Coagulase-negative staphylococci (CoNS) are a common etiology of serious and recurrent infections in immunocompromised patients. Although most isolates appear susceptible to vancomycin, a single strain might have a subpopulation of resistant bacteria. This phenomenon is termed heteroresistance and may adversely affect the response to treatment. A retrospective cohort study was performed of pediatric patients with leukemia treated at St. Jude Children’s Research Hospital who developed CoNS central line-associated bloodstream infection (CLABSI).
KEYWORDS: staphylococci, Staphylococcus epidermidis, child, heteroresistance, leukemia, vancomycin
ABSTRACT
Coagulase-negative staphylococci (CoNS) are a common etiology of serious and recurrent infections in immunocompromised patients. Although most isolates appear susceptible to vancomycin, a single strain might have a subpopulation of resistant bacteria. This phenomenon is termed heteroresistance and may adversely affect the response to treatment. A retrospective cohort study was performed of pediatric patients with leukemia treated at St. Jude Children’s Research Hospital who developed CoNS central line-associated bloodstream infection (CLABSI). Available isolates were sequenced and tested for vancomycin heteroresistance by population analysis profiling. Risk factors for heteroresistance and the association of heteroresistance with treatment failure (death or relapse of infection) or poor clinical response to vancomycin therapy (treatment failure or persistent bacteremia after vancomycin initiation) were evaluated. For 65 participants with CoNS CLABSI, 62 initial isolates were evaluable, of which 24 (39%) were vancomycin heteroresistant. All heteroresistant isolates were of Staphylococcus epidermidis and comprised multiple sequence types. Participants with heteroresistant bacteria had more exposure to vancomycin prophylaxis (P = 0.026) during the 60 days prior to infection. Of the 40 participants evaluable for clinical outcomes, heteroresistance increased the risk of treatment failure (P = 0.012) and poor clinical response (P = 0.001). This effect persisted after controlling for identified confounders. These data indicate that vancomycin heteroresistance is common in CoNS isolates from CLABSIs in pediatric patients with leukemia and is associated with poor clinical outcomes. Validation of these findings in an independent cohort and evaluation of alternative antibiotic therapy in patients with heteroresistant infections have the potential to improve care for serious CoNS infections.
INTRODUCTION
Vancomycin is the definitive therapy for many serious Gram-positive infections; however, although most such infections remain susceptible to vancomycin, therapeutic failure is common. One potential explanation for the poor outcomes is heteroresistance, that is, the presence of a small subpopulation of resistant organisms recalcitrant to antibiotic therapy. This phenomenon has been well-described in some bacteria, including Enterobacteriaceae and Staphylococcus aureus (1, 2). Heteroresistance does appear to be associated with poor treatment outcomes in methicillin-resistant S. aureus infections (1, 3); however, the clinical significance of heteroresistance in coagulase-negative staphylococci (CoNS) is unknown (4–10).
CoNS are Gram-positive bacteria that colonize human skin and mucosa. They are a frequent etiology of central line-associated bloodstream infection (CLABSI), especially in immunocompromised patients (11–13). Although often thought of as benign, CoNS CLABSI in vulnerable populations can cause prolonged hospitalization and serious illness, including organ dysfunction and neurocognitive damage (14, 15). These bacteria typically appear susceptible to vancomycin, and vancomycin is recommended as first-line therapy; however, antibiotic treatment failure or relapse is common (16, 17). The possibility that heteroresistance contributes to poor clinical response in CoNS infections has not been evaluated. Clinical susceptibility testing, which uses an inoculum of approximately 106 CFU/ml, typically cannot detect a resistant subpopulation at a frequency of 10−6 or lower. Since the genetic basis for vancomycin heteroresistance remains unknown, molecular testing is unavailable. Therefore, vancomycin heteroresistance might be present in CoNS causing bloodstream infections in immunocompromised patients and might impair the clinical response to vancomycin therapy. We undertook a retrospective study to evaluate the frequency, risk factors, and clinical significance of vancomycin heteroresistance in CoNS CLABSI in pediatric patients undergoing treatment for leukemia at St. Jude Children’s Research Hospital (St. Jude).
RESULTS
Incidence of heteroresistance.
A total of 74 isolates from 65 study participants were available; 7 participants had had 2 episodes of CoNS CLABSI and 1 had had 3 episodes. Two of the 65 participants were excluded because their isolates were not identified as CoNS following sequencing, and 1 was excluded because the heteroresistance testing was unsuccessful due to poor in vitro growth. The characteristics of the 62 included participants are presented in Table 1. Analysis of the first isolates from all participants revealed that 24 (39%) were classified as heteroresistant, suggesting that heteroresistance is common in CoNS isolated from bloodstream infections in the pediatric leukemia population (see Table S1 in the supplemental material).
TABLE 1.
