Abstract
Background:
The primary objective of this study was to compare the characteristics of culture positive and negative status in septic patients. We also determined whether culture status is associated with mortality and whether unique variables are associated with mortality in culture positive and culture negative patients separately.
Methods:
Utilizing patient records from intensive care units, the emergency department and general care wards in a large academic medical center, we identified adult patients with 2 or more systemic inflammatory response syndrome (SIRS) criteria between January 1, 2007 and May 31, 2014. We compared the characteristics between culture positive and culture negative patients and used binary logistic regression to identify variables independently associated with culture status and mortality.
Results:
The study population included 9,288 (89%) culture negative patients and 1,105 (11%) culture positive patients. Culture negative patients received more antibiotics during the 48 hours preceding diagnosis, but otherwise demonstrated similar characteristics as culture positive patients. After adjusting for illness severity, a positive culture was not independently associated with mortality (OR = 1.01 [95% C.I. 0.81, 1.26], p = 0.945). The models predicting mortality separately in culture negative and culture positive patients demonstrated very good and excellent discrimination, (c-statistic ± SD: 0.87 ± 0.01 and 0.92 ± 0.01), respectively. After adjusting for multiple comparisons, vasopressor use, renal replacement therapy and plasma transfusion before presentation was associated with mortality in culture negative patients but not culture positive patients.
Conclusions:
Culture negative and culture positive patients with sepsis demonstrate similar characteristics and, after adjusting for severity of illness, similar mortality. Despite this, obtaining cultures are important as, when positive, the sensitivities will affect the class of antibiotic and treatment duration.
The most important factor associated with negative cultures is receipt of antibiotics during the preceding 48 hours. Negative cultures should not give the clinician a false sense of reassurance - patients are still at risk of death, with the risks being dependent on severity of the physiologic derangements before and on the day of sepsis.
Keywords: sepsis, SIRS, culture negative, culture positive, infection
Introduction
Sepsis is one of the most common causes of mortality in hospitalized patients and until recently was defined on the basis of having systemic inflammatory response syndrome (SIRS) with an identified (or presumed) infection.1-3 While the risk of mortality increases with severity of SIRS, a large portion of patients (30 – 60%) fail to demonstrate a culture proven source of infection (culture negative sepsis).4-11
A few small studies suggest that mortality is not impacted by identification of an infective pathogen and that many clinical characteristics are similar between patients with or without microbiologically documented infection.5-10 However, these studies were limited by only studying patients with septic shock,7,10 patients with acute respiratory distress syndrome,8 emergency department patients,5 or by being more than two decades old.6,9 While most of these studies found few differences in patient characteristics and no difference in survival between culture positive and culture negative patients, two recent studies reached very different conclusions. A single center study of 1,000 medical intensive care unit patients in Asia found that culture negative patients had fewer comorbidities and a lower risk of death.7 After controlling for other factors, having a positive culture was not significantly associated with mortality.7 In contrast, a second study utilizing administrative data from the Nationwide Inpatient Sample found that culture negative patients had higher burden of comorbidity and higher risk of death.11 Given the contrasting methodology and results, we sought to further evaluate this topic using a large clinical database that contained patient-level data.
The primary objective of this study was to compare the characteristics of culture positive and negative status in septic patients. The secondary objectives were to determine whether culture status is associated with mortality and to determine whether unique variables are associated with mortality in culture positive and culture negative patients separately.
Methods
Data source
The University of Michigan Institutional Review Board reviewed and approved this retrospective study (HUM00052066) and waived consent. No funding sources were used for this study. All demographic, vital signs, laboratory and outcomes data were obtained from the University of Michigan Health System’s electronic health record system (Centricity, General Electric, Waukegan, IL).
Study population
We studied adult patients (≥ 18 years of age) who presented between January 1, 2007 and May 31, 2014 and developed 2 or more SIRS criteria with suspected source of infection. We used the original American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference Definition for SIRS: temperature < 36°C or > 38°C, heart rate > 90 beats/min, respiratory rate > 20 breaths/min or PaCO2 < 32 mmHg, or white blood cell count > 12,000/mm3, < 4000/mm3 or > 10% immature (band) forms.3
Patients were included in the study if (1) a blood sample was obtained and sent for culture and (2) they met 2 or more SIRS criteria within 12 hours before or after the blood culture (24-hour window). We included patients who were in any adult intensive care units, emergency department and general care units. To ensure best culture practices, the microbiology lab is Clinical Laboratory Improvement Amendments (CLIA) and College of American (CAP) certified. It maintains strict standards for ensuring the correct amount of blood is added to the culture bottles. Bottles with lesser amounts are rejected and redrawn.
In order to evaluate clinical data, vital signs were captured during the three hours before and three hours after meeting SIRS criteria (six-hour window). Laboratory values recorded were closest to the time of blood culture order, six hours before or six hours after (twelve-hour window) presentation. Blood cultures are typically ordered by house officers, nurse practitioners, or physician assistants based on vital signs, physical exam (e.g., wound appearance, productive cough), and clinical history (e.g., recent surgery, comorbidities placing the patient at risk for infections).
In order to identify culture status, we evaluated whether the blood sample obtained subsequently became positive, at any time in the future. We also included all other samples (urine, wound, respiratory or additional blood) taken during the 24 hours after initial blood culture and determined culture results. Patients who had any positive culture were referred to as “culture positive” and those who did not were “culture negative.” We reviewed the medical records of twenty culture negative septic shock patients to confirm that they were actually culture negative.
To examine the liberality of obtaining blood cultures, i.e., having a too-low threshold leading to excessive cases of culture negative sepsis, we conducted a post hoc analysis of a random sample of hospital adult admissions (40% of all admissions) weighted to match the temporal distribution of patients in the study. Using vital signs, white blood cell count, and PaCO2, we calculated hourly SIRS scores for each patient. Patients with 2 or more SIRS points had SIRS; if they had a blood culture obtained within the 12-hour window, they were deemed to have sepsis.
To determine the impact of including prior and subsequent infections in our dataset, we performed three sensitivity analyses by including patients with samples taken (1) only prior to presentation, (2) only subsequent to presentation (greater than 24 hours), and (3) those with both. We defined “presentation” as the 24 hours after initial blood culture required for inclusion in the study. We also performed a subgroup analysis of patients with septic shock, defined as requiring a vasopressor or inotrope at the time of presentation.
