Abstract
We have recently reported on a series of Lys-covalent agents targeting the BIR3 domain of the X-linked Inhibitor of Apoptosis Protein (XIAP) using a benzamide-sulfonyl fluoride warhead. Using XIAP as a model system, we further investigated a variety of additional warheads that can be easily incorporated into binding peptides, and analyzed their ability to form covalent adducts with lysine and other amino acids, including tyrosine, histidine, serine, and threonine, using biochemical and biophysical assays. Moreover, we tested aqueous, plasma stability, cell permeability, and cellular efficacy of the most effective agents. These studies identified aryl-fluoro sulfates as likely the most suitable electrophiles to effectively form covalent adducts with Lys, Tyr, and His residues, given that these agents were cell permeable, and stable in aqueous buffer and in plasma. Our studies contain a number of general findings that open new possible avenues for the design of potent covalent PPIs antagonists.
Keywords: Drug discovery, lysine covalent, tyrosine covalent, histidine covalent, PPIs, protein-protein interactions, XIAP, IAP, covalent inhibitors, sulfonyl fluorides, fluoro sulfates
Graphical Abstract
Introduction
The design of effective inhibitors of protein-protein interactions (PPIs) for therapeutic use has been notoriously an arduous and challenging task; hence, PPIs remain a largely untapped target space for new therapeutics. While several factors can contribute to this challenge, one main issue in targeting PPIs is the large surface area at the site of interaction that is less prone to be effectively inhibited by small molecule compounds that are typically populating libraries for high-throughput screening (HTS) campaigns. During the past two decades, applications of more rational biophysical approaches, combined with fragment- and/or structure-based design strategies have resulted in the design of a variety of novel PPIs antagonists, culminating in 2016 in the approval of Venetoclax (ABT199),1 a Bcl-2 antagonist derived using a combination of NMR-based fragment screening2–6 and iterative structure-based optimizations.1 Alternative strategies to this successful yet still relatively convoluted approach have been proposed, including the use of short peptides derived from structural studies of the intervening binding partners, or from peptide library screening approaches. For example, the Inhibitors of Apoptosis Proteins (IAPs)7–9 are tightly regulated in normal cells by a natural protein inhibitor, namely the second mitochondria-derived activator of caspases (SMAC). Structural and cellular studies identified that the activation of SMAC after mitochondrial release into the cytosol exposes an N-terminal tetrapeptide of sequence Ala-Val-Pro-Phe (AVPF, or also the peptide AVPI) that mediates its interactions with various members of the IAP family, including XIAP.3, 10–12 Using structure-based approaches, aimed at increasing drug-likeness of this tetrapeptide, including increasing their affinity, stability, and cell permeability, several drug-like agents that could mimic SMAC AVPF peptide have been developed as potential therapeutic agents5, 6, 13–30 and a few have entered clinical trials. This example clearly demonstrates that short peptide sequences could provide valuable starting point for the design of potential therapeutic agents against PPIs. One major challenge while working with short peptides is optimizing their affinity, selectivity, and drug-likeness. We and others have recently demonstrated that potency and selectivity of binding in peptides can be accomplished by introducing mild electrophiles such as an acrylamides or chloroacetamides at side chain amines (including Lysine, or Lys; Ornithine, or Orn; di-amino-propionic acid, or Dap; di-amino butyric acid, or Dab) if these are in proximity of a cysteine residue.31, 32 More recently, we have also demonstrated that coupling sulfonyl fluoride benzoic acids on the same amines on a peptide ligand can result in peptides that can covalently target binding site Lys residues.33 Because these approaches can target covalently a surface amino acid at the binding site, the resulting agents can acquire remarkable potency for the intended target. While reactivity, selectivity, and stability of acrylamides and chloroacetamides in targeting Cys residues have been more extensively studied, similar studies characterizing the introduction of aryl-sulfonyl fluorides34–36 or aryl-fluoro sulfates37, 38 warheads into binding peptides have not been fully investigated, which is the topic of our studies. Hence, using the BIR3 domain of XIAP, we probed the potential reactivity of these electrophiles against binding site side chains, including lysine, tyrosine, histidine, serine, and threonine. Our studies provide a number of practical considerations and simple methodologies that are useful in optimizing binding peptides into covalent agents targeting lysine, tyrosine, or histidine residues that we envision can be particularly useful in targeting PPIs.
Results
Synthesis of peptide mimetics containing aryl-sulfonyl fluorides or aryl-fluoro sulfates.
To incorporate aryl-sulfonyl fluorides or aryl-fluoro sulfates into linear peptides we used the general schemes reported in Figure 1. The synthesis of aryl-sulfonyl fluorides peptides was easily accomplished using orthogonal deprotection of ivDde-protected primary amines on the side chains of Dap, Dab, Orn, or Lys. As the ivDde protecting group is stable in 20% piperidine, but is cleaved with 2–4% hydrazine in DMF, it allows the amino groups of ivDde-protected residues to be selectively unmasked on the solid phase without affecting the side-chain protecting groups of other residues, facilitating subsequent site-specific modifications. Hence, after selective deprotection of these primary amines, subsequent reaction of the free amine with sulfonyl fluoride benzoic acids will afford agents as reported in Figure 1A. Likewise, reaction with bromomethyl-benzensulfonyl fluorides will result in agents as reported in Figure 1B. The synthesis of aryl-fluoro sulfates followed a recently developed approach that takes advantage of the [4-(acetylamino)phenyl]imidodisulfuryl difluoride (AISF)37 reagent for the insertion of the fluoro sulfate functional group into Tyr or homo-Tyr (Figure 1C). Alternatively, substituted or unsubstituted phenol carboxylic acids can be introduced in the side chains of Dap, Dab, Orn, or Lys, and subsequently the phenols can be converted to their corresponding aryl-fluoro sulfates (Figure 1D) using the same procedure as reported in Figure 1C. Hence, these four simple strategies can inexpensively and effectively incorporate a variety of aryl-sulfonyl fluorides or aryl-fluoro sulfates into peptides and peptide mimetics. When applied to tetrapeptides of sequence Ala-X-Pro-Phe-NH2, these strategies generated a small library of sulfonyl fluorides and fluoro sulfates listed in Tables 1 and 2. These agents were subsequently used to assess their reactivity with surface Lys, Tyr, Ser, Thr, or His amino acids, using biochemical and biophysical assays.
Figure 1. General schemes for the incorporation of aryl-sulfonyl fluorides (A,B) or aryl-fluoro sulfates (C,D) into peptides.