Characteristics of evaluable participants (N = 62)a
| Characteristic | Nonheteroresistant (n = 38) |
Heteroresistant (n = 24) |
All participants (N = 62) |
|---|---|---|---|
| Age in yrs, mean (SD) | 10.6 (6.2) | 7.5 (6.3) | 9.4 (6.4) |
| Sex, no. (%) | |||
| Female | 12 (31.6) | 14 (58.3) | 26 (41.9) |
| Male | 26 (68.4) | 10 (41.7) | 36 (58.1) |
| CVC type, no. (%) | |||
| Tunneled CVC | 14 (36.8) | 16 (66.7) | 30 (48.4) |
| Port | 15 (39.5) | 4 (16.7) | 19 (30.6) |
| PICC | 2 (5.3) | 0 (0) | 2 (3.2) |
| Other | 7 (18.4) | 4 (16.7) | 11 (17.7) |
| Leukemia type, no. (%) | |||
| ALL | 26 (68.4) | 13 (54.2) | 39 (62.9) |
| AML | 12 (31.6) | 11 (45.8) | 23 (37.1) |
| History of HCT | 7 (18.4) | 6 (25) | 13 (21) |
| Race, no. (%) | |||
| White | 28 (73.7) | 16 (66.7) | 44 (71) |
| Black | 7 (18.4) | 4 (16.7) | 11 (17.7) |
| Other | 3 (7.9) | 4 (16.7) | 7 (11.3) |
SD, standard deviation; CVC, central venous catheter; PICC, peripherally inserted central catheter; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; HCT, hematopoietic cell transplantation.
Vancomycin exposure.
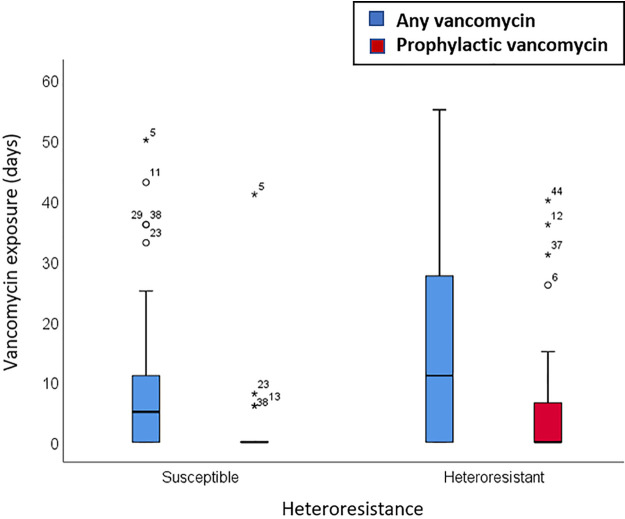
We next tested whether vancomycin exposure before infection increased the risk of a heteroresistant organism being recovered. In the 60 days before infection, participants with heteroresistant CoNS had received more days of vancomycin therapy (P = 0.11) and more days of vancomycin prophylaxis (P = 0.026) than had participants without heteroresistant CoNS (Fig. 1). These data suggest that prolonged exposure to intravenous vancomycin at the subtherapeutic doses used for prophylaxis may contribute to the emergence or selection of heteroresistant bacterial populations.
FIG 1.
Vancomycin exposure in the 60 days before infection for heteroresistant and nonheteroresistant isolates (N = 62). Box plots show the differences between heteroresistant and nonheteroresistant infections with respect to vancomycin exposure (P = 0.11) and prophylactic vancomycin exposure (P = 0.026). The boxes represent the 25th to 75th percentiles, the whiskers extend up to 1.5 times the box height, and the circles and stars represent outliers and extreme outliers, respectively.
Effect of heteroresistance on clinical outcomes.
We evaluated the effect of vancomycin heteroresistance on the clinical response to vancomycin treatment. Only 40 episodes were included in this analysis; the other 22 were excluded as being possibly attributable to blood culture contamination (see exclusion criteria in Materials and Methods). Seven (18%) of these 40 participants experienced treatment failure and 15 (38%) experienced poor clinical response. Of 6 participants with relapse of infection, sequence typing was available for 3 pairs of isolates; in all cases, the sequence types (STs) of the initial and subsequent isolates were identical. Participants with heteroresistant infections had higher cumulative incidences of treatment failure (P = 0.012) (Fig. 2a) and poor clinical response (P = 0.001) (Fig. 2b) compared to participants without heteroresistant infections.
FIG 2.
