Abstract
Over the past decades, several antiviral drugs have been developed to treat a range of infections. Yet the number of treatable viral infections is still limited, and resistance to current drug regimens is an ever-growing problem. Therefore, additional strategies are needed to provide a rapid cure for infected individuals. An interesting target for antiviral drugs is the process of viral attachment and entry into the cell. Although most viruses use distinct host receptors for attachment to the target cell, some viruses share receptors, of which sialic acids are a common example. This review aims to give an update on entry inhibitors for a range of sialic-acid-targeting viruses and provides insight into the prospects for those with broad-spectrum potential.
Introduction
Antiviral drugs are highly valuable for slowing down or clearing established viral infections. In addition, most of these drugs can be given prophylactically, which is beneficial for high-risk groups. However, the actual number of treatable viral infections is limited. Of the 219 viruses currently known to be infectious to humans [1], only nine are treatable with antiviral drugs [2]. For this reason, there is an urgent need to investigate further antiviral strategies.
Antiviral drugs can interfere with any step of the viral life cycle. An interesting target for antiviral therapy is the process of viral attachment and entry. However, entry inhibitors have seen limited clinical application, with only seven compounds being approved by the US Food and Drug Administration (FDA) for four virus types: human immunodeficiency virus, varicella zoster virus, herpes simplex virus and respiratory syncytial virus [2]. Moreover, these drugs are not commonly prescribed owing to their expense and the availability of more cost-effective alternatives. Therefore, novel ways of targeting viral entry processes should be urgently explored.
Viruses deploy several mechanisms for entry into the host cell, using distinct moieties on the cell membrane as a receptor. Such moieties include sialic acids, which are used as attachment receptor by several viruses. Sialic acids are a family of monosaccharides composed of a nine-carbon backbone. They are typically located at the terminal end of carbohydrate chains, attached to several glycoproteins and glycolipids on the eukaryotic cell membrane. For this reason, sialic acids are highly accessible for protein–ligand and receptor–ligand interactions [3]. Because of their ubiquitous expression on a wide variety of cell types, sialic acids play a part in many physiological processes [4, 5, 6, 7, 8, 9, 10,16]. Although they are widely found on mucosal tissue and appear in soluble form in mucosal secretions, the exact tissue distribution of sialic acids in the human body remains largely unknown, and the distribution might vary depending on the physiological conditions and gene expression profile of the individual [11].
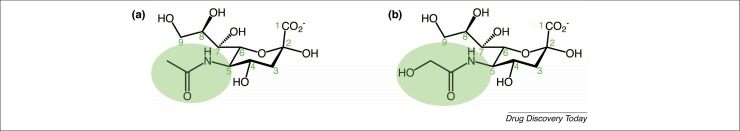
Sialic acids exist in different isoforms, depending on chemical substitutions and on the way they are linked to the terminal end of the abovementioned glycoconjugates 12, 13, 14. The most common isoform is neuraminic acid. The two main types of sialic acid found in mammals are two modifications of neuraminic acid: N-glycolylneuraminic acid (Neu5Gc) and N-acetylneuraminic acid (Neu5Ac) [4]. In humans, only Neu5Ac is present because humans lack the ability to produce the enzyme CMP-N-acetylneuraminic acid hydroxylase, which converts the acetyl group into a glycolyl group (Fig. 1 ) [15].
Figure 1.
Chemical structures of the two sialic acids predominantly present in mammals: (a) N-acetylneuraminic acid (Neu5Ac) and (b) N-glycolylneuraminic acid (Neu5Gc). The green ellipse highlights the only difference between the two: the substitution on the carbon atom at position 5. Neu5Gc is not present in humans because of a mutation in the gene encoding the enzyme CMP‐N‐acetylneuraminic acid hydroxylase.
In addition to their role in normal physiology, sialic acids are involved in the interaction of many pathogens with their host cells. Along with parasites and bacteria, a large number of viruses have been shown to interact with sialic acids for cellular attachment and entry [3]. However, the specific sialic acid derivative that serves as the attachment receptor differs according to the virus strain and depends on the receptor binding site of the viral attachment protein. For this reason, viral tropism is mostly dependent on the location of the attachment receptor. Interestingly, several viruses have been described to be not strictly dependent on sialic acids for attachment and entry, but rather use sialic acids as a co-receptor or enhancer of attachment. At least 16 viral genera have been identified to interact with sialic acids in some way for their entry process, as reviewed elsewhere [11].
Because of the profound role of sialic acids in the viral entry process, an entry inhibitor that prevents virus attachment to sialic acids would have the potential to serve as a broad-spectrum antiviral drug and might therefore be of high therapeutic relevance. This review provides an overview of the epidemiology of a selection of viruses that use sialic acids as their main cellular receptor to initiate entry, and we discuss the current development status of entry inhibitors targeting these viruses. Both sialic-acid-mimicking and sialic-acid-targeting drugs are discussed to highlight the most promising entry inhibitors, which are those with broad-spectrum activity.
Sialic-acid-targeting viruses and the development of entry inhibitors
The following sections provide an overview of the most clinically relevant sialic-acid-targeting viruses for which effective antiviral therapy is urgently needed. We discuss current modes of treatment for these viruses and highlight advances in the development of entry inhibitors, if applicable. The chemical structures of the entry inhibitors described in this review are depicted in Table 1 . Because virus–sialic acid binding characteristics at the molecular level have been extensively reviewed elsewhere 13, 17, they are not described in detail here.
Table 1.
Structures of compounds discussed in this review
| Compound | Structure | In vitro activity |
|---|---|---|
| Umifenovir (Arbidol) | 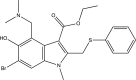 |
Influenza [67] EC50: 12.9 μM ± 1.4–30.9 μM ± 0.7a Zika 158, 191 EC50 (vero cells): 12.09 μM ± 0.77 10.57 μM ± 0.74; IC50 (A549 cells): 11 μM |
| DAS-181 (Fludase) | NA | Influenza 62, 192, 193 IC50: 0.25 nM to 1.0 nMa; EC50: 0.02 μM to 0.75 μMa |
| LSTc-bearing liposomesb | 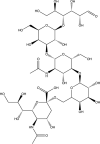 |
Influenza [181] IC90: 0.04 μM to 0.98 μMa |
| 6′SL-PAMAM conjugates | 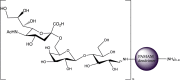 |
Influenza [182] IC50 range (for different compound variations): 3.4 μM to 220 μM |
| Linear polyglycerol sialosides | 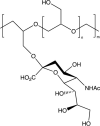 |
Influenza [183] IC50: 2.35 nM + 0.83 |
| BCX-2798 derivative | 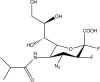 |
HPIV-1 [82] IC50: 0.39 μM |
| PAC-3066 | 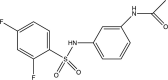 |
HPIV-3 [84] IC50: 37 μM |
| 17a | 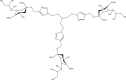 |
Adenovirus-D37 [103] IC50 (HCE cell-binding assay): 1.4 nM; IC50 (infection assay): 2.9 nM |
| AY4 | 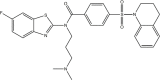 |
JC-polyomavirus [123]: dose-dependent inhibition from 100 μM to 1 mM |
| EK1 | 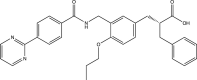 |
MERS-CoV IC50: 0.11 μM; OC43-CoV IC50: 0.62 μM [134] |
Range for several strains of influenza A and/or B.
6′SL-PAMAM, 6′-sialyllactose-polyamidoamine; HPIV, human parainfluenza virus; LSTc, sialylneolacto-N-tetraose c.
Influenza viruses
Belonging to the Orthomyxoviridae family, the influenza A virus is one of the most prevalent viral pathogens of our time, frequently causing infections in various mammalian and avian species. Seasonal human influenza epidemics, which are associated with the viruses’ susceptibility to undergo antigenic drift [18], are still a major threat to high-risk groups such as the elderly and immunocompromised, accounting for 3 million–5 million cases of severe illness and approximately 500,000 deaths every year [19]. Occasionally, the emergence of new influenza A virus strains can cause even more fatal pandemics [20].
The genome of influenza A and B viruses comprises eight segments of negative-sense, single-stranded RNA, and is enclosed by a lipid envelope [21]. The viral envelope is composed of mainly two proteins, haemagglutinin and neuraminidase, which are both crucial for infection. The primary criterion for the classification of influenza A viruses is based on the subtype of haemagglutinin (H1–H18) and neuraminidase (N1–N11), with the virus being named accordingly (e.g., H1N1) [19]. The trimeric haemagglutinin consists of two smaller subunits: HA-1 and HA-2. HA-1 contains a part of the stalk and forms the globular head domain, which carries the receptor-binding sites for sialic acid and thereby is of utmost importance for viral attachment to the host cell [23]. HA-2 forms the main part of the stalk, including the transmembrane region and cytosolic tail of the protein, and harbours the 20–23-amino-acid-long fusion peptide (104).
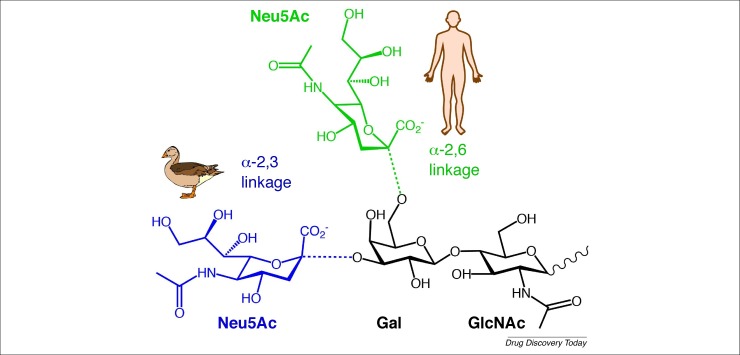
The haemagglutinin protein of human-adapted influenza strains initiates cell entry by binding to the trisaccharide sequence Neu5Ac-α(2-6)-Gal-β(1-4)-GlcNAc, also known as 6′-sialyllactosamine (6′-SLN). This sequence, linking Neu5Ac at the α-2,6 position to galactose, is the predominant sialic-acid-containing sequence on human tracheal epithelium and therefore serves as the primary binding site for human influenza A and B strains [24]. By contrast, avian influenza strains preferentially bind to Neu5Ac, linked at the α-2,3 position to galactose, which is present on the intestinal epithelium of (aquatic) birds (Fig. 2 ) 25, 26. These differences in linkage specificity between human-adapted and avian-adapted influenza A strains are an important determinant for their host cell tropism. The binding preference of haemagglutinin from avian and mammalian influenza virus strains for differentially linked sialic-acid subtypes, together with the distribution of these sialic acids and their pH stability, serve as a restrictive factor for zoonotic transmission of influenza and determine which species can be infected successfully [27].