Statistical analysis
To compare characteristics by culture status, we used Pearson Chi-Square tests to compare categorical variables, Student’s t tests to compare normally distributed continuous variables and Mann–Whitney U tests to compare non-parametric continuous variables. Because patients had varying collections of laboratory test and vital signs and these missing data were not missing at random, but were obtained (or not obtained) based on perceived clinical need, we adjusted for missing data by binning these variables. These continuous variables were binned into deciles and the missing data became an 11th bin and treated as an 11-level categorical variable.12 This adjusts for both the presence or absence of a test and the numerical value of the result when obtained. For these variables, the number of patients with available data are listed in parentheses in the relevant tables.
To determine the variables associated with culture status and mortality, we created logistic regression models using Akaike Information Criteria (AIC) to select the final variables, except culture status, which was always included. We used all demographic; APACHE II scores; vital signs; laboratory values; antibiotic use; transfusions; use of mechanical ventilation, renal replacement therapy, and vasopressors; and culture results (Table 1) as covariates in all regression models. We also used this method to determine variables associated with mortality in culture negative and culture positive patients separately.
Table 1.
Univariate analysis comparing culture negative to culture positive patients
| Characteristic | Negative Cultures | Positive Cultures | |
|---|---|---|---|
| n = 9,288 | n = 1,105 | p-value | |
| mean ± sd frequency (%) |
mean ± sd frequency (%) |
||
| Age, years | 55 ± 17 | 57 ± 17 | <0.001 |
| Male sex | 5,573 (60) | 574 (52) | <0.001 |
| Race | 0.941 | ||
| African American | 1,004 (11) | 114 (10) | |
| Caucasian | 7,280 (78) | 875 (79) | |
| Other | 343 (4) | 40 (4) | |
| Physiological data and lab values on presentation | |||
| Temperature maximum, °C (8,945; 1,065) | 37.8 ± 0.9 | 37.9 ± 1.0 | 0.017 |
| Temperature minimum, °C (8,945; 1,065) | 36.6 ± 0.7 | 36.5 ± 0.6 | 0.109 |
| Median heart rate, min−1 (9,270; 1,101) | 98 ± 17 | 101 ± 19 | <0.001 |
| Median respiratory rate, min−1 (9,168; 1,105) | 21 ± 5 | 21 ± 6 | 0.235 |
| Median systolic blood pressure, mmHg (9,248; 1,098) | 122 ± 22 | 120 ± 23 | 0.043 |
| Median diastolic blood pressure, mmHg (9,239; 1,098) | 64 ± 12 | 63 ± 12 | 0.226 |
| Oxygen saturation maximum, % (9,222; 1,096) | 98 ± 2 | 99 ± 2 | 0.022 |
| Oxygen saturation minimum, % (9,222; 1,096) | 91 ± 10 | 90 ± 11 | 0.012 |
| White blood cell count, K/uL (7,050; 875) | 13.7 ± 13.0 | 14.0 ± 16.2 | 0.701 |
| Hematocrit, % (7,071; 881) | 30.8 ± 6.2 | 30.7 ± 5.9 | 0.461 |
| Platelets, K/uL (7,074; 877) | 200 ± 131 | 206 ± 140 | 0.538 |
| Bicarbonate, mmol/L (7,011; 862) | 25.4 ± 4.7 | 24.9 ± 5.2 | 0.013 |
| Blood urea nitrogen, mg/dL (6,986; 854) | 27 ± 21 | 29 ± 22 | 0.007 |
| Creatinine, mg/dL (7,064; 869) | 1.41 ± 1.40 | 1.37 ± 1.23 | 0.785 |
| Glasgow Coma Scale maximum (5,007; 614) | 13 ± 4 | 12 ± 4 | 0.377 |
| Glasgow Coma Scale minimum (5,007; 614) | 11 ± 4 | 11 ± 4 | 0.460 |
| APACHE II score (9,288, 1,105) | 12 ± 8 | 13 ± 8 | <0.001 |
| Antibiotic use | |||
| None | 3,899 (42) | 632 (57) | <0.001 |
| Received only on day 3 before presentation | 496 (5) | 119 (11) | <0.001 |
| Received on days 2 & 3 before presentation | 219 (2) | 14 (1) | 0.018 |
| Received on days 1, 2 & 3 before presentation | 1,715 (18) | 146 (13) | <0.001 |
| Received on days 1 & 3 before presentation | 111 (1) | 16 (1) | 0.468 |
| Received only on day 2 before presentation | 159 (2) | 15 (1) | 0.457 |
| Received on days 1 & 2 before presentation | 955 (10) | 31 (3) | <0.001 |
| Received only on day 1 before presentation | 1,734 (19) | 132 (12) | <0.001 |
| Before presentation | |||
| Mechanical ventilation | 1,939 (21) | 167 (15) | <0.001 |
| Renal replacement therapy | 142 (2) | 15 (1) | 0.794 |
| Plasma transfusion | 1,032 (11) | 154 (14) | 0.007 |
| Red blood cell transfusion | 1,245 (13) | 168 (15) | 0.104 |
| Platelet transfusion | 309 (3) | 51 (5) | 0.899 |
| Receiving vasopressors | 1,242 (13) | 156 (14) | 0.485 |
| Therapies initiated within 24 hours of presentation | |||
| Mechanical ventilation | 2,074 (22) | 239 (22) | 0.619 |
| Renal replacement therapy | 242 (3) | 37 (3) | 0.167 |
| Plasma transfusion | 309 (3) | 51 (5) | 0.030 |
| Red blood cell transfusion | 823 (9) | 97 (9) | >0.999 |
| Platelet transfusion | 420 (5) | 70 (6) | 0.011 |
| Outcomes | |||
| Hospital Survival | 8,137 (88) | 926 (84) | <0.001 |
| Post sepsis hospital length of stay, days (9,283; 1,105) | 3.5 ± 5.1 | 4.2 ± 5.9 | <0.001 |
Data are presented as mean ± or frequency (percentage). *If patients are missing values, the numbers of patients with values for that variable are shown in parentheses after the variable (n for negative cultures; n for positive cultures). Bonferroni adjusted p-value for statistical significance p<0.008.