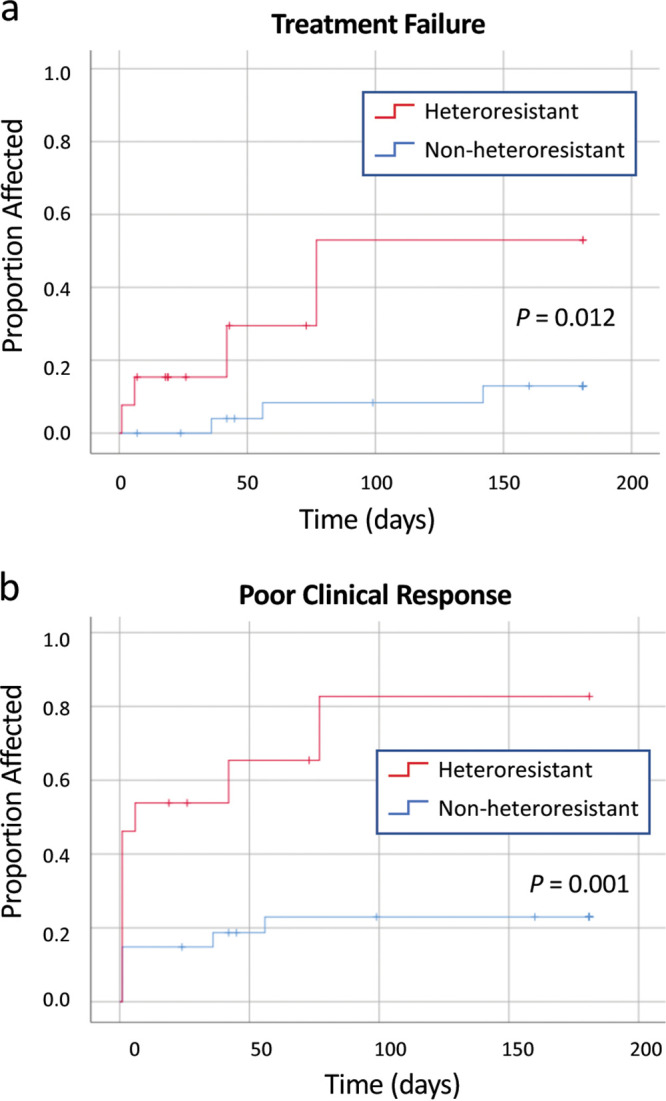
(a) Cumulative incidence of treatment failure in patients with CoNS bloodstream infection (N = 40). (b) Cumulative incidence of poor clinical response in patients with CoNS bloodstream infection (N = 40).
Of the other variables evaluated, only vancomycin MIC and age of the participant were considered to be potential confounders. Vancomycin MIC was significantly associated with poor clinical response (P = 0.037) but not with treatment failure (P = 0.669). Age group was also significantly associated with poor clinical response (P = 0.023), but its apparent association with treatment failure was not statistically significant (P = 0.054). Variables that were not associated with treatment failure or poor clinical response were central venous catheter (CVC) type (P = 0.727 or P = 0.377, respectively), leukemia type (P = 0.476 or P = 0.939, respectively), sex (P = 0.800 or P = 0.601, respectively), and self-reported race (P = 0.802 or P = 0.964, respectively). Therefore, vancomycin MIC and participant age were included in stratified analyses. These analyses showed that poor clinical response and treatment failure remained associated with heteroresistance after controlling for vancomycin MIC (P = 0.008 and P = 0.021, respectively) and for age group (P = 0.014 and P = 0 0.12, respectively). An additional subgroup analysis restricted to S. epidermidis isolates also showed that heteroresistance was significantly associated with treatment failure (P = 0.028) and poor clinical response (P = 0.006).
Genetic characterization of bacterial isolates.
Most isolates were identified as S. epidermidis by sequencing (Table 2). All heteroresistant isolates were S. epidermidis (P = 0.005), but they represented several sequence types. The most common sequence type was ST2, a well-known hospital pathogen (18, 19), and all S. epidermidis isolates were classified as members of genetic clusters (GCs) associated with hospital sources (GC1, GC5, and GC6) (20, 21). Despite all heteroresistant isolates being identified as S. epidermidis, there was no evidence of clonal outbreaks.
TABLE 2.
Species and multilocus sequence type for all first included CoNS isolates (N = 62)a
| Species/sequence type | No. (%) nonheteroresistant (n = 38) |
No. (%) heteroresistant (n = 24) |
|---|---|---|
| Staphylococcus epidermidis | 28 (53.8) | 24 (46.2) |
| ST2 | 7 (58.3) | 5 (41.7) |
| ST5 | 3 (33.3) | 6 (66.7) |
| ST16 | 2 (100) | 0 (0) |
| ST22 | 0 (0) | 1 (100) |
| ST23 | 0 (0) | 1 (100) |
| ST57 | 0 (0) | 1 (100) |
| ST73 | 0 (0) | 1 (100) |
| ST83 | 3 (60) | 2 (40) |
| ST89 | 1 (100) | 0 (0) |
| ST173 | 0 (0) | 1 (100) |
| ST218 | 1 (50) | 1 (50) |
| ST291 | 0 (0) | 1 (100) |
| NT | 11 (73.3) | 4 (26.7) |
| Staphylococcus haemolyticus | 4 (100) | 0 (0) |
| ST1 | 1 (100) | 0 (0) |
| ST3 | 1 (100) | 0 (0) |
| NT | 2 (100) | 0 (0) |
| Staphylococcus hominis | 6 (100) | 0 (0) |
| ST18 | 2 (100) | 0 (0) |
| NT | 4 (100) | 0 (0) |
| Total | 38 (61.3) | 24 (38.7) |
ST, sequence type; %, percentage of species/sequence type; NT, nontypeable.