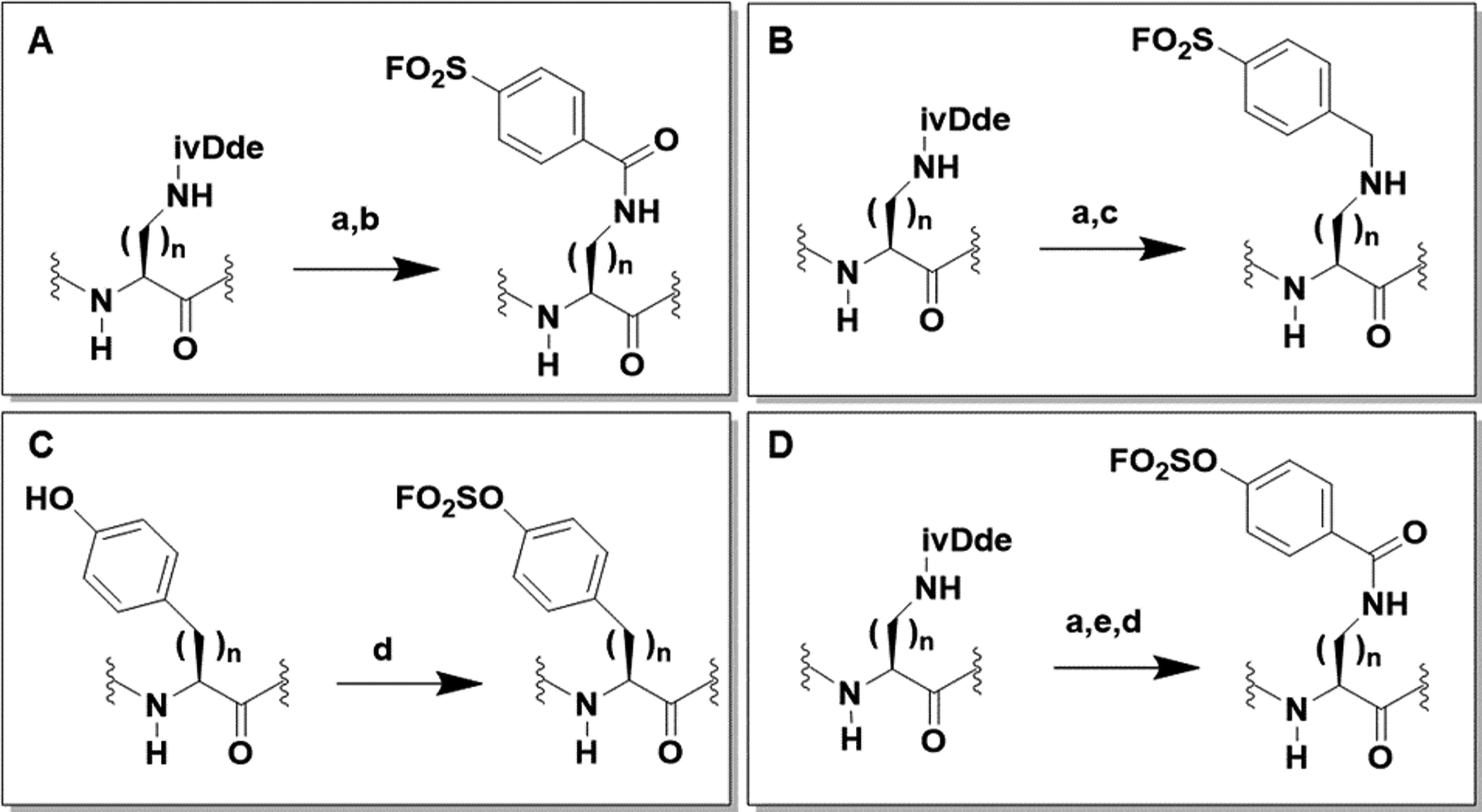
Reaction conditions: a) 4% N2H2 in DMF (3×5ml), rt; b) FO2SPhCOOH, HATU, DIPEA, DMF, rt; c) FO2SPhCH2Br, DIPEA, DMF, rt; d) AISF, DBU, THF, rt.; e) OHPhCOOH, HATU, DIPEA, DMF, rt.
Table 1.
Displacement assays, thermal shift data, and chemical stability data for aryl-sulfonyl fluorides and aryl-fluoro sulfates.
 | ||||
|---|---|---|---|---|
| ID | X | IC50 XIAP-BIR3 [nM]a | ΔTm [°C]b | Chemical stabilityc |
| AVPF | 108 ± 2 56 ± 6 64 ± 10 |
5.5 17 |
> 5h | |
| 1 | 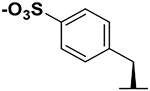 |
47 ± 6 47 ± 0.1 39 ± 0.4 |
13.5 17 |
> 5h |
| 2 | 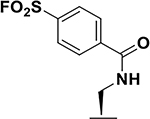 |
88 ± 1 11 ± 1 13 ± 1 |
31 37 |
~ 30 min |
| 3 | 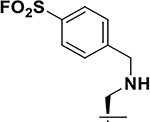 |
95 ± 5 17 ± 0.1 8 ± 0.1 |
35.5 35 |
~ 2h 30 min |
| 4 | 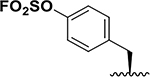 |
160 ± 60 71 ± 7 24 ± 1 |
8.5 33 |
> 5h |
IC50 values represent dose-response curves obtained after incubating protein and ligand for 15 min, 2 h, or 8 h at room temperature.
Thermal shift (ΔTm) data were obtained incubating protein and ligand for 2 h at room temperature or 6 h at 37 °C.
Chemical stability (half-life) was assessed by collecting 1D 1H NMR spectra at various times for ligands dissolved in phosphate buffer pH 7.5 at 37°C (Figure 2C).
Table 2.
Displacement assays, thermal shift data for aryl-sulfonyl fluorides and aryl-fluoro sulfates tested against several XIAP BIR3 mutants.
 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agent | BIR3wt | Lys311Ala | Lys311Tyr | Lys311His | Lys311Thr | Lys311Ser | |||||||
| ID | X | IC50a [nM] |
ΔTm [°C]b |
IC50 [nM]a |
ΔTm [°C]b |
IC50 [nM]a |
ΔTm [°C]b |
IC50 [nM]a |
ΔTm [°C]b |
IC50 [nM]a |
ΔTm [°C]b |
IC50 [nM]a |
ΔTm [°C]b |
| 2 | 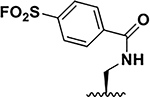 |
11 ± 1 13 ± 1 |
31 37 |
86 ± 2 59 ± 6 |
1.5 8.5 |
12 ± 2 10 ± 0.8 |
37.5 34.5 |
26 ± 0.1 21 ± 1 |
35.5 28.5 |
201 ± 1 72 ± 2 |
4.0 6.5 |
160 ± 8 80 ± 9 |
5 11 |
| 3 | 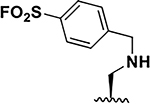 |
17 ± 0.1 8 ± 0.1 |
35.5 35 |
61 ± 4 27 ± 0.1 |
5 7.5d |
6.4 ± 0.1 3.5 ±0.1 |
36 33 |
62 ± 14 16 ± 1 |
31 28.5 |
161 ± 9 45 ± 2 |
5 6d |
145 ± 7 39 ± 1 |
5 7.5d |
| 4 | 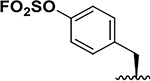 |
71 ± 7 24 ± 1 |
8.5 33.0 |
65 ± 1 55 ± 0.2 |
11 13.5 |
37 ± 5 21 ± 2 |
8.5 35.5 |
113± 22 63 ± 4 |
11 12.5 |
140±20 120±10 |
9.5 11.5 |
177 ± 4 126 ± 2 |
10.5 14.5 |
| 5 | 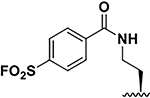 |
190± 20 105± 10 |
11.5 16 |
86 ± 9 95 ± 4 |
10.5 11.5 |
55± 16 71 ± 8 |
10.5 11.5 |
137 ± 4 110 ± 3 |
8.5 8.5 |
231±20 220±10 |
8.5 9.5 |
241 ± 1 230± 10 |
6.5 13.5 |
| 6 | 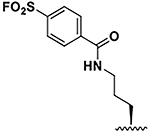 |
140 ± 1 100 ± 5 |
11.5 15.0 |
89 ± 0.2 108 ± 6 |
10.5 11.5 |
63 ± 10 85 ± 4 |
10.5 11.5 |
140± 24 130± 10 |
11.5 12 |
240±12 220±20 |
7.5 9.5 |
250±1 276±14 |
8 13.5 |
| 7 | 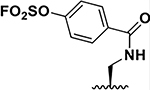 |
590± 65 100 ± 7 |
4.5 31c |
218 ±8 270± 25 |
8 8.5 |
113 ± 2 48 ± 6 |
5.5 32 |
270±70 260±20 |
36.5e 38e |
620±40 630±20 |
5.5 6.5 |
713± 3 646± 32 |
8.5 10.5 |
| 8 | 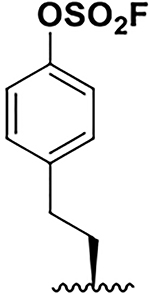 |
2000± 20 90 ± 1 |
11.5 28.5c |
70 ± 2 70 ± 7 |
11 12.5 |
80 ± 2 86 ± 13 |
10.5 29.5c |
126± 14 98 ± 1 |
11 29.5c |
180± 40 165± 3 |
8.5 10 |
250± 1 237± 13 |
10.5 10.5 |
IC50 values represent dose-response curves obtained after incubating protein and ligand for 2 h or 8 h at room temperature.