For variable selection in building our logistic regression model, we used stepwise AIC variable selection to find reduced models as follows. Beginning with the full model, each variable was removed, one at a time, and the AIC calculated for a model including only the remaining variables. The model with the minimum AIC was selected and once again, each variable was removed one at a time, and the model with the minimum AIC was selected. From here, each remaining variable was removed and the AIC calculated for the model including the remaining variables, and each already-removed variable was re-entered to the model, one at a time, with the AIC calculated, and the model with the minimum AIC was selected. This stepwise procedure - removing each remaining variable as well as adding each removed variable and choosing the model with the minimum AIC - continued until the current model had a lesser AIC than any of the models removing a remaining variable or re-entering a removed variable. Culture status was forced to remain in the model and was not eligible to be removed, no matter what AIC removing it would produce. We used this model as our reduced model.
Discrimination of the models is presented as c-statistics ± standard deviation. Hosmer - Lemeshow test for goodness of fit and calibration plots were used to assess calibration for each regression.
We used the Bonferroni method to adjust for multiple outcomes. We had 6 analyses for the primary and secondary aims, 3 for the sensitivity analyses, and 3 for septic shock. Hence the adjusted p-values for statistical significance are .008, .017, and .017, respectively. We present both the unadjusted and adjusted p-values in a footnote for each table in the results. All analyses were performed using R version 3.2.1 (R Foundation for Statistical Computing, Vienna, Austria).13 This manuscript adheres to the applicable STROBE reporting guidelines.
Power analysis
Recent studies of sepsis have been powered to find a 20% difference in relative risk of mortality.13,14 For us to have 90% power to detect a 20% difference in mortality of 2.5% (12.5 v. 10%) between the culture positive and culture negative groups, with alpha = 0.05, required 8,000 patients (3,200 and 4,800 in each group). However, after collecting our data, our population was more than 8,000 patients but fewer than 3,200 in the culture positive group. We therefore did a post hoc power calculation using our actual numbers. With respective mortality of 16.2% and 12.4% with 1,105 culture positive and 9,208 culture negative patients with alpha = 0.05, we had 92% power to detect the actual 3.8% difference.
Results
Of the 10,393 patients who met inclusion criteria (Figure 1), 9,288 (89%) were culture negative and 1,105 (11%) were culture positive (666 – blood, 432 – urine, 42 – wound, 19 – respiratory; some patients had more than one positive site). By univariate analysis, culture negative patients were more likely to be younger, male, have a lower Acute Physiology and Chronic Health Evaluation II – Acute Physiology Score (APACHE II), and to have received antibiotics within the preceding 48 hours (Table 1). Culture positive patients were more likely to receive transfusion of plasma prior to presentation. However, presenting vital signs and laboratory values were fairly similar between culture negative and culture positive patients.
Figure 1:
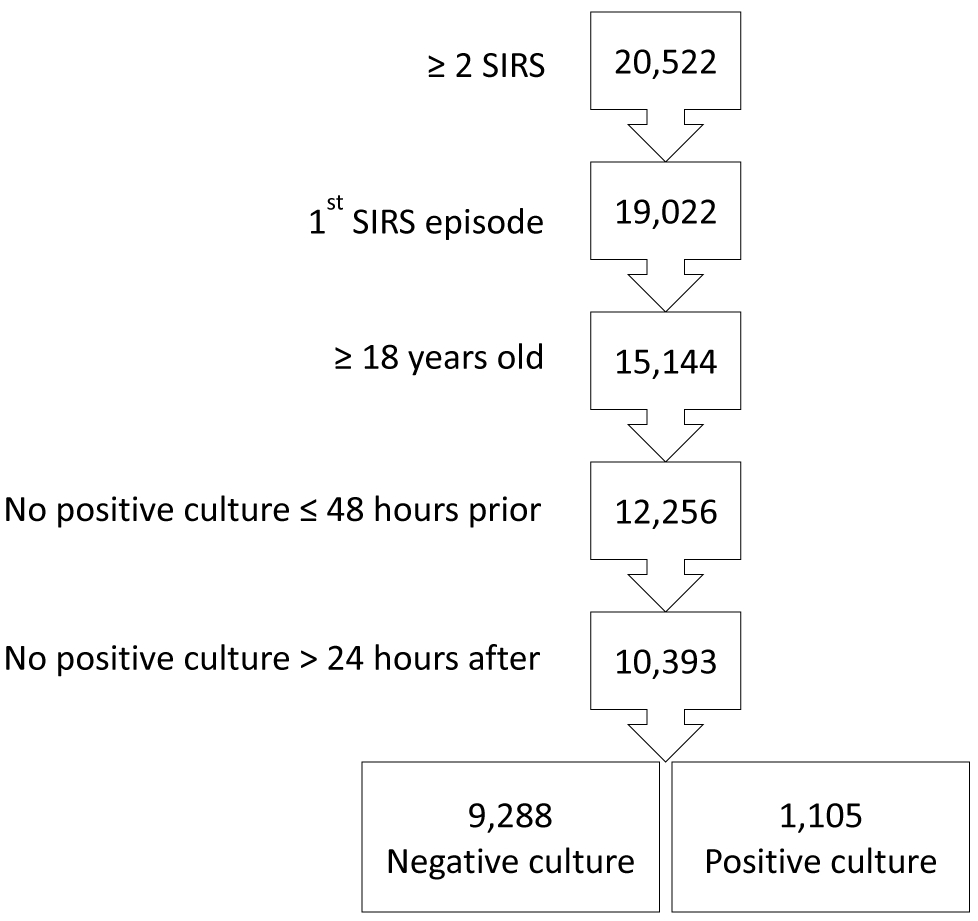
A total of 20,522 patients with 2 or more SIRS criteria were identified. Patients younger than 18 years of age (3,878) and subsequent sepsis episodes during hospitalization (1,500) were excluded, resulting in 15,144 patients. Then, patents were further excluded if they had grown positive cultures from samples taken 0 to 48 hours prior to presentation (2,888) or beyond 24 hours after presentation (1,863) resulting in 10,393 patients of whom 9,288 were culture negative and 1,105 were culture positive. SIRS = Systemic Inflammatory Response Syndrome; Culture negative = patients who had samples taken during the first 24 hours of presentation that did not grow a pathogen; Culture positive = patients who had samples taken during the first 24 hours of presentation that did grow a pathogen.