We next looked for mutations in genes known to confer vancomycin resistance in S. epidermidis. Specifically, D471E and I527M in rpoB cause cross-resistance to rifampin and vancomycin, and V500F in walK causes cross-resistance to daptomycin and vancomycin (18, 19). Various nonsynonymous mutations in rpoB were identified in 8 S. epidermidis strains, of which 3 were heteroresistant (Table S2). Only one strain of ST23 had the dual resistance alleles in rpoB (E471, M527); it was resistant to rifampin (MIC > 32) and heteroresistant to vancomycin. Two nonsynonymous mutations in walK were identified in 2 heteroresistant S. epidermidis strains, but neither mutation had been previously linked to resistance (Table S2). No nonsynonymous mutations occurred in the walK gene’s cognate response regulator, walR. These results indicate that mutations in rpoB and walK are not necessary for vancomycin heteroresistance in the studied strains and also highlight a knowledge gap in our understanding of the genetic basis for this phenotype in S. epidermidis.
DISCUSSION
In this study, vancomycin heteroresistance was common in CoNS isolates from CLABSIs in children with leukemia. Data on the incidence of heteroresistance in CoNS are limited, but the high rate found in this study is similar to that reported for various CoNS species in various disease contexts in previous studies (4–10, 22).
Clonal spread of heteroresistant CoNS has been described previously, especially in neonatal intensive care units (9, 23). However, in this study, there was no single genetic marker for heteroresistance and a wide range of sequence types was involved. This suggests that the phenomenon of heteroresistance likely emerged on multiple occasions and by different mechanisms. Gene amplification may be a key genetic mechanism of heteroresistance in Gram-negative bacteria (24). However, heteroresistance to vancomycin may differ from heteroresistance to other antibiotics; because Gram-positive organisms commonly have high MICs within the susceptible range (the result of “MIC creep”) (25), heteroresistance might emerge from minor changes in these near-resistant subpopulations, rather than from the dramatic increases in MIC seen in Gram-negative species.
The study had some important limitations. The retrospective nature of the data collection means that some information might have been missed or misclassified or some confounding variables excluded. Additional variables, such as CVC removal and concomitant antibiotic therapy, were not included in the analysis, but were not expected to vary by heteroresistance status. The finding of poorer clinical outcomes in patients with heteroresistant infections might also be attributable to host factors, linkage disequilibrium, or another factor that affects heteroresistance testing. Because the method of detecting heteroresistance is not standardized, alternative laboratory approaches might yield different results and interpretations. Using a sensitive but nonstandard definition of poor clinical response improved our ability to evaluate the effect of heteroresistance on clinical outcomes, but the clinical relevance of these outcomes is uncertain. A major challenge to incorporating heteroresistance measurement into clinical practice and diagnosis is the absence of a widely accepted clinical test for the heteroresistance phenotype. In cases of infection for which rapid diagnosis and susceptibility testing are paramount, the time- and resource-intensive methodology used in this study is likely to be impractical. To maximize our ability to detect a difference between outcomes for heteroresistant and susceptible organisms, and to allow stratified analyses to control for potential confounders, we used both a standard definition of treatment failure and a more sensitive measure of poor clinical response to initial therapy. Although this approach identified a larger number of episodes in which clinical resolution of bacteremia did not occur immediately after initiation of vancomycin, the classification is nonstandard, and as more information becomes available, multivariable analyses using standard definitions of treatment failure will become important to confirm these findings.
The study also had several strengths that support our findings. We were able to connect information obtained from laboratory experiments to clinical data for a large cohort of patients with CoNS bloodstream infection, which enabled us to demonstrate the differences in clinical outcomes associated with heteroresistance. The 180-day duration of follow-up was appropriate because relapse of CLABSI after that time is uncommon (26, 27). Inclusion of isolates collected during the same time period and from the same population helped to exclude confounding by secular or cohort effects.
Similarly to other mechanisms of resistance, prior exposure to antibiotics increased the risk of heteroresistance for this study population (28, 29). Intriguingly, exposure to intravenous vancomycin administered as prophylaxis appeared to be more strongly associated with risk of heteroresistance than were other intravenous exposures. Subtherapeutic antibiotic levels over an extended period may provide strong selective pressure by encouraging the emergence of heteroresistance or helping to maintain the transient heteroresistance that emerges as a result of unstable gene amplifications or mutations (30). The immunocompromised state of the study participants might also have contributed to the evolution of heteroresistance by reducing the rate of clearance of heteroresistant bacteria (31). Concerns about contributing to the rise of antibiotic resistance have limited the implementation of glycopeptide prophylaxis (32, 33). A previous study found that vancomycin prophylaxis increased the risk of vancomycin-resistant enterococcal (VRE) infections (33); therefore, the finding that vancomycin prophylaxis also appears to contribute to heteroresistance in CoNS is important.
