Abstract
Background:
During HIV infection distinct mechanisms drive immune activation of the CD4 and CD8 T cells leading to CD4 T-cell depletion and expansion of the CD8 T-cell pool. This immune activation is polyclonal and extends beyond HIV-specific T cells. One consequence of this immune activation is a profound decrease in IL-7Rα (CD127) expression on memory CD8 T cells. The mechanisms leading to this are unknown and because of the potential impact of reduced IL-7 signaling in memory T cells specific to HIV and other pathogens, in the present study we examined the molecular mechanisms implicated in this downregulation of CD127.
Methods:
Membrane bound (mIL7RA) and soluble (sIL7RA) mRNA expression was determined by qRT-PCR. CD127, Eomesodermin (Eomes) and T-bet expression in healthy controls and HIV-infected patients were studied by flow cytometry.
Results:
CD127 downregulation occurs at the transcriptional level for both mIL7RA and sIL7RA alternative spliced forms in the CD127low memory CD8 T cells. CD127low memory CD8 T cells exhibited increased Eomes expression and an ‘effector-like’ gene profile. These changes were associated with higher HIV-RNA levels. Following combination antiretroviral therapy (cART), there was an increase in CD127 expression over an extended period of time (>5 months) which was associated with decreased Eomes expression.
Conclusion:
CD127 is downregulated at a transcriptional level in memory CD8 T cells in association with upregulation of Eomes expression.
Keywords: CD127, Eomes, HIV, T-bet, T-box transcription factors
Introduction
The hallmark of the pathogenesis of HIV infection is the depletion of CD4 T cells and a global immune activation involving most elements of the innate and adaptive immune systems including both the CD4 and CD8 T-cell pools. This immune activation extends beyond HIV-specific response and T cells with other specificities also show an activated phenotype [1]. There is substantial evidence that immune activation leads to immune dysfunction and ultimately the development of AIDS, which is associated with opportunistic infections and poor vaccine responses [2].
Activation of the CD4 and CD8 T-cell pools is manifested in a variety of ways such as an increased proportion of ‘memory-like’ phenotype T cells, increased expression of surface activation markers such as HLA-DR and CD38 [3] and increased proportion of proliferating CD4 and CD8 T cells in vitro and in vivo [4–10]. The mechanisms driving activation of CD4 and CD8 T cells are distinct and are associated with differences in the homeostatic regulation of these pools. Proliferation of CD4 T-cell subsets is driven by a combination of the homeostatic response to the CD4 T-cell depletion and the inflammatory environment driven by viremia. These environments are reflected by increased expression of mRNA transcripts of genes associated with signaling of both common gamma-chain (γc) cytokines and Type-I IFN in naive (CD45RA+CD27+) and memory (CD45RA−CD27+) phenotype CD4 T cells [11–13]. In contrast, the main force driving immune activation of CD8 T cells appears to be levels of HIV-RNA and the associated environment as reflected by increased mRNA transcripts related to Type-I IFN signaling [10,11,14].
Maintenance and survival of memory CD8 T cells is primarily mediated by the homeostatic cytokines IL-7 and IL-15 [15,16]. IL-7 signals through a heterodimeric receptor composed of the common γ-chain (CD132), which is shared with other cytokines such as IL-2, IL-4, IL-9, IL-15 and IL-21, and the IL-7-specific α-chain (IL-7Rα or CD127) [17–19]. Although IL-7 signaling is primarily regulated by membrane-bound CD127 (mCD127), recent data have also suggested that a soluble form of CD127 (sCD127) may serve as a decoy receptor and suppress IL-7 signaling. Both an alternative spliced isoform, encoding only the ectodomain of CD127 as well as the generation of a proteolytically cleaved CD127 from the cell surface, have been suggested as pathways leading to sCD127 production [20–26]. Several groups have reported that the immune activation of memory CD8 T cells during HIV infection is associated with a loss of CD127 expression, leading to a poor response to in vitro stimulation with IL-7 [27–32].
CD127 can be downregulated by T-cell receptor (TcR) stimulation. The differentiation of naive T cells into effector and memory T cells is orchestrated by a partially defined network of signals from TcR, costimulation and cytokines [33]. Expression of the T-box transcription factors, T-bet and Eomes have been described as critical master regulators for the differentiation of CD8 T cells into effectors, driving the expression of cytotoxic molecules [34–36]. Studies in animal models have shown that despite the overlapping roles of T-bet and Eomes in effector differentiation [35,37,38], they have reciprocal functions in promoting effector versus memory differentiation [39–43]. Animals lacking T-bet showed an enhanced differentiation of central memory CD8 T cells [36,38]. Conversely, memory CD8 T cells from animals lacking Eomes have diminished long-term persistence and secondary expansion after challenge [39].
In humans, the roles of Eomes and T-bet expression have been studied in the context of CD8 T-cell effector differentiation and function. CD8 T cells from patients infected with human cytomegalovirus (CMV) have higher mRNA expression of the transcription factors Eomes and T-bet on cells with a ‘terminal effector memory (TEMRA) phenotype’ (CD45RA+CD27−) [44]. HIV-specific CD8 T cells from elite controllers have been reported to have higher levels of T-bet after stimulation than CD8 T cells of HIV-infected progressors [45].
Because of the potential impact of the loss of IL-7 signaling in immunity against HIV, other pathogens and vaccine responses in HIV-infected patients, in the present study we examined the mechanisms that could be involved in the regulation of CD127 expression in the memory CD8 T-cell pool irrespective of any specificity. We found that mRNA transcription of both membrane bound – and soluble splice form – IL7RA were downregulated in the memory CD127low CD8 T cells that accumulate in HIV-infected patients with poorly controlled viremia. This downregulation was associated with an ‘effector-like’ differentiated gene profile and higher levels of Eomes. Moreover, recovery of CD127 expression on the memory CD8 T cells from patients undergoing combination antiretroviral therapy (cART) was strongly associated with a decrease in Eomes expression that continued to evolve even after 5 months of therapy and suppressed viremia. These data suggest that HIV-driven immune activation leads to increased expression of Eomes in association with CD8 T-cell activation and downregulation of CD127.
Materials and methods
Patient and healthy volunteers
Patients and healthy controls were consented and studied in NIAID/CCMD intramural program IRB approved HIV clinical research studies. The patients analyzed in the cross-sectional cohort and patients used to study the effect of cART on the expression of Eomes, T-bet and CD127 longitudinally are described in supplementary material and methods, http://links.lww.com/QAD/A346.