Figure 2.
Schematic representation of the difference between avian (blue) and human (green) sialic-acid receptors. The only difference is the linkage between N-acetylneuraminic acid (Neu5Ac) and galactose (Gal). In humans, Neu5Ac is linked through an α-2,6 linkage to galactose, whereas for birds, Neu5Ac is linked to galactose via an α-2,3 linkage. In humans, the sequence Neu5Ac-α-(2-6)-Gal-β-(1-4)-GlcNAc is also known as 6′-sialyl-N-acetyllactosamine (6′-SLN).
After the haemagglutinin binds to sialic acids on the host’s respiratory epithelial cells, the virus is internalized by receptor-mediated endocytosis into endosomes. Under acidic conditions, the HA protein undergoes conformational changes that lead to the exposure of the HA-2 fusion peptide, which facilitates the low-pH-induced fusion of the viral envelope with the endosomal membrane. This consequently causes an influx of H+ ions via the M2 ion channel, which in turn leads to the release of the viral ribonucleoprotein into the cytoplasm 28, 29. After replication of the virus has taken place in the host cell, neuraminidase (also known as sialidase) is responsible for the enzymatic cleavage of sialic acid residues, with which it facilitates the release of virions into the extracellular milieu [30].
Prevention of influenza virus infections primarily relies on vaccination. However, vaccines are only moderately effective when they match circulating strains, and they can be ineffective if this is not the case [31]. This has led to interest in the development of influenza-specific antiviral drugs. For the 2019–20 influenza season, four antiviral drugs approved by the FDA were recommended for the treatment of persistent influenza infection in the United States: orally administered oseltamivir phosphate, inhalable zanamivir, intravenously administered peramivir (all of which are neuraminidase inhibitors) and oral baloxavir (which blocks viral RNA transcription) (https://cdc.gov/flu/professionals/antivirals/summary-clinicians.htm#Table1). In the European Union and the European Economic Area, only oseltamivir and zanamivir, sold under their respective trade names Tamiflu and Relenza, have been approved for individuals at a high risk of experiencing complications [33]. Although these drugs have been effective in treating the disease, the emergence of resistance against the currently used antivirals is a major concern for effective treatment and prophylaxis of influenza in the future 34, 35, 36, 37. Therefore, the development of novel drugs or combination therapies that target multiple pathways of the viral replication cycle remains necessary [38].
In this light, entry inhibitors might be a promising alternative. Entry inhibitors for influenza can be generally divided into two categories on the basis of their mechanism of action. The first category includes compounds that interfere with the initial attachment process of the virus. This can be done by directly targeting host sialic acids or by targeting the receptor-binding domain of the virus, using sialic acid analogues as a decoy receptor. Both of these mechanisms disable the ability of influenza-virus particles to bind sialic-acid molecules on the host cell. The other category includes compounds that bind to the haemagglutinin stalk, affecting the fusion of the viral envelope with the endosomal membrane. Although fusion inhibitors do not block the initial attachment, they do block the release of the viral genome into the cytoplasm and thereby interfere with the last step of the viral entry process.
In the past five years, a plethora of novel influenza entry inhibitors have shown potency in vitro 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58. An overview of compounds that have been further evaluated in the preclinical or clinical phase is given in Table 2, Table 3 . Interestingly, only a few of the highlighted entry inhibitors interfere with the initial attachment process of the influenza virus (i.e., with the receptor-binding site of haemagglutinin or with sialic acids). The primary reason for this is that compounds that target the receptor-binding site need to bind the haemagglutinin protein in such a way that they do not lose efficacy on seasonal mutation of the virus, as is the case with neutralizing antibodies from the host. In this context, it should be remarked that haemagglutinin is found in a trimeric form [59]. This implies that it ideally binds to three sialic-acid moieties on the cell surface. Such a concept (i.e., multiple ligands binding to multiple targets) is known as multivalency and is highly important in several biological and pathogenic interactions (Box 1 ).
Table 2.
Influenza entry inhibitors in the clinical phase
| Compound | Type | Sponsor | Proposed target region | Clinical status | Trial identifier | Refs |
|---|---|---|---|---|---|---|
| DAS-181 | Sialidase | Ansun Biopharma | Host sialic acid | Phase IIb study completed assessing safety and efficacy in otherwise healthy individuals infected with influenza A | Phase I: NCT00527865; NCT01173224; NCT01651494 Phase II: NCT01037205; NCT01740063; NCT04298060 | 61, 62, 159, 192 |
| CR6261 | Monoclonal antibody | US National Institute of Allergy and Infectious Diseases | HAa stalk region | Phase II study completed assessing efficacy of the drug in healthy individuals challenged with H1N1 compared to placebo | Phase I: NCT01406418 Phase II: NCT02371668 | 194, 195 |
| Umifenovir | Indole derivative | Pharmstandard | HA stalk region | Phase III study recruiting in China assessing the safety and efficacy of the drug in combination with oseltamivir in hospitalized influenza patients; phase IV study with unknown status | Phase III: NCT03787459 Phase IV: NCT01651663 | 67, 69, 196 |
| CT-P27 | Monoclonal antibody | Celltrion | HA stalk region | Phase IIb study recruiting assessing the safety and efficacy of the drug in influenza-A-infected patients compared to placebo | Phase I: no data posted Phase II: NCT02071914; NCT01740063; NCT03511066; KCT0002211 | NA |
| MEDI8852 | Monoclonal antibody | MedImmune LLC | HA stalk region | Phase IIb withdrawn (owing to delay in site enrolment timelines) | Phase I: NCT02350751 Phase II: NCT02603952; NCT03903718 | 70, 197, 198 |
| MHAA4549A | Monoclonal antibody | Genentech | HA stalk region | Phase II study completed assessing efficacy of the drug in hospitalized influenza-A-infected patients compared with oseltamivir | Phase I: NCT01877785; NCT02284607 Phase 2: NCT01980966; NCT02623322; NCT02293863 | 71, 199, 200 |
| VIS-410 | Monoclonal antibody | Visterra | HA stalk region | Phase II study recruiting to assess safety and efficacy of the drug in hospitalized influenza-A-infected patients compared with oseltamivir | Phase I: NCT02045472 Phase II: NCT02989194; NCT02468115; NCT03040141 | 72, 201, 202, 203 |
aHA, haemagglutinin.
Table 3.
Influenza entry inhibitors in the preclinical phase
| Compound | Type | Proposed target region | Preclinical relevance | Refs |
|---|---|---|---|---|
| Sialic acid-functionalized Qβ-bacteriophage capsids | Sialic-acid-functionalized bacteriophage capsid | Receptor binding site of HAa | In vitro and in vivo protection against two H3N2 strains | [184] |
| IY7640 | Small molecule inhibitor | HA stalk region | In vitro protection against H3N2, H1N1 and influenza B; in vivo efficacy against H1N1 | [204] |
| JNJ4796 | Small molecule inhibitor | HA stalk region | Protected mice against lethal and sublethal H1N1 influenza challenge after oral administration | [205] |
| Carbinoxamine maleate; chlorpheniramine maleate | Histamine antagonist | Endocytic pathway | Showed in vivo protection after challenge with H7N9 influenza A virus | [206] |
| 1428A33/1; 1428B5/1; F3A19 | Monoclonal antibody | Receptor binding site of HA | In vivo efficacy against the A(H1N1)pdm09 strain | [60] |
| Dialtizem | Calcium channel blocker | Voltage-gated Ca2+ channel Cav1.2 | In vivo efficacy against H1N1 | [207] |
| S-KKWK | Lipopeptide | HA stalk region | Prevented HA-2 rearrangements and subsequent membrane fusion of several H1N1 strains and H3N2 in vitro, and protected mice from lethal infection with H1N1 | [208] |
| Linear polyglycerol sialosides | Linear polyglycerol sialosides | Receptor binding site of HA | In vitro protection against H3N2 and both avian H3N2 and avian H7N1; in vivo protection against H3N2 | [183] |
| Urumin | Frog-derived peptide | HA stalk region of H1-type HA | In vitro efficacy against several H1N1 strains; in vivo protection against lethal H1N1 influenza A virus infection | [209] |
| 3′-SL- and 6′-SL-linked PAMAM dendrimers | PAMAM conjugates | Receptor binding site of HA | In vivo protection against lethal H1N1 | 41, 182 |
| Multivalent carbohydrate-binding modules | Sialidase derivatives | Host sialic acid | In vivo protection against lethal H1N1 | [63] |
aHA, haemagglutinin; PAMAM, polyamidoamine; SL, sialyllactose.
Box 1. The concept of multivalency in biological systems.
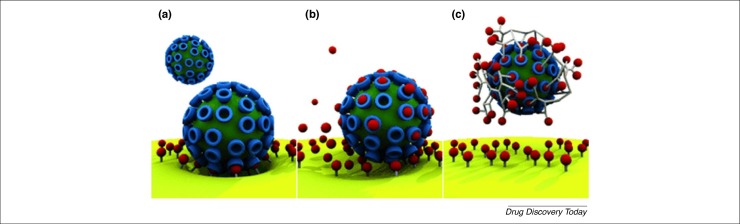
To the best of our knowledge, the concept of multivalency has been observed with all viral attachment proteins that have been structurally resolved to date. Multivalency is in fact common in biological systems [187]. It is understood that nature uses it to increase binding strength and consequently selectivity 188, 189. On the basis of this, many receptor analogues have been designed to be multivalent: that is, they were designed as macromolecules containing many repetitive units of the viral receptor to better bind to the virus (Fig. 3 ). Another advantage of multivalent drugs over monovalent compounds is that a lower dose can be used to elicit the required effect [188]. The design of these compounds further requires the fine-tuning of several parameters such as scaffold geometry and the flexibility of the linkers, their length and their relative distance 182, 188, 190.
Figure 3.
Schematic representation of multivalency. (a) Multivalent interaction of a virus with its receptors on the host cell. (b) Monovalent interaction of a drug with the viral attachment protein. (c) Multivalent interaction of a drug with the viral attachment protein. Reproduced, with permission, from [188].
Alt-text: Box 1
The concept of multivalency is widely used in the development of potentially broad-spectrum sialic-acid-based receptor analogues, which are described in further detail later in this review. Apart from using multivalent drugs to block the receptor-binding site, another strategy is to use monoclonal antibodies that bind to more conserved parts of the HA1 head domain [60]. However, none of the abovementioned compounds has been evaluated in clinical trials.
Besides targeting haemagglutinin to block the attachment process, another way is to directly target host sialic acids. Arguably the most promising influenza entry inhibitor using this method is DAS-181, which is a recombinant fusion protein that can enzymatically cleave terminal sialic acids from carbohydrate chains on the host-cell membrane 61, 62. Because DAS-181 has shown promising results as a broad-spectrum drug, we discuss it later on in this review.