Modeling of heteroresistance in mammalian models has suggested that it can contribute to antibiotic treatment failure in patients with S. aureus, Enterobacter species, or Klebsiella species infections (3, 34, 35). Moreover, vancomycin heteroresistance in S. aureus infections appeared to contribute to treatment failure in multiple studies (36, 37). However, little information is available about the effect of heteroresistance on outcomes in CoNS infections. One case-control study that included CoNS and S. aureus found increased mortality in patients with heteroresistant infections (10), and a recent case report described a neonate with a heteroresistant Staphylococcus capitis infection that responded poorly to treatment with vancomycin (38). To our knowledge, the present cohort study was the first to have shown that the clinical response to vancomycin is demonstrably worse in infections caused by heteroresistant CoNS.
Conclusions.
Heteroresistance is common in isolates of S. epidermidis from bloodstream infections in children with cancer; it may also occur in other species, but it was not detected in this study. Systemic vancomycin exposure, especially at the subtherapeutic levels used for prophylaxis, is associated with subsequent heteroresistance. Heteroresistance appears to increase the risk of poor clinical outcomes, including poor initial response to treatment and recurrence of the infection after the completion of therapy. If these findings are validated in subsequent studies, development of a clinical test for heteroresistance and development of alternative treatment strategies for heteroresistant staphylococci could improve treatment outcomes for vulnerable patients.
MATERIALS AND METHODS
Clinical data.
This was a retrospective cohort study comprising all patients who developed CoNS CLABSI during treatment for leukemia at St. Jude between 1 January 2010, and 28 March 2016. The study was approved by the St. Jude institutional review board (approval XPD16-036). Participants were identified from the electronic medical record, and data on demographics, malignancy, vancomycin exposure, and infection outcomes were abstracted from the electronic medical record. Episodes were excluded if the isolate was not available for testing, was not identified as CoNS on sequencing, or was not successfully tested for heteroresistance.
Bacterial isolates.
CoNS isolates were obtained from the clinical microbiology laboratory, where they were stored in glycerol at −80°C for quality assurance purposes until tested. A prototypical vancomycin-heteroresistant S. aureus strain (Mu3) was obtained from ATCC (Manassas, VA) for use as a positive control. All isolates were classified as susceptible to vancomycin by conventional testing (with Vitek 2 [bioMérieux, Inc.]; MIC range, <0.5 to 2 μg/ml) (39).
Evaluation of vancomycin heteroresistance.
Population analysis profiling (PAP) was used to identify vancomycin heteroresistance in clinical isolates. PAP was performed in accordance with standard published techniques for detecting heterogeneous vancomycin-intermediate S. aureus (hVISA) (40, 41). Each isolate was grown in 10 ml of tryptic soy broth (BD Sciences, catalog no. 211825) overnight at 37°C. Of the 74 available clinical isolates, 30 were analyzed in 2016 and 44 were analyzed in 2017. The latter set of isolates required the addition of 0.2% yeast extract (BD Biosciences, catalog no. 212750) for growth. Overnight stationary-phase cultures were serially diluted 1:100 in 1× phosphate-buffered saline (PBS) to give dilutions ranging from 10−2 to 10−8, and 100 μl of each dilution was spread on brain heart infusion (BHI) agar plates (made from Bacto brain heart infusion and Bacto agar from BD Biosciences) containing 0, 1, 2, 4, 6, or 8 μg/ml of vancomycin (vancomycin hydrochloride; Sigma-Aldrich, St. Louis, MO). Each strain at each dilution was inoculated onto duplicate plates for each antibiotic concentration. After a 48-h incubation at 37°C, colonies were enumerated to estimate the bacterial survival, expressed as CFU per milliliter (CFU/ml), at each vancomycin concentration. The average number of colonies on duplicate plates was used to calculate the CFU/ml for each vancomycin concentration; CFU/ml was plotted as a function of vancomycin concentration, and the area under the curve (AUC) was calculated using GraphPad Prism 7. The AUC for each clinical isolate was then compared by a simple ratio to the average AUC for heteroresistant reference strain Mu3, obtained using the identical technique, and if the AUC ratio was 0.9 or greater, the isolate was classified as heteroresistant (40). PAP values for each isolate are shown in Table S1 and Fig. S1 and 2 in the supplemental material.
Genomic sequencing.
