Abstract
Building on our recent report of an active H2 production catalyst [Ni(PPh2NProp-peptide)2]2+ (Prop = para-phenylpropionic acid, peptide (R10) = WIpPRWTGPR-NH2, p = D-proline and P2N = 1-aza-3,6-diphosphacycloheptane) that contains structured β-hairpin peptides, here we investigate how H2 production is effected by: (1) the length of the hairpin (eight or ten residues) and (2) limiting the flexibility between the peptide and the core complex by altering the length of the linker: para-phenylpropionic acid (three carbons) or para-benzoic acid (one carbon). Reduction of the peptide chain length from ten to eight residues increases or maintains the catalytic current for H2 production for all complexes, suggesting a non-productive steric interaction at longer peptide lengths. While the structure of the hairpin appears largely intact for the complexes, NMR data are consistent with differences in dynamic behavior which may contribute to the observed differences in catalytic activity. Molecular dynamics simulations demonstrate that complexes with a one-carbon linker have the desired effect of restricting the motion of the hairpin relative to the complex; however, the catalytic currents are significantly reduced compared to complexes containing a three-carbon linker as a result of the electron withdrawing nature of the -COOH group. These results demonstrate the complexity and interrelated nature of the outer coordination sphere on catalysis.
Keywords: Hydrogen production, Electrocatalysis, Peptide catalyst, Outer coordination sphere, Renewable energy, Enzyme mimic
1. Introduction
The superior catalytic activity of enzymes compared to synthetic catalysts suggests that features of the enzyme scaffold are critical to their function. Enzymes utilize the protein scaffold to create a pocket for the reaction, and to move substrates such as gases, protons, and electrons into and out of the active site [1, 2]. A more subtle feature, but equally important, is the finely tuned environment of this well-structured scaffold. This enzymatic scaffold is defined by precise spatial placement of natural amino acids that provide atomic control over the hydrophobicity, hydrophilicity, and dielectric environment around the active site [1, 3].
Hydrogenases efficiently interconvert hydrogen and protons, and mimics of these catalysts have been studied extensively [4]. Due to the enhanced performance of the enzymes compared to synthetic catalysts, several research groups have focused on incorporating features of the enzyme scaffold into molecular mimics to achieve the enhanced activity, specificity, selectivity, and/or stability observed in enzymes [5–13]. For hydrogenase mimics, several approaches have been undertaken to introduce features found in the outer coordination spheres of enzymes, from using viscous media and binding to resin beads [14–16], to attaching solubilized polymers [17–19] and amino acids/peptides [20–28]. A peptide-based outer coordination sphere allows for the emulation of a minimal biomimetic scaffold around the active site that has the potential to offer all of the advantages of the enzyme framework; an essential step is to synthetically incorporate structured peptides around the active molecular catalysts. Our laboratory has been focusing on the incorporation of protein motifs onto some of the most efficient molecular electrocatalysts for H2 production reported to date: the [Ni(PPh2NX2)2]2+ and [Ni(7PPh2NX)2]2+ complexes (P2N2 = 1,5-diaza-3,7-diphosphacyclooctane and 7P2N = 1-aza-3,6-diphosphacycloheptane) [20]. These complexes have a well-understood first coordination sphere, making them ideal platforms on which to test the influence of a peptide-based outer coordination sphere on catalytic reactivity. We recently reported the incorporation of a β-hairpin peptide (R10) onto [Ni(PPh2NProp)2]2+ (Prop) to produce [Ni(PPh2NProp-R10)2]2+ (Prop-R10), where Prop = para-phenylpropionic acid and R10 = WIpPRWTGPR-NH2 (p = D-proline) [27]. This β-hairpin peptide is an ideal peptide motif because it has a very stable secondary structure and the primary sequence can be readily modified to examine different features of the outer coordination sphere [29, 30]. Additionally, the hairpin’s β-turn directs the peptide’s C-terminus toward the complex, the desired location for it to have an impact on catalytic activity.
In the seminal work reported for this complex, R10 largely maintained its structure within the Prop-R10 complex and improved the catalytic current relative to the parent complex (Prop) demonstrating the impact of the outer coordination sphere on electrochemical hydrogen production [27]. However, the propionic acid linker allows a great deal of structural flexibility of the peptide relative to the catalyst core, limiting the R10 peptide from creating a precisely positioned outer coordination sphere around the active site. In an effort to reduce the mobility of the outer coordination sphere relative to the core complex, and to understand the role of specific residues on activity, we have investigated a series of complexes spectroscopically and computationally. This series of complexes has two varying features: the linker length (one and three carbons) and the length of the peptide chain. The resulting Prop-peptide complexes described in this study (figure 1) have higher catalytic currents than the parent complex. The Meth-peptide complexes have restricted motion, as designed; however, this linker negatively influences the catalytic performance. The catalytic currents for the Meth parent complex are 50-times lower than for the Prop parent complex, and the catalytic currents for the Meth-peptide complexes are the same or lower than the Meth parent complex, possibly due to structural restriction.
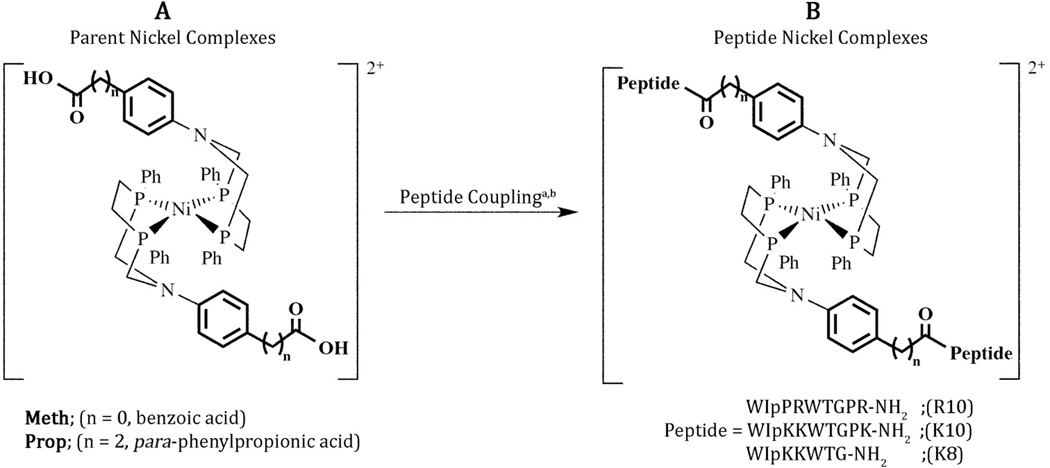
Figure 1.
Schematic of (A) the NiII-parent complexes ([Ni(PPh2NX)2]2+) and (B) the NiII-peptide complexes ([Ni(PPh2NX-peptide)2]2+) referred to with the short-hand notation given in table 1. The amino acid “p” is D-proline. aAny given complex was prepared with only one of the peptides shown: R10, K10, or K8. b See experimental section for the coupling procedure.
2. Results
2.1. Synthesis
Five metal complexes were prepared for this study (figure 1 and table 1). The NiII-parent complexes ([Ni(PPh2NX)2]2+) with X = para-phenylpropionic acid and para-benzoic acid are referred to as Prop and Meth, respectively. After coupling to the N-terminal of the β-hairpin, three (Prop) and one (Meth) carbons link the phenyl ring to the peptide (figure 1). Both ligands and NiII-parent complexes were prepared following previously reported procedures [27, 31, 32]. The ligands were isolated as white powders containing the meso- and rac-isomers and the NiII-parent complexes were isolated as red to orange crystalline products containing the nickel bound only to the meso isomer [31]. Each of the parent ligands was characterized by elemental analysis, and 31P{1H} and 1H NMR spectroscopy. The NiII-parent complexes were characterized by 31P{1H} and 1H NMR spectroscopy, mass spectrometry, cyclic voltammetry, and X-ray crystallography (for Prop [27] and Meth; figure S1, tables S1–S3).
Table 1.
Shorthand notation for the two parent [Ni(PPh2NX)2]2+ and the five [Ni(PPh2NX-peptide)2]2+ complexes prepared for these studies. Peptides: R10 = WIpPRWTGPR-NH2, K10 = WIpKKWTGPK-NH2, K8 = WIpKKWTG-NH2; p = D-proline.
| Linker X | Peptide | Shorthand notation |
|---|---|---|
| para-benzoic acid | None | Meth |
| K8 | Meth-K8 | |
| K10 | Meth-K10 | |
| para-phenylpropionic acid | None | Propa |
| K8 | Prop-K8 | |
| K10 | Prop-K10 | |
| R10 | Prop-R10a |
Reported previously [27]
The synthesis of the NiII-peptide complexes, [Ni(PPh2NX-peptide)2]2+, defined in figure 1 and table 1, were prepared on resin as previously reported by coupling the unprotected carboxylic acid of the parent PPh2NX ligand to the N-terminal W1 amine to create an amide bond [27]. The ligands were then metallated, followed by cleavage of the NiII-peptide complexes from the resin, precipitation from diethyl ether, dissolution into water, and isolation after lyophilization as light red fluffy powders [27]. Each complex was purified by HPLC to a single peak (figure S2 and table S4). Each of the NiII-peptide complexes was characterized by 1D and 2D 1H NMR spectroscopy, 31P{1H} NMR spectroscopy, mass spectrometry, cyclic voltammetry, and circular dichroism (CD) spectroscopy.
