Abstract
Objective
To delineate the phenotype associated with biallelic ATAD1 variants.
Methods
We describe 2 new patients with ATAD1-related disorder diagnosed by whole-exome sequencing and compare their phenotype to 6 previous patients.
Results
Patients 1 and 2 had a similar distinctive phenotype comprising congenital stiffness of limbs, absent spontaneous movements, weak sucking, and hypoventilation. Both had absent brainstem evoked auditory responses (BEARs). Patient 1 carried the homozygous p.(His357Argfs*15) variant in ATAD1. In the light of the finding in patient 1, a second reading of exome data for patient 2 revealed the novel homozygous p.(Gly128Val) variant.
Conclusions
Analysis of the phenotypes of these 2 patients and of the 6 previous cases showed that biallelic ATAD1 mutations are responsible for a unique congenital encephalopathy likely comprising absent BEAR, different from hyperekplexia and other conditions with neonatal hypertonia.
Neonatal unexplained muscular stiffness is a relatively rare condition. Various genetic syndromes/etiologies such as hyperekplexia,1,2 encephalopathies due to BRAT13,4 or GRIA45 genes, and Crisponi syndrome6 are causes of persistent neonatal stiffness. Paroxysmal stiffness is observed in newborns with SCN4A-related severe neonatal episodic laryngospasm (SNEL),7 and facial paroxysms are a hallmark of Crisponi syndrome. Features other than hypertonia may be part of these phenotypes: developmental delay with or without epilepsy (BRAT1 and GRIA4), laryngeal spasms (SCN4A and Crisponi), permanent (BRAT1) or paroxysmal (SCN4A and Crisponi) sucking difficulties, and exaggerated startles (hyperekplexia, GRIA4, and Crisponi).
Biallelic variants in the ATAD1 gene are the most recently described cause of persistent neonatal stiffness.8–10 The phenotype ascribed to these genotypes is referred to as hyperekplexia 4 (MIM 618011). Six patients belonging to 3 different families have been reported to date in the literature. We describe here the ATAD1-related disorder in 2 additional patients of unrelated families, diagnosed by whole-exome sequencing (WES).
Methods
DNA was extracted from parent and proband samples. Trio WES was performed for each patient on a NextSeq 500 Sequencing System (Illumina, San Diego, CA), with a 2× 150 bp high-output sequencing kit after a 12-plex enrichment with the SeqCap EZ MedExome kit (Roche, Basel, Switzerland), according to the manufacturer's specifications. Sequence quality was assessed with FastQC 0.11.5; then, the reads were mapped using BWA-MEM (version 0.7.13), sorted, and indexed in a bam file (samtools 1.4.1), duplicates were flagged (sambamba 0.6.6), and coverage was calculated (picard-tools 2.10.10). Variant calling was performed with GATK 3.7 Haplotype Caller. Variants were then annotated with SnpEff 4.3, dbNSFP 2.9.3, Genome Aggregation Database (gnomAD), ClinVar, Human Gene Mutation Database, Variome Great Middle East, and an internal database. Coverage for these patients was at least 93% at a 20× depth threshold. This work was approved by the local ethics committee (Comité de Protection des Personnes).
Standard protocol approvals, registrations, and patient consents
The parents of both patients gave their consent for genetic studies and for the publication of the results and videos and have been given the opportunity to review the manuscript.
Data availability
Anonymized data not published within this article will be made available by request from qualified investigators.
Patient description and results
A full description is available in supplemental data 1 (links.lww.com/NXG/A322).