Each clinical isolate was inoculated in TSB and grown overnight at 37°C for DNA extraction. The entire overnight culture of each isolate was pelleted at 6,000 rpm for 10 min, and the pellets were resuspended in TSM solution (50 mM Tris [pH 7.5], 0.5 M sucrose, 10 mM MgCl2) before the addition of lysostaphin at 2 mg/ml. The mixture was then incubated at 37°C for 10 min. Once the bacterial cells had lysed, DNA was extracted using a Wizard genomic DNA purification kit (Promega, Madison, WI) in accordance with the manufacturer’s instructions. DNA libraries for next-generation sequencing were prepared using a Nextera DNA Flex library kit (Illumina, San Diego, CA) and sequenced on an Illumina Hi-Seq 2000 sequencing system.
Sequencing and typing.
Sequence reads for 74 clinical strains and Mu3 were adapter trimmed (minimum length of 15 bp) and then quality trimmed (minimum base quality of 12) by using BBMap (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbmap-guide/). Sequences were assembled de novo by using SPAdes v3.11.1. Sequence types (STs) from species-specific multilocus sequence typing (MLST) schemes were extracted from the assemblies by using mlst v2.8 (https://github.com/tseemann/mlst). For Staphylococcus epidermidis strains with typeable STs, genetic clusters were assigned according to the method of Tolo et al. (20). Sequences were aligned by the use of the Burrows-Wheeler Alignment tool bwa v0.7.17, using S. epidermidis DAR1907 (GenBank accession no. CP013943.1) as a reference strain. Sequences were subjected to coordinate sorting by using Picard v1.141 and realigned around short insertion-deletion polymorphisms (indels) with Genome Analysis Toolkit (GATK) v2.8-1. Variants were called by GATK and filtered for a minimum base quality of 30 and the presence of the variant in all strains. The functional effects of the variants were determined using SnpEff v4.
Statistical analysis.
The first available episode for each participant was used to evaluate associations between preceding vancomycin exposure and heteroresistance. Preceding vancomycin exposure was calculated as the number of days in the preceding 60 days on which intravenous vancomycin was administered. The Mann-Whitney U test was used to evaluate the significance of differences between groups. An unadjusted P value of less than 0.05 was considered to indicate statistical significance.
To evaluate associations between heteroresistance and clinical outcomes, the first episode for each participant was again selected, but episodes that were probably the result of contaminants, defined as episodes with single positive blood cultures and at least 1 simultaneously collected negative blood culture, were excluded. Treatment failure was defined as death during the CLABSI episode or recurrence of BSI with the same organism within 180 days; poor clinical response was defined as persistent positive blood cultures collected at least 1 day after the initiation of vancomycin or treatment failure as defined above. Poor clinical response was designed to be a more sensitive measure of decreased initial response to vancomycin therapy, as death and recurrence are relatively uncommon in this cohort.
The log rank test was used to compare data representing the cumulative incidence of each clinical outcome with heteroresistant and nonheteroresistant bacteria. Cases were censored at unrelated death, completion of cancer therapy, permanent discharge from the institution, 180 days after onset of BSI, or completion of the study period. Potential confounders were each evaluated for their association with outcome and were included in stratified survival analyses, pooled over all strata, if the P value was 0.1 or less. Statistical analyses were performed with SPSS (IBM SPSS Statistics for Windows, Version 26.0: IBM Corporation, Armonk, NY).
Supplementary Material
ACKNOWLEDGMENTS
We thank Bruce Levin and Elaine Tuomanen for lively discussion on heteroresistance and encouragement, Kris Branum for regulatory support, and Keith A. Laycock for scientific editing of the manuscript.
No specific funding was obtained for this project.