Another study focused on using multivalent proteins derived from the binding domains of bacterial sialidases to target host sialic acids. However, the compounds, which mask the receptor rather than cleaving it, only worked prophylactically, indicating that the virus successfully competed for sialic-acid binding when the same dose was administered therapeutically [63].
Most of the compounds that have been evaluated in preclinical or clinical settings target the stalk domain, the part of the haemagglutinin protein that is involved in the fusion process and is generally more conserved [64]. One interesting example is umifenovir (trade name: Arbidol), a drug that has been licensed in Russia and China and is claimed to have activity against a broad variety of influenza strains 65, 66, 67, 68. The proposed working mechanism is that by interacting with the upper region of the stalk, the drug prevents the low-pH-induced protein rearrangements that are necessary to elicit fusion of the virus with the cell membrane [69]. Other clinically relevant compounds that target the stalk include monoclonal antibodies 70, 71, 72. Of these, five are currently in clinical evaluation (Table 2).
Parainfluenza virus
The Paramyxoviridae family consists of enveloped negative-sense, single-stranded RNA viruses, among which are the parainfluenza viruses. Human parainfluenza viruses (HPIVs) are a common cause of respiratory disease in infants, young children, elderly people and immunocompromised individuals [73]. HPIV infections are the second most prevalent cause of respiratory-disease-related hospitalizations in children under five years of age, after respiratory syncytial virus infection [74]. Symptoms range from mild and cold-like to more severe, including pneumonia and croup. These symptoms can be worse in patients who are already suffering from chronic airway diseases such as asthma and chronic obstructive pulmonary disease [75].
Four types of HPIV have been described: HPIV-1–HPIV-4. Their main outer proteins are haemagglutinin-neuraminidase (HN), which is responsible for attachment to sialic acids, and the fusion protein, which mediates fusion of the virus with the host cell membrane [75]. All HPIV strains are known to bind to sialic acids, preceding cell entry. HPIV-1 and HPIV-3 have been shown to use α-2,3-linked sialic acids, and HPIV-3 also binds to α-2,6-linked sialic acids [76]. This contrasts with avian influenza viruses, which also bind to α-2,3-linked sialic acids but are very rarely transmitted from human to human because they have a different linkage specificity [77]. To our knowledge, the linkage specificity of HPIV-2- and HPIV-4-binding sialic acids has not yet been described.
Because vaccination strategies have not yet proven to be effective in eliminating the disease [78], patients suffering from HPIV infection could benefit from antiviral therapy. Currently, no drugs have been approved for treating HPIV infections. Because HPIV needs sialic acids for entry, research has been done regarding the development of suitable antivirals for blocking this mechanism. The HN inhibitor and sialic-acid derivative BCX-2798 has been proven to effectively protect mice against challenge with a 90% lethal dose of a chimeric strain of HPIV-1, in which the fusion and HN proteins of Sendai viruses were replaced by those of HPIV. Moreover, the compound had prophylactic potential against chimeric HPIV-3 79, 80, 81. BCX-2798 was chemically modified by Eveno et al. to bind covalently to a key catalytic component of the HN protein of HPIV-1. Hereafter, HPIV-1 replication in vitro was reduced by 13-fold compared with its parent compound [82]. Research into the compound is still ongoing.
Recently, the small molecule CM9 was found to interact with the HN protein in vitro and in vivo, causing the fusion protein to undergo its conformational change prior to the actual attachment of HPIV-3 to cells, which led to a block in viral infectivity [83]. It was found that the molecule interacts with the second sialic-acid-binding site of the HN protein without disturbing the receptor-binding capacity of the virus. This proof-of-concept study led to the design of another compound, named premature activating compound-3066 (PAC-3066), which targets the same second sialic-acid-binding site, but is 100 times more effective in vitro than its predecessor [84]. Aside from these studies, only the recombinant sialidase DAS-181 has made it into clinical trials.
Adenovirus
Human adenoviruses (AdVs) are non-enveloped double-stranded DNA viruses with a size range of 90–100 nm. They belong to the family of Adenoviridae and are divided into seven different species, AdV-A to AdV-G, which can be further classified into more than 60 different subtypes [85]. Young children are the most susceptible to AdV infection, and account for roughly 80% of cases. This is mainly due to their lack of humoral immunity against the virus [86].
A wide variety of symptoms can arise following adenoviral infection, including gastrointestinal and ocular inflammation. Most commonly, however, symptoms manifest in the respiratory tract, and they vary from common cold-like symptoms, such as sneezing and coughing, to pneumonia and bronchitis [87]. One of the less common symptoms is epidemic keratoconjunctivitis (EKC), or inflammation of the ocular surface tissue, which is caused by AdV-B1, AdV-E and, in the more severe form, by AdV-D subtypes. EKC is mostly characterized by eye redness, itchiness, soreness and excessive tearing [85]. Although some antiviral drugs, such as the DNA polymerase inhibitor cidofovir, have been reported to be helpful in treating AdV-related disease in people who have received an organ transplant 88, 89, no drug has been made available to the clinic because the virus is mostly self-limiting and can be cleared within 3 weeks [85]. However, susceptible patient groups might benefit from broad-spectrum antiviral drugs.
The two main receptors for adenovirus attachment are the coxsackievirus-adenovirus receptor (CAR) on epithelial cells [90] and the ubiquitously expressed membrane cofactor protein (or CD46) [91]. After binding, a second interaction with αV-integrin is necessary for initiation of endocytosis 92, 93. Of all adenovirus strains that have been described to cause EKC, AdV-D37, AdV-D8 and AdV-D19a account for most severe disease [85]. Interestingly, all of these strains have been reported to interact with sialic-acid residues for binding to the host cell 94, 95, 96. The reported adenoviruses all share a prevalence for α-2,3-linked sialic acids, which contrasts with the requirement of human influenza viruses to solely bind α-2,6-linked sialic acids [97]. This distinction can be explained by AdVs having a tropism for ocular tissue, where α-2,3-linked sialic acids are abundant, rather than respiratory epithelium. The ocular tropism of the above adenoviruses also explains the onset of symptoms affecting the eyes [98]. The AdV-D37 spike protein knob domain has been found to contain three sialic-acid-binding sites, all of which are involved in the interaction with sialic-acid residues that are attached to a GD1a glycan motif on host corneal epithelial cells [99]. The importance of this interaction was highlighted by the fact that after pretreatment of these cells with GD1a-based monoclonal antibodies and soluble GD1a, inoculation with AdV-D37 did not lead to infection. The implications of these findings for the in vivo setting have not yet been fully elucidated [99].
In 2005, Johansson and colleagues were the first to design multiple multivalent 3′-sialyllactose derivatives, conjugated to human serum albumin, which were able to effectively bind to AdV and inhibit infection in a model of human corneal epithelial cells [100]. Furthermore, they found that the multivalent molecules yielded a 1000-fold higher inhibitory effect than the monovalent form, which can be explained by the fact that AdV uses several of its fibre proteins for entry into the host cell. The same group confirmed their findings by using sialic-acid conjugates instead of 3′-siallylactose and showed that treatment with this compound led to virus aggregation [101]. On the basis of these results, Spjut et al. designed trivalent sialic acid-based inhibitors that bound to all three of the AdV-D37 sialic-acid-binding sites on its knob domain. The most promising compound, ME0322, was a potent inhibitor of AdV-D37 in vitro [102]. Based on this, an even more potent trivalent compound was designed, which was highly effective in cell-attachment and -infection assays [103]. In addition to binding CAR, the gastroenteritis-causing AdV-G52 was recently found to bind α-2,8-linked polysialic acids, which are a unique type of post-translationally modified sialic acid that is usually found in brain tissue [104]. Also, AdV-D26 has recently been shown to rely on sialic acids for cell entry [105]. Future insights into the binding mechanisms of these viruses might pave the way for new types of potentially broad-spectrum entry inhibitors for sialic-acid-dependent AdV subtypes.
Polyomavirus
Polyomaviruses include a variety of non-enveloped double-stranded DNA viruses [106]. Their icosahedral-shaped capsid consists of three proteins, namely VP1, which is the viral attachment protein and makes up most of the viral capsid [107], VP2 and VP3, which are probably involved in insertion of the viral genome into the host cell nucleus [108]. Around 14 different human polyomaviruses have been identified [109]. The two strains that are most infectious to humans are BK virus (BKV) and JC virus (JCV), both of which can be further divided into different subtypes [107]. The newly discovered Merkel cell polyomavirus was recently added to this list [110].
Polyomavirus infections are highly prevalent among the global population. An estimated 80% of humans are infected in their lifetime, with the first infection usually occurring in early childhood [107]. Mostly, the virus latently resides in the kidneys and peripheral blood, making the infection asymptomatic or mildly symptomatic for the majority of infected individuals [111]. However, in immune-deficient individuals, the virus might start to replicate again and cause specific disease. In this context, BKV is a common cause of nephropathy, mostly occurring in people who have undergone a kidney transplant 112, 113. By contrast, recurring infections with JCV can lead to progressive multifocal leukoencephalopathy. It is thought that JCV infiltrates the brain tissue via B cells in the blood, from where it can cause a lytic infection in myelin-producing oligodendrocytes and in astrocytes 114, 115. In humans, Merkel cell polyomavirus has been found to be the primary cause of Merkel cell carcinoma, which is a highly aggressive form of skin cancer 110, 116.
Different types of human polyomavirus interact with different sialic-acid residues on host cells. JCV has been found to interact with α-2,6-linked sialic acids 117, 118, as well as with α-2,3-linked sialic acids [119]. BKV was found to interact with α-2,3-linked sialic acids only [120]. Merkel cell polyomavirus has also been found to rely on α-2,3-linked sialic acids for attachment, although it can use α-2,6-linked sialic acids as well 121, 122.
With knowledge of the receptors used for attachment, it might be possible to design antiviral drugs based on the viral entry mechanism. Using computational screening, Yatawara et al. identified four different compounds that blocked JCV infectivity in an in vitro setting involving the astrocyte cell line SVGA. Of those, the small-molecule inhibitor AY4 was the most effective inhibitor of infection, albeit with low affinity. AY4 was shown to directly bind to the VP1 attachment protein to block viral attachment [123]. Because the options for treating polyomavirus infections and related diseases are limited, the development of novel antiviral compounds is important.
