ABSTRACT
Small interfering RNA (siRNA) is a critical loss-of-function tool for elucidating the role of genes in biomedical studies. The effective use of siRNA needs transfection technology that delivers siRNA into the correct location of target cells, especially those which are extremely difficult to transfect. Macrophages, which play an important role in the pathogenesis of many diseases, are known to be extremely hard to transfect. Thus, to elucidate the functions of genes in human macrophage biology, it is essential to devise technology for efficient siRNA transfection. However, a fast and efficient method for siRNA transfection in primary human macrophages has not been reported. The siRNA transfection is a tug-of-war between transfection rate and cytotoxicity. A higher transfection rate is generally accompanied with increased cytotoxicity, therefore, choosing a transfection reagent that limits cell death while maintain a desirable transfection rate is important. In this study, we employed auto-analysis function of the IncuCyte® to devise a fast and cost-saving technology for efficient transfection of adherent cells and particularly human macrophages. We show that DharmaFECT3 transfection reagent from Dharmacon was the most efficient in transfecting primary human monocyte-derived macrophages and PMA-differentiated U937 cells, whereas other transfection reagents tested were cytotoxic. This method exhibited approximately 85% transfection efficiency in human macrophages. Moreover, siRNA silencing of Bax with this technique effectively protected primary human macrophages and PMA-differentiated U937 cells against Resveratrol-induced cell death. In addition, this method inherently takes the balance between transfection rate and cytotoxicity of siRNA transfection reagents into consideration.
KEYWORDS: siRNA transfection, primary human macrophages, Bax, Resveratrol, apoptosis
Introduction
Macrophages, the multifunctional phagocytes, play crucial roles in many physiological activities, such as immune regulation, cleaning up exogenous antigens, clearance of endogenous cell debris, tissue repair, regeneration, and fibrosis [1–4]. In addition, macrophages are long-lived cells distributed all over the human body with a lifespan of months to years [5,6]. Thus, these cells play critical roles in innate immunity and in the pathogeneses of several chronic and inflammatory diseases through the induction or resolution of inflammation and tissue repair [7]. Recent evidence obtained by way of microarray analysis and conventional approaches of in vivo studies has suggested the involvement of M1 proinflammatory macrophages in gastrointestinal typhoid fever, tuberculoid leprosy and active tuberculosis. Anti–inflammatory M2 macrophages, however, are associated with lepromatous leprosy and chronic rhinosinusitis [7–13]. In addition, it has been shown that macrophages can be altered by the tumour microenvironment and can promote solid tumour progression and metastasis [14–17]. Moreover, macrophages have been shown to play a major role in HIV reservoir formation [18]. Targeting macrophages as a potential therapeutic strategy via induced apoptosis has been suggested, such as downregulating pro-inflammatory pathways and targeting tumour-associated or HIV-infected macrophages [16,19–21]. Small interfering RNA (siRNA) silencing has been identified as a critical tool in inducing apoptosis in infected or neoplastic cells [22,23], but effective use of siRNA needs transfection technology to deliver siRNA into the correct subcellular location of target cells [24].
Primary human macrophages are well known to be extremely hard to transfect [25–27], primarily due to their recognition of foreign nucleic acids, and their initiation of immune responses to exogenous siRNA molecules [26,27]. Moreover, in some infections, such as HIV, the infection rate of human macrophages in vitro or in vivo is usually low [28–31]. As a result, ambiguous results may be obtained if the transfection rate is also very low when studying specific killing of HIV-infected macrophages by siRNA silencing. These challenges may be overcome by optimizing the transfection technology for primary human macrophages. Transfection is a tug-of-war between transfection rate and cytotoxicity, however, a higher transfection rate is generally accompanied with increased cytotoxicity, which may non-specifically activate certain genes and unfavourably affect experimental data [32]. Cellular toxicity is correlated with the transfection reagent and cell type [32–34], but an optimized method for siRNA transfection in primary human macrophages, which takes into consideration the balance between transfection rate and cytotoxicity, has not been reported.
In 2011, Guha et al reported that an anti-oxidant drug, Resveratrol (RESV), killed a monocytic cancer cell line, U937, by upregulating the gene expression of Bax, a component of the intrinsic apoptosis signalling pathway [35]. We hypothesize that an ideal siRNA transfection reagent can effectively deliver Bax siRNA into human macrophages and protect cells against apoptosis induced by RESV. The IncuCyte® has been widely used in recent years to study cell viability without removing cells from incubators [36], as it automatically analyzes and generates presentation-ready graphs. When dyes suitable for staining dead cells are applied, this technology can automatically report the time course of cell death induced by drug treatments [37], allowing for a time and cost-effective way of selecting the optimal transfection reagent for adherent cells. Herein, we combined the protective effects of Bax siRNA to RESV–induced cell death and the auto-analysis function of the IncuCyte® for a fast selection of optimal siRNA transfection reagents for primary human macrophages. This novel method may be universally applied for a rapid optimization of siRNA transfection technology for adherent cells. Moreover, this method provides the comparison of transfection efficiency between multiple reagents that is simple, intuitive, visual, and objective. Most importantly, this strategy takes into consideration the balance between transfection rate and cytotoxicity.
Results
RESV induces cell death in PMA-differentiated U937 and primary human MDMs
RESV has been shown to induce apoptosis in U937 cells [35]. To determine if RESV induces apoptosis in primary human macrophages, we first determined the optimal dose of RESV to induce cell death in PMA-differentiated U937 cells and primary human monocyte-derived macrophages (MDMs). We have previously used a second mitochondria-derived activator of caspase (SMAC) mimetics dimer, AEG40730, to study apoptosis in human MDMs [38]. In this study, AEG40730 was used as a reference drug to determine the optimal dose of RESV to moderately induce apoptosis of differentiated U937 and human MDMs. The optimal dose of RESV should be able to kill target cells effectively, but the killing effect should not exceed the rate observed with 10.0 µM AEG40730. Propidium iodide (PI), which specifically stains the RNA and DNA of dead cells, was employed to track cell death, and the IncuCyte® was used for real-time imaging, data analyses, and generating time-course graphs of cell death.