We report that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 2.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MR, Perdreau-Remington F, Chambers HF. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob Agents Chemother 47:1262–1266. doi: 10.1128/AAC.47.4.1262-1266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong J, Caya C, Levesque S, Quach C. 2016. Heteroresistant vancomycin intermediate coagulase negative Staphylococcus in the NICU: a systematic review. PLoS One 11:e0164136. doi: 10.1371/journal.pone.0164136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Mello D, Daley AJ, Rahman MS, Qu Y, Garland S, Pearce C, Deighton MA. 2008. Vancomycin heteroresistance in bloodstream isolates of Staphylococcus capitis. J Clin Microbiol 46:3124–3126. doi: 10.1128/JCM.00592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashaly GE-S, El-Mahdy RH. 2017. Vancomycin heteroresistance in coagulase negative Staphylococcus blood stream infections from patients of intensive care units in Mansoura University Hospitals, Egypt. Ann Clin Microbiol Antimicrob 16:63–63. doi: 10.1186/s12941-017-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunes AP, Schuenck RP, Bastos CC, Magnanini MM, Long JB, Iorio NL, Santos KR. 2007. Heterogeneous resistance to vancomycin and teicoplanin among Staphylococcus spp. isolated from bacteremia. Braz J Infect Dis 11:345–350. doi: 10.1590/s1413-86702007000300009. [DOI] [PubMed] [Google Scholar]
- 8.Nunes A, Teixeira L, Iorio N, Bastos C, Fonseca L, Soutopadron T, Dossantos K. 2006. Heterogeneous resistance to vancomycin in Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus warneri clinical strains: characterisation of glycopeptide susceptibility profiles and cell wall thickening. Int J Antimicrob Agents 27:307–315. doi: 10.1016/j.ijantimicag.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Van Der Zwet WC, Debets-Ossenkopp YJ, Reinders E, Kapi M, Savelkoul PH, Van Elburg RM, Hiramatsu K, Vandenbroucke-Grauls CM. 2002. Nosocomial spread of a Staphylococcus capitis strain with heteroresistance to vancomycin in a neonatal intensive care unit. J Clin Microbiol 40:2520–2525. doi: 10.1128/jcm.40.7.2520-2525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong SS, Ho PL, Woo PC, Yuen KY. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin Infect Dis 29:760–767. doi: 10.1086/520429. [DOI] [PubMed] [Google Scholar]
- 11.Zakhour R, Chaftari AM, Raad II. 2016. Catheter-related infections in patients with haematological malignancies: novel preventive and therapeutic strategies. Lancet Infect Dis 16:e241–e250. doi: 10.1016/S1473-3099(16)30213-4. [DOI] [PubMed] [Google Scholar]
- 12.van Pelt C, Nouwen J, Lugtenburg E, van der Schee C, de Marie S, Schuijff P, Verbrugh H, Löwenberg B, van Belkum A, Vos M. 2003. Strict infection control measures do not prevent clonal spread of coagulase negative staphylococci colonizing central venous catheters in neutropenic hemato-oncologic patients. FEMS Immunol Med Microbiol 38:153–158. doi: 10.1016/S0928-8244(03)00114-7. [DOI] [PubMed] [Google Scholar]
- 13.Celebi S, Sezgin ME, Cakir D, Baytan B, Demirkaya M, Sevinir B, Bozdemir SE, Gunes AM, Hacimustafaoglu M. 2013. Catheter-associated bloodstream infections in pediatric hematology-oncology patients. Pediatr Hematol Oncol 30:187–194. doi: 10.3109/08880018.2013.772683. [DOI] [PubMed] [Google Scholar]
- 14.Rosa RG, Dos Santos RP, Goldani LZ. 2014. Mortality related to coagulase-negative staphylococcal bacteremia in febrile neutropenia: a cohort study. Can J Infect Dis Med Microbiol 25:e14–e17. doi: 10.1155/2014/702621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedroso S, Sandes SHC, Filho RAT, Nunes AC, Serufo JC, Farias LM, Carvalho MAR, Bomfim MRQ, Santos SG. 2018. Coagulase-negative staphylococci isolated from human bloodstream infections showed multidrug resistance profile. Microb Drug Resist 24:635–647. doi: 10.1089/mdr.2017.0309. [DOI] [PubMed] [Google Scholar]
- 16.Garrett DO, Jochimsen E, Murfitt K, Hill B, McAllister S, Nelson P, Spera RV, Sall RK, Tenover FC, Johnston J, Zimmer B, Jarvis WR. 1999. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect Control Hosp Epidemiol 20:167–170. doi: 10.1086/501605. [DOI] [PubMed] [Google Scholar]
- 17.Sieradzki K, Roberts RB, Serur D, Hargrave J, Tomasz A. 1999. Heterogeneously vancomycin-resistant Staphylococcus epidermidis strain causing recurrent peritonitis in a dialysis patient during vancomycin therapy. J Clin Microbiol 37:39–44. doi: 10.1128/JCM.37.1.39-44.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang J-H, Dexter C, Cameron DR, Monk IR, Baines SL, Abbott IJ, Spelman DW, Kostoulias X, Nethercott C, Howden BP, Peleg AY. 2019. Evolution of daptomycin resistance in coagulase-negative staphylococci involves mutations of the essential two-component regulator WalKR. Antimicrob Agents Chemother 63:e01926-18. doi: 10.1128/AAC.01926-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JYH, Monk IR, Gonçalves da Silva A, Seemann T, Chua KYL, Kearns A, Hill R, Woodford N, Bartels MD, Strommenger B, Laurent F, Dodémont M, Deplano A, Patel R, Larsen AR, Korman TM, Stinear TP, Howden BP. 2018. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat Microbiol 3:1175–1185. doi: 10.1038/s41564-018-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolo I, Thomas JC, Fischer RSB, Brown EL, Gray BM, Robinson DA. 2016. Do Staphylococcus epidermidis genetic clusters predict isolation sources? J Clin Microbiol 54:1711–1719. doi: 10.1128/JCM.03345-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas JC, Zhang L, Robinson DA. 2014. Differing lifestyles of Staphylococcus epidermidis as revealed through Bayesian clustering of multilocus sequence types. Infect Genet Evol 22:257–264. doi: 10.1016/j.meegid.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieradzki K, Villari P, Tomasz A. 1998. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother 42:100–107. doi: 10.1128/AAC.42.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasigade JP, Raulin O, Picaud JC, Tellini C, Bes M, Grando J, Ben Said M, Claris O, Etienne J, Tigaud S, Laurent F. 2012. Methicillin-resistant Staphylococcus capitis with reduced vancomycin susceptibility causes late-onset sepsis in intensive care neonates. PLoS One 7:e31548. doi: 10.1371/journal.pone.0031548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicoloff H, Hjort K, Levin BR, Andersson DI. 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 25.Steinkraus G, White R, Friedrich L. 2007. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–05. J Antimicrob Chemother 60:788–794. doi: 10.1093/jac/dkm258. [DOI] [PubMed] [Google Scholar]
- 26.Wolf J, Allison KJ, Tang L, Sun Y, Hayden RT, Flynn PM. 2014. No evidence of benefit from antibiotic lock therapy in pediatric oncology patients with central line-related bloodstream infection: results of a retrospective matched cohort study and review of the literature. Pediatr Blood Cancer 61:1811–1815. doi: 10.1002/pbc.25101. [DOI] [PubMed] [Google Scholar]
- 27.Wolf J, Connell TG, Allison KJ, Tang L, Richardson J, Branum K, Borello E, Rubnitz JE, Gaur AH, Hakim H, Su Y, Federico SM, Mechinaud F, Hayden RT, Monagle P, Worth LJ, Curtis N, Flynn PM. 2018. Treatment and secondary prophylaxis with ethanol lock therapy for central line-associated bloodstream infection in paediatric cancer: a randomised, double-blind, controlled trial. Lancet Infect Dis 18:854–863. doi: 10.1016/S1473-3099(18)30224-X. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee A, Modarai M, Naylor NR, Boyd SE, Atun R, Barlow J, Holmes AH, Johnson A, Robotham JV. 2018. Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect Dis 18:e368–e378. doi: 10.1016/S1473-3099(18)30296-2. [DOI] [PubMed] [Google Scholar]
- 29.Sakoulas G, Gold HS, Cohen RA, Venkataraman L, Moellering RC, Eliopoulos GM. 2006. Effects of prolonged vancomycin administration on methicillin-resistant Staphylococcus aureus (MRSA) in a patient with recurrent bacteraemia. J Antimicrob Chemother 57:699–704. doi: 10.1093/jac/dkl030. [DOI] [PubMed] [Google Scholar]
- 30.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 31.Margolis E, Rosch JW. 2018. Fitness landscape of the immune compromised favors the emergence of antibiotic resistance. ACS Infect Dis 4:1275–1277. doi: 10.1021/acsinfecdis.8b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boztug H, Muhlegger N, Potschger U, Attarbaschi A, Peters C, Mann G, Dworzak M. 2017. Antibiotic prophylaxis with teicoplanin on alternate days reduces rate of viridans sepsis and febrile neutropenia in pediatric patients with acute myeloid leukemia. Ann Hematol 96:99–106. doi: 10.1007/s00277-016-2833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inaba H, Gaur AH, Cao X, Flynn PM, Pounds SB, Avutu V, Marszal LN, Howard SC, Pui CH, Ribeiro RC, Hayden RT, Rubnitz JE. 2014. Feasibility, efficacy, and adverse effects of outpatient antibacterial prophylaxis in children with acute myeloid leukemia. Cancer 120:1985–1992. doi: 10.1002/cncr.28688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, Burd EM, Weiss DS. 2016. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1:16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS. 2018. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9:e02448-17. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes DM, Ward KE, LaPlante KL. 2015. Clinical implications of vancomycin heteroresistant and intermediately susceptible Staphylococcus aureus. Pharmacotherapy 35:424–432. doi: 10.1002/phar.1577. [DOI] [PubMed] [Google Scholar]
- 37.Koh YR, Kim KH, Chang CL, Yi J. 2016. Prevalence and clinical impact of heterogeneous vancomycin-intermediate Staphylococcus aureus isolated from hospitalized patients. Ann Lab Med 36:235–243. doi: 10.3343/alm.2016.36.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butin M, Claris O, Laurent F. 2019. Clinical impact of vancomycin heteroresistance in staphylococcal strains involved in neonatal sepsis: discussion of a case report. Arch Pediatr 26:236–237. doi: 10.1016/j.arcped.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement (document M100-S24). CLSI, Wayne, PA. [Google Scholar]
- 40.Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47:399–403. doi: 10.1093/jac/47.4.399. [DOI] [PubMed] [Google Scholar]
- 41.Wootton M, MacGowan AP, Walsh TR, Howe RA. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J Clin Microbiol 45:329–332. doi: 10.1128/JCM.01508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.