibiotic concentrations reflecting resistance of S. aureus RN4220 expressing vga(A) was tested (Fig. 3A).
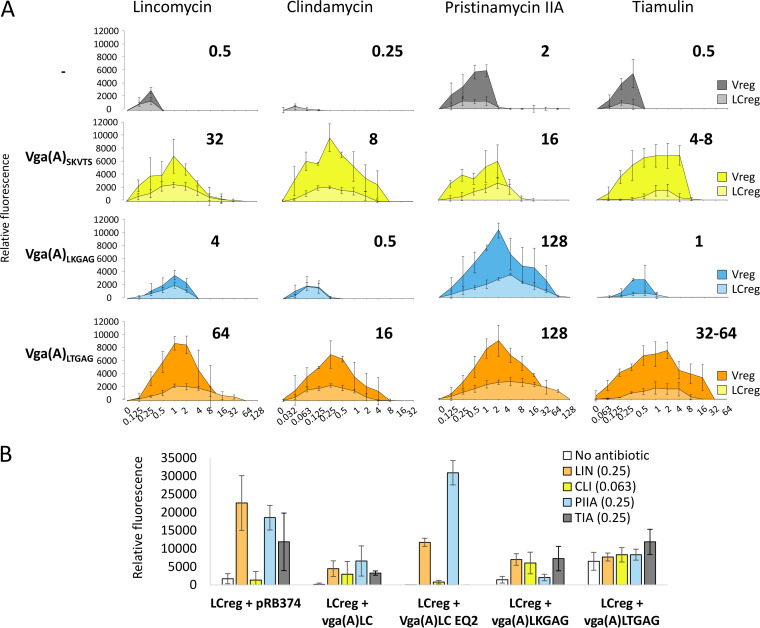
FIG 3.
Expression of vga(A) in response to antibiotics is shaped by the resistance activity of Vga(A). (A) Effect of constitutive expression of mutated vga(A) (10) with different specificity of resistance to lincosamides, streptogramin A, and pleuromutilins on the expression of eqFP650 reporter under the control of the vga(A)LC 5′ UTR (LCreg) in S. aureus RN4220. Relative fluorescence intensity of cells grown in the presence of different concentrations of lincomycin (LIN), clindamycin (CLI), pristinamycin IIA (PIIA), and tiamulin (TIA) overnight in 96-well plates are shown. MIC values of S. aureus RN4220 expressing different Vga(A) mutants are indicated above each graph. (B) Comparison of the activity of LCreg reporter in the absence or the presence of a subinhibitory concentration of LIN, CLI, PIIA, or TIA in S. aureus RN4220 expressing active Vga(A), Vga(A)LC, Vga(A)LKGAG, Vga(A)LTGAG, or ATPase-deficient Vga(A)LC EQ2 forms. Fluorescence intensity of cells grown in 3-ml cultures in the absence or the presence of antibiotics until an optical density of 1is reached is shown. (A, B) Averages and standard deviations of three independent measurements are shown. The constitutive production of all Vga(A) variants was checked by Western blot analysis (see Fig. S8 in the supplemental material). Growth of RN4220 expressing different Vga(A) variants in the presence of antibiotics is shown in Fig. S9. Fig. S10 shows that the expression of vga(A)LCEQ2 does not inhibit the growth of RN4220.
As shown in Fig. 3A, the expression of fluorescent reporters LCreg and Vreg in response to antibiotics was substantially improved when they were coexpressed with Vga(A). Moreover, the expression of the reporters correlated with the level of resistance conferred by a particular Vga(A) mutant (Fig. 3A), indicating that Vga(A) drives the antibiotic specificity of its induction. However, coexpression of the 5′ UTR regulatory regions with Vga(A) mutants expressed constitutively in trans may not reflect the situation in clinical isolates, where the presence of antibiotics fine tunes the expression of Vga(A). Therefore, we fused LCreg with full-length vga(A)LC and eqFP650 to create LCreg-vga (A)LC-eqFP650 translation fusion construct, and we compared its expression in RN4220 with that in strains in which LCreg was expressed alone or in combination with constitutive vga(A)LC (see Fig. S7 in the supplemental material). Similarly, as in the previous experiment, the induction pattern of LCreg-vga(A)LC–eqFP650 corresponded to the specificity of Vga(A)LC, but it was even more efficient in terms of both the level of induction (Fig. S7A) and the conferred resistance (Fig. S7B). Collectively, these results demonstrate that Vga(A) indeed shapes the specificity of its expression in response to antibiotics.