Flow cytometry
Frozen PBMCs from patients and healthy controls were thawed and rested overnight before staining (see supplementary material and methods, http://links.lww.com/QAD/A346).
Sorting and quantitative real-time PCR
Sorted CD8+ T-cell subsets from HIV-infected patients with HIV-RNA levels of more than 50 copies/ml, less than 50 copies/ml and from healthy controls were analyzed for mRNA expression (see supplementary material and methods, http://links.lww.com/QAD/A346).
Statistical analysis
See supplementary material and methods, http://links.lww.com/QAD/A346.
Results
Decreases in membrane-bound and soluble CD127 in memory CD8 T cells from patients with chronic HIV infection are due to decreased IL7RA transcription
Memory CD8 T cells from patients with HIV infection and viral loads more than 50 copies/ml show a profound decrease in the expression of CD127 (Figure S1, http://links.lww.com/QAD/A346) [28,29].
The mechanisms for downregulation of CD127 could be accounted by decreased transcription of the CD127 gene, an increased transcription of the alternative spliced CD127 isoform that encodes the soluble CD127 [20], and or shedding of the membrane-bound CD127 [22].
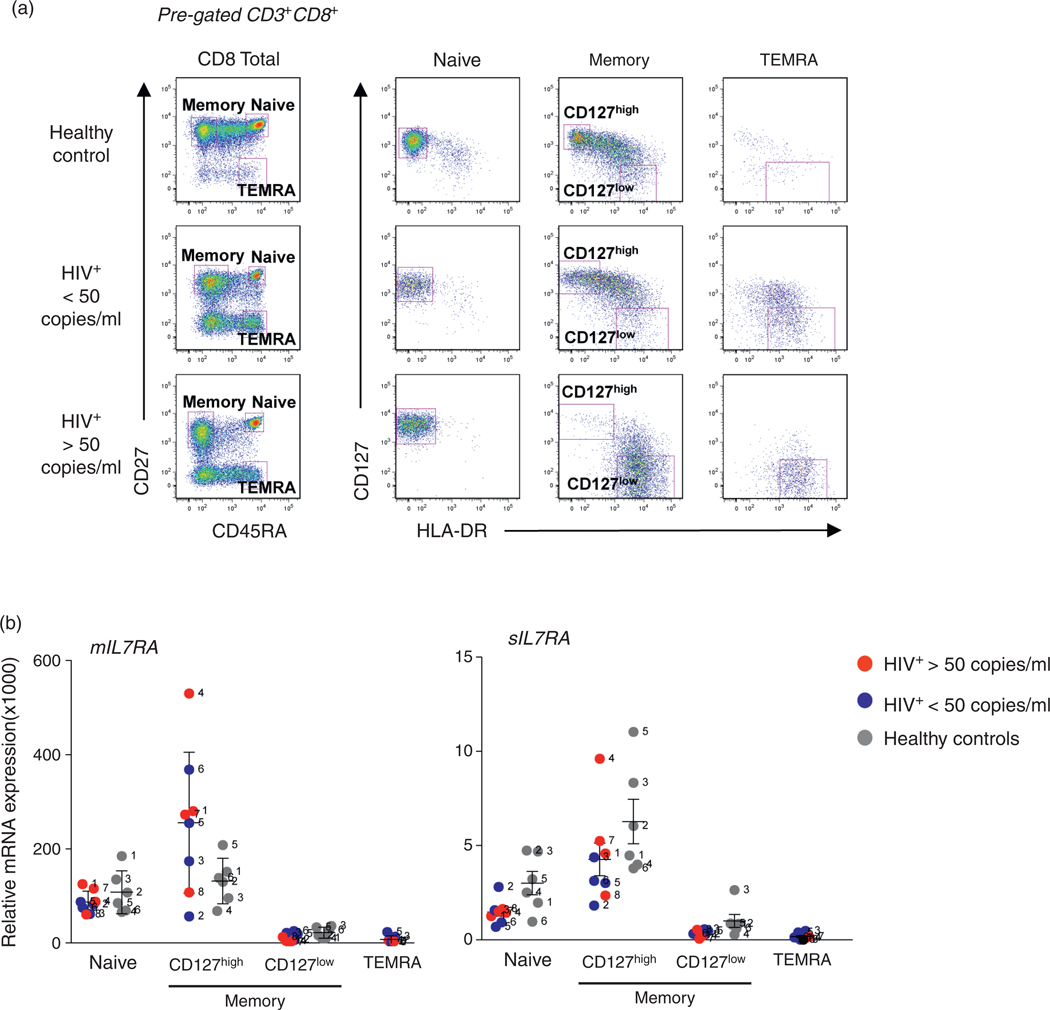
We analyzed expression of IL7RA mRNA transcripts corresponding to the membrane bound (mIL7RA) and soluble forms (sIL7RA) in sorted CD8 T-cell subsets, as defined in supplementary materials and methods, http://links.lww.com/QAD/A346 [44,46,47]. Because of the immune activation observed during HIV infection we included the activation marker HLA-DR as a marker of activation and cell-cycle progression [48] to clearly distinguish resting from activated CD8 T-cell subsets. Of note, the memory CD127low phenotypic subset (CD45RA−CD27+CD127lowHLA-DR+) was highly enriched in patients with HIV infection in comparison with healthy controls (Fig. 1a). However, the TEMRA subset could not be studied in healthy controls due to low number of cells (Fig. 1a). Expression levels of IL7RA mRNA transcripts in CD8 T-cell subsets from healthy controls and HIV-infected individuals are shown in Fig. 1b. The highest levels of both membrane and soluble forms of CD127 mRNA were found in the naive and memory CD127high subsets for both HIV-infected patients and healthy controls (Fig. 1b). We found no evidence in healthy volunteers or patients that overexpression of the mRNA corresponding to the soluble form of CD127 could account for the decreased levels of the membrane bound form. As shown in Fig. 1, the lack of CD127 expression on the memory CD127low cells and TEMRA cells was clearly due to decreased CD127 transcription.
Fig. 1. Expression of IL7RA membrane and soluble transcripts in sorted CD8 T cells.
