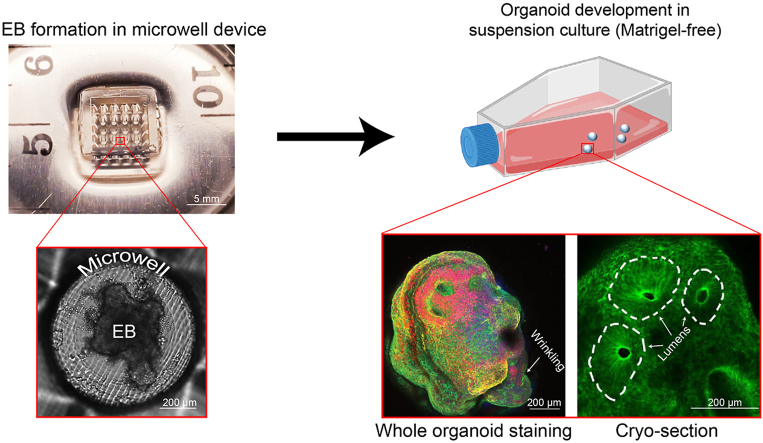
Abstract
The current methods of generating human cerebral organoids rely excessively on the use of Matrigel or other external extracellular matrices (ECM) for cell micro-environmental modulation. Matrigel embedding is problematic for long-term culture and clinical applications due to high inconsistency and other limitations. In this study, we developed a novel microwell culture platform based on 3D printing. This platform, without using Matrigel or external signaling molecules (i.e., SMAD and Wnt inhibitors), successfully generated matured human cerebral organoids with robust formation of high-level features (i.e., wrinkling/folding, lumens, neuronal layers). The formation and timing were comparable or superior to the current Matrigel methods, yet with improved consistency. The effect of microwell geometries (curvature and resolution) and coating materials (i.e., mPEG, Lipidure, BSA) was studied, showing that mPEG outperformed all other coating materials, while curved-bottom microwells outperformed flat-bottom ones. In addition, high-resolution printing outperformed low-resolution printing by creating faithful, isotropically-shaped microwells. The trend of these effects was consistent across all developmental characteristics, including EB formation efficiency and sphericity, organoid size, wrinkling index, lumen size and thickness, and neuronal layer thickness. Overall, the microwell device that was mPEG-coated, high-resolution printed, and bottom curved demonstrated the highest efficacy in promoting organoid development. This platform provided a promising strategy for generating uniform and mature human cerebral organoids as an alternative to Matrigel/ECM-embedding methods.
Keywords: Cortical wrinkling, Cortical folding, 3D culture, Biomanufacturing, Embryoid body spheroid, Microfabrication
Graphical abstract
Highlights
-
•
Mature cerebral organoids were generated in ECM-free conditions.
-
•
3D printing techniques produced microtopography, dependent on printing modality.
-
•
Surface coating was shown to influence organoid formation, with mPEG performing best.
-
•
Curved geometry produced larger lumens and thicker neuronal layers in organoids.
1. Introduction
Recent advances in the generation of human cerebral organoids have augmented our understanding of early human brain development and diseases [[1], [2], [3], [4], [5], [6], [7], [8]]. Human cerebral organoids recapitulate critical high-level physiological features (e.g., lumen formation [9], cortical layer development [9], and folding/wrinkling [10]), which emerge during early brain morphogenesis and successive maturation [3,4]. While various methods were used for unguided human cerebral organoids generation, virtually all require embedding in Matrigel, a complex basement membrane matrix that modulates the stem cell micro-environment and cell signaling [11]. However, Matrigel suffers from inconsistency in manufacturing and complexity in composition [12]; this variability influences the organoid formation and thus limits clinical applications of human cerebral organoids [12,13]. Long-term culturing in Matrigel was also shown to lead to pre-mature organoids [14] and induce undesired neurite outgrowth [15]. Although synthetic extracellular matrix (ECM) substitutes were recently demonstrated [16], removing these materials (necessary for cell or imaging analysis) is still problematic because it unavoidably inflicts mechanical damage to the organoids [17,18]. Thus, finding alternative ECM-free methods continues to be one major topic of this field.
Microwell platforms [17] and other microfluidic 3D culturing platforms [19,20] have been successfully employed for the generation of spheroids [[21], [22], [23]], which are similar to organoids but lack high-level physiological features. When forming spheroid cultures, microwell platforms provide a geometrically-controlled micro-environment that defines spheroid biology (e.g., shape, size, and differentiation) [[24], [25], [26]]. Besides, microwell platforms also feature high-scalability, cost-effectiveness, ease of operation, and uniform spheroid generation [21,23,[27], [28], [29], [30], [31], [32], [33]].
Microwell platforms have been used in facilitating the rudimentary aggregation of embryoid bodies (EBs), which are spheroids of stem cells and the precursor of cerebral organoids [2,15,17,34]. Microwell platforms aided the initial self-organization of EBs, by providing a confined space (such as reverse-pyramid-shaped AggreWell [2] and curved surface [24,32,35]), enabling cell aggregation, and controlling initial cell densities [32]. Lately, microwell-derived EBs were further cultured to develop into organoids [17,34], however, still relying on Matrigel-embedding for modulating the organoid micro-environment [13,17,34]. In addition, most of the microwell generated spheroids (structureless aggregation) [36,37] or spheroid-like organoids (e.g. Islet organoids [21,38]) have much lower structural and developmental complexity compared to brain organoids. Oftentimes the studies that employed a microwell platform for the brain organoid generation relied on Matrigel embedding for organoid maturation. Worthy of mentioning, a recent study generated uniquely structured spheroids using AggreWell, and then cultured in an ECM-free environment [2]. The resulting spheroids showed putative lumen formation, similar to early-stage cerebral organoids [2]. However, no other cerebral organoid characteristic structures were observed. In an ECM-free system, the effect of the microwell platform will be a major determining factor in modulating cell-fate, which provides an opportunity for engineering. However, the impact of microwell material properties (e.g., geometry, surface coating) on the subsequent maturation and structure of human cerebral organoids has been largely understudied.
In this study, we evaluated the long-term effects of a 3D-printed microwell in unguided organoid formation without introducing Matrigel or another ECM. Our results showed the microwell-derived organoids robustly developed mature physiological features (e.g., lumen structures, cortical layers, and folding/wrinkling), which were not seen with any other unguided (ECM-free) or microwell methods. With physiological development similar or superior to the traditional Matrigel-embedded methods, our micro-well platform presented a simple, scalable, and ECM-free alternative for the engineering and fabrication of human cerebral organoids.
Furthermore, unlike the existing microwell devices fabricated through vacuum-based photolithography [31], injection molding (Aggrewell) [2], and other complex fabrication process [21,29], our microwell devices were made through 3D-printing and soft lithography and thus offer an unprecedented capability in rapid prototyping. Thus, we were able to optimize our 3D-printed microwell platform and found that the microwell geometry (e.g., printing resolution and well curvature) and surface coating have a profound effect on the early-stage development of human cerebral organoids. As a cell-repulsive surface coating, mPEG outperformed Lipidure and bovine serum albumin (BSA) and generated wrinkling and lumen structures with significantly higher consistency and efficiency. Additionally, we demonstrated that high-resolution curved microwell-devices significantly promoted neuronal differentiation and maturation.
2. Materials and methods
2.1. 3D printing of reverse mold
Devices were designed in SolidWorks (Dassault Systems SolidWorks Corporation, USA). Each microdevice is composed of a five by five array of microwells, allowing for high throughput. Two CAD files were prepared: (1) flat microwell geometry and (2) curved microwell geometry. In both cases, the microwell diameter was kept constant at 800 microns and the resulting arrays fit comfortably within a six-well plate format.
The molds were printed by two methods; high-resolution direct light processing (DLP) 3D prints were supplied by ResinWorks3D (Ontario, Canada) and low-resolution FLX-S85 material inkjet 3D prints were obtained from an Objet 260 Connex 2 (Stratasys Ltd.).
After printing, all molds were cleaned by sonication in 200-proof ethanol for 10 min and then dried thoroughly. The molds were placed under vacuum with two drops of a silanizing agent (97%, 1H,1H,2H, 2H-Perfluorooctyl-trichlorosilane, Alfa Aesar, USA) for three hours to improve subsequent polydimethylsiloxane (PDMS) molding.
2.2. PDMS device preparation
PDMS (Sylgard 184 silicone elastomer; Dow Silicon Corporation, USA) was prepared at a ratio of 10:1 base to the initiator and degassed thoroughly. Molds were pre-wet in PDMS, then placed feature-down for reverse molding (Fig. 1A). After overnight curing at 50 °C, the molds were peeled from PDMS, forming the microwell platforms for EB aggregation. For sterilization, devices were sonicated in 200-proof ethanol (3*10 min), and deionized (DI) water (2*10 min). After drying, devices were autoclaved (Autoclave, Tuttnauer, USA).
