Abstract
Malic Enzyme 1 (ME1) is a cytosolic protein that catalyzes the conversion of malate to pyruvate while concomitantly generating NADPH from NADP. Early studies identified ME1 as a mediator of intermediary metabolism primarily through its participatory roles in lipid and cholesterol biosynthesis. ME1 was one of the first identified insulin-regulated genes in liver and adipose and is a transcriptional target of thyroxine. Multiple studies have since documented that ME1 is pro-oncogenic in numerous epithelial cancers. In tumor cells, the reduction of ME1 gene expression or the inhibition of its activity resulted in decreases in proliferation, epithelial-to-mesenchymal transition and in vitro migration, and conversely, in promotion of oxidative stress, apoptosis and/or cellular senescence. Here, we integrate recent findings to highlight ME1’s role in oncogenesis, provide a rationale for its nexus with metabolic syndrome and diabetes, and raise the prospects of targeting the cytosolic NADPH network to improve therapeutic approaches against multiple cancers.
Keywords: malic enzyme, glutathione, thioredoxin, NADPH, hyperinsulinemia, cancer
Introduction
Malic Enzyme 1 (ME1) is a cytosolic protein that catalyzes the conversion of malate to pyruvate while simultaneously regenerating NADPH from NADP. ME1 (along with its cousins, malic enzyme 2 (ME2) and malic enzyme 3 (ME3)) have been well studied for their roles in intermediary metabolism (Fig. 1). ME1 is a major participant in lipid and cholesterol biosynthesis, since it generates NADPH, a required cofactor for fatty acid and cholesterol biosynthesis, and it regulates the reversible oxidative decarboxylation of malate to pyruvate, thus linking the glycolytic and citric acid pathways (Fig. 1). ME1 is also an indirect participant in many other NADPH-dependent metabolic pathways, by virtue of its contribution to the cytosol NADPH pool. Recent studies have demonstrated that ME1 is pro-oncogenic in a diverse array of epithelial cancers. This review focuses on this new role of ME1 in cancer and potentially at the interface of cancer, diabetes and obesity (Lega & Lipscombe 2020).
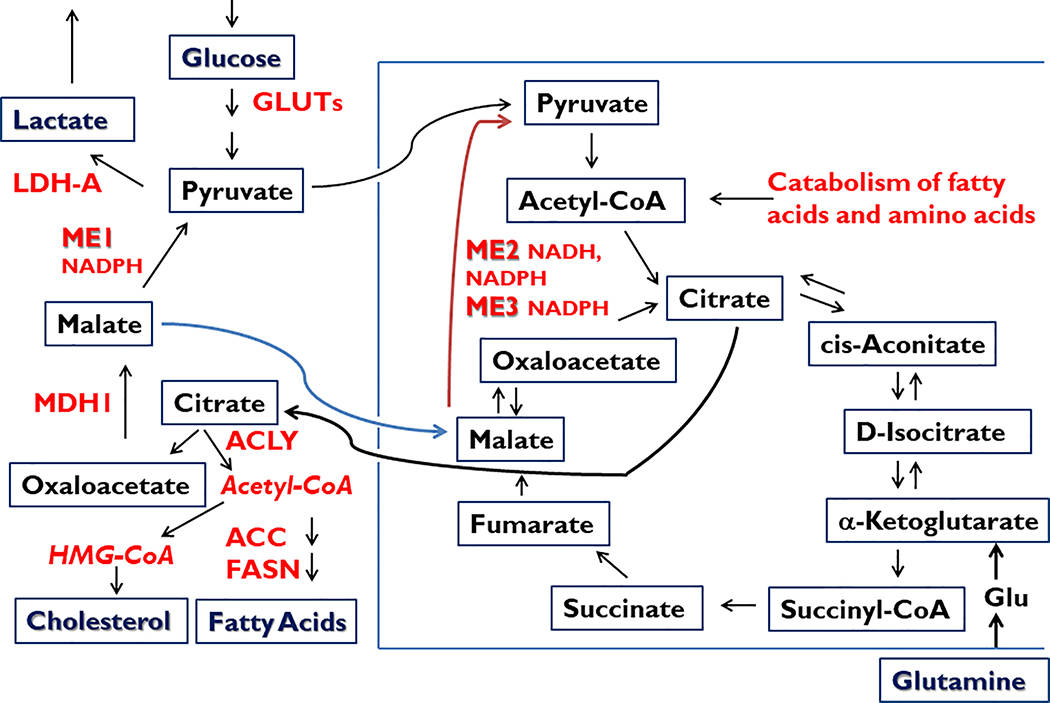
Figure 1.
Central role of malic enzymes (ME) in normal and cancer cell metabolism. Malic enzymes located within cytoplasmic (ME1) and mitochondrial (ME2, ME3) compartments catalyze the oxidative decarboxylation of malate to pyruvate, while concomitantly storing reducing equivalents in the form of NADPH (ME1, ME2, ME3) or NADH (ME2). NADPH serves in multiple biochemical capacities such as in countering ROS (via glutathione and thioredoxin pathways), contributing to ROS signaling (via NOX/DUOX family members), and providing reductant capacity for the biosynthesis of fatty acids and cholesterol, among many other metabolic pathways (Goodman et al. 2018). Besides being a major source of cytosolic NADPH, ME1 participates in a pathway by which excess mitochondrial Acetyl-CoA is transferred to cytosol (in the form of citrate; via the pyruvate-malate cycle) for incorporation into cholesterol and fatty acids. ME1 also participates in the Warburg effect (aerobic glycolysis, secretion of lactate) that is characteristic of many cancer cells and tumors. Lactate may serve as a paracrine energy source for tumor cells. ACC, acetyl-CoA carboxylase; ACLY, ATP citrate lyase; FASN, fatty acid synthase; Glu, Glutamate; GLUTs, glucose transporters; LDH-A, lactate dehydrogenase A; MDH1, malate dehydrogenase 1.
ME1 and the cytosolic NADPH pool
NADPH is a universal electron donor in all organisms and is a required cofactor in the biosynthesis of major cell components. NADPH is also a required participant in other biological processes such as anti-oxidative stress pathways, detoxification pathways, and the generation of reactive oxygen species (ROS). To help maintain an adequate pool of NADPH, regeneration of the oxidized form (NADP+) occurs. Within the cell cytoplasm, there are three major enzymes/pathways that contribute to NADP-NADPH recycling; these are ME1, isocitrate dehydrogenase 1 (IDH1), and two enzymes of the pentose phosphate pathway (glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (PGD)). Published work indicates that these enzymes comprise a regulatory network in maintaining the cytosolic NADPH pool, which remains not well understood (Merritt et al. 2009, Rzezniczak & Merritt 2012, Goodman et al. 2018).
ME1: the early years
ME1 was an active focus of research during the 1980s and 1990s, serving as a model for nutritional and hormonal regulation of gene expression/transcription and in so doing, contributed to the emergence of the field of molecular endocrinology. Insulin and thyroxine were found to induce ME1 gene expression (transcriptional rate and mRNA abundance) in rat liver and adipose (Drake & Mucenski 1985, McHugh & Drake 1989, Garcia-Jiménez et al. 1994). Consistent with ME1 as a lipogenic enzyme, a high carbohydrate diet up-regulated liver and adipose ME1 expression (Mariash et al. 1980). ME1 gene expression was increased in the epididymal fat of rats fed a cafeteria diet, high in fat and carbohydrates, coincident with increased lipid synthesis and storage (López et al. 2003). Dietary effects on ME1 gene expression were not limited to liver and adipose tissues. In C57BL/6J mice, short-term feeding of a high-fat diet led to inductions (~18-fold) in ME1 expression in the small intestine, coincident with increased cell proliferation in the small intestine crypts, resulting in greater villus length and cell number per villus (de Wit et al. 2008). Similarly, C57BL/6N mice fed a Western-type diet high in fat had increased (~3-fold) small intestine ME1 transcript levels (Desmarchelier et al. 2012). Support for the direct involvement of ME1 in the proliferation of colon stem-progenitor cells came from studies using mice null for ME1 protein (Al-Dwairi et al. 2012). In this study, average colon crypt depth of wild-type (WT) C57BL/6J mice was found to be greater than that for counterpart mice that lacked ME1 protein, when both groups were fed a high-fat diet. Conversely, enhancement of gastrointestinal expression of ME1 in C57BL/6J mice (via germline integration of a villin-ME1 cDNA transgene) conferred, in jejunum, a phenotype of increased intracellular NADPH, deeper crypts, and greater incorporation of BrdU into crypt stem-progenitor cells (Al-Dwairi et al. 2014). These collective findings led to an emergent picture of ME1 as a nutritionally- and hormonally-regulated metabolic gene/protein with lipogenic actions in liver and adipose and with NADPH-elevating and pro-proliferative actions within the gastrointestinal tract. These observations likewise support the contribution (direct or indirect) of ME1 in the development of obesity and type 2 diabetes (Yang et al. 2009; Zhong et al. 2010), via its integrated actions in multiple tissues (Al-Dwairi et al. 2014).