(a) Cell sorting of CD8 T-cell subsets from healthy controls (n = 6, gray symbol) and HIV-infected patients with viral load more than 50 copies/ml (n = 4, red symbol) and viral load less than 50 copies/ml (n = 4, blue symbol) was performed based on surface expression of CD45RA and CD27. The naive CD8 T-cell subset was CD45RA+CD27+CD127highHLADR−; the memory CD8 T-cell subset was divided by expression of CD127 into memory CD127high (CD45RA−CD27+CD127highHLADR−), and memory CD127low (CD45RA−CD27+CD127lowHLADR+). The terminal effector memory (TEMRA) subset was defined as CD45RA+CD27−CD127lowHLADR+. The figure shows a representative example of the sorted cohorts pregated on live CD3+CD8+ cells. (b) Relative fold increase of membrane bound (mIL7RA) and soluble (sIL7RA) forms of the IL7RA as determined by qRT-PCR. The results were normalized to RPL13 expression. No significant differences were noted between the groups at the P ≤ 0.01 level.
CD8 memory CD127low T cells from patients with HIV infection exhibit an ‘effector-like’ transcription profile
The observation that the patterns of CD127 surface expression correlate with those observed at the mRNA level, confirmed that CD127 expression is highly regulated at the level of transcription [25,26]. If such regulation is a consequence of HIV-associated immune activation, one would predict that it would correlate with either the homeostatic forces associated with CD4 T-cell depletion or the level of the inflammatory environment driven by HIV-RNA [28]. When assessing the first possibility, we found that CD4 T-cell counts had no significant contribution to the loss of CD127 in CD8 memory T cells (R = −0.230, P = 0.248). Conversely, there was a strong inverse correlation between CD127 expression and HIV-RNA levels (R = −0.719, P < 0.001). Similar relationships were observed with the activation markers HLA-DR (R = −0.696, P < 0.001) and combined HLA-DR/CD38 (R = −0.807, P < 0.001).
To determine if the loss of CD127 by memory CD8 T cells was associated with differentiation into effector cells, we analyzed the mRNA expression of genes related to cytokine expression, signaling and regulation. This analysis revealed that the memory CD127low phenotype cells showed a gene expression profile most closely related to that observed in the ‘effector-like’ TEMRA CD8 T cells (Table S1, http://links.lww.com/QAD/A346). In addition, a positive association was noted between CD27 and CD127 expression (R = 0.599, P = 0.002).
Taken together, these data suggest that the in vivo loss of CD127 by the memory phenotype CD8 T cells during HIV infection is driven by differentiation into ‘effector-like’ T cells.
Increased expression of eomesodermin in the memory CD8 T cells from HIV-infected patients is associated with loss of CD127
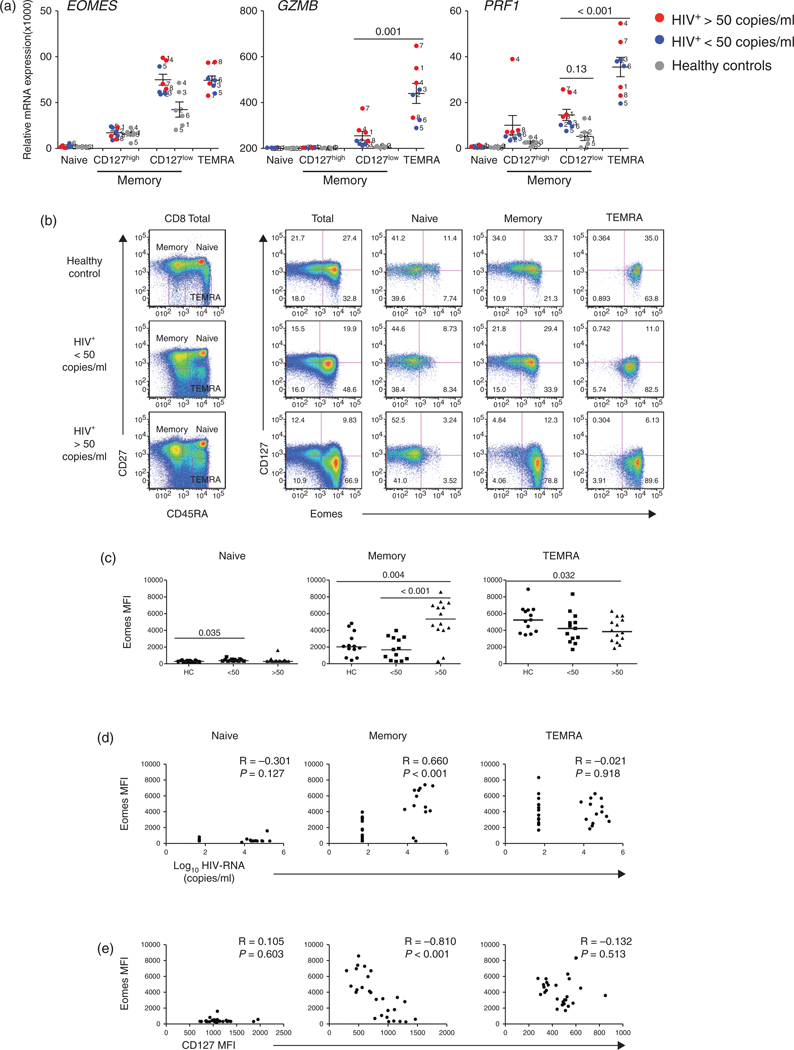
The above data suggested that CD127low memory CD8 T cells from patients with HIV infection exhibit an ‘effector-like’ gene expression profile. CD8 T-cell effector differentiation is governed at least in part by two T-box transcription factors, Eomes and T-bet [36,38,49]. In addition to the well defined role of Eomes in promoting effector function, animal models have suggested a unique role for Eomes in the maintenance and fitness of memory CD8 T cells [39]. Yet its role in humans has not been defined. To determine if the changes in CD127 expression were associated with changes in Eomes, we examined its expression in CD8 T-cell subsets in healthy controls and patients with HIV infection. In healthy controls, EOMES mRNA expression was restricted to the antigen-experienced subsets and was highest in memory CD127low T cells (Fig. 2a). A similar pattern was seen in the HIV-infected cohort. In addition, the memory CD127low subset showed similar levels of expression to that observed in the TEMRA, this was consistent with the ‘effector-like’ nature of this subset. Of note, EOMES, GZMB (Granzyme B) and PRF1 (Perforin) transcripts tended to be higher in the memory CD127low phenotype cells of the HIV-infected patients compared with healthy controls. This was particularly true for the patients with HIV-RNA levels of more than 50 copies/ml (Fig. 2a).