The resistance activity of Vga(A) is the main factor affecting the specificity of the vga(A) induction.
There are two possibilities for how Vga(A) can affect the specificity of self-induction. The first is that ribosome protection by Vga(A) allows growth at higher antibiotic concentrations, which results in higher vga(A) expression. However, the observation that the ATP hydrolysis-deficient Vga(A)LCEQ2 mutant inhibits transpeptidation in vitro (11) suggests that Vga(A) can by itself induce ribosome stalling. Therefore, the second possibility is that the Vga(A) protein can be directly involved in the induction. To find out whether Vga(A) contributes to the activation of its expression, we evaluated an LCreg reporter in the presence of ATPase-deficient Vga(A)LCEQ2 or functional Vga(A) variants in the absence or the presence of antibiotics. At subinhibitory concentrations of lincomycin (0.25 mg/liter) and pristinamycin IIA (0.25 mg/liter), the fluorescence of the reporter was significantly higher in the absence than in the presence of functional Vga(A) (Fig. 3B). In contrast, the presence of Vga(A)LCEQ2, which does not confer resistance (see Table S6 and Fig. S9 in the supplemental material), resulted in a similar induction pattern as that of LCreg expressed alone (Fig. 3B). The exception was tiamulin, which did not induce expression in the presence of Vga(A)LCEQ2 because tiamulin substantially inhibited the growth of the respective strain (Fig. S9B). These observations suggest that active Vga(A) variants at low concentrations of antibiotics clear the antibiotic from the ribosome, thereby reducing reporter expression in a feedback manner. Contrary to our expectations, Vga(A)LTGAG, but not Vga(A)LCEQ2, activated LCreg reporter expression in the absence of antibiotics (Fig. 3B). This indicates that only this Vga(A) variant can induce reporter expression similarly to an antibiotic. Taken together, our results suggest that the induction of vga(A) expression is predominantly fine-tuned indirectly through the resistance activity of Vga(A) variants.
DISCUSSION
In this work, we show that induction of vga(A)LC expression is mediated by an attenuation mechanism that involves a short ORF encoding the MIN peptide and requires antibiotics to bind to the ribosome (Fig. 2B). We dissected the differential ability of clindamycin to induce vga(A) expression in S. haemolyticus isolates (Fig. 1B) by analyzing the antibiotic specificity of attenuators located upstream of several vga(A) variants in S. aureus. We have shown that differential expression of vga(A) is not due to the discriminatory ability of the attenuators toward clindamycin but depends on the ability of vga(A) to confer resistance (Fig. 2C and Fig. 3A). Induction of reporters with clindamycin in the presence of Vga(A)LC resulted in maximum values even at a concentration approximately four times lower than that for lincomycin (see Fig. S7A in the supplemental material). Therefore, clindamycin, which inhibits S. aureus ribosomes more effectively than lincomycin (35), seems to be a more effective inducer than lincomycin in the presence of a vga(A) variant that confers resistance to clindamycin. The antibiotic responsiveness of the attenuator, together with the ability of Vga(A) to confer resistance, results in the tuned expression of vga(A) that reflects both the amount of antibiotic and the resistance specificity of the protein (Fig. 4). In other words, mutations leading to more effective forms of Vga(A) can be immediately phenotypically manifested without the need to modify the specificity of the attenuator. On the other hand, a Vga(A) variant which is not effective in protecting the ribosome will not be produced. From a clinical perspective, more potent Vga(A) variants active against new LSAP antibiotic derivatives can emerge more easily.