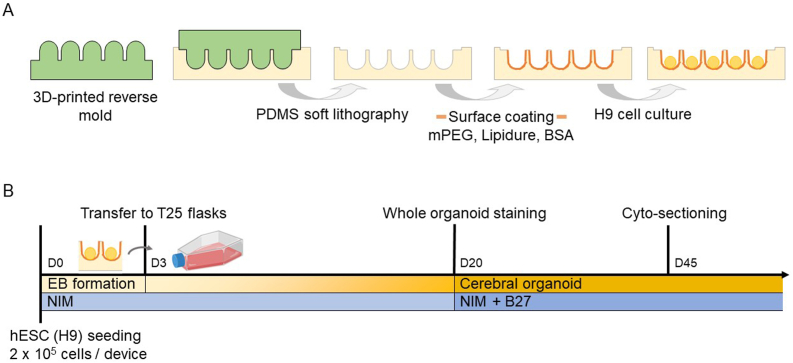
Fig. 1.
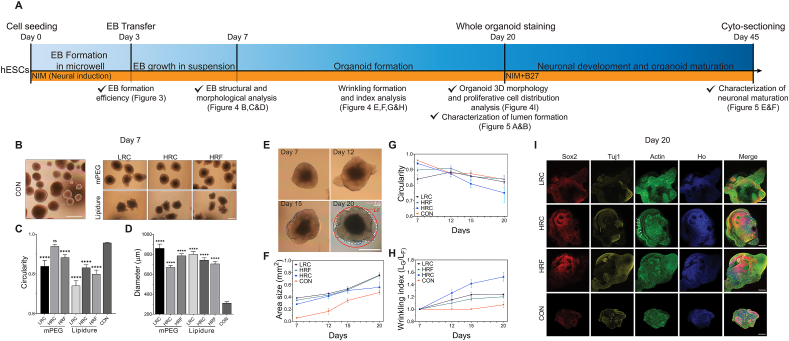
Microwell fabrication and use in human cerebral organoid culture in this study. A) Schematic of microwell fabrication and post-treatment. The reverse mold (micropillars) was 3D-printed. By molding in PDMS through soft lithography, the microwell platform was created, followed by a surface coating of mPEG, Lipidure, or BSA, and then the embryonic stem cell (H9) culturing. B) Timeline of the microwell culturing and human cerebral organoid generation. hESCs (H9) were seeded at 0.2 million cells per microwell device and cultured for three days for EB formation. The formed EBs were then transferred to T25 flasks for prolonged culture and neural induction. Cerebral organoid formation and maturation were then assessed on day 20 and day 45 by whole organoid staining and cyto-sectioning, respectively. NIM medium was used throughout the entire culture process, and B27 was added since day 20.
2.3. Scanning electron microscopy (SEM)
SEM images were taken with FEI Quanta FEG 650 Scanning Electron Microscope (SEM) (ThermoFischer, USA).
2.4. Surface coating
Devices were coated with mPEG (mPEG-silane, 5k Da, Creative PEGWorks, USA), Lipidure (NOF Corporation, Japan), or BSA (Fischer Scientific, USA). Coating concentrations and the modified coating protocol were based on previous reports [mPEG [39]; Lipidure [11]; BSA [21]].
For mPEG coated surfaces, 4 mM mPEG was prepared in 95% ethanol solution at room temperature. Then, devices were treated with handheld plasma (Electro Technic Products, USA) for approximately thirty seconds each. 120 μL of mPEG solution was immediately pipetted into the device and allowed to incubate at room temperature for two hours. All remaining solution was removed, and devices were stored at 4 °C until further use.
For Lipidure coated surfaces, Lipidure was prepared at 0.5 wt% in 200-proof ethanol. 120 μL of Lipidure solution was then pipetted into each device and allowed to incubate at room temperature for two hours. Afterward, all remaining solution was removed, and devices were stored at 4 °C until further use.
For BSA coated surfaces, devices were coated in a 3% BSA solution at 37 °C overnight. Afterward, the devices were washed with cell culture medium (3* 5 min) before cell-seeding. BSA had been diluted in DI water and sterile filtered before use.
2.5. hESC maintenance and passage
Human embryonic stem cells (hESCs) (H9, passages 35–45, WiCell) were maintained in a feeder-free environment on vitronectin-coated six-well plates (Life Technologies, USA). A standard protocol using the Essential 8 basal medium (Life Technologies, USA) was followed (www.wicell.com). hESC colonies were passaged by using EDTA (Lonza Inc., USA) when they became 60–70% confluent.
2.6. Cell culture in microwell devices
When cells became 60–70% confluent in the six-well plates, hESC colonies were treated with EDTA for 2 min. Cells were washed with DMEM/F12 1:1 (Life Technologies, USA) and dissociated into single cells. Cells were counted under a hemocytometer. ~2 × 105 cells were prepared in 100 μL Neural Induction Medium (NIM; Neural Induction Medium:1% NEAA (Life Technologies, USA), 1% N2 supplement (Life Technologies, USA), 98% DMEM/F12 1:1) and then seeded to each microwell device. After 24 h, an additional 4 ml of NIM was added. On day 3, EBs were transferred to T25 flasks. The medium was changed every other day. Beginning on day 20, 1:50 B27 supplement minus Vitamin A (Life Technologies, USA) was added to NIM (Fig. 1B).
2.7. The control group (CON)
The control group did not use microwell devices. Instead, the EBs were formed in a T25 flask since day 0. The rest of the culture procedure was identical to that in microwell devices in section 2.6. For the EB formation, 60–70% confluent hESC colonies in the six-well plates were treated with 1 U/mL dispase (Life Technologies, USA) at 37 °C for 1–2 min, followed by DMEM/F12 1:1 washing, as previously reported [40]. After detachment in each well of the well plates, cells were suspended in NIM in a T25 flask.
2.8. Whole organoid immunostaining
On day 20, organoids were fixed with 4% paraformaldehyde (PFA, Sigma Aldrich, USA) overnight at 4 °C. Then organoids were washed with phosphate-buffered saline (PBS) for 2 h at 4 °C. After being washed with PBS 3 times, samples were processed with 1% Triton X-100 (VWR, USA) and 5% donkey serum (Millipore, USA) overnight at 4 °C. The next day, samples were incubated at 4 °C for 24–48 h (depending on the size of the organoids) with primary antibodies (Table S1), which were suspended in 1% Triton X-100 and 5% donkey serum. Then the organoids were washed with PBS for 2 h at 4 °C. After being washed with PBS 3 times, organoids were incubated with secondary antibodies, prepared in 5% donkey serum, overnight at 4 °C. The nuclei were counterstained with Hoechst 33258. Then, samples were washed with PBS for 2 h before fluorescent imaging.
2.9. Cryo-sectioning and immunostaining
On day 45, cerebral organoids were collected and fixed with 4% PFA overnight at 4 °C. Then cerebral organoids were washed with PBS for 2 h at 4 °C. After being washed with PBS for 3 times, samples were cryoprotected in 30% sucrose in PBS overnight at 4 °C. The next day, organoids were transferred to the cryomold and filled with optimal cutting temperature (OCT) compound (Sakura Finetek, USA). Samples were then stored at −80 °C. Frozen samples were sliced in a Leica microtome. The organoids were sliced into 30 μm thickness, and the sliced samples were transferred to PDL-coated slides for immunostaining.
The OCT compound was removed by PBS washing for 20 min. The tissue was outlined by a water-resistant marker. Samples were treated with 1% Triton X-100 and 5% donkey serum for 1 h at room temperature. Then the samples were incubated overnight at 4 °C with primary antibodies suspended in 1% Triton X-100 and 5% donkey serum. The next day, samples were washed with PBS for 10 min at room temperature. After being washed with PBS 3 times, samples were then incubated in secondary antibody, prepared from 5% donkey serum, for 30 min at room temperature. The nuclei were counterstained with Hoechst 33258. Then, samples were washed with PBS for 30 min and captured with fluorescent imaging.
2.10. Image acquisition
Tissue samples were imaged using bright-field (Julistage Real Time Cell History Recorder, NanoEntek Inc or AmScope IN300TC-FL optical microscope with a Canon EOS Rebel T5 digital camera) and confocal imaging (Carl Zeiss LSM-710 live cell imaging system, Zeiss). On day 2, EBs were imaged within the microwell devices, and formation efficiency was manually calculated. Day 7 organoids were analyzed for circularity and diameter from bright-field images using ImageJ (NIH). The wrinkling index of the organoids was measured from day 7 to day 20, following a protocol defined earlier [11]. On day 7 to day 20, organoid area size was also measured using ImageJ. Day 45 immunostaining images were similarly processed and measured for lumen diameter, thickness, and quantity.