Coronavirus
Coronaviruses (CoVs) belong to the Coronaviridae family and are enveloped, positive-sense, single-stranded RNA viruses. The abundant spike (S) glycoprotein functions as the viral attachment protein. The S protein is composed of two subunits, S1 and S2: S1 contains the receptor-binding site, whereas S2 contains the fusion machinery [124]. CoVs are divided into four subtypes (α, β, γ and δ). Of these, only certain α-CoVs (CoV-229E and CoV-NL63) and β-CoV (OC43-CoV, HKU1-CoV, SARS-CoV, MERS-CoV and the recently emerged SARS-CoV-2) infect humans, and are all thought to be of zoonotic origin 124, 125, 126. CoVs have emerged unexpectedly three times over the past two decades, with the most recent example being the SARS-CoV-2 pandemic, which causes respiratory disease designated as COVID-19 [126]. Overall, most CoVs cause mild respiratory disease varying from cough to fever. However, symptoms can be worse, especially in susceptible groups such as elderly people and individuals with an underlying illness. To manage the current pandemic and prevent future epidemics or pandemics, the development of novel antiviral compounds has become urgent.
Many clinical trials of potential antiviral compounds such as nucleoside analogues and corticosteroids have failed [127]; thus, other strategies for treating patients are needed to prevent future outbreaks. In that respect, entry inhibitors that target the viral receptor or receptor-binding site of the CoV S protein might be of therapeutic value. Several of the β-CoVs have been shown to interact with sialic acid moieties for cell entry processes. Both OC43-CoV and HKU1-CoV specifically target 9-O-acetylated sialic acids 124, 128, 129, which are modified sialic acids that are thought to have an important role in many biological and pathological processes 130, 131. Attachment occurs via the receptor-binding site in the S1 domain of the S protein 124, [132], 133. Recently, the small molecule EK1, derived from the HR2 domain of the OC43-CoV spike protein, was found to elicit high fusion-inhibitory activity against various human CoVs, underlining the potential for its development as a broad-spectrum drug [134]. MERS-CoV interacts with dipeptidyl peptidase 4 as its receptor to enter the cell 135, 136, using α-2,3-linked sialic acids as a co-receptor for attachment 137, 138. Although monoclonal antibodies that target the sialic-acid-binding S1a domain might provide a synergistic effect in combination treatment regimens, sialic acids do not seem to be essential for MERS-CoV infectiveness [139].
Enterovirus D68
Enterovirus D68 (EV-D68) is a member of the Picornaviridae family, which is a large family consisting of non-enveloped, single-stranded RNA viruses [140]. Unlike most other enteroviruses, EV-D68 infects the upper respiratory tract, primarily causing disease in the paediatric population [141]. In most patients, the virus causes symptoms similar to those of the common cold. However, in some cases, infections are accompanied by acute flaccid myelitis (AFM), a polio-like disorder that is characterized by lesions in the spinal cord, which can eventually cause paralysis 142, 143. Most of these patients suffer from underlying respiratory disease, have a history of organ transplantation or are immunocompromised in another way [144]. However, whether there is a causal relationship between the virus and AFM pathogenesis is still a matter of debate 141, 145, 146. The emergence of more severe cases, together with recent outbreaks, have led to an increased interest in the virus and in disease pathogenicity.
In recent years, EV-D68 has been found to exploit sialic acids for attachment and for cell entry [147]. It was found that these sialic acid molecules are terminally linked via α-2,6-linkers [148] to intercellular adhesion molecule 5 (ICAM5), which is found in the upper respiratory tract as well as on neurons [149], with the latter providing a possible link to the pathogenesis of AFM. Interestingly, it was found that EV-D68 can also exploit α-2,3-linked sialic acid in the lower respiratory tract for infection, increasing the probability of the virus successfully infecting its host [150]. Owing to this, interest in the development of entry inhibitors for the currently untreatable infection has increased. Rhoden et al. were among the first to test existing antivirals for the treatment of EV-D68 in a cell-based assay, one of which was DAS-181. They found that the drug was able to reduce the viral cytopathic effect on infection with three EV-D68 strains isolated from the 2014 outbreak, and one prototype strain [151]. However, it is unclear whether these results can be translated to an in vivo system, partly owing to the lack of suitable animal models.
Zika virus
The Zika virus (ZIKV) is a single-stranded, enveloped RNA virus in the Flaviviridae family. It is transmitted via mosquitoes, but once it has infected a host, it can be transmitted through the congenital and perinatal route, and it can be sexually transmitted [152]. Infection with ZIKV can affect all age groups and is usually asymptomatic or mildly symptomatic, generally resolving within 2 weeks. However, several reports have indicated that ZIKV infection of pregnant women can cause congenital brain abnormalities of the foetus such as microcephaly, in which the foetus has an abnormally small brain, leading to severe cognitive and motor deficits 153, 154. Although the virus has caused infections only sporadically in the past, several outbreaks have emerged since 2007, which shows the need for preventive measures [152]. People are usually advised to use mosquito nets or DEET-containing insect repellents to prevent initial transmission, but pregnant women who have become infected might benefit from antiviral therapy.
ZIKV has been shown to infect a range of cells from the reproductive tract, as well as certain brain cell types, including neuronal cells from the foetal brain [155]. The primary surface protein of ZIKV is the envelope (E) protein. It is thought that the E protein interacts with several cellular attachment factors before cell entry, with each individual interaction contributing to its binding avidity to the host cell [156]. Once the binding strength has reached the threshold, the virus is internalized via clathrin-mediated endocytosis.
ZIKV was recently found to use α-2,3-sialic acids for cell entry, supposedly as a factor contributing to ZIKV internalization after attachment has taken place [157]. In a study conducted by Fink et al., it was found that the anti-influenza drug umifenovir caused inhibition of ZIKV infection in a range of cell types, including vaginal and cervical epithelial cells. Although the authors propose that the drug inhibits viral entry, the exact mechanisms of inhibition are unknown [158]. Additional insights into the entry mechanism and specific attachment factors of ZIKV might lead to specific therapy for pregnant women at risk of infection.
Antiviral drugs with broad-spectrum potential
Although most studies on antiviral therapies focus on virus-specific pathways, it might be more promising to search for drugs that affect mechanisms common to multiple viruses: for example, the mechanism of viral entry into the host cell. To develop such broad-spectrum entry inhibitors in the context of sialic acid-targeting viruses, they should either target the sialic-acid residues or mimic them by making use of drugs that function as decoy receptors to block the virus from binding to the host cell. Below, both strategies are discussed.
Sialic-acid-targeting drugs
One of the potentially broad-spectrum antiviral drugs that target the entry pathway is DAS-181, a recombinant neuraminidase analogue that cleaves sialic-acid residues from carbohydrate chains on host epithelia. In this way, virus binding to the host cell can be indirectly inhibited. More specifically, DAS-181 is a fusion protein that contains both the catalytic domain of the sialidase enzyme of Actinomyces viscosus and the anchoring domain of human amphiregulin [61]. Because the drug is administered via inhalation, it prevents entry of the virus directly at the airway epithelium, which is thought to keep adverse systemic side effects to a minimum [159]. DAS-181 is considered to be a promising candidate for the treatment of infections with sialic-acid-targeting viruses that are transmitted via the respiratory tract because it targets the host receptor rather than the virus, and thereby circumvents any losses of efficacy related to viral mutations.
Recently, a randomized double-blinded phase IIb clinical trial has been completed regarding the safety and therapeutic efficacy of DAS-181 for treatment of influenza (ClinicalTrials.gov identifier: NCT01740063). Results so far have indicated that the inhaled drug is generally well tolerated for up to 7 days 159, 160. Longer treatment periods have shown to lead to symptoms related to systemic reabsorption and to induce DAS-181-specific antibodies. A new phase IIb clinical trial has been scheduled to assess the efficacy, safety and pharmacokinetics of DAS-181 in people who have been hospitalized with severe influenza and require supplemental oxygen (NCT04298060). In a sub-cohort of the study, DAS-181 efficacy will also be tested in hospitalized patients who have been infected with other sialic-acid-dependent viruses.
In addition, DAS-181 has shown to be effective for HPIV infections. In preclinical studies, administration of the drug has been shown to lead to a significant reduction in the number of HPIV-infected cells in vitro and in vivo [161]. Furthermore, the drug has successfully passed a phase II clinical trial in which its efficacy was investigated in immunocompromised patients with parainfluenza infection (NCT01644877) [162]. DAS-181 was also shown to have therapeutic potential in the treatment of HPIV-infected individuals who recently underwent haematopoietic stem cell transplantation [163]. A phase III clinical trial has started recruiting participants for evaluating the treatment of human parainfluenza infections in hospitalized, immunocompromised patients (NCT03808922). These developments have led the FDA to designate DAS-181 as both a fast-track and breakthrough therapy.
Aside from the promising advances regarding its use for the treatment of influenza and parainfluenza infections, DAS-181 has shown high therapeutic efficacy against EV-D68 at concentrations in the nanomolar range [151]. However, concerns have been raised owing to the finding that isolated strains of EV-D68 might evade the drug because they are capable of infecting desialylated cells, although it is not known whether these strains actively circulate in the human population [150].
The cleavage of viral attachment receptors seems like a promising strategy, but it is uncertain whether the removal of sialic-acid residues from the respiratory epithelium leads to adverse side effects. Concerns over the use of neuraminidase analogues are primarily based on the hypothesis that neuraminidase treatment makes the patient more prone to secondary infections by bacteria such as Streptococcus pneumoniae 164, 165, 166. This possibility was strengthened after treatment with the neuraminidase inhibitor oseltamivir in influenza-infected mice was reported to lead to a decrease in cases of secondary bacterial pneumonia [165].
Additional concerns were raised over the possibility that cleavage of sialic-acid residues by neuraminidase might expose cryptic receptors that are necessary for the opportunistic bacteria to adhere 167, 168. This hypothesis has been tested in in vivo colonization studies with S. pneumoniae in influenza-infected mice, which showed that DAS-181 had no effect on bacterial growth or might even reduce the risk of acquiring a secondary bacterial infection [169].
However, because sialic acids are necessary for several physiological processes, the long-term effects of their removal should be considered carefully. Moreover, another limitation might be that the removal of sialic acids by sialidases could lead to the emergence of highly resistant escape mutants that do not strictly depend on the sialic-acid interaction.
Notwithstanding these concerns, DAS-181 is currently the only compound in advanced clinical development that tackles sialic-acid-targeting viruses by targeting the cellular receptor itself.
Sialic-acid analogues
Another strategy for broad-spectrum targeting is to mimic the cellular receptor. This can be done by using compounds that function as a decoy, thereby competing with the cellular receptor to bind the virus (i.e., competitive-binding inhibitors). A number of studies have investigated this possibility for treating sialic-acid-targeting-virus infections, especially infections with influenza viruses.