Thermal shift (ΔTm) data were obtained incubating protein and ligand for 2 h at room temperature or 6 h at 37°C. The data reported represent the major peak observed (see supplementary information for individual curves).
In this experiment, the peak corresponding to the non-covalent ΔTm is still present.
An additional smaller peak that corresponds to a ΔTm ~30 °C is observed perhaps due to a covalent interaction with Lys322.
In this experiment the shift reported corresponds to a minor peak.
Targeting Lys311 of the BIR3 domain of XIAP.
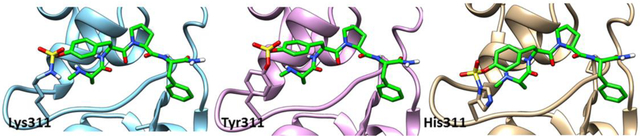
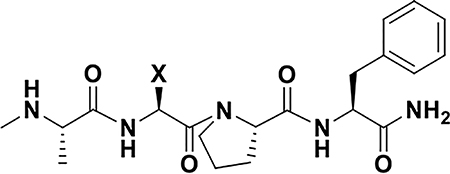
In order to assess the potential of the proposed agents in targeting XIAP BIR3 Lys311, we prepared a number of N-methyl-Ala-X-Pro-Phe-NH2 mimetics as listed in Table 1, which, based on structural considerations, were predicted to juxtapose each of the listed X electrophiles with the side chain of residue Lys311 in XIAP (Figure 2A).33 Subsequently, we characterized the interactions between each agent and the BIR3 domain of XIAP using biochemical and biophysical assays. First, we used a Dissociation-Enhanced Lanthanide Fluorescence Immunoassay (DELFIA) displacement assay platform, as we have recently described39 in which IC50 values represented the ability of the agents to displace the binding of the given test agent from a biotinylated AVPI reference peptide. IC50 values were obtained from dose responses curves measured after 15 min, 2 h or 8 h incubation. A large increase in affinity at longer incubation times was interpreted as possible covalent interaction. Subsequently, each agent was used to measure their induced denaturation thermal shifts (ΔTm) on the BIR3 domain of XIAP. ΔTm values were measured after different ligand/protein incubation times. Non-covalent agents, such as AVPF or compound 1, displayed a ΔTm < 20 °C (after 2 or 6 h incubation), while putative covalent compounds showed significantly larger shifts (ΔTm of > 30 °C; Table 1, and supplementary Figure S1). Finally, presumed covalent agents were verified by SDS gel electrophoresis (Figure 2B) and the rate of reactivity of the agents was assessed by collecting samples at various incubation times. These data clearly suggested that compounds 2, 3, and 4 formed a stable covalent adduct with the BIR3 domain, with a reactivity order: compound 2 > compound 3 > compound 4. Subsequently, to assess their chemical stability, the integrity of each agent was monitored using solution 1H 1D NMR measured over time, at different temperatures and pH values (Figure 2C, Table 1). Of note is that all agents were very stable at low pH (supplementary Figure S2), while the benzamide-sulfonyl fluoride (compound 2) was the least stable at physiological pH = 7.2 (t1/2 ~ 30 min). The aryl-fluoro sulfate (compound 4) and the benzyl-sulfonyl fluoride (compound 3) presented perhaps the best compromise between reactivity and stability in aqueous solution at physiological pH. To further confirm that compounds 2, 3, and 4 targeted Lys311 selectively, we conducted SDS gel electrophoresis measurements with a single point mutant Lys311Ala (Supplementary Figure S3). Moreover, thermal shift data with these agents and the Lys311Ala revealed ΔTm values < 20 °C, also suggesting non-covalent binding (Table 2). In addition, 2D [15N,1H] NMR analyses with a 15N-Lys labeled BIR3 sample and compound 4 revealed a time-dependent chemical shift changes for the backbone amide of Lys 311, presumably due to the covalent bond formation with its side chain over time (Figure 2D). These data collectively demonstrated that compounds 2, 3, and 4 were effective Lys-covalent agents for the BIR3 domain of XIAP targeting Lys 311.
Figure 2. Covalent agents targeting XIAP BIR3 Lys311.
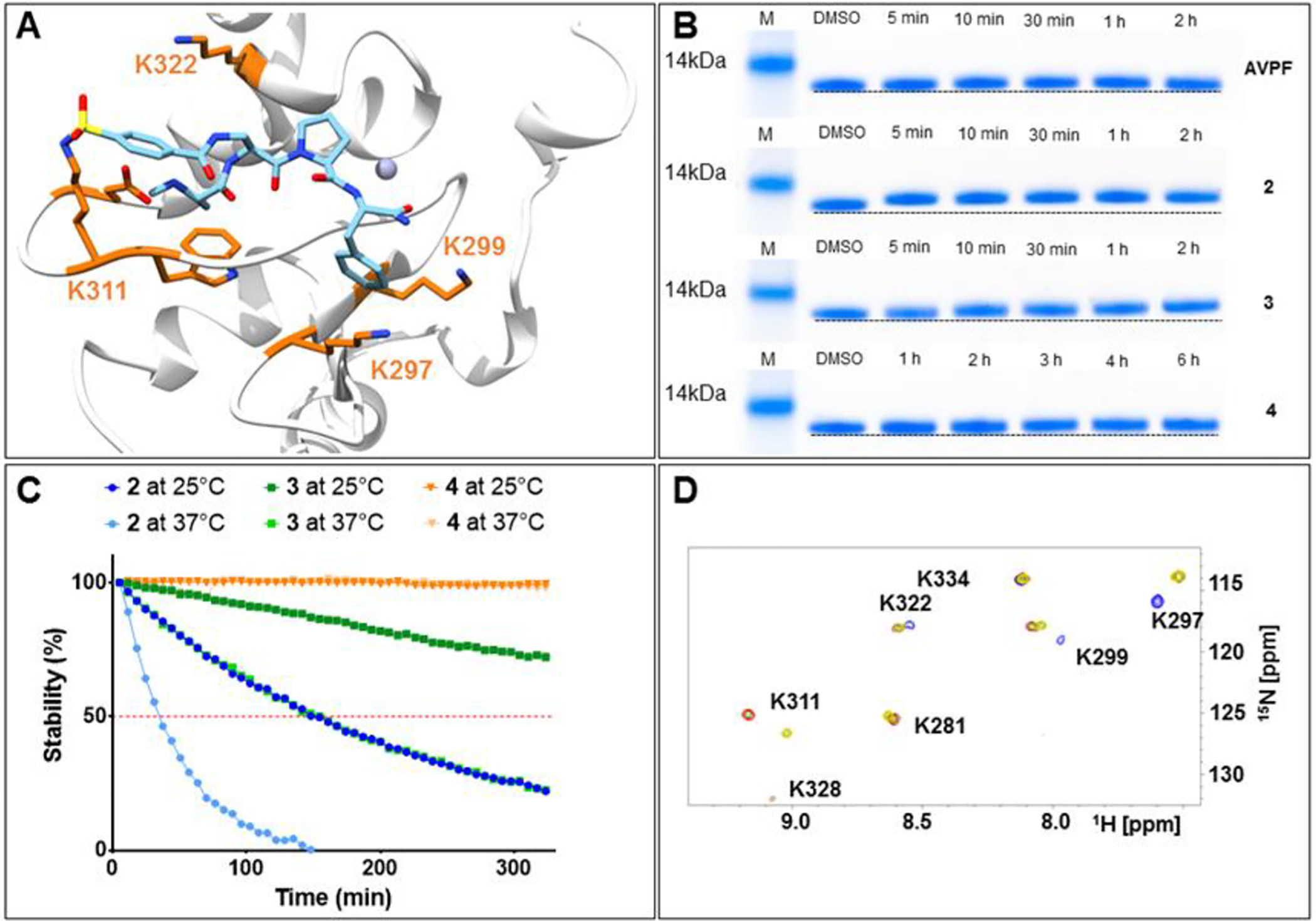
A) Covalent docking pose of compound 2 into the binding pocket of the BIR3 domain of XIAP (PDB ID 2OPZ). B) SDS-PAGE gel electrophoresis followed by Coomassie staining of the BIR3 domain of XIAP in the absence and presence of compounds AVPF, 2, 3, and 4 incubated at different time points. For compounds AVPF, 2, and 3 the incubation was carried out at room temperature, while compound 4 was incubated at 37°C. C) Aqueous stability of compounds 2, 3, and 4 measured at 25°C in 25mM TRIS buffer pH 7.5, 150 mM NaCl, and at 37°C in 50 mM phosphate buffer pH 7.5, 150 mM NaCl. The stability has been measured by NMR spectroscopy by measuring the decrease in peak intensity in the aromatic region of the spectrum. D) [1H, 15N]-HSQC spectra of 20 μM BIR3 domain of XIAP selectively labeled with 15N-Lysine (blue spectrum) in the presence of 40 μM of compound 4, recorded at different incubation times: after 30 min (red), after 4 h (green), and after 7 h 30 min (yellow). The spectra were recorded in 25 mM TRIS pH 8, 150 mM NaCl at 25°C.