PMA-differentiated U937 and human MDMs were treated with various doses of RESV and PI was added to the complete medium simultaneously to track cell death. Cells were incubated, imaged every 2 hr by IncuCyte®, and cell death was analysed by IncuCyte® Zoom 2016B software to generate kill curves of AEG40730 and RESV. Our results show that cell death was detectable by the 10 hr and peaked at 30–36 hr post-RESV treatment (Fig. 1). RESV at a concentration of 300 µM induced reproducible and consistent levels of cell death in PMA-differentiated U937 cells (Fig. 1A). However, primary human MDMs were more sensitive to RESV–induced cell death than PMA-differentiated U937 cells (Fig. 1B). Moreover, the sensitivity of primary human MDMs to RESV–induced cell death varied from donor to donor. RESV at a concentration of 300 µM was highly toxic to primary human MDMs from some donors, causing extensive cell death. Therefore, we selected 250 µM RESV as the optimal dose to induce cell death in primary human MDMs.
Figure 1.
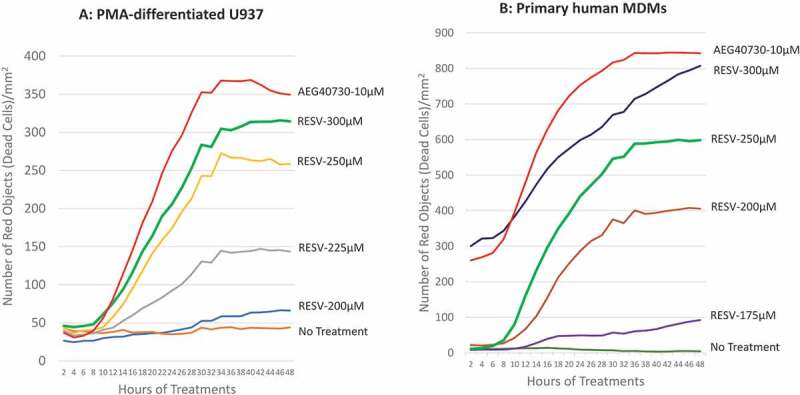
Determining optimal RESV dosage inducing apoptosis of U937 and primary human MDMs. Representative graphs for time-course cell death of PMA-differentiated U937 cells (A) and primary human MDMs (B) induced by AEG40730 and RESV.
Selection of appropriate siRNA transfection reagent for PMA-differentiated U937 cells and primary human MDMs
To determine the protective effect of Bax siRNA, PMA-differentiated U937 cells and primary human MDMs were treated with 20 nM Bax siRNA and each of the 10 transfection reagents obtained from various biotechnology companies (DharmaFECT 3 and DharmaFECT 4 from Dharmacon, HiPerFect from Qiagen, Lipofectamine RNAiMAX, Lipofectamine 2000 and Lipofectamine 3000 from Invitrogen, PureFection™ from SBI System Bioscience, X-tremeGENE from Roche, siRNA Transfection Reagent from Santa Cruz, and Ambion siPORT Amine siRNA Transfection Reagent from ThermoFisher) for 24 hr as per the manufactures’ protocols suggested. Subsequently, the transfected U937 cells and MDMs were treated with 300 µM and 250 µM RESV respectively, followed by incubation in the IncuCyte® for 48 hr. PI was added to the complete medium simultaneously. To achieve efficient transfection, we fixed the amount of siRNA at 20 nM/well but varied the amount of transfection reagents by ±25% of the dose recommended by the manufacturers. For example, Dharmacon recommended 0.8 µl/well DharmaFECT 3 transfection reagent for macrophages seeded in a 12-well plate. Therefore, 20 nM Bax siRNA was transfected with 0.6 µl, 0.8 µl, or 1.0 µl/well DharmaFECT 3 transfection reagent, respectively, for 24 hr before RESV treatment. Cells were imaged and analysed by IncuCyte® at various time points post-RESV treatment, and kill curves were generated to compare the protective effects of Bax siRNA with different transfection reagents. Using this approach, we were able to decipher, via IncuCyte, the level of cell death caused at each concentration of the reagents. The kill curves closest to the negative control (no RESV treatment, no siRNA, and no transfection reagent) would indicate the optimal transfection reagent with least cytotoxicity, as more efficient transfection should result in a greater protective effect of Bax siRNA with least cytotoxic effect of the transfection reagent (Fig. 2A and 2B). The results show that of all the 10 transfection reagents examined, Bax siRNA transfected with DharmaFECT 3 transfection reagent prevented maximally RESV–induced cell death in both PMA-differentiated U937 cells and primary human MDMs. For clarity and ease of presentation, only the two best kill curves, which were closer to the ‘no treatment curve’ for a transfection reagent, are shown. The kill curve with the concentration causing the most cell death was not shown. However, if the best two curves selected for a transfection reagent were very close to each other, only one curve was shown in the figure (e.g. Santa Cruz transfection reagent in Fig. 2A). The protective effect of Bax siRNA was detectable as early as 12 hr post-RESV treatment and the maximal protective effect was observed by 24 hr post-RESV treatment. In contrast, Bax siRNA transfected with X-tremeGENE from Roche caused the highest level of cell death. The transfection reagents from Santa Cruz, Lipofectamine RNAiMAX, and HiPerFect containing Bax siRNA moderately prevented RESV–induced cell death. Fig. 2B shows that for primary human MDMs, Lipofectamine 2000 was less toxic than other reagents but relatively more toxic than DharmaFECT 3 reagent. This was further confirmed via fluorescence microscopy. For this, 20 nM BLOCK-iT™ Alexa Fluor® Red Fluorescent Control siRNA was transfected with either 1.0 μl DharmaFECT 3 or 6.0 μl Lipofectamine 2000 for 16 hr following which the cells were imaged under fluorescence microscope. The results revealed altered cell morphology and extensive granulation in cells treated with Lipofectamine 2000 compared to the cells treated with DharmaFECT 3 transfection reagent (Fig. 2C), indicating Lipofectamine 2000 is more toxic than DharmaFECT 3 for primary human MDMs. These results suggest that DharmaFECT 3 is the most appropriate transfection reagent for primary human MDMs. Therefore, in the next series of experiments, we primarily focused on DharmaFECT 3 transfection reagent.
Figure 2.