Fig. 2. Expression of Eomes and CD127 in CD8 T-cell subsets from healthy controls and HIV-infected patients.
(a) The analysis of mRNA expression of the transcription factor EOMES and the effector molecules granzyme B (GZMB) and perforin (PRF1) were measured as described in Fig. 1. (b) The protein expression of Eomes and CD127 in CD8 T-cell subsets were analyzed by flow cytometry in PBMCs from healthy controls (n = 13), HIV-infected patients with viral load less than 50 copies/ml (n = 13) and HIV-infected patients with viral load more than 50 copies/ml (n = 14). Median fluorescence intensities for CD127 and Eomes were calculated using FlowJo. A representative example from each of the three cohorts is displayed (gating strategy is shown in Figure S2). (c) Comparisons of Eomes expression in CD8 T-cell subsets between HIV-infected groups and healthy controls. (d) Correlations between Eomes expression and HIV-RNA levels and (e) Correlations between CD127 and Eomes expression.
To determine if the increased mRNA transcript levels for Eomes were associated with increased levels of protein expression, we next measured intracellular levels of Eomes protein in ex vivo T-cell subsets from healthy controls, HIV-infected patients with HIV-RNA levels of more than 50 copies/ml and HIV-treated patients with HIV-RNA levels of less than 50 copies/ml (Fig. 2b). The patterns of Eomes protein expression in healthy controls matched that observed at the mRNA expression level. There was increased expression of Eomes in association with increased differentiation with TEMRA > memory > naive (Fig. 2b and c). Of note, HIV-infected patients with HIV-RNA levels of more than 50 copies/ml demonstrated increased Eomes protein expression in the memory phenotype T cells compared with healthy controls and patients with HIV-RNA levels of less than 50 copies/ml (P = 0.004 and P < 0.001, respectively). As reported in other systems [41], Eomes expression was associated with the transcription of cytotoxic molecules such as Perforin. Consistent with previous reports Perforin expression was mainly restricted to the more differentiated subset (TEMRA) and was marginally increased in viremic HIV-infected patients [50], when compared with healthy controls (Figure S2, http://links.lww.com/QAD/A346). We next assessed the association of Eomes expression with IFNγ producing memory CD8 T cells after stimulation with mAbs anti-CD3/CD28 or HIV Gag-peptides. In patients with HIV-RNA levels of more than 50 copies/ml, IFNγ expressing memory CD8 T cells showed increased Eomes expression when compared with healthy controls. These differences were more pronounced in the Gag stimulated T cells from patients with HIV-RNA levels of more than 50 copies/ml compared to those patients with suppressed viremia (P = 0.015) (Figure S3, http://links.lww.com/QAD/A346).
Expression of Eomes in the memory CD8 T cells from HIV-infected patients was directly correlated with HIV-RNA levels (R = 0.660, P < 0.001) and inversely associated (R = −0.810, P < 0.001) with CD127 expression (Fig. 2d and e). IFNγ producing cells were CD127low and showed increased expression of Eomes reflecting the ‘effector-like’ nature of this population (Fig. S3, http://links.lww.com/QAD/A346). These results demonstrate that in humans, Eomes can play a role in the regulation of both memory and effector CD8 T cells.
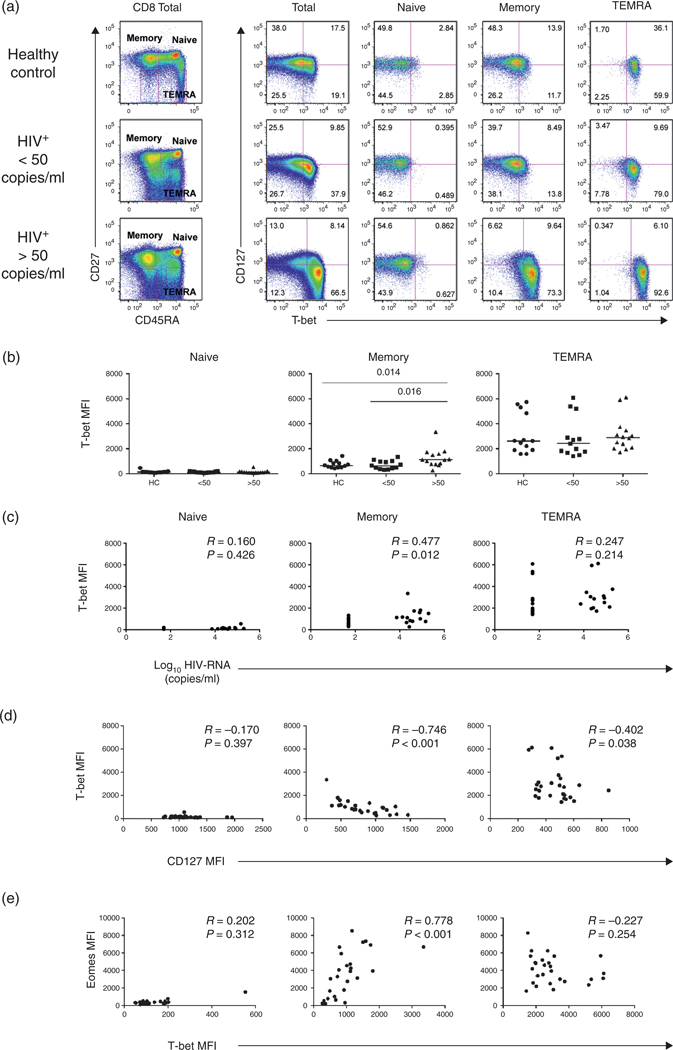
Upregulation of T-bet in the memory CD127low CD8 T cells from HIV-infected patients
The results above suggested that increased levels of Eomes may drive CD8 T cells to differentiate into an ‘effector-like’ profile in association with downregulation of CD127. Recent studies in animal models have suggested overlapping roles for Eomes and T-bet in T-cell effector differentiation [39]. In humans, T-bet expression has been described to be restricted to the more differentiated phenotypes and its expression to be important in CD8 T-cell effector function [44,45,49]. Examination of CD8 T-cell subsets from healthy controls showed that, contrary to Eomes, expression of T-bet was restricted to the TEMRA, consistent with the suggested role of this transcription factor in effector function (Fig. 3a and b). As in the case of Eomes, a positive correlation was noted between T-bet expression and HIV-RNA levels (R = 0.447, P = 0.012; Fig. 3c). Increased expression of T-bet was associated with decreased CD127 expression (R = −0.746, P < 0.001; Fig. 3d) and increased Eomes (R = 0.778, P < 0.001; Fig. 3e). These data suggest that the immune activation leading to the loss of CD127 in the memory CD8 T-cell pool may occur via a transcriptional programming involving increased expression of both T-box transcription factors Eomes and T-bet.