ME1 in cancer biology
Pioneering work in the 1990’s showed that ME1 ‘activity’ was relatively high in human tumor cells, and that the tumor-expressed ME1 protein was the same as that expressed in normal liver and adipose (Loeber et al. 1994). Many clinical studies of more recent vintage have indeed described the elevated ME1 expression in tumors when compared to adjacent non-tumor tissues (Table 1). These studies serve to highlight the linkage of ME1 over-expression with multiple solid tumors (e.g., gastrointestinal tract, hepatobiliary system and the aerodigestive tract, in particular) and importantly, argue for the potential prognostic value of tumor ME1 expression in adverse patient outcomes (Table 1). From the standpoint of human tumor genomic landscapes, ME1 mutations are relatively rare (1–2% frequency). Exceptions to the latter are prostate and endometrial cancers, where deletions, missense mutations and amplifications occur at frequencies between 3–6%. The underlying molecular basis for cancer-related ME1 over-expression remains an important question, given the limited studies to date. In human colorectal cancers, mutations in TP53 and KRAS elicited induction of steady-state ME1 mRNA levels (Jiang et al. 2013, Chakrabarti 2015, Shen et al. 2017). The contribution of microRNAs (miRNAs) in the regulation of ME1 expression is gaining ground, with specific miRNAs negatively influencing ME1 expression (Table 2). A decline in abundance of miRNA(s) that interact with ME1 mRNA in tumor cells resulted in an imbalance in redox potential favoring oxidative stress and increased accumulation of intracellular reactive oxygen species (ROS) and consequently, promotion of DNA damage (Table 2). The ME1 gene is richly endowed with diverse transcriptional regulatory motifs as well as larger multipartite regions for transcription factor interactions, including those that mediate its transcriptional regulation by insulin, thyroxine, nuclear receptors (e.g., PPARs) and NRF2, the latter a master regulator of anti-oxidative stress pathways. However, the potential contributions of these cis- and trans-acting motifs/factors to tumor ME1 over-expression remain largely unexplored.
Table 1:
ME1 as a Human Cancer Biomarker
| Cancer (tumor type) | Approach | Reported Associations/Findings | References |
|---|---|---|---|
| Human gastric cancer (adenocarcinoma) | Immunohistochemistry (IHC) of tumors. | ME1 was more highly expressed in tumors than adjacent normal tissue; ME1 was more highly expressed in metastases than primary tumor; tumor ME1 protein abundance (IHC signal) was correlated with shorter overall survival and disease-free survival of patients. | Lu et al. (2018b) |
| Human gastric cancer (adenocarcinoma) | IHC of tissue multi array (TMA); independent scoring by several pathologists. | No difference in IHC staining score between tumor and adjacent normal (differs from Lu et al. 2018); no significant correlation between ME1 expression and clinico-pathological parameters; however, moderate or high ME1 expression predicted lower cumulative patient survival. | Shi et al. (2019) |
| Human breast cancer | Mining of gene expression databases; Western blots of tumor lysates. | ME1 was more highly expressed in basal-like breast cancer (BLBC) than luminal breast cancer. ME1 copy number amplification was evident for many BLBCs; larger tumors expressed more ME1; Grade 3 tumors expressed more ME1 than Grade 1 and 2 tumors; patients with tumors with higher ME1-expression had worse overall and relapse-free survival and higher frequency of chemotherapy resistance. | Liao et al. (2018) |
| Human colon (mucinous adenocarcinoma) | Mining of gene expression databases. | ME1 mRNA expression was greater in tumor than adjacent non-tumor tissue. | Jiang et al. (2013) |
| Human colon and rectal cancers (adenocarcinoma) | IHC of TMA. | ME1 protein content was positively correlated with tumor pathological stage and invasion. | Gdynia et al. (2016) |
| Human colon and rectal cancers (adenocarcinoma) | Quantitative reverse transcriptase-PCR (qRT-PCR), Western blot analysis. | ME1 expression was higher in tumor than adjacent non-tumor tissue; KRAS-mutant tumors had more ME1 than KRASwt tumors. | Shen et al. (2017) |
| Human colon and rectal cancers (adenocarcinoma) | IHC of TMA. | ME1 expression was robust in tumor epithelial but not stromal cells; however, not all tumors were of the high ME1-expressing type. | Fernandes et al. (2018) |
| Human hepatocellular carcinoma (HCC) (adenocarcinoma) | IHC of tumor and paired adjacent non-tumor; segregation of patients/tumors into ME1 high vs. ME1 low-expressing types. | HCC had more ME1 staining than did adjacent non-tumor tissue; patients with an “ME1 high” tumor had lower overall survival and progression-free survival than did those with an “ME1 low” tumor. | Wen et al. (2015) |
| Human hepatocellular cholangiocarcinoma, subtype with stem cell features, intermediate-cell type (INT) | Microarrays and IHC. | INT tumor had greater expression of ME1 mRNA and protein than did HCC or intrahepatic cholangiocarcinoma tumors. | Mihara et al. (2019) |
| Human non-small cell lung cancer (NSCLC) | IHC of TMA (tumors and including some non-tumor tissues). | ME1 protein abundance was greater in NSCLC than non-tumor lung tissue; within NSCLC, ME1 expression was greater for squamous cell carcinomas than for adenocarcinomas; ME1 expression was greater in tumors of smokers than non-smokers; ME1 expression in primary tumor was positively associated with metastasis to lymph nodes; tumor ME1 expression predicted longer overall survival and shorter overall survival in younger and older patients, respectively (median cut-off of 65 years of age). | Csanadi et al. (2015) |
| Human non-small cell lung cancer (NSCLC) | Mining of gene expression databases. | Patients with elevated tumoral ME1 expression had worse overall survival after radiotherapy, but this effect was not observed with chemotherapy alone. | Chakrabarti (2015) |
| Human hereditary clear cell renal cell carcinoma | Mining of gene expression databases. | ME1 expression was greater in tumor than non-tumor tissue. | Jiang et al. (2013) |
| Human oral squamous cell carcinoma | IHC of TMA. | ME1 expression was greater in tumor than non-tumor tissue; ME1 expression was positively associated with cancer progression; patients with moderate to high tumor ME1 had worse prognosis. | Nakashima et al. (2018) |
| Human laryngeal squamous cell carcinoma | Microarrays of tumors and paired nonmalignant mucosa. | ME1 mRNA expression was greater in tumor than non-tumor tissue. | Nicolau-Neto et al. (2020) |
Table 2.
Functional Identification of ME1 mRNA-interacting microRNAs
| MicroRNA | Cells/tumors | Associations/effects | References |
|---|---|---|---|
| miR-30a | KRAS-mutant colorectal cancers; HCT116 and DLD1 human colorectal cancer cell lines | Over-expression (OE) of miR-30a suppressed ME1 mRNA and protein; miR-30a was down regulated in KRAS-mutant colorectal tumors compared to KRASwt tumors; miR-30a inhibited the growth of HCT116 and DLD1 cells in vitro; miR-30 attenuated proliferation, migration and invasion of HCT116 cells in vitro, as well as growth of HCT116 xenografts in nude mice. | Shen et al. (2017) |
| miR-612 | Human T24 bladder cancer cell line | miR-612 expression was decreased with bladder tumor progression; miR-612 depressed the expression of ME1; OE of miR-612 inhibited bladder cancer cell growth, colony formation, migration, invasion and epithelial-mesenchymal transition. | Liu et al. (2018) |
| miR-30c-5p | Mouse aorta smooth muscle cells | OE of miR-30c-5p caused down-regulation of ME1 and increased ROS in vitro; in vivo silencing of miR-30c-5p led to increase in ME1 and reduction in ROS. | Zhu et al. (2018) |
The positive association between ME1 and cell migration (Table 2) is strongly supported by recent studies that directly targeted ME1 in a diverse array of tumor cell models (Table 3). Knock-down of ME1 mRNA or inhibition of ME1 activity, in tumor cell lines and mouse models consistently abrogated proliferation, epithelial-to-mesenchymal transition (EMT) and in vitro migration, and promoted oxidative stress, apoptosis and/or senescence (Table 3). While the mechanistic underpinnings of ME1 to these cancer cell phenotypes remain unknown, elevations in intracellular NADPH levels can lead to suppression of apoptosis (Yang et al. 2010). However, documentation of this putative association in tumor cells remains lacking. Similarly, increased metabolic fluxes involving the biosynthesis of membrane components such as fatty acids and cholesterol and the pyruvate-malate cycle, provide essential support for cancer cell proliferation and migration (Fig. 1).
Table 3.