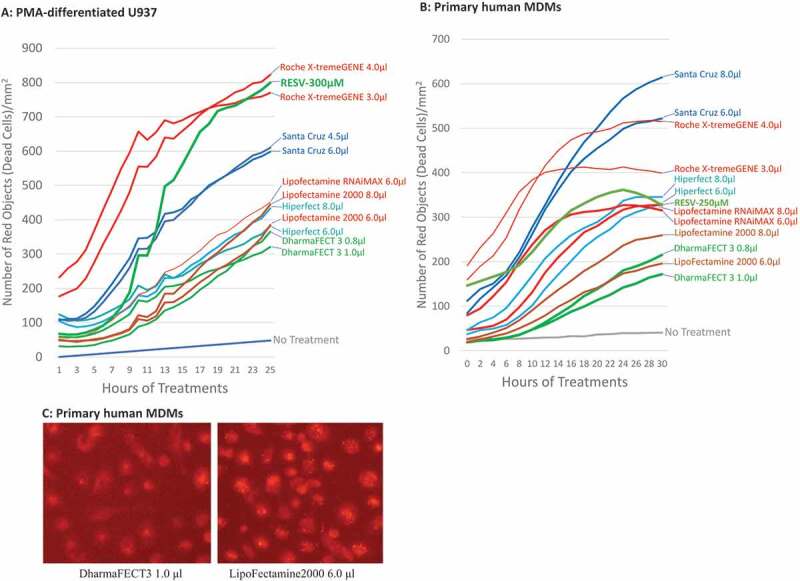
Transfection reagent selection for U937 and human macrophages. 20 nM Bax siRNA was transfected with 3 different doses of each transfection reagent (the amount recommended by the manufacturer, +25% and −25% of the dose recommended by the manufacturer). For clarity and ease of presentation, the best two kill curves for the transfection reagents closest to the ‘no treatment curve’ are shown. If the best two curves selected for a transfection reagent were very close to each other, however, only one curve is shown. Representative graph for the time-course cell death of PMA-differentiated U937 cells (A) and primary human MDMs (B) pretreated with Bax siRNA 24 hr before treatment with RESV. (C) The cytotoxicity comparison between DharmaFECT 3 and Lipofectamine 2000 in primary human MDMs. 20 nM BLOCK-iT™ Alexa Fluor® Red Fluorescent control siRNA was transfected with either 1.0 μl DharmaFECT 3 or 6.0 μl Lipofectamine 2000 for 16 hr following which the cells were imaged under fluorescence microscope.
Selection of optimal dose of siRNA and the DharmaFECT 3 transfection reagent for primary human MDMs
To determine the optimal dose of siRNA and DharmaFECT 3 transfection reagent for human macrophages, various amounts of Bax siRNA (20–30 nM) and DharmaFECT 3 transfection reagent (1.0 to 2.0 μl) were combined to pretreat primary human MDMs for 24 hr followed by treatment with 250 µM RESV. 1.0 μl/ml PI was added into the complete medium simultaneously to track cell viability. Cell death was monitored by IncuCyte® and kill curves were generated (Fig. 3A). Our results show that the most protective effect to RESV–induced cell death of human MDMs was achieved when the combination of 1.0 μl/well of DharmaFECT 3 transfection reagent and 20 nM/well of Bax siRNA was applied. In contrast, 30 nM/well of Bax siRNA along with 1.5 or 2.0 µl/well of DharmaFECT 3 transfection reagent killed cells similar to positive control (250 µM RESV without siRNA pretreatment). However, the combination of 20 nM Bax siRNA along with 1.2 or 1.5 μl DharmaFECT 3 transfection reagent exhibited modest protective effect against RESV–induced cell death, indicating that high dose of siRNA (30 nM) and/or transfection reagent (2.0 μl) were very toxic to primary human MDMs.
Figure 3.
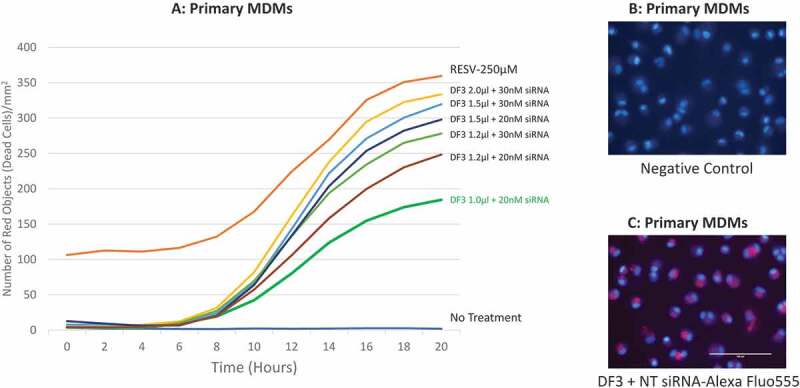
Dosage combination of DharmaFECT 3 and siRNA, and localization of siRNA. (A): time-course cell death of dosage combination of DharmaFECT 3 and Bax siRNA. (B): MDMs stained with Hoechst 33342 for 15 min without siRNA transfection. (C): MDMs transfected with the BLOCK-iT™ Alexa Fluor Red Fluorescent Control siRNA for 6 hr and then stained with Hoechst 33342 for 15 min. DF3 refers to DharmaFECT 3 transfection reagent; NT siRNA refers to the BLOCK-iTTM Alexa Fluor Red Fluorescent Control siRNA.
To verify if siRNA was delivered into the correct location of target cells, we tracked siRNA in macrophages by transfecting 20 nM/well the BLOCK-iT™ Alexa Fluor® Red Fluorescent Control siRNA with 1.0 μl DharmaFECT 3 transfection reagent. Cells were stained with Hoechst 33342 for 15 min, as a nuclear counterstain, and imaged under a fluorescence microscope. We found that after 6 hr of transfection, red fluorescence (indicates location of transfected siRNA) was observed in the cytoplasm of primary MDMs (Fig. 3B and 3C), suggesting that the DharmaFECT 3 transfection reagent we selected, and the dose we determined, 1.0 µl/well, successfully delivered siRNA into the cytoplasm of primary human MDMs.