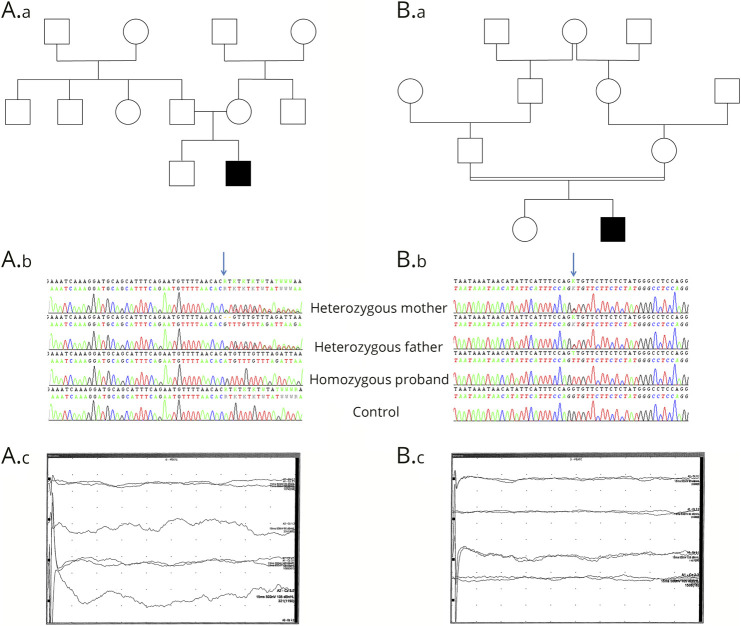
Patient 1 (video 1, links.lww.com/NXG/A323, figure, A) was the second child of healthy parents. He was born at 36 weeks of gestation with normal parameters. Respiratory distress and hypotonia occurred at 20 minutes of life requiring oxygen therapy and noninvasive ventilation (NIV). At 3 hours of life, the patient's weak sucking required parenteral nutrition. The day after, physical examination revealed marked generalized stiffness, brisk tendon reflexes, truncal hypotonia, tremor of limbs, absent spontaneous movements and absent eye contact, weak respiratory movements, and a constant moaning. These clinical findings remained the same from that point on. During the following weeks, patient 1 needed a nasogastric tube. After NIV removal, he required oxygen supply and experienced many episodes of apnea and desaturation. Two brain MRIs were normal. Electroencephalographic (EEG) recordings were initially normal (at days 2, 4, 11, and 21), then showed a slow, monotone and nonreactive activity without spatial organization, amplitude and frequency were asymmetric with scarce spikes without epileptiform brain discharges. Brainstem evoked auditory responses (BEARs) were absent on both ears despite stimulation up to 105 dB (figure, A). EMG performed at 2 months of life showed a persistent contraction without myotonia. Patient 1 died at 4 months of cardiorespiratory arrest. WES revealed the homozygous NM_001321967.1:c.1070_1071del p.(His357Argfs*15) variant in ATAD1 confirmed by Sanger sequencing. This variant has been previously reported in ATAD1-related disorder8 with thorough functional studies. Analysis of the parents showed that they were heterozygous carriers.
Figure. Pedigrees, electrophoregrams, and BEAR.
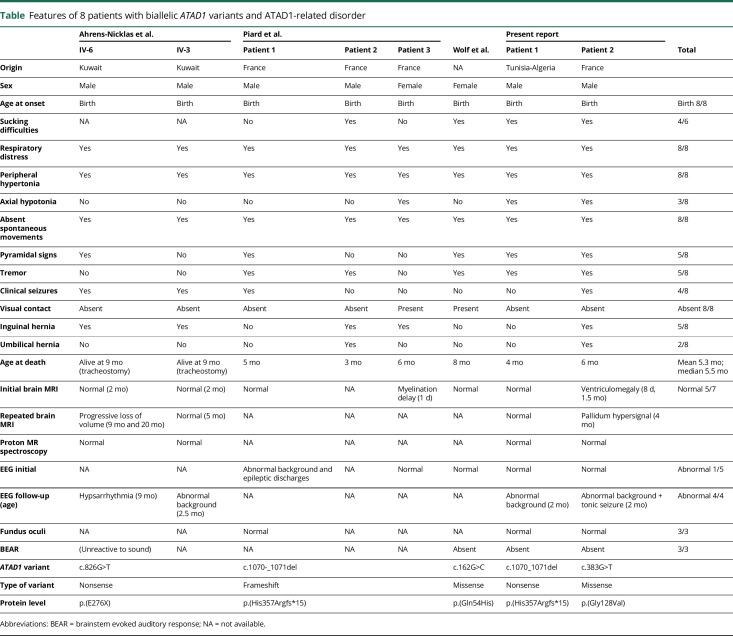
(A) Pedigree (A.a) and electrophoregrams (A.b) showing the homozygous p.(His357Argfs*15) variant in the proband and the heterozygous state of his parents. BEARs were absent with stimulation up to 90 and 105 dB (A.c). (B) Pedigree (B.a) of patient 2 and electrophoregrams (B.b) showing the homozygous p.(Gly128Val) variant in the proband and the heterozygous state of his parents. BEARs were also absent in patient 2 with stimulation up to 90 and 105 dB (B.c). BEAR = brainstem evoked auditory response.