Effects of Knock-down (KD), Knock-out (KO) or Over-expression (OE) of ME1 in Human and Mouse Tumor Cells
| Cell line(s)/Models | Reagent/Approach | Major Effect(s) of ME1 Perturbation | References |
|---|---|---|---|
| HCT116 human colorectal cancer | siRNA KD | Decreased cell proliferation, decreased colony formation in agar, decreased xenograft growth in nude mice, reduced glutamine consumption, increased number of senescent cells, no effect on apoptosis or intracellular ROS level. | Jiang et al. (2013) |
| HCT116 human colorectal cancer | siRNA KD | Increased number of senescent cells, decreased cell growth, decreased colony formation in vitro, increased rate of glycolysis, increased flux through the pentose phosphate pathway, oxidative stress state, decreased extracellular glucose levels, increased extracellular lactate levels. | Murai et al. (2017) |
| HCT116, DLD1 human colorectal cancer | shRNA KD | Decreased colony formation in vitro, decreased content of intracellular NADPH and triglycerides in KRAS-mutant but not KRASwt cell lines, decreased xenograft growth in nude mice. | Shen et al. (2017) |
| HCT116, HT29 human colorectal cancer | Small molecule inhibitor of ME1 | Suppressed growth (fewer cell numbers) in vitro, decreased cell viability, decreased colony-formation in vitro. | Fernandes et al. (2018) |
| HCT116 human colorectal cancer | CRISPR deletions | ME1 KO: no effect on in vitro
growth or fatty acid synthesis. ME1/G6PD KO: major cell growth inhibition. |
Chen et al. (2019) |
| ApcMin mouse model of Familial Adenomatous Polyposis | ME1 OE (villin promoter-enhancer driving rat ME1 transgene) | Increased number (and overall size distribution) of adenomas in the small intestine, increased expression of Sp5, a Wnt-inducible transcription factor of intestinal stem-progenitor cells, no effect on EMT-related gene expression, trend to decreased apoptosis. | Fernandes et al. (2018) |
| AOM/DSS-induced mouse colorectal cancer model | ME1 OE (adenoviral ME1 construct delivered intraperitoneal) | Mice that received adenoviral ME1 exhibited increased number and size of carcinomas in the colo-rectum. | Zhu et al. (2020) |
| SGC7901, MGC803 human gastric cancer | shRNA KD, low glucose medium | ME1 KD: decreased NADPH level, increased apoptosis, increased intracellular level of ROS. | Lu et al. (2018b) |
| SGC7901, MGC803 human gastric cancer | shRNA KD, anoikis condition | ME1 KD: decreased NADPH level, increased intracellular H2O2, decreased colony-formation, increased apoptosis. | Lu et al. (2018b) |
| SGC7901 human gastric cancer | shRNA KD | ME1 KD: less cell growth, cell cycle arrest (in G2), decreased cell migration (transwell assay). | Shi et al. (2019) |
| MHCC97H, HCCLM3 human hepatocellular carcinoma | shRNA KD | ME1 KD: decreased NADPH level, increased ROS, decreased cell migration and invasion (transwell assay), reversion of EMT. | Wen et al. (2015) |
| SUM159, T47D human breast cancer | shRNA KD, OE | ME1 enhanced glucose uptake and lactate production by cells, depressed OXPHOS, and promoted aerobic glycolysis; perturbations in ME1 levels caused minimal growth effects under normoxic conditions, however, KD of ME1 under hypoxic conditions caused inhibition of proliferation, ME1 promoted colony formation under normoxic conditions. | Liao et al. (2018) |
| BT474 human ERBB2-positive breast cancer | shRNA KD | ME1 KD: decreased cell viability. | Kourtidis et al. (2010) |
| H522 (KRASwt), HCC44 (KRASmut) human non-small cell lung cancer (NSCLC) cell lines | siRNA KD | ME1 KD had no effect on clonogenic survival (colony number) after radiation treatment; whereas KD of HCC44 cells eliminated colony formation after radiation treatment. | Chakrabarti (2015) |
| A549 human lung cancer U2OS Human osteosarcoma | siRNA KD of wild-type ME1 coupled with OE of enzyme-inactive (mutant) ME1 | OE of enzymatically dead ME1 augmented proliferation and colony formation of tumor cells (A549, U2OS) that were devoid of wild-type ME1. | Yao et al. (2017) |
| T24 human bladder cancer | siRNA KD | ME1 KD: decreases in cell proliferation, colony formation, migration, invasion, reversion of EMT. | Liu et al. (2018) |
| CNE-2 human nasopharyngeal cancer | shRNA KD | ME1 KD: inductions in expression of the PPP enzymes G6PD and PGD, NADPH level was suppressed only in a low glucose medium, reduced level of GSH, increased intracellular ROS, decreased levels of FASN and SNAIL, decreased cell migration and invasion in vitro. | Zheng et al. (2012) |
| HN31 human head and neck squamous cell carcinoma (HNSCC) | shRNA KD | ME1 KD: no effect on NADPH level, increased cell sensitivity to metformin or ionizing radiation in the colony formation assay, enhanced senescence with metformin treatment, increased intracellular ROS level. | Woo et al. (2016) |
| HSC3 human tongue squamous cell carcinoma | ME1 antisense oligonucleotide | ME1 KD: decreased cell proliferation, reversion of EMT, less “stemness”, decreased lactate production, decreased GSH content, increased oxidative stress state. | Nakashima et al. (2018) |
Our examination of gene expression databases, established from diverse cancer cell lines, revealed consistent co-expression of ME1 mRNA with mRNAs encoding two cytosolic NADPH generating enzymes (G6PD and IDH1), other cytosolic enzymes that utilize NADPH as an electron donor (e.g., prostaglandin reductase 1 (PTGR1), NAD(P)H quinone dehydrogenase 1 (NQO1) and proteins that may serve anti-oxidative stress functions (e.g., tripartite motif containing 16 (TRIM16)). Similarly, perusal of expression databases for human tumors show robust co-expression of ME1 mRNA with mRNAs encoding reductases (e.g., PTGR1) that utilize NADPH, as well as those encoding anti-oxidative stress pathway protein components. The expression correlations noted between ME1, G6PD and IDH1 warrant further interest. G6PD is a rate-limiting enzyme of the pentose phosphate pathway and its over-expression has been noted for multiple human tumor types (Chen et al. 2018, Lu et al. 2018a). Similar to ME1, knock-down of G6PD abrogated tumor cell growth and enhanced intracellular levels of ROS (Chen et al. 2018) and inhibited in vitro cell migration and invasion (Lu et al. 2018a). IDH1 mutations are drivers of several human malignancies (reviewed in Bergaggio & Piva 2019). Overexpression of wild-type IDH1 has also been observed for multiple cancers (e.g., non-small cell lung carcinoma (NSCLC), glioblastoma multiforme (GBM)) whereas its KD abrogated in vitro and in vivo tumor cell growth, NADPH/NADP ratio, and resistance to oxidative stress (Bergaggio & Piva 2019), similar to that observed for ME1 KD (Table 3).
ME1 stimulates the pentose phosphate pathway (PPP) flux via a physical interaction with and resultant activation of 6-phosphogluconate dehydrogenase (6PGD), the second PPP enzyme that also provides reduced NADP in cytosol (Yao et al. 2017). 6PGD itself is implicated in tumorigenesis (Sarfraz et al. 2020). Notably, ME1 enhanced 6PGD levels, functioning independent of its enzymatic activity, as evidenced from studies using constructs encoding enzymatically inactive, mutant ME1. Remarkably, when these same mutant ME1 proteins were expressed in cells in which wild-type ME1 was suppressed, augmented cell growth and colony formation in vitro were found, supporting the conclusion that ME1’s pro-proliferative effects are primarily mediated by interaction with and activation of 6PGD (Yao et al. 2017; Table 3). This new mechanism may explain some of the observed effects of KD and over-expression of ME1 in tumor cells (Table 3); however, they are not easily reconciled with the cellular growth-inhibitory effects of ME1 active site-inhibitors (Zhang et al. 2006; Saha et al. 2014, Fernandes et al. 2018). The reciprocal and dynamic regulation of ME1 activity (manifested as increased NADPH/NADP ratio and increased production of fatty acids) by acetylation at amino acid K337 (positive regulation) and by phosphorylation at amino acid S336 (negative regulation) (Zhu et al. 2020) raises the important question of whether post-translational modification(s) of ME1 affect its interactions with 6PGD, in differing tumor types and tissue contexts.