2.11. Data analysis
Statistical significance between samples' means was determined using a student's unpaired t-test and between groups was determined by one-way ANOVA followed by Tukey's post-hoc tests. Statistical tests were performed either in SPSS (IBM, USA) or in GraphPad (GraphPad, USA).
3. Results and discussion
3.1. Device fabrication
Previously, microwell platforms were used for the culturing of 3D spheroids, which often lack high-level structures. Three characteristics of microwells were believed critical for the formation of cell aggregates: spatial confinement for defined cell numbers [26], an anti-fouling smooth surface for non-adhesive cell contacts [17], and a concave curvature to facilitate spherical aggregation [21,31,35,41]. While the seeding cell density can help determine the former function, the latter two characteristics are mostly regulated through device fabrication and treatment. We sought to extend microwell platforms to cerebral organoid generation, by first investigating how smoothness, curvature, and anti-fouling surface were realized in our 3D printed platform.
To demonstrate different smoothness and curvature, we employed various resolutions and curvatures using different printing methods (i.e., printing technique and resin) and designs, respectively. Three devices were fabricated to include high resolution flat—HRF, high resolution curved—HRC, and low resolution curved—LRC. The PDMS stamps inversely replicated the 3D-printed molds with high-fidelity after soft lithography (Fig. S1A&B), with no discernible deviation (Fig. S1C), and were used for topographic assessment of the printed 3D molds. As shown in the SEM images (Fig. 2A&B), high-resolution printing (HRF and HRC) yielded 3D structures faithful to the CAD design, which consists of 800 μm diameter wells with either flat or concave bottom surfaces. The slanted side-walls which connect to the wells (Fig. 2A&B, Top-View of HRF and HRC) were designed to accelerate cell movement to the bottom of the wells.
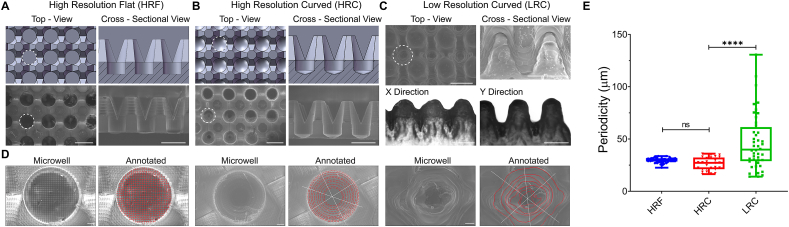
Fig. 2.
Characterization of shape and microtopography of the PDMS microwell devices. A) High-resolution flat (HRF) device. Top and cross-sectional views of CAD models (upper) and corresponding SEM micrographs (lower) are shown. White dash lines were used to outline a single microwell culturing space. B) High-resolution curve (HRC) device. Top and cross-sectional views of CAD models (upper) and corresponding SEM micrographs (lower) are shown. White dash lines were used to outline a single microwell culturing space. C) Low-resolution curved (LRC) devices showed distorted morphology (SEM micrographs, upper), and anisotropic shape in the x- and y-directions (inspection microscopy sectional views, lower). D) Representative top-view high-magnification SEM images and corresponding microtopography analysis of a microwell for HRF (left two), HRC (middle two), and LRC (right two) devices. SEM images of the microwell bottom revealed microtopography, contours of which were analyzed and annotated in red lines. The microtopography was measured as a distance between two adjacent contours along the white lines. The contour analysis in HRC and HRF showed a regularly-spaced grid and concentric circles, whereas that in LRC showed irregular concentric circles. E) Distribution of microtopography periodicity as a function of the device. Periodicity was measured as a distance between two adjacent contours along the white lines, which are the vertical/horizontal axis for HRF and diagonal axis for LRC and HRC. Statistical analysis: One-way ANOVA with Tukey's post-hoc analysis. Scale bars: (A, B and C): 1 mm; (D) 100 μm.
Further, as seen from the cross-sectional view of the CAD models and corresponding SEM images, the V-shaped converging pillar created a network between the wells and medium above, for better nutrient exchange. In the HRC design, the contacting angle between the microwell edge and wall was designed to be 450 (Fig. S3A). The bottom-curved surface was to aid cell aggregation. In contrast to the high-resolution devices, the low-resolution printing (LRC) resulted in substantial deviation. In fact, the LRC devices shown in Fig. 2C share the same flat-bottom design as the HRF devices in Fig. 2A, but deviated far enough to render a curved bottom. This discrepancy, we believe, was due to the resolution limit of the printing method. The size of the LRC device features were smaller than the minimum resolution of the inkjet printer, resulting in altered and smoothed dimensions. We also observed anisotropic printing deviation with the LRC devices, where the wells were shallower in the x-direction and deeper in the y-direction (Fig. 2C). This deviation may be due to the motion of the printhead during printing, as ink material was deposited in parallel lines before curing [42]. Nevertheless, the LRC device, which was the most similar to a V-shaped well amongst our devices, maintained all essential features. Namely, it had a slanted wall and a narrow well, which helped in cell aggregation, and a network between wells for nutrient exchange.
Other studies producing microwells for cell aggregation have generally created smooth-surfaced structures. In contrast, 3D printing unavoidably created surface texture and micro-topography due to its intrinsic mechanism of polymer fiber deposition [[43], [44], [45], [46]] (Fig. 2). Thus, we observed that the printing technique's resolution determined the pattern and periodicity of these micro-topographic features (Fig. 2D&E). High-resolution devices (HRC and HRF) had significantly lower and consistent periodicity compared to low-resolution devices (LRC) (Fig. 2E). In the HRF devices, a grid-like pattern made up of approximately 29 μm squares was apparent. The HRC devices had concentric ridges, repeating, on average, every 26 μm. The LRC devices' microtopography was less regular; we measured distances between concentric ridges ranging from 14 to 130 μm. Collectively, although the curved devices (LRC and HRC) may functionally be similar, their differences in periodicity and topography provided different 3D micro-environments during cell aggregation, which may further impact the organoid-maturation process.
3.2. Effect of geometry and surface coating on EB formation efficiency
We sought to test the influence of each device on EB formation, which is the first developmental step towards organoid formation [21]. During EB formation, cells establish cell-cell connections that lead to self-assembly and differentiation [2]. The initial size of EBs has been shown to affect the subsequent maturation of organoids [47]. In microwell platforms, an EB's initial cell aggregation is mainly affected by microwell's geometry [48] and surface coating [17], similar to the spheroid formation. Hence, to establish a method that reproducibly generates human cerebral organoids in ECM-free conditions, optimization of initial EB formation parameters is crucial.
First, three coating materials (i.e., mPEG, Lipidure, and BSA), which are commonly used in spheroid culture, were chosen to test the effect of surface coating on EB aggregation. We verified the efficacy of these coating by measuring the change in the surface property (water-contact angle) and stem cell adhesiveness on flat PDMS surfaces. For all coatings, the hydrophilicity of the coated surfaces increased significantly compared to an uncoated surface (Fig. S2A). These results are consistent with the anti-fouling mechanisms of the chosen passivation materials [39,49,50]. The inherent hydrophobicity of PDMS promotes protein adsorption and cell attachment [49,51,52]. Passivation coatings generally render a PDMS surface hydrophilic, preventing protein adsorption and cell-adhesion [53]. For BSA, albumin protein adsorbs to the PDMS surface like any protein, which renders the surface hydrophilic [49,54,55], creating an anti-fouling layer that inhibits other protein adsorption. However, protein-based anti-fouling coatings are temporary because the PDMS regains its hydrophobicity over time. For mPEG coating, a self-assembled monolayer of the PEG moieties [39,56], covalently attached to the surface of the substrate, forms an anti-fouling surface by exerting steric repulsion to protein absorption [57,58] (Fig. S2B). Lipidure, a zwitterionic based coating, is made of a biocompatible polymer, phosphorylcholine, which has equal amounts of negative and positive charges [57,58]. This equal charge distribution makes the surface electrically neutral, which prevents protein adsorption. It is noteworthy that Lipidure is used in many commercial products for EB and other 3D spheroid generation [50]. We also established the efficacy of each surface coating in a stem cell adhesiveness assay, where cell attachment was negligible in all samples but substantial in the positive control (vitronectin coated) (Fig. S2C).
Then, we evaluated the synergistic effect of geometries (i.e., LRC, HRC, HRF) and coating materials on EB formation. Devices in all geometries, coated with mPEG, Lipidure, BSA, and uncoated, were seeded with hESCs and cultured for three days for EB formation. As seen in Fig. 3A, both surface coating and device geometry influenced cell morphology and aggregation. To compare between groups, we quantified EB formation efficiency, which we defined as the percent of wells that formed a single aggregate with a clear border, a hallmark of EB development (Fig. 3B: Top).