Fig. 3. Protein expression of T-bet and CD127 in CD8 T-cell subsets from healthy controls and HIV-infected patients.
(a) Representative dot plots from the three cohorts studied (Fig. 2 for T-bet and CD127 expression). (b) T-bet median fluorescence intensity in CD8 T-cell subsets (as above). (c) Correlations between T-bet expression and HIV-RNA levels; and (d) Correlations between CD127 and T-bet expression. (e) Correlations between Eomes and T-bet expression in CD8 T-cell subsets.
Combination antiretroviral therapy leads to progressive decreases in Eomes in association with increased expression of CD127 on memory CD8 T cells
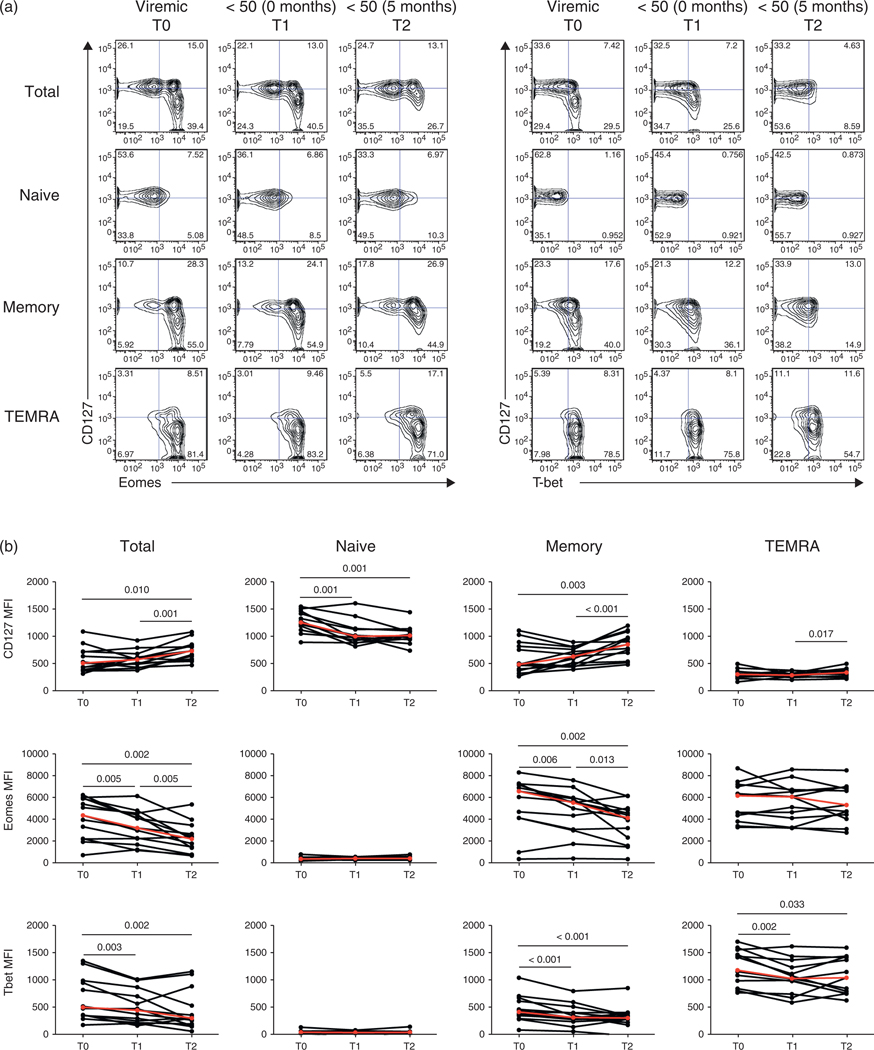
The above results suggest that HIV driven immune activation may lead to the differentiation of memory CD8 T cells into ‘effector-like’ cells through increased expression of the T-box transcription factors Eomes and T-bet. We next studied the relationships between cART and the expression of Eomes, T-bet and CD127 in CD8 T cells. To do so, we analyzed the effects of cART in a cohort of HIV-infected patients followed over the course of their treatment (Fig. 4, Table 1). Significant changes in CD4 T-cell counts (median cells/μl: 267 at T0, 337 at T1 and 504 at T2; P = 0.010, P = 0.002, respectively) but not CD8 T-cell counts were observed following initiation of cART (Figure S4, http://links.lww.com/QAD/A346). In addition, significant decreases in Eomes (P = 0.006) and T-bet (P < 0.001) occurred early after treatment in the memory subset (Fig. 4b) without significant changes in CD127 expression. Following 5–8 months of treatment and HIV-RNA levels persistently less than 50 copies/ml (T2), there was a continued decrease in Eomes expression concomitant with a significant increase in CD127 expression (P < 0.001). These observations suggest that HIV-RNA levels in serum may not accurately reflect the degree of ongoing viral replication as evidenced by residual immune activation in the CD8 T cells. Changes of Eomes expression in memory CD8 T cells between T0 and T2 were strongly associated with changes in CD127 expression (R = −0.830, P = 0.001; Table 1).
Fig. 4. Effect of cART on T-box transcription factors T-bet and Eomes and CD127 expression in CD8 T-cell subsets.
(a) Representative contour plots for CD127, Eomes and T-bet expression over three time points: more than 50 copies/ml (T0), first month less than 50 copies/ml (T1), and 5–8 months later (T2). (b) Changes over time of the MFI of Eomes, T-bet and CD127 in CD8 T-cell subsets. Median change denoted by red line.
Table 1.