Efficiency of siRNA transfection into primary human MDMs
Next, we determined the efficiency of siRNA transfection in primary human MDMs using the BLOCK-iT™ Alexa Fluor® Red Fluorescent Control siRNA (Alexa Fluor control siRNA) with DharmaFECT 3 transfection reagent and compared it with the transfection reagents from other manufacturers. The Alexa Fluor control siRNA (20 nM) was transfected into human MDMs with DharmaFECT 3 or X-tremeGENE transfection reagent for 16 hr. Cells were imaged (Fig. 4A1-Fig. 4D1), trypsinized, and the percentages of Alexa Fluor555 positive cells were analysed by flow cytometry. The results show that without any treatment, none of the cells were stained (Fig. 4A1), whereas cells treated with Alexa Fluor control siRNA in the absence of transfection reagent showed only few deeply stained macrophages (Fig. 4B1). However, with DharmaFECT 3 and X-tremeGENE transfection reagents, most of the macrophages were deeply stained (Fig. 4C1, 4D1). The lightly stained cells might be the macrophages in which the Alexa Fluor control siRNA is bound on the cell membranes but are not transfected. The quantification of cells transfected with the Alexa Fluor control siRNA by flow cytometric analysis indicated that ~83.9% of human MDMs were effectively transfected with 1.0 µl DharmaFECT 3 transfection reagent (Fig. 4C2) compared to the 0% in untreated and un-transfected cells (Fig. 4A2) and 15% in cells treated with Alexa Fluor control siRNA alone without transfection reagent (Fig. 4B2). Similarly, approximately 95% of cells were transfected with Alexa Fluor control siRNA and X-tremeGENE transfection reagent (Fig. 4D2). Although the transfection rate of X-tremeGENE was as high as 95% (Fig. 4D2), this reagent exhibited significantly higher number of cell death (Fig. 2A and 2B), suggesting that the X-tremeGENE transfection reagent from Roche was very toxic to primary human MDMs. In addition, the siRNA transfection efficiency of Lipofectamine 2000 was in the range of 85-90% (data not shown), but the high transfection rate was offset by its higher cytotoxicity compared to DharmaFECT 3 transfection reagent (Fig. 2B and 2C). Thus, DharmaFECT 3 was identified as the optimal transfection reagent for primary human MDMs.
Figure 4.
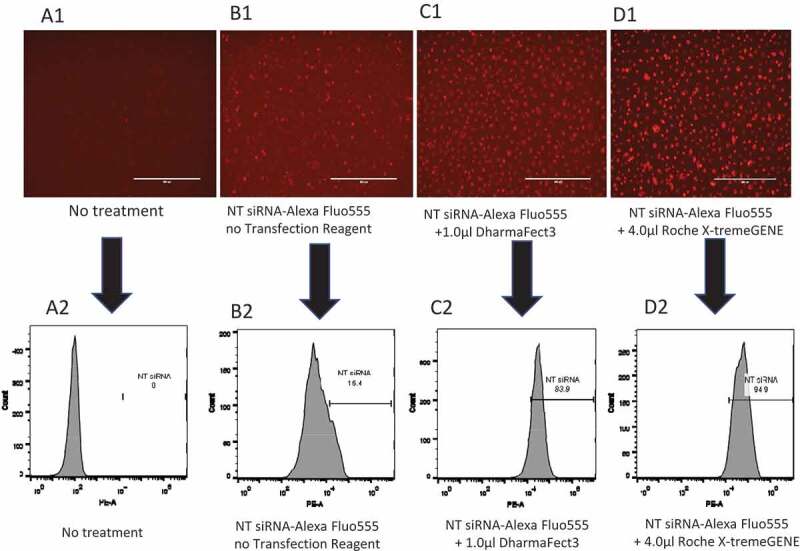
siRNA transfection efficiency in primary MDMs. Primary human MDMs were transfected with 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control siRNA and various transfection reagents for 16 hr. A1: No siRNA, no transfection reagent; B1: 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control without transfection reagent; C1: 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control with 1.0 µl DharmaFECT 3; D1: 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control with 4.0 µl Roche X-tremeGENE. A2: Percentage of control siRNA+ at A1; B2: Percentage of control siRNA+ at B1; C2: Percentage of control siRNA+ at C1; D2: Percentage of control siRNA+ at D1.
Quantification of the protective effect of Bax siRNA with DharmaFECT 3 transfection reagent for PMA-differentiated U937 and primary human MDMs
To quantify the protective effects of Bax siRNA transfected with DharmaFECT 3 transfection reagent for PMA-differentiated U937 cells against RESV–induced cell death, cells were treated with 20 nM Bax siRNA and 1.0 μl DharmaFECT 3 transfection reagent as before followed by RESV treatment along with PI for 24 or 48 hr. The images were retrieved from IncuCyte® for quantitative analysis. RESV treatment alone for 24 hr (Fig. 5A1) and 48 hr (Fig. 5A2) resulted in a large number of dead cells as indicated by red dots. In contrast, there was a significant reduction in the number of dead cells in PMA-differentiated U937 cells pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 transfection reagent followed by RESV treatment for 24 hr (Fig. 5C1) and 48 hr (Fig. 5C2) as revealed by staining with PI (red dots), suggesting that Bax siRNA transfection effectively protected U937 cells against RESV–induced cell death. As expected, very few dead cells were observed in negative controls without RESV treatment and siRNA transfection (Fig. 5B1 and 5B2). To quantify dead cells, we retrieved the data on ‘Red Object Counts’ (dead cells stained with PI) recorded by IncuCyte® from the same images (Fig. 5A1 to 5C1, and Fig. 5A2 to 5C2), and converted the data into a bar diagram from one representative experiment (Fig. 5D). Fig. 5E shows the percentage of dead cells calculated from three independent experiments. In addition, our Western blot and densitometry analysis revealed that 20 nM Bax siRNA along with 1.0 µl DharmaFECT 3 transfection reagent significantly silenced the expression of Bax gene in PMA-differentiated U937 cells (Fig. 5F and 5G).
Figure 5.
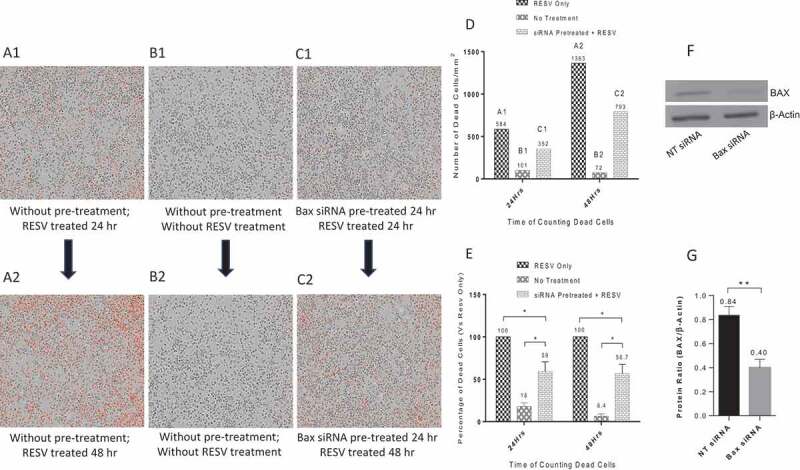
Protective effects of Bax siRNA in PMA-differentiated U937 cells. A1: RESV treated for 24 hr; A2: RESV treated for 48 hr; B1: No RESV treatment 24 hr; B2: No RESV treatment 48 hr; C1: Pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 24 hr then RESV treated for 24 hr; C2: Pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 24 hr then RESV treated for 48 hr. D: Number of dead cells in A1, B1, C1 and A2, B2, C2, information directly retrieved from Incucyte®. E: Percentage of dead cells compared to RESV treated; F: Representative Western blot analysis of PMA-differentiated U937 cells transfected with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 48 hr. G: Densitometry analysis of BAX and β-Actin protein bands from three independent Western blot experiments with results shown as mean ± SD (n = 3). * indicates p ≤ 0.05. ** indicates p ≤ 0.01.