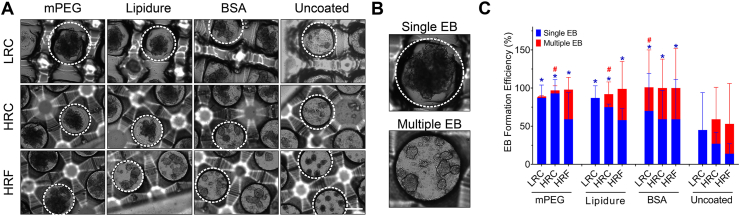
Fig. 3.
EB formation efficiency of the microwell devices. A) Representative bright-field images of EB formation in microwells on day 2, with respect to various surface coating materials and device geometries. The white line indicates a single microwell culture space. EBs were visible as the dark-phased cell aggregates. B) Examples of single and multiple EB formation. C) Quantification of single and multiple EB formation efficiency with respect to surface coatings and microwell geometries, including the uncoated groups. All surface coated groups formed more EBs than the uncoated ones. Within each group, curved devices (LRC and HRC) had higher efficiency than flat devices (HRF) in single EB formation. Statistical analysis: One-way ANOVA with Tukey's post-hoc analysis. Stars and pound signs indicate the significance of single and multiple EB formation, respectively, compared to uncoated devices. Scale bar (A, B): 500 μm. All data were presented as mean ± SD.
With respect to surface coating, EB formation efficiency was the highest and most consistent in mPEG-coated devices, followed by Lipidure, BSA, and then uncoated (Fig. 3C). In fact, EB formation efficiency in coated devices was significantly higher (p < 0.001) compared to uncoated devices, demonstrating the crucial role of a surface coating. Because of this result, we excluded uncoated surfaces from further experiments.
In regard to geometry, we observed a general trend that curved devices (LRC and HRC) had significantly higher EB formation efficiency than flat devices (HRF) (p < 0.05) (except for BSA coated devices). This trend was observed irrespective of surface coating or device resolution. Our observations are consistent with previous studies, which have found that curved microwell environments promote better cell aggregation in spheroids [25,41].
We also quantified multiple EB formation, where a single well formed two or more EBs (Fig. 3B: Bottom). Geometry-wise, flat wells generated significantly more multiple EBs than curved devices (LRC and HRC) (p < 0.001) (except for BSA coated devices). This observation is consistent with other studies [59]. With regard to coating, BSA coated wells produced significantly more multiple EBs than Lipidure or mPEG coated wells (p < 0.01). Although interesting, multiple EB formation is undesirable, as it increases variability in the generated EBs [59]. Hence, we excluded BSA coated microwells from further experiments.
The 3D micro-environment described here allowed us to control single EB formation through geometry and surface coating. As shown in previous studies [17], we observed that anti-fouling coating was necessary to achieve high and consistent EB formation. Besides, uncoated devices had less than half EB formation efficiency compared to the most efficient conditions (mPEG and Lipidure). Additionally, the prevalent formation of multiple EBs in BSA coated devices is likely due to the stability of BSA coating, which is vulnerable to culture conditions and is likely to crack into multiple small coated areas [54,55]. Furthermore, device curvature did not seem to salvage BSA coated devices, as BSA-coated HRC (curved) and HRF (flat) devices showed an insignificant difference in EB formation efficiency.
Similarly, no surface coating salvaged any flat-curvature condition, as HRF devices showed similar single-EB formation in all surface-coated groups. In summary, these results proved that the surface coating and geometry plays a vital role in initial EB formation in our platform. Among all coating materials, mPEG showed the highest percentage of single-EB formation. However, as there were no statistically significant differences between mPEG and Lipidure coated curved devices, both coatings were used in further studies.
3.3. The effect of micro-environment on EB and organoid morphology
The timeline of human cerebral organoid generation using our method is similar to that of a typical unguided one, including EB formation, EB growth, organoid formation, and organoid maturation (Fig. 4A). Development of brain organoids often consists of multiple stages, each involving different developmental milestones. For example, during EB formation by day 7, it is vital to generate cell aggregates of high sphericity that can be surrounded by a well-formed basement membrane. In contrast, during organoid development after day 7, it is desired to generate surface wrinkle/folding, which creates irregular non-spherical shapes. Hitherto, we explored the effect of geometry and surface coating on single-EB and multiple EB formation efficiency as a key milestone during EB formation (day 0–3). In previous studies of EB cultures, these microwell properties had a prolonged effect in regulating the stem cell proliferation and subsequent differentiation long after cells were transferred out of the microwell [33]. Herein, we explored whether such a prolonged effect existed in our organoid culture platform by evaluating the developmental milestones in each stage. Various microwell geometries and surface coatings were used to evaluate their long-term effect. The result was also compared to that of the control group (CON; never cultured within the microwell device), introduced in methods 2.7.
Fig. 4.
Characterization of EB and organoid formation. A) Timeline illustrated a multi-stage culturing process, including EB formation in microwells, EB growth in suspension, organoid formation, and organoid maturation. Major experiments and procedures were labeled above the timeline, along with the days. Major analyses and associated figures in this paper were labeled beneath the timeline. The cell culture medium condition was tagged in the yellow stripe. B) Representative bright-field images of EBs on day 7 of the control group (CON) and microwell groups (with respect to three geometries and two coating conditions). C) EB circularity on day 7. The mPEG-HRC was not significantly different from CON, while other microwell groups were significantly lower. D) EB diameter on day 7. All groups were significantly larger than CON group in diameter. E) Bright-field images of HRC-derived organoids from day 7 to day 20 exemplified the organoid growth and wrinkling/folding formation. The bottom right image illustrated how LG and LF were measured for the determination of the wrinkling index. F) Area size as a function of time in LRC, HRC, HRF, and CON. All showed an increasing trend, with all microwell groups significantly larger than CON. G) Circularity as a function of time in LRC, HRC, HRF, and CON. The circularity of all groups declined after day 7 or 12 at various speeds, with CON being the slowest and HRC being the fastest. The HRC group achieved the lowest circularity on day 20. H) Wrinkling index as a function of time in LRC, HRC, HRF, and CON. Wrinkling index of all microwell groups increased since day 7 at various speeds, with HRC group being the fastest and the highest. In contrast, CON group stayed un-wrinkled (wrinkle index ~ 1.0) until day 20, when the wrinkle index increased slightly. I) Immunocytochemistry of whole-mount organoids on day 20 revealed the overall cell organization for each microwell group and CON. Sox2, Tuj1, Actin, and Hoechst (Ho) were used to mark stem cells, differentiated neurons, cytoskeleton (neurites), and nuclei (all cell types). The location of wrinkling grooves was marked with white arrows. Note, all microwells used in E)-I) were coated with mPEG for maximum performance. Scale Bars (B, E): 500 μm; (I) 200 μm. (C, D) Every condition was statistically compared to CON group using Student's t-test ****P < 0.0001; ns, no significance. All data were presented as the mean ± SEM.
Day 7 is often considered to be the timing of mature EB formation. In traditional organoid generation methods, EBs are then embedded in Matrigel for neural differentiation and develop into organoids. The size and circularity of EBs are vital for this transition [1]. Hence, it is necessary to determine these characteristics of EBs on day 7. In our system, on day 7, as a whole, EBs generated in all tested conditions (all geometries; control group, mPEG, and Lipidure) showed characteristic EB morphology (i.e., round, with an optically clear smooth boundary) (Fig. 4B). Within each device geometry, EBs derived from mPEG coated devices had higher circularity than those derived from Lipidure coated devices (Fig. 4C). Additionally, EBs derived from high-resolution devices (HRC, HRF) showed a higher circularity than those from low-resolution devices, regardless of surface coating. Compared with the control group, most microwell devices (both geometry and coating) produced significantly reduced circularity (Fig. 4C) on day 7. Notably, however, the mPEG-coated HRC group produced EBs of a comparably high circularity, which was not significantly different from that of the control group.
Within all device groups, EB diameter showed an inverse relation to the circularity on day 7, i.e., the most circular groups were also the smallest in diameter (Fig. 4C&D). In addition, the device-generated EBs were all significantly larger (2–3 folds higher diameter) than the control group (Fig. 4D), suggesting that microwell devices promoted proliferation.
Although the LRC and HRC groups showed an insignificant difference in EB formation efficiency, the LRC group yielded a much lower circularity. This result can be attributed to the LRC device's anisotropic shape, as seen from the SEM images (Fig. 2C&D). The prolonged effect of this anisotropic point-of-contact surface might lead to anisotropic EB shape, thus leading to lower circularity. In addition, the mPEG derived EBs showed higher circularity than the Lipidure derived EBs; this is likely because mPEG coating, as a self-assembled monolayer, forms a permanent, single-molecular smooth and compliant surface (Fig. S2B) [60,61]. This surface can facilitate higher cell aggregation and prolonged anti-fouling effect. In contrast, EBs derived from Lipidure coated surface showed lower circularity and less desirable morphology. Hence, we only selected mPEG generated EBs for differentiation and maturation analysis due to better resulting EB morphology.
