Abstract
Objective
To further clarify the molecular pathogenesis of RNA polymerase III (Pol III)-related leukodystrophy caused by biallelic POLR1C variants at a cellular level and potential effects on its downstream genes.
Methods
Exome analysis and molecular functional studies using cell expression and long-read sequencing analyses were performed on 1 family with hypomyelinating leukodystrophy showing no clinical and MRI findings characteristic of Pol III–related leukodystrophy other than hypomyelination.
Results
Biallelic novel POLR1C alterations, c.167T>A, p.M56K and c.595A>T, p.I199F, were identified as causal variants. Functional analyses showed that these variants not only resulted in altered protein subcellular localization and decreased protein expression but also caused abnormal inclusion of introns in 85% of the POLR1C transcripts in patient cells. Unexpectedly, allelic segregation analysis in each carrier parent revealed that each heterozygous variant also caused the inclusion of introns on both mutant and wild-type alleles. These findings suggest that the abnormal splicing is not direct consequences of the variants, but rather reflect the downstream effect of the variants in dysregulating splicing of POLR1C, and potentially other target genes.
Conclusions
The lack of characteristic clinical findings in this family confirmed the broad clinical spectrum of Pol III–related leukodystrophy. Molecular studies suggested that dysregulation of splicing is the potential downstream pathomechanism for POLR1C variants.
RNA polymerase III (Pol III)-related leukodystrophy is characterized by hypomyelination in the CNS with various additional manifestations such as hypogonadotropic hypogonadism, hypodontia, cerebellar ataxia, and atrophy of the corpus callosum. After the proposal of multiple clinical entities,1,2 the discovery of pathogenic variants in genes encoding 2 major subunits of Pol III, POLR3A and POLR3B,3,4 in the majority of patients led to the concept of Pol III–related leukodystrophy being emerged.5 Recently, variants in yet another gene coding for Pol III complex, POLR1C, were identified in patients who were negative for, but showed clinical features similar to those with, POLR3A and POLR3B variants.6
Here, we report a patient with novel POLR1C pathogenic variants, who showed clinical and imaging features compatible with hypomyelinating leukodystrophy without additional features characteristic of Pol III–related leukodystrophy. We also propose a potential molecular mechanism of POLR1C variants involving dysregulation of splicing.
Methods
This study was approved by the Institutional Review Board of the National Center of Neurology and Psychiatry. Genomic DNA and total RNA were extracted from the peripheral blood of the patient and parents. For DNA diagnostic testing, we performed quantitative PCR for the screening of PLP1 duplication, followed by exome sequencing for Mendelian disease panel (TruSight One, Illumina), as we previously performed according to the manufacturer's protocol.7 POLR1C complementary DNAs were obtained by reverse transcriptase (RT)-PCR, which were cloned into an expression vector, pcDNA3.1 (Invitrogen) with FLAG-tag at the N-terminus, for subsequent Sanger sequencing and transient expression studies in HeLa cells for Western blotting and fluorescent immunostaining. For long-read next-generation sequencing, barcoded RT-PCR products (control, father, mother, and patient) were sequenced on a single MinION R9.4 flow cell (Nanopore).
Data availability
Any data not published within the article will be shared by request from any qualified investigator.
Case report
The patient was a Japanese boy without a family history of neuromuscular diseases and had normal neurodevelopment during infancy. At age 2 years, he developed action tremor of his fingers, had difficulty in writing, and showed early signs of motor dyspraxia. At age 3 years, he developed amblyopia secondary to hypermetropia and astigmatism. Myopia was not noted. At age 3 years and 10 months, he presented with action tremors in fingers, but there were no other neurologic abnormalities. He showed a developmental quotient of 105 (Enjoji analytical developmental test for infants and toddlers). Subsequently, he became neurodevelopmental abilities stagnated and regressed in his daily activities and nystagmus became apparent. On his visit at age 5 years and 9 months, he had a short stature with 101.4 cm (−2.1 SD) tall without apparent microcephaly, facial abnormalities, or ambiguous genitalia. No delay or abnormal order in dentation was noted. He exhibited lateral nystagmus, action tremors, and slurred speech. The finger-to-nose, pronosupination, and tandem walk tests showed mild dysmetria and ataxia. Deep tendon reflexes of the lower limbs were increased. He exhibited a staggering wide-based gait and was unable to stand on 1 leg for more than 2 seconds. Both parents were intellectually and physically normal with no neurologic findings.