Early studies of the use of sialic-acid-containing compounds to inhibit viral infection primarily investigated naturally occurring soluble substances that contain sialic-acid molecules, such as egg white, serum and respiratory mucus [170]. These substances were shown to be able to compete with cell-associated sialic acids for the binding of influenza virus. It should be noted that the inhibitory capacity of these molecules was found to be highly dependent on several parameters such as size, rigidity and accessibility to haemagglutinin, as well as on the susceptibility of the substance to being neutralized by neuraminidase molecules [170]. Other interesting examples of molecules that are thought to contribute to the defence mechanism of the host are mucins 171, 172 and surfactant proteins 173, 174, 175, 176, which are produced by cells of the respiratory tract as a component of mucus and contain a wide variety of carbohydrates, including sialic acid. These compounds, which are part of the innate immune system, can function as naturally occurring decoy receptors against intruding sialic-acid-targeting pathogens, leading to aggregation of the virus particles and an enhanced ability of phagocytic cells to recognize such particles, as has been shown recently [176].
As well as naturally occurring molecules, several synthetic sialic-acid analogues have been tested against influenza, albeit with limited success. The efficacy of these drugs increased markedly with the development of the first multivalent inhibitors, although this success varied for different strains of influenza [170]. To overcome the problem of strain-dependent efficacy of the inhibitors, research focused on moieties that are essential to most influenza strains, such as 6′-SLN (Fig. 2). Synthetic compounds containing 6′-SLN epitopes were shown to effectively inhibit influenza A virus infection in mice, both prophylactically and therapeutically 177, 178. In a series of studies conducted by Papp et al., sialic-acid-functionalized nanoparticles were shown to effectively inhibit influenza virus infection in vitro by binding to viral haemagglutinin in a multivalent manner 179, 180. Another application of multivalent receptor analogues was introduced by Wang and colleagues, who functionalized liposomes with sialylneolacto-N-tetraose c (LSTc) glycans. The compounds, which acted like polymeric receptor decoys, were able to inhibit infection with several influenza strains, both in vitro and in vivo [181].
To optimize drugs that target the receptor-binding site of influenza, Kwon and colleagues designed multivalent 6′-sialyllactose-polyamidoamine (6′SL-PAMAM) conjugates with well-defined linker spacings to match the spacing of HA molecules on the viral envelope, and were therefore able to inhibit infection of mice with a lethal dose of H1N1 [182]. On the basis of this, Günther et al. recently fine-tuned the design of these conjugates to bear more functional groups, and were able to confirm the efficacy of the compounds against various human and avian influenza strains [41].
Other interesting compounds that target the receptor-binding site are linear polyglycerol sialosides. By optimizing the ligand densities, these compounds were shown to prevent influenza infection in mice in a superior fashion compared with dendritic polyglycerol sialosides. The authors state that the efficacy of dendritic polyglycerol sialosides was probably affected by steric shielding [183]. Another interesting application of multivalent receptor decoys for the treatment of influenza was introduced recently by Lauster and colleagues, who used bacteriophage capsids functionalized with sialic-acid moieties as a decoy for the virus. With ligand arrangements matching the distance of the receptor-binding sites of trimeric influenza haemagglutinin, the systems were able to multivalently bind to influenza virions, thereby efficiently blocking viral attachment in vitro, ex vivo and in vivo [184].
Although the concept of sialic-acid-receptor analogues is promising, not many studies have been conducted on this idea in recent years. This lack of interest could be partially attributed to the poor translatability of in vitro results to the clinical setting. The low in vivo effectiveness of sialic-acid-receptor analogues is intrinsic to their mechanism of action; such compounds are competitive binding inhibitors that interact with the virus, thus preventing the infection just above a certain concentration. On dilution, a usual condition in the in vivo setting, the reversible interaction is lost, and the drug releases an intact virion that can restart an infection cycle, which explains the lack of efficacy in vivo. As such, the drug has a so-called virustatic effect.
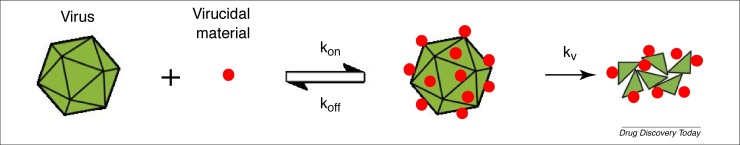
Arguably, a superior approach would be based on a virucidal mechanism of action. A virucidal drug is able to irreversibly inhibit a virus, so that the inhibition is retained even on dilution (Fig. 4 ). Compounds that are based on a virucidal mechanism of action enhance the properties of a standard competitive inhibitor because they permanently inactivate the virus. Examples of virucidal compounds are bleach, disinfectants and strong acids, which are all used to deactivate viruses in vitro but are also extremely toxic. Therefore, they are not suitable as drug candidates.
Figure 4.
A schematic representation of the virucidal mechanism of action. The virucidal drug first interacts with the virus with kinetic constants Kon and Koff. If the compound used binds with an irreversible mechanism, a local increase in pressure on the virus leads to its inactivation, driven by a kinetic constant Kv.
One way of designing non-toxic virucidal drugs is to couple decoy receptors to nanoparticles via long, flexible and hydrophobic linkers. The validity of this concept has been shown for heparan sulfate proteoglycans (HSPGs), which are common receptors for many viruses. The modified compounds were shown to bind to several HSPG-targeting viruses in a multivalent manner, leading to irreversible viral deformation, probably owing to a local increase in pressure [185].
Recently, this approach has been successfully translated to sialic-acid-targeting viruses using natural molecules as a core. Specifically, β-cyclodextrins were modified with long and hydrophobic linkers, bearing either 6′-SLN or 3′-SLN as functional group. These compounds were able to irreversibly inhibit different strains of human and avian influenza in the nanomolar range. The results were confirmed ex vivo and validated in vivo [186].
Taken together, sialic-acid-based receptor analogues might hold promise as entry inhibitors for sialic-acid-dependent viruses, especially for influenza. The possibility of using monosaccharides (such as Neu5Ac) could pave the way for broad-spectrum antiviral drugs with a virucidal mechanism of action.
Concluding remarks and perspectives
Because the number of approved antiviral drugs is limited and new virus strains continue to emerge, the urgency of developing novel therapeutic strategies remains high. Despite numerous attempts to find efficient antiviral drugs, in the end, many of them will fail in phase II or III studies: a frequent occurrence in drug development. Therefore, more effort should be put into the development of potent, preferably broad-spectrum antivirals.
In addition, the administration route should be considered. For example, respiratory-virus infections are likely to be treated most optimally with antivirals that are administered via the respiratory tract. Because several viruses share a common attachment receptor, we think that focusing on the mechanism of viral entry could open doors to the development of compounds with broad-spectrum activity. Many viruses use sialic-acid residues as their receptor, so this review has focused on the process of viral entry of these sialic-acid-targeting viruses and the current status of entry-inhibitor development, including those which might have broad-spectrum activity. Although numerous studies have been conducted on entry inhibitors targeting this group of viruses, most of the compounds studied have strain-specific targets. Therefore, we think that receptor-targeting compounds that directly interfere with the attachment of the virus to sialic acids, as well as multivalent sialic acid receptor analogues, deserve more attention, because they might have the potential to broadly combat a wide variety of sialic-acid-targeting viruses.
Competing interests
F.S. is the inventor on patent number WO 2018/015465 A1—Virucidal compounds and uses thereof. The author declares no other competing interests.
Acknowledgements
This work was supported by the Swiss National Science Foundation (via a Sinergia grant CRSII5_180323 to F.S. and H.W.F.). M.G. was supported by NCCR Bio-Inspired Materials.
Biographies

Rick Heida received his master’s degree in biomedical sciences at the University of Groningen in the Netherlands in 2017, with a main interest in microbiology and immunology. Currently, he is pursuing his PhD at the department of Pharmaceutical Technology and Biopharmacy at the University of Groningen under the supervision of Henderik W. Frijlink, Anke L.W. Huckriede and Wouter L.J. Hinrichs. His research focuses mainly on the evaluation of antiviral entry inhibitors with broad-spectrum potential and their suitability to be formulated as dry-powder formulations. In line with this, he works on optimizing methods for pulmonary administration of dry-powder formulations in small laboratory animals.

Yoshita C. Bhide completed her PhD in virology and immunology in 2018 at the University of Groningen under the supervision of Anke L.W. Huckriede. Her PhD research focused on improved and cross-protective influenza vaccine evaluation in vivo. She is now working as a postdoctoral researcher with Henderik W. Frijlink. Her current research focuses on in vitro and in vivo evaluation of novel entry inhibitors against influenza viruses. Her research has resulted in several peer-reviewed publications. Being a trained virologist, her research interests are in infectious diseases, especially viral pathogenesis and immune modulation, and the development of vaccines and antivirals.

Matteo Gasbarri received his master’s degree in materials science at the Tor Vergata University of Rome. His master thesis focused on flexible perovskite solar cells and was carried out at the Center for Hybrid and Organic Solar Energy (CHOSE). In 2017, he joined Saule Technologies, developing new generation perovskite photovoltaic cells. Since September 2017, he has been a PhD student in the Supramolecular Nanomaterials and Interfaces Laboratory (SuNMIL) at École Polytechnique Fédérale de Lausanne (EPFL), under the supervision of Francesco Stellacci. His research is primarily focused on the development and physicochemical understanding of non-toxic broad-spectrum antiviral nanomaterials.
