Abstract
Guide RNA design for CRISPR genome editing of gene families is a challenging task as usually good candidate sgRNAs are tagged with low scores precisely because they match several locations in the genome, thus time-consuming manual evaluation of targets is required. To address this issues, I have developed ARES-GT, a Python local command line tool compatible with any operative system. ARES-GT allows the selection of candidate sgRNAs that match multiple input query sequences, in addition of candidate sgRNAs that specifically match each query sequence. It also contemplates the use of unmapped contigs apart from complete genomes thus allowing the use of any genome provided by user and being able to handle intraspecies allelic variability and individual polymorphisms. ARES-GT is available at GitHub (https://github.com/eugomin/ARES-GT.git).
Introduction
The design of optimal single guide RNAs (sgRNAs) is a critical step in CRISPR/Cas genome editing, and it must ensure specificity and minimize the possibility of offtarget mutations. Although good online tools are available for identification of CRISPR DNA targets, which have popularized genome editing, their use is limited to a restricted list of genomes [1–6], sometimes corresponding to less than ten species [7, 8]. Even Breaking-Cas [9], a free online tool which currently offers more than 1600 genomes, lacks the flexibility to easily incorporate unpublished genomes or contemplate genomes of populations with allelic variants -an issue partially addressed by AlleleAnalyzer for the human genome [10]. Several command-line tools present more flexibility incorporating any genome provided by users, like sgRNA-cas9 [11] or CRISPRseek [12]. However, an additional problem posed by the design of sgRNAs targeting gene families is that good candidate sgRNAs can be tagged with low scores precisely because they match several locations in the genome, thus time-consuming manual evaluation of targets is required. To address this issue, I have developed ARES-GT, a local command line tool in Python programming language.
Methods
ARES-GT
ARES-GT is written in python programming language (https://www.python.org/) so it can be runned in any operative system. The software is available at GitHub (https://github.com/eugomin/ARES-GT.git): version 2.0 is a python2.7 version while version 2.0.1 is updated to python3.8. In addition of sys and re modules, ARES-GT also requires the third-party regex module (https://pypi.org/project/regex/).
Complete analysis presented in this work were performed in minimum 3 hours and maximum 12 hours, depending of the analysis, in a Linux server running Ubuntu 18.04 LTS with Intel Xeon 2.0 GHz processor and 32 GB RAM. When option “OR” is selected (so only analysis of candidates matching several query sequences), the same analysis were performed in 15 min or less. Running time directly depends on the number of query sequences, genome size and selected parameters.
Genome sequences
Arabidopsis reference genome (Col-0) were obtained from TAIR (www.arabidopsis.org). Good quality genome assemblies of seven A. thaliana accessions (An-1, C24, Cvi, Eri, Kyo, Ler and Sha) [13] were downloaded from Arabidopsis 1001 genomes project (https://1001genomes.org/), and Cardamine hirsuta genome from its genetic and genomic resource (http://chi.mpipz.mpg.de/index.html). All sequences of CBF genes are available in S1 File.
CBF genes
Genomic sequences of Arabidopsis thaliana CBF genes (AtCBFs) were obtained from TAIR (https://www.arabidopsis.org/), corresponding to Col-0 TAIR v10. Genomic sequences of AtCBFs homologs in C. hirsuta were identified by BLAST in the C. hirsuta genetic and genomic resource (http://chi.mpipz.mpg.de/index.html) using the AtCBFs protein sequences and supported by alignment with ClustalX2 [14]. Ecotype specific genomic sequence of each CBF gene were retrived using the genomic coordinates from ARES-GT results using AtCBFs (Col-0).
Results
Identification of CRISPR targets candidates
The high sequence similarity shared in gene families increase the possibility of also sharing CRISPR targets, both with perfect match or with few mismatches. While this is especially interesting for targeting multiple members of the same family, they are usually discarded or evaluated with low scores. Similarly to other available software, ARES-GT starts with the identification of all candidate guide RNAs in query sequences and then the reference genome is used to find possible offtargets, but an additional step is added to evaluate which guide sequences match several query sequences.
Offtargets evaluation is based in a mismatch criteria. It has been reported that the specificity of both Cas9 and Cas12a is particularly sensitive to mismatches in the PAM proximal sequence (on an 11- and 8-nucleotide stretch for Cas9 and Cas12a, respectively), named “seed” [15–18]. Mismatches in the seed sequence has a critical impact into cleavage efficiency on DNA target, and it is unlikely that seed sequences with 2 or more mismatches cause real offtargets in vivo. Sequence composition and the number and distribution of mismatches also affects cleavage efficiency [15]. Therefore the ARES-GT algorithm discards possible offtargets using as criterium the presence of 2 or more mismatches in the seed sequence, while the user defines the threshold criterium out of seed sequence. In addition, the user must also indicate whether a “NAG” PAM, which Cas9 can recognise though with lower efficiency [15], must be taken into account when evaluating possible Cas9 offtargets.