We further analyzed the organoid formation phase from day 7 to day 20 by monitoring the variation in area size, wrinkling index, and circularity (Fig. 4E–H). The area size of the device-derived organoids was larger than the control group (Fig. 4F). Notably, the area size of HRC derived organoids was the closest to the control group on day 20. This was our first implication of differences between the long-term effects of the microwells on organoid formation.
Furthermore, beginning on day 12, we observed the surface wrinkling or folding in organoids derived from mPEG coated devices. Wrinkling or folding emerges in the neural tube, which defines different regions of the brain [5,11] and promotes the expansion of the three-dimensional brain surface area [[62], [63], [64]] in a confined space. Early-stage organoid wrinkling was previously found in a 2D model to be remarkably analogous to cortical gyrification in folding morphology and temporal dynamics [11]. To characterize the folding extent, the wrinkling index was measured as the contour length LG normalized by the outer convex contour length LF (Fig. 4E&H). A wrinkling index larger than 1 indicates the formation of wrinkling. In microwell-derived organoids, the wrinkling index became larger than 1 on day 12 and continued to increase after that (Fig. 4E&H). In contrast, wrinkling did not emerge in the control group until day 20. In addition, wrinkling increased at a faster rate in microwell derived organoids, as compared to the control group, with surface wrinkling being highly expressed in the HRC generated group. Among the microwell generated organoids, the wrinkling dynamics started to plateau on day 20 at 1.24, 1.69, and 1.20 for LRC, HRC, and HRF, respectively.
The emergence of wrinkling created an irregular-shaped organoid edge, which intuitively decreased the circularity. In our system, the LRC and HRF group's circularity exhibited a peak on day 12 and then decreased. In contrast, the control and HRC groups' circularity consistently showed a decreasing trend since day 7 (Fig. 4G), in which the circularity of the HRC derived organoids was lower compared to the control group. On day 20, the HRC group had the lowest circularity and highest wrinkling index compared to all other groups (Fig. 4H). The circularity in the LRC, HRF, and control groups was close, but the LRC and HRF still behaved better in wrinkling formation than the control group. This variation of circularity can be attributed to surface folding or winkling formation. Folding increased the 3D surface area size of the organoids, however, due to folding on the surface, the circularity decreased. Previous studies, using ECM embedding or confining EBs in laminated devices, demonstrated that wrinkling formation is induced and modulated by the mechanical micro-environment of the outer layer cells [11,65,66]. We believe our microwell device played a similar role by providing spatial confinement to the growing EBs. In a novel way, our results showed that these effects were long-term, as the HRC-derived organoids formed higher amounts of folding. The HRC's strong confinement effect was evidenced by its superior EB circularity on day 7, as we described in previous sections, which can be correlated to the increased folding during organoid formation. From our results, microwell devices promoted organoid early-stage growth and wrinkling/folding formation, which were tunable through the 3D geometry of the well.
Cell differentiation status and organization are also vital for the proper development of organoids [67]. We used whole-mount staining to assess the 3D morphology and cell distribution in our early-stage human cerebral organoids. Stemness marker Sox2, β-Tubulin III marker Tuj1, and cytoskeletal marker actin were used to label stem cells, neurons, and actin filaments, respectively (Fig. 4I). Sox2 signals were preferentially located in the center of organoids in microwell-cultured organoids, especially those in the high resolution printed microwell devices (HRF and HRC). This result suggested the formation of well-organized stem cell structures, such as lumens. In contrast, the distribution of Sox2 was random and not well organized for lumen formation for the control group. The expression of Tuj1 was observed in all microwell-devices generated organoids, which indicated proper neuronal differentiation. In the control group, the signal of Tuj1 was discontinuous and scattered. Additionally, more enriched neuronal axons (actin-labeled) were observed on the surface of HRC- and HRF-generated organoids, which suggested enhanced neuronal differentiation and the development of neuronal layers. These primary characteristics indicated that microwell devices, especially those of higher resolution, promoted the self-organization of human cerebral organoids. In contrast, we rarely observed a well-defined cell organization in the control group.
With the whole mount staining of the HRC group, we observed an interesting wrinkling pattern of several long and deep grooves (exemplified in Figs. 4I and S3B white arrows). These grooves were parallel to one other and equally spaced with a periodicity of 30–40 μm on day 20 (Fig. S3B). Their position and spacing matched the microtopography trace found in HRC microwell-devices (spaced by 10–26 μm) (Fig. S3A), suggesting the grooves grew from the imprint of the microwells in proportion to the entire organoid growth. On day 3, the average diameter of transferred microwell-derived EBs was 430 μm. As the organoids grew from day 20 to day 45, the microtopography trace was also increased from 30 μm to 60 μm (Fig. S3C). This fold change is consistent with the fold change of cell diameter, which was ~1018 μm on day 20 and ~1950 μm on day 45. Detailed monitoring of the mechanisms of this imprinting is currently under investigation for our follow up study.
3.4. Lumen structure and matured neuronal layer formation of the human cerebral organoids
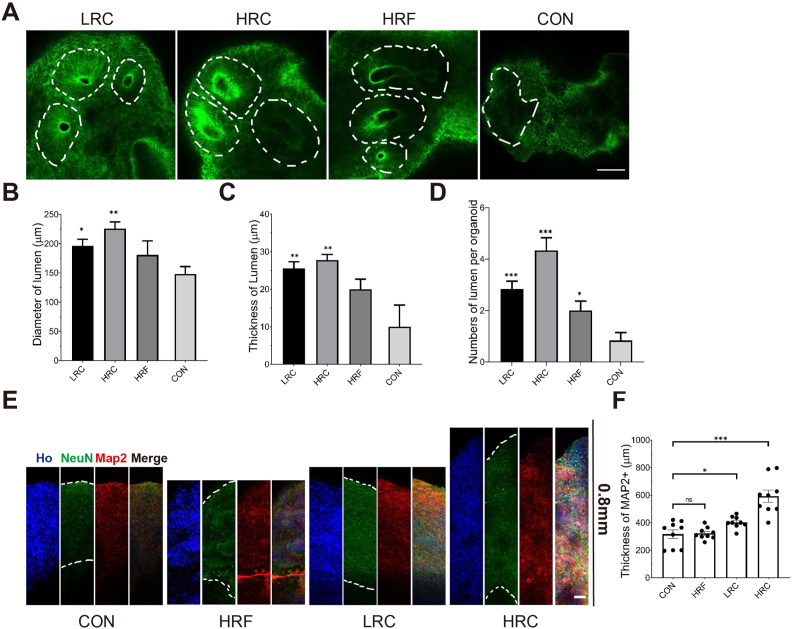
We performed cryogenic sectioning on organoids and further characterized the lumen formation and neuronal maturation as two key features for the early development of the human cerebral organoids. Following neural induction, fluid-filled lumens appear within the organoids [68]. These ventricle-like lumen formation is a hallmark of healthy developed organoids in early stages [69,70]. Lumens supply Sox2+ cells that differentiate and migrate to support neurogenesis, ventricle expansion, and cortical development [68,[71], [72], [73]]. Thus, lumen properties (size, thickness, and number) may be critical to the organoids' subsequent cortical layer development [74].
In our system, on day 20, multiple ventricle-like lumens were formed in all device-derived organoids (Fig. 5A). These lumens exhibited a healthy, spherical shape and radiating cell arrangement, similar to those generated through Matrigel-embedded methods [11]. In contrast, the control group formed none or very few, ill-shaped lumens. Quantification of lumen size, thickness, and number per organoid confirmed these qualitative observations (Fig. 5B–D). In all measures, the two curved groups were significantly better than the control. Among all device groups, HRC was once again the best choice in promoting lumen formation, producing significantly more, larger, and thicker lumens than the LRC and HRF groups (Fig. 5B–D). Taken together, our results suggest that both curvature (perhaps more significantly) and the resolution of 3D printed microwells played an important role in promoting lumen formation. In addition, the variance of lumen characteristics for each device group was small: only 5–10% of the average values. This variance was significantly lower compared to Matrigel-embedded traditional methods (which we estimate to be 30–60% based on our previous culturing experience). Thus, our method presents a robust biomanufacturing strategy to generate consistent lumen models.
Fig. 5.