Targeting Tyr, Ser, Thr, and His residues.
In order to assess whether any aryl-sulfonyl fluorides or aryl-fluoro sulfates were capable to react irreversibly with other potential nucleophilic side chains in the protein target, we used the BIR3 domain of XIAP as a model system and prepared several single point mutants. These mutants introduced Tyr, Ser, Thr, or His in lieu of Lys311, and we tested each mutant against a small library of peptides (Table 2). Hence, each agent was tested against each mutant in the DELFIA assay after 2 h and 8 h incubation. In addition, ligand-induced thermal shift measurements were collected after 2 h and 6 h incubation times for each agent against each mutant (Table 2).
Time-dependent decreases in IC50 values and concomitant large ΔTm values (> 20 °C) were interpreted as caused by possible covalent interactions. These criteria identified aryl-sulfonyl fluorides compounds 2, 3 (Figure 3A), and aryl-fluoro sulfate 4 as putative covalent agents for mutant Lys311Tyr (Table 2). In addition, compounds 2 and 3 (Figure 3B) also exhibited potential covalent interactions with mutant Lys311His and, albeit it to a much lesser extent, also Lys311Ser and Lys311Thr (Table 2, supplementary Figure S1). Interestingly, IC50 and ΔTm values for compounds 5 and 6 did not suggest significant covalent interactions with any of the mutants, while compounds 7 and 8 displayed some possible covalent interactions with wt-BIR3, with the Lys311Tyr mutant, and with the Lys311His mutant (Table 2), indicating that proper juxtaposition of the electrophile with the targeting amino acid is essential for the reaction. Subsequently, to further confirm possible covalent interactions with the Lys311Tyr and with Lys311His mutants, respectively, SDS gel electrophoresis and mass spectrometry analyses were conducted (Figures 3C, D). Excluding compound 2 that seemed particular unstable in aqueous buffer (Figure 1), these data concluded that benzyl-sulfonyl fluorides (such as compound 3) can be effective in targeting Lys, Tyr, or His residues, while aryl-fluoro sulfates (such as compounds 4, 7, 8) can target Lys, Tyr, and perhaps also His residues (albeit at a slower rate). Hence, these electrophiles could be properly incorporated into peptides (Figure 1) to target Lys, Tyr, or His residues.
Figure 3. Covalent agents targeting XIAP BIR3 Lys311Tyr and Lys311His.
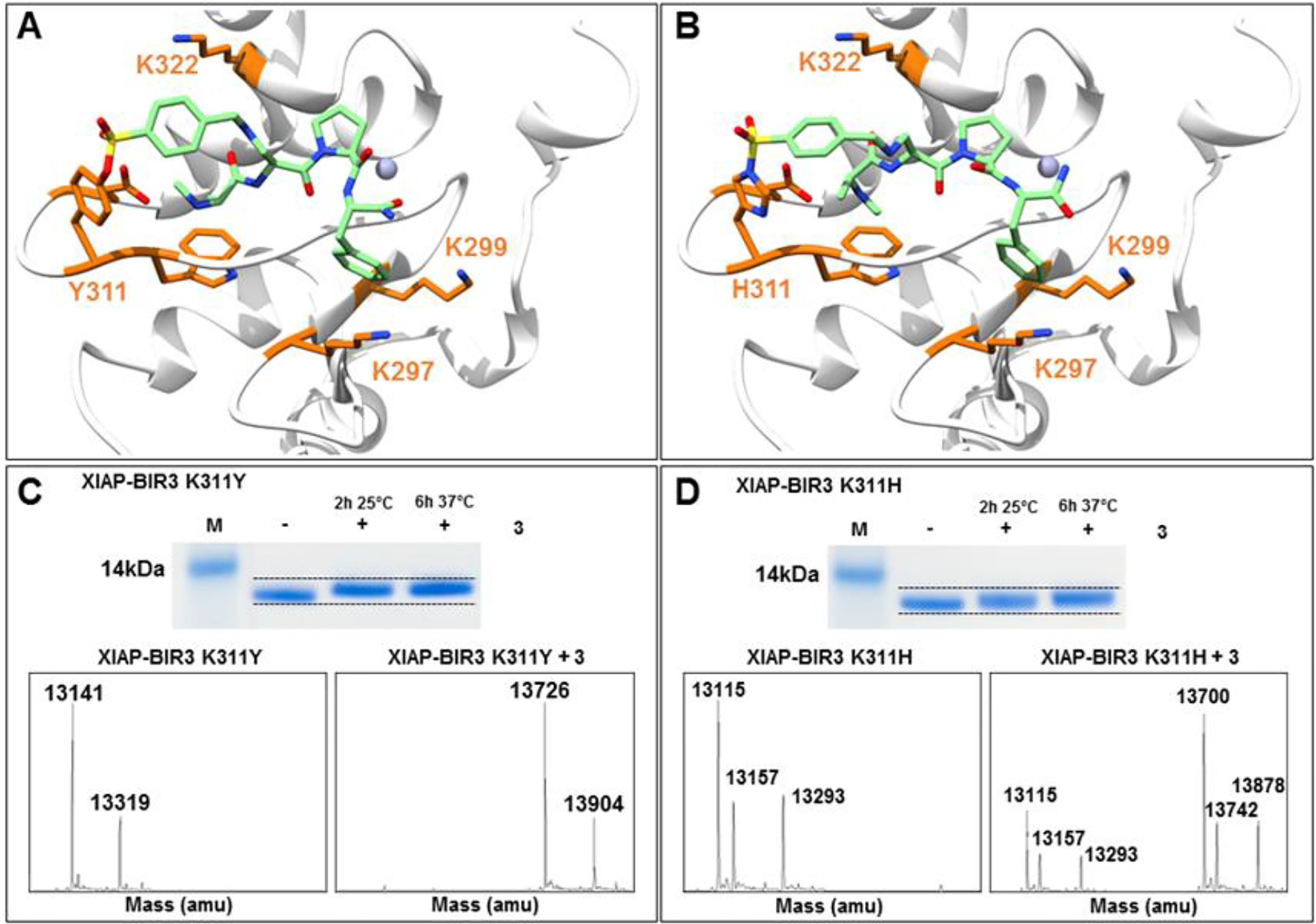
A) Covalent docking pose of compound 3 into the binding pocket of the Lys311Tyr mutant of BIR3 domain of XIAP (PDB ID 2OPZ). B) Covalent docking pose of compound 3 into the binding pocket of the Lys311His mutant of BIR3 domain of XIAP (PDB ID 2OPZ). C) SDS-PAGE gel electrophoresis followed by Coomassie staining of the Lys311Tyr mutant BIR3 domain of XIAP in the absence and presence of compound 3 incubated for 2 h at room temperature, and for 6h at 37°C. The bottom panel reports the LC-MS spectra of the Lys311Tyr mutant BIR3 domain of XIAP in the absence (left) and presence (right) of compound 3 incubated at room temperature. D) SDS-PAGE gel electrophoresis followed by Coomassie staining of the Lys311His mutant BIR3 domain of XIAP in the absence and presence of compound 3 incubated for 2 h at room temperature, and for 6 h at 37°C. The bottom panel reports the LC-MS spectra of the Lys311His mutant BIR3 domain of XIAP in the absence (left) and presence (right) of compound 3 incubated at 37°C.