ARES-GT can identify targets of the two most widely used CRISPR enzymes (Cas9 and Cas12a/Cpf1) and evaluates possible offtargets in a user-provided reference genome, including non assembled contigs and unpublished genomes from any species. A list is generated with the best candidates (those with no offtargets based on parameters selected by user) and, if multiple query genes from the same family are targeted, the list includes sgRNAs that match more than one of them. Detailed information for each possible target is also provided, including an alignment with the possible offtargets. ARES-GT have been already used successfully in Arabidopsis, tomato and rice while under development [19, 20].
Design of guide RNA matching multiple CBF genes
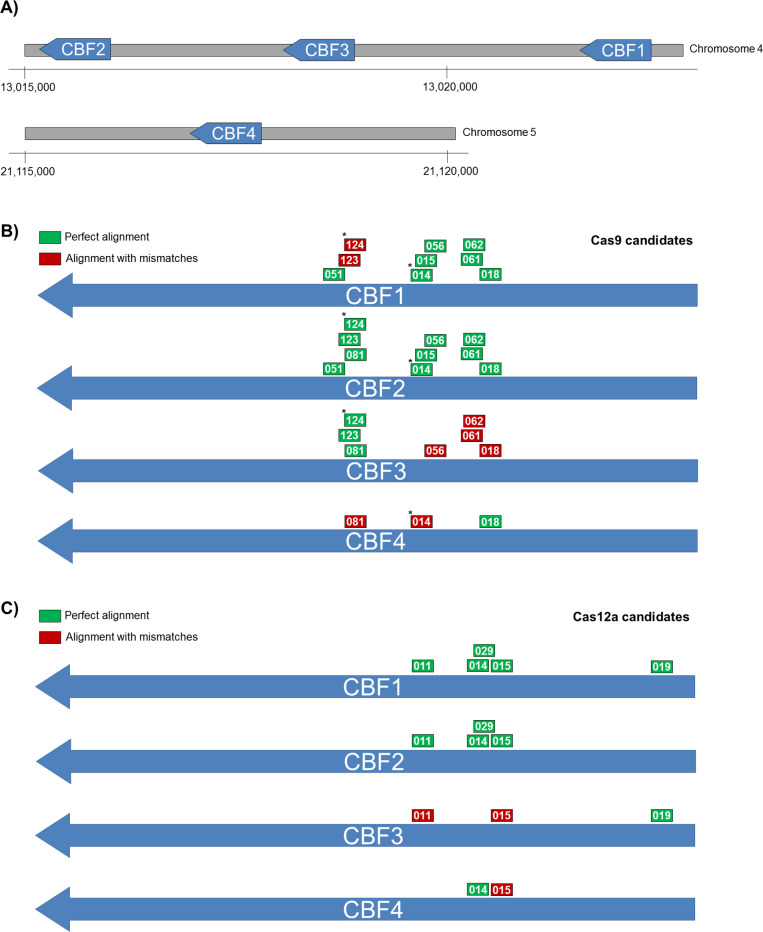
As a proof of concept, I have choosen the C-repeat/DRE-Binding Factor (CBF) gene family of plant transcription factors to test the various novelties implemented in ARES-GT. Among the four members identified in Arabidopsis thaliana, three of them–AtCBF1, AtCBF2 and AtCBF3–, have been implicated in the response to cold temperatures, while AtCBF4 has been implicated in the response to drought [21, 22]. The first three members of this family are closely located in less than 8 Kb in chromosome 4 (Fig 1A), making extremely difficult to obtain a triple mutant by classical crossing strategy. This has been recently achieved by CRISPR/Cas9-induced mutagenesis [23] using two sgRNAs that the authors selected by manual evaluation of sequence alignments, manual selection of candidates, and specificity verification with CRISPR-P [1]. I used the A. thaliana genomic coding sequences (TAIR v10) of the four CBF genes as a multiple query in ARES-GT, to search for candidate sgRNAs using both Cas9 and Cas12a. A total of 96 and 34 unique specific targets matching only one location in the genome and with no predicted offtargets were found for each the four genes, using Cas9 and Cas12a, respectively. More interestingly, the program also listed 13 candidates for Cas9 and 10 candidates for Cas12a that match multiple CBF genes (Tables 1 and 2). In total, 10 Cas9 and 5 Cas12a candidates were identified that match more than one CBF gene and did not present any offtarget outside CBF genes (Fig 1B and 1C). Among them were included the two sequences previously reported [23], corresponding to Cas9CBF1_015 and Cas9CBF2_124 in this work.
Fig 1. sgRNS targets in CBF genes.
A) Genomic distribution of CBF genes in Arabiopsis thaliana chromosomes 4 and 5. Location of Cas9 (B) and Cas12a (C) candidates with multiple CBF gene targets. (*) Asterisk marks candidates corresponding with previously reported sgRNAs (Cho et al., 2017).
Table 1. Multiple targets Cas9 candidates for AtCBF genes.