Characterization of early-stage maturation of organoids. A) Immunocytochemistry of actin in LRC, HRC, HRF and CON revealed detailed lumen structures and cell organization, as outlined by white dashed lines. B) The diameter of lumens on day 20 was significantly larger in LRC and HRC group than CON, with the HRC group having the largest lumens. C) The thickness of lumens on day 20 shared a similar trend with the lumen diameter. D) The number of lumens on day 20 was significantly larger in all microwell groups than CON, while the HRC group generated the highest lumen numbers. E) Immunocytochemistry of organoid cryogenic sections on day 45 revealed neuronal cell layers, as labeled by mature neuron related markers NeuN and Map2. F) The thickness of Map2 positive cells in CON, HRF, LRC and HRC averaged over at least 3 organoids. The thickness of the mature layer was significantly increased in LRC and HRC group. The HRC group has the maximum average value of the thickness of mature neurons. The data are presented as the mean ± SEM. Every microwell condition was compared to control using Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance. Scale bars (A: 100 μm, E: 50 μm). Note, all data were acquired with at least 3 organoids for each condition.
In human cerebral organoids, the lumen is the source of undifferentiated stem cells, which is critical for cortical development. These stem cells differentiate batch by batch into neurons, migrate to the organoid edge, and form matured layers [71]. Thus, the success of the following corticogenesis in the organoids depends on the quality of lumen structures and mature neuronal layer. To evaluate these structures, we evaluated organoid maturation through immunocytochemistry on day 45 (Fig. 5E). Mature neurons and dendrites were labeled using the neuronal nuclei marker NeuN and microtubule-associated protein 2 marker Map2, respectively. The neuronal dendrites were enriched and continuous in the HRC group, in contrast to the thin and non-homogenously scattered organization in the control and HRF groups (Fig. 5E). The LRC group showed continuous organization, slightly thicker than the control and HRF groups (Fig. 5E). The intensity of NeuN was also much higher in the HRC group than other conditions, taken together, indicating that the HRC microwell-devices promoted the layer maturation.
The subsequent thickness quantification of the matured neuronal layer based on Map2 staining found a device-dependent trend similar to that of the lumen characteristics. Specifically, both bottom-curved device groups yielded a significantly thicker neuronal layer than the bottom-flat device group and non-device control. The HRC group yielded the thickest layer (592.88 μm), doubling that of the control (Fig. 5E&F). Taken together, our results suggest that both curvature (much more significantly) and the resolution of 3D printed microwells played an essential role in promoting cell differentiation and maturation. Because lumen formation and maturation followed similar trends, our results imply that these processes are related, and perhaps synergistic. We plan to investigate this connection in future work thoroughly.
Our study showed that microwell devices, especially with high-resolution concave surfaces, were able to facilitate the process of organoid development, from EB formation and wrinkling/folding to lumen formation and neuronal layer maturation. Such a process demands a series of micro-environment modulations, which is usually achieved by Matrigel-embedding in traditional unguided methods. For instance, in previous reports, lumens and other key organoid features were not observed until ECM embedding [11]. However, the side effects of Matrigel and other ECM materials should not be neglected, as we discussed in the introduction. Strikingly, our microwell platform produced results similar to or better than the Matrigel-embedded methods, suggesting a viable substitute for Matrigel embedding. This is likely attributed to our novel microwell designs combining several features that may modulate the early-stage micro-environment. These features were individually found to promote 3D culture systems in previous studies. Specifically, the provided concave spherical curvature and high spatial confinement were found to promote 3D cell aggregation in spheroids [25,30,38], very similar to the vital influence of the skull in the brain development [[75], [76], [77], [78]]. In EB formation, these two features were further shown to increase the cell packing density [37], form tighter junctions, and modulate ECM networks [36,37]. Previous studies also established that microwell device's 3D topography and surface coating could affect cell behaviors (e.g., proliferation, differentiation) [17,41,79,80] and tissue morphogenesis (e.g., surface wrinkling) [10,81]. Furthermore, the provided deep and highly-networked (as discussed in section 3.1) microwells were previously found to enhance cell aggregation, nutrient exchange, and physiological functions of spheroids [21,38].
EBs in our devices were only cultured for 3 days before transferring to culture flasks but still showed that the effect of the micro-environmental modulation went far beyond the treatment period (until day 45, the longest time we explored). Unlike a real skull that grows along with the brain, our devices are fixed in size and shape. Thus, they could only hold the culture for up to three days before nutrient/waste exchange became challenging. Yet, the formed human cerebral organoids showed superb development in all tested characteristics, equivalent to or better than the Matrigel-embedding method. Although cultured in microwells for only 3 days, these organoids demonstrated superb maturation and quality in the long term. This prolonged effect was likely attributed to the three-germ layer formation pre-defined during the EB formation. The three-germ layer formation, especially ectoderm layer's promotion, is essential to organoid development [82]. This spherical confinement was shown to lead to consistent EBs with distinct germ layers and enriched SOX2 (ectoderm marker) [11,83]. Besides, the higher cell packing density in the microwell derived EBs (like the effect of the skull and meninges [84]) intensifies the cell-cell tension that drives the folding process [11].
Previous work from other researchers used microwells for ECM-free EB formation [2]; the resulting spheroids achieved putative organoid features (e.g., lumen), but failed to develop other vital features (e.g., wrinkling/folding, neuronal layer). In contrast, the human cerebral organoids developed in our study have exhibited a much more comprehensive range of features, including wrinkling/folding and neuronal layer besides lumen. Another advantage of our approach is that it is unguided and Matrigel-free. We attribute this substantial advancement of our platform to the likely synergistic effect of our microwell designs that innovatively integrated high curvature, networked-space, and deep pocket, as well as treatment time. Yoon et al. allowed cell aggregation in the microwell device for only 18 h [2]. This treatment time was only ~25% of that in our study, possibly insufficient in thoroughly modulating the micro-environment during EB formation. Additionally, we have shown that the curvature providing compliant contact was critical for the EB formation and had long-term effects on organoid development. In that sense, the reverse-pyramid shape used in their study was likely to further reduce the modulation ability of the device. Growing long-term brain organoids is time- and supply-consuming, with a high risk of necrosis. In our microwells, we observed better-organized development and more mature neuronal layers, as compared to the organoids at the same age in other methods. In other words, the development is faster with our system, which could be a more efficient model for the late-stage development: superficial layer and whole-tissue level maturation.
4. Conclusion
We demonstrated, for the first time, a 3D printed microwell platform to generate human cerebral organoids with mature physiological features, without the use of Matrigel/ECM embedding or external signaling molecules (i.e., SMAD and Wnt inhibitors). Our results confirmed that surface coating influences the EB generation efficiency, in which, we found mPEG is better for PDMS microwell coatings, compared to Lipidure or BSA. We also showed that microwell device geometry is vital for EB formation efficiency: curved geometry is better for organoid generation than flat geometry. In addition, we found that the 3D printing modality affected the fidelity of the microwell system, which led to isotropic (HRC) and anisotropic (LRC) curved geometry. Higher-fidelity HRC devices led to better organoid formation and maturation, compared to the lower-fidelity, anisotropic LRC devices. Furthermore, we showed that the device geometry and microtopography had a long-term impact on the organoid growth, even after cells had been removed from the devices for over two months. Even without ECM-embedding or chemical guidance, we still observed high-level structures like wrinkling/folding and lumen. This is a significant step forward, helping to expand our knowledge of organoid generation methods. Specifically, the micro-environment modulated by high-resolution printed, bottom-curved devices resulted in the highest lumen formation and the thickest layers of mature neurons, which modeled early organoid development effectively in an ECM-free condition.
CRediT authorship contribution statement
Cheng Chen: Methodology, Validation, Investigation, Writing - original draft, Writing - review & editing. Venkatakrishnan Rengarajan: Formal analysis, Validation, Investigation, Writing - original draft, Writing - review & editing. Andrew Kjar: Formal analysis, Validation, Investigation, Writing - original draft, Writing - review & editing. Yu Huang: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Resources, Supervision.
Declaration of competing interest
None.