Studies of gastrointestinal cancers have illuminated the pro-proliferative role of ME1. Intercrossing of a villin-ME1 transgene into the ApcMin mouse background led to increased numbers as well as overall sizes of adenomas within the small intestine (Fernandes et al. 2018; Table 3). Also of interest were the noted changes in ME1 expression in the outwardly “normal” mucosa adjacent to adenomas. Fernandes et al. 2018 demonstrated that normal-appearing small intestine villi bordering individual adenomas displayed increased ME1 immunoreactivity as well as increased numbers of mucin-producing goblet cells compared to more distant, and presumably more normal, villi. These “transitional” villi, presumably lying within the cancer field, had deeper crypts and were longer than their counterparts more distant from the adenoma. In this regard, a previous report described an increase in lipid content in the epithelial-like borders of ApcMin mouse adenomas (Mutoh et al. 2009). We speculate that the ME1 phenotype for cancers of the gastrointestinal tract includes a significant lipogenesis component, as well as paracrine communication between tumors and adjacent non-tumor tissue, to induce production of lipids and mucins in the latter for tumor growth.
The ME1-related protein, ME2 also is implicated in tumorigenesis. Recent work has documented the over-expression of ME2 and its functional contributions to tumor phenotypes of erythroleukemia (Ren et al. 2010), glioblastoma multiforme (GBM) (Cheng et al. 2016), and head and neck squamous cell carcinoma (Woo et al. 2016). In the non-small cell lung cancer (NSCLC) cell line A549, ME2 KD resulted in growth cessation, increased cell death, higher levels of ROS, lower ATP and an increased sensitivity to cisplatin (Ren et al. 2014). Interestingly, in the same study, KD of ME2 also led to an induction in PTEN levels and a down-regulation of AKT signaling. This reciprocal regulation between ME2 and tumor suppressor (PTEN) vs. tumor promoter (AKT) is reminiscent of the inverse relationship found for ME1 and TP53 in colon cancer cells (Jiang et al. 2013). Further, the interesting connections between the malic enzymes and major tumor suppressive and tumor promoting pathways suggest their more extensive linkages in various cancers and are worthy of further exploration.
ME1 and lung cancer subtypes: a paradigm for parsing ME1-dependent tumor pathways
Human lung cancers express higher levels of ME1 (gene and protein) than does normal lung tissue, and tumor ME1 over-expression may confer a worse patient prognosis (Table 1). Within lung cancer subtypes, squamous cell carcinomas express more ME1 than do adenocarcinomas (Table 1). Perusal of gene expression databases for lung tumors revealed robust and relatively consistent associations of ME1 mRNA with mRNAs encoding multiple NADPH-dependent reductases (Fig. 2). As a class, NADP/NADPH-dependent reductases are involved in the detoxification or activation of xenobiotics, metabolism of endogenous biochemical pathway intermediates, regeneration of endogenous antioxidants, and overall mediation of cellular redox state. These enzymes also influence susceptibility of cells to exogenous drugs and carcinogens. One example of this is prostaglandin reductase 1 (PTGR1), a cytosolic, NADP-dependent, alkenal/one oxidoreductase that inactivates prostaglandins and related eicosanoids (Sánchez-Rodriguez et al. 2017) and which is highly co-expressed with ME1 (Fig. 2). This protein has significant anti-oxidative actions by virtue of its reduction of 4-hydroxy-2-nonenal (4HNE) and fatty acid nitroalkenes. Recent studies have pointed to the potential targeting of PTGR1 in treatment of prostate cancer (Xue et al. 2016) and hepatocellular carcinoma (Sánchez-Rodriguez et al. 2017). In particular, KD of PTGR1 led to decreased cell proliferation, increased apoptosis and decreased protection against hydrogen peroxide and 4-HNE-induced cell death. Yet another example of an ME1 co-expressed mRNA in lung tumors (Fig. 2) is the mRNA for thioredoxin reductase 1 (TXNRD1), an enzyme which utilizes NADPH to regenerate reduced thioredoxin and thereby, increase cellular reducing capacity and protection against oxidative stress. TXNRD1 KD in radiation-resistant lung cancer cells sensitized these cells to radiation treatment (Hao et al. 2017). A third example (Fig. 2) is the mRNA that encodes aldo-keto reductase family 1 member C1 (AKR1C1), a NADP-dependent cytosolic enzyme involved in intermediary metabolism and for which multiple studies have identified contributory roles in therapy resistance, EMT and metastasis of cancer cells (Tian et al. 2016, Zhu et al. 2018).
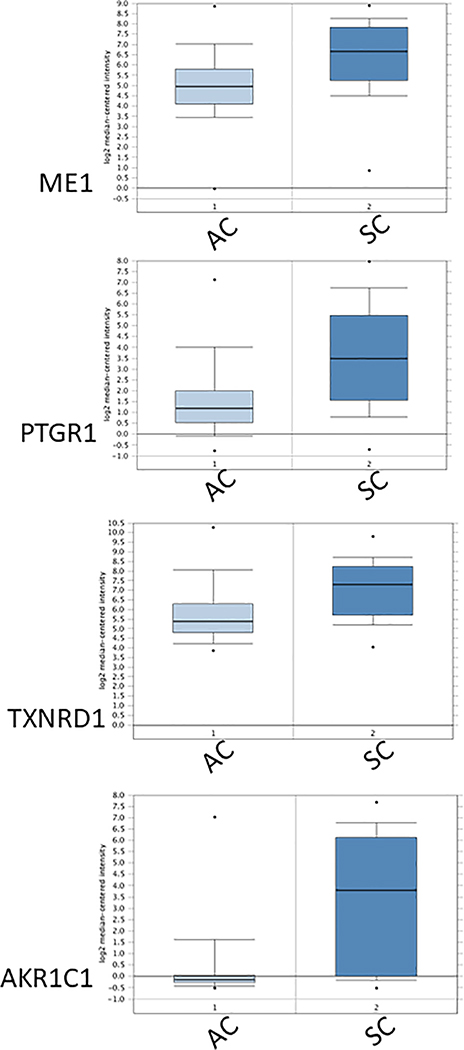
Figure 2.
Elevated abundance and significant co-expression of mRNAs encoding ME1 and the NADPH-dependent proteins PTGR1, TXNRD1 and AKR1C1 in human lung squamous cell carcinomas (SC) compared to human lung adenocarcinomas (AC). Co-expression analysis identified many NADPH-dependent reductases that are highly co-expressed with ME1 in lung tumors; shown are expression data for three such genes. Box plots present normalized expression data for 63 individual AC and 75 individual SC (obtained from Oncomine Research Edition ((http://www.oncomine.org; Rhodes et al. 2004); gene expression data are originally from Lee et al. 2008). Dots represent maximum and minimum values; error bars represent 10th and 90th percentiles; boxes span 25th to 75th percentiles; thick line is the median. Fold changes (SC compared to AC) were ME1, 2.677; PTGR1, 3.818; TXNRD1, 2.405; AKR1C1, 8.188. All fold-differences were highly significant (P <0.01).
Of interest are the findings that mutations in TP53 and PI3KCA are more frequent in lung squamous cell carcinomas than in lung adenocarcinomas (de Sousa & Carvalho 2018). Inference from other cancers such as those of the colo-rectum suggests that mutations in both genes, along with those of KRAS, constitute potential drivers of enhanced ME1 expression in squamous cell carcinomas. Lung adenocarcinomas exhibit similar, as well as distinct, metabolic characteristics from lung squamous cell carcinomas (Rocha et al. 2015). Both lung cancer subtypes manifest respectively, lower glucose and higher lactate levels, when compared to non-tumor tissue; however, squamous carcinomas have a more exaggerated glycolytic phenotype and have greater reliance on glutaminolysis (Rocha et al. 2015), highlighting known functional linkages of over-expressed ME1 with increased glutaminolytic flux. Interestingly, squamous adenocarcinomas also have a greater amount of reduced glutathione (GSH, γ-Glu-Cys-Gly) (Rocha et al. 2015), consistent with more ME1 available to supply the NADPH used for GSH regeneration. Published work of others (Lee et al. 2008, Chakrabarti 2015, Csanadi et al. 2015, Hao et al. 2017, Yan et al. 2019) support the concept of targeting ME1 and downstream NADPH-dependent pathways as an adjunct approach to lung cancer treatment.
ME1 at the nexus of hyperinsulinemia and cancer initiation and progression: NOX/DUOX family
Obesity and type 2 diabetes are associated with elevated risks for numerous cancers (Lega & Lipscombe 2020). Sequela of obesity and diabetes that are likely contributory to this elevated cancer risk include hyperinsulinemia, hyperglycemia, inflammation and oxidative stress. Based on the current literature, oxidative stress is a probable point of convergence of ME1, obesity and type 2 diabetes, and cancer development. Perhaps, increased oxidative stress (via induced ME1) provides one mode by which hyperinsulinemia increases cancer risk.