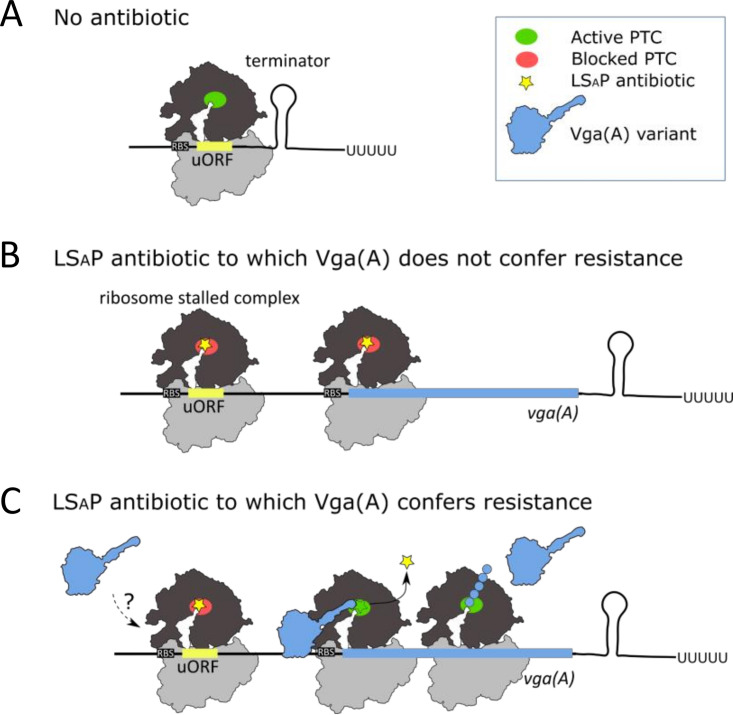
FIG 4.
Proposed model for fine-tuned vga(A) expression in response to antibiotics. (A) In the absence of antibiotic, transcription of vga(A) is prematurely terminated by the 5′ UTR secondary structure. (B) Ribosome stalled complex formed at uORF in the presence of lincosamide, streptogramin A, or pleuromutilin (LSAP) antibiotic to which a particular vga (A) variant does not confer resistance releases the transcription-permissive 5′ UTR secondary structure. Gene is transcribed; however, Vga(A) is not effectively translated at higher antibiotic concentrations, since Vga(A) cannot rescue the ribosome from inhibition. (C) In the presence of an LSAP antibiotic to which a particular Vga(A) variant confers resistance, ribosomes are protected, and Vga(A) is effectively translated proportionally to the amount of antibiotic. We speculate that direct modulation of the stalled ribosome complex by Vga(A) is part of regulation.
Although we have shown that the ability of Vga(A) to confer resistance is an important prerequisite for antibiotic-induced vga(A) expression, we have also observed that Vga(A)LTGAG, which is the most efficient Vga(A) form, activated reporter in the absence of antibiotic (Fig. 3B). This observation indicates that Vga(A) may additionally modify its expression directly by interacting with the ribosome during ribosome stalling. Structures of the related antibiotic resistance ABCF proteins VmlR and Msr(E) in complex with the ribosome (27, 36) revealed that linkers reach the PTC of the ribosome where, consistently with this hypothesis, they change conformations of the same 23S rRNA residues that are important for antibiotic-induced ribosome stalling at uORFs upstream of erm resistance genes (37–39). For further understanding of the involvement of Vga(A) proteins in the direct regulation of their expression, it will be necessary to separate proposed regulatory activity from antibiotic resistance activity. However, this could be challenging, namely, because both antibiotic removal and modulation of ribosome stalling might involve the same Vga(A)-induced conformational changes of the PTC.
Lincomycin, pristinamycin IIA, and tiamulin represent the three antibiotic classes with an overlapping binding site on the ribosome to which vga(A) confers resistance (10). Although LSAP antibiotics differ substantially in their structure (40), we have demonstrated here for the first time that they all induce vga(A) expression, whereas a macrolide antibiotic, erythromycin, to which vga(A) does not confer the resistance, does not (Fig. 2C). Contrary to the broad inducing specificity of vga(A), expression of erm genes conferring resistance to macrolide-lincosamide-streptogramin B antibiotics are induced only by some macrolides (41). Whether a macrolide is an inducer is determined by the specific motifs within the uORF sequence (42–44). On the other hand, uORFs of vga(A) (MIN) and lmo0919 (MKF) (28) do not share sequence similarity, but both are only 3 amino acids long. The short size of the uORF may correspond to the fact that translation in the presence of lincomycin stops after 1 or 2 cycles (45). It is therefore reasonable to think that the short length of the vga(A) uORF broadly but clearly defines the specificity of the vga(A) attenuator to structurally diverse LSAP antibiotics with the overlapping binding sites.