Association between changes in the levels of Eomes, T-bet and CD127 expression in CD8 T cells.
| Total | Naive | Memory | TEMRA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Change betweena | R | P | R | P | R | P | R | P | |
| T0 and T1 | Eomes | −0.742 | 0.005 | −0.484 | 0.097 | −0.644 | 0.018 | −0.011 | 0.972 |
| T-bet | 0.113 | 0.714 | −0.269 | 0.373 | 0.006 | 0.986 | 0.094 | 0.761 | |
| T0 and T2 | Eomes | − 0.797 | 0.002 | −0.687 | 0.012 | −0.830 | 0.001 | −0.124 | 0.687 |
| T-bet | 0.140 | 0.648 | 0.275 | 0.358 | 0.126 | 0.676 | 0 | 1 | |
| T1 and T2 | Eomes | −0.615 | 0.029 | −0.019 | 0.950 | −0.445 | 0.130 | −0.396 | 0.182 |
| T-bet | −0.275 | 0.363 | −0.017 | 0.957 | −0.203 | 0.505 | −0.176 | 0.566 | |
TEMRA, terminal effector memory CD45RA+.
change: measured at the later time point minus that at earlier time point.
In addition, as observed in the cross-sectional cohort, changes in CD127 were not associated with changes in CD4 T cells counts (Table S2, http://links.lww.com/QAD/A346). However, as IL-7 signaling regulates the expression of CD127 at a transcriptional level [25], we tested whether or not serum levels of IL-7 were associated with levels of CD127 expression and/or levels of the T-box transcription factors in CD8 T cells (Table S3, http://links.lww.com/QAD/A346). IL-7 serum levels were inversely associated with CD127 expression (R = −0.61, P = 0.03) at T0 (Table S3, http://links.lww.com/QAD/A346, http://links.lww.com/QAD/A346, Figure S4, http://links.lww.com/QAD/A346) and positively associated with Eomes expression (R = 0.760, P < 0.01). However, in vitro, IL-7 was not sufficient to modulate levels of Eomes or T-bet expression suggesting that other factors or a combination of factors are involved in the modulation of these transcription factors in patients HIV infection (Figure S5, http://links.lww.com/QAD/A346).
Discussion
The present study has clearly demonstrated a role for the T-box transcription factors T-bet and Eomes in the regulation of the immune activation seen in the CD8 T cells of patients with HIV infection. This activation has its most profound effect on the memory subset of CD8 T cells in the setting of higher levels of HIV-RNA. In addition, it is associated with downregulation of CD127 mRNA and protein, and upregulation of effector molecules such as IFNγ. The immune activation seen in the setting of HIV infection plays a major role in disease pathogenesis ranging from susceptibility to opportunistic infections and neoplasia to serious non-AIDS defining diseases such as atherosclerosis, osteoporosis and cancer [51,52]. Although the precise mechanisms are poorly understood, it is clear that this is a multifactorial process that differentially affects CD4 and CD8 T-cell subsets and may be driven by forces from T-cell homeostasis, to Type-I IFN, to microbial translocation [3,8,10,53]. The level of immune activation of the CD8 T-cell pool is highly associated with HIV-RNA levels [11,28,31,54], an inefficient immune response against HIV and other opportunistic pathogens, and poor responses to vaccines [55–57]. CD8 T-cell-mediated immunity relies on the survival and self-renewal characteristics of memory T cells in which IL-7 signaling plays a critical role [31,32,58,59]. Our present data demonstrated that HIV driven immune activation leads to transcriptional downregulation of both forms of the IL7RA, membrane associated and soluble forms.
The transcriptional downregulation of CD127 in CD8 memory T cells in the present study, was associated with differentiation of CD8 T cells to an ‘effector-like’ gene profile with upregulation of the transcription factors Eomes and T-bet. Modulation of CD127 expression by memory CD8 T cells has also been observed in chronic infections such as CMV in which CMV-specific memory CD8 T cells are CD127low, suggesting continuous TcR engagement of these cells in vivo [15,60]. In HIV-infected patients this phenomenon appears to be more generalized and not restricted to HIV-specific CD8 T cells [1]. The clinical relevance of the impact oflosing CD127 expression becomes evident from the recent studies that showed that early treatment is beneficial to the preservation of functional CD127high memory T cells [61,62].
The differences in CD127 expression between healthy controls and patients with HIV infection were most pronounced in the memory CD8 T-cell subset [10,63]. The upregulation of Eomes and T-bet in the memory CD8 T cells was associated with higher HIV-RNA levels. The apparent transitional and reversible nature of this activation was evident by the regained expression of CD127 by the memory CD8 T-cell subset following successful suppression of HIV replication by cART.
In the present study we found that the expression of Eomes was increased in patients with HIV infection and HIV-RNA levels of more than 50 copies/ml suggesting a role for this transcription factor in HIV-associated immune activation. Consistent with this is the observation that Type-I IFN is capable of inducing Eomes expression in a murine system [64]. A recent report had demonstrated a reciprocal expression of Eomes and T-bet in hepatitis C virus (HCV)-specific T cells isolated from liver tissue of chronically infected patients when compared to cells isolated from patients with recently resolved infection [65]. In that study, HCV-specific T cells from the peripheral blood of infected and treated patients showed no significant differences in their expression of T-bet and Eomes. These findings suggest differences in the host response to viruses and antigen-specific T cells from peripheral blood might not reflect those antigen-specific CD8 T cells that transit/reside in peripheral tissues. Therefore, more studies analyzing the dynamics of the modulation of the T-box transcription factors in HIV infection are needed to understand the nature of the immune activation of these cells.
The changes observed in the context of cART and viral loads suppressed to less than 50 copies/ml suggest that these markers of immune activation of CD8 T cells could be a surrogate for ongoing viral replication. This is consistent with evidence from other studies suggesting ongoing viral replication despite several months of cART treatment and viremia suppressed to less than 50 copies/ ml [66,67]. The increased levels of biomarkers for inflammation and coagulation such as IL-6, sCD14 and D-dimer seen despite HIV-RNA levels of less than 50 copies/ml is in support of this possibility. The clinical significance of this activation/low level of replication is reflected in the increased risks of all-cause mortality observed in patients with high levels of these markers [68].
In the present study, patients undergoing cART treatment demonstrated continued immunological changes even after viral loads had declined to less than 50 copies/ml for at least 5 months, implying that despite ‘undetectable’ viral loads there was ongoing viral replication and virus associated immune activation [66,67]. These data are in agreement with other studies that reported similar observations in the context of treatment [54,62].