Reactive oxygen species (e.g., .NO, H2O2, OH and O2-) are chemically reactive molecules that contain oxygen and are important participants in extracellular and intracellular signaling elicited by various hormones and growth factors (Bauer 2014). Depending on their steady-state levels and cell/tissue context, they can act as inducers of cell proliferation and of cell death. At low levels, ROS may act as signaling molecules that activate proliferation. ROS at intermediate levels can promote DNA damage and mutagenesis while at high levels, they can cause oxidative stress that may result in apoptosis or senescence. Since cancer cells, which are highly proliferative, produce relatively high levels of ROS, they must buffer excess ROS to maintain proliferation, while avoiding apoptosis and senescence. This intracellular redox state is maintained via the buffering capacity of the reduced glutathione (GSH)-oxidized glutathione (GSSG) system and the thioredoxin system, in support of proliferation.
One mechanism by which NADPH (and indirectly ME1) can contribute to oxidative damage is via its direct coupling to NADPH oxidases (NOX/DUOX family members) that catalyze the formation of ROS. This pathway also provides a plausible means by which insulin can induce DNA damage in cells during pre-neoplastic and neoplastic stages. Studies have implicated NOX/DUOX family members in the pathway(s) by which elevations in circulating insulin lead to inductions in ROS and ROS-mediated DNA damage (Xiao et al. 2008, Meng et al. 2012, Othman et al. 2014, Mahmoud et al. 2017, Steinhorn et al. 2017). Moreover, suppression of ROS generation by inhibition of NOX/DUOX member(s) can inhibit insulin signaling (Sypowska & Burgering 2011).
One tissue whose cancer risk is known to be positively associated with obesity, diabetes and hyperinsulinemia is the colo-rectum. Certain members of the NOX/DUOX family were found to be over-expressed and likely growth promoting in tumors of the human colon and rectum (Juhasz et al. 2009, Juhasz et al. 2017, Lin et al. 2017, Cho et al. 2018, Kang et al. 2018). NOX1 overexpression (similar to that of ME1) in human colorectal cancers is associated with activating mutations in KRAS (Laurent et al. 2008). Interestingly, individual NOX/DUOX members differ with respect to their apparent contributions to clinical outlook and tumor therapeutic response (Lin et al. 2017, Cho et al. 2018). Cancer cells, compared to normal cells, are generally more susceptible to the cytotoxic effects of glucose deprivation, effects that are mediated, in part, by their increased production of ROS via NOX/DUOX (Aykin-Burns et al. 2009). In this regard, human colon cancer cells (HCT116) grown in a glucose-restricted medium were extra-sensitive to the growth-inhibition elicited by ME1 KD (Murai et al. 2017). Screening of human colon and rectal cancers for KRAS mutations coupled with determination of relative co-expression of ME1 and NOX/DUOX genes/proteins may allow dissection of these potential functional linkages and if confirmed, enable therapeutic targeting of tumor cells via ME1 and NOX/DUOX inhibitors.
Future directions
The work to date suggests several avenues to refine our understanding of ME1 as pro-oncogenic in multiple tissues and to apply this emerging knowledge to future cancer treatments. Foremost to consider is that ME1 is a participant in a network of cancer promoters, rather than as a single, independent entity (Fig. 3). We infer that ME1 is a major contributor to: a) cancer cell biosynthesis of lipids and cholesterol, b) generation of ROS via NOX/DUOX proteins for pro-proliferative (and perhaps for pro-migration/EMT) signaling, and c) buffering of ROS via NADPH-dependent recycling of glutathione and thioredoxin pathways, working in concert with other NADP-recycling cytosolic enzymes G6PD, PGD and IDH1 (Fig. 3). We further hypothesize that insulin (i.e., hyperinsulinemia) via its coincident induction of ME1 expression and of NOX/DUOX activity in pre-neoplastic cells, can tip the balance towards oxidative stress/genotoxic state while countering ROS-induced apoptosis and senescence (Fig. 3).
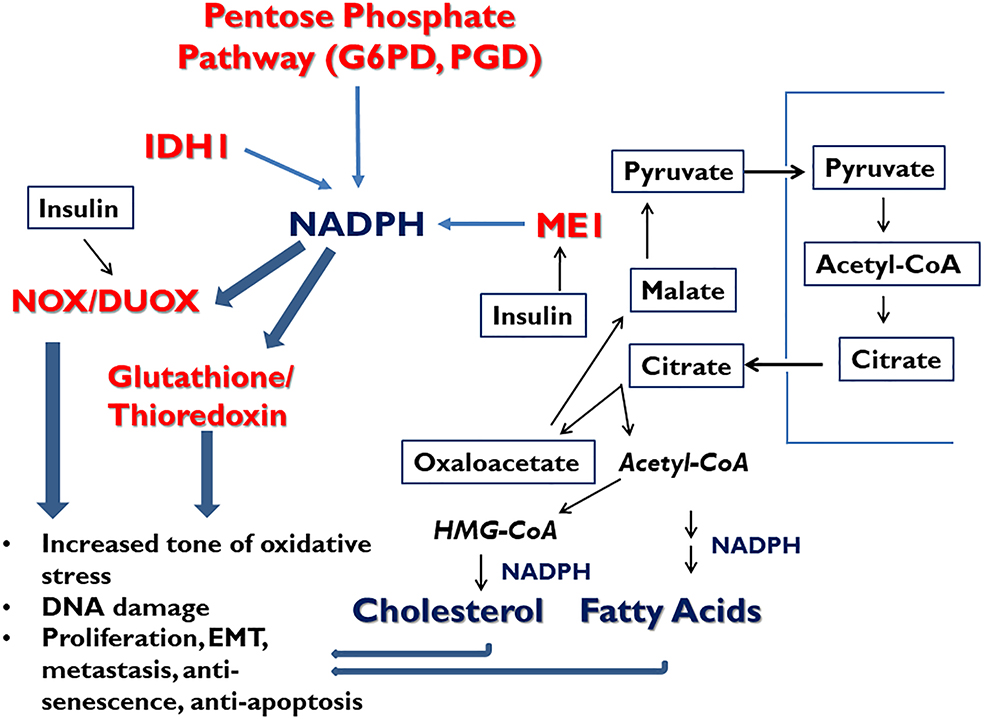
Figure 3.
Integrative model for involvement of ME1 and the other cytosolic NADP-reducing enzymes in the initiation and progression of cancer. The model posits that ME1, IDH1, and pentose phosphate pathway function collaboratively within a network to maintain required levels of NADPH. In pre-cancerous and tumor cells, one or more of these enzymes/pathways becomes over-expressed, or augmented in activity, thereby favoring increased steady state levels of NADPH. This elevation in NADPH promotes NOX/DUOX signaling leading to increased oxidative stress and DNA damage, and increased glutathione and thioredoxin recycling; these latter pathways protect tumor cells from the apoptosis-inducing effects of increased oxidative stress. The NADPH-dependent pathways favor proliferation, DNA damage, epithelial-mesenchymal transition (EMT), cell migration and metastasis, anti-senescence and anti-apoptosis. We further posit that hyperinsulinemia drives a) the increased expression of ME1 favoring increased activity of the pyruvate-malate cycle and thereby, leading to increased synthesis of fatty acids and cholesterol in support of biosynthesis, proliferation and anti-apoptosis (Xiao et al. 2008), and b) the increased activity of NOX/DUOX members. The collective actions contribute to a heightened genotoxic and proliferative state, ultimately contributing to tumor genesis.
In Drosophila flies and human cancer cells, the major cytosolic NADP-reducing enzymes (ME1, G6PD, PGD, IDH1) behave as a network, with changes of some members occurring in response to diminution in expression/activity of another (Merritt et al. 2009, Rzezniczak & Merritt 2012, Chen et al. 2019). Interestingly, not all such changes in this network are compensatory, suggesting an unappreciated degree of complexity. A key to understanding the nature of these relationships and their cooperation during tumor genesis will require further discernment of their network regulation, of which little is presently known. Additional knowledge of these enzyme’s interactions, both physical and temporal, within normal, pre-cancerous and cancerous cells is clearly of interest. Further molecular dissection of the NADP/NADPH flux as well as identification of the functioning NADP/NADPHome (Goodman et al. 2018) in tumor cells will be important for the selective targeting of NADPH regeneration leading to tumor progression. In this regard, ROS-induced signaling can occur in defined signaling complexes (Xi et al. 2013, Bauer 2014). Thus, a defined molecular picture of the spatiotemporal organization of ME1, G6PD, PGD, IDH1, other NADPHome partners, and NADPH is necessary to understand how these proteins form complex tumor-promoting networks. The role of ME1 and its immediate network partners (and the expanded NADPHome) in oxidative stress during aging (Hecker et al. 2014), and how these pathways influence cancer development in older individuals is ripe for further exploration.
Glutathione and thioredoxin anti-oxidant pathways buffer oxidative stress in cancer cells and appear to be synergistic with respect to tumor initiation and progression (Harris et al. 2015, Lien et al. 2016, Yan et al. 2019). Combined inhibition of both anti-oxidant pathways leads to cancer cell death in vitro and in vivo (Palomares et al. 2009). Targeting ME1 and its network of associated NADPH-regenerating enzymes may phenocopy or may be additive or even synergistic with the co-targeting of the glutathione and thioredoxin pathways to counter cancer cell growth, survival, metastasis and therapy resistance.