Acknowledgments
We thank Hemdeep Patel from the ResinWorks3D company for providing us the high-resolution 3D printed mold devices as a gift. We also thank Angela Clyde for the initial discussion on device design. We thank Utah State University's College of Engineering Undergraduate Research Program (EURP) for supporting Andrew Kjar. We also thank Utah State University Research Catalyst (RC) program for supporting Cheng Chen.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.10.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon S.-J., Elahi L.S., Pașca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., Fan H.C. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sloan S.A., Andersen J., Pașca A.M., Birey F., Pașca S.P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 2018;13:2062–2085. doi: 10.1038/s41596-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krieger, T.G.; Tirier, S.M.; Park, J.; Jechow, K.; Eisemann, T.; Peterziel, H.; Angel, P.; Eils, R.; Conrad, C. Modeling glioblastoma invasion using human brain organoids and single-cell transcriptomics. Neuro Oncol., doi:10.1093/neuonc/noaa091. [DOI] [PMC free article] [PubMed]
- 5.Tallinen T., Chung J.Y., Rousseau F., Girard N., Lefèvre J., Mahadevan L. On the growth and form of cortical convolutions. Nat. Phys. 2016;12:588–593. doi: 10.1038/nphys3632. [DOI] [Google Scholar]
- 6.Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Yang S.M., Berger D., Maria N., Scholvin J., Goldman M., Kinney J. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paşca S.P. Assembling human brain organoids. Science. 2019;363:126–127. doi: 10.1126/science.aau5729. [DOI] [PubMed] [Google Scholar]
- 9.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G., Jacob F., Zhong C. Brain region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Muffat J., Omer A., Bosch I., Lancaster M.A., Sur M., Gehrke L., Knoblich J.A., Jaenisch R. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20:385–396. doi: 10.1016/j.stem.2016.11.017. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karzbrun E., Kshirsagar A., Cohen S.R., Hanna J.H., Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y., Eglen R.M. Three-dimensional cell cultures in drug discovery and development. SLAS discov. Adv. Sci. Drug Discov. 2017;22:456–472. doi: 10.1177/1087057117696795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nawy T. Move over. Matrigel. Nat. Methods. 2017;14 doi: 10.1038/nmeth.4138. 10–10. [DOI] [Google Scholar]
- 14.Mansour A.A., Gonçalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tieng V., Stoppini L., Villy S., Fathi M., Dubois-Dauphin M., Krause K.-H. Engineering of midbrain organoids containing long-lived dopaminergic neurons. Stem Cell. Dev. 2014;23:1535–1547. doi: 10.1089/scd.2013.0442. [DOI] [PubMed] [Google Scholar]
- 16.Jabaji Z., Brinkley G.J., Khalil H.A., Sears C.M., Lei N.Y., Lewis M., Stelzner M., Martín M.G., Dunn J.C.Y. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivitilli A.A., Gosio J.T., Ghoshal B., Evstratova A., Trcka D., Ghiasi P., Hernandez J.J., Beaulieu J.M., Wrana J.L., Attisano L. Robust production of uniform human cerebral organoids from pluripotent stem cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., Ming G. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ao Z., Cai H., Havert D.J., Wu Z., Gong Z., Beggs J.M., Mackie K., Guo F. One-stop microfluidic assembly of human brain organoids to model prenatal cannabis exposure. Anal. Chem. 2020;92:4630–4638. doi: 10.1021/acs.analchem.0c00205. [DOI] [PubMed] [Google Scholar]
- 20.Pedro R., Marc I., Gaurav Singh R., Patrick A.-K., Oliver Knights M., Guido B., Julie L., Kristoffer Lihme E., Dylan M.R., Malin P. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee G.H., Lee J.S., Lee G.-H., Joung W.Y., Kim S.H., Lee S.H., Park J.Y., Kim D.-H. Networked concave microwell arrays for constructing 3D cell spheroids. Biofabrication. 2017;10 doi: 10.1088/1758-5090/aa9876. [DOI] [PubMed] [Google Scholar]
- 22.Patra B., Chen Y.-H., Peng C.-C., Lin S.-C., Lee C.-H., Tung Y.-C. A microfluidic device for uniform-sized cell spheroids formation, culture, harvesting and flow cytometry analysis. Biomicrofluidics. 2013;7 doi: 10.1063/1.4824480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeon S.-E., No D.Y., Lee S.-H., Nam S.W., Oh I.-H., Lee J., Kuh H.-J. Application of concave microwells to pancreatic tumor spheroids enabling anticancer drug evaluation in a clinically relevant drug resistance model. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hribar K.C., Chen S., Finlay D., Vuori K., Xuanyi M. 2019. Method for Fabrication of Microwells for Controlled Formation of 3-dimensional Multicellular-Shapes. [Google Scholar]
- 25.Kim K., Kim S.-H., Lee G.-H., Park J.Y. Fabrication of omega-shaped microwell arrays for a spheroid culture platform using pins of a commercial CPU to minimize cell loss and crosstalk. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aad7d3. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y., Wang L., Yu H., Yin F., Wang Y., Liu H., Jiang L., Qin J. In situ generation of human brain organoids on a micropillar array. Lab Chip. 2017;17:2941–2950. doi: 10.1039/C7LC00682A. [DOI] [PubMed] [Google Scholar]
- 27.Moeller H.-C., Mian M., Shrivastava S., Chung B.G., Khademhosseini A. A microwell array system for stem cell culture. Biomaterials. 2008;29:752–763. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr J.C., de Pablo J.J., Palecek S.P. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen D., Sa S., Pegan J.D., Rich B., Xiang G., McCloskey K.E., Manilay J.O., Khine M. Tunable shrink-induced honeycomb microwell arrays for uniform embryoid bodies. Lab Chip. 2009;9:3338–3344. doi: 10.1039/B914091C. [DOI] [PubMed] [Google Scholar]
- 30.Pettinato G., Wen X., Zhang N. formation of well-defined embryoid bodies from dissociated human induced pluripotent stem cells using microfabricated cell-repellent microwell Arrays. Sci. Rep. 2014;4:7402. doi: 10.1038/srep07402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi Y.Y., Chung B.G., Lee D.H., Khademhosseini A., Kim J.-H., Lee S.-H. Controlled-size embryoid body formation in concave microwell arrays. Biomaterials. 2010;31:4296–4303. doi: 10.1016/j.biomaterials.2010.01.115. [DOI] [PubMed] [Google Scholar]
- 32.Karp J.M., Yeh J., Eng G., Fukuda J., Blumling J., Suh K.-Y., Cheng J., Mahdavi A., Borenstein J., Langer R. Controlling size, shape and homogeneity of embryoid bodies using poly(ethylene glycol) microwells. Lab Chip. 2007;7:786–794. doi: 10.1039/B705085M. [DOI] [PubMed] [Google Scholar]
- 33.Hwang Y.-S., Chung B.G., Ortmann D., Hattori N., Moeller H.-C., Khademhosseini A. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y., Wang L., Yin F., Yu Y., Wang Y., Liu H., Wang H., Sun N., Liu H., Qin J. A hollow fiber system for simple generation of human brain organoids. Integr. Biol. Quant. Biosci. Nano Macro. 2017;9:774–781. doi: 10.1039/c7ib00080d. [DOI] [PubMed] [Google Scholar]
- 35.Jeong G.S., Song J.H., Kang A.R., Jun Y., Kim J.H., Chang J.Y., Lee S.-H. Surface tension-mediated, concave-microwell Arrays for large-scale, simultaneous production of homogeneously sized embryoid bodies. Adv. Healthc. Mater. 2013;2:119–125. doi: 10.1002/adhm.201200070. [DOI] [PubMed] [Google Scholar]
- 36.Huang C.-K., Paylaga G.J., Bupphathong S., Lin K.-H. Spherical microwell arrays for studying single cells and microtissues in 3D confinement. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab6eda. [DOI] [PubMed] [Google Scholar]
- 37.Jeong G.S., Jun Y., Song J.H., Shin S.H., Lee S.-H. Meniscus induced self organization of multiple deep concave wells in a microchannel for embryoid bodies generation. Lab Chip. 2012;12:159–166. doi: 10.1039/c1lc20619b. [DOI] [PubMed] [Google Scholar]
- 38.Lee G., Jun Y., Jang H., Yoon J., Lee J., Hong M., Chung S., Kim D.-H., Lee S. Enhanced oxygen permeability in membrane-bottomed concave microwells for the formation of pancreatic islet spheroids. Acta Biomater. 2018;65:185–196. doi: 10.1016/j.actbio.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 39.Sharma V., Dhayal M., Shivaprasad S., Jain S. Surface characterization of plasma-treated and PEG-grafted PDMS for micro fluidic applications. Vacuum. 2007;81:1094–1100. [Google Scholar]
- 40.Chen C., Dong X., Fang K.-H., Yuan F., Hu Y., Xu M., Huang Y., Zhang X., Fang D., Liu Y. Develop a 3D neurological disease model of human cortical glutamatergic neurons using micropillar-based scaffolds. Acta Pharm. Sin. B. 2019;9:557–564. doi: 10.1016/j.apsb.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong S.F., No D.Y., Choi Y.Y., Kim D.S., Chung B.G., Lee S.-H. Concave microwell based size-controllable hepatosphere as a three-dimensional liver tissue model. Biomaterials. 2011;32:8087–8096. doi: 10.1016/j.biomaterials.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 42.Kjar A., Huang Y. Application of micro-scale 3D printing in pharmaceutics. Pharmaceutics. 2019;11:390. doi: 10.3390/pharmaceutics11080390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ligon S.C., Liska R., Stampfl J., Gurr M., Mülhaupt R. Polymers for 3D printing and customized additive manufacturing. Chem. Rev. 2017;117:10212–10290. doi: 10.1021/acs.chemrev.7b00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magnien J., Cosemans P., Nutal N., Kairet T. Current surface issues in additive manufacturing. Plasma Process. Polym. 2020;17:1900154. [Google Scholar]
- 45.Maurya N.K., Rastogi V., Singh P. Comparative study and measurement of form errors for the component printed by FDM and PolyJet process. J. Homepage Httpiieta Orgjournalsi2m. 2019;18:353–359. [Google Scholar]
- 46.Derby B. Inkjet printing of functional and structural materials: fluid property requirements, feature stability, and resolution. Annu. Rev. Mater. Res. 2010;40:395–414. [Google Scholar]
- 47.Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Limraksasin P., Okawa H., Zhang M., Kondo T., Osathanon T., Pavasant P., Egusa H. Size-optimized microspace culture facilitates differentiation of mouse induced pluripotent stem cells into osteoid-rich bone constructs. Stem Cell. Int. 2020:2020. doi: 10.1155/2020/7082679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan D.-H., Yuan S.-W., Shen Y.-M. Surface modification with BSA blocking based on in situ synthesized gold nanoparticles in poly(dimethylsiloxane) microchip. Colloids Surf. B Biointerfaces. 2010;75:608–611. doi: 10.1016/j.colsurfb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 50.Plate, R.-C.; Dish, R.-C. LIPIDURE® COAT PLATE A-F96 (Flat Bottom).