The protective effects of Bax siRNA against RESV–induced cell death for primary human MDMs were also studied. Similar to the PMA-differentiated U937 cells, we pretreated primary human MDMs with 20 nM Bax siRNA and 1.0 μl DharmaFECT 3 transfection reagent for 24 hr, followed by RESV treatment along with PI for 24 or 48 hr. The images retrieved from IncuCyte® were analysed. RESV treatment alone for 24 hr (Fig. 6A1) and 48 hr (Fig. 6A2) resulted in a large number of dead cells (red dots). In contrast, there was a significant reduction in the number of dead cells in MDMs pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 transfection reagent for 24 hr followed by RESV treatment for 24 hr (Fig. 6C1) and 48 hr (Fig. 6C2), indicating that Bax siRNA effectively protected MDMs against RESV–induced cell death. As expected, very few dead cells were observed in negative controls without RESV treatment and Bax siRNA transfection (Fig. 6B1 and 6B2). To quantify dead cells, we retrieved the data of MDMs on ‘Red Object Counts’ recorded by IncuCyte® from the same images (Fig. 6A1 to 6C1, and Fig. 6A2 to 6C2), and converted the data into a bar diagram from one representative experiment (Fig. 6D). Fig. 6E shows the percentage of dead cells calculated from three different experiments. These results suggest that Bax siRNA significantly protected MDMs from RESV–induced cell death. In addition, Western blot (Fig. 6F) and densitometry analysis (Fig. 6G) revealed that 20 nM Bax siRNA along with 1.0 µl DharmaFECT 3 transfection reagent significantly silenced the expression of Bax gene in primary human MDMs. Overall, pretreating PMA-differentiated U937 and primary human MDMs with Bax siRNA and DharmaFECT 3 transfection reagent significantly reduced both the number and percentage of dead cells induced by RESV.
Figure 6.
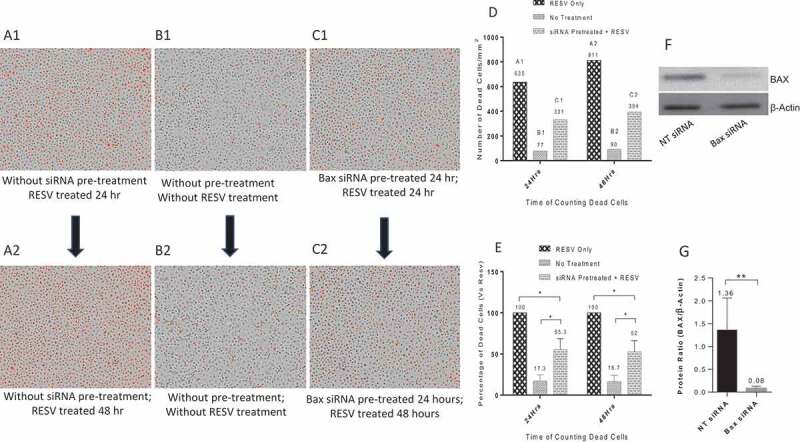
Protective effects of Bax siRNA in primary human MDMs. A1: RESV treated for 24 hr; A2: RESV treated for 48 hr; B1: No RESV treatment 24 hr; B2: No RESV treatment 48 hr; C1: Pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 24 hr then RESV treated for 24 hr; C2: Pretreated with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 24 hr then RESV treated for 48 hr. D: Number of dead cells in A1, B1, C1 and A2, B2, C2; information was directly retrieved from IncuCyte®. E: Percentage of dead cells compared to RESV treated; the mean value and standard deviations were calculated using Microsoft Excel (n = 3). F: Representative Western blot analysis of primary human MDMs transfected with 20 nM Bax siRNA and 1.0 µl DharmaFECT 3 for 48 hr. G: Densitometry analysis of BAX and β-Actin protein bands from three independent Western blot experiments. The results shown are mean ± SD (n = 3). * indicates p ≤ 0.05. ** indicates p ≤ 0.01.
Discussion
siRNA is a critical loss-of-function tool for elucidating the roles of genes in experimental in vitro studies. Primary human macrophages are well known to be hard-to-transfect cells with siRNA [25–27]. Currently, a fast, cost-saving, and easy-to-do method which can be used to select efficient siRNA transfection reagents for various adherent cell types, and in particular, primary human macrophages, have not been reported. In this study, we took advantage of the protective effect of Bax siRNA and the auto-analysis function of IncuCyte® to select a transfection reagent that efficiently transfects siRNA into primary human macrophages and PMA-differentiated U937 cells. Our results suggest that DharmaFECT 3, a transfection reagent from Dharmacon, was the most efficient to transfect siRNA into both cell types, whereas most of the other transfection reagents were cytotoxic. Furthermore, ~85% of primary human MDMs can be transfected by this selected reagent. We also show that Bax siRNA effectively prevented RESV–induced cell death in primary human macrophages and PMA-differentiated U937 cells.
We observed that primary human MDMs from many individuals were not sensitive to siRNA transfection. The mechanisms responsible for resistance to transfection are not well understood. There are at least two factors that determine resistant phenotype of macrophages to siRNA transfection [25–27]. First, macrophages are professional immune cells; they have evolved to recognize foreign nucleic acids and may initiate immune responses to exogenous siRNA molecules [26,27]. Second, macrophages possess more restriction factors than other cell types which may interfere with intracellular machinery for siRNA silencing. For example, Li et al identified 114 restriction factors which significantly impact HIV replication in macrophages [39]. SamHD1, Apobec3, Tetherin, and Viperin are well-known restriction factors that affect multiple viruses at distinct stages of their life cycles [40]. SamHD1 has RNase activity which may degrade exogenous siRNA [41], directly causing the failure of siRNA transfection into macrophages.