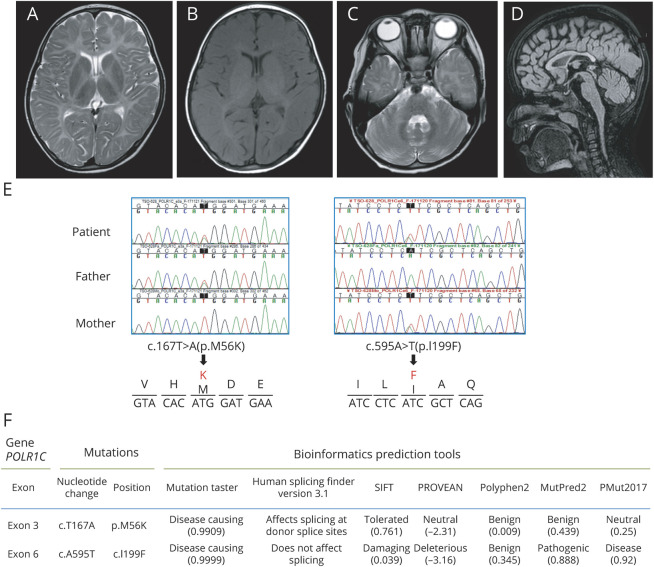
We performed several tests at age 5–6 years. Laboratory tests revealed prepubertal patterns of pituitary gonadotropins and testosterone. The Wechsler Intelligence Scale for Children, fourth edition, showed regression with a full-scale intelligence quotient of 70. Peripheral nerve conduction velocities and auditory brainstem responses were normal. EEG was normal. MRI showed diffuse T2 hyper- and T1 iso-intensities in the white matter, indicating hypomyelination (figure 1, A and B). T1 and T2 shortening in the optic radiation, the ventrolateral thalamus, and the dentate nucleus was noted, as typically observed in Pol III–related leukodystrophy (figure 1, A–C).8 Cerebellar atrophy or thinning of the corpus callosum was not evident (figure 1, C and D). MRIs of the parents were not available.
Figure 1. MRI and POLR1C variants.
Brain MRI of the present patient examined at age 5 years 9 months. (A) T2-weighted axial image shows diffusely elevated white matter signal. T2 shortening in the optic radiation and the ventrolateral thalamus is noted. (B) T1-weighted axial shows high to iso-signal, consistent with hypomyelination. T1 shortening in the optic radiation and the ventrolateral thalamus is noted. (C) T2-weighted axial image shows no sign of cerebellar atrophy. (D) FLAIR sagittal image demonstrates no thinning of the corpus callosum or cerebellar atrophy. (E) POLR1C Sanger sequence chromatograms showing compound heterozygous c.T167A:p.M56K and c.A595T:p.I199F variants in the patient (top) inherited from each parent (middle and bottom). Lower panels show the deduced amino acid change in each allele. (F) In silico prediction of pathogenicity. FLAIR = fluid-attenuated inversion recovery.