Examination of patient #1 aged 4.5 months.Download Supplementary Video 1 (19.2MB, mp4) via http://dx.doi.org/10.1212/000520_Video_1
Patient 2 (video 2, links.lww.com/NXG/A324, figure, B) was a boy born 5 years before patient 1 to healthy consanguineous parents. He was born full term by spontaneous vaginal delivery with normal parameters. Tremor, weak sucking, and continuous crying were noticed during the first hours of life. Patient 2 was hospitalized because of repeated episodes of oxygen desaturation at the 10th hour of life. Clinical examination showed generalized stiffness, truncal hypotonia, absent spontaneous movements, and brisk tendon reflexes. He needed nasogastric tube feeding as well as oxygen therapy and intermittent NIV because of persistent hypoventilation. Tonic seizures began when he was 3.5 months. He died at age 6 months of a cardiorespiratory arrest. Three brain MRIs showed a moderate left ventriculomegaly. EEG recordings were initially normal (at days 3 and 29) and then showed an unstable, slow background activity without organization from the age of 3.5 months. BAER showed no responses on both ears despite stimulation up to 105 dB (figure, B). WES analysis revealed the homozygous Chr10(GRCh37):g.89544427C>A, NM_001321967.1:c.383G>T, p.(Gly128Val) variant in ATAD1 confirmed by Sanger sequencing. This variant is present on all coding refseq transcripts, is absent from control databases (Exome Variant Server, Exome Aggregation Consortium, and gnomAD), affects a highly conserved amino acid of the protein in the AAA+ ATPase domain, and is considered as disease causing with all the prediction algorithms we have tested. Both parents were heterozygous carriers of the variant.
Examination of patient #2 aged 3 months. Note in both patients the absence of spontaneous movements, stiff limbs, axial hypotonia, and limited facial expression. Induced tremor is also visible in patient #1.Download Supplementary Video 2 (7.2MB, mp4) via http://dx.doi.org/10.1212/000520_Video_2
Discussion
ATAD1 encodes for thorase, an AAA+ ATPase protein discovered in 2010 in primary rat cortical neurons.11 Thorase plays a role in mitochondria and peroxisomes as a quality control of membrane proteins and regulates surface expression of brain α-amino-3-hydroxy-5-methylisoxazole-4-propionate (AMPA) receptors. At the synaptic level, thorase induces the internalization of AMPA receptor by disassembling the subunits GLUR2 and GRIP1. Thus, thorase modulates central neurotransmission, which explains the neurologic phenotype of individuals unable to produce normal thorase species.
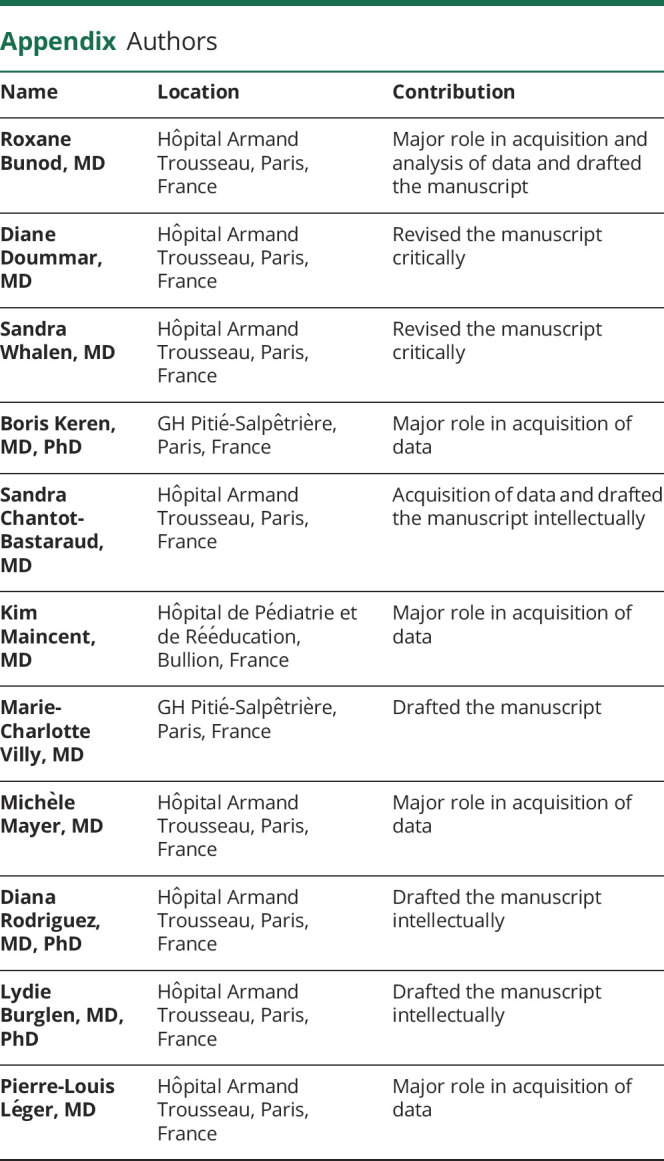
Six patients with biallelic ATAD1 mutations belonging to 3 different families have been reported in the literature to date (table).8–10 As for patient 2 of the present report, one of these diagnoses was made retrospectively, based on the identification of the distinctive ATAD1-related neurologic syndrome,9 which demonstrates the uniqueness of the condition. The phenotype is characterized by a neonatal onset of stiffness of limbs associated with truncal hypotonia or hypertonia. Unlike other causes of neonatal hypertonia, including hyperekplexia, these newborns have no spontaneous movements and display poor facial expression. Clinical examination induces limb tremor and exacerbation of hypertonia, but no exaggerated startle response to sensory stimuli. These newborns need intensive care from the first days of life because of poor sucking and respiratory distress/hypoventilation. This vital distress is the main cause of their early death (3–8 months). Of note, the only 2 patients alive at age 9 months underwent tracheostomy.10
Table.