Interestingly, the 3-amino-acid uORFs seem to be shared by attenuators of many vga(A) homologs that confer resistance to LSAP antibiotics (28) but not by attenuators of other lincomycin resistance gene classes (46, 47). Therefore, the size of the short uORF does not seem to be a prerequisite for the attenuation induced by lincosamides in general but could be important for the LSAP antibiotic resistance proteins of the ABCF family, where it may reflect direct involvement of these proteins in the induction.
Among vga(A) variants found in S. haemolyticus, the dominant variant, especially in clinical isolates, was vga(A)LC, which is the only vga(A) variant that conferred resistance to clindamycin (see Table S3 in the supplemental material). Despite the acquired knowledge of vga(A) regulation gained by this study, we were unable to explain many of the observed findings (Fig. 1B). Although the sequences of the 5′ UTRs and the vga(A)LC genes were identical, some strains did not produce Vga(A)LC in response to antibiotics, while in others, the production was constitutive (Fig. 1B). These inconsistencies indicate that the vga(A) regulation is complex, perhaps involving co-occurring macrolide-lincosamide-streptogramin-pleuromutilin resistance proteins or reflecting strain-specific polymorphism. Notably, five LSAP sensitive strains with silenced vga(A)LC (see Table S4 in the supplemental material) represent a potential threat to the therapeutic efficacy of these antibiotics, since these strains may revert to antibiotic-resistance during treatment (48). Further studies are required to understand the mechanism of vga(A) induction and the direct involvement of Vga(A) and other cellular factors in this process.
MATERIALS AND METHODS
Strains.
The following S. haemolyticus strains from two previously described whole-genome sequenced collections were used: 134 invasive S. haemolyticus isolates collected from geographically diverse origins across Europe, North America, and Japan, and 48 S. haemolyticus isolates collected from the skin of healthy volunteers (22, 30). Sequences of natural vga(A) variants and mutants are provided in the File S2 in the supplemental material. Staphylococcus aureus RN4220 was used as a host for eqFP650 reporter constructs. Staphylococcus aureus JE2 and its insertion mutant NE1858 with ermB inserted into the ABC transporter ATP-binding protein (JE2::ermB) (49) were used to test the effect of ermB expression on the induction of vga(A) by lincomycin. Staphylococcus aureus ATCC 29213 was used as a reference strain for susceptibility testing. Escherichia coli strains XL1-Blue and BL21 were used for cloning and heterologous expression of Vga(A)LC. Escherichia coli IM08B was used to prepare plasmids for highly efficient electroporation into staphylococci (50).
Antibiotics.
Chloramphenicol, lincomycin, clindamycin, tiamulin, and erythromycin were purchased from Sigma-Aldrich; carbenicillin was purchased from Duchefa Biochemie, and pristinamycin IIA was kindly provided by Aventis Pharma (Vitry-sur-Seine, France).
Plasmids.
All plasmids used in the study are described in Table S1 in the supplemental material. Oligonucleotides used for cloning are listed in the Table S2. eqFP650 reporter constructs were assembled using seamless ligation cloning extract (SLiCE) (51). All created plasmids were sequenced and then transformed into Escherichia coli IM08B (50) to mimic adenine methylation patterns of recipient staphylococcal strains, purified, and electroporated into S. aureus strains using a MicroPulser electroporation apparatus (Bio-Rad) according to the instrument application guide. Site-directed mutagenesis was performed using two complementary primers, according to the QuikChange site-directed mutagenesis kit protocol (Agilent Technologies).
MICs.
MICs for S. haemolyticus were determined by the broth microdilution method according to ISO standard 20776-1. MICs for S. aureus RN4220 carrying plasmids were determined in the presence of 25 μg/ml chloramphenicol and that of 25 μg/ml kanamycin to maintain pRMC2 and pRB374-based constructs, respectively. All measurements were performed in triplicates two times. Susceptible S. aureus ATCC 29213 was used as a control.
Production of the recombinant Vga(A)LC protein and anti-Vga(A) antibody preparation.
Production of Vga(A)LC (plasmid pJL040) in E. coli BL21 (DE3, Novagen) in the presence of coexpressed chaperonins GroES and GroEL was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when optical density at 600 nm (OD600) reached 0.6, then further incubated for 16 h at 18°C.