All possible genome targets and offtargets (with ARES-GT thresholds: L0 = 4 and L1 = 3) of each candidate are listed with indication of genome coordinates (TAIR v10) and whether it corresponds to a CBF gene. In alignments, black boxes mark mismatches and a space separates PAM (NGG or NAG) from sequence. Differences in the “N” position in the PAM are not marked.
| Candidate ID | Targets + Offtargets (L0 = 4, L1 = 3) | |||||
|---|---|---|---|---|---|---|
| A. thaliana | Gene | chrom | start | end | sense | sequence |
| Cas9AtCBF1_014 | AtCBF2 | 4 | 13015820 | 13015842 | + | AGCACGAGCTGCCATCTCAG CGG |
| AtCBF1 | 4 | 13022305 | 13022327 | + | AGCACGAGCTGCCATCTCAG CGG | |
| AtCBF3 | 4 | 13018737 | 13018759 | + | AGCTCGAGCTGCCATCTCAG CGG | |
| Cas9AtCBF1_015 | AtCBF2 | 4 | 13015825 | 13015847 | + | GAGCTGCCATCTCAGCGGTT TGG |
| AtCBF1 | 4 | 13022310 | 13022332 | + | GAGCTGCCATCTCAGCGGTT TGG | |
| Cas9AtCBF1_018 | AtCBF2 | 4 | 13015920 | 13015942 | + | TGACGAACTCCTCTGTAAAT TGG |
| AtCBF1 | 4 | 13022405 | 13022427 | + | TGACGAACTCCTCTGTAAAT TGG | |
| AtCBF4 | 5 | 21117612 | 21117634 | + | TGACGAACTCCTCTGTAAAT CGG | |
| AtCBF3 | 4 | 13018837 | 13018859 | + | CGACGAACTCCTCTGTATAT TGG | |
| Cas9AtCBF1_019 | AtCBF2 | 4 | 13015921 | 13015943 | + | GACGAACTCCTCTGTAAATT GGG |
| AtCBF1 | 4 | 13022406 | 13022428 | + | GACGAACTCCTCTGTAAATT GGG | |
| ---- | 1 | 1597274 | 1597296 | + | CACAATCTCCTCTGTAAATT CAG | |
| AtCBF3 | 4 | 13018838 | 13018860 | + | GACGAACTCCTCTGTATATT GGG | |
| Cas9AtCBF1_051 | AtCBF2 | 4 | 13015738 | 13015760 | - | CCG GGATTCGTAGCCGCCAAGCC |
| AtCBF1 | 4 | 13022223 | 13022245 | - | CCG GGATTCGTAGCCGCCAAGCC | |
| Cas9AtCBF1_056 | AtCBF2 | 4 | 13015831 | 13015853 | - | CCA TCTCAGCGGTTTGGAAAGTC |
| AtCBF1 | 4 | 13022316 | 13022338 | - | CCA TCTCAGCGGTTTGGAAAGTC | |
| AtCBF3 | 4 | 13018748 | 13018770 | - | CCA TCTCAGCGGTTTGAAATGTT | |
| Cas9AtCBF1_061 | AtCBF2 | 4 | 13015900 | 13015922 | - | CCC ACTTACCGGAGTTTCTTTGA |
| AtCBF1 | 4 | 13022385 | 13022407 | - | CCC ACTTACCGGAGTTTCTTTGA | |
| AtCBF3 | 4 | 13018817 | 13018839 | - | CCC ACTTACCGGAGTTTCTCCGA | |
| Cas9AtCBF1_062 | AtCBF2 | 4 | 13015901 | 13015923 | - | CCA CTTACCGGAGTTTCTTTGAC |
| AtCBF1 | 4 | 13022386 | 13022408 | - | CCA CTTACCGGAGTTTCTTTGAC | |
| AtCBF3 | 4 | 13018818 | 13018840 | - | CCA CTTACCGGAGTTTCTCCGAC | |
| Cas9AtCBF1_063 | AtCBF2 | 4 | 13015908 | 13015930 | - | CCG GAGTTTCTTTGACGAACTCC |
| AtCBF1 | 4 | 13022393 | 13022415 | - | CCG GAGTTTCTTTGACGAACTCC | |
| ---- | 2 | 6123419 | 6123441 | - | CCC GACTTTCTTTGAAGAACTCC | |
| Cas9AtCBF1_064 | AtCBF2 | 4 | 13015929 | 13015951 | - | CCT CTGTAAATTGGGTGACGAGT |
| AtCBF1 | 4 | 13022414 | 13022436 | - | CCT CTGTAAATTGGGTGACGAGT | |
| AtCBF3 | 4 | 13018846 | 13018868 | - | CCT CTGTATATTGGGTGACGAGT | |
| ---- | 1 | 4290740 | 4290762 | - | CCT CTGTAAACTGGGTGACGTGT | |
| ---- | 1 | 23368054 | 23368076 | - | CCT CTGTAGATTGGGTGACGTGT | |
| AtCBF4 | 5 | 21117621 | 21117643 | - | CCT CTGTAAATCGGATGACGTGT | |
| Cas9AtCBF2_081 | AtCBF2 | 4 | 13015760 | 13015782 | + | CGAGTCAGCGAAATTGAGAC AGG |
| AtCBF3 | 4 | 13018677 | 13018699 | + | CGAGTCAGCGAAATTGAGAC AGG | |
| AtCBF4 | 5 | 21117452 | 21117474 | + | AGAATCAGCGAAATTGAGAC AAG | |
| Cas9AtCBF2_123 | AtCBF2 | 4 | 13015754 | 13015776 | - | CCA AGCCGAGTCAGCGAAATTGA |
| AtCBF3 | 4 | 13018671 | 13018693 | - | CCA AGCCGAGTCAGCGAAATTGA | |
| AtCBF1 | 4 | 13022239 | 13022261 | - | CCA AGCCGAGTCAGCGAAGTTGA | |
| Cas9AtCBF2_124 | AtCBF2 | 4 | 13015759 | 13015781 | - | CCG AGTCAGCGAAATTGAGACAG |
| AtCBF3 | 4 | 13018676 | 13018698 | - | CCG AGTCAGCGAAATTGAGACAG | |
| AtCBF1 | 4 | 13022244 | 13022266 | - | CCG AGTCAGCGAAGTTGAGACAT | |
Table 2. Multiple targets Cas12a candidates for AtCBF genes.