The nature (and the counter balance) of downstream oncogenic and tumor suppressive proteins/pathways that are activated by NOX/DUOX-elicited ROS in different tissues and tumors is an important question in cancer biology (Xi et al. 2013, Chen et al. 2018). Review of literature suggests that NADPH/NOX/DUOX signaling may be particularly relevant to initiation, progression and treatment of cancers of the colon and rectum. In this regard, Komiya et al. 2015 documented a suppressive effect of apocynin, an NADPH oxidase-inhibitor, on adenoma occurrence in the ApcMin mouse model.
Pyruvate, the oxidative product of ME1, is also consequential to any discussion of ME1 in cancer biology. One route that ME1-derived pyruvate can follow is its conversion to lactate by the enzyme lactate dehydrogenase A (LDH-A) during the process of anaerobic glycolysis (Warbug effect) (Fig. 1). LDH-A is itself a molecular target for cancer therapies (Miao et al. 2013).
While the studies summarized in Tables 2 and 3 have implicated ME1 in epithelial-mesenchymal transition (EMT) and tumor metastasis, many of the molecular details of how these actions are promoted by ME1 (and/or its network) are missing. However, one candidate pathway may involve the induction of EMT by DUOX2-generated ROS in 5-flourouracil-resistant human colon cancer cells in vitro (Kang et al. 2018).
Conclusions
Malic Enzyme 1 is an old enzyme, both figuratively and evolutionarily, with an illustrious history. It is an important player in intermediary metabolism in animals and plants where it is subject to complex multifactorial transcriptional and post-transcriptional regulation. As a multifunctional enzyme, ME1 contributes to adipogenesis, lipogenesis and steroidogenesis. Variations in human ME1 sequence/expression have been linked to propensity for developing type 2 diabetes and for fat deposition (Yang et al. 2009, Zhong et al. 2010). In the plant kingdom, ME1 has been explored as a marker/tool to increase stress resistance of plants. In its newest fascinating iteration, ME1 and its interacting network may constitute important drivers of cancer development and metastasis, in part, by inducing oxidative stress resistance, somewhat akin to its role in plant physiology. Further development and refinement of small molecule inhibitors of ME1 (Zhang et al. 2006, Saha et al. 2014, Fernandes et al. 2018) may prove useful as adjuncts to therapies for primary and metastatic cancers and particularly for cancers associated with obesity, type 2 diabetes and hyperinsulinemia.
Acknowledgments
Funding
Research from our laboratories was supported by the National Institutes of Health (R01CA136493; Translational Research Institute grant U54 TR001629) and UAMS intramural grant programs (Medical Research Endowment; Winthrop P. Rockefeller Cancer Institute Cancer Prevention and Population Science Program; Chancellor’s Research Development Award). The content is solely the responsibility of the authors.
Footnotes
Declaration of interest
The authors declare that there are no conflicts of interest that could be perceived as prejudicing the impartiality of this review.
References
- Al-Dwairi A, Pabona JM, Simmen RC, Simmen FA. Cytosolic malic enzyme 1 (ME1) mediates high fat diet-induced adiposity, endocrine profile, and gastrointestinal tract proliferation-associated biomarkers in male mice. PLoS One. 2012;7(10):e46716. doi: 10.1371/journal.pone.0046716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dwairi A, Brown AR, Pabona JM, Van TH, Hamdan H, Mercado CP, Quick CM, Wight PA, Simmen RC, Simmen FA. Enhanced gastrointestinal expression of cytosolic malic enzyme (ME1) induces intestinal and liver lipogenic gene expression and intestinal cell proliferation in mice. PLoS One. 2014;9(11):e113058 Published 2014 Nov 17. doi: 10.1371/journal.pone.0113058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykin-Burns N, Ahmad IM, Zhu Y, Oberley LW, Spitz DR. Increased levels of superoxide and H2O2 mediate the differential susceptibility of cancer cells versus normal cells to glucose deprivation. Biochem J. 2009;418(1):29–37. doi: 10.1042/BJ20081258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G Targeting extracellular ROS signaling of tumor cells. Anticancer Res. 2014;34(4):1467–1482. [PubMed] [Google Scholar]
- Bergaggio E, Piva R. Wild-Type IDH enzymes as actionable targets for cancer therapy. Cancers (Basel). 2019;11(4):563 Published 2019 Apr 19. doi: 10.3390/cancers11040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti G Mutant KRAS associated malic enzyme 1 expression is a predictive marker for radiation therapy response in non-small cell lung cancer. Radiat Oncol. 2015;10:145 Published 2015 Jul 16. doi: 10.1186/s13014-015-0457-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, Rabinowitz JD. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–415. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu Z, Zhu Z, Chen A, Fu G, Wang Y, Pan H, Jin B. Modulation of G6PD affects bladder cancer via ROS accumulation and the AKT pathway in vitro. Int J Oncol. 2018;53(4):1703–1712. doi: 10.3892/ijo.2018.4501 [DOI] [PubMed] [Google Scholar]
- Cheng CP, Huang LC, Chang YL, Hsieh CH, Huang SM, Hueng DY. The mechanisms of malic enzyme 2 in the tumorigenesis of human gliomas. Oncotarget. 2016;7(27):41460–41472. doi: 10.18632/oncotarget.9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SY, Kim JS, Eun HS, Kang SH, Lee ES, Kim SH, Sung JK, Lee BS, Jeong HY, Moon HS. Expression of NOX family genes and their clinical significance in colorectal cancer. Dig Dis Sci. 2018;63(9):2332–2340. doi: 10.1007/s10620-018-5121-5 [DOI] [PubMed] [Google Scholar]
- Csanadi A, Kayser C, Donauer M, Gumpp V, Aumann K, Rawluk J, Prasse A, zur Hausen A, Wiesemann S, Werner M, et al. Prognostic value of Malic Enzyme and ATP-Citrate Lyase in non-small cell lung cancer of the young and the elderly. PLoS One. 2015;10(5):e0126357 Published 2015 May 11. doi: 10.1371/journal.pone.0126357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarchelier C, Dahlhoff C, Keller S, Sailer M, Jahreis G, Daniel H. C57Bl/6 N mice on a western diet display reduced intestinal and hepatic cholesterol levels despite a plasma hypercholesterolemia. BMC Genomics. 2012;13:84 Published 2012 Mar 6. doi: 10.1186/1471-2164-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa VML, Carvalho L. Heterogeneity in lung cancer. Pathobiology. 2018;85(1–2):96–107. doi: 10.1159/000487440 [DOI] [PubMed] [Google Scholar]
- de Wit NJ, Bosch-Vermeulen H, de Groot PJ, Hooiveld GJ, Bromhaar MG, Jansen J, Muller M, van der Meer R. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med Genomics. 2008;1:14 Published 2008 May 6. doi: 10.1186/1755-8794-1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RL, Mucenski CG. Insulin mediates the asynchronous accumulation of hepatic albumin and malic enzyme messenger RNAs. Biochem Biophys Res Commun. 1985;130(1):317–324. doi: 10.1016/0006-291x(85)90420-6 [DOI] [PubMed] [Google Scholar]
- Fernandes LM, Al-Dwairi A, Simmen RCM, Marji M, Brown DM, Jewell SW, Simmen FA. Malic Enzyme 1 (ME1) is pro-oncogenic in ApcMin/+ mice. Sci Rep. 2018;8(1):14268 Published 2018 Sep 24. doi: 10.1038/s41598-018-32532-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Jiménez C, Benito B, Jolin T, Santisteban P. Insulin regulation of malic enzyme gene expression in rat liver: evidence for nuclear proteins that bind to two putative insulin response elements. Mol Endocrinol. 1994;8(10):1361–1369. doi: 10.1210/mend.8.10.7854353 [DOI] [PubMed] [Google Scholar]