Vga(A)LC was purified as described previously (7) with the following exceptions: cell extract was loaded onto 1-ml home-packed Ni Sepharose high-performance (HP) (GE Healthcare Life Sciences) columns, washed with 50 mM NaH2PO4/K2HPO4, 2 M NaCl, and 50 mM imidazole (pH 7.5), and eluted with 250 mM imidazole. Fractions containing Vga(A)LC were mixed and concentrated on an Amicon Ultra 10K device (Merck Millipore) to a final volume of 1 ml and loaded onto an ENrich SEC 650 column (Bio-Rad) equilibrated by 50 mM Tris-Cl, 200 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol, and 10% glycerol (pH 8). Protein was further stored at −80°C.
Polyclonal anti-Vga(A) antibody was produced by BioGenes GmBH (Berlin, Germany) by immunizing rabbits with purified Vga(A)LC-6×His protein. Immunized serum was then affinity purified against agarose-bound Vga(A)LC-6×His and then against Msr(A)-6×His to remove the His tag-reacting antibody fraction. The specificity of the anti-Vga(A) antibody was tested by Western blot analysis of S. aureus RN4220 with an empty pRMC2 vector and with vectors expressing Vga(A)LC or Msr(A)-6×His (see Fig. S1 in the supplemental material).
Western blot analysis of Vga(A) expression.
Vga(A) expression was tested in S. haemolyticus cells harvested from cultures grown to an optical density at 600 nm (OD600) of 0.7 to 1.2 in 1 ml brain heart infusion (BHI) in the absence or the presence of subinhibitory concentrations of lincomycin (0.25 μg/ml), clindamycin (0.125 μg/ml), erythromycin (0.125 μg/ml), pristinamycin IIA (0.25 μg/ml), tiamulin (0.25 μg/ml), and carbenicillin (0.25 μg/ml). Harvested cells were washed twice in 1× phosphate-buffered saline (PBS) and lysed by lysostaphin (1 μg per 100 μl of resuspended cells in 1× PBS) for 15 min at 37°C. 6× SDS loading buffer (20 μl; BioLabs) was added to the lysed cell suspension, and the overall mixture was incubated for 15 min at 96°C. Furthermore, 10 μl of the mixture was loaded and separated on a 12% SDS-PAGE gel. After separation, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Merck Millipore, United States) using the Bio-Rad SemiDry blotting system (10 min at 15 V). Vga(A) was detected by an anti-Vga(A) polyclonal antibody (1:1,000) and by secondary goat anti-rabbit IgG antibody horseradish peroxidase (HRP) conjugate (1:2,000) (Sigma-Aldrich, Germany). Protein abundance was measured using Immobilon Western HRP substrate (Merck Millipore, United States), and the signal was developed using the Bio-Rad ChemiDoc MP imaging system.
eqFP650 reporter fluorescence measurements.
Colonies grown overnight at 37˚C on BHI agar plates with appropriate antibiotics were resuspended in 0.9% NaCl to a McFarland turbidity standard of 0.5. Suspension (5 μl) was inoculated in 200 μl BHI per well in black 96-well plates (Thermo Fisher Scientific, Germany) in the absence or presence of antibiotics at a range of concentrations. For strains without vga(A) expression, the antibiotics and ranges were as follows: lincomycin (0.008 to 1 μg/ml), clindamycin (0.002 to 0.25 μg/ml), and pristinamycin IIA (0.15 to 16 μg/ml). For strains expressing mutated vga(A), the antibiotics and ranges were as follows: lincomycin (0.125 to 128 μg/ml), clindamycin (0.031 to 32 μg/ml), and pristinamycin IIA (0.125 to 128 μg/ml). Bacteria were grown for 24 h at 37°C and fluorescence (excitation at 590 nm [Ex590]/emission at 590 nm [Em650]), and absorbance at 600 nm (A600) of the bacterial cultures were measured at the end of cultivation using a Tecan Infinite 200 Pro plate reader. Autofluorescence values of the strains without eqFP650 were subtracted from the fluorescence values of the strains that encoded eqFP650, and they were normalized to the absorbance. The experiment was repeated three times, each time with duplicates.