References
- 1.Woolhouse M., et al. Human viruses: discovery and emergence. Philos. Trans. R. Soc. B. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 5.Gorog P., et al. Effect of removing sialic acids from endothelium on the adherence of circulating platelets in arteries in vivo. Proc. R. Soc. Lond. B. 1982;214:471–480. doi: 10.1098/rspb.1982.0022. [DOI] [PubMed] [Google Scholar]
- 6.Gelberg H., et al. In vivo enzymatic removal of alpha 2-->6-linked sialic acid from the glomerular filtration barrier results in podocyte charge alteration and glomerular injury. Lab. Invest. 1996;74:907–920. [PubMed] [Google Scholar]
- 7.Born G.V., Palinski W. Unusually high concentrations of sialic acids on the surface of vascular endothelia. Br. J. Exp. Pathol. 1985;66:543–549. [PMC free article] [PubMed] [Google Scholar]
- 8.Strilić B., et al. Electrostatic cell-surface repulsion initiates lumen formation in developing blood vessels. Curr. Biol. 2010;20:2003–2009. doi: 10.1016/j.cub.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 9.Wang B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv. Nutr. 2012;3 doi: 10.3945/an.112.001875. 465S–472S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker C.G., et al. The polysialic acid modification of the neural cell adhesion molecule is involved in spatial learning and hippocampal long-term potentiation. J. Neurosci. Res. 1996;45:143–152. doi: 10.1002/(SICI)1097-4547(19960715)45:2<143::AID-JNR6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 11.Wasik B.R., et al. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 2016;24:991–1001. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angata T., Varki A. Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 13.Blaum B.S., Stehle T. Sialic acids in nonenveloped virus infections. Adv. Carbohydr. Chem. Biochem. 2019;76:65–111. doi: 10.1016/bs.accb.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Langereis M.A., et al. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varki A. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am. J. Phys. Anthropol. 2001;116:54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B. Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 2009;29:177–222. doi: 10.1146/annurev.nutr.28.061807.155515. [DOI] [PubMed] [Google Scholar]
- 17.Stencel-Baerenwald J.E., et al. The sweet spot: defining virus–sialic acid interactions. Nat. Rev. Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster R.G., et al. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 19.Davidson S. Treating InfluenzaInfection, From Now and Into the Future. Front. Immunol. 2018;9:1946. doi: 10.3389/fimmu.2018.01946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taubenberger J.K., et al. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taubenberger J.K., Morens D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. Mech. Dis. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasparini R., et al. Compounds with anti-influenza activity: present and future of strategies for the optimal treatment and management of influenza. Part I: Influenza life-cycle and currently available drugs. J. Prev. Med. Hyg. 2014;55:69–85. [PMC free article] [PubMed] [Google Scholar]
- 24.Gambaryan A.S., et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-ace. Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 25.Ito T., et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers G.N., Paulson J.C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 27.Ito T. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol. Immunol. 2000;44:423–430. doi: 10.1111/j.1348-0421.2000.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 28.Dou D., et al. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 2018;9:1581. doi: 10.3389/fimmu.2018.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edinger T.O., et al. Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 30.Seto J.T., Rott R. Functional significance of sialidose during influenza virus multiplication. Virology. 1966;30:731–737. doi: 10.1016/0042-6822(66)90178-4. [DOI] [PubMed] [Google Scholar]
- 31.Lewnard J., Cobey S. Immune history and influenza vaccine effectiveness. Vaccines. 2018;6:28. doi: 10.3390/vaccines6020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.European Centre for Disease Prevention and Control . ECDC; 2017. Expert Opinion on Neuraminidase Inhibitors for the Prevention and Treatment of Influenza. Review of Recent Systematic Reviews and Meta-Analyses. [Google Scholar]
- 34.Imai M., et al. Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets. Nat. Microbiol. 2020;5:27–33. doi: 10.1038/s41564-019-0609-0. [DOI] [PubMed] [Google Scholar]
- 35.Kormuth K.A., Lakdawala S.S. Emerging antiviral resistance. Nat. Microbiol. 2020;5:4–5. doi: 10.1038/s41564-019-0639-7. [DOI] [PubMed] [Google Scholar]
- 36.Uehara T., et al. Treatment-emergent influenza variant viruses with reduced baloxavir susceptibility: impact on clinical and virologic outcomes in uncomplicated influenza. J. Infect. Dis. 2019;221:346–355. doi: 10.1093/infdis/jiz244. [DOI] [PubMed] [Google Scholar]
- 37.Chesnokov A., et al. Replicative fitness of seasonal influenza A viruses with decreased susceptibility to baloxavir. J. Infect. Dis. 2019;221:367–371. doi: 10.1093/infdis/jiz472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitley R.J., Monto A.S. Resistance of influenza virus to antiviral medications. Clin. Infect. Dis. 2019;71:1092–1094. doi: 10.1093/cid/ciz911. [DOI] [PubMed] [Google Scholar]
- 39.Luganini A., et al. The cranberry extract Oximacro® exerts in vitro virucidal activity against influenza virus by interfering with hemagglutinin. Front. Microbiol. 2018;9:1826. doi: 10.3389/fmicb.2018.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadam R.U., Wilson I.A. A small-molecule fragment that emulates binding of receptor and broadly neutralizing antibodies to influenza A hemagglutinin. Proc. Natl Acad. Sci. USA. 2018;115:4240–4245. doi: 10.1073/pnas.1801999115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Günther S.C., et al. Antiviral potential of 3′-sialyllactose- and 6′-sialyllactose-conjugated dendritic polymers against human and avian influenza viruses. Sci. Rep. 2020;10:768. doi: 10.1038/s41598-020-57608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., et al. Synthesis of a hexavalent betulinic acid derivative as a hemagglutinin-targeted influenza virus entry inhibitor. Mol. Pharm. 2020;17:2546–2554. doi: 10.1021/acs.molpharmaceut.0c00244. [DOI] [PubMed] [Google Scholar]
- 43.Hussein A.F.A., et al. Identification of entry inhibitors with 4-aminopiperidine scaffold targeting group 1 influenza A virus. Antiviral Res. 2020;177:104782. doi: 10.1016/j.antiviral.2020.104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaisina I.N., et al. Optimization of 4-aminopiperidines as inhibitors of influenza A viral entry that are synergistic with oseltamivir. J. Med. Chem. 2020;63:3120–3130. doi: 10.1021/acs.jmedchem.9b01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Castro S., et al. N-benzyl 4,4-disubstituted piperidines as a potent class of influenza H1N1 virus inhibitors showing a novel mechanism of hemagglutinin fusion peptide interaction. Eur. J. Med. Chem. 2020;194:112223. doi: 10.1016/j.ejmech.2020.112223. [DOI] [PubMed] [Google Scholar]
- 46.Ye M., et al. An oleanolic acid derivative inhibits hemagglutinin-mediated entry of influenza A virus. Viruses. 2020;12:225. doi: 10.3390/v12020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cihan-Üstündağ G., et al. Superior inhibition of influenza virus hemagglutinin-mediated fusion by indole-substituted spirothiazolidinones. Bioorg. Med. Chem. 2020;28:115130. doi: 10.1016/j.bmc.2019.115130. [DOI] [PubMed] [Google Scholar]
- 48.Memczak H., et al. Anti-hemagglutinin antibody derived lead peptides for inhibitors of influenza virus binding. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatia S., et al. Adaptive flexible sialylated nanogels as highly potent influenza A virus inhibitors. Angew. Chem. Int. Ed. 2020;59:12417–12422. doi: 10.1002/anie.202006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie C., et al. Topology‐matching design of an influenza‐neutralizing spiky nanoparticle‐based inhibitor with a dual mode of action. Angew. Chem. Int. Ed. 2020;59:15532–15536. doi: 10.1002/anie.202004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leiva R., et al. Aniline-based inhibitors of influenza H1N1 virus acting on hemagglutinin-mediated fusion. J. Med. Chem. 2018;61:98–118. doi: 10.1021/acs.jmedchem.7b00908. [DOI] [PubMed] [Google Scholar]
- 52.Li S., et al. Structure-activity relationships of 3-O-β-chacotriosyl oleanic acid derivatives as entry inhibitors for highly pathogenic H5N1 influenza virus. Bioorg. Med. Chem. 2017;25:4384–4396. doi: 10.1016/j.bmc.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 53.Wang H., et al. Design, synthesis and biological evaluation of novel l-ascorbic acid-conjugated pentacyclic triterpene derivatives as potential influenza virus entry inhibitors. Eur. J. Med. Chem. 2016;110:376–388. doi: 10.1016/j.ejmech.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Wu W., et al. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses. 2015;8:6. doi: 10.3390/v8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu W., et al. New influenza a virus entry inhibitors derived from the viral fusion peptides. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W., et al. Super short membrane-active lipopeptides inhibiting the entry of influenza A virus. Biochim. Biophys. Acta. 2015;1848:2344–2350. doi: 10.1016/j.bbamem.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., et al. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur. J. Med. Chem. 2015;93:182–195. doi: 10.1016/j.ejmech.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Sriwilaijaroen N., et al. 6SLN-lipo PGA specifically catches (coats) human influenza virus and synergizes neuraminidase-targeting drugs for human influenza therapeutic potential. J. Antimicrob. Chemother. 2015;70:2797–2809. doi: 10.1093/jac/dkv193. [DOI] [PubMed] [Google Scholar]
- 59.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 60.Yasuhara A., et al. Isolation and characterization of human monoclonal antibodies that recognize the influenza A(H1N1)pdm09 virus hemagglutinin receptor-binding site and rarely yield escape mutant viruses. Front. Microbiol. 2018;9:2660. doi: 10.3389/fmicb.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malakhov M.P., et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob. Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Triana-Baltzer G.B., et al. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connaris H., et al. Prevention of influenza by targeting host receptors using engineered proteins. Proc. Natl Acad. Sci. USA. 2014;111:6401–6406. doi: 10.1073/pnas.1404205111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krammer F., Palese P. Universal influenza virus vaccines that target the conserved hemagglutinin stalk and conserved sites in the head domain. J. Infect. Dis. 2019;219:S62–S67. doi: 10.1093/infdis/jiy711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leneva I.A., et al. Characteristics of arbidol-resistant mutants of influenza virus: Implications for the mechanism of anti-influenza action of arbidol. Antiviral Res. 2009;81:132–140. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 66.Leneva I.A., et al. Sensitivity of various influenza virus strains to arbidol. Influence of arbidol combination with different antiviral drugs on reproduction of influenza virus A. Ter Arkh. 2005;77:84–88. (in Russian) [PubMed] [Google Scholar]
- 67.Leneva I.A., et al. Umifenovir susceptibility monitoring and characterization of influenza viruses isolated during ARBITR clinical study. J. Med. Virol. 2019;91:588–597. doi: 10.1002/jmv.25358. [DOI] [PubMed] [Google Scholar]
- 68.Wang M., et al. Efficacy and safety of arbidol in treatment of naturally acquired influenza. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2004;26:289–293. (in Chinese) [PubMed] [Google Scholar]
- 69.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl Acad. Sci. USA. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali S.O., et al. Evaluation of MEDI8852, an anti-influenza a monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00694-18. e00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng R., et al. Pharmacokinetics of MHAA4549A, an anti-influenza a monoclonal antibody, in healthy subjects challenged with influenza A virus in a phase IIa randomized trial. Clin. Pharmacokinet. 2018;57:367–377. doi: 10.1007/s40262-017-0564-y. [DOI] [PubMed] [Google Scholar]