Results
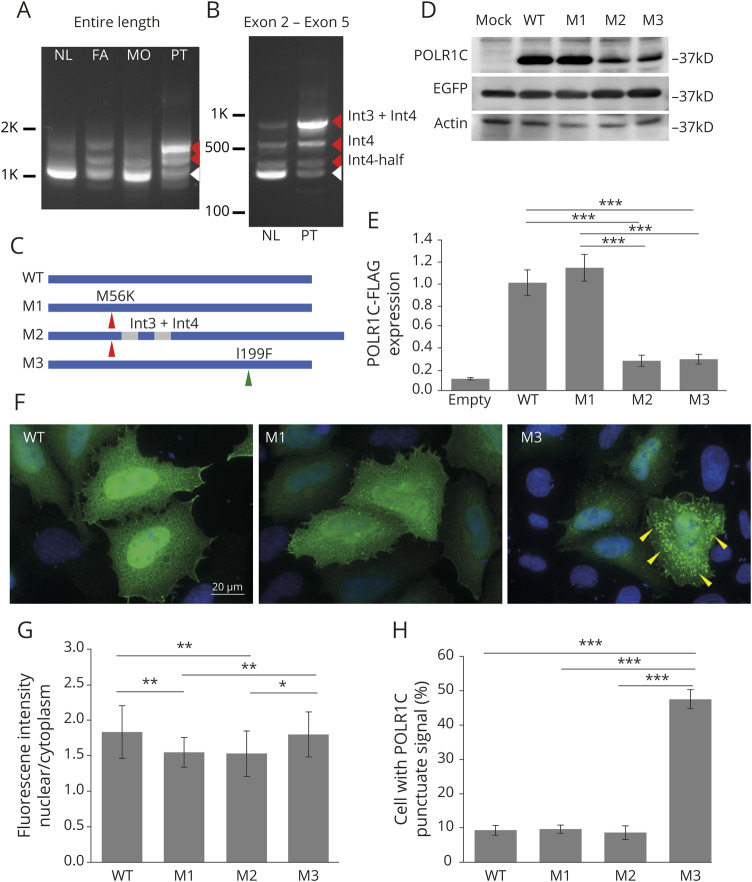
After PLP1 duplication was excluded, the panel exome sequencing identified 2 novel heterozygous missense variants in exon 3 and exon 6 of the POLR1C gene (NM_203290.3:c.167T>A, p.M56K and c.595A>T, p.I199F, respectively; figure 1E). Parental segregation analysis confirmed compound heterozygosity. In silico prediction analyses revealed both variants to be pathogenic at different levels (figure 1F). RT-PCR using the patient's sample showed increased proportion of splicing variants with a combination of full intron 3 and/or half/full intron 4 inclusions, all of which are presumably nonfunctioning variants with premature termination codons on sequence validation (figure 2, A and B). The patient's major transcript was the variant including both intron 3 and intron 4. Three representative variants expressed in HeLa cells showed that p.M56K alone did not change the protein stability, but the nuclear localization was modestly diminished (figure 2, C–G). p.M56K with intron 3 and intron 4 inclusion significantly decreased the protein level. Meanwhile, p.I199F caused cytosolic punctation and reduced protein expression (figure 2, C–H). The punctation did not overlap with lysosomal marker Lamp1, an autophagosome marker LC3, or with the proteosome marker, ubiquitin (data not shown).
Figure 2. Molecular effects of POLR1C variants.