2.2. Core structure
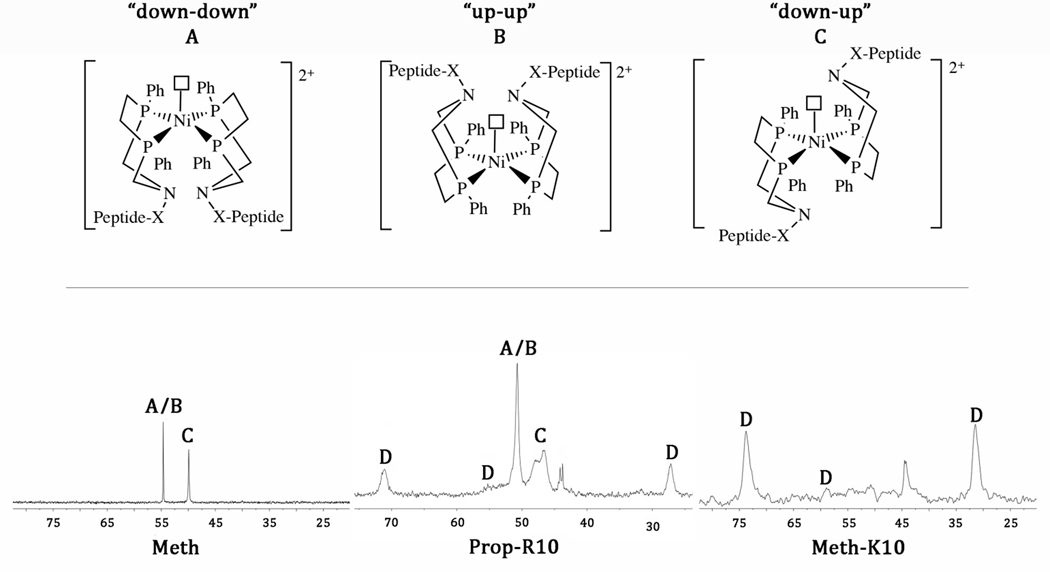
The 31P{1H} NMR spectra of the NiII-parent complexes, Prop and Meth, in CD3CN are similar to analogous complexes of this type [31, 32] and show two resonances in a ~1:1 ratio, previously assigned to a five-coordinate “down-up” isomer and a combination of “up-up” and “down-down” isomers [32], where a solvent molecule (acetonitrile in this case) is the fifth ligand (figure 2, where the terms “up” and “down” refer to the position of the pendant nitrogen atom in relation to the solvent molecule).
Figure 2.
Top: Three isomers (A, B, C) are observed for the NiII-peptide complexes, which contain a fifth ligand, a coordinated solvent molecule (acetonitrile) designated by an open square. Bottom: 31P{1H} NMR spectra of the NiII-parent complexes show only three isomers as shown for Meth. The Prop-peptide complexes also show all three isomers plus a down-up isomer with restricted structural mobility (D) as shown for Prop-R10. The Meth-peptide complexes show only the D isomer as shown for Meth-K10. The unlabeled resonances at ~45 ppm are oxidized phosphine.
The 31P{1H} NMR (95:5 CD3CN:H2O) spectra of the NiII-peptide complexes are considerably more complex than their parent compounds, indicating slower exchange between the different isomers (figure 2). The Prop-K10 and Prop-K8 complexes each show all three isomers (A, B, C), with the addition of new resonances (D) consistent with motional restriction of the “down-up” isomer, as previously reported for the Prop-R10 [27] complex but in different ratios (table 2). The less fluxional “down-up” five-coordinate species is thought to have less structural flexibility due to intra- or intermolecular interactions between the peptide chains. Restricted mobility is observed at low temperatures in the related [Ni(PPh2NPh-X)2]2+ complexes (figure S3) [31, 33], and the assignment to the “down-up” isomers for the β-hairpin peptide complexes here is based on the similarity of the lineshape and chemical shift difference of the structurally restricted resonances. Structural restriction is also observed at room temperature for similar dipeptide substituted [Ni(PPh2Ndipeptide2)2]2+ complexes [22, 23]. The 31P{1H} spectra of the Meth-K8 and Meth-K10 complexes indicate that the main isomer is the less fluxional “down-up” five coordinate species (table 2). We recently confirmed exchange between the two resonances for complex D for the Prop-R10 complex using a 31P-31P EXSY experiment, suggesting they are unique phosphorous atoms in the same molecule [27].
Table 2.
Population of each isomer based on the 31P{1H} NMR spectra, excluding the percentage of the free oxidized phosphine ligand (unlabeled resonances in spectra, figure 2).
| Nickel peptide complex | Up-up (A) and down-down (B) (%) | Down-up (C) (%) | Restricted down-up (D) (%) |
|---|---|---|---|
| Meth-K8 | <1 | <1 | >99 |
| Meth-K10 | <1 | <1 | >99 |
| Prop-K8 | 16 | 23 | 61 |
| Prop-K10 | 29 | 31 | 40 |
| Prop-R10a | 34 | 41 | 25 |
Reported previously [27]
2.3. Peptide structure
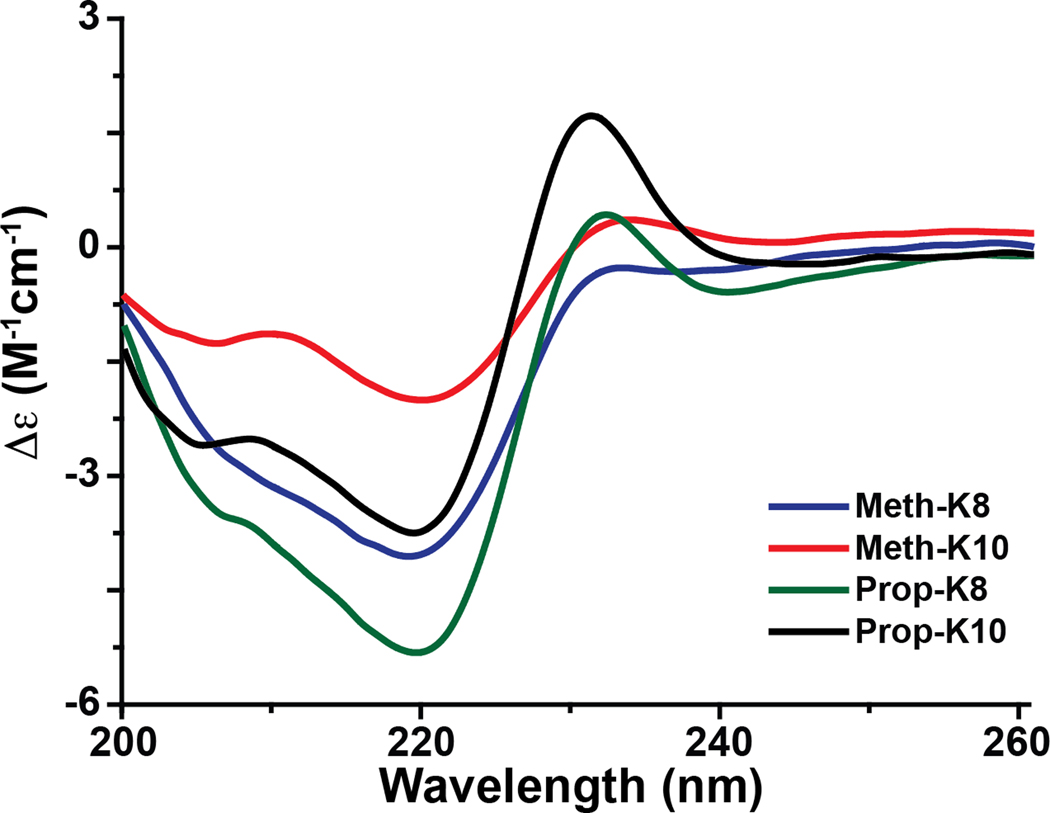
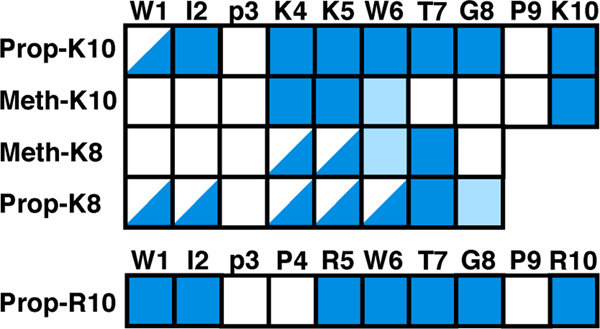
The peptide structure of each complex was evaluated by circular dichroism (CD) (95:5 acetonitrile:water) and NMR spectroscopy (90:10 acetonitrile:water). As illustrated in figure 3, the CD spectrum for each compound contains an exciton couplet with a maximum at ~228 nm characteristic of a face-to-edge Trp-Trp interaction in a β-hairpin fold [29]. While the intensity of the ~228 nm band can be indicative of the degree of peptide folding, it cannot be used to quantitate the relative amount of folding between compounds because it is sensitive to the precise orientation and rotamer preference of the two Trp residues, particularly in the presence of bulky substituents in the linker regions of the NiII-peptide complexes [34]. Quantitation of the ~228 nm band is especially difficult if there are multiple alignments of the Trp rings or if the ring-ring orientation is dynamic. Dynamics/conformational heterogeneity were suggested for many of the NiII-peptide complexes based on the 1H and 31P NMR data. For example, while the natural abundance 1H-15N HSQC spectra for the compounds showed chemical shift dispersion in both the 1H and 15N dimension characteristic of a structured peptide, weak/broadened, missing or multiple backbone amide cross peaks were reported for W1 and/or W6 in many of the compounds (figure 4 and tables S5–S6). A consequence of the multiple conformations/dynamics is that the NOESY spectra were not rich in information for many of the compounds. At a minimum however, the long-range W1 (1Hα) to W6 (1Hα) NOE, unambiguously indicative of the β-hairpin folded peptides, was observed for each of the complexes, corroborating the CD data that suggested all these compounds adopted a β-hairpin fold. The trend observed for the I2 amide (table S5) is consistent with Prop-R10 (8.83 ppm) being most folded and Prop-K8 (8.19 ppm) least folded based on the chemical shift values, with Prop-K10 (8.3 ppm) and Meth-K10 (8.53 ppm) being intermediately folded [30, 34].