- 72.Hershberger E., et al. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine. 2019;40:574–582. doi: 10.1016/j.ebiom.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henrickson K.J. Parainfluenza viruses. Clin. Microbiol. Rev. 2003;16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall C.B. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 2001;344:1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 75.Pawełczyk M., Kowalski M.L. The role of human parainfluenza virus infections in the immunopathology of the respiratory tract. Curr. Allergy Asthma Rep. 2017;17:16. doi: 10.1007/s11882-017-0685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukushima K., et al. Terminal sialic acid linkages determine different cell infectivities of human parainfluenza virus type 1 and type 3. Virology. 2014;464–465:424–431. doi: 10.1016/j.virol.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 77.Amonsen M., et al. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 2007;81:8341–8345. doi: 10.1128/JVI.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmidt A.C., et al. Progress in the development of human parainfluenza virus vaccines. Expert Rev. Respir. Med. 2011;5:515–526. doi: 10.1586/ers.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alymova I.V., et al. Efficacy of novel hemagglutinin-neuraminidase inhibitors BCX 2798 and BCX 2855 against human parainfluenza viruses in vitro and in vivo. Antimicrob. Agents Chemother. 2004;48:1495–1502. doi: 10.1128/AAC.48.5.1495-1502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alymova I.V., et al. Efficacy of the novel parainfluenza virus haemagglutinin-neuraminidase inhibitor BCX 2798 in mice – further evaluation. Antivir. Ther. 2009;14:891–898. doi: 10.3851/IMP1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe M., et al. Effect of hemagglutinin-neuraminidase inhibitors BCX 2798 and BCX 2855 on growth and pathogenicity of Sendai/human parainfluenza type 3 chimera virus in mice. Antimicrob. Agents Chemother. 2009;53:3942–3951. doi: 10.1128/AAC.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eveno T., et al. Targeting human parainfluenza virus type-1 haemagglutinin-neuraminidase with mechanism-based inhibitors. Viruses. 2019;11:417. doi: 10.3390/v11050417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bottom-Tanzer S.F., et al. Inhibiting human parainfluenza virus infection by preactivating the cell entry mechanism. mBio. 2019;10 doi: 10.1128/mBio.02900-18. e02900-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marcink T.C., et al. Hijacking the fusion complex of human parainfluenza virus as an antiviral strategy. mBio. 2020;11 doi: 10.1128/mBio.03203-19. e03203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghebremedhin B. Human adenovirus: viral pathogen with increasing importance. Eur. J. Microbiol. Immunol. 2014;4:26–33. doi: 10.1556/EuJMI.4.2014.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koirala B., Wang J.P., et al. In: Viruses and the Lung. Fraire A., editor. Springer; 2014. Adenovirus; pp. 35–41. [Google Scholar]
- 87.Khanal S., et al. The repertoire of adenovirus in human disease: the innocuous to the deadly. Biomedicines. 2018;6:30. doi: 10.3390/biomedicines6010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Oliveira C.B.R., et al. Evaluation of Cidofovir (HPMPC, GS-504) against adenovirus type 5 infection in vitro and in a New Zealand rabbit ocular model. Antiviral Res. 1996;31:165–172. doi: 10.1016/0166-3542(95)00962-0. [DOI] [PubMed] [Google Scholar]
- 89.Legrand F., et al. Early diagnosis of adenovirus infection and treatment with cidofovir after bone marrow transplantation in children. Bone Marrow Transplant. 2001;27:621–626. doi: 10.1038/sj.bmt.1702820. [DOI] [PubMed] [Google Scholar]
- 90.Bergelson J.M., et al. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J. Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaggar A., et al. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 92.Mathias P., et al. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arnberg N. Adenovirus receptors: implications for tropism, treatment and targeting. Rev. Med. Virol. 2009;19:165–178. doi: 10.1002/rmv.612. [DOI] [PubMed] [Google Scholar]
- 94.Arnberg N., et al. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 95.Arnberg N., et al. Initial interactions of subgenus D adenoviruses with A549 cellular receptors: sialic acid versus alpha(v) integrins. J. Virol. 2000;74:7691–7693. doi: 10.1128/jvi.74.16.7691-7693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arnberg N., et al. Adenovirus type 37 binds to cell surface sialic acid through a charge-dependent interaction. Virology. 2002;302:33–43. doi: 10.1006/viro.2002.1503. [DOI] [PubMed] [Google Scholar]
- 97.Gagneux P., et al. Human-specific regulation of α2-6-linked sialic acids. J. Biol. Chem. 2003;278:48245–48250. doi: 10.1074/jbc.M309813200. [DOI] [PubMed] [Google Scholar]
- 98.Kumlin U., et al. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respi. Viruses. 2008;2:147–154. doi: 10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nilsson E.C., et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat. Med. 2011;17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]
- 100.Johansson S.M.C., et al. Multivalent HSA conjugates of 3′-sialyllactose are potent inhibitors of adenoviral cell attachment and infection. ChemBioChem. 2005;6:358–364. doi: 10.1002/cbic.200400227. [DOI] [PubMed] [Google Scholar]
- 101.Johansson S.M.C., et al. Multivalent sialic acid conjugates inhibit adenovirus type 37 from binding to and infecting human corneal epithelial cells. Antiviral Res. 2007;73:92–100. doi: 10.1016/j.antiviral.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 102.Spjut S., et al. A potent trivalent sialic acid inhibitor of adenovirus type 37 infection of human corneal cells. Angew. Chem. Int. Ed. 2011;50:6519–6521. doi: 10.1002/anie.201101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Caraballo R., et al. Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells. Org. Biomol. Chem. 2015;13:9194–9205. doi: 10.1039/c5ob01025j. [DOI] [PubMed] [Google Scholar]
- 104.Lenman A., et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc. Natl Acad. Sci. USA. 2018;115:E4264–E4273. doi: 10.1073/pnas.1716900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baker A.T., et al. Human adenovirus type 26 uses sialic acid–bearing glycans as a primary cell entry receptor. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aax3567. eaax3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pinto M., Dobson S. BK and JC virus: a review. J. Infect. 2014;68:S2–S8. doi: 10.1016/j.jinf.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 107.Boothpur R., Brennan D.C. Human polyoma viruses and disease with emphasis on clinical BK and JC. J. Clin. Virol. 2010;47:306–312. doi: 10.1016/j.jcv.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bennett S.M., et al. Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry. Virology. 2015;474:110–116. doi: 10.1016/j.virol.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prado J., et al. Human polyomaviruses and cancer: an overview. Clinics. 2018;73 doi: 10.6061/clinics/2018/e558s. e558s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng H., et al. Clonal Integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eash S., et al. The human polyomaviruses. Cell Mol. Life Sci. 2006;63:865–876. doi: 10.1007/s00018-005-5454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nickeleit V., et al. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J. Am. Soc. Nephrol. 1999;10:1080–1089. doi: 10.1681/ASN.V1051080. [DOI] [PubMed] [Google Scholar]
- 113.Drachenberg C.B., et al. Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: a prospective evaluation. Transplantation. 2007;84:323–330. doi: 10.1097/01.tp.0000269706.59977.a5. [DOI] [PubMed] [Google Scholar]
- 114.Tan C.S., Koralnik I.J. Progressive multifocal leukoencephalopathy and other disorders caused by JC virus: clinical features and pathogenesis. Lancet Neurol. 2010;9:425–437. doi: 10.1016/S1474-4422(10)70040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sabath B.F., Major E.O. Traffic of JC virus from sites of initial infection to the brain: the path to progressive multifocal leukoencephalopathy. J. Infect. Dis. 2002;186:S180–S186. doi: 10.1086/344280. [DOI] [PubMed] [Google Scholar]
- 116.Becker M., et al. Infectious entry of Merkel cell polyomavirus. J. Virol. 2019;93 doi: 10.1128/JVI.02004-18. e02004-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu C.K., et al. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal alpha(2-6)-linked sialic acids. J. Virol. 1998;72:4643–4649. doi: 10.1128/jvi.72.6.4643-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Neu U., et al. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dugan A.S., et al. Direct correlation between sialic acid binding and infection of cells by two human polyomaviruses (JC virus and BK virus) J. Virol. 2008;82:2560–2564. doi: 10.1128/JVI.02123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dugan A.S., et al. An N-linked glycoprotein with α(2,3)-linked sialic acid is a receptor for BK virus. J. Virol. 2005;79:14442–14445. doi: 10.1128/JVI.79.22.14442-14445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neu U., et al. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bauer P.H., et al. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 1999;73:5826–5832. doi: 10.1128/jvi.73.7.5826-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yatawara A., et al. Small-molecule inhibitors of JC polyomavirus infection. J. Pept. Sci. 2015;21:236–242. doi: 10.1002/psc.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tortorici M.A., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zumla A., et al. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang X., et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vlasak R., et al. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl Acad. Sci. USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Klein A., Roussel P. O-Acetylation of sialic acids. Biochimie. 1998;80:49–57. doi: 10.1016/s0300-9084(98)80056-4. [DOI] [PubMed] [Google Scholar]
- 131.Park S.S. Post-glycosylation modification of sialic acid and its role in virus pathogenesis. Vaccines. 2019;7:171. doi: 10.3390/vaccines7040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hulswit R.J.G., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl Acad. Sci. USA. 2019;116:2681–2890. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peng G., et al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl Acad. Sci. USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xia S., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. eaav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Raj V.S., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mou H., et al. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li W., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl Acad. Sci. USA. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Park Y.-J., et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Widjaja I., et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes Infect. 2019;8:516–530. doi: 10.1080/22221751.2019.1597644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zell R. Picornaviridae-the ever-growing virus family. Arch. Virol. 2018;163:299–317. doi: 10.1007/s00705-017-3614-8. [DOI] [PubMed] [Google Scholar]