Agarose gel electrophoresis images of RT-PCR products using total RNA from blood cells, amplifying (A) the entire length and (B) exon 2–exon 5 of POLR1C cDNA. (A) Although a normal control (NL) shows a single 1-Kb band (white arrowhead), the patient (PT) showed longer abnormal bands (red arrowheads) along with a fainter normal band. Both the father (FA) and the mother (MO) also showed abnormal bands with different intensities. (B) In addition to the expected 340-bp normal band (white arrowhead), 3 additional bands were observed in both the normal and patient samples (red arrowheads). Band isolation and sequencing confirmed that the strongest amplicon in the patient (842 bp band) included both intron 3 and intron 4 (int3 + int4). The others contain either the entire or the first half of intron 4 (int4 or int4-half, respectively). (C) A scheme of each variant transcript cloned into an expression vector, pcDNA3.1. Arrowheads indicate each variant. M2 includes intron 3 and intron 4 (int3 + int4) flanking exon 3. (D) Western blot of POLR1C variants transiently expressed in HeLa cells. Upper panel: POLR1C; middle panel: enhanced green fluorescent protein (EGFP) (cotransfected as an inner control for normalization of transfection efficiency); lower panel: actin (loading control). Cells were harvested after 24 hours of transfection. An anti-FLAG antibody was used to visualize exogenous POLR1C. The sizes of the M2 band appear to be the same as WT, suggesting that these introns are partially spliced out before translation. Truncated protein was not observed, presumably due to the removal by nonsense-mediated messenger RNA decay before translation. (E) Quantification of the POLR1C protein level. Experiments were performed in triplicates. POLR1C was normalized to EGFP. The y-axis indicates relative value to the average of wild type. (F) Fluorescent immunostaining of HeLa cells transiently expressing wild-type and mutant POLR1C. Subcellular localization of exogenous protein was determined using anti-FLAG antibody. Bar indicates 20 µm. Wild-type POLR1C showed strong nuclear expression (WT). The M56K mutant showed reduced nuclear staining (M1). The I199F mutant showed prominent cytosolic punctations (yellow arrowheads; M3). DAPI nuclear staining shows blue signals. Images were obtained using Keyence BZ-X710 fluorescence microscope (Keyence, Japan). (G) Quantification of nuclear localization. Using imaging software (Keyence), the ratio of signal intensities of the nucleus and cytosol was measured (n = 30 cells per group). (H) The proportion of cells with cytosolic punctations was calculated in more than 300 cells (10 fields [30–40 cells per field] at 200× magnification). Error bars indicated standard errors. *p < 0.05, **p < 0.01, ***p < 0.001. One-way analysis of variance.
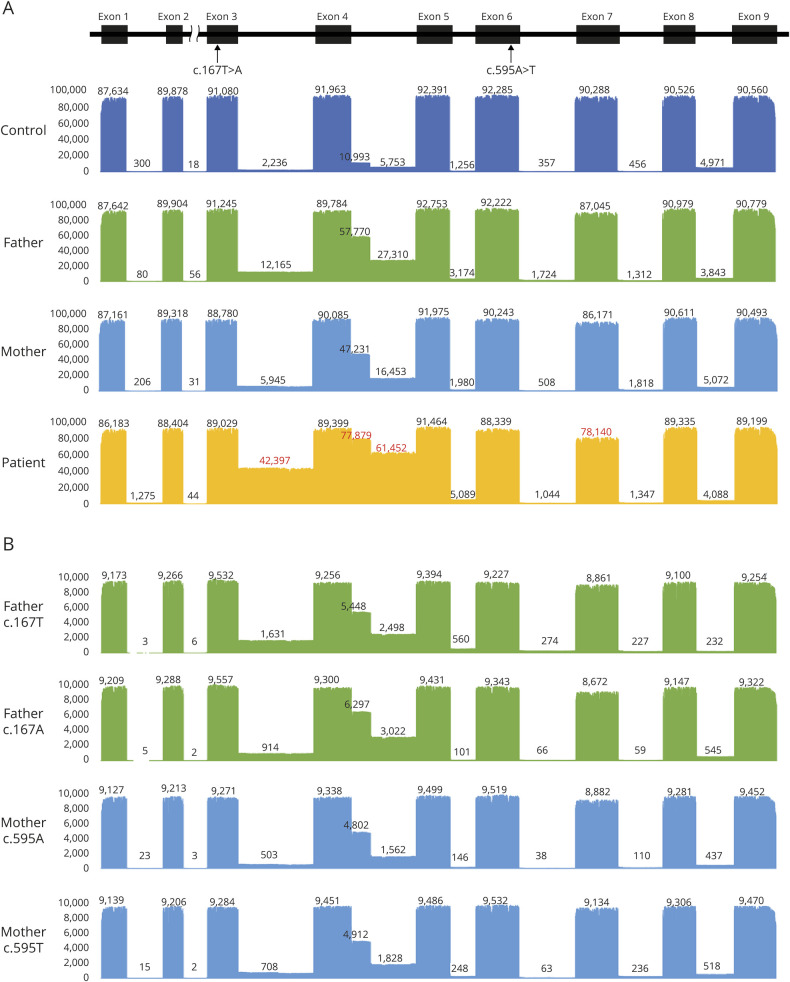
To our surprise, both parents also showed increased proportion of the intron 3/4 inclusion variant (figure 2A), which prompted us to use long-read next-generation sequencing to obtain deep reads of all variants with allelic segregation. Mapping patient POLR1C transcripts on genome demonstrated that they were biallelic, and more than 85% of correctly mapped transcripts were intron-containing variants (figure 3A). Both parents also showed apparently increased proportion of intron-containing variants (64% in the father and 52% in the mother). These findings suggested 2 possible mechanisms: (1) both c.167T>A and c.595A>T variants directly affected the splicing to properly remove intron 3/4, or (2) splicing abnormality resulted from impaired function of Pol III target genes that play roles in the maintenance of splicing. To delineate these 2 possibilities, the parental reads of each allele were remapped to determine whether each variant affected splicing in cis or trans. Surprisingly, the proportion of intron-containing variants was equivalent between wild-type and mutant alleles in both parents, indicating that both variants affect splicing in trans (figure 3B). Because it is unlikely that the variant in one allele directly affects the splicing of the other, this trans effect is probably driven by the latter hypothesis.