Plasma stability, cell permeability, and cellular activity of covalent agents.
While in vitro studies with purified target indicated that electrophiles in the compounds 3 and 4 could be effective Lys-, Tyr-, or His-covalent agents, their use as pharmacological tools or future therapeutics requires the agents to be stable not only in buffer (our studies suggested that compound 2 was deemed too unstable in aqueous buffer to be used in further studies), but also in plasma, and to be cell permeable. Hence, to further make these critical determinations we first synthesized compounds 9, 10, and 11 listed in Table 3, in which the Phe residue in the Ala-X-Pro-Phe-NH2 peptides was replaced by amino-indanes, that together with the methylation of the N-terminal Ala, increased drug-likeness and cell-permeability of the peptide mimetics. Table 3 reports their structures together with the IC50 values against XIAP BIR3 domain after 2 h and 8 h incubation, as well as thermal shift values measured after 2 h and 6 h incubation times. As references, the antagonists LCL161 (Novartis) and GDC0152 (Genentech)15 were also tested.
Table 3.
Displacement assays, thermal shift wdata, and plasma stability and cellular activity of aryl-sulfonyl fluorides and aryl-fluoro sulfates.
| ID | Structure | IC50 XIAP-BIR3 [nM]a | EC50 (SKOV3) [nM]b | Plasma Stabilityc | ΔTm [°C]d |
|---|---|---|---|---|---|
| LCL161 | 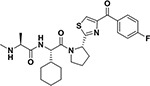 |
48 ± 5 53 ± 3 |
3 ± 5 1 ± 0.5 |
> 2h | 14 18.5 |
| GDC-0152 | 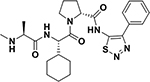 |
21 ± 1 21 ± 2 |
3 ± 6 2.5 ± 7 |
~1 h | 15 19.5 |
| 9 | 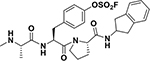 |
239 ± 27 65 ± 19 |
400 ± 2 300 ± 1 |
> 2h | 7.5 31.0 |
| 10 | 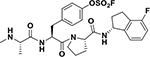 |
63 ± 6 28 ± 6 |
7 ± 2.5 4 ± 2.5 |
> 2h | 9.5 33.5 |
| 11 |  |
24 ± 1 3.5 ± 0.1 |
43 ± 2 50 ±1 |
~15 min | 35 34.5 |
IC50 values were calculated from dose-response curves obtained after incubating protein and ligand for 2 h or 8 h at room temperature.
EC50 values estimated form the apoptosis assay measured after 24 h (top value) or 48 hr (bottom value; Figure 4C).
Plasma stability refers to measurements of compounds’ integrity (LC/MS method) at various time points after incubation with mouse plasma.
Thermal shift (ΔTm) data were obtained incubating protein and ligand for 2 h at room temperature or 6 h at 37°C. The data reported represent the major peak observed (see supplementary information for individual curves).
All tested agents were relatively plasma stable, with the exception of compound 11 that was completely degraded in plasma after 40 minutes (Table 3). Next, to assess cell permeability of these agents, we obtained a cell line that is stably transfected with HA-BIR3 of XIAP.40 Exposure of compounds to this cell line, followed by western blot analyses of cell lysates using an anti-HA antibody was used to assess cell permeability of our agents. This could be appreciated by a small shift in molecular weight (band shift) similar to what we observed in the SDS gel electrophoresis in vitro. However, we noticed that the unbound BIR3 domain of XIAP is fairly unstable at 37 °C, as revealed by 1D 1H NMR measurements (Figure 4A), while the domain gets stabilized by ligand binding, as indicated by both 1D 1H NMR (Figure 4A) and by our thermal shift studies (Tables 1–3). Hence, exposing the HA-BIR3 expressing HEK293 cell line to cell permeable compound GDC0152 resulted in the stabilization of the BIR3 domain that resulted in an increase in band intensity in the western blot. On the contrary, exposure of cells to cell impermeable AVPF did not result in a visible band. Similarly, compounds 9 and 10 and 11 stabilized HA-BIR3 in this experiment and also induced an appreciable gel shift, both consistent with cell permeability of the agents and covalent adduct formation (Figure 4B).
Figure 4. Cellular permeability and efficacy of selected agents.
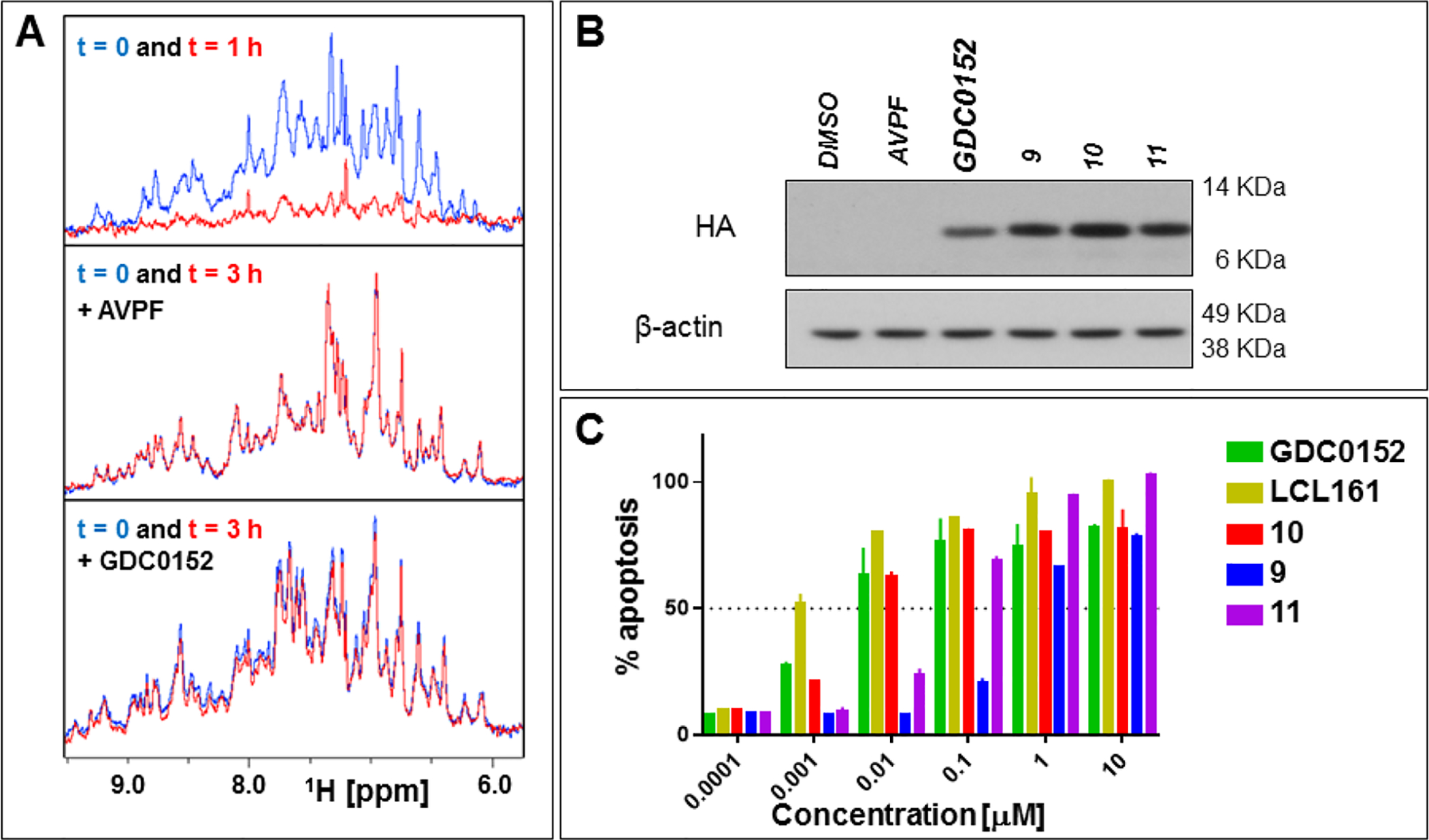
A) 1H-1D NMR spectra of the amide/aromatic region of 20 μM BIR3 domain of XIAP in the apo form (top panel), in the presence of 40 μM of AVPF (middle panel), or in the presence of 40 μM of GDC0152 (bottom panel). The blue spectra were collected at time 0, the red the spectra were collected after 1 h at 37°C for the apo protein, and after 3h at 37°C in presence of AVPF, or GDC0152. B) Immunoblot of HEK293 expressing HA-tagged BIR3 domain of XIAP. The lysates were collected from cells that were exposed for 6 h to 10 μM of the indicated compounds. C) SKOV3 cells were exposed to different compounds at various concentrations. Subsequently, the plate was imaged by IncuCyte S3 live-cell analysis system and images were collected every 2 h for 48 h and data were analyzed using the IncuCyte S3 basic analyzer. The histogram represents data collected at 48 h.