All possible genome targets and offtargets (with ARES-GT thresholds: L0 = 4 and L1 = 3) of each candidate are listed with indication of genome coordinates (TAIR v10) and whether it corresponds to a CBF gene. In alignments, black boxes mark mismatches and a space separates PAM (TTTN) from sequence. Differences in the “N” position in the PAM are not marked.
| Candidate ID | Targets + Offtargets (L0 = 4, L1 = 3) | |||||
|---|---|---|---|---|---|---|
| A. thaliana | Gene | chrom | start | end | sense | sequence |
| Cas12aAtCBF1_011 | AtCBF2 | 4 | 13015814 | 13015837 | - | GCTGCCATCTCAGCGGTTTG GAAA |
| AtCBF1 | 4 | 13022299 | 13022322 | - | GCTGCCATCTCAGCGGTTTG GAAA | |
| Cas12aAtCBF1_012 | AtCBF2 | 4 | 13015827 | 13015850 | - | CGGTTTGGAAAGTCCCGAGC CAAA |
| AtCBF1 | 4 | 13022312 | 13022335 | - | CGGTTTGGAAAGTCCCGAGC CAAA | |
| ---- | 1 | 27242286 | 27242310 | + | TTTG GCTCGGGACTTTCAACACAG | |
| ---- | 3 | 8296023 | 8296047 | + | TTTG GCTCGGGACGTTCGAAAGCG | |
| ---- | 5 | 17806910 | 17806934 | + | TTTG GCTCGGGACATTCGACACGG | |
| ---- | 5 | 21618544 | 21618567 | - | CCGTCTCAAAAGTCCCGAGC CAAA | |
| ---- | 4 | 7932903 | 7932927 | + | TTTG GCTCGGCACTTTTGAAACCG | |
| ---- | 4 | 10190722 | 10190745 | - | CAGTTTGGAACGTTCCGAGC CAAA | |
| AtCBF3 | 4 | 13018744 | 13018767 | - | CGGTTTGAAATGTTCCGAGC CAAA | |
| Cas12aAtCBF1_014 | AtCBF2 | 4 | 13015902 | 13015925 | - | TTCTTTGACGAACTCCTCTG TAAA |
| AtCBF1 | 4 | 13022387 | 13022410 | - | TTCTTTGACGAACTCCTCTG TAAA | |
| AtCBF4 | 5 | 21117594 | 21117617 | - | TCCTCTGACGAACTCCTCTG TAAA | |
| Cas12aAtCBF1_015 | AtCBF2 | 4 | 13015924 | 13015947 | - | AATTGGGTGACGAGTCTCAC GAAA |
| AtCBF1 | 4 | 13022409 | 13022432 | - | AATTGGGTGACGAGTCTCAC GAAA | |
| AtCBF3 | 4 | 13018841 | 13018864 | - | TATTGGGTGACGAGTCTCAC GAAA | |
| AtCBF4 | 5 | 21117616 | 21117639 | - | AATCGGATGACGTGTCTCAC GAAA | |
| Cas12aAtCBF1_017 | AtCBF2 | 4 | 13016031 | 13016054 | - | AATCGGAGCCAAACATTTCA GAAA |
| AtCBF3 | 4 | 13018948 | 13018971 | - | AATCGGAGCCAAACATTTCA GAAA | |
| AtCBF1 | 4 | 13022507 | 13022530 | - | AATCGGAGCCAAACATTTCA GAAA | |
| ---- | 1 | 8279033 | 8279056 | - | AATCAGAGCCTAACACTTCA AAAA | |
| ---- | 3 | 9399469 | 9399493 | + | TTTA TGAAGTGTTTGGTTCCTATT | |
| Cas12aAtCBF1_018 | AtCBF2 | 4 | 13016032 | 13016055 | - | ATCGGAGCCAAACATTTCAG AAAA |
| AtCBF3 | 4 | 13018949 | 13018972 | - | ATCGGAGCCAAACATTTCAG AAAA | |
| AtCBF1 | 4 | 13022508 | 13022531 | - | ATCGGAGCCAAACATTTCAG AAAA | |
| ---- | 1 | 9505057 | 9505081 | + | TTTG CTGAAATGGTTGCCTCTAAT | |
| Cas12aAtCBF1_019 | AtCBF3 | 4 | 13018950 | 13018973 | - | TCGGAGCCAAACATTTCAGA AAAA |
| AtCBF1 | 4 | 13022509 | 13022532 | - | TCGGAGCCAAACATTTCAGA AAAA | |
| Cas12aAtCBF1_024 | AtCBF2 | 4 | 13015842 | 13015865 | + | TTTG GAAAGTCCCGAGCCAAATCC |
| AtCBF1 | 4 | 13022327 | 13022350 | + | TTTG GAAAGTCCCGAGCCAAATCC | |
| ---- | 3 | 8296020 | 8296043 | - | GGGTTTGGCTCGGGACGTTC GAAA | |
| Cas12aAtCBF1_028 | AtCBF2 | 4 | 13015913 | 13015936 | + | TTTC TTTGACGAACTCCTCTGTAA |
| AtCBF1 | 4 | 13022398 | 13022421 | + | TTTC TTTGACGAACTCCTCTGTAA | |
| ---- | 5 | 16311156 | 16311179 | + | TTTT TTTGACGAATTTCTCTGTGG | |
| Cas12aAtCBF1_029 | AtCBF2 | 4 | 13015917 | 13015940 | + | TTTG ACGAACTCCTCTGTAAATTG |
| AtCBF1 | 4 | 13022402 | 13022425 | + | TTTG ACGAACTCCTCTGTAAATTG | |