One thousand, three hundred eight patients (13%) did not survive to discharge. Patients who survived were more likely to be young, African American, and have a lower APACHE II (Table 2). Those who died were more likely to be culture positive, have abnormal vital signs and laboratory values, and have received transfusion of plasma and platelets, mechanical ventilation and renal replacement therapy (Table 2). Of those who died, 133 of 1,329 (10%) cultures obtained became positive compared to 533 of 9,9045 (6%) in those who lived, p < 0.001.
Table 2.
Univariate analysis comparing in-hospital survival to mortality
| Characteristic | No Hospital Mortality n = 9,063 |
Hospital Mortality n = 1,330 |
p-value |
|---|---|---|---|
| Age, years | 55 ± 17 | 62 ± 16 | <0.001 |
| Male sex | 5351 (59) | 796 (60) | 0.591 |
| Race | <0.001 | ||
| African American | 1002 (11) | 116 (9) | |
| Caucasian | 7122 (79) | 1033 (78) | |
| Other race | 343 (4) | 40 (3) | |
| Physiological data and lab values on presentation | |||
| Temperature maximum, °C (8,739; 1,271) | 37.8 ± 0.9 | 37.7 ± 1.2 | <0.001 |
| Temperature minimum, °C (8,739; 1,271) | 36.6 ± 0.6 | 36.3 ± 1.2 | <0.001 |
| Median heart rate, min−1 (9,047; 1,324) | 98 ± 17 | 100 ± 21 | <0.001 |
| Median respiratory rate, min−1 (8,974; 1,286) | 20 ± 5 | 23 ± 7 | <0.001 |
| Median systolic blood pressure, mmHg (9,030; 1,316) | 122 ± 22 | 114 ± 24 | <0.001 |
| Median diastolic blood pressure, mmHg (9,024; 1,313) | 65 ± 12 | 60 ± 13 | <0.001 |
| Oxygen saturation maximum, % (9,012; 1,306) | 98 ± 2 | 99 ± 3 | 0.005 |
| Oxygen saturation minimum, % (9,012; 1,306) | 91 ± 8 | 85 ± 16 | <0.001 |
| White blood cell count, K/uL (6,764; 1,161) | 13.1 ± 10.2 | 17.5 ± 24.7 | <0.001 |
| Hematocrit, % (6,786; 1,166) | 30.9 ± 6.1 | 30.6 ± 6.4 | 0.265 |
| Platelets, K/uL (6,785; 1,166) | 206 ± 133 | 168 ± 119 | <0.001 |
| Bicarbonate, mmol/L (6,731; 1,142) | 25.6 ± 4.4 | 23.5 ± 6.2 | <0.001 |
| Blood urea nitrogen, mg/dL (6,678; 1,162) | 25 ± 19 | 39 ± 26 | <0.001 |
| Creatinine, mg/dL (6,769; 1,164) | 1.33 ± 1.37 | 1.83 ± 1.35 | <0.001 |
| Glasgow Coma Scale maximum (4,895; 726) | 13 ± 3 | 9 ± 5 | <0.001 |
| Glasgow Coma Scale minimum (4,895; 726) | 12 ± 4 | 8 ± 5 | <0.001 |
| APACHE II score (9,063, 1,330) | 11 ± 7 | 19 ± 9 | <0.001 |
| Positive Culture | 926 (10) | 179 (13) | <0.001 |
| Antibiotic use | |||
| None | 3912 (43) | 619 (47) | <0.001 |
| Received only on day 3 before presentation | 568 (6) | 47 (4) | |
| Received on days 2 & 3 before presentation | 221 (2) | 12 (1) | |
| Received on days 1, 2 & 3 before presentation | 1595 (18) | 266 (20) | |
| Received on days 1 & 3 before presentation | 104 (1) | 23 (2) | |
| Received only on day 2 before presentation | 165 (2) | 9 (1) | |
| Received on days 1 & 2 before presentation | 879 (10) | 107 (8) | |
| Received only on day 1 before presentation | 1619 (18) | 247 (19) | |
| Before presentation | |||
| Mechanical ventilation | 1789 (20) | 317 (24) | <0.001 |
| Renal replacement therapy | 100 (1) | 57 (4) | <0.001 |
| Plasma transfusion | 821 (9) | 365 (27) | <0.001 |
| Red blood cell transfusion | 1215 (13) | 198 (15) | 0.153 |
| Platelet transfusion | 575 (6) | 123 (9) | <0.001 |
| Receiving vasopressors | 881 (10) | 517 (39) | <0.001 |
| Therapies initiated within 24 hours of presentation | |||
| Mechanical ventilation | 1735 (19) | 578 (43) | <0.001 |
| Renal replacement therapy | 149 (2) | 130 (10) | <0.001 |
| Plasma transfusion | 213 (2) | 147 (11) | <0.001 |
| Red blood cell transfusion | 715 (8) | 205 (15) | <0.001 |
| Platelet transfusion | 339 (4) | 151 (11) | <0.001 |
Data are presented as mean ± or frequency (percentage). * If patients are missing values, the numbers of patients with values for that variable are shown in parentheses after the variable (n for negative cultures; n for positive cultures). Bonferroni adjusted p-value for statistical significance p<0.008.
After adjustment for other factors using logistic regression, older age, worse vital signs, more abnormal laboratory values, mechanical ventilation, renal replacement therapy, plasma transfusion, and vasopressors at the time of presentation were associated with mortality (Table 3). However, having a positive culture itself was not significantly associated with mortality after adjusting for severity of illness and other factors (OR = 1.01 [95% C.I. 0.81, 1.26], p = 0.945), (Bonferroni significance p < 0.008) (Table 3). The model predicting mortality demonstrated very good discrimination (c-statistic = 0.87 ± 0.01) and good calibration with a line close to 45° (Supplemental Figure 1).
Table 3.