To further assess if these surrogate cell permeability data translated into cellular efficacy, we tested the ability of these agents to induce caspase-3 activation in the ovarian cancer cell line SKOV3 that has been reported previously to be very sensitive to XIAP antagonists (Table 3, Figure 4C).41 EC50 values obtained with various agents are reported in Table 3 and parallel the in vitro IC50 values, as well as the plasma stability of the compounds.
Discussion and conclusions
While targeted therapy strategies have been increasingly more successful in bringing new agents to the clinic, the design of effective therapeutics targeting protein-protein interactions (PPIs) is lagging behind despite the fact that PPIs represent potentially a large class of viable therapeutic targets. While several potentially effective strategies to tackle PPIs have emerged in the past decade, including fragment-, structure-, and/or NMR-based approaches, phage display, and DNA encoded libraries,42–44 these strategies still struggle to effectively optimize initial binding agents into potent, selective, and cell permeable ligands for continued target validation and lead optimizations. Recent years have also witnessed a resurgence of targeted covalent therapeutics with several newly FDA approved covalent agents targeting surface Cys residues.45–47 More recently, Lys covalent agents have also emerged, using a variety of possible warheads.35 Few reports have appeared that successfully demonstrate covalent targeting of Lys residues in active sites of proteins by introduction of appropriately placed electrophiles on an existing ligand.35, 48, 49 Most excitingly, similar studies revealed that it is possible to target surface Lys residues located at protein-protein interfaces, including the recent examples of a covalent Mcl-1 inhibitor,50 a covalent inhibitor of MDM2/P53 interactions,51 and our recent studies targeting Lys311 of XIAP.33 Here, we further investigated the use of aryl-sulfonyl fluorides or aryl-fluoro sulfates as possible warheads to target not only Lys but also other surface aminoacids, such as Tyr, His, Ser, and Thr. As mentioned, we have recently reported on the first XIAP covalent agent incorporating a benzamide-sulfonyl fluoride in lieu of the valine residue in its tetra-peptide ligand of sequence Ala-Val-Pro-Phe-NH2.33 Because simple N-methylation of the amino terminal residue and replacements of C-terminal Phe in the tetra-peptide with a variety of aryl compounds led to clinical candidates, one could envision that simple strategies aimed at identifying short linear binding peptides (perhaps using the HTS by NMR approach, phage display, or guided by structural studies of the PPI as in the case of the XIAP/SMAC complex), followed by peptide derivatization with proper electrophiles to target Lys, Tyr, His, Ser, or Thr, could provide an effective platform for the design of therapeutically viable covalent PPIs antagonists. Hence, to extend previous findings on targeting surface Cys residues located in proximity of binding sites in PPIs,31, 32 we sought here to investigate whether aryl-sulfonyl fluorides or aryl-fluoro sulfates, which could be easily incorporated into binding peptides (Figure 1) could be deployed to target Lys, Tyr, His, Ser, or Thr. In addition to measuring their basic pharmacological properties to evaluate their suitability as chemical probes or even therapeutics.
Our recent NMR and enthalpy measurements with Ala-X-Pro-Phe-NH2 agents indicated that compound 1 (with X= L-Phe(p-sulfone) could form a salt bridge with Lys311,39 and that when X = L-Dap(benzamide-psulfonyl fluoride), the resulting agent (compound 2) was effective in targeting covalently Lys311.33 Hence, we further incorporated a L-Dap(benzyl-p-sulfonyl fluoride) (compound 3) and, aided by a simple scheme reported in Figure 1C, L-Phe(p-fluoro sulfate) (compound 4) was also obtained. Additional compounds incorporating chains of various lengths between the backbone and the electrophiles (compounds 5–8) were also obtained using the general schemes of Figure 1 and are reported in Table 2. Each agent was subsequently tested against wt-BIR3 domain of XIAP (hence probing their interaction with Lys311; Figure 2) and against a variety of Lys311 mutants, including Lys311Ala, Lys311Tyr, Lys311His, Lys311Ser, and Lys311Thr (Table 2). Two general measurements were conducted: first, IC50 values using a displacement DELFIA assay were measured after incubation of ligand and protein at various times (Tables 1 and 2); second, thermal shift data with each ligand/protein pairs were obtained, and also measured after different incubation times (Tables 1 and 2). Ligands/protein complexes that displayed a time-dependent decrease in IC50 values and concomitantly increased ΔTm values (> 20 °C), compared to those observed with reference non-covalent agents, were interpreted as potential covalent complexes. The isolated BIR3 domain of XIAP in its unbound form is relatively unstable at 37 °C. Indeed, thermal shift measurements indicated a relatively large effect by non-covalent agents such as the peptide AVPF (ΔTm = 17 °C), the Genentech agent GDG0152 (ΔTm = 19.5 °C), or the Novartis clinical candidate (https://clinicaltrials.gov/ct2/show/NCT01934634) LCL161 (ΔTm = 18.5 °C), after 6 h incubation (Tables 1 and 3). In agreement with the covalent nature of the interactions provided by compounds 2, 3, and 4 against wt-BIR3, their thermal shifts were dramatically larger under the same experimental conditions (ΔTm = 37 °C, 35 °C, and 33 °C, respectively, after 6 h incubation; Table 1). Interestingly, the thermal shifts for the aryl-fluoro sulfate compound 4 after 2 h incubation are closer to those of non-covalent agents, suggesting a slower reaction rate. These observations were further corroborated by SDS gel electrophoresis collected on compounds 2, 3, and 4 in complex with wt-BIR3 after various incubation times (Figure 2B). These studies indicated the relative Lysreactivity order: benzamide-sulfonyl fluoride (compound 2) > benzyl-sulfonyl fluoride (compound 3) > arylfluoro sulfate (compound 4), that paralleled the relative aqueous reactivity of these agents (Figure 2C). ΔTm values with these agents were also collected against the Lys311Ala mutant (Table 2). Of note is that a minor peak in the thermal shift spectra of the Lys311Ala in complex with compounds 3, after 6 h incubation, is visible corresponding to a ΔTm of about 29 °C (supplementary Figure S1). It is possible that at longer incubation times, these agents could reach a nearby Lys322 (Figure 1A). This is consistent with a SDS gel shift assay conducted with agents 2, 3 and 4, performed against a Lys311Ala mutant, where a broader band is observed only for compound 3 (Supplementary Figure S4). Albeit more speculative, 2D [15N,1H] HSQC NMR data (using 15N-Lys selectively labeled wt-BIR3 domain of XIAP) was also consistent with covalent interactions, with covalent agents inducing time-dependent chemical shift perturbations of the backbone amide of Lys311 compared to non-covalent agents (Figure 2D).