Figure 3.
Circular dichroism spectra of the NiII-peptide complexes in 95:5 acetonitrile:water contain a band with a maximum ellipticity at ~228 nm that is characteristic of a Trp-Trp exciton due to adoption of a β-hairpin fold by the peptide.
Figure 4.
Amide cross peak patterns observed in the 1H-15N HSQC spectra for each of the NiII-peptide complexes. Each square represents an amide cross peak from the 1H-15N HSQC: solid blue = present, light blue = present but weak, half blue = dual resonances and white = not present (no cross peak is expected for proline). Cross peaks for the first two residues have the most sensitivity to the linker identity, while residues 4–6 are the most sensitive to peptide length.
2.4. Peptide orientation relative to the core complex
Intramolecular 1H–1H NOEs were not observed between the peptide and the peptide-metal linker region for any of the complexes. The absence of such NOEs could be due to dynamics (consistent with NMR observations reported above) or the distance of the peptide from the linker of > 6 Å, the maximum expected distance measurable from an NOE. In order to gain an understanding of the relative position of the peptide to the metal core, replica exchange molecular dynamic (REMD) simulations were performed on Meth-K8, Meth-K10, Prop-K8 and Prop-R10 using the NMR and CD observations to restrain the peptide to a β-hairpin. These calculations showed that the phenyl group is within 6 Å of several of the amino acid side chains in the hairpin (residues 1, 5, and 6 for all five complexes, residue 2 for Meth-K10 and Prop-R10, and residue 4 for Meth-K8, Meth-K10 and Prop-K8; figure S4). This suggests that the hairpin is positioned in close proximity to the core and the lack of NOEs is a result of mobility of the peptide relative to the core complex.
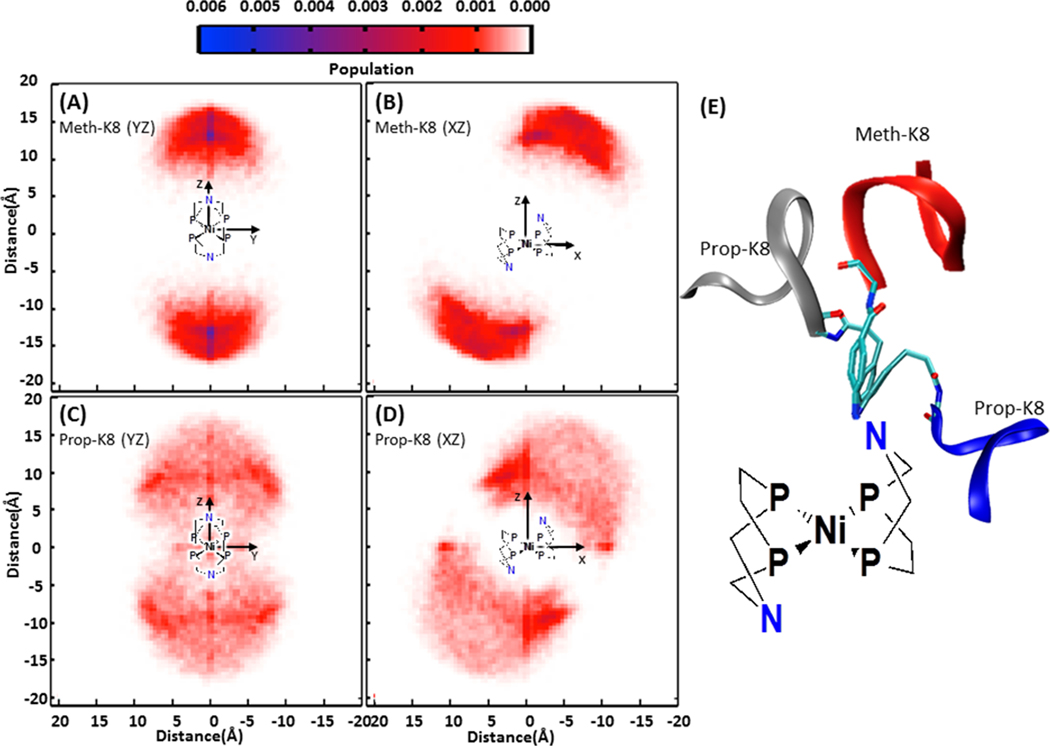
While the hairpin relative to the core has variable orientation in all of the complexes studied, the REMD simulations show that the range of motion for the hairpin relative to the linker is reduced for the Meth-peptide complexes compared to the Prop-peptide complexes. This is shown by projecting the tip of the hairpin (Cα of the D-Pro residue) into three planes (described in detail in the Methods section) as illustrated in figure 5. The projection in the YZ plane (figure 5A and 5C) and the XZ plane (figure 5B and 5D) show a significantly smaller and more localized region for each hairpin of Meth-K8 (figure 5A and 5B) compared to the hairpins on Prop-K8 (figure 5C and 5D). The projection of the full hairpin backbone is shown in figure S5.
Figure 5.
Probability distribution of the projection of the hairpin peptide from Meth-K8 onto the YZ plane (A) and XZ plane (B), and Prop-K8 onto the YZ plane (C) and XZ plane (D), as defined in the Methods section. Meth-K8 shows the β-hairpins have a smaller range of motion and one shallow minima (dark blue in the YZ plane), whereas Prop-K8 has a larger range of motion and two preferred orientations as indicated by the dark red regions. (E) Representative orientations of the preferred structures are shown on the right (red ribbon: Meth-K8; blue and gray ribbons: Prop-K8).
The projections in figure 5 show that for Meth-K8 there is a single, preferred orientation of the hairpin relative to the core (blue region), with the N-(D)Pro vector being perpendicular to the YZ plane (~90 degrees) (figure 5A), although the well is shallow and accommodates a distribution of structures. For Prop-K8, two orientations are favored, the dark red regions in figure 5C and 5D, on either side of the N. Representative structures from the most highly populated regions of the Meth-K8 and Prop-K8 complex are shown in figure 5E. The REMD data are qualitatively consistent with the 1H NMR HSQC results which show two conformers of the peptide in residues 1 and sometimes 2 for the Prop-peptide complexes, while the Meth-peptide complexes have no resonance, suggestive of slowly interconverting species (restricted mobility).
The mobility between the peptide and the core in the Meth-peptide complexes is limited to rotations about the phenyl ring axis. For the Prop-peptide systems, rotations about the phenyl ring axis are combined with additional degrees of freedom due to the three-carbon chain linker that allows a hinge-like motion. The 10-mer peptide for both Meth-K10 and Prop-R10 also increase the space covered by the peptide compared to the 8-mer peptide (figure S6). Interestingly, inter-hairpin hydrogen bonding within the complex is observed only for the Prop-peptide complexes, where many hydrogen bonds are observed for both Prop-K8 and Prop-R10, figure S7. The lack of interaction between chains in the Meth-peptide complexes is due to the limited degrees of freedom afforded by the shorter linker. No correlation was found between the motions of the two hairpins for any complex.
2.5. Electrochemistry
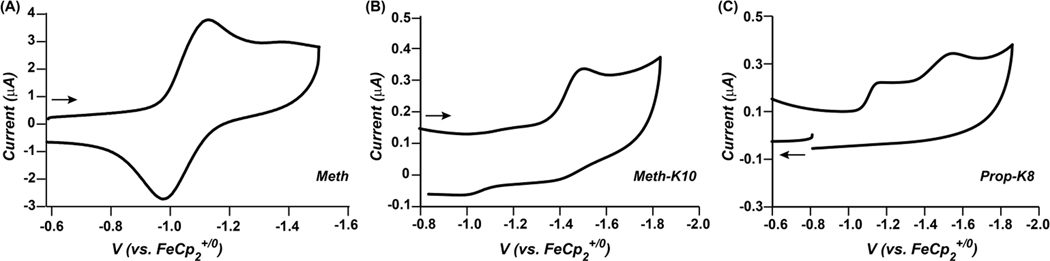
The cyclic voltammograms of all of the NiII-parent complexes indicate two overlapping one-electron, reversible redox couples at −1.10 V (Meth) and −1.12 V (Prop) versus the Cp2Fe+/0 couple in 100% acetonitrile, consistent with observations for similar complexes [31, 32]. Similar potentials are observed for the NiII-peptide complexes in acetonitrile with 2–3% water, although the waves are irreversible, consistent with the previously reported Prop-R10 complex [27] (figure 6). Adding a small amount of the base triethyl amine resulted in a reversible wave (figure S8), suggesting that residual acid was already performing H2 production. In the NiII-peptide complexes, a second wave at −1.4 V was also observed. The latter wave is irreversible and is more pronounced for the Meth-peptide complexes than for the Prop-peptide complexes (figure 6), and could be consistent with association of a fifth ligand, either solvent or a functional group from the peptide, but could also be consistent with restricted motion due to the hairpins. The negative shift of the NiII/I wave upon association of a fifth ligand for these complexes has been reported previously [35]. Both interpretations are consistent with the restricted mobility observed by 31P NMR spectroscopy (figure 2).