To test that AREST-GT can work with any user-provided genome, including unmapped contigs, I selected the first version of the genome of Cardamine hirsuta [24]. The available genome sequence spans over its 8 chromosomes, but also contains 622 unmapped contigs in addition to chloroplast and mithocondria genomes. The sequence information was downloaded and used locally with ARES-GT for searching CRISPR targets in the four C. hirsuta CBF homologous genes. In addition to unique specific targets (86 for Cas9 and 28 for Cas12a), 10 candidate sgRNAs for Cas9 and 3 for Cas12a were identified that perfectly match ChCBF1 and ChCBF2 (Table 3). Taking into account possible offtargets, only 5 and 3 sequences for Cas9 and Cas12a, respectively, are relyable candidate sgRNAs targeting only ChCBF family genes. For instance, Cas9ChCBF1_044 perfectly matches ChCBF1 and ChCBF2, and it also matches ChCBF3 with one mismatch.
Table 3. Multiple targets Cas9 and Cas12a candidates for ChCBF genes.
All possible genome targets and offtargets (with ARES-GT thresholds: L0 = 4 and L1 = 3) of each candidate are listed with indication of genome coordinates (Cardamine hirsuta v1.0) and whether it corresponds to a CBF gene. In alignments, black boxes mark mismatches and a space separates PAM (NGG/NAG or TTTN) from sequence. Differences in the “N” position in the PAM are not marked.
| Candidate ID | Targets + Offtargets (L0 = 4, L1 =3) | |||||
|---|---|---|---|---|---|---|
| C. hirsuta | Gene | chrom | start | end | sense | sequence |
| Cas9ChCBF1_004 | ChCBF2 | 4 | 6514798 | 6514820 | + | AGCTGTCCCAAGAAACCAGC TGG |
| ChCBF1 | 7 | 17908883 | 17908905 | - | CCG GCTGGTTTCTTGGGACAGCT | |
| Cas9ChCBF1_010 | ChCBF2 | 4 | 6514878 | 6514900 | + | CTCCGGTAAGTGGGTGTGTG AGG |
| ChCBF1 | 7 | 17908803 | 17908825 | - | CCT CACACACCCACTTACCGGAGE | |
| Cas9ChCBF1_018 | ChCBF2 | 4 | 6514910 | 6514932 | + | CAAACAAGAAATCTAGGATT TGG |
| ChCBF1 | 7 | 17908771 | 17908793 | - | CCA AATCCTAGATTTCTTGTTTG | |
| ChCBF3 | 8 | 13812274 | 13812296 | - | CCA AATCCTCGATTTCTTGTTAG | |
| ---- | 5 | 18638271 | 18638293 | - | CTT AATCCTACATTTGTAGTTTG | |
| ---- | 5 | 21152837 | 21152859 | - | CTT AATCCTACATTTCTGGTTTT | |
| Cas9ChCBF1_013 | ChCBF2 | 4 | 6514915 | 6514937 | + | AAGAAATCTAGGATTTGGCT TGG |
| ChCBF1 | 7 | 17908766 | 17908788 | - | CCG AGCCAAATCCTAGATTTCTT | |
| ---- | 8 | 18333140 | 18333162 | - | CCA AGCCAAATCCTAGAACCCTT | |
| ---- | 1 | 5556241 | 5556263 | + | AGGAAACGGAGGATTTGGCT TGG | |
| ---- | 1 | 370416 | 370438 | + | AAAAAATCTCGGATTTGGCT CGG | |
| ChCBF3 | 8 | 13812269 | 13812291 | - | CCT AACCAAATCCTCGATTTCTT | |
| Cas9ChCBF1_033 | ChCBF2 | 4 | 6515264 | 6515286 | + | TGCCGCCTCCGTCCGTACAA TGG |
| ChCBF1 | 7 | 17908390 | 17908412 | - | CCA TTGTACGGACGGAGGCGGCA | |
| NSCAFA. | 444 | 2316 | 2338 | + | CGCCGCCACCGTCCGTACAC CGG | |
| Cas9ChCBF1_036 | ChCBF2 | 4 | 6514793 | 6514815 | - | CCG TGAGCTGTCCCAAGAAACCA |
| ChCBF1 | 7 | 17908888 | 17908910 | + | TGGTTTCTTGGGACAGCTCA CGG | |
| Cas9ChCBF1_043 | ChCBF2 | 4 | 6514880 | 6514902 | - | CCG GTAAGTGGGTGTGTGAGGTA |
| ChCBF1 | 7 | 17908801 | 17908823 | + | TACCTCACACACCCACTTAC CGG | |
| Cas9ChCBF1_044 | ChCBF2 | 4 | 6514909 | 6514931 | - | CCA AACAAGAAATCTAGGATTTG |
| ChCBF1 | 7 | 17908772 | 17908794 | + | CAAATCCTAGATTTCTTGTT TGG | |