Analysis of IC50 and ΔTm values with the compound library tested against the various mutants revealed that compounds 2, 3, and 4 could be putative covalent agents for Lys311, but also Tyr311, or His 311. SDS gel collected with the Lys311Tyr mutant after exposure to putative covalent compound 3 (Table 2) revealed that this agent forms potentially a covalent adduct with this mutant, as also confirmed by mass spectrometry analyses (Figure 3A, B). Likewise, data with agents 2, 3, 7, 8 (Table 2) suggested possible covalent interaction with the mutant Lys311His, as corroborated by the SDS gel shift and mass spectrometry data with compound 3 (Figure 3D), albeit the reactions required longer incubation times to be completed, as compared to those observed with Lys and Tyr. Hence, collectively these data identified compounds 2, 3, 4, 7, and 8 as putative Lys/Tyr/His covalent agents.
To further ascertain if these agents could be used as pharmacological tools and eventually be moved along into lead optimization studies, we first probed their aqueous stability at various temperatures and pH values. Not surprisingly, all agents were very stable at low pH, while various degrees of instability were observed at physiological pH, with the benzamide-sulfonyl fluoride (compound 2) being the least stable, the aryl-fluoro sulfate (compound 4) being the most stable (Table 1), and the benzyl-sulfonyl fluoride (compound 3) displaying intermediate stability. These chemical stability data also paralleled the rate of reactivity of the agents for Lys311 (Figure 2B). On balance, it seemed that the aryl-fluoro sulfates and the benzyl-sulfonyl fluorides may possess the best compromise between aqueous stability and reactivity with the binding site nucleophiles among the agents tested. To further investigate plasma stability and cell permeability of these agents, we further incorporated Phe mimetics39 into the N-Me-Ala-X-Pro-Phe-NH2 agents 3 and 4, resulting in agents 9, 10, and 11 (Table 3), and compared their pharmacological properties with LCL161 and GDC0152. Not surprisingly, all tested agents were found to be plasma-stable under the same experimental conditions, with the notable exception of 11 that still displayed a relatively short plasma half-life (Table 3). Next, we obtained an HA tagged BIR3 construct and transfected HEK293 cells. The isolated BIR3 domain of XIAP, in its unbound form, seemed particularly unstable at 37 °C as revealed by our thermal shift studies (Tables 1–3) and by 1D 1H NMR (Figure 4A). In agreement, western blot analysis using anti-HA antibody of HEK293 cell lysates that express HA-BIR3, revealed only a fainted band (if any) for HA. Exposing the cells to GDC0152, but not to Ala-Val-Pro-Phe-NH2 which is not cell permeable, resulted in the stabilization of the BIR3 domain (again, as also observed in vitro by NMR and by thermal shift assays) as it is manifested in an intense band in the western blot (Figure 4B). Similarly, exposing cells to covalent agents 9, 10 and 11, resulted in intense bands that are also appreciably shifted, suggesting that each agent was cell permeable and covalently interacted with the target in cell. Finally, to further corroborate these data, we preliminarily assessed cellular efficacy of these agents by measuring their ability to induce cell apoptosis in the ovarian cancer cell line SKOV3 that has been reported to be very sensitive to XIAP antagonists (Table 3, Figure 4C).41 Using the IncuCyte® Cytotox Green apoptosis assay with different compounds in Table 3 at various concentrations, we observed nanomolar efficacy with all agents, with EC50 values that all in all paralleled the observed in vitro data. For example, compound 11, despite being most active in vitro, is also the least stable in plasma, and thus was less active in cell. Compound 9 was less potent in vitro and in cell, while compound 10 seemed to possess the best properties in terms of affinity for XIAP and plasma stability, resulting in the most efficacious agent (Table 3, Figure 4C). More extensive studies with a battery of cell lines are ongoing and fall outside the scope of this manuscript. Hence, the full potential of these agents as therapeutics in oncology and for other indications such as pulmonary fibrosis,52 has yet to be fully determined.
In conclusion, taken together our studies identified possible simple avenues to derive covalent ligands to PPIs starting from short binding peptides of modest affinity (single digit micromolar). These initial binding peptides could be obtained by structural studies of the binding partners, by phage display, or by other approaches such as our recently proposed HTS by NMR, for example.53–58 Structural studies of the identified peptide in complex with the target could suggest possible derivatizations with benzyl-sulfonyl fluorides or aryl-fluoro sulfates to reach Lys, Tyr, or His, or Lys of Tyr, respectively.59 Given the ease of synthesis in incorporating these electrophiles into short peptides, absent of structural studies, one can also envision introducing these residues systematically at various positions of a binding peptide and use simple follow up characterizations, including thermal shift measurements, and/or SDS gel electrophoresis, and/or mass spectrometry, and/or IC50 measurements after various incubation times, to investigate if any of the agents can form a covalent adduct with the target.37 However, caution must be taken when incorporating aryl-sulfonyl fluorides, despite it may be appealing to use these electrophiles to target both Lys33 or Tyr,60, 61 as these are too unstable to be used as chemical probes, let alone for therapeutics in current forms. Benzamide sulfonyl fluorides were very unstable in buffer, hence our data suggest that these electrophiles should be avoided. The benzyl-sulfonyl fluorides seemed significantly more stable in buffer, but it resulted still fairly unstable in plasma. Perhaps introduction of deactivating groups in the phenyl ring could ameliorate this problem. Therefore, all in all, the aryl-fluoro sulfates should be preferred when targeting Lys or Tyr residues, and potentially also His residues, given their increased stability in buffer and plasma. Hence, absent of structural information on the complex, a systematic approach introducing aryl-fluoro sulfates reported in Figure 1C,D on a short binding peptide seems entirely feasible, entailing the synthesis of a few dozen agents. However, available SAR studies and other considerations can also be used to narrow down the number of residues where the electrophiles could be placed. For example, binding peptides containing glutamic or aspartic residues (or other residues for which there are reasons to believe that are solvent-exposed) are likely the best candidates for substitution with electrophiles targeting Lys. Likewise, Tyr or Phe residues in binding peptides could be directly replaced with aryl-fluoro sulfates in the attempt to reach possible binding site Tyr or His residues in their proximity. Our data also suggest that measurements at various times of 2D [15N,1H] NMR correlation spectra of 15N-Lys-specific labeled spectra of a given protein target could be used as a rapid screening method to test libraries of aryl-fluoro sulfated containing peptides or peptide mimetics, perhaps arranged in a positional scanning fashion.53–58 This approach could be easily adapted to detect Tyr, via 13C labeling, or His, via direct NMR measurements of their side chains,56 and we are currently investigating these possible applications.
With the resurgence and the success of covalent drugs, and the paucity of effective PPIs antagonists in the clinic, we are confident that our studies provide novel and practical insights into the optimization and derivation of effective covalent peptides that widen the target space from the Cysteinome45–47 to other more abundant residues such as Lys, Tyr, or even His. Applied to PPIs, we are confident that such covalent agents could represent significant stepping-stones in the development of novel pharmacological tools or even drug candidates against such a large class of therapeutic targets.
Experimental Section
General Chemistry.