For better sensitivity, testing of the strains with constitutive vga(A) expression in the presence of subinhibitory concentrations of antibiotics (Fig. 3B) was performed on 3 ml of culture inoculated with 150 μl from the 0.5 McFarland suspension and cultivated until an OD600 of 1 at 37°C. Cells were harvested by centrifugation (5,000 rpm, 10 min), resuspended in 200 μl of 1× PBS, and 100 μl was transferred to black 96-well plates (Thermo Fisher Scientific, Germany) to measure the fluorescence, while 100 μl was subjected to cell lysis and Western blot analysis to control the presence of Vga(A) proteins.
Supplementary Material
ACKNOWLEDGMENTS
Research on this project was supported by Czech Science Foundation grant number 15-16225Y, Grant Agency of Charles University grant number 772214 (granted to J.L.), by the Ministry of Education, Youth and Sport of the Czech Republic within the National Sustainability Program II (project BIOCEV-FAR; LQ1604), and by the project BIOCEV (CZ.1.05/1.1.00/02.0109) from the European Regional Development Fund in the Czech Republic.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dar D, Sorek R. 2017. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Curr Opin Microbiol 36:111–117. doi: 10.1016/j.mib.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Sharkey LKR, O’Neill AJ. 2018. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infect Dis 4:239–246. doi: 10.1021/acsinfecdis.7b00251. [DOI] [PubMed] [Google Scholar]
- 3.Ero R, Kumar V, Su W, Gao YG. 2019. Ribosome protection by ABC-F proteins—molecular mechanism and potential drug design. Protein Sci 28:684–693. doi: 10.1002/pro.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ousalem F, Singh S, Chesneau O, Hunt JF, Boël G. 2019. ABC-F proteins in mRNA translation and antibiotic resistance. Res Microbiol 170:435–447. doi: 10.1016/j.resmic.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DN, Hauryliuk V, Atkinson GC, O’Neill AJ. 2020. Target protection as a key antibiotic resistance mechanism. Nat Rev Microbiol doi: 10.1038/s41579-020-0386-z. [DOI] [PubMed] [Google Scholar]
- 6.Murina V, Kasari M, Takada H, Hinnu M, Saha CK, Grimshaw JW, Seki T, Reith M, Putrinš M, Tenson T, Strahl H, Hauryliuk V, Atkinson GC. 2019. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J Mol Biol 431:3568–3590. doi: 10.1016/j.jmb.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquet E, Girard J-MM, Ramaen O, Pamlard O, Lévaique H, Betton J-MM, Dassa E, Chesneau O, Levaique H. 2008. ATP hydrolysis and pristinamycin IIA inhibition of the Staphylococcus aureus Vga(A), a dual ABC protein involved in streptogramin A resistance. J Biol Chem 283:25332–25339. doi: 10.1074/jbc.M800418200. [DOI] [PubMed] [Google Scholar]
- 8.Chesneau O, Ligeret H, Hosan-Aghaie N, Morvan A, Dassa E. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob Agents Chemother 49:973–980. doi: 10.1128/AAC.49.3.973-980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother 50:4070–4076. doi: 10.1128/AAC.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenart J, Vimberg V, Vesela L, Janata J, Novotna GB. 2015. Detailed mutational analysis of Vga(A) interdomain linker: tmplication for antibiotic resistance specificity and mechanism. Antimicrob Agents Chemother 59:1360–1364. doi: 10.1128/AAC.04468-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murina V, Kasari M, Hauryliuk V, Atkinson GC. 2018. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res 46:3753–3763. doi: 10.1093/nar/gky050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allignet J, Loncle V, el Sohl N. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45–51. doi: 10.1016/0378-1119(92)90488-B. [DOI] [PubMed] [Google Scholar]
- 13.Qin X, Poon B, Kwong J, Niles D, Schmidt BZ, Rajagopal L, Gantt S. 2011. Two paediatric cases of skin and soft-tissue infections due to clindamycin-resistant Staphylococcus aureus carrying a plasmid-encoded vga(A) allelic variant for a putative efflux pump. Int J Antimicrob Agents 38:81–83. doi: 10.1016/j.ijantimicag.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Tessé S, Trueba F, Berthet N, Hot C, Chesneau O. 2013. Resistance genes underlying the LSA phenotype of staphylococcal isolates from France. Antimicrob Agents Chemother 57:4543–4546. doi: 10.1128/AAC.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozano C, Aspiroz C, Rezusta A, Gómez-Sanz E, Simon C, Gómez P, Ortega C, Revillo MJ, Zarazaga M, Torres C, José M. 2012. Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in meticillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int J Antimicrob Agents 40:306–312. doi: 10.1016/j.ijantimicag.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Mendes RE, Smith TC, Deshpande L, Diekema DJ, Sader HS, Jones RN. 2011. Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn Microbiol Infect Dis 71:177–180. doi: 10.1016/j.diagmicrobio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob Agents Chemother 52:4507–4509. doi: 10.1128/AAC.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadlec K, Pomba CF, Couto N, Schwarz S. 2010. Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J Antimicrob Chemother 65:2692–2693. doi: 10.1093/jac/dkq365. [DOI] [PubMed] [Google Scholar]
- 19.Petrelli D, Repetto A, D’Ercole S, Rombini S, Ripa S, Prenna M, Vitali LA. 2008. Analysis of meticillin-susceptible and meticillin-resistant biofilm-forming Staphylococcus aureus from catheter infections isolated in a large Italian hospital. J Med Microbiol 57:364–372. doi: 10.1099/jmm.0.47621-0. [DOI] [PubMed] [Google Scholar]
- 20.Monecke S, Ehricht R. 2005. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturised oligonucleotide arrays. Clin Microbiol Infect 11:825–833. doi: 10.1111/j.1469-0691.2005.01243.x. [DOI] [PubMed] [Google Scholar]
- 21.McNeil JC, Hulten KG, Kaplan SL, Mason EO. 2014. Decreased susceptibilities to retapamulin, mupirocin, and chlorhexidine among Staphylococcus aureus isolates causing skin and soft tissue infections in otherwise healthy children. Antimicrob Agents Chemother 58:2878–2883. doi: 10.1128/AAC.02707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavanagh JP, Hjerde E, Holden MTG, Kahlke T, Klingenberg C, Flaegstad T, Parkhill J, Bentley SD, Ericson Sollid JU, Flægstad T, Parkhill J, Bentley SD, Sollid JUE. 2014. Whole-genome sequencing reveals clonal expansion of multiresistant Staphylococcus haemolyticus in European hospitals. J Antimicrob Chemother 69:2920–2927. doi: 10.1093/jac/dku271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Sun H, Pan Y, Zhai Y, Cai T, Yuan X, Gao Y, He D, Liu J, Yuan L, Hu G. 2018. Prevalence, resistance pattern, and molecular characterization of Staphylococcus aureus isolates from healthy animals and sick populations in Henan Province, China. Gut Pathog 10:31. doi: 10.1186/s13099-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fessler AT, Kadlec K, Hassel M, Hauschild T, Eidam C, Ehricht R, Monecke S, Schwarz S. 2011. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl Environ Microbiol 77:7151–7157. doi: 10.1128/AEM.00561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes E, Conceição T, Poirel L, de Lencastre H, Aires-de-Sousa M. 2019. Epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus isolates colonizing pigs with different exposure to antibiotics. PLoS One 14:e0225497. doi: 10.1371/journal.pone.0225497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Duijkeren E, Greko C, Pringle M, Baptiste KE, Catry B, Jukes H, Moreno MA, Pomba MCMF, Pyörälä S, Rantala M, Ružauskas M, Sanders P, Teale C, Threlfall EJ, Torren-Edo J, Törneke K. 2014. Pleuromutilins: use in food-producing animals in the European Union, development of resistance and impact on human and animal health. J Antimicrob Chemother 69:2022–2031. doi: 10.1093/jac/dku123. [DOI] [PubMed] [Google Scholar]
- 27.Crowe-McAuliffe C, Graf M, Huter P, Takada H, Abdelshahid M, Nováček J, Murina V, Atkinson GC, Hauryliuk V, Wilson DN. 2018. Structural basis for antibiotic resistance mediated by the Bacillus subtilis ABCF ATPase VmlR. Proc Natl Acad Sci U S A 115:8978–8983. doi: 10.1073/pnas.1808535115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. 2016. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pain M, Hjerde E, Klingenberg C, Cavanagh JP. 2019. Comparative genomic analysis of Staphylococcus haemolyticus reveals key to hospital adaptation and pathogenicity. Front Microbiol 10:2096. doi: 10.3389/fmicb.2019.02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanagh JP, Pain M, Askarian F, Bruun JA, Urbarova I, Wai SN, Schmidt F, Johannessen M. 2019. Comparative exoproteome profiling of an invasive and a commensal Staphylococcus haemolyticus isolate. J Proteomics 197:106–114. doi: 10.1016/j.jprot.