- Gdynia G, Sauer SW, Kopitz J, Fuchs D, Duglova K, Ruppert T, Miler M, Pahl J, Cerwenka A, Enders M, et al. The HMGB1 protein induces a metabolic type of tumour cell death by blocking aerobic respiration. Nat Commun. 2016;7:10764 Published 2016 Mar 7. doi: 10.1038/ncomms10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RP, Calvo SE, Mootha VK. Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J Biol Chem. 2018;293(20):7508–7516. doi: 10.1074/jbc.TM117.000258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Xu X, Ma J, Xia J, Dai B, Liu L, Ma Y. MicroRNA-124 regulates the radiosensitivity of non-small cell lung cancer cells by targeting TXNRD1. Oncol Lett. 2017;13(4):2071–2078. doi: 10.3892/ol.2017.5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D Knobbe-Thomsen CB, Cox MA, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression [published correction appears in Cancer Cell. 2015 Feb 9;27(2):314]. Cancer Cell. 2015;27(2):211–222. doi: 10.1016/j.ccell.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, Meldrum E, Sanders YY, Thannickal VJ. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci Transl Med. 2014;6(231):231ra47. doi: 10.1126/scitranslmed.3008182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493(7434):689–693. doi: 10.1038/nature11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz A, Ge Y, Markel S, Chiu A, Matsumoto L, van Balgooy J, Roy K, Doroshow JH. Expression of NADPH oxidase homologues and accessory genes in human cancer cell lines, tumours and adjacent normal tissues. Free Radic Res. 2009;43(6):523–532. doi: 10.1080/10715760902918683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz A, Markel S, Gaur S, Liu H, Lu J, Jiang G, Wu X, Antony S, Wu Y, Melillo G, et al. NADPH oxidase 1 supports proliferation of colon cancer cells by modulating reactive oxygen species-dependent signal transduction. J Biol Chem. 2017;292(19):7866–7887. doi: 10.1074/jbc.M116.768283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KA, Ryu YS, Piao MJ, Shilnikova K, Kang HK, Yi JM, Boulanger M, Paolillo R, Bossis G, Yoon SY, et al. DUOX2-mediated production of reactive oxygen species induces epithelial mesenchymal transition in 5-fluorouracil resistant human colon cancer cells. Redox Biol. 2018;17:224–235. doi: 10.1016/j.redox.2018.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya M, Fujii G, Miyamoto S, Takahasi M, Ishigamori R, Onuma W, Ishino K, Totsuka Y, Fujimoto K, Mutoh M. Suppressive effects of the NADPH oxidase inhibitor apocynin on intestinal tumorigenesis in obese KK-A(y) and Apc mutant Min mice. Cancer Sci. 2015;106(11):1499–1505. doi: 10.1111/cas.12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70(5):1783–1792. doi: 10.1158/0008-5472.CAN-09-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent E, McCoy JW 3rd, Macina RA, Liu W, Cheng G, Robine S, Papkoff J, Lambeth JD. Nox1 is over-expressed in human colon cancers and correlates with activating mutations in K-Ras. Int J Cancer. 2008;123(1):100–107. doi: 10.1002/ijc.23423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, Kim H, Lee HJ, Choi HY, Jung Y, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14(22):7397–7404. doi: 10.1158/1078-0432.CCR-07-4937 [DOI] [PubMed] [Google Scholar]
- Lega IC, Lipscombe LL. Review: diabetes, obesity, and cancer-pathophysiology and clinical implications. Endocr Rev. 2020;41(1):bnz014. doi: 10.1210/endrev/bnz014 [DOI] [PubMed] [Google Scholar]
- Liao R, Ren G, Liu H, Chen X, Cao Q, Wu X, Li J, Dong C. ME1 promotes basal-like breast cancer progression and associates with poor prognosis. Sci Rep. 2018;8(1):16743 Published 2018 Nov 13. doi: 10.1038/s41598-018-35106-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, Toker A. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol. 2016;18(5):572–578. doi: 10.1038/ncb3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Chang IW, Hsieh PL, Lin CY, Sun DP, Sheu MJ, Yang CC, Lin LC, He HL, Tian YF. High immunoreactivity of DUOX2 is associated with poor response to preoperative chemoradiation therapy and worse prognosis in rectal cancers. J Cancer. 2017;8(14):2756–2764. Published 2017 Aug 23. doi: 10.7150/jca.19545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Chen Y, Huang B, Mao S, Cai K, Wang L, Yao X. Tumor-suppressing effects of microRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. Int J Oncol. 2018;52(6):1923–1933. doi: 10.3892/ijo.2018.4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber G, Dworkin MB, Infante A, Ahorn H. Characterization of cytosolic malic enzyme in human tumor cells. FEBS Letts. 1994;344:181–186 [DOI] [PubMed] [Google Scholar]
- López IP, Marti A, Milagro FI, Md MAZ, Moreno-Aliaga MJ, Martinez JA, Miguel CD. DNA microarray analysis of genes differentially expressed in diet-induced (cafeteria) obese rats. Obes Res. 2003;11(2):188–194. doi: 10.1038/oby.2003.30 [DOI] [PubMed] [Google Scholar]
- Lu M, Lu L, Dong Q, Yu G, Chen J, Qin L, Wang L, Zhu W, Jia H. Elevated G6PD expression contributes to migration and invasion of hepatocellular carcinoma cells by inducing epithelial-mesenchymal transition. Acta Biochim Biophys Sin (Shanghai). 2018a;50(4):370–380. doi: 10.1093/abbs/gmy009 [DOI] [PubMed] [Google Scholar]
- Lu YX, Ju HQ, Liu ZX, Chen DL, Wang Y, Zhao Q, Wu QN, Zeng ZL, Qiu HB, Hu PS, et al. ME1 regulates NADPH homeostasis to promote gastric cancer growth and metastasis [published correction appears in Cancer Res. 2019 Jul 15;79(14):3789]. Cancer Res. 2018b;78(8):1972–1985. doi: 10.1158/0008-5472.CAN-17-3155 [DOI] [PubMed] [Google Scholar]
- Mahmoud AM, Ali MM, Miranda ER, Mey JT, Blackburn BK, Haus JM, Phillips SA. Nox2 contributes to hyperinsulinemia-induced redox imbalance and impaired vascular function. Redox Biol. 2017;13:288–300. doi: 10.1016/j.redox.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariash CN, Kaiser FE, Schwartz HL, Towle HC, Oppenheimer JH. Synergism of thyroid hormone and high carbohydrate diet in the induction of lipogenic enzymes in the rat. Mechanisms and implications. J Clin Invest. 1980;65(5):1126–1134. doi: 10.1172/JCI109766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh KM, Drake RL. Insulin-mediated regulation of epididymal fat pad malic enzyme. Mol Cell Endocrinol. 1989;62(2):227–233. doi: 10.1016/0303-7207(89)90009-9 [DOI] [PubMed] [Google Scholar]
- Meng D, Mei A, Liu J, Kang X, Shi X, Qian R, Chen S. NADPH oxidase 4 mediates insulin-stimulated HIF-1α and VEGF expression, and angiogenesis in vitro. PLoS One. 2012;7(10):e48393. doi: 10.1371/journal.pone.0048393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt TJ, Kuczynski C, Sezgin E, Zhu CT, Kumagai S, Eanes WF. Quantifying interactions within the NADP(H) enzyme network in Drosophila melanogaster. Genetics. 2009;182(2):565–574. doi: 10.1534/genetics.109.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013;65(11):904–910. doi: 10.1002/iub.1216 [DOI] [PubMed] [Google Scholar]
- Mihara Y, Akiba J, Ogasawara S, Kondo R, Fukushima H, Itadani H, Obara H, Kakuma T, Kusano H, Naito Y, et al. Malic enzyme 1 is a potential marker of combined hepatocellular cholangiocarcinoma, subtype with stem-cell features, intermediate-cell type. Hepatol Res. 2019;49(9):1066–1075. doi: 10.1111/hepr.13365 [DOI] [PubMed] [Google Scholar]
- Murai S, Ando A, Ebara S, Hirayama M, Satomi Y, Hara T. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis. 2017;6(5):e329 Published 2017 May 8. doi: 10.1038/oncsis.2017.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh M, Komiya M, Teraoka N, Ueno T, Takahashi M, Kitahashi T, Sugimura T, Wakabayashi K. Overexpression of low-density lipoprotein receptor and lipid accumulation in intestinal polyps in Min mice. Int J Cancer. 2009;125(11):2505–2510. doi: 10.1002/ijc.24667 [DOI] [PubMed] [Google Scholar]
- Nakashima C, Yamamoto K, Fujiwara-Tani R, Luo Y, Matsushima S, Fujii K, Ohmori H, Sasahira T, Sasaki T, Kitadai Y, et al. Expression of cytosolic malic enzyme (ME1) is associated with disease progression in human oral squamous cell carcinoma. Cancer Sci. 2018;109(6):2036–2045. doi: 10.1111/cas.13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolau-Neto P, de Souza-Santos PT, Ramundo MS, Valverde P, Martins I, Santos IC, Dias F, Simao TA, Pinto LFR. Transcriptome analysis identifies ALCAM overexpression as a prognosis biomarker in laryngeal squamous cell carcinoma. Cancers (Basel). 2020;12(2):E470 Published 2020 Feb 18. doi: 10.3390/cancers12020470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman EM, Hintzsche H, Stopper H. Signaling steps in the induction of genomic damage by insulin in colon and kidney cells. Free Radic Biol Med. 2014;68:247–257. doi: 10.1016/j.freeradbiomed.2013.12.010 [DOI] [PubMed] [Google Scholar]