Thus, monitoring changes of this type may be of value in the evaluation of patients in whom strategies aimed to the eradication of HIV are studied.
Supplementary Material
Acknowledgements
We would like to thank the patients of the NIAID HIV-Clinic, for their participation in this study and the healthy controls of the NIH Blood bank. We would like to thank Dr Stephen A. Migueles for his useful discussions and critical reading of the manuscript. We would like to thank Chris Wilhelm for his contribution setting up some of the panels used in the flow cytometry assays. We would like to thank Dr Anthony Fauci for his guidance and support.
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Conflicts of interest
The authors report no conflict of interest.
References
- 1.Doisne JM, Urrutia A, Lacabaratz-Porret C, Goujard C, Meyer L, Chaix ML, et al. CD8+ T cells specific for EBV, cytomegalovirus, and influenza virus are activated during primary HIV infection. J Immunol 2004; 173:2410–2418. [DOI] [PubMed] [Google Scholar]
- 2.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis 1999; 179:859–870. [DOI] [PubMed] [Google Scholar]
- 3.Ho HN, Hultin LE, Mitsuyasu RT, Matud JL, Hausner MA, Bockstoce D, et al. Circulating HIV-specific CD8+ cytotoxic T cells express CD38 and HLA-DR antigens. J Immunol 1993; 150:3070–3079. [PubMed] [Google Scholar]
- 4.Sachsenberg N, Perelson AS, Yerly S, Schockmel GA, Leduc D, Hirschel B, et al. Turnover of CD4+ and CD8+ T lymphocytes in HIV-1 infection as measured by Ki-67 antigen. J Exp Med 1998; 187:1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang ZQ, Notermans DW, Sedgewick G, Cavert W, Wietgrefe S, Zupancic M, et al. Kinetics of CD4+ T cell repopulation of lymphoid tissues after treatment of HIV-1 infection. Proc Natl Acad Sci U S A 1998; 95:1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 1995; 373:123–126. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs JA, Lempicki RA, Sidorov IA, Adelsberger JW, Herpin B, Metcalf JA, et al. Identification of dynamically distinct subpopulations of T lymphocytes that are differentially affected by HIV. J Exp Med 2001; 194:1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lempicki RA, Kovacs JA, Baseler MW, Adelsberger JW, Dewar RL, Natarajan V, et al. Impact of HIV-1 infection and highly active antiretroviral therapy on the kinetics of CD4+ and CD8+ T cell turnover in HIV-infected patients. Proc Natl Acad Sci U S A 2000; 97:13778–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieg SF, Rodriguez B, Asaad R, Jiang W, Bazdar DA, Lederman MM. Peripheral S-phase T cells in HIV disease have a central memory phenotype and rarely have evidence of recent T cell receptor engagement. J Infect Dis 2005; 192:62–70. [DOI] [PubMed] [Google Scholar]
- 10.Catalfamo M, Di Mascio M, Hu Z, Srinivasula S, Thaker V, Adelsberger J, et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc Natl Acad Sci U S A 2008; 105:19851–19856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalfamo M, Wilhelm C, Tcheung L, Proschan M, Friesen T, Park JH, et al. CD4 and CD8 T Cell Immune Activation during Chronic HIV Infection: Roles of Homeostasis, HIV, Type I IFN, and IL-7. J Immunol 2011; 186:2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyrcza MD, Kovacs C, Loutfy M, Halpenny R, Heisler L, Yang S, et al. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J Virol 2007; 81:3477–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, et al. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest 2011; 121:2391–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane HC. Pathogenesis of HIV infection: total CD4+ T-cell pool, immune activation, and inflammation. Top HIV Med 2010; 18:2–6. [PubMed] [Google Scholar]
- 15.van Leeuwen EM, de Bree GJ, Remmerswaal EB, Yong SL, Tesselaar K, ten Berge IJ, et al. IL-7 receptor alpha chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood 2005; 106:2091–2098. [DOI] [PubMed] [Google Scholar]
- 16.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol 2004; 78:5535–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol 2001; 1:200–208. [DOI] [PubMed] [Google Scholar]
- 18.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol 2007; 7:144–154. [DOI] [PubMed] [Google Scholar]
- 19.Gadina M, Hilton D, Johnston JA, Morinobu A, Lighvani A, Zhou YJ, et al. Signaling by type I and II cytokine receptors: ten years after. Curr Opin Immunol 2001; 13:363–373. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin RG, Friend D, Ziegler SF, Jerzy R, Falk BA, Gimpel S, et al. Cloning of the human and murine interleukin-7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell 1990; 60:941–951. [DOI] [PubMed] [Google Scholar]
- 21.Rose T, Lambotte O, Pallier C, Delfraissy JF, Colle JH. Identification and biochemical characterization of human plasma soluble IL-7R: lower concentrations in HIV-1-infected patients. J Immunol 2009; 182:7389–7397. [DOI] [PubMed] [Google Scholar]
- 22.Crawley AM, Faucher S, Angel JB. Soluble IL-7R alpha (sCD127) inhibits IL-7 activity and is increased in HIV infection. J Immunol 2010; 184:4679–4687. [DOI] [PubMed] [Google Scholar]
- 23.Blom-Potar MC, Bugault F, Lambotte O, Delfraissy JF, Theze J. Soluble IL-7Ralpha (sCD127) and measurement of IL-7 in the plasma of HIV patients. J Acquir Immune Defic Syndr 2009; 51:104–105. [DOI] [PubMed] [Google Scholar]
- 24.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol 2009; 10:1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JH, Yu Q, Erman B, Appelbaum JS, Montoya-Durango D, Grimes HL, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity 2004; 21:289–302. [DOI] [PubMed] [Google Scholar]
- 26.Alves NL, van Leeuwen EM, Derks IA, van Lier RA. Differential regulation of human IL-7 receptor alpha expression by IL-7 and TCR signaling. J Immunol 2008; 180:5201–5210. [DOI] [PubMed] [Google Scholar]
- 27.Miller JD, Masopust D, Wherry EJ, Kaech S, Silvestri G, Ahmed R. Differentiation of CD8 T cells in response to acute and chronic viral infections: implications for HIV vaccine development. Curr Drug Targets Infect Disord 2005; 5:121–129. [DOI] [PubMed] [Google Scholar]
- 28.Paiardini M, Cervasi B, Albrecht H, Muthukumar A, Dunham R, Gordon S, et al. Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J Immunol 2005; 174:2900–2909. [DOI] [PubMed] [Google Scholar]