Efficiency of siRNA transfection may vary with the cell type and more so with the adherent cells depending upon the transfection reagents used. Reynolds emphasized the importance of selecting a transfection reagent for target cells [34], but a fast, cost-saving, and easy-to-do method for human macrophages has yet to be reported. Therefore, we analysed 10 transfection reagents commercially available from 7 biotechnology companies to select the most suitable reagent for siRNA transfection in primary human MDMs. By employing Bax siRNA, we successfully selected the optimal transfection reagents for two hard-to-transfect cells, primary human macrophages [25–27] and PMA-differentiated U937 cells [42], suggesting that this novel method may be widely applied for the selection of an optimal siRNA transfection reagent for various adherent cells. Most importantly, the time-course graphs auto-generated by IncuCyte® software are an integrated readout of both transfection rate and cytotoxicity of the transfection reagents. Therefore, this strategy inherently takes the balance between transfection rate and cytotoxicity of the transfection reagents into consideration. The BLOCK-iT™ Alexa Fluor™ Red Fluorescent Non-targeting Control siRNA has been reported to be a useful reagent for assessing the transfection efficiency of siRNA [43]. This reagent alone, however, does not ascertain the cytotoxicity of the transfection reagents, and the higher transfection rate may be offset by the cytotoxicity of the transfection reagents. In addition, it also reports false-positive siRNA signals caused by siRNA attaching to the outer cell membrane, without actually reaching the cytoplasm. Thus, this reagent can be used to track the location of siRNA, but does not convey the optimal transfection reagent for target cells.
In summary, taking advantage of the protective effects of Bax siRNA to RESV–induced cell death and the auto-analysis function of IncuCyte®, we report that DharmaFECT 3 is the optimal transfection reagent for both PMA-differentiated U937 and primary human MDMs. Although DharmaFECT 3 has been used for siRNA transfection of cell lines LNCaP (Prostate carcinoma) and SK-OV-3 (Ovarian adenocarcinoma), MCF7 [44], A2780 [45], SW480 and HT29 [46], the novel method we describe herein is indeed a quick and effective strategy to select the optimal transfection reagent for adherent cells, and in particular, hard-to-transfect primary human macrophages. The kill curves are automatically generated by the IncuCyte®, avoiding human bias. Thus, it is easy to determine the optimal siRNA transfection reagent directly from the graphs auto-generated by the software. Moreover, the comparison of the transfection efficiency between multiple reagents is simple, intuitive, visual, and objective. This strategy takes into consideration the balance between transfection rate and cytotoxicity of the transfection reagents and can be widely applied to a fast and cost-saving selection of optimal siRNA transfection reagent for adherent cells. The limitations of this method are that the application requires the IncuCyte® Imaging System, familiarity with IncuCyte® data analysis, and high quality of cell monolayer seeded in culture plates.
Material and methods
Cell culture and differentiation of U937
U937 (ATCC Cat. CRL-1593.2™) was maintained in complete medium (DMEM from Wisent, Cat.: 319-105-CL; 10% Foetal Bovine Serum (FBS) from Sigma, SKU: F1051; 100U/ml Penicillin G from Sigma, SKU: P3032; 100μg/ml Streptomycin from Sigma, SKU: S9137) at 37°C, 5% CO2 at the density of 105 ~ 106 cells/ml. For differentiated U937, 4.5 × 105 cells/well were seeded in co-star 12-well plates (Sigma, SKU: CLS3513) with 1 ml complete medium supplemented with 50 ng Phorbol 12-myristate 13-acetate (PMA, Sigma, SKU: P1585) for 3 days; cells were washed with PBS (Wisent, Cat.: 311-425-CL) and maintained in complete medium with 50 ng PMA for 2 more days before further experiments.
Preparation of primary human MDMs
Peripheral Blood Mononuclear Cells (PBMCs) were prepared following the protocol of Lymphoprep Density Gradient (StemCell, Cat.: 07851). 1.5 ~ 2.0 x 106 cells/well were seeded in co-star 12-well plates in DMEM medium without supplement at 37°C, 5% CO2. Three hours later, supernatant was removed, and the cell monolayer was washed twice with PBS. Subsequently, cells were maintained in complete medium supplemented with 10 ng/ml Macrophage-Colony Stimulating Factor (M-CSF, R&D, Cat.: 216-MC-025) at 37°C, 5% CO2. After 3 days, the cells were washed once with PBS and maintained in complete medium with M-CSF for 4 more days before further experiments.
siRNA transfection
Two hours before siRNA transfection, cells were washed with PBS and maintained in 0.6 ml/well of antibiotics-free DMEM medium supplemented with 10% FBS, and incubated at 37°C, 5% CO2. 20 nM Bax siRNA (Dharmacon, Accell Human Bax siRNA, Cat.: E-003308-00-0005) or BLOCK-iT™ Alexa Fluor™ Red Fluorescent Non-targeting Control siRNA (ThermoFisher Scientific Invitrogen Cat.: 14,750,100) in 200 µl transfection medium was used in all siRNA transfections unless specifically annotated. Differentiated U937 and MDMs were pretreated with 20 nM Bax siRNA and transfection reagents for 24 hr in a volume of 0.8 ml/well and a total of 10 transfection reagents were tested following the protocols provided by the manufacturers (Lipofectamine RNAiMax, Invitrogen, Cat.: 13,778,075; Lipofectamine 2000, Invitrogen, Cat.: 11,668–019; Lipofectamine 2000, Invitrogen, Cat.: L3000-008; siRNA Transfection Reagent, Santa Cruz, Cat.: sc-29,528; DharmaFECT 3, Dharmacon, Cat.: T-2003-03; DharmaFECT 4, Dharmacon, Cat.: T-2004-01; HiPerFect, Qiagen, Cat.: 301,704; X-tremeGENE siRNA Transfection Reagent, Roche, Cat.: 04 476 093 001; PureFection™, SBI System Bioscience, Cat.: LV750A-1; Ambion siPORT Transfection Agent, ThermoFisher, P/N: AM4510).