| ChCBF3 | 8 | 13812275 | 13812297 | + | CAAATCCTCGATTTCTTGTT AGG | |
| Cas9ChCBF1_056 | ChCBF2 | 4 | 6515266 | 6515288 | - | CCG CCTCCGTCCGTACAATGGAA |
| ChCBF1 | 7 | 17908388 | 17908410 | + | TTCCATTGTACGGACGGAGG CGG | |
| ---- | 2 | 8347578 | 8347600 | + | GGCCAGAGTACGGACGGAGG AGG | |
| Cas9ChCBF1_057 | ChCBF2 | 4 | 6515269 | 6515291 | - | CCT CCGTCCGTACAATGGAATCA |
| ChCBF1 | 7 | 17908385 | 17908407 | + | TGATTCCATTGTACGGACGG AGG | |
| ---- | 1 | 17089187 | 17089209 | + | TGGTCCGGTTGTACGGACGG CGG | |
| ---- | 5 | 5225681 | 5225703 | - | CCA CCGTCCGTACACTGGATTAT | |
| Cas21aChCBF1_018 | ChCBF2 | 4 | 6514830 | 6514853 | + | TTTC GTGAGACTCGTCACCCAATT |
| ChCBF1 | 7 | 17908848 | 17908871 | - | AATTGGGTGACGAGTCTCAC GAAA | |
| ChCBF3 | 8 | 13812351 | 13812374 | - | AATCGGATGACGTGTCTCAC GAAA | |
| Cas21aChCBF1_029 | ChCBF2 | 4 | 6515260 | 6515283 | + | TTTT GCCGCCTCCGTCCGTACAAT |
| ChCBF1 | 7 | 17908391 | 17908414 | - | ATTGTACGGACGGAGGCGGC AAAA | |
| Cas21aChCBF1_030 | ChCBF2 | 4 | 6515261 | 6515284 | + | TTTG CCGCCTCCGTCCGTACAATG |
| ChCBF1 | 7 | 17908390 | 17908413 | - | CATTGTACGGACGGAGGCGG CAAA | |
Finally, to contemplate intraspecific allelic variability in the design of sgRNAs for genome editing, I used ARES-GT in combination with the genome sequences available through the Arabidopsis 1001 genomes project (https://1001genomes.org/). ARES-GT can be used to design ecotype-specific targets taking advantage of polymorphic sequences in the different accessions. Good quality genome assemblies of seven A. thaliana accessions (An-1, C24, Cvi, Eri, Kyo, Ler and Sha) [13] were downloaded, and ARES-GT was used to design sgRNAs targeting CBF genes in each accession. As reflected in Table 4, the SNPs in CBF genes between the different accessions are responsible of the identification of different number of candidate sgRNAs that match several genes of the family, from 18 Cas9 candidates with CBF genes from Kyo genome to 11 Cas9 candidates with CBF genes from Cvi genome. The selection of CRISPR candidates with specific unique target (without offtargets) also varied between accessions (Table 4). I used each accession CBF genes as query for ARES-GT but using either the standar Col-0 reference or the corresponding accession genome. Candidates only listed when Col-0 is used as reference (Col-0 exclusive) are false positives, as they have offtargets in the corresponding accession genome. The accession`s exclusive candidates would be false negatives, as they are discarded if Col-0 is used but do not have offtargets in the corresponding accession genome (Table 4). Differences in the identification of offtargets also affects the selection of efficient candidates matching several CBF genes. For instance, candidate C24_CBF1_019 perfectly match C24_CBF1, C24_CBF2 and C24_CBF3 but has a possible offtarget (4 mismatches in distal sequence) in the chromosome 3 of C24 genome, which is above offtarget thresholds in Col-0 genome because of an extra mismatch in the proximal sequence (Table 5). In the other sense, Eri_Cas12aCBF1_017 is a candidate that perfectly match Eri_CBF1, Eri_CBF2 and Eri_CBF3 without offtargets in Eri genome, however it would be discarded because two offtargets are detected if Col-0 genome is used (Table 5).