Figure 3. Depth of coverage of aligned reads of POLR1C cDNA.
(A) Top: schematic representation of POLR1C consisting of 9 exons (black rectangles). We examined transcript variant 1 (NM_203290.3). Bottom: the bar graph shows the sequence depth at each position, and the number shows the average depth of exons and introns. Red numbers highlight aberrant transcripts. In addition to intron 3 and intron 4 inclusion, skipping of exon 7 was evident in a small proportion of transcripts. From the top: control (blue), father (green), mother (light blue), and patient (yellow). The proportion of aberrant splicing variants was obtained by dividing the highest read count of intron 4 by the highest read count of all exons (e.g., 77879/91464 in the patient). (B) Alignment of each allelic reads. The c.167T and c.167A allele reads were selected from sequence reads of the father, and the c.595A and c.595T allele reads were selected from those of the mother. Ten thousand reads of each allele were aligned. There was no obvious difference in the proportion of variants with intron inclusions between the 2 alleles in each parent.
Discussion
In this study, we reported 1 family with novel POLR1C compound heterozygous variants. Clinically, our patient so far had no characteristic features of Pol III–related leukodystrophy, such as dental abnormality and hypogonadism, and no cerebellar atrophy and thinning of the corpus callosum. Although these features are common in patients with POLR3A or POLR3B variants,9 thereby serving as diagnostic key features, findings other than hypomyelination are not necessarily present in all cases.10 Previously reported 32 cases with POLR1C variants presented with at least one of these features.6,11,12 Thus, the present case also suggested that lacking characteristic features of Pol III–related leukodystrophy does not exclude the presence of POLR1C variants.
The molecular mechanisms underlying POLR1C variants causing hypomyelination remain unknown. We showed that the POLR1C variant on each allele does not only affect the subcellular localization and/or amount of the protein but also affects the splicing that removes the intron 3/4. Allelic segregation studies using a long-read sequencing technology in the patient revealed a large proportion of abnormal splicing variants. Moreover, the analyses in the parental samples showed 2 unexpected findings. First, in addition to the exon 3 variant, the exon 6 variant also affected splicing even in a heterozygous status, indicating that haploinsufficiency of POLR1C caused molecular deficits despite the autosomal recessive mode of inheritance. Second, each allelic variant caused the inclusion of introns on both alleles. This most likely resulted from altered transcription of Pol III target genes that play a role in the regulation of splicing of POLR1C. As one such candidate, we examined the expression of U6 snRNA, which plays a central role in spliceosome,13 but was not altered in either the patient nor his parents (data not shown). Although the exact effectors downstream of the POLR1C variants remain unknown, our findings provide a potential molecular mechanism for POLR1C variants that affect the activity of Pol III and transcription of its target genes, which may lead to dysregulation of splicing of genes including POLR1C.