Factors associated with mortality by multivariable logistic regression
| Characteristic | odds ratio | 95% CI | p-value |
|---|---|---|---|
| Positive culture | 1.01 | (0.81, 1.26) | 0.945 |
| Age, 10 years | 1.397 | (1.331, 1.553) | < 0.001 |
| Physiological data and lab values on presentation | |||
| Oxygen saturation maximum < 96% | 1.68 | (1.23, 2.31) | 0.001 |
| Oxygen saturation unknown | 7.46 | (3.36, 16.56) | < 0.001 |
| Median respiratory rate, min−1 | 1.03 | (1.02, 1.05) | < 0.001 |
| Median heart rate, min−1 | 1.01 | (1.00, 1.01) | 0.007 |
| Median systolic blood pressure, mmHg | 0.99 | (0.99, 1.00) | 0.001 |
| Glasgow Coma Scale maximum, 8 - 10 | 1.65 | (1.13, 2.41) | 0.009 |
| Glasgow Coma Scale maximum, < 7 | 3.54 | (2.44, 5.14) | < 0.001 |
| Glasgow Coma Scale, unknown | 3.29 | (2.30, 4.70) | < 0.001 |
| Bicarbonate < 19 mmol/L | 1.41 | (1.05, 1.90) | 0.021 |
| APACHE II score | 1.11 | (1.09, 1.12) | < 0.001 |
| Before presentation | |||
| Renal replacement therapy | 2.21 | (1.38, 3.53) | 0.001 |
| Plasma transfusion | 1.94 | (1.60, 2.36) | < 0.001 |
| Red blood cell transfusion | 0.62 | (0.49, 0.78) | < 0.001 |
| Receiving vasopressors | 1.85 | (1.54, 2.23) | < 0.001 |
| Therapies initiated within 24 hours of presentation | |||
| Mechanical ventilation | 1.31 | (1.09, 1.59) | 0.005 |
95% CI = 95% confidence interval. Bonferroni adjusted p-value for statistical significance p<0.008.
After adjusting for other factors, we found that older and female patients were more likely to be culture positive, while the use of antibiotics within the preceding 48 hours was associated with culture negativity (Table 4). Higher heart rate and transfusion of plasma prior to presentation were also significantly associated with culture positivity. Notably, race, APACHE II, vital signs, and laboratory values were not discriminators between culture positive and negative sepsis, (c-statistic = 0.67 ± 0.01).
Table 4.
Factors associated with positive cultures by multivariable logistic regression
| Characteristic | odds ratio |
95% CI | p-value |
|---|---|---|---|
| Age, 10 years | 1.105 | (1.062, 1.149) | < 0.001 |
| Male sex | 0.71 | (0.62, 0.81) | < 0.001 |
| Median systolic blood pressure, mmHg | 1.00 | (0.99, 1.00) | 0.111 |
| Median heart rate, min−1 | 1.01 | (1.00, 1.01) | < 0.001 |
| APACHE II score | 1.01 | (1.00, 1.02) | 0.014 |
| Before presentation | |||
| Mechanical ventilation | 0.75 | (0.62, 0.91) | 0.004 |
| Plasma transfusion | 1.23 | (1.01, 1.50) | 0.043 |
| Red blood cell transfusion | 1.26 | (1.03, 1.53) | 0.026 |
| Antibiotic use | |||
| None (ref) | 1.00 | ||
| Received only on day 3 before presentation | 1.57 | (1.25, 1.97) | < 0.001 |
| Received on days 2 & 3 before presentation | 0.43 | (0.25, 0.75) | 0.003 |
| Received on days 1, 2 & 3 before presentation | 0.54 | (0.44, 0.66) | < 0.001 |
| Received on days 1 & 3 before presentation | 0.87 | (0.51, 1.49) | 0.602 |
| Received only on day 2 before presentation | 0.58 | (0.34, 1.00) | 0.051 |
| Received on days 1 & 2 before presentation | 0.22 | (0.15, 0.31) | < 0.001 |
| Received only on day 1 before presentation | 0.47 | (0.39, 0.58) | < 0.001 |
95% CI = 95% confidence interval. Bonferroni adjusted p-value for statistical significance p<0.008.
Culture positive patients had a longer post sepsis length of stay (4.2 ± 5.9 days vs 3.5 ± 5.1, p < 0.001) and greater mortality (179 of 1,105 (16%) vs 1,151 of 9,288 (12%), p < 0.001). Vasopressor use, renal replacement therapy and plasma transfusion within 24 hours prior to presentation was significantly associated with mortality in culture negative patients compared to culture positive patients. Initiation of renal replacement therapy after presentation was significantly associated with mortality in culture positive patients. Other factors associated with death were similar between culture negative and culture positive patients (Table 5). The models predicting mortality in each group showed very good and excellent discrimination (culture negative c-statistic = 0.87 ± 0.01 and culture positive c-statistic 0.92 ± 0.01), respectively, along with good calibration (Supplemental Figure 2).
Table 5.
Factors associated with mortality by multivariable logistic regression
| Culture Negative (n = 9,134) | Culture Positive (n = 1,087) | |||||
|---|---|---|---|---|---|---|
| Characteristic |
odds ratio |
95% CI | p-value | odds ratio | 95% CI | p-value |
| Age, 10 years | 1.384 | (1.305, 1.452) | < 0.001 | 1.397 | (1.184, 1.629) | < 0.001 |
| Physiological data and lab values on presentation | ||||||
| Oxygen saturation maximum, < 96% | 1.778 | (1.273, 2.482) | 0.001 | - | - | - |
| Oxygen saturation unknown | 7.101 | (3.115, 16.189) | < 0.001 | - | - | - |
| Median respiratory rate, min−1 | 1.033 | (1.021, 1.045) | < 0.001 | 1.04 | (1.001, 1.08) | 0.046 |
| Median heart rate, min−1 | 1.005 | (1.001, 1.009) | 0.022 | - | - | - |
| Median systolic blood pressure, mmHg | 0.995 | (0.991, 0.998) | 0.002 | 0.991 | (0.981, 1.001) | 0.068 |
| Diastolic blood pressure, mmHg | - | - | - | - | - | - |
| Glasgow Coma Scale maximum, 8 - 10 | 1.747 | (1.163, 2.624) | 0.007 | 1.855 | (0.559, 6.156) | 0.313 |
| Glasgow Coma Scale maximum, < 7 | 3.477 | (2.33, 5.187) | < 0.001 | 5.651 | (1.672, 19.101) | 0.005 |
| Glasgow Coma Scale unknown | 3.499 | (2.381, 5.14) | < 0.001 | 3.153 | (1.045, 9.513) | 0.042 |
| Platelets < 54 K/uL | - | - | - | 4.866 | (1.618, 14.634) | 0.005 |
| Bicarbonate > 31 mmol/L | 1.118 | (0.773, 1.616) | 0.554 | 2.22 | (0.785, 6.277) | 0.133 |
| Bicarbonate < 19 mmol/L | 1.389 | (1.01, 1.912) | 0.043 | 1.878 | (0.739, 4.772) | 0.185 |
| Bicarbonate unknown | 1.834 | (0.833, 4.034) | 0.132 | 0.639 | (0.212, 1.93) | 0.427 |
| BUN 25 – 30 mg/dL | 1.64 | (1.06, 2.55) | < 0.001 | - | - | - |
| BUN 31 – 38 mg/dL | 1.87 | (1.212, 2.885) | 0.005 | - | - | - |
| BUN 39 – 54 mg/dL | 2.641 | (1.703, 4.095) | < 0.001 | - | - | - |
| BUN > 54 mg/dL | 3.082 | (1.945, 4.886) | < 0.001 | - | - | - |
| APACHE II score | 1.106 | (1.091, 1.122) | < 0.001 | 1.155 | (1.108, 1.203) | < 0.001 |
| Before presentation | ||||||
| Mechanical ventilation | 0.838 | (0.665, 1.057) | 0.135 | - | - | - |
| Renal replacement therapy | 2.141 | (1.295, 3.539) | 0.003 | - | - | - |
| Plasma transfusion | 1.936 | (1.565, 2.396) | < 0.001 | 1.693 | (0.916, 3.127) | 0.093 |
| Red blood cell transfusion | 0.622 | (0.475, 0.814) | 0.001 | 0.526 | (0.265, 1.044) | 0.066 |
| Receiving vasopressors | 1.848 | (1.511, 2.261) | < 0.001 | - | - | - |
| Therapies initiated within 24 hours of presentation | ||||||
| Mechanical ventilation | 1.339 | (1.077, 1.663) | 0.008 | - | - | - |
| Renal replacement therapy on day of presentation | - | - | - | 5.3 | (1.815, 15.482) | 0.002 |
95% CI = 95% confidence interval. Bonferroni adjusted p-value for statistical significance p<0.008.