Figure 6.
(A) Cyclic voltammogram of 0.25 mM Meth in 0.1 M [Et4N][BF4]/acetonitrile with a scan rate 1.0 Vs−1 at 25 °C. This data is representative of all of the NiII-parent complexes. (B and C) Cyclic voltammogram of 0.25 mM Meth-K10 (B) and 0.25 mM Prop-K8 (C) in 0.1 M [Et4N][BF4]/acetonitrile/2% water with a scan rate 0.05 Vs−1 at 25 °C. The arrows indicate initial scanning direction. All data collected with a 1 mm glassy carbon working electrode.
2.6. Catalysis
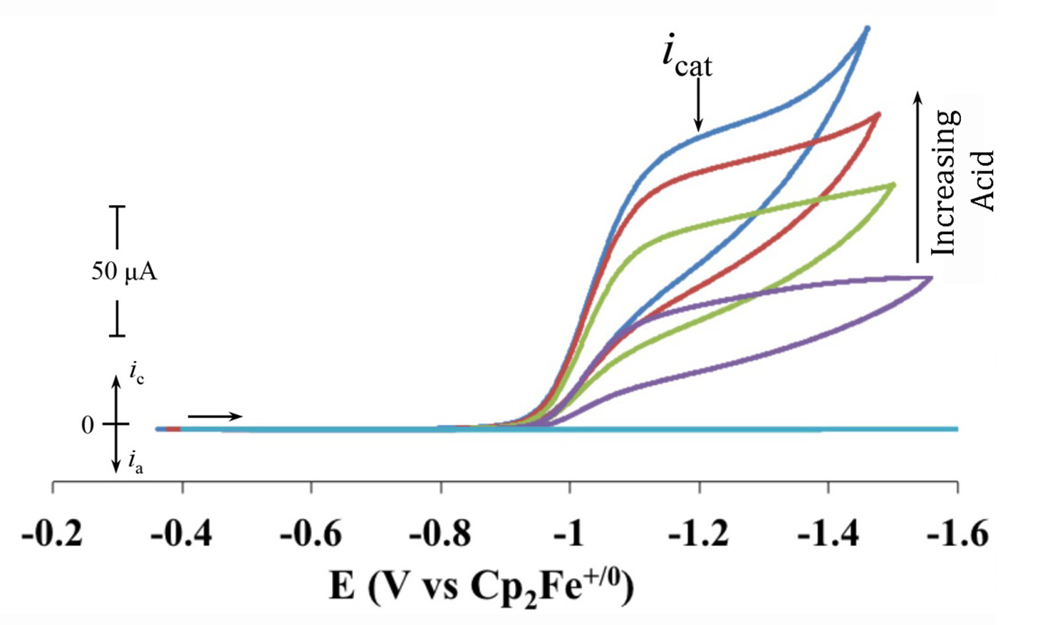
Each of the NiII-peptide complexes requires a small percentage of water to become soluble in acetonitrile. Due to the previously reported high sensitivity of the catalytic TOF to water concentration [23, 36], water content was optimized for electrochemical measurements and catalytic studies were carried out in initial solution conditions containing either 100% acetonitrile (Meth and Prop); 97:3 (v:v) acetonitrile:water (Meth-K10 and Meth-K8); or 98:2 (v:v) acetonitrile:water (Prop-K10, Prop-R10 and Prop-K8) (table 3). Upon the addition of acid (protonated dimethylformamide triflate, ([(DMF)H]+), each complex shows an increase in the cathodic current (icat), consistent with catalytic hydrogen production (figure 7). The catalytic production of H2 was previously confirmed by quantitative gas chromatography during a controlled potential electrolysis of Prop-R10 with a current efficiency of 96% [27]. After the maximum current was reached with the addition of acid (acid independence), water was added to realize any further enhancements [22, 36].
Table 3.
Summary of icat and overpotential determined for 0.25 mM catalyst at maximum acid (0.5 – 1.3 M) and optimal water (0.4 to 2.0 M) concentrations for each of the parent and peptide complexes studied.
| Complex | OP (mV) (±10%) | icat x 105(A) (±25%) | TOF (s−1) (±25%) |
|---|---|---|---|
| Meth | 470 | 2 | 1350 |
| Meth-K8 | 520 | 2 | NA |
| Meth-K10 | 480 | 0.5 | NA |
| Propa | 550 | 6 | 80,000a [37] |
| Prop-K8 | 540 | 14.5 | NA |
| Prop-K10 | 560 | 7 | NA |
| Prop-R10a | 540 | 10 | NA |
Reported previously [27]
Figure 7.
Cyclic voltammograms of 0.25 mM Prop-K8 in 0.1 M [Et4N]+[BF4]-/acetonitrile/2% water with subsequent additions of [(DMF)H]+; representative of all the peptide complexes. Data collected at 25 °C using a 1 mm glassy carbon working electrode with a scan rate 0.05 Vs−1. The icat values were taken at −1.2 V, as indicated.
The icat was measured at the potential where the current first reaches its plateau (−1.2 V as shown in figure 7 by the arrow labeled icat), and for the parent complexes, the ratio of icat/ip used in equation 4 to calculate the TOF (see Materials and Methods). The maximum TOFs for each of the complexes are listed in table 3 and found to be 1,350 s−1 for Meth and 80,000 s−1 for Prop [27, 37]. We were unable to obtain TOF values for any of the peptide complexes due to the second observed wave (~−1.4 to −1.5 V) and the resulting difficulty in determining ip. Instead we report only the icat measured at −1.2 V, values that can be compared to one another to give insight into relative catalytic activity, given that similar concentrations are used and that scan rate independence is observed (figure S9).
The icat for H2 production of the parent and Prop-peptide complexes decreases in the following order: Prop-K8 > Prop-R10 > Prop-K10 ≈ Prop. The icat for Prop-K8 is about ~2.5 times higher than the parent Prop complex, while Prop-K10 is 20% higher (table 3). The icat for the Meth complex is about three times lower than for the Prop complex, an observation attributed to pKa, described below. Each of the Meth-peptide complexes has the same or less current enhancement than the parent complex, and similar overpotentials are obtained (table 3). The overpotentials for the complexes (table 3) are insensitive to the presence or absence of the peptide and are similar to previously reported values [27, 31, 32].
Each of the NiII-peptide complexes was tested for catalytic hydrogen production in 100% water under various conditions (see Materials and Methods). Unfortunately, the addition of different acids (1.0 M H2SO4 (2 μM), 1.0 M HClO4 (2 μM) or 1.0 M HTFSI (2 μM)) to a 0.25 mM solution of each of the NiII-peptide complexes in 0.1 M Na2SO4/water/1–5% acetonitrile resulted in instant precipitation of the complexes, therefore electrocatalytic studies in 100% water were not possible. Previous work of the [Ni(PPh2NPh2)2]2+ was reported in 100% water when it was associated with Photosystem I [38], suggesting that a larger peptide scaffold may be needed to attain full water solubility.
2.7. Pendant amine pKa values for the parent complexes
The pKa values for the parent complexes of each protonated pendant amine were determined using the linear free energy relationships we recently proposed for [Ni(PR2NR’2)2]2+ complexes [39]. These correlations can also be used for [Ni(7PR2NR’)2]2+ complexes without losing accuracy (unpublished observations), and were found to vary as a function of the substituent on N ([Ni(PPh2NX)(PPh2NXH)]2+) in 100% acetonitrile: Ph = 5.9, Meth = 2.5, Prop = 5.7. They correlate the pKa value of a protonated pendant amine in the complex with the NiI/Ni0 redox potential of the non-protonated complex and the pKa value of the parent primary aminium from which the complex is derived. While the absolute values are likely inaccurate, the relative values are reliable. The experimental redox potential was used in the linear free energy correlations.
3. Discussion
Enzymes use the protein scaffold to position functional groups in specific locations to achieve the desired reactivity. A recent report using a “top-down” approach [40], i.e., binding the catalyst to a preformed protein, demonstrated that assembling [Ni(PPh2NPh2)2]2+ with photosystem I and its electron transfer partners can modify the redox potentials for H2 production [38]. Using a “bottom-up” approach [40], i.e., starting with the catalyst and building a protein-like framework onto it, we recently demonstrated that incorporating a structured peptide (R10) onto the outer coordination sphere of Prop to result in Prop-R10 increased the catalytic activity without affecting the overpotential [27, 41]. Unfortunately, the para-phenylpropionic acid linker used to attach the peptide to the complex allowed significant flexibility of the hairpin relative to the core complex, limiting the ability to control precise interactions around the metal center such as those achieved with the enzyme scaffold. The work reported here is an attempt to further reduce the range of motion of the peptide about the metal center in order to have a structurally well-defined environment with which we can start systematically probing the role of the peptide in catalysis.