Table 4. Intraspecies variability effect in the number of Cas9 and Cas12a candidates targeting multiple or unique AtCBF genes.
Sequence variability in the CBF genes from different Arabidopsis thaliana accessions change the number of candidates that can match multiple targets due to SNPs in the 20 nucleotides of the guide but also SNPs affecting PAM sequence. The use of the standard Col-0 genome reference (TAIR v10) or the corresponding accession genome affects the identification of offtargets thus the correct identification of specific (unique) candidates matching only one CBF gene. The column “exclusive” indicates the number of specific candidates that are only listed when the corresponding reference genome is used.
|
CBF genes accession |
Multiple Targets Candidates | Reference Genome |
Unique Cas9 Candidates | Unique Cas12a Candidates | |||
|---|---|---|---|---|---|---|---|
| Cas9 | Cas12a | Total | Exclusive | Total | Exclusive | ||
| Col | 13 | 10 | Col | 96 | - | 34 | - |
| An-1 | 13 | 9 | Col | 100 | 3 | 37 | 2 |
| An-1 | 105 | 8 | 41 | 6 | |||
| C24 | 13 | 10 | Col | 100 | 4 | 33 | 2 |
| C24 | 101 | 5 | 31 | 0 | |||
| Cvi | 11 | 9 | Col | 102 | 6 | 34 | 3 |
| Cvi | 107 | 11 | 37 | 6 | |||
| Eri | 13 | 10 | Col | 101 | 2 | 32 | 1 |
| Eri | 101 | 2 | 31 | 0 | |||
| Kyo | 18 | 6 | Col | 99 | 8 | 32 | 2 |
| Kyo | 103 | 12 | 33 | 3 | |||
| Ler | 13 | 10 | Col | 102 | 3 | 32 | 0 |
| Ler | 105 | 6 | 34 | 2 | |||
| Sha | 13 | 10 | Col | 101 | 6 | 31 | 2 |
| Sha | 102 | 7 | 31 | 2 | |||
Table 5. Intraspecies variability effect in the identification of targets and possible offtargets.
For each example, upper file shows the targets and offtargets listed by ARES-GT (with thresholds L0 = 4 and L1 = 3) for each reference genome. SNPs differences between genomes that explain why some targets or offtargets are not detected are shown in lower file (separated by discontinuous line) as red boxes. Black boxes mark mismatches with candidates sequence.
| Candidate ID | ||||||
|---|---|---|---|---|---|---|
| A. thaliana | Gene | chrom | start | end | sense | sequence |
| C24_Cas21aCBF1_019 | C24CBF2 | C24_4 | 13745457 | 13745480 | - | TCGGAGCCAAACATTTCAGA AAAA |
| C24CBF3 | C24_4 | 13748381 | 13748404 | - | TCGGAGCCAAACATTTCAGA AAAA | |
| C24CBF1 | C24_4 | 13751940 | 13751963 | - | TCGGAGCCAAACATTTCAGA AAAA | |
| ---- | C24_3 | 4670219 | 4670243 | + | TTTG TCTGAAATGTGCAGTTCCGA | |
| ColCBF3 | Col_4 | 13018950 | 13018973 | - | TCGGAGCCAAACATTTCAGA AAAA | |
| ColCBF1 | Col_4 | 13022509 | 13022532 | - | TCGGAGCCAAACATTTCAGA AAAA | |
| ColCBF2 | Col_4 | 13016046 | 13016068 | - | TCGGAGCCAAACATTTCAGA AAAG | |
| ---- | Col_3 | 4673610 | 4673633 | + | TTTG TCTGAAAGGTGCAGTTCCGA | |
| Eri_Cas12aCBF1_017 | EriCBF2 | Eri_4 | 12981374 | 12981397 | - | AATCGGAGCCAAACATTTCA GAAA |
| EriCBF3 | Eri_4 | 12984307 | 12984330 | - | AATCGGAGCCAAACATTTCA GAAA | |
| EriCBF1 | Eri_4 | 12987866 | 12987889 | - | AATCGGAGCCAAACATTTCA GAAA | |
| ColCBF2 | Col_4 | 13016031 | 13016054 | - | AATCGGAGCCAAACATTTCA GAAA | |
| ColCBF3 | Col_4 | 13018948 | 13018971 | - | AATCGGAGCCAAACATTTCA GAAA | |
| ColCBF1 | Col_4 | 13022507 | 13022530 | - | AATCGGAGCCAAACATTTCA GAAA | |
| ---- | Col_1 | 8279033 | 8279056 | - | AATCAGAGCCTAACACTTCA AAAA | |
| ---- | Col_3 | 9399469 | 9399493 | + | TTTA TGAAGTGTTTGGTTCCTATT | |
| ---- | Eri_1 | 8194484 | 8194507 | - | AATTAGGGCCTAACACTTCA AAAA | |
| ---- | Eri_3 | 9400735 | 9400758 | + | TTTA TGAAGTGTTTGGTTCCTTTT | |
Discussion
Sequence similarity in gene families usually difficults the identification of CRISPR target candidates matching several member of the family and it requires manual time-consuming task. ARES-GT in addition of gene specific guide RNAs also evaluates which candidates match several query sequences. By selection of which sequences are included in the query file user has the maximal flexibility for working with complete families, subfamilies or a particular set of genes to find candidates specifically matching those genes. I have also shown how using ecotype-specific genomes can prevent the identification of false positive/negative candidates, which also apply to individual genomes taking into account polymorphisms.