Examination of the liberality of obtaining blood cultures
Of the 57,415 patients, admitted on average twice in the 7 years, in our 40% random sample of adult hospital patients, 16,156 (28%) had one or more SIRS episodes. Of these 16,156 SIRS patients, 6,547 (41%) had blood cultures obtained within the 12-hour window of SIRS. Thus 11% (6,547/57,415) of patients had sepsis, or ~5.6% of hospital admissions.
Sensitivity Analyses
We performed sensitivity analyses by sequentially re-including patients who had cultures before or after the initial sepsis episode and found similar results. Culture positivity was not significantly associated with mortality in these sensitivity analyses: adjusted odds ratio (aOR) = 0.98 ([95% C.I. 0.81,1.18], p = 0.829), 1.01 ([95% C.I. 0.84, 1.21], p = 0.913) and 1.05 ([95% C.I. 0.90, 1.22], p = 0.562) (Bonferroni significance p < 0.017) for patients with prior infections, subsequent infections, or either, respectively.
Septic Shock
Of the 1,398 patients who presented with septic shock, 1,242 (89%) were culture negative and 156 (11%) were culture positive. Culture positive patients with septic shock had higher APACHE II (21 ± 8 vs 20 ± 9, p = 0.035) and mortality rate (46% vs 36%, p = 0.014) compared to culture negative patients.(Supplemental Table 1) After adjusting for other factors, age (aOR = 1.01 ([95% C.I. 1.002 – 1.03], p = 0.026) and initiation of renal replacement therapy (aOR = 2.04 ([95% C.I. 1.25 – 3.34], p = 0.004) was associated with culture positivity, while the use of antibiotics within the preceding 48 hours was associated with culture negativity, (c-statistic = 0.56 ± 0.02). (Supplemental Table 2)
Eight hundred eighty-one septic shock patients (63%) survived to discharge and 517 (37%) died. Patients who survived were more likely to be younger and have a lower APACHE II (Supplemental Table 3). Patients who died were more likely to be blood culture positive (72/5517 (14%) versus 84/881 (10%), p = 0.014), have worse vital signs and laboratory values, and have received transfusion, mechanical ventilation and renal replacement therapy (Supplemental Table 3). After adjustment for other factors using logistic regression, older age, lower oxygen saturation, lower Glasgow coma score, more abnormal laboratory values, transfusion of plasma, and renal replacement therapy were associated with mortality. Yet, having a positive culture was not significantly associated with mortality (OR = 1.36 [95% C.I. 0.88, 2.09]), p = 0.165 (Bonferroni significance p < 0.017), (Supplemental Table 4). The model predicting mortality in patients with septic shock demonstrated very good discrimination (c-statistic= 0.84 ± 0.01), along with good calibration (Supplemental Figure 3).
Discussion
We found that culture negative and culture positive patients had similar mortality after correcting for greater severity of illness and other factors. We also found that the clinical picture of laboratory values and vital signs had only fair discrimination between culture positive and culture negative patients, and that culture positive and culture negative patients had mostly similar risk factors for death. The most important predictor of culture negativity was receipt of antibiotics within the preceding 48 hours.
Patients with culture positive sepsis accounted for 11% of our patient population, a smaller percentage than reported in most previously published studies that ranged between 40-70%.6,8,16,17,18 This may be a result of including patients from all hospital units in our study, rather than restricting to the ICU. Furthermore, prior studies included culture results from samples collected prior to presentation or prospectively beyond 24 hours, and results may be impacted by pre-existing or subsequent hospital acquired infection. A strength of our study is that cultures were restricted to samples taken during the first 24 hours after meeting sepsis criteria, which better isolates the effect of an episode of sepsis. It is also possible that the higher culture negative rate in our study represents institutional practices reflecting a more liberal culture strategy or antibiotic administration prior to obtaining cultures. In contrast to one study that demonstrated a step-wise increase in rate of pathogen identification with severity of sepsis – culture positive rates with sepsis, severe sepsis and septic shock were 17%, 25% and 69%, respectively9 – we found 11% and 11% for sepsis and septic shock patients, respectively. This may be a result restricting culture analysis to samples obtained during the first 24 hours of presentation, changes in critical care over the past 20 years, or the large percentage of patients receiving antibiotics prior to presentation. Our goal was to analyze sepsis based on what was known to the physician at the time of presentation or within a short time thereafter. Our rate of sepsis, 5.6% of admissions, is similar to a large US study of 409 hospitals Rhee et al. that found a 6% rate.19 Given the same rate of sepsis as found by Rhee et al., further study is needed to assess the lower culture positive rate.19
We found culture negative patients had a lower APACHE II compared to culture positive patients. This may reflect a lower bacterial load or milder biological insult resulting in decreased sensitivity of laboratory culture testing. A negative culture could also be a result of sampling error or testing error (false-negative), but this is rare.20 Negative culture results may also reflect issues unique to the microorganism, viral or fungal origin, non-infectious source of symptoms, or genetic differences. Studies have shown that only about 1% of environmental bacteria are currently culturable,21,22 only half of bacterial species inhabiting the human mouth have been characterized, and the colonic flora is suspected to be mostly unidentified.23,24 Infection by any of these non-culturable organisms would result in culture negative sepsis. Similar to other studies, as prior antibiotic use was associated with culture negative sepsis (Tables 1 and 4), antibiotics may have sterilized the cultures.7,25 This suggests that presepsis antibiotics may either sterilize the cultures or select for a non-culturable infection. Another possibility is toxemia with very low grade or intermittent bacteremia. Further study is needed to determine why most septic patients have negative cultures and how best to treat them. Despite these reasons for negative cultures, obtaining cultures are important as, when positive, the sensitivities will affect the class of antibiotic and treatment duration.