In conclusion, we reported 1 family with hypomyelinating leukodystrophy caused by novel POLR1C variants. Both pathogenic variants resulted in changes in subcellular localization and reduction in protein levels, as well as inclusion of introns, which presumably resulted in loss of function. Allelic segregation analyses of full-length transcripts in both patient and parents revealed that the aberrant splicing variants are not direct consequences of the coding variants, but rather reflect the downstream effect of the variants in dysregulating splicing of POLR1C, and potentially other target genes.
Acknowledgment
The authors thank the patient and his family for their co-operation. They are grateful to Drs. Hitoshi Osaka (Jichi Medical University, Japan) and Toshiyuki Yamamoto (Tokyo Women's Medical University, Japan) for their critical review of the clinical information.
Glossary
- Pol III
polymerase III
Appendix. Authors
Study funding
This study was supported in part by grants from the Japan Agency of Medical Research and Development, AMED, Practical Research Project for Rare/Intractable Diseases (K. Inoue, 16ek0109016 and 17ek0109270), and Grant-in-aid for Research on Measures for Intractable Diseases, No. H30-Nanchi-Ippan-008, by the Ministry of Health, Labour and Welfare, Japan (J. Takanashi, K. Kurosawa, H. Saitsu, and K. Inoue).
Disclosure
The authors report no disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Sasaki M, Takanashi J, Tada H, Sakuma H, Furushima W, Sato N. Diffuse cerebral hypomyelination with cerebellar atrophy and hypoplasia of the corpus callosum. Brain Dev 2009;31:582–587. [DOI] [PubMed] [Google Scholar]
- 2.Timmons M, Tsokos M, Asab MA, et al. Peripheral and central hypomyelination with hypogonadotropic hypogonadism and hypodontia. Neurology 2006;67:2066–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard G, Chouery E, Putorti ML, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet 2011;89:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saitsu H, Osaka H, Sasaki M, et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet 2011;89:644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard G, Vanderver A. POLR3-Related leukodystrophy. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReview® [online]. Seattle: University of Washington, Seattle; 2012:1993-2020. Available at: ncbi.nlm.nih.gov/books/NBK99167/. Accessed April 24, 2020. [PubMed] [Google Scholar]
- 6.Thiffault I, Wolf NI, Forget D, et al. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis if RNA polymerase III. Nat Commun 2015;6:7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami H, Enomoto Y, Tsurusaki Y, Sugio Y, Kurosawa K. A female patient with x-linked Ohdo syndrome of the Maat-Kievit-Brunner phenotype caused by a novel variant of MED12. Congenit Anom (Kyoto) 2020;60:91–93. [DOI] [PubMed] [Google Scholar]
- 8.La Piana R, Tonduti D, Gordish Dressman H, et al. Brain magnetic resonance imaging (MRI) pattern recognition in pol III-related leukodystrophies. J Child Neurol 2014;29:214–220. [DOI] [PubMed] [Google Scholar]
- 9.Wolf NI, Vanderver A, van Spaendonk RM, et al. Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology 2014;83:1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daoud H, Tétreault M, Gibson W, et al. Mutations in POLR3A and POLR3B are a major cause of hypomyelinating leukodystrophies with or without dental abnormalities and/or hypogonadotropic hypogonadism. J Med Genet 2013;50:194–197. [DOI] [PubMed] [Google Scholar]
- 11.Gauquelin L, Cayami FK, Sztriha L, et al. Clinical spectrum of POLR3-related leukodystrophy caused by biallelic POLR1C pathogenic variants. Neurol Genet 2019;5:e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraoua I, Karkar A, Drissi C, et al. Novel POLR1C mutation in RNA polymerase III-related leukodystrophy with severe myoclonus and dystonia. Mol Genet Genomic Med 2019;7:e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didychuk AL, Butcher SE, Brow DA. The life of U6 small nuclear RNA, from cradle to grave. RNA 2018;24:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Any data not published within the article will be shared by request from any qualified investigator.