3.1. Catalytic activity of parent complexes
To reduce the degrees of freedom of the peptide about the pendant amine phenyl group in our original compound, Prop-R10, complexes containing a one-carbon linker (Meth) were synthesized and studied. For the parent complex, a significant reduction in TOF is observed for Meth (1,350 s−1) compared to Prop (80,000 s−1) [27]. We correlate this loss in activity to an increase in the electron-withdrawing nature of the immediately bound -COOH group that lowers the pKa of the pendant amine (table 3). Recent studies have shown that the relative pKa of the pendant amine of [Ni(PPh2NC6H4X)2]2+ complexes with respect to the proton source is a determining factor for the TOF of H2 production [32, 39]. For example, electron-withdrawing groups on [Ni(PPh2NC6H4X)(PPh2NC6H4XH)]2+ (where X = CF3, Cl or Br) result in lower pKa values of the pendant amine with respect to the proton source (in this case [(DMF)H]+, pKa = 6.1 in acetonitrile). The mismatched pKa values for the solvent acid ([(DMF)H]+) and the pendant amine result in slower or incomplete protonation at the pendant amine, thus slower TOFs are observed [32, 39].
3.2. Effect of linker length on catalytic activity
The shorter linker was introduced to limit the mobility of the β-hairpin peptide relative to the core complex so that ultimately the positioning of the amino acids relative to the core complex could be controlled. Evaluation of the structural and dynamic features of the peptide complexes suggests that the role of limiting mobility was achieved. First, the REMD calculations suggest that introducing the shorter linker produced the desired result of limiting the mobility in the Meth-peptide complexes. Second, the 31P NMR show restricted motion in the core catalyst for the Meth-peptide complexes beyond that which was observed for the Prop-peptide complexes, a feature which could also be beneficial to catalytic activity by controlling the location of the pendant amines relative to the metal [32, 42, 43]. Third, consistent with the REMD analysis, 1H NMR data suggest differing local dynamics and β-hairpin populations for the Meth-peptide complexes and Prop-peptide complexes, particularly at W1 and I2, based on missing, weak, or dual amide resonances in the 1H-15N HSQC spectra as a function of linker (figure 4). While the limited mobility of the shorter linker could be completely responsible for the change in dynamics as suggested by REMD, contributions from binding a fifth ligand from one of the peptide functional groups cannot be ruled out. The 31P NMR spectra of five-coordinate Ni(PR2NR’2)2X (X = acetonitrile) complexes have been shown to have similar spectra to that shown in figure 2 [22, 44]. Additionally, the binding of a fifth ligand shifts the electrochemistry to more reducing potentials, consistent with that observed by the presence of the second wave in the CV (figure 6). While a more thorough investigation would be needed to resolve these questions, the drawbacks of this system are significant enough that the time needed to perform this evaluation is not warranted.
3.3. Effect of chain length on catalytic activity
The catalytic rates, as inferred from the icat values, were greater for corresponding complexes with an 8-residue peptide (K8) than a 10-residue peptide (R10 or K10). This observation is surprising and could be due to several factors, including a structural change in the core catalyst, an influence of the peptide functional groups, steric interactions of the longer chain, or peptide structural changes. A structural change in the core is an unlikely explanation based on the identical 31P{1H} NMR spectra within a given linker family showing insensitivity to chain length (figure 2 and table 2). The REMD simulations indicate a slightly higher propensity for the hairpin motif to be located in the plane of the active site for Prop-R10 than for Prop-K8 (figure S6), suggesting that the longer chain could be hindering access of the acid to the active site and consequently, slowing catalysis for Prop-R10. Structural changes in the peptide are also observed. While the 1H NMR and CD data suggest that the peptides adopt a β-hairpin structure in all of the complexes, different residue-specific spectral features (split, weak, or absent amide resonances) are observed in the 1H-15N HSQC spectrum for each complex (figure 4), suggesting differences in local dynamics or β-hairpin populations. Specifically, spectral changes in residues 4–6 have a strong correlation to chain length; the 10-mer hairpin complexes all have a single conformer for each residue, while the 8-mer hairpin complexes exhibit two conformers based on the split peaks observed for most of these residues in Meth-K8 and Prop-K8 (figure 4). The sensitivity of residues 4–6 to chain length is not clear, but these differences in local dynamics and β-hairpin populations may be influencing catalysis. The REMD results also show that complexes containing a 10-residue peptide have residue 2 closer to the core a higher percentage of the time, while residue 4 is closer to the core in complexes with an 8-residue peptide. The potential role of these modeled structural differences requires further investigation to fully understand any mechanistic implications.
Other work focused on introducing peptides into the outer coordination sphere of molecular complexes have seen enhancements in rates or water solubility due to the incorporation of a peptide [38, 45, 46]. Complementary work where metals have been incorporated into stable proteins have also resulted in active H2 production catalysts [47]. In some cases, a structured outer coordination sphere has been added, without resulting function [21, 28]. Thorough functional and structural characterization of these systems to allow structure-function relationships has yet to be reported, and when determined, will significantly enhance our understanding of the role of the outer coordination sphere on catalytic performance. In an elegant study of hydrogenase itself, incorporating a molecular diiron complex into the apo-protein results in function identical to the native protein [48]. The ease of synthesis of this derivative provides a route to investigate the role of residues around the active site and their impact on catalysis. Ultimately, the combination of these three approaches is needed to provide a detailed understanding of the mechanistic function of the outer coordination sphere.
4. Summary
The goals of this work were to evaluate the role of a peptide outer coordination sphere on catalytic performance by limiting the mobility of the peptide relative to the core complex using a one (Meth) and three (Prop) carbon linker and to understand the role of peptide chain length using 8-residue and 10-residue peptides. The incorporation of structured peptides into active H2 production electrocatalysts enhances catalytic current for H2 production, with the shorter peptide (8-mer) having a larger increase than complexes with a longer peptide (10-mer). In all cases, the overpotential was not influenced by the peptide. Attempts to limit the flexibility of the peptide relative to the core by introducing a shorter linker were successful, based on both experimental and computational studies. However, as shown for the parent complexes, the linker alone decreased catalytic activity, likely due to changes in the pKa of the pendant amine. These results demonstrate the importance of both the second and outer coordination spheres in having a positive and negative impact upon catalytic performance and the importance of maintaining some structural flexibility while positioning amino acids in a defined way around the catalytic core. Perhaps more importantly, these results highlight the interrelated and complex nature of contributions of the outer coordination sphere in catalysis that must be understood in order to rationally enhance the performance of designed molecular catalysts.
5. Materials and methods
5.1. Materials
All reactions were performed under an inert atmosphere of nitrogen using standard Schlenk techniques except for cleaving the complex from the resin. Solvents were deoxygenated and purified using an Innovative Technology, Inc. PureSolv™ solvent purification system. Acetonitrile-d3 (Cambridge Isotope Laboratories, 99.5%D) was vacuum-distilled from P2O5. Chloroform-d (Cambridge Isotope Laboratories, 99.5%D) and methylene chloride-d2 (Cambridge Isotope Laboratories, 99.5%D) were degassed and stored over molecular sieves. Water was dispensed from a Millipore MilliQ purifier at 18 MΩ and sparged with nitrogen. 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), 1-hydroxybenzotriazole (HOBT), 3-(4-aminophenyl)propionic acid, 2-(4-aminophenyl)acetic acid and 4-aminobenzoic acid (Aldrich) were used as received. Diisopropylethyl amine (DIPEA) (Aldrich) was degassed prior to use by the freeze-pump-thaw method. The peptides (R10 = WIpPRWTGPR-NH2, K10 = WIpKKWTGPK-NH2 and K8 = WIpKKWTG-NH2) were synthesized with an amidated C-terminus and left on the resin with the N-terminal FMOC group until ready for use. Synthesis was performed on the polystyrene-based Rink-Amide MBHA resin at the Protein Chemistry Technology Center, University of Texas, Dallas, TX. Tetraethylammoniumtetrafluoroborate (Et4N+BF4-) (Alfa-Aesar) was recrystallized twice from hot ethanol; the crystals obtained were dried under vacuum. Sodium sulfate (Aldrich) was used as received. Ferrocene and (hydroxymethyl) ferrocene (Aldrich) were sublimed under vacuum before use. Dimethylformamide-trifluoromethanesulfonic acid, [(DMF)H]+, was prepared by the method of Favier and Duñach [49]. [Ni(MeCN)6](BF4)2 and PPh2NProp (where Prop = 3-(4-aminophenyl)propionic acid) and PPh2NMeth (where Meth = 4-aminobenzoic acid) were prepared following literature methods [27, 31, 50].
5.2. Syntheses
meso/rac PPh2NX. To 1,2-bis(hydroxymethylphenylphosphino)ethane (2.49 g, 8.13 mmol) in 100 mL ethanol at 75 °C was added the aniline reagent (8.13 mmol) (~0.5 g every 25 min while stirring) and the mixture was stirred at 75 °C overnight. The ethanol was removed leaving an off white powder that was washed with 60 mL of ethanol followed by diethyl ether, dried under vacuum and isolated as a 50/50 mixture of the meso- and rac-isomers. PPh2NProp: Yield: 2.98 g, 84.1%. 31P {1H} NMR (CD3CN, ppm); δ −26.2 (s) and −25.5 (s).1H NMR (CD3CN, ppm): δ 7.58 – 6.73 (m, 14H, P-C6H5 and N-C6H4-CH2); 4.33 – 3.68 (m, 4H, PCH2N); 2.84 – 2.48 (m, 4H, PCH2CH2P); 2.46 – 2.17 (m, 4H, CH2CH2-C(O)); elemental analysis (%) calcd: C 68.96, H 6.25, N 3.22; found: C 69.74, H 6.37, N 3.10. PPh2NMeth: Yield: 3.1 g, 93.1%. 31P{1H} NMR (CD2Cl2, ppm); δ −24.19 (s) and −25.55 (s). 1H NMR (CD2Cl2, ppm): δ 8.04 – 6.74 (m, 14H, P-C6H5 and N-C6H4-CH2); 4.51 – 3.69 (m, 4H, PCH2N); 2.51 – 2.25 (m, 4H, PCH2CH2P); elemental analysis (%) calcd: C 67.81, H 5.69, N 3.44; found: C 68.07, H 5.72, N 3.49.