Our results of no difference in mortality by culture status in both all septic patients and only septic shock patients is similar to many other studies.7,9-10,16-18 In contrast, Gupta et al. found a higher mortality in culture negative patients.11 They used the Nationwide Inpatient Sample and International Classification of Diseases (ICD) codes to identify hospital associated infections and deemed patients culture negative when billing records did not include coding for an infectious organism.11 However, the accuracy of ICD coding used to identify sepsis and hospital associated infection was not evaluated. Other studies have suggested the use of ICD codes to diagnose severe sepsis has limited accuracy19,26 and low sensitivity for identifying the organism.27,28 In addition, identified infections, co-morbidities and mortality were associated with the entire hospitalization and may be confounded by previously existing condition or hospital acquired conditions that occurred after or were not related to sepsis. Unlike our study, actual culture results were not used.
Compared to other studies, 9,10,16-18 our study demonstrated a much lower rate of hospital mortality - 13% overall, 12% for culture negative and 16% for culture positive patients. This may be explained by the lower mean APACHE II in our study (culture negative 12 ± 8 and culture positive 13 ± 8), inclusion of non-ICU and non-shock patients, and a generalized improvement in sepsis survival since the earlier studies.29 Although culture negative and culture positive patients demonstrated similar risk factors for mortality in our study, the mortality we observed in culture positive patients is likely attributed to the greater occurrence of risk factors, such as older age and higher APACHE II.
While the protective association between survival and pre-sepsis red blood cell transfusion that we found may be a result of increased oxygen carrying capacity or improvement in intravascular volume, prospective studies showed no improved survival with red blood cell transfusion in ICU and septic shock patients.30-32 Our findings may simply reflect that patients receiving red blood cell transfusion develop SIRS as a result of a transfusion reaction that is not a true infection, and therefore are more likely to survive. Further study is needed on this important point.
Our study has several limitations. First, patient and culture data were collected retrospectively; therefore, our findings represent associations only and prospective studies are needed to infer causality. Furthermore, retrospective data collection may be subject to missing data, and therefore less accurate than information collected prospectively.33 Second, our study was conducted at a single institution, a large academic medical center, and results may not be generalizable to other settings. However, our study is the largest study using clinical variables comparing the differences between culture negative and culture positive septic patients and included patients presenting from a broad clinical setting (ICUs, emergency department and general hospital wards). Finally, we used a SIRS based definition of sepsis. While a new organ dysfunction definition of sepsis has been proposed, it has not been fully accepted by some professional societies34 and our SIRS based definition of sepsis is consistent with the Centers for Medicare and Medicaid Services Quality Measure SEP-1: The Early Management Bundle for Severe Sepsis/Septic Shock. Additionally, voluminous literature uses SIRS based criteria permitting more generalizability of our results.
As culture negative sepsis is the most common type of sepsis in both our study and in the case-based study of 409 United States hospitals19, further study is needed to determine the effect of the duration and spectrum (broad v. narrow) of antibiotics in culture negative patients on outcome.
Conclusions
Culture negative and culture positive patients with sepsis demonstrate similar characteristics. After adjusting for severity of illness, having a positive culture is not significantly associated with mortality. Despite this, obtaining cultures are important as, when positive, the sensitivities will affect the class of antibiotic and treatment duration.
Antibiotic treatment within the prior 48 hours is the most important factor predicting culture negativity. However, culture negative and culture positive patients demonstrate similar mortality risk factors. Negative cultures should not give the clinician a false sense of reassurance – patients are still at risk of death, with the risks being dependent on severity of the physiologic derangements before and on the day of sepsis.
Supplementary Material
Supplemental Figure 1:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit
Supplemental Figure 2:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit
Supplemental Figure 3:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit.
Key Points.
Question: Our goal was to compare the characteristics between culture positive and negative patients, determine whether culture status is associated with mortality, and develop models predicting mortality in culture positive and culture negative patients separately.
Findings: We found that (1) patients with sepsis have similar characteristics regardless of culture status, (2) culture negativity is associated with receipt of antibiotics during the 48 hours preceding presentation, (3) after adjusting for disease severity, culture status itself is not associated with mortality and (4) vasopressor use, renal replacement therapy and plasma transfusion are significantly associated with morality in culture negative patients.
Meaning: Culture negative and culture positive sepsis patients demonstrate largely similar characteristics and mortality, and receipt of antibiotics prior to presentation is associated with culture negative sepsis.
Footnotes
The above authors have no conflicts of interest and no financial disclosures.