AEG40730 and RESV treatments with PI staining
The concentration of SMAC Mimetics AEG40730 (Tocris Bioscience, Cat.: 5330) was fixed to be 10 µM in this study. Resveratrol (RESV, Sigma, SKU: R5010-100MG) were diluted as suggested by the manufacturer. The concentration of Propidium Iodide (PI, Sigma, SKU: P4170-10MG) was also fixed to be 1.0 µl/ml in all experiments, and it was added to the master-mix with AEG40730 or RESV when it was used to track cell death in the IncuCyte®. The amount of AEG40730 or RESV and PI was calculated and added to 200 µl/well complete medium without antibiotics, and directly loaded to cells pretreated with siRNA, making the final volume to be 1.0 ml/well. Cell death was monitored immediately with IncuCyte®, and maintained at 37°C, 5% CO2 for further experiments.
Flow cytometry analysis of siRNA transfection rates
Primary human MDMs were transfected with 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control siRNA and transfection reagents for 16 hr, washed with PBS, trypsinized with 0.25% Trypsin-1.0 mM EDTA (StemCell, Cat.: 07901) for 30 min. Cells were pipetted up and down 16 times, harvested, centrifuged, washed with PBS, re-suspended in 500 µl of PBS with 0.5% BSA (Bovine Serum Albumin, Sigma-Aldrich, SKU: A2153-100G) and the percentages of Alexa Fluor555-positive cells were analysed by flow cytometer (BD LSRFortessaTM X-20) in the PE Channel.
Monitoring cell death by incucyte®
Immediately after the drugs (RESV and AEG40730 with PI) were added to the supernatant, cell culture plates were incubated at 37°C, 5% CO2 in the IncuCyte® (Essen Bioscience IncuCyteTM Zoom). The 10x objective lens was selected to monitor cell death at both phase-contrast and red channels, and cells were scanned every 2 hr. Image collection and processing definition were completed following the IncuCyte® manual, and only the objective area between 50 µm2 and 2000 µm2 were counted to be single cells. For image editing purposes, the original data after auto-analysis were exported to Microsoft Excel for the generation of time-course cell death graphs.
Localization of siRNA in primary human MDMs
Primary human MDMs were transfected with 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control siRNA and 1.0 µl DharmaFECT 3 for 6 hr. Subsequently, the supernatant was removed, and cells were washed with PBS. The Hoechst 33342 (ThermoFisher Scientific Invitrogen, Cat.: H1399) stock solution (10 mg/ml) was diluted 1:2,000 in PBS, and 500 µl was added to cover all cells for 15 min. The staining solution was removed, cells were washed 3 times in PBS and imaged at DAPI Channel of EVOS FL Cell Imaging System.
Microscopy
Images of cells treated with AEG40730 and RESV were retrieved directly from IncuCyte®. For the transfection efficiency of siRNA in primary human MDMs, cells were transfected with 20 nM BLOCK-iT Alexa Fluor Red Fluorescent Control with various transfection reagents for 16 hr and imaged under DAPI channel of EVOS FL Cell Imaging System.
Western blotting
Forty-eight hours after Bax siRNA transfection, cells were harvested and lysed in cell lysis buffer (Cell Signalling, Product #: 9803S). Proteins were quantitated following the Bio-Rad Protein Assay (Bio-Rad, Cat.: 5,000,006) manual, and 50 µg of each sample was analysed in 15% SDS-Polyacrylamide Gel Electrophoresis (PAGE). Subsequently, proteins were transferred onto PVDF membranes (Bio-Rad, Cat.: 1,620,177) and blocked with 5% non-fat dried milk in PBS at 4ºC overnight. Membranes were probed with Anti-BAX Antibody (Santa Cruz, Cat.: sc-20,067) in PBS with 1% BSA for 1 hr at room temperature, followed by Rat Anti-mouse IgG conjugated with Horseradish Peroxidase (Bio-Rad, Cat.: MCA152) in 1% BSA. Immunoblots were visualized using Clarity Max™ Western ECL Blotting Substrates (Bio-Rad, Cat.: 1,705,060) and imaged with Chemigenius Bio-imaging System (Syngene) and GeneSnap Software (Syngene). ImageJ Software (developed by Wayne Rasband from National Institute of Health of USA) was used for the densitometric analysis of BAX and β-Actin protein bands, and the protein ratio (BAX/β-Actin) of each experiment was calculated independently. The results were summarized into bar diagrams.
Statistical analysis
Microsoft Excel 2016 was applied to compute Mean Value, Standard Deviation (SD) and plot kill curves; P-value was calculated using Student t-test. * indicates significant (p ≤ 0.05); ** indicates p ≤ 0.01. GraphPad Prism 6.0 software was used to create bar diagrams. Each experiment was repeated three times independently (n = 3).
Ethics statement
Healthy participants involved in the study gave informed written consent and the protocol for obtaining blood samples was approved by the Review Ethics Board of the Ottawa General Hospital and the Children’s Hospital of Eastern Ontario, Ottawa, ON, Canada.
Acknowledgments
We acknowledge the generous help provided by the blood donors and the nurses in collecting blood sample. We would like to thank Stephen Baird for his daily helps with computer work and data backup. Many thanks to Xiang Xiao, who was very informative to the progress of this study. We would also like to thank Katelynn J. Rowe, who devoted her time to editing this manuscript.
Funding Statement
This work was supported by grants from the Canadian Institute of Health Research (CIHR) under Grant to A.K. HOP98830 and by The Canadian HIV Cure Enterprise Team Grant HIG-133050 to A.K. from the CIHR in partnership with CANFAR and IAS.