[Ni(PPh2NX)2]2+. A mixture of the meso/rac PPh2NX ligand (0.34 mmol) was combined with [Ni(CH3CN)6](BF4)2 (0.17 mmol) in 20 mL acetonitrile forming a dark red/orange solution that was stirred at room temperature for two hours. The solution was filtered through celite and the solvent removed, followed by the addition of 3 mL of dichloromethane. The reaction mixture was then added to diethyl ether (273 K) with stirring to precipitate orange to red crystals. Only the meso isomer reacts with Ni (confirmed by 31P NMR), resulting in a pure [Ni(PPh2NX)2]2+ complex. The product was isolated by filtration, washed two times with 5 mL diethyl ether (273 K) and dried under vacuum. Prop: Yield: 68.7 mg, 36.4%. 31P {1H} NMR (CD3CN, ppm); δ 52.0 (s) and 48.1 (s). 1H NMR (CD3CN, ppm): δ 7.82 – 7.11 (m, 28H, P-C6H5 and N-C6H4-CH2); 4.86 (d, 4H, PCH2N); 3.96 (d, 4H, PCH2N); 2.89 (m, 4H, CH2C(O)); 2.70 (m, 4H, N-PhCH2); 2.34 (m, 4H, PCH2CH2P); 2.02 (m, 4H, PCH2CH2P).MALDI MS: m/z calcd for [Ni(PPh2NProp)2]2+: 928.24; found 928.24. Meth: Yield: 72.5 mg, 45.1%. 31P {1H} NMR (CD3CN, ppm); δ 54.63 (s) and 49.91 (s). 1H NMR (CD3CN, ppm): δ 8.05 – 7.07 (m, 28H, P-C6H5 and N-C6H4); 4.35 – 4.02 (m, 4H, PCH2N); 3.96 (m, 4H, PCH2N); 2.70 – 2.54 (m, 8H, PCH2CH2P). MALDI MS: m/z calcd for [Ni(PPh2NMeth)2]2+: 872.18; found 872.13.
meso/rac PPh2NX-Peptide. Coupling of meso/rac PPh2NX to the peptide to make the PPh2NX-Peptide ligand was performed on the Rink Amide MBHA resin. The fluorenylmethoxy carbonyl (FMOC) group was removed by stirring the resin bound peptide (0.08 mmol) in 20 mL of 20% piperidine in N-methylpyrrolidone for 10 min, filtered and repeated. The resin was then rinsed with 30 mL N-methylpyrrolidone, stirred in 20 mL of N-methylpyrrolidone for 5 min and rinsed in a medium frit with an additional 30 mL of N-methylpyrrolidone. The resin was then rinsed with 200 mL dichloromethane, stirred in 20 mL dichloromethane for 5 min and then rinsed in a medium frit with 200 mL dichloromethane. The resin was then rinsed with 30 mL dimethylformamide, stirred for 5 min in 20 mL dimethylformamide and then rinsed in a medium frit with 30 mL dimethylformamide. The meso/rac PPh2NX was activated in a stirred solution of 10 mL of a dimethylformamide solution containing PPh2NX (0.092 mmol), HATU (0.092 mmol), HOBT (0.092 mmol) and DIPEA (0.36 mmol). The mixture was allowed to stir for 5 minutes and then the FMOC deprotected peptide-resin was added along with an additional 10 mL of dimethylformamide. The solution was stirred at room temperature for 3 h (Prop-peptide complexes) to 12 h (Meth-peptide complexes), at which time a ninhydrin test showed complete coupling. The solution was filtered using a medium frit and the resin rinsed with 30 mL dimethylformamide stirred in 20 mL dimethylformamide for 5 min and then rinsed with 30 mL of dimethylformamide. This protocol was repeated with dichloromethane and then acetonitrile. The product was left attached to the resin for incorporation of nickel.
NiII-peptide complexes: The final resin above was added to a 5 mL acetonitrile solution containing 0.5 equivalents of [Ni(CH3CN)6](BF4)2. After 5 min an additional 15 mL of dimethylformamide was added to this reaction mixture and stirred overnight to yield a deep red/orange resin.
To cleave the NiII-peptide complexes from the resin, the resin was suspended in a 1 mL mixture of 50% triisopropylsilane/50% water (v:v) for 5 minutes followed by the addition of 4 mL of trifluoroacetic acid and stirred for 3 h at room temperature. The resulting reaction mixture was filtered and the volume of the filtrate reduced to less than 1 mL under vacuum. The resulting red solution was flash precipitated by addition of this solution into 60 mL diethyl ether (273 K). The product was dissolved in 100% water and purified by HPLC (figure S2 and table S4). Prop-R10: Yield: 7.3 mg, 5.1%. 31P{1H} NMR (10% H2O in CD3CN); “down-down and “up-up” (47.9, 46.7 ppm); “down-up” (71.3, 50.7, 27.4 ppm); “oxidized ligand” (44.1, 43.8 ppm). MALDI MS: m/z calcd for [Ni(PPh2NR10)2]2+: 3422; found 1714: [O2PPh2NR10 + 2H+]. Prop-K10: Yield: 7.9 mg, 9.3%. 31P {1H} NMR (10% H2O in CD3CN, ppm); “down-down” and “up-up” (46.54); “down-up” (70.26, 49.95, 26.36); “oxidized ligand” (43.34, 42.83). MALDI MS: m/z calcd for [Ni(PPh2NProp-K10)2]2+: 3369.68; found 1678.33: [PPh2NProp-K10 + Na+]. Prop-K8: Yield: 5.8 mg, 4.7%. 31P {1H} NMR (10% H2O in CD3CN, ppm); “down-down” and “up-up” (46.83); “down-up” (70.97, 50.15, 27.07); “oxidized ligand” (43.80). MALDI MS: m/z calcd for [Ni(PPh2NProp-K8)2]2+: 2921.35; found 1485.45: [O2PPh2NProp-K8 + Na+]. Meth-K10: Yield: 8.3 mg, 7.4%. 31P {1H} NMR (10% H2O in CD3CN); “down-up” (73.84, 31.56); “oxidized ligand” (44.49).MALDI MS: m/z calcd for [Ni(PPh2NMeth-K10)2]2+: 3313.16; found 1682.37: [O2PPh2NMeth-K10 + Na+]. Meth-K8: Yield: 4.4 mg, 5.3%. 31P {1H} NMR (10% H2O in CD3CN, ppm); “down-up” (72.78, 30.38); “oxidized ligand” (43.74). MALDI MS: m/z calcd for [Ni(PPh2NMeth-K8)2]2+: 2865.29; found 1425.60: [PPh2NMeth-K8 + Na+].
5.3. Spectroscopy and chromatography
Nuclear Magnetic Resonance (NMR).
All NMR spectra were recorded on Varian VNMRS or Inova spectrometers operating at proton resonance frequencies of 500, 600, 750 or 800 MHz. Proton chemical shifts were internally calibrated to residual solvent protons and 31P{1H} chemical shifts were externally referenced to phosphoric acid. Two-dimensional 1H-1H TOCSY, 1H-1H NOESY, and natural abundance 1H-15N HSQC spectra were collected at 25 °C for all peptide nickel complexes.
Circular Dichroism (CD).
All the Ni-peptide complexes were prepared in an acetonitrile:water (95:5) (v:v) mixture. The concentrations of the samples were measured using the predicted molar extinction coefficient (ε) at 280 nm, calculated using the four tryptophan residues in each complex (4 × εW280 nm (5600 M−1cm−1) = 22400 M−1cm−1). The sample concentrations were between 26 to 45 μM. The circular dichroism (CD) data [51, 52] were collected from 200 to 260 nm on a Aviv model 410 spectropolarimeter (Lakewood, NJ) in a quartz cell of 0.1 cm path length. The raw data were recorded as ellipticity ([θ]raw), which was later converted to molar ellipticity ([θ]M) using equation 1:
| (1) |
where c = concentration of the sample and l is the path length. When [θ]raw, c and l are represented in units of degree (deg), Molar (M), and cm, respectively, the units of [θ]M become deg cm2/dmol. The calculated molar ellipticity ([θ]M) data was further divided by the number of amide bonds in the complex to further normalize it to molar ellipticity per residue ([θ]r) (equation 2). The number of amide bond residues for the complexes containing 8-mer and 10-mer peptides are 14 and 18, respectively.
| (2) |
Finally the molar ellipticity per residue ([θ]r) data was converted to decadic molar circular dichroism (Δε) unit by using equation 3.
| (3) |
Spectra were processed by subtracting a blank spectrum from the sample spectrum followed by automatic line smoothing using the Aviv software.
Mass spectrometry.
A Bruker Autoflex II MALDI-TOF-MS was operated in reflectron mode with delayed extraction at 100 ns and an accelerating voltage of 19.8 kV. Spectra were acquired over m/z ranges of 1060–4460 or 300–2240. Samples containing 0.5 mg of analyte were suspended into 500 μL of 10% 18 cm/MΩ water and 90% acetonitrile. The sample was mixed 1:10 (sample:matrix) with matrix consisting of 10 mg of α-cyano-4-hydroxycinnamic acid in 33.3% acetonitrile, 33.3% ethanol (100%) and 33.3% 18 MΩ·cm water (0.1% trifluoroacetic acid) and deposited in multiple layers directly onto a stainless steel plate. Identification was based on the mass assignment match to the expected m/z matching the theoretical m/z within 100 ppm mass error. Data were collected using a minimum of 1000 shots in positive ion mode.