The publisher’s final edited version of this article is available at Anesth Analg
References:
- 1.Kumar G, Kumar N, Taneja A, Kaleekal T, Tarima S, McGinley E et al. : Nationwide trends of severe sepsis in the 21st century (2000-2007). Chest. 2011;140:1223–31. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. : The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016;315:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone RC, Balk RA, Cerra FB: Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. [DOI] [PubMed] [Google Scholar]
- 4.Kaukonen KM, Bailey M, Pilcher D, Cooper DJ, Bellomo R. Systemic inflammatory response syndrome criteria in defining severe sepsis. N Engl J Med. 2015;372:1629–38. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong-Briley D, Hozhabri NS, Armstrong K, Puthottile J, Benavides R, Beal S: Comparison of length of stay and outcomes of patients with positive versus negative blood culture results. Proc (Bayl Univ Med Cent). 2015;28:10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peduzzi P, Shatney C, Sheagren J, Sprung C: Predictors of bacteremia and gram-negative bacteremia in patients with sepsis. The Veterans Affairs Systemic Sepsis Cooperative Study Group. Arch Intern Med. 1992;152:529–535. [PubMed] [Google Scholar]
- 7.Phua J, Ngerng W, See K, et al. : Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit Care. 2013;17:R202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SC, Liao KM, Chen CW, Lin WC: Positive blood culture is not associated with increased mortality in patients with sepsis-induced acute respiratory distress syndrome. Respirology. 2013;18:1210–1216. [DOI] [PubMed] [Google Scholar]
- 9.Rangel-Frausto MS, Pittet D, Costigan M, Hwang T, Davis CS, Wenzel RP: The natural history of the systemic inflammatory response syndrome (SIRS). A prospective study. JAMA. 1995;273:117–123. [PubMed] [Google Scholar]
- 10.Brun-Buisson C, Doyon F, Carlet J, Dellamonica P, Gouin F, Lepoutre A: Incidence, risk factors, and outcome of severe sepsis and septic shock in adults. A multicenter prospective study in intensive care units. French ICU Group for Severe Sepsis. JAMA. 1995;274:968–974. [PubMed] [Google Scholar]
- 11.Gupta S, Sakhuja A, Kumar G, Mcgrath E, Nanchal RS, Kashani KB: Culture Negative Severe Sepsis - Nationwide Trends and Outcomes. Chest. 2016150:1251–1259. [DOI] [PubMed] [Google Scholar]
- 12.Engoren M Does erythrocyte blood transfusion prevent acute kidney injury? Propensity-matched case control analysis. Anesthesiology. 2010;113:1126–33. [DOI] [PubMed] [Google Scholar]
- 13.R Development Core Team (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Available at: http://www.R-project.org. Accessed October 4, 2016. [Google Scholar]
- 14.Mouncey PR, Osborn TM, Power GS, et al. Trial of early, goal-directed resuscitation for septic shock. N Engl J Med. 2015;372:1301–1311. [DOI] [PubMed] [Google Scholar]
- 15.Peake SL, Delaney A, Bailey M, et al. Goal-directed resuscitation for patients with early septic shock. N Engl J Med. 2014;371:1496–506. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Roberts D, Wood KE, et al. : Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. [DOI] [PubMed] [Google Scholar]
- 17.Vincent JL, Sakr Y, Sprung CL, et al. : Sepsis in European intensive care units: results of the SOAP study. Crit Care Med. 2006;34:344–353. [DOI] [PubMed] [Google Scholar]
- 18.Kethireddy S, Bilgili B, Sees A, et al. Culture-negative septic shock compared with culture-positive septic shock: a retrospective cohort study. Crit Care Med. 2018;46:506–12. [DOI] [PubMed] [Google Scholar]
- 19.Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA. 2017;318:1241–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peretz A, Isakovich N, Pastukh N, Koifman A, Glyatman T, Brodsky D: Performance of Gram staining on blood cultures flagged negative by an automated blood culture system. Eur J Clin Microbiol Infect Dis. 2015;34:1539–41. [DOI] [PubMed] [Google Scholar]
- 21.Ward DM, Bateson MM, Weller R, Ruff-Roberts AL: Ribosomal analysis of microorganisms as they occur in nature In: Marshall KC (Ed). Advances in Microbial Ecology. New York, Plenum Press, 1992, pp 219–286. [Google Scholar]
- 22.Amann RI, Ludwig W, Schleifer KH: Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wade W: Unculturable bacteria - the uncharacterized organisms that cause oral infections. J R Soc Med. 2002;95:81–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siqueira JF, Rôças IN: As-yet-uncultivated oral bacteria: breadth and association with oral and extra-oral diseases. J Oral Microbiol. 2013;5:21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Prost N, Razazi K, Brun-Buisson C: Unrevealing culture-negative severe sepsis. Crit Care. 2013;17:1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwashyna TJ1, Odden A, Rohde J, et al. : Identifying patients with severe sepsis using administrative claims: patient-level validation of the angus implementation of the international consensus conference definition of severe sepsis. Med Care. 2014;52:e39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones G, Taright N, Boelle PY, et al. : Accuracy of ICD-10 codes for surveillance of Clostridium difficile infections, France. Emerging Infect Dis. 2012;18:979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guevara RE, Butler JC, Marston BJ, Plouffe JF, File TM Jr, Breiman RF: Accuracy of ICD-9-CM codes in detecting community-acquired pneumococcal pneumonia for incidence and vaccine efficacy studies. Am J Epidemiol. 1999:149:282–89. [DOI] [PubMed] [Google Scholar]
- 29.Kaukonen KM, Bailey M, Suzuki S, Pilcher D, Bellomo R: Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000-2012. JAMA. 2014;311:1308–1316. [DOI] [PubMed] [Google Scholar]
- 30.Sadaka F, Aggu-Sher R, Krause K, O’Brien J, Armbrecht ES, Taylor RW: The effect of red blood cell transfusion on tissue oxygenation and microcirculation in severe septic patients. Ann Intensive Care. 2011;1:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hébert PC, Wells G, Blajchman MA, et al. : A multicenter, randomized, controlled clinical trial of transfusion requirements in critical care. Transfusion requirements in critical care investigators, Canadian Critical Care Trials Group. N Engl J Med. 1999;340:409–17. [DOI] [PubMed] [Google Scholar]
- 32.Holst LB, Haase N, Wetterslev J, et al. : Lower versus higher hemoglobin threshold for transfusion in septic shock. N Engl J Med. 2014;371:1381–91. [DOI] [PubMed] [Google Scholar]
- 33.Sedgwick P. Retrospective cohort studies: advantages and disadvantages. BMJ. 2014;348:g1072. [DOI] [PubMed] [Google Scholar]
- 34.Simpson SQ. New Sepsis Criteria: A Change We Should Not Make. Chest. 2016;149:1117–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit
Supplemental Figure 2:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit
Supplemental Figure 3:
The red line shows perfect calibration (45-degree line), the black line with the beige envelope shows the actual calibration with 95% confidence intervals. Hosmer - Lemeshow test was used to assess goodness of fit.