Features of 8 patients with biallelic ATAD1 variants and ATAD1-related disorder
Most of these patients (6/8) do not have eye contact despite normal ophthalmologic examination and normal electroretinogram/visual evoked potential recordings (present report). This persists throughout their lifetime and is likely related to the severity of their developmental encephalopathy. Their poor reactions to stimuli may be aggravated by hearing impairment, as suggested by absent BEAR in our 2 patients and in a previous one.9 This may be related to a deficit of AMPA receptor–dependent neurotransmission of the signals arising from cochlear hair cells.12
Repeated clinical examinations of these newborns show severe encephalopathy during their entire lifetime. All these features make a strong difference with other conditions associated with neonatal stiffness, i.e., hyperekplexia,1,2 Crisponi syndrome,6 SCN4A-related SNEL syndrome,7 and GRIA4-related encephalopathy.5 Only the BRAT1-related encephalopathy coined as “lethal neonatal rigidity and multifocal seizure syndrome” bears comparison with the ATAD1-related phenotype.3,4,13 Refractory epilepsy is usual in the former, and although seizures are not rare in the latter (4/8), the epilepsy is severe enough to suggest epileptic encephalopathy in a minority of them (2/8). Thus, the term “hyperekplexia 4” phenotype ascribed to ATAD1 mutations seems to be inaccurate.
The 8 patients reported so far belong to 4 families carrying 4 different ATAD1 variants. All of these variants are homozygous, and the only recurrent variant is the p.(His357Argfs*15) frameshift identified in patient 1 with unrelated parents and in Piard et al.8 All 4 parents with this frameshift in the heterozygous state are of Tunisian descent, with those reported in Piard et al. being related. This variant is very rare, reported 3 times at the heterozygous state in the gnomAD database (allele frequency 1.2e-5), but the origin of the carriers is not mentioned in the database. This variant is either a true recurrent variant or a variant found in 2 patients with an unidentified common Tunisian ancestry. We also report the novel p.(Gly128Val) missense variant. This variant is the second missense causing ATAD1-related disorder, together with p.(Gln54His).9 The exact consequence of this change is unknown for now, but its pathogenicity is highly likely according to all tested prediction algorithms and given the uniqueness of the syndrome. The first pathogenic variant ever reported is a nonsense [p.(E276X)10], which suggested that the loss of thorase function is the cause of ATAD1-related disorder. This was substantiated by studies showing that the in vitro overexpression of thorase results in a decreased expression of cell surface AMPA receptors, whereas the loss of thorase expression induces the surface expression of these receptors and an excess of AMPA-mediated current.14 These data are in line with the clinical involvement compatible with glutamate overload. One cannot exclude, however, peroxisomal dysfunction in patients with ATAD1-related disorder. Other functional studies showed that the mutant thorase with the Tunisian variant impairs the recycling of glutamate receptors at the cell surface, acting as a gain-of-function–inducing variant.8 Further investigations are needed to clarify this issue, especially since Ahrens-Nicklas et al.10 recommended the use of perampanel, an AMPA receptor antagonist, in these patients.
Acknowledgment
The authors thank the patients' parents for consenting to the publication of this report.