- 141.Cassidy H., et al. Enterovirus D68 – the new polio? Front. Microbiol. 2018;9:2677. doi: 10.3389/fmicb.2018.02677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sejvar J.J., et al. Acute flaccid myelitis in the United States, August–December 2014: results of nationwide surveillance. Clin. Infect. Dis. 2016;63:737–745. doi: 10.1093/cid/ciw372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Knoester M., et al. Upsurge of enterovirus D68, the Netherlands, 2016. Emerg. Infect. Dis. 2017;23:140–143. doi: 10.3201/eid2301.161313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Holm-Hansen C.C., et al. Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis. 2016;16:e64–e75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 145.Messacar K., et al. Enterovirus D68 and acute flaccid myelitis—evaluating the evidence for causality. Lancet Infect. Dis. 2018;18:e239–e247. doi: 10.1016/S1473-3099(18)30094-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brown D.M., et al. Contemporary circulating enterovirus D68 strains have acquired the capacity for viral entry and replication in human neuronal cells. mBio. 2018;9 doi: 10.1128/mBio.01954-18. e01954-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu Y., et al. Sialic acid-dependent cell entry of human enterovirus D68. Nat. Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Imamura T., et al. Antigenic and receptor binding properties of enterovirus 68. J. Virol. 2014;88:2374–2384. doi: 10.1128/JVI.03070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei W., et al. ICAM-5/telencephalin is a functional entry receptor for enterovirus D68. Cell Host Microbe. 2016;20:631–641. doi: 10.1016/j.chom.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 150.Baggen J., et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl Acad. Sci. USA. 2016;113:1399–1404. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rhoden E., et al. In vitro efficacy of antiviral compounds against enterovirus D68. Antimicrob. Agents Chemother. 2015;59:7779–7781. doi: 10.1128/AAC.00766-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Plourde A.R., Bloch E.M. A literature review of Zika virus. Emerg. Infect. Dis. 2016;22:1185–1192. doi: 10.3201/eid2207.151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mlakar J., et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 154.Schuler-Faccini L., et al. Possible association between Zika virus infection and microcephaly — Brazil, 2015. MMWR Morb. Mortal. Wkly Rep. 2016;65:59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- 155.Sirohi D., Kuhn R.J. Zika virus structure, maturation, and receptors. J. Infect. Dis. 2017;216:S935–S944. doi: 10.1093/infdis/jix515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hamel R., et al. Biology of Zika virus infection in human skin cells. J. Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tan C.W., et al. Cell surface α2,3-linked sialic acid facilitates Zika virus internalization. Emerg. Microbes Infect. 2019;8:426–437. doi: 10.1080/22221751.2019.1590130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fink S.L., et al. The antiviral drug arbidol inhibits Zika virus. Sci. Rep. 2018;8:8989. doi: 10.1038/s41598-018-27224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zenilman J.M., et al. Phase 1 clinical trials of DAS181, an inhaled sialidase, in healthy adults. Antiviral Res. 2015;123:114–119. doi: 10.1016/j.antiviral.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Moss R.B., et al. A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J. Infect. Dis. 2012;206:1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moscona A., et al. A recombinant sialidase fusion protein effectively inhibits human parainfluenza viral infection in vitro and in vivo. J. Infect. Dis. 2010;202:234–241. doi: 10.1086/653621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chemaly R.F., et al. 1715. A randomized, double-blind, placebo-controlled multicenter phase 2 trial to examine the effects of DAS181 in immunocompromised (IC) patients with parainfluenza virus (PIV) lower respiratory tract infection (LRTI) on supplemental oxygen (SO) Open Forum Infect. Dis. 2018;5:S50–S51. [Google Scholar]
- 163.Salvatore M., et al. DAS181 for treatment of parainfluenza virus infections in hematopoietic stem cell transplant recipients at a single center. Biol. Blood Marrow Transplant. 2016;22:965–970. doi: 10.1016/j.bbmt.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 164.Peltola V.T., et al. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 2005;192:249–257. doi: 10.1086/430954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McCullers J.A. Effect of antiviral treatment on the outcome of secondary bacterial pneumonia after influenza. J. Infect. Dis. 2004;190:519–526. doi: 10.1086/421525. [DOI] [PubMed] [Google Scholar]
- 166.Zhang H. Concerns of using sialidase fusion protein as an experimental drug to combat seasonal and pandemic influenza. J. Antimicrob. Chemother. 2008;62:219–223. doi: 10.1093/jac/dkn026. [DOI] [PubMed] [Google Scholar]
- 167.Nicholls J.M., et al. Comment on: Concerns of using sialidase fusion protein as an experimental drug to combat seasonal and pandemic influenza. J. Antimicrob. Chemother. 2008;62:426–428. doi: 10.1093/jac/dkn167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duez J.-M., et al. Influenza virus infection: don’t forget the role of the mucociliary system! J. Antimicrob. Chemother. 2008;63:421–422. doi: 10.1093/jac/dkn468. [DOI] [PubMed] [Google Scholar]
- 169.Hedlund M., et al. Sialidase‐based anti–influenza virus therapy protects against secondary pneumococcal infection. J. Infect. Dis. 2010;201:1007–1015. doi: 10.1086/651170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Matrosovich M., Klenk H.-D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev. Med. Virol. 2003;13:85–97. doi: 10.1002/rmv.372. [DOI] [PubMed] [Google Scholar]
- 171.Lamblin G., Roussel P. Airway mucins and their role in defence against micro-organisms. Respir. Med. 1993;87:421–426. doi: 10.1016/0954-6111(93)90067-a. [DOI] [PubMed] [Google Scholar]
- 172.Scharfman A., et al. Interactions between human respiratory mucins and pathogens. Biochem. Soc. Trans. 1995;23:836–839. doi: 10.1042/bst0230836. [DOI] [PubMed] [Google Scholar]
- 173.Benne C.A., et al. Interactions of surfactant protein A with influenza A viruses: binding and neutralization. J. Infect. Dis. 1995;171:335–341. doi: 10.1093/infdis/171.2.335. [DOI] [PubMed] [Google Scholar]
- 174.Hartshorn K.L., et al. Mechanisms of anti-influenza activity of surfactant proteins A and D: comparison with serum collectins. Am. J. Physiol. Lung Cell. Mol. Physiol. 1997;273:L1156–L1166. doi: 10.1152/ajplung.1997.273.6.L1156. [DOI] [PubMed] [Google Scholar]
- 175.LeVine A.M., et al. Absence of SP-A modulates innate and adaptive defense responses to pulmonary influenza infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L563–L572. doi: 10.1152/ajplung.00280.2001. [DOI] [PubMed] [Google Scholar]
- 176.van Eijk M., et al. Enhanced antiviral activity of human surfactant protein D by site-specific engineering of the carbohydrate recognition domain. Front. Immunol. 2019;10:2476. doi: 10.3389/fimmu.2019.02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gambaryan A., et al. Polymeric inhibitor of influenza virus attachment protects mice from experimental influenza infection. Antiviral Res. 2002;55:201–205. doi: 10.1016/s0166-3542(02)00020-7. [DOI] [PubMed] [Google Scholar]
- 178.Gambaryan A.S., et al. Polymer-bound 6′ sialyl-N-acetyllactosamine protects mice infected by influenza virus. Antiviral Res. 2005;68:116–123. doi: 10.1016/j.antiviral.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 179.Papp I., et al. Inhibition of influenza virus infection by multivalent sialic-acid-functionalized gold nanoparticles. Small. 2010;6:2900–2906. doi: 10.1002/smll.201001349. [DOI] [PubMed] [Google Scholar]
- 180.Papp I., et al. Inhibition of influenza virus activity by multivalent glycoarchitectures with matched sizes. ChemBioChem. 2011;12:887–895. doi: 10.1002/cbic.201000776. [DOI] [PubMed] [Google Scholar]
- 181.Hendricks G.L., et al. Sialylneolacto-N-tetraose c (LSTc)-bearing liposomal decoys capture influenza a virus. J. Biol. Chem. 2013;288:8061–8073. doi: 10.1074/jbc.M112.437202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kwon S.J., et al. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nat. Nanotechnol. 2017;12:48–54. doi: 10.1038/nnano.2016.181. [DOI] [PubMed] [Google Scholar]
- 183.Bhatia S., et al. Linear polysialoside outperforms dendritic analogs for inhibition of influenza virus infection in vitro and in vivo. Biomaterials. 2017;138:22–34. doi: 10.1016/j.biomaterials.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 184.Lauster D., et al. Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nat. Nanotechnol. 2020;15:373–379. doi: 10.1038/s41565-020-0660-2. [DOI] [PubMed] [Google Scholar]
- 185.Cagno V., et al. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 2018;17:195–203. doi: 10.1038/nmat5053. [DOI] [PubMed] [Google Scholar]
- 186.Kocabiyik O., et al. Non-toxic virucidal macromolecules show high efficacy against influenza virus ex vivo and in vivo. bioRxiv. 2020 doi: 10.1101/2020.03.18.996678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mammen M., et al. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 188.Fasting C., et al. Multivalency as a chemical organization and action principle. Angew. Chem. Int. Ed. 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 189.Müller M., et al. Mobility-based quantification of multivalent virus-receptor interactions: new insights into influenza A virus binding mode. Nano. Lett. 2019;19:1875–1882. doi: 10.1021/acs.nanolett.8b04969. [DOI] [PubMed] [Google Scholar]
- 190.Liese S., Netz R.R. Influence of length and flexibility of spacers on the binding affinity of divalent ligands. Beilstein J. Org. Chem. 2015;11:804–816. doi: 10.3762/bjoc.11.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Haviernik J., et al. Arbidol (umifenovir): a broad-spectrum antiviral drug that inhibits medically important arthropod-borne flaviviruses. Viruses. 2018;10:184. doi: 10.3390/v10040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Marjuki H., et al. An investigational antiviral drug, DAS181, effectively inhibits replication of zoonotic influenza a virus subtype H7N9 and protects mice from lethality. J. Infect. Dis. 2014;210:435–440. doi: 10.1093/infdis/jiu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Triana-Baltzer G.B., et al. Novel pandemic influenza A(H1N1) viruses are potently inhibited by DAS181, a sialidase fusion protein. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Koudstaal W., et al. Pre‐ and postexposure use of human monoclonal antibody against H5N1 and H1N1 influenza virus in mice: viable alternative to oseltamivir. J. Infect. Dis. 2009;200:1870–1873. doi: 10.1086/648378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Friesen R.H.E., et al. New class of monoclonal antibodies against severe influenza: prophylactic and therapeutic efficacy in ferrets. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Boriskin Y., Leneva I., Pecheur E.-I., Polyak S. Arbidol: A Broad-Spectrum Antiviral Compound that Blocks Viral Fusion. Curr Med Chem. 2008;15:997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- 197.Kallewaard N.L., et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell. 2016;166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Paules C.I., et al. The hemagglutinin A stem antibody MEDI8852 prevents and controls disease and limits transmission of pandemic influenza viruses. J. Infect. Dis. 2017;216:356–365. doi: 10.1093/infdis/jix292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nakamura G., et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 200.McBride J.M., et al. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza a virus challenge model. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01154-17. e01154-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wollacott A.M., et al. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBioMedicine. 2016;5:147–155. doi: 10.1016/j.ebiom.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baranovich T., et al. The hemagglutinin stem-binding monoclonal antibody VIS410 controls influenza virus-induced acute respiratory distress syndrome. Antimicrob. Agents Chemother. 2016;60:2118–2131. doi: 10.1128/AAC.02457-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tharakaraman K., et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc. Natl Acad. Sci. USA. 2015;112:10890–10895. doi: 10.1073/pnas.1502374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kim Jl., et al. Novel small molecule targeting the hemagglutinin stalk of influenza viruses. J. Virol. 2019;93 doi: 10.1128/JVI.00878-19. e00878-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.van Dongen M.J.P., et al. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science. 2019:363. doi: 10.1126/science.aar6221. eaar6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xu W., et al. The antihistamine drugs carbinoxamine maleate and chlorpheniramine maleate exhibit potent antiviral activity against a broad spectrum of influenza viruses. Front. Microbiol. 2018;9:2643. doi: 10.3389/fmicb.2018.02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fujioka Y., et al. A sialylated voltage-dependent Ca2+ channel binds hemagglutinin and mediates influenza A virus entry into mammalian cells. Cell Host Microbe. 2018;23:809–818. doi: 10.1016/j.chom.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 208.Lin D., et al. Potent influenza A virus entry inhibitors targeting a conserved region of hemagglutinin. Biochem. Pharmacol. 2017;144:35–51. doi: 10.1016/j.bcp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 209.Holthausen D.J., et al. An amphibian host defense peptide is virucidal for human H1 hemagglutinin-bearing influenza viruses. Immunity. 2017;46:587–595. doi: 10.1016/j.immuni.2017.03.018. [DOI] [PubMed] [Google Scholar]