- 29.Vranjkovic A, Crawley AM, Patey A, Angel JB. IL-7-dependent STAT-5 activation and CD8+ T cell proliferation are impaired in HIV infection. J Leukoc Biol 2011; 89:499–506. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Imamichi H, Sukura A, Kovacs JA. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J Infect Dis 2001; 184:1358–1362. [DOI] [PubMed] [Google Scholar]
- 31.Colle JH, Moreau JL, Fontanet A, Lambotte O, Joussemet M, Delfraissy JF, et al. CD127 expression and regulation are altered in the memory CD8 T cells of HIV-infected patients–reversal by highly active antiretroviral therapy (HAART). Clin Exp Immunol 2006; 143:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirskyj D, Theze J, Kumar A, Kryworuchko M. Disruption of the gamma c cytokine network in T cells during HIV infection. Cytokine 2008; 43:1–14. [DOI] [PubMed] [Google Scholar]
- 33.Obar JJ, Lefrancois L. Early events governing memory CD8+ T-cell differentiation. Int Immunol 2010; 22:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med 2007; 204:2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 2007; 27:281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 2003; 302:1041–1043. [DOI] [PubMed] [Google Scholar]
- 37.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 2010; 32:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 2005; 6:1236–1244. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, et al. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 2010; 185:4988–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton SE, Jameson SC. CD8(+) T cell differentiation: choosing a path through T-bet. Immunity 2007; 27:180–182. [DOI] [PubMed] [Google Scholar]
- 41.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 2009; 206:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol 2010; 11:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002; 111:621–633. [DOI] [PubMed] [Google Scholar]
- 44.Hertoghs KM, Moerland PD, van Stijn A, Remmerswaal EB, Yong SL, van de Berg PJ, et al. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J Clin Invest 2010; 120:4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood 2011; 117:3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A 2008; 73:975–983. [DOI] [PubMed] [Google Scholar]
- 47.Takata H, Takiguchi M. Three memory subsets of human CD8+ T cells differently expressing three cytolytic effector molecules. J Immunol 2006; 177:4330–4340. [DOI] [PubMed] [Google Scholar]
- 48.Imamichi H, Lempicki RA, Adelsberger JW, Hasley RB, Rosenberg A, Roby G, et al. The CD8+ HLA-DR+ T cells expanded in HIV-1 infection are qualitatively identical to those from healthy controls. Eur J Immunol 2012; 42:2608–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flatz L, Roychoudhuri R, Honda M, Filali-Mouhim A, Goulet JP, Kettaf N, et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc Natl Acad Sci U S A 2011; 108:5724–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 2008; 29:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duprez DA, Neuhaus J, Kuller LH, Tracy R, Belloso W, De Wit S, et al. Inflammation, coagulation and cardiovascular disease in HIV-infected individuals. PloS One 2012; 7:e44454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boasso A, Shearer GM. Chronic innate immune activation as a cause of HIV-1 immunopathogenesis. Clin Immunol 2008; 126:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douek D. HIV disease progression: immune activation, microbes, and a leaky gut. Top HIV Med 2007; 15:114–117. [PubMed] [Google Scholar]
- 54.Sharma TS, Hughes J, Murillo A, Riley J, Soares A, Little F, et al. CD8+ T-cell interleukin-7 receptor alpha expression as a potential indicator of disease status in HIV-infected children. PloS One 2008; 3:e3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Far M, Halwani R, Said E, Trautmann L, Doroudchi M, Janbazian L, et al. T-cell exhaustion in HIV infection. Curr HIV/AIDS Rep 2008; 5:13–19. [DOI] [PubMed] [Google Scholar]
- 56.Migueles SA, Weeks KA, Nou E, Berkley AM, Rood JE, Osborne CM, et al. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol 2009; 83:11876–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reuter MA, Pombo C, Betts MR. Cytokine production and dysregulation in HIV pathogenesis: lessons for development of therapeutics and vaccines. Cytokine Growth Factor Rev 2012; 23:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacPherson PA, Fex C, Sanchez-Dardon J, Hawley-Foss N, Angel JB. Interleukin-7 receptor expression on CD8(+) T cells is reduced in HIV infection and partially restored with effective antiretroviral therapy. J Acquir Immune Defic Syndr 2001; 28:454–457. [DOI] [PubMed] [Google Scholar]
- 59.Landires I, Bugault F, Lambotte O, de Truchis P, Slama L, Danckaert A, et al. HIV infection perturbs interleukin-7 signaling at the step of STAT5 nuclear relocalization. AIDS 2011; 25:1843–1853. [DOI] [PubMed] [Google Scholar]
- 60.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U S A 2004; 101:16004–16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabbaj S, Heath SL, Bansal A, Vohra S, Kilby JM, Zajac AJ, et al. Functionally competent antigen-specific CD127(hi) memory CD8+ T cells are preserved only in HIV-infected individuals receiving early treatment. J Infect Dis 2007; 195:108–117. [DOI] [PubMed] [Google Scholar]
- 62.Lecuroux C, Girault I, Boutboul F, Urrutia A, Goujard C, Meyer L, et al. Antiretroviral therapy initiation during primary HIV infection enhances both CD127 expression and the proliferative capacity of HIV-specific CD8+ T cells. AIDS 2009; 23:1649–1658. [DOI] [PubMed] [Google Scholar]
- 63.Sasson SC, Zaunders JJ, Zanetti G, King EM, Merlin KM, Smith DE, et al. Increased plasma interleukin-7 level correlates with decreased CD127 and Increased CD132 extracellular expression on T cell subsets in patients with HIV-1 infection. J Infect Dis 2006; 193:505–514. [DOI] [PubMed] [Google Scholar]
- 64.Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM, Bohjanen PR, Mueller DL, et al. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol 2009; 183:1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 2012; 338:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon V, Ho DD. HIV-1 dynamics in vivo: implications for therapy. nature reviews. Microbiology 2003; 1:181–190. [DOI] [PubMed] [Google Scholar]
- 67.Vandergeeten C, Fromentin R, Chomont N. The role of cytokines in the establishment, persistence and eradication of the HIV reservoir. Cytokine Growth Factor Rev 2012; 23:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 2008; 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.