High Pressure Liquid Chromatography (HPLC).
Reverse phase chromatography was performed using a Waters XBridge C-18 (5 μm ODB) 19 × 250 mm prep column with a 20% to 80% gradient of solvent B over 15 minutes at a flow rate of 20 mL/min. Solvent A: 0.1% trifluoroacetic acid in water; Solvent B: 0.1% trifluoroacetic acid in acetonitrile.
5.4. Electrochemistry
General.
All electrochemical studies were done in an N2 glove box in 100% acetonitrile with 2–3% water (0.1 M Et4N+BF4-) at the ambient temperature of the glovebox, ~27 °C, using ferrocene as an internal standard. All electrochemical studies in water were done in an N2 glove box in water with 1–5% acetonitrile (0.1 M Na2SO4) at the ambient temperature of the glovebox, ~27 °C, using (hydroxymethyl)ferrocene as an internal standard. Cyclic voltammetry experiments were performed on a CH Instruments 1100A or 600D electrochemical analyzer using a standard three-electrode configuration. The working electrode was a 1 mm diameter glassy carbon disk coated in PEEK (Cypress Systems EE040), polished between scans using 0.25 μMMetaDi® II diamond polishing paste (Buehler) on a Buehler MicroCloth®. The counter electrode was a 3 mm glassy carbon rod (Alfa-Aesar) and the pseudo reference electrode was a silver wire suspended in a solution of Et4N+BF4- (0.1 M) in acetonitrile and separated from the analyte solution by a Vycor frit (Alfa-Aesar).
Electrocatalysis.
NiII-parent complexes: A stock solution of known concentration was made in acetonitrile for Meth and aliquots of this were further diluted with 0.1 M Et4N+BF4- in acetonitrile to 0.25 mM in a 4 mL glass vial. A small amount of ferrocene was added as an internal standard and the solution was stirred for 5 min. A voltammogram was measured for the analyte solution. The analyte solution was then titrated with 20 μL aliquots of 5.0 M [(DMF)H]+ in acetonitrile using a 50 μL syringe. The working electrode was polished before measuring the cyclic voltammogram after each addition of acid. Acid was added to the analyte solution until no further increase in catalytic current was observed. This was then followed by a titration with water to reach the maximum catalytic activity. Scan rate independence was observed for this complex from 0.05 V/s to 5 V/s (figure S9). The ip was most reversible at 1 Vs−1, therefore, all studies were performed using a scan rate of 1.0 Vs-1. The Prop complex was studied previously and was collected at 200 mV/s [27].
NiII-peptide complexes.
A stock solution of known concentration was made in acetonitrile with 8% water for each of the NiII-peptide complexes and aliquots of this were further diluted to 0.25 mM in a 4 mL glass vial with 0.1 M Et4N+BF4- in acetonitrile. Small aliquots (1 μL) of water were added to obtain a final solvent percentage of 2–3% water. The electrocatalytic evaluation proceeded as for the parent complexes. Prop-R10, Meth-K8 and Meth-K10 were collected at 0.2 Vs−1 and Prop-K8 and Prop-K10 were collected at 0.05 Vs-1. A typical example of the results from the acid addition is shown in figure S10. Scan rate independence was observed from 200 mV/s to 1 V/s (figure S9) with 50 mV/s being scan rate dependent but within the error of our measurements.
NiII-peptide complexes in water.
A stock solution of known concentration was made in water with 5% acetonitrile and aliquots of this were further diluted to 0.25 mM in a 4 mL glass vial with 0.1 M Na2SO4 in water. Small aliquots (1 μL) of acetonitrile were then added to obtain a final solvent percentage of 1–5% acetonitrile. A small amount of (hydroxymethyl)ferrocene was added as an internal standard and the solution was stirred for 5 min. A voltammogram was measured for the analyte solution at 0.05 Vs−1. The analyte solution was then titrated with 5 μL aliquots of 1.0 M H2SO4, 1.0 M HClO4 or 1.0 M HTFSI in water using a 10 μL syringe. The working electrode was polished before measuring the cyclic voltammogram after each addition of acid. Acid was added to the analyte solution until no further increase in catalytic current was observed.
TOF and overpotential calculations.
The turnover frequency or TOF (kobs, s−1) was calculated for Meth using equation 4 from the ratio of the maximum catalytic current (icat), measured at −1.2 V (figure 7) to the current obtained with the catalyst in the absence of acid (ip). The measured ip value was divided by 1.7 to account for the overlapping NiII/I and NiI/0 waves [32].
| (4) |
In equation 4, R is the gas constant, T is the temperature in kelvin, F is the Faraday constant, and ν is the scan rate (Vs−1) [53–55]. To determine icat/ip, the cathodic current (ip) was corrected for dilution. Each measurement was repeated at least three times for statistical purposes. Overpotentials were calculated using Ecat/2 and the method of Evans et al. (equation 5) [56], with E° H+= −0.14V and the pKa of [(DMF)H]+ = 6.1.
| (5) |
5.5. Molecular dynamic simulations
Replica Exchange Molecular Dynamic (REMD) simulations were performed on the [Ni(PPh2NX-peptide2)2]2+ complexes using the software GROMACS in order to explore the conformational space spanned by the molecules. Restraints were imposed to reproduce the experimentally observed NOEs and W1-W6 interactions, resulting in stable β-hairpin structures for the peptides. The acetonitrile solvent environment was modeled using the Generalized Born implicit solvation model. We used force field parameters for [Ni(PPh2NX-peptide2)2]2+ as reported previously [27]. Parameters for the linkers and peptide were taken from the Generalized Amber Force Field (GAFF) [49] andAmber03 Force Field [57], respectively. The RESP charges for the linkers were calculated using the standard Amber procedure [58]. The solvent ligand, acetonitrile, was not included in these calculations though we anticipate that this will not influence the relative results.
The REMD simulations were carried out with 20 replicas, spanning a range of temperatures from 281 K to 931 K. The temperatures were taken from an exponential distribution. Each replica was equilibrated for ~200 ps, after which a 20 ns REMD simulation was started. Exchange between replicas were attempted every 20000 steps resulting in an acceptance ratio of ~40%. The reported analysis is for the trajectory at 300 K. For all simulations a time step for the integration of the equation of motion of 0.5 fs was used, ensuring stability at all temperatures.
The conformational space covered by each complex was analyzed in terms of probability distributions of the projection of the position of the backbone atoms of the peptide onto three orthogonal planes passing through the Ni position. The planes are defined in terms of the following three vectors: (1) NiPX, where PX is the midpoint between the position of two P atoms on a ligand, (2) NPY where PY is the midpoint between the two adjacent P atoms on different ligands, and (3) the vector NiPZ perpendicular to the NiPX and NiPY. The plane defined by NiPX and NiPY is referred to as the XY plane, the plane defined by NiPX and NiPZ is referred to as the XZ plane, and the plane defined by NiPY and NiPZ is referred to as the YZ plane.
5.6. Calculations for pKa
The pKa values were determined using the linear free energy relationships recently proposed for [Ni(PR2NR’2)2]2+ complexes [39]. All calculations for the parent complex were obtained from DFT calculations using an isodesmic reaction scheme. Computational details are provided in reference [39]. The experimental redox potential was used in the linear free energy correlations.
5.7. X-ray crystallography
Single crystals of Meth (C100H98B4Cl4F16N7Ni2O8P8) were grown by slow ether diffusion into a saturated acetonitrile solution of the compound. A suitable crystal was selected and mounted on a nylon loop using Paratone-N oil, and the data collected on a Bruker APEX-II CCD diffractometer. The crystal was kept at 100(2) K during data collection. Using Olex2 [59], the structure was solved with the ShelXS [60] structure solution program using Direct Methods and refined with the ShelXL [60] refinement package using Least Squares minimization.
Supplementary Material
Acknowledgements
This work was funded by the U.S. Department of Energy (US DOE) Basic Energy Sciences (BES), Physical Bioscience program (MLR, SR), the Office of Science Early Career Research Program through the US DOE BES (WJS, AD, GWB, BG), the Center for Molecular Electrocatalysis, an Energy Frontier Research Center funded by the US DOE, Office of Science, Office of BES (MLH) and the National Institutes of Health Grant: GM099889 (BLK). Part of the research was conducted at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. DOE’s Office of Biological and Environmental Research (BER) program located at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the US DOE.
Footnotes
Supporting information
Crystallographic data; HPLC data; NMR data; Molecular Dynamics data; Electrochemistry controls.