Author contributions
S.X.M.D conceived and designed the study, reviewed literatures, conducted the lab work, developed protocols, edited figures, performed data analyses and drafted the manuscript. R.E provided daily assistances, such as preparation of primary human MDMs and Western blot. D.L.F.R. provided cell plates and helped to polish figures. H.A. helped computational methods and performed bioinformatics analysis. E.C. and A.K. supervised and coordinated the study and finalized the manuscript. All authors have read and approved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Wijburg OLC, van den Dobbelsteen GPJM, Vadolas J, et al. The role of macrophages in the induction and regulation of immunity elicited by exogenous antigens. Eur J Immunol. 1998;28:479–487. [DOI] [PubMed] [Google Scholar]
- [2].Gordon S. The role of the macrophage in immune regulation. Research in Immunology. 1998;149:685–688. [DOI] [PubMed] [Google Scholar]
- [3].Park S-Y, Jung M-Y, Lee S-J, et al. Stabilin-1 mediates phosphatidylserine-dependent clearance of cell corpses in alternatively activated macrophages. Journal of Cell Science. 2009;122:3365–3373. [DOI] [PubMed] [Google Scholar]
- [4].Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Perelson AS, Neumann AU, Markowitz M, et al. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. [DOI] [PubMed] [Google Scholar]
- [6].Crowe S, Zhu T, Muller WA. The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. Journal of Leukocyte Biology. 2003;74:635–641. [DOI] [PubMed] [Google Scholar]
- [7].Chen Y, Zhang X. Pivotal regulators of tissue homeostasis and cancer: macrophages. Exp Hematol Oncol. 2017;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krysko O, Holtappels G, Zhang N, et al. Alternatively activated macrophages and impaired phagocytosis of S aureus in chronic rhinosinusitis. Allergy. 2011;66:396–403. [DOI] [PubMed] [Google Scholar]
- [9].Firszt R, Vickery B. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Pediatrics. 2011;128:S145–S146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Montoya D, Cruz D, Teles RMB, et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe. 2009;6:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Quiding-Järbrink M, Raghavan S, Sundquist M. Enhanced M1 macrophage polarization in human helicobacter pylori-associated atrophic gastritis and in vaccinated mice. PLoS One. 2010;5:e15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mège J-L, Mehraj V, Capo C. Macrophage polarization and bacterial infections. Current Opinion in Infectious Diseases. 2011;24:230–234. [DOI] [PubMed] [Google Scholar]
- [13].Thompson LJ, Dunstan SJ, Dolecek C, et al. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proceedings of the National Academy of Sciences. 2009;106:22433–22438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Parisi L, Gini E, Baci D, et al. Macrophage polarization in chronic inflammatory diseases: killers or builders? Journal of Immunology Research. 2018;2018:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. [DOI] [PubMed] [Google Scholar]
- [16].Ponzoni M, Pastorino F, Di Paolo D, et al. Targeting macrophages as a potential therapeutic intervention: impact on inflammatory diseases and cancer. International Journal of Molecular Sciences. 2018;19:1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hadji P, Coleman R, Gnant M, et al. The impact of menopause on bone, zoledronic acid, and implications for breast cancer growth and metastasis. Annals of Oncology. 2012;23:2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dahl V, Josefsson L, Palmer S. HIV reservoirs, latency, and reactivation: prospects for eradication. Antiviral Research. 2010;85:286–294. [DOI] [PubMed] [Google Scholar]
- [19].Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Koren-Gluzer M, Rosenblat M, Hayek T. Paraoxonase 2 induces a phenotypic switch in macrophage polarization favoring an M2 anti-inflammatory state. International Journal of Endocrinology. 2015;2015:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Porcheray F, Viaud S, Rimaniol A-C, et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ovcharenko D, Kelnar K, Johnson C, et al. Genome-scale MicroRNA and small interfering RNA screens identify Small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. [DOI] [PubMed] [Google Scholar]
- [24].Xia Y, Gates B, Yin Y. Building complex structures from monodisperse spherical colloids. Aust J Chem. 2001;54:287–290. [Google Scholar]
- [25].Stroh T, Erben U, Kühl AA, et al. Combined pulse electroporation – a novel strategy for highly efficient transfection of human and mouse cells. PLoS One. 2010;5:e9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dokka S, Toledo D, Shi X, et al. High-efficiency gene transfection of macrophages by lipoplexes. International Journal of Pharmaceutics. 2000;206:97–104. [DOI] [PubMed] [Google Scholar]
- [27].Keller -A-A, Maeß MB, Schnoor M, et al. Transfecting macrophages. Methods in Mol Biol. 2018;1784:187–195. [DOI] [PubMed] [Google Scholar]
- [28].Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology. 2012;9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].DiNapoli SR, Hirsch VM, Brenchley JM. Macrophages in progressive human immunodeficiency virus/simian immunodeficiency virus infections. J Virol. 2016;90:7596–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Clayton KL, Garcia V, Clements JE, et al. HIV infection of macrophages: implications for pathogenesis and cure. Pathogens and Immunity. 2017;2:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aquaro S, Caliò R, Balzarini J, et al. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antiviral Research. 2002;55:209–225. [DOI] [PubMed] [Google Scholar]
- [32].Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. [DOI] [PubMed] [Google Scholar]
- [33].Breunig M, Lungwitz U, Liebl R, et al. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proceedings of the National Academy of Sciences. 2007;104:14454–14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Reynolds A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guha P, Dey A, Sen R, et al. Intracellular GSH depletion triggered mitochondrial bax translocation to accomplish Resveratrol-induced apoptosis in the U937 cell line. J Pharmacol Exp Ther. 2011;336:206–214. [DOI] [PubMed] [Google Scholar]
- [36].Single A, Beetham H, Telford BJ, et al. A comparison of real-time and endpoint cell viability assays for improved synthetic lethal drug validation. J Biomol Screen. 2015;20:1286–1293. [DOI] [PubMed] [Google Scholar]
- [37].Li Y, Yuan J. Systematic metrics depicting cell death kinetics. Cell Chem Biol. 2017;24:785–786. [DOI] [PubMed] [Google Scholar]
- [38].Busca A, Saxena M, Kumar A. Critical role for antiapoptotic Bcl-xL and Mcl-1 in human macrophage survival and cellular IAP1/2 (cIAP1/2) in resistance to HIV-Vpr-induced apoptosis. J Biol Chem. 2012;287:15118–15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu L, Oliveira NMM, Cheney KM, et al. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chemudupati M, Kenney AD, Bonifati S, et al. From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochimica Et Biophysica Acta (BBA) - Molecular Cell Research. 2019;1866:382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ryoo J, Choi J, Oh C, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tietz SM, Berghoff M. Gene silencing of MK2 in hard-to-transfect human U937 cells. J Biomol Tech. 2012;23:47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mehrotra S, Lee I, Chan C. Multilayer mediated forward and patterned siRNA transfection using linear-PEI at extended N/P ratios. Acta Biomater. 2009;5:1474–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Iorns E, Lord CJ, Ashworth A. Parallel RNAi and compound screens identify the PDK1 pathway as a target for tamoxifen sensitization. Biochemical Journal. 2009;417:361–371. [DOI] [PubMed] [Google Scholar]
- [45].Pink RC, Samuel P, Massa D, et al. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecologic Oncology. 2015;137:143–151. [DOI] [PubMed] [Google Scholar]
- [46].Tsofack SP, Garand C, Sereduk C, et al. NONO and RALY proteins are required for YB-1 oxaliplatin induced resistance in colon adenocarcinoma cell lines. Mol Cancer. 2011;10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]