- 51.Gökaltun A., Kang Y.B.A., Yarmush M.L., Usta O.B., Asatekin A. Simple surface modification of poly (dimethylsiloxane) via surface segregating smart polymers for biomicrofluidics. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-43625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chumbimuni-Torres K.Y., Coronado R.E., Mfuh A.M., Castro-Guerrero C., Silva M.F., Negrete G.R., Bizios R., Garcia C.D. Adsorption of proteins to thin-films of PDMS and its effect on the adhesion of human endothelial cells. RSC Adv. 2011;1:706–714. doi: 10.1039/C1RA00198A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boxshall K., Wu M.-H., Cui Z., Cui Z., Watts J.F., Baker M.A. Simple surface treatments to modify protein adsorption and cell attachment properties within a poly(dimethylsiloxane) micro-bioreactor. Surf. Interface Anal. 2006;38:198–201. doi: 10.1002/sia.2274. [DOI] [Google Scholar]
- 54.Park J.-W., Kurosawa S., Aizawa H., Wakida S., Yamada S., Ishihara K. Comparison of stabilizing effect of stabilizers for immobilized antibodies on QCM immunosensors. Sensor. Actuator. B Chem. 2003;91:158–162. [Google Scholar]
- 55.Ma G.J., Ferhan A.R., Jackman J.A., Cho N.-J. Quantitative assessment of bovine serum albumin proteins for blocking applications. bioRxiv. 2019:869677. doi: 10.1101/869677. [DOI] [Google Scholar]
- 56.Sarvi F., Yue Z., Hourigan K., Thompson M.C., Chan P.P.Y. Surface-functionalization of PDMS for potential micro-bioreactor and embryonic stem cell culture applications. J. Mater. Chem. B. 2013;1:987–996. doi: 10.1039/C2TB00019A. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H., Chiao M. Anti-fouling coatings of poly (dimethylsiloxane) devices for biological and biomedical applications. J. Med. Biol. Eng. 2015;35:143–155. doi: 10.1007/s40846-015-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S., Li L., Zhao C., Zheng J. Surface hydration: principles and applications toward low-fouling/nonfouling biomaterials. Polymer. 2010;51:5283–5293. [Google Scholar]
- 59.Hribar K.C., Finlay D., Ma X., Qu X., Ondeck M.G., Chung P.H., Zanella F., Engler A.J., Sheikh F., Vuori K. Nonlinear 3D projection printing of concave hydrogel microstructures for long-term multicellular spheroid and embryoid body culture. Lab Chip. 2015;15:2412–2418. doi: 10.1039/c5lc00159e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plegue T.J., Kovach K.M., Thompson A.J., Potkay J.A. Stability of polyethylene glycol and zwitterionic surface modifications in PDMS microfluidic flow chambers. Langmuir. 2018;34:492–502. doi: 10.1021/acs.langmuir.7b03095. [DOI] [PubMed] [Google Scholar]
- 61.Kovach K.M., Capadona J.R., Gupta A.S., Potkay J.A. The effects of PEG‐based surface modification of PDMS microchannels on long‐term hemocompatibility. J. Biomed. Mater. Res. A. 2014;102:4195–4205. doi: 10.1002/jbm.a.35090. [DOI] [PubMed] [Google Scholar]
- 62.Sun T., Hevner R.F. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Florio M., Huttner W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 64.Mota B., Herculano-Houzel S. Cortical folding scales universally with surface area and thickness, not number of neurons. Science. 2015;349:74–77. doi: 10.1126/science.aaa9101. [DOI] [PubMed] [Google Scholar]
- 65.Karzbrun E., Tshuva R.Y., Reiner O. An on-chip method for long-term growth and real-time imaging of brain organoids. Curr. Protoc. Cell Biol. 2018;81:e62. doi: 10.1002/cpcb.62. [DOI] [PubMed] [Google Scholar]
- 66.Karzbrun E., Reiner O. Brain organoids—a bottom-up approach for studying human neurodevelopment. Bioengineering. 2019;6 doi: 10.3390/bioengineering6010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cimadamore F., Amador-Arjona A., Chen C., Huang C.-T., Terskikh A.V. SOX2–LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossant J., Tam P.P.L. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 70.Shao Y., Sang J., Fu J. On human pluripotent stem cell control: the rise of 3D bioengineering and mechanobiology. Biomaterials. 2015;52:26–43. doi: 10.1016/j.biomaterials.2015.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryan A.Q., Chan C.J., Graner F., Hiiragi T. Lumen expansion facilitates epiblast-primitive endoderm fate specification during mouse blastocyst formation. Dev. Cell. 2019;51:684–697. doi: 10.1016/j.devcel.2019.10.011. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Compagnucci C., Petrini S., Higuraschi N., Trivisano M., Specchio N., Hirose S., Bertini E., Terracciano A. Characterizing PCDH19 in human induced pluripotent stem cells (iPSCs) and iPSC-derived developing neurons: emerging role of a protein involved in controlling polarity during neurogenesis. Oncotarget. 2015;6:26804. doi: 10.18632/oncotarget.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lv X., Ren S.-Q., Zhang X.-J., Shen Z., Ghosh T., Xianyu A., Gao P., Li Z., Lin S., Yu Y. TBR2 coordinates neurogenesis expansion and precise microcircuit organization via Protocadherin 19 in the mammalian cortex. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-019-11854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Noctor S.C., Martínez-Cerdeño V., Kriegstein A.R. Contribution of intermediate progenitor cells to cortical histogenesis. Arch. Neurol. 2007;64:639–642. doi: 10.1001/archneur.64.5.639. [DOI] [PubMed] [Google Scholar]
- 75.Kelava I., Lancaster M.A. Stem cell models of human brain development. Cell Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 76.Zilles K., Palomero-Gallagher N., Amunts K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 77.Baer M.J. Patterns of growth of the skull as revealed by vital staining. Hum. Biol. 1954;26:80–126. [PubMed] [Google Scholar]
- 78.Welker W. Cerebral Cortex. Springer; 1990. Why does cerebral cortex fissure and fold? pp. 3–136. [Google Scholar]
- 79.Jäger M., Zilkens C., Zanger K., Krauspe R. Significance of nano- and microtopography for cell-surface interactions in orthopaedic implants. J. Biomed. Biotechnol. 2007;2007 doi: 10.1155/2007/69036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shin H.-S., Hong H.J., Koh W.-G., Lim J.-Y. Organotypic 3D culture in nanoscaffold microwells supports salivary gland stem-cell-based organization. ACS Biomater. Sci. Eng. 2018;4:4311–4320. doi: 10.1021/acsbiomaterials.8b00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schulte C., Lamanna J., Moro A.S., Piazzoni C., Borghi F., Chighizola M., Ortoleva S., Racchetti G., Lenardi C., Podestà A. Neuronal cells confinement by micropatterned cluster-assembled dots with mechanotransductive nanotopography. ACS Biomater. Sci. Eng. 2018;4:4062–4075. doi: 10.1021/acsbiomaterials.8b00916. [DOI] [PubMed] [Google Scholar]
- 82.Metzger J.J., Simunovic M., Brivanlou A.H. Synthetic embryology: controlling geometry to model early mammalian development. Curr. Opin. Genet. Dev. 2018;52:86–91. doi: 10.1016/j.gde.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tallinen T., Chung J.Y., Biggins J.S., Mahadevan L. Gyrification from constrained cortical expansion. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:12667–12672. doi: 10.1073/pnas.1406015111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee C.-T., Bendriem R.M., Wu W.W., Shen R.-F. 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017;24:59. doi: 10.1186/s12929-017-0362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.