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Haroche J, Allignet J, Buchrieser C, El Solh N. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob Agents Chemother 44:2271–2275. doi: 10.1128/aac.44.9.2271-2275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohki R, Tateno K, Takizawa T, Aiso T, Murata M. 2005. Transcriptional termination control of a novel ABC transporter gene involved in antibiotic resistance in Bacillus subtilis. J Bacteriol 187:5946–5954. doi: 10.1128/JB.187.17.5946-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shcherbo D, Shemiakina II, Ryabova AV, Luker KE, Schmidt BT, Souslova EA, Gorodnicheva TV, Strukova L, Shidlovskiy KM, Britanova OV, Zaraisky AG, Lukyanov KA, Loschenov VB, Luker GD, Chudakov DM. 2010. Near-infrared fluorescent proteins. Nat Methods 7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae T, Glass EM, Schneewind O, Missiakas D. 2008. Generating a collection of insertion mutations in the Staphylococcus aureus genome using bursa aurealis. Methods Mol Biol 416:103–116. doi: 10.1007/978-1-59745-321-9_7. [DOI] [PubMed] [Google Scholar]
- 35.Matzov D, Eyal Z, Benhamou RI, Shalev-Benami M, Halfon Y, Krupkin M, Zimmerman E, Rozenberg H, Bashan A, Fridman M, Yonath A. 2017. Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus. Nucleic Acids Res 45:10284–10292. doi: 10.1093/nar/gkx658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su W, Kumar V, Ding Y, Ero R, Serra A, Lee BST, Wong ASW, Shi J, Sze SK, Yang L, Gao Y-G. 2018. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc Natl Acad Sci U S A 115:5157–5162. doi: 10.1073/pnas.1803313115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DN, Arenz S, Beckmann R. 2016. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol 37:123–133. doi: 10.1016/j.sbi.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Koch M, Willi J, Pradere U, Hall J, Polacek N. 2017. Critical 23S rRNA interactions for macrolide-dependent ribosome stalling on the ErmCL nascent peptide chain. Nucleic Acids Res 45:6717–6728. doi: 10.1093/nar/gkx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. 2018. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem 87:451–478. doi: 10.1146/annurev-biochem-062917-011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen NE. 1977. Macrolide resistance in Staphylococcus aureus: inducers of macrolide resistance. Antimicrob Agents Chemother 11:669–674. doi: 10.1128/aac.11.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, Mankin AS, Vázquez-Laslop N. 2016. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol 12:153–158. doi: 10.1038/nchembio.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min YH, Jeong JH, Choi YJ, Yun HJ, Lee K, Shim MJ, Kwak JH, Choi EC. 2003. Heterogeneity of macrolide-lincosamide-streptogramin B resistance phenotypes in enterococci. Antimicrob Agents Chemother 47:3415–3420. doi: 10.1128/aac.47.11.3415-3420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayford M, Weisblum B. 1990. The ermC leader peptide: amino acid alterations leading to differential efficiency of induction by macrolide-lincosamide-streptogramin B antibiotics. J Bacteriol 172:3772–3779. doi: 10.1128/jb.172.7.3772-3779.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol 330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 46.Duval M, Dar D, Carvalho F, Rocha EPC, Sorek R, Cossart P. 2018. HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance. Proc Natl Acad Sci U S A 115:13359–13364. doi: 10.1073/pnas.1810555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reilman E, Mars RAT, Van Dijl JM, Denham EL. 2014. The multidrug ABC transporter BmrC/BmrD of Bacillus subtilis is regulated via a ribosome-mediated transcriptional attenuation mechanism. Nucleic Acids Res 42:11393–11407. doi: 10.1093/nar/gku832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kime L, Randall CP, Banda FI, Coll F, Wright J, Richardson J, Empel J, Parkhill J, O’Neill AJ. 2019. Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. mBio 10:e01755-19. doi: 10.1128/mBio.01755-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:1–8. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. 2015. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15–e00315. doi: 10.1128/mBio.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Werling U, Edelmann W. 2012. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res 40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.