Glossary
- AMPA
α-amino-3-hydroxy-5-methylisoxazole-4-propionate
- BEAR
brainstem evoked auditory response
- gnomAD
Genome Aggregation Database
- NIV
noninvasive ventilation
- SNEL
severe neonatal episodic laryngospasm
- WES
whole-exome sequencing
Appendix. Authors
Study funding
No targeted funding reported.
Disclosure
The authors report no disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Andermann F, Keene DL, Andermann E, Quesney LF. Startle disease or hyperekplexia: further delineation of the syndrome. Brain 1980;103:985–997. 10.1093/brain/103.4.985. [DOI] [PubMed] [Google Scholar]
- 2.Praveen V, Patole SK, Whitehall JS. Hyperekplexia in neonates. Postgrad Med J 2001;77:570–572. 10.1136/pmj.77.911.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Ommeren RH, Gao AF, Blaser SI, Chitayat DA, Hazrati LN. BRAT1 mutation: the first reported case of Chinese origin and review of the literature. J Neuropathol Exp Neurol 2018;77:1071–1078. 10.1093/jnen/nly093. [DOI] [PubMed] [Google Scholar]
- 4.Hanes I, Kozenko M, Callen DJA. Lethal neonatal rigidity and multifocal seizure syndrome—a misnamed disorder? Pediatr Neurol 2015;53:535–540. 10.1016/j.pediatrneurol.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Martin S, Chamberlin A, Shinde DN, et al. . De novo variants in GRIA4 lead to intellectual disability with or without seizures and gait abnormalities. Am J Hum Genet 2017;101:1013–1020. 10.1016/j.ajhg.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buers I, Persico I, Schöning L, et al. . Crisponi/cold-induced sweating syndrome: differential diagnosis, pathogenesis and treatment concepts. Clin Genet 2020;97:209–221. 10.1111/cge.13639. [DOI] [PubMed] [Google Scholar]
- 7.Lion-Francois L, Mignot C, Vicart S, et al. . Severe neonatal episodic laryngospasm due to de novo SCN4A mutations: a new treatable disorder. Neurology 2010;75:641–645. 10.1212/WNL.0b013e3181ed9e96. [DOI] [PubMed] [Google Scholar]
- 8.Piard J, Umanah GKE, Harms FL, et al. . A homozygous ATAD1 mutation impairs postsynaptic AMPA receptor trafficking and causes a lethal encephalopathy. Brain 2018;141:651–661. 10.1093/brain/awx377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf NI, Zschocke J, Jakobs C, Rating D, Hoffmann GF. ATAD1 encephalopathy and stiff baby syndrome: a recognizable clinical presentation. Brain 2018;141:e49 10.1093/brain/awy095. [DOI] [PubMed] [Google Scholar]
- 10.Ahrens-Nicklas RC, Umanah GKE, Sondheimer N, et al. . Precision therapy for a new disorder of AMPA receptor recycling due to mutations in ATAD1. Neurol Genet 2017;3:e130 doi:10.1212/NXG.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai C, Liang D, Li H, Sasaki M, Dawson TM, Dawson VL. Functional identification of neuroprotective molecules. PLoS One 2010;5:e15008 10.1371/journal.pone.0015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ottersen OP, Takumi Y, Matsubara A, Landsend AS, Laake JH, Usami S. Molecular organization of a type of peripheral glutamate synapse: the afferent synapses of hair cells in the inner ear. Prog Neurobiol 1998;54:127–148. 10.1016/s0301-0082(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava S, Olson HE, Cohen JS, et al. . BRAT1 mutations present with a spectrum of clinical severity: BRAT1 mutations present with a spectrum of clinical. Am J Med Genet 2016;170:2265–2273. 10.1002/ajmg.a.37783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Wang Y, Chi Z, et al. . The AAA+ ATPase thorase regulates AMPA receptor-dependent synaptic plasticity and behavior. Cell 2011;145:284–299. 10.1016/j.cell.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examination of patient #1 aged 4.5 months.Download Supplementary Video 1 (19.2MB, mp4) via http://dx.doi.org/10.1212/000520_Video_1
Examination of patient #2 aged 3 months. Note in both patients the absence of spontaneous movements, stiff limbs, axial hypotonia, and limited facial expression. Induced tremor is also visible in patient #1.Download Supplementary Video 2 (7.2MB, mp4) via http://dx.doi.org/10.1212/000520_Video_2
Data Availability Statement
Anonymized data not published within this article will be made available by request from qualified investigators.