ARES-GT is written in Python so can be used in any operative system and it has not high computational complexity so it is expected to work without problems with any processor. ARES-GT also has an option for working only with candidates matching several query sequences (option “–OR”) which reduce computer time to 15 min.
Conclusion
In summary, I have shown how the architecture of the ARES-GT tool (i) allows the selection of candidate sgRNAs that match multiple input query sequences for simultaneous editing of several members of gene families; (ii) contemplates the use of unmapped contigs apart from complete genomes; and (iii) can be used for the design of ecotype-specific CRISPR targets. ARES-GT is available at GitHub (https://github.com/eugomin/ARES-GT.git).
Supporting information
DNA sequences of all CBF genes used in this work.
(ZIP)
Acknowledgments
I thank Prof. Miguel A. Blazquez for edition and comments on the manuscript.
Data Availability
ARES-GT is available from GitHub (https://github.com/eugomin/ARES-GT.git). All further relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7(9):1494–6. Epub 2014/04/11. 10.1093/mp/ssu044 . [DOI] [PubMed] [Google Scholar]
- 2.Bae S, Park J, Kim J-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–5. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic acids research. 2019;47(W1):W171–W4. 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome biology. 2016;17(1):148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nature methods. 2014;11(2):122–3. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen L-L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Molecular Plant. 2017;10(3):530–2. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 7.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology. 2016;34(2):184–91. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pliatsika V, Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biology Direct. 2015;10(1):4 10.1186/s13062-015-0035-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveros JC, Franch M, Tabas-Madrid D, San-Leon D, Montoliu L, Cubas P, et al. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic acids research. 2016;44(W1):W267–71. 10.1093/nar/gkw407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keough KC, Lyalina S, Olvera MP, Whalen S, Conklin BR, Pollard KS. AlleleAnalyzer: a tool for personalized and allele-specific sgRNA design. Genome biology. 2019;20(1):167 10.1186/s13059-019-1783-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: A Software Package for Designing CRISPR sgRNA and Evaluating Potential Off-Target Cleavage Sites. PloS one. 2014;9(6):e100448 10.1371/journal.pone.0100448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PloS one. 2014;9(9):e108424–e. 10.1371/journal.pone.0108424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao W-B, Schneeberger K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. bioRxiv. 2019:738880 10.1101/738880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. 10.1093/bioinformatics/btm404 . [DOI] [PubMed] [Google Scholar]
- 15.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827–32. 10.1038/nbt.2647 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swarts DC, van der Oost J, Jinek M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Molecular cell. 2017;66(2):221–33 e4. 10.1016/j.molcel.2017.03.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aliaga-Franco N, Zhang C, Presa S, Srivastava AK, Granell A, Alabadí D, et al. Identification of Transgene-Free CRISPR-Edited Plants of Rice, Tomato, and Arabidopsis by Monitoring DsRED Fluorescence in Dry Seeds. Frontiers in plant science. 2019;10(1150). 10.3389/fpls.2019.01150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernabé-Orts JM, Casas-Rodrigo I, Minguet EG, Landolfi V, Garcia-Carpintero V, Gianoglio S, et al. Assessment of Cas12a-mediated gene editing efficiency in plants. Plant biotechnology journal. 2019;17(10):1971–84. 10.1111/pbi.13113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual review of plant biology. 2006;57:781–803. Epub 2006/05/04. 10.1146/annurev.arplant.57.032905.105444 . [DOI] [PubMed] [Google Scholar]
- 22.Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant physiology. 2002;130(2):639–48. Epub 2002/10/12. 10.1104/pp.006478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho S, Yu S-i, Park J, Mao Y, Zhu J-K, Yun D-J, et al. Accession-Dependent CBF Gene Deletion by CRISPR/Cas System in Arabidopsis. Frontiers in plant science. 2017;8(1910). 10.3389/fpls.2017.01910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan X, Hay A, Kwantes M, Haberer G, Hallab A, Ioio RD, et al. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants. 2016;2(11):16167 10.1038/nplants.2016.167 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences of all CBF genes used in this work.
(ZIP)
Data Availability Statement
ARES-GT is available from GitHub (https://github.com/eugomin/ARES-GT.git). All further relevant data are within the manuscript and its Supporting Information files.