- Palomares T, Caramés M, García-Alonso I, Alonso-Varona A. Glutathione modulation reverses the growth-promoting effect of growth factors, improving the 5-fluorouracil antitumour response in WiDr colon cancer cells. Anticancer Res. 2009;29(10):3957–3965. [PubMed] [Google Scholar]
- Ren JG, Seth P, Everett P, Clish CB, Sukhatme VP. Induction of erythroid differentiation in human erythroleukemia cells by depletion of malic enzyme 2. PLoS One. 2010;5(9):e12520 Published 2010 Sep 2. doi: 10.1371/journal.pone.0012520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JG, Seth P, Clish CB, Lorkiewicz PK, Higasi RM, Lane AN, Fan TWM, Sukhatme VP. Knockdown of malic enzyme 2 suppresses lung tumor growth, induces differentiation and impacts PI3K/AKT signaling. Sci Rep. 2014;4:5414 Published 2014 Jun 24. doi: 10.1038/srep05414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha CM, Barros AS, Goodfellow BJ, Carreira IM, Gomes A, Sousa V, Bernardo J, Carvalho L, Gil AM, Duarte IF. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis. 2015;36(1):68–75. doi: 10.1093/carcin/bgu226 [DOI] [PubMed] [Google Scholar]
- Rzezniczak TZ, Merritt TJ. Interactions of NADP-reducing enzymes across varying environmental conditions: a model of biological complexity. G3 (Bethesda). 2012;2(12):1613–1623. doi: 10.1534/g3.112.003715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Thakkar S, Al-Dwairi A, Gokulan K, Simmen FA, Varughese KI. Malic enzyme is a target for drug design to combat obesity and cancer. Acta Crystallographica Section A Foundations and Advances. 2014;70 C846–C846. 10.1107/S2053273314091530. [DOI] [Google Scholar]
- Sánchez-Rodríguez R, Torres-Mena JE, Quintanar-Jurado V, Chagoya-Hazas V, Del Castillo ER, Yauner LDP, Villa-Trevino S, Perez-Carreon JI. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic Biol Med. 2017;102:87–99. doi: 10.1016/j.freeradbiomed.2016.11.027 [DOI] [PubMed] [Google Scholar]
- Sarfraz I, Rasul A, Hussain G, Shah MA, Zahoor AF, Asrar M, Selamoglu Z, Ji XY, Adem S, Sarker SD. 6-Phosphogluconate dehydrogenase fuels multiple aspects of cancer cells: From cancer initiation to metastasis and chemoresistance [published online ahead of print, 2020 Feb 10]. Biofactors. 2020;10.1002/biof.1624. doi: 10.1002/biof.1624 [DOI] [PubMed] [Google Scholar]
- Shen H, Xing C, Cui K, Li Y, Zhang J, Du R, Zhang X, Li Y. MicroRNA-30a attenuates mutant KRAS-driven colorectal tumorigenesis via direct suppression of ME1. Cell Death Differ. 2017;24(7):1253–1262. doi: 10.1038/cdd.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhou S, Wang P, Guo Y, Xie B, Ding S. Malic enzyme 1 (ME1) is a potential oncogene in gastric cancer cells and is associated with poor survival of gastric cancer patients. Onco Targets Ther. 2019;12:5589–5599. Published 2019 Jul 11. doi: 10.2147/OTT.S203228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorn B, Sartoretto JL, Sorrentino A, Romero N, Kalwa H, Abel ED, Michel T. Insulin-dependent metabolic and inotropic responses in the heart are modulated by hydrogen peroxide from NADPH-oxidase isoforms NOX2 and NOX4. Free Radic Biol Med. 2017;113:16–25. doi: 10.1016/j.freeradbiomed.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szypowska AA, Burgering BM. The peroxide dilemma: opposing and mediating insulin action. Antioxid Redox Signal. 2011;15(1):219–232. doi: 10.1089/ars.2010.3794 [DOI] [PubMed] [Google Scholar]
- Tian H, Li X, Jiang W, Lv C, Sun W, Huang C, Chen R. High expression of AKR1C1 is associated with proliferation and migration of small-cell lung cancer cells. Lung Cancer (Auckl). 2016;7:53–61. Published 2016 May 2. doi: 10.2147/LCTT.S90694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Liu D, Tang J, Dong L, Liu Y, Tao Z, Wan J, Gao D, Wang L, Sun H, et al. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumour Biol. 2015;36(8):6211–6221. doi: 10.1007/s13277-015-3306-5 [DOI] [PubMed] [Google Scholar]
- Woo SH, Yang LP, Chuang HC, Fitzgerald A, Lee HY, Pickering C, Myers JN, Skinner HD. Down-regulation of malic enzyme 1 and 2: Sensitizing head and neck squamous cell carcinoma cells to therapy-induced senescence. Head Neck. 2016;38 Suppl 1:E934–E940. doi: 10.1002/hed.24129 [DOI] [PubMed] [Google Scholar]
- Xi G, Shen XC, Wai C, Clemmons DR. Recruitment of Nox4 to a plasma membrane scaffold is required for localized reactive oxygen species generation and sustained Src activation in response to insulin-like growth factor-I. J Biol Chem. 2013;288(22):15641–15653. doi: 10.1074/jbc.M113.456046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Su Y, Simmen RC, Simmen FA. Dietary soy protein inhibits DNA damage and cell survival of colon epithelial cells through attenuated expression of fatty acid synthase. Am J Physiol Gastrointest Liver Physiol. 2008;294(4):G868–G876. doi: 10.1152/ajpgi.00515.2007 [DOI] [PubMed] [Google Scholar]
- Xue L, Zhu Z, Wang Z, Li H, Zhang P, Wang Z, Chen Q, Chen H, Chong T. Knockdown of prostaglandin reductase 1 (PTGR1) suppresses prostate cancer cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Trends. 2016;10(2):133–139. doi: 10.5582/bst.2016.01045 [DOI] [PubMed] [Google Scholar]
- Yan X, Zhang X, Wang L, Zhang R, Pu X, Wu S, Li L, Tong P, Wang J, Meng QH, et al. Inhibition of Thioredoxin/Thioredoxin Reductase induces synthetic lethality in lung cancers with compromised glutathione homeostasis. Cancer Res. 2019;79(1):125–132. doi: 10.1158/0008-5472.CAN-18-1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Thomenius MJ, Gan EC, Tang W, Freel CD, Merritt TJS, Nutt LK, Kornbluth S. Metabolic regulation of Drosophila apoptosis through inhibitory phosphorylation of Dronc. EMBO J. 2010;29(18):3196–3207. doi: 10.1038/emboj.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41(4):415–423. doi: 10.1038/ng.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Sun H, Xu C, Chen T, Zou B, Jiang P, Du W. Evidence for a direct cross-talk between malic enzyme and the pentose phosphate pathway via structural interactions. J Biol Chem. 2017;292(41):17113–17120. doi: 10.1074/jbc.M117.810309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Wang Z, Sprous D, Nabioullin R. In silico design and synthesis of piperazine-1-pyrrolidine-2,5-dione scaffold-based novel malic enzyme inhibitors. Bioorg Med Chem Lett. 2006;16(3):525–528. doi: 10.1016/j.bmcl.2005.10.065 [DOI] [PubMed] [Google Scholar]
- Zheng FJ, Ye HB, Wu MS, Lian YF, Qian CN, Zeng YX. Repressing malic enzyme 1 redirects glucose metabolism, unbalances the redox state, and attenuates migratory and invasive abilities in nasopharyngeal carcinoma cell lines. Chin J Cancer. 2012;31(11):519–531. doi: 10.5732/cjc.012.10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6(5):e1000932 Published 2010 May 6. doi: 10.1371/journal.pgen.1000932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Chang LL, Yan FJ, Hu Y, Zeng CM, Zhou TY, Yuan T, Ying MD, Cao J, He QJ, et al. AKR1C1 activates STAT3 to promote the metastasis of non-small cell lung cancer. Theranostics. 2018;8(3):676–692. Published 2018 Jan 1. doi: 10.7150/thno.21463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wang H, Wei J, Sartor GC, Bao MM, Pierce CT, Wahlestedt CR, Dykxhoorn DM, Dong C. Cocaine exposure increases blood pressure and aortic stiffness via the miR-30c-5p-Malic Enzyme 1-reactive oxygen species pathway. Hypertension. 2018; 71(4):752–760. doi: 10.1161/HYPERTENSIONAHA.117.10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Gu L, Lin X, Liu C, Lu B, Cui K, Zhou F, Zhao Q, Prochownik EV, Fan C, et al. Dynamic regulation of ME1 phosphorylation and acetylation affects lipid metabolism and colorectal tumorigenesis. Mol Cell. 2020;77(1):138–149.e5. doi: 10.1016/j.molcel.2019.10.015 [DOI] [PubMed] [Google Scholar]