References
- [1].Fontecilla-Camps JC, Volbeda A, Cavazza C, Y. Nicolet. Chem. Rev, 107, 4273 (2007). [DOI] [PubMed] [Google Scholar]
- [2].Creighton TE, Proteins: Structure and Molecular Properties, W.H. Freeman and Company, New York: (1996). [Google Scholar]
- [3].Armstrong RN. Chem. Rev, 90, 1309 (1990). [Google Scholar]
- [4].Gloaguen F, Rauchfuss TB. Chem. Soc. Rev, 38, 100 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kohler V, Wilson YM, Lo C, Sardo A, Ward TR. Curr. Opin. Biotechnol, 21, 744 (2010). [DOI] [PubMed] [Google Scholar]
- [6].Lu Y, Yeung N, Sieracki N, Marshall NM. Nature, 460, 855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nanda V, Koder RL. Nat. Chem, 2, 15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koder RL, Dutton PL. Dalton Trans., 3045 (2006). [DOI] [PubMed] [Google Scholar]
- [9].Zaykov AN, Popp BV, Ball ZT. Chem.-Eur. J, 16, 6651 (2010). [DOI] [PubMed] [Google Scholar]
- [10].Kaplan J, DeGrado WF. Proc. Natl. Acad. Sci. U.S.A, 101, 11566 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gilbertson SR, Chen GH, Kao J, Beatty A, Campana CF. J. Org. Chem, 62, 5557 (1997). [Google Scholar]
- [12].Gilbertson SR, Wang XF, Hoge GS, Klug CA, Schaefer J. Organometallics, 15, 4678 (1996). [Google Scholar]
- [13].Maglio O, Nastri F, de Rosales RTM, Faiella M, Pavone V, DeGrado WF, Lombardi A. C.R. Chim, 10, 703 (2007). [Google Scholar]
- [14].Green KN, Hess JL, Thomas CM, Darensbourg MY. Dalton Trans., 4344 (2009). [DOI] [PubMed] [Google Scholar]
- [15].Frederix PWJM, Kania F, Wright JA, Lamprou DA, Ulijn RV, Pickett CJ, Hunt NT. Dalton Trans., 41, 13112 (2012). [DOI] [PubMed] [Google Scholar]
- [16].Pool DH, Stewart MP, O’Hagan M, Shaw WJ, Roberts JAS, Bullock RM, DuBois DL. Proc. Natl. Acad. Sci. U.S.A, 109, 15634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Singleton ML, Beibenspies JH, Darensbourg MY. J. Am. Chem. Soc, 132, 8870 (2010). [DOI] [PubMed] [Google Scholar]
- [18].Ibrahim S, Woi PM, Alias Y, Pickett CJ. Chem. Commun, 46, 8189 (2010). [DOI] [PubMed] [Google Scholar]
- [19].Pullen S, Fei H, Orthaber A, Cohen SM, Ott S. J. Am. Chem. Soc, 135, 16997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shaw WJ, Helm ML, DuBois DL. Biochim. Biophys. Acta, 1827, 1123 (2013). [DOI] [PubMed] [Google Scholar]
- [21].Jones AK, Lichtenstein BR, Dutta A, Gordon G, Dutton PL. J. Am. Chem. Soc, 129, 14844 (2007). [DOI] [PubMed] [Google Scholar]
- [22].Jain A, Lense S, Linehan JC, Raugei S, Cho H, DuBois DL, Shaw WJ. Inorg. Chem, 50, 4073 (2011). [DOI] [PubMed] [Google Scholar]
- [23].Jain A, Reback ML, Lindstrom ML, Thogerson CE, Helm ML, Appel AM, Shaw WJ. Inorg. Chem, 51, 6592 (2012). [DOI] [PubMed] [Google Scholar]
- [24].Reback ML, Ginovska-Pangovska B, Ho M-H, Jain A, Squier TC, Raugie S, Roberst JAS, Shaw WJ. Chem.-Eur. J, 19, 1928 (2013). [DOI] [PubMed] [Google Scholar]
- [25].Dutta A, Lense S, Hou J, Engelhard MH, Roberts JAS, Shaw WJ. J. Am. Chem. Soc, 135, 18490 (2013). [DOI] [PubMed] [Google Scholar]
- [26].Lense S, Ho MH, Chen ST, Jain A, Raugei S, Linehan JC, Roberts JAS, Appel AM, Shaw W. Organometallics, 31, 6719 (2012). [Google Scholar]
- [27].Reback ML, Buchko GW, Kier BL, Ginovska-Pangovska B, Xiong Y, Lense S, Hou J, Roberts JAS, Sorensen CM, Raugei S, Squier TC, Shaw WJ. Chem.-Eur. J, 20, 1510 (2014). [DOI] [PubMed] [Google Scholar]
- [28].Roy S, Shinde S, Hamilton GA, Hartnett HE, Jones AK. Eur. J. Inorg. Chem, 1050 (2011). [Google Scholar]
- [29].Kier BL, Andersen NH. J. Am. Chem. Soc, 130, 14675 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kier BL, Shu I, Eidenschink LA, Andersen NH. Proc. Natl. Acad. Sci. U.S.A, 107, 10466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Helm ML, Stewart MP, Bullock RM, DuBois MR, DuBois DL. Science, 333, 863 (2011). [DOI] [PubMed] [Google Scholar]
- [32].Stewart MP, Ho MH, Wiese S, Lindstrom ML, Thogerson CE, Raugei S, Bullock RM, Helm ML. J. Am. Chem. Soc, 135, 6033 (2013). [DOI] [PubMed] [Google Scholar]
- [33].Stewart MP, Ho M-H, Wiese S, Lindstrom ML, thogerson CE, Raugei S, Bullock RM, Helm ML. J. Am. Chem. Soc, 135, 6033 (2013). [DOI] [PubMed] [Google Scholar]
- [34].Kier BL, Andersen NH. J. Peptide Sci, 20, 704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Seu CS, Appel AM, Doud MD, DuBois DL, Kubiak CP. Energy Environ. Sci, 5, 6480 (2012). [Google Scholar]
- [36].Kilgore UJ, Roberts JAS, Pool DH, Appel AM, Stewart MP, Rakowski DuBois M, Dougherty WG, Kassel WS, Bullock RM, DuBois DL. J. Am. Chem. Soc, 133, 5861 (2011). [DOI] [PubMed] [Google Scholar]
- [37].When originally reported, the contribution of two electrons was not taken into account. The data was reevaluated here and the ip divided by 1.7, as reported in the Methods section.
- [38].Silver SC, Niklas J, Du P, Poluektov OG, Tiede DM, Utschig LM. J. Am. Chem. Soc, 135, 13246 (2013). [DOI] [PubMed] [Google Scholar]
- [39].Chen S, Ho M-H, Bullock RM, DuBois DL, Dupuis M, Rousseau R, Raugei S. ACS Catal., 4, 229 (2014). [Google Scholar]
- [40].Shaw WJ. Catal. Rev. Sci. Eng, 54, 489 (2012). [Google Scholar]
- [41].In the original publication, we reported TOFs for the Prop-R10 complex. At that time we were unaware of the second irreversible wave. With our new information, the previous rates may not be accurate, but the reported icat values are accurate and are consistent with increased catalytic activity for the peptide-based catalyst.
- [42].O’Hagan M, Ho MH, Yang JY, Appel AM, Rawkowski DuBois M, Raugei S, Shaw WJ, DuBois DL, Bullock RM. J. Am. Chem. Soc, 134, 19409 (2012). [DOI] [PubMed] [Google Scholar]
- [43].O’Hagan M, Shaw WJ, Raugei S, Chen S, Yang JY, Kilgore UJ, DuBois DL, Bullock RM. J. Am. Chem. Soc, 133, 14304 (2011). [DOI] [PubMed] [Google Scholar]
- [44].Franz JA, O’Hagan M, Ho M-H, Liu T, Helm ML, Lense S, DuBois DL, Shaw WJ, Appel AM, Raugei S, Bullock RM. Organometallics, 32, 7034 (2013). [Google Scholar]
- [45].Kleingardner JG, Kandemir B, Bren KL. J. Am. Chem. Soc, 136, 4 (2014). [DOI] [PubMed] [Google Scholar]
- [46].Sano Y, Onoda A, T. Hayashi. Chem. Commun, 47, 8229 (2011). [DOI] [PubMed] [Google Scholar]
- [47].Slater JW, Shafaat HS. J. Phys. Chem. Lett, 6, 3731 (2015). [DOI] [PubMed] [Google Scholar]
- [48].Berggren G, Adamska A, Lambertz C, Simmons TR, Esselborn J, Atta M, Bambarelli S, Mouesca J-M, Reijerse E, Lubitz W, Happe T, Artero V, Fontecave M. Nature, 499, 66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Favier I, Duñach E. Tetrahedron Lett., 45, 3393 (2004). [Google Scholar]
- [50].Le Goff A, Artero V, Jousselme B, Tran PD, Guillet N, Metaye R, Fihri A, Palacin S, Fontecave M. Science, 326, 1384 (2009). [DOI] [PubMed] [Google Scholar]
- [51].Woody RW. Methods Enzymol., 246, 34 (1995). [DOI] [PubMed] [Google Scholar]
- [52].Greenfield NJ. Nat. Protoc, 1, 2876 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Savéant JM, E. Vianello. Electrochim. Acta, 10, 905 (1965). [Google Scholar]
- [54].Savéant JM, E. Vianello. Electrochim. Acta, 12, 629 (1967). [Google Scholar]
- [55].Pool DH, DuBois DL. J. Organomet. Chem, 694, 2858 (2009). [Google Scholar]
- [56].Felton GAN, Glass RS, Lichtenberger DL, Evans DH. Inorg. Chem, 45, 9181 (2006). [DOI] [PubMed] [Google Scholar]
- [57].Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. J. Comput. Chem, 24, 1999 (2003). [DOI] [PubMed] [Google Scholar]
- [58].Bayly CI, Cieplak P, Cornell W, Kollman PA. J. Phys. Chem, 97, 10269 (1993). [Google Scholar]
- [59].Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H. J. Appl. Cryst, 42, 339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sheldrick GM. Acta Cryst., A64, 112 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.