Solvent and reagents were commercially obtained and used without further purification. NMR spectra used to check concentration were recorded on Bruker Avance III 700MHz. High-resolution mass spectral data were acquired on an Agilent LC-TOF instrument. RP-HPLC purifications were performed on a JASCO preparative system equipped with a PDA detector and a fraction collector controlled by a ChromNAV system (JASCO) on a Luna C18 10μ 10 × 250mm (Phenomenex) to > 95% purity. LCL161 and GDG0152 were obtained from MedChem Express. 2-Chlorotritylchloride and Rink amide resins were purchased from Novabiochem. Fmoc-amino acids were purchased from Chem-Impex and Novabiochem. The AISF reagent was obtained by Paloza Molecule. Peptides were synthesized by using standard solid phase protocols or using standard microwave-assisted Fmoc peptide synthesis protocols with a Liberty Blue Peptide Synthesizer (CEM). For each coupling reaction, 6 eq. of Fmoc-AA, 3 eq. of DIC and 1 eq. of OximaPure in 4.5 ml of DMF were used. The coupling reaction was allowed to proceed for 5 min at 90°C. ivDde deprotection was performed using 4% N2H2 in DMF (3×5ml), at room temperture. Fmoc deprotection was performed by treating the resin-bound peptide with 20% 4-methylpiperidine in DMF (2×3ml) for 3 min at 90°C. Peptides were cleaved from Rink amide resin with a cleavage cocktail containing TFA/TIS/water/phenol (94:2:2:2) for 3 h. The cleaving solution was filtered from the resin, evaporated under reduced pressure and the peptides precipitated in Et2O, centrifuged and dried in high vacuum. The crude peptide was purified by preparative RP-HPLC using a Luna C18 column (Phenomenex) and water/acetonitrile gradient (5% to 100%) containing 0.1% TFA. The final compounds were characterized by HRMS. Detailed experimental procedures for key compounds are reported below.
Compound 9: 4-((S)-3-((S)-2-((2,3-dihydro-1H-inden-2-yl)carbamoyl)pyrrolidin-1-yl)-2-((S)-2(methylamino)propanamido)-3-oxopropyl)phenyl fluoro-sulfate.
2-Chlorotrityl chloride resin was used as solid-phase support (0.05 mmol scale), and the previously described coupling conditions were used to obtain the peptidic part of the agent. Aryl-floro sulfate incorporation was performed on resin, using [4-(acetylamino)phenyl]imidodisulfuryl difluoride (AISF) reagent (1.2 eq., 2.2 eq. of DBU in THF, overnight reaction at room temperature). The protected sequence was then cleaved from the resin using a 2% TFA solution in DCM for 1h then purified by preparative RP-HPLC using a Luna C18 column (Phenomenex) and water/acetonitrile gradient (5% to 100%) containing 0.1% TFA. The purified material was coupled with 2aminoindane (3eq., 3 eq. HATU and 5 eq. DIPEA in DMF, overnight reaction at room temperature), then completely deprotected using HCl 4N in dioxane for 30 min, and finally purified using the previous described method to obtain a white powder (12.6 mg, 42.8%). HRMS: calcd 560.2107 (M+H)+; obs 561.2179 (M+H)+, 583.1993 (M+Na)+.
Compound 10: 4-((S)-3-((S)-2-(((R)-4-fluoro-2,3-dihydro-1H-inden-1-yl)carbamoyl)pyrrolidin-1-yl)-2-((S)-2(methylamino)propanamido)-3-oxopropyl)phenyl fluoro-sulfate.
2-Chlorotrityl chloride resin was used as solid-phase support (0.05 mmol scale), and the previously described coupling conditions were used to obtain the peptidic part of the agent. Aryl-floro sulfate incorporation was performed on resin, using [4-(acetylamino)phenyl]imidodisulfuryl difluoride (AISF)37 reagent (1.2 eq., 2.2 eq. of DBU in THF, overnight reaction at room temperature). The protected sequence was then cleaved from the resin using a 2% TFA solution in DCM for 1h then purified by preparative RP-HPLC using a Luna C18 column (Phenomenex) and water/acetonitrile gradient (5% to 100%) containing 0.1% TFA. The purified material was coupled with 4-Fluoro1R-aminoindane (3eq., 3 eq. HATU and 5 eq. DIPEA in DMF, overnight reaction at room temperature), then completely deprotected using HCl 4N in dioxane for 30 min, and finally purified using the previous described method to obtain a white powder (15.3 mg, 52.9%). HRMS: calcd 578.2011 (M+H)+; obs 601.2416 (M+Na)+.
Compound 11: 4-((((S)-3-((S)-2-(((R)-4-fluoro-2,3-dihydro-1H-inden-1-yl)carbamoyl)pyrrolidin-1-yl)-2-((S)-2(methylamino)propanamido)-3-oxopropyl)amino)methyl)benzenesulfonyl fluoride.
2-Chlorotrityl chloride resin was used as solid-phase support (0.05 mmol scale), and the previously described coupling conditions were used to obtain the peptidic part of the agent. ivDde protecting group was removed as previously described, and 4(bromomethyl)benzenesulfonyl fluoride (1.2 eq., 3 eq. of DIPEA in DMF, overnight reaction at room temperature) was added to the reactor. The protected sequence was then cleaved from the resin using a 2% TFA solution in DCM for 1h then purified by preparative RP-HPLC using a Luna C18 column (Phenomenex) and water/acetonitrile gradient (5% to 100%) containing 0.1% TFA. The purified material was coupled with 4-Fluoro1R-aminoindane (3eq., 3 eq. HATU and 5 eq. DIPEA in DMF, overnight reaction at room temperature), then completely deprotected using HCl 4N in dioxane for 30 min, and finally purified using the previous described method to obtain a white powder (9.9 mg, 33.5%). HRMS: calcd 591.2327 (M+H)+; obs 592.2403 (M+H)+, 614.221 (M+Na)+.
Protein expression and purification
For the expression of XIAP BIR3, a pET15b vector encoding for the human BIR3 domain of XIAP fragment (residues 253–347) and an N-terminal His tag was transformed into E. coli BL21(DE3) Gold cells and expressed as reported previously.33 For the expression of 15N-Lys labeled BIR3, 200 mg of U-15N Lys (SIGMA) were added to 1L of minimal media just prior induction with 1mM IPTG overnight at 25°C, as recently published.62 Bacteria were collected and lysed by sonication at 4°C and proteins were purified using Ni2+ affinity chromatography. The final buffer used for the eluted protein was finally exchanged using a desalting column into an aqueous buffer composed of 25 mM Tris pH = 8.0, 150 mM NaCl, 50 μM Zn(Ac)2, and 1 mM DTT. The BIR3 domain of XIAP where the Lys 311 was mutated to either Ala, Tyr, His, Ser, or Thr, were expressed in the same way described above.
Thermal Shift Assay
Thermal shift assays for BIR3 construct/inhibitor complexes were conducted using a BioRad CFX Connect Real-Time PCR Detection System, with studies on each inhibitor/protein complex being conducted in triplicate. Incubation of BIR3 protein with inhibitor followed one of two parameters, either 37°C for 6 hours or 25°C for 2 hours. Protein/inhibitor complexes and 5000x SYPRO Orange dye (Sigma) were diluted using reaction buffer, 50 mM Tris pH 8.0, 150 mM NaCl, 50 μM Zinc acetate, to obtain final concentrations of 5 μM BIR3, 10 μM inhibitor, and 60x SYPRO Orange. Sample plates were heated from 10°C to 95°C with heating increments of 0.5°C, over 30 min. Fluorescence intensity was measured within the excitation/emission ranges 470–505/540–700 nm.
Gel Electrophoresis
10 μM of each protein were incubated at various times with 20 μM of compounds in 25 mM TRIS at pH 8, 150 mM NaCl, 50 μM zinc acetate, and 1 mM DTT buffer. Samples were subjected to gel electrophoresis with SDS-PAGE gel using the NuPAGE 12% bis-tris mini gels (Life Technologies), and MES as running buffer. Gels were subsequently stained with SimplyBlue SafeStain (Life Technologies) according to the manufacture’s protocol.
Immunoblot study
The HA-XIAP-BIR3 plasmid was a gift from Dr. Colin Duckett (Addgene plasmid # 25689). One million HEK293 cells were plated in 6-well plates and left to attach overnight. The following day, cells were transfected with 1 μg of HA-XIAP-BIR3 plasmid using Lipofectamine 2000 (ThermoFisher) in complete DMEM media. 18 h post-transfection, media was replaced with serum-free DMEM containing 10 μM of compounds (1% of DMSO) and incubated for an additional 6 h. Finally, cells were lysed with lysis buffer (20 mM Tris, pH 7.4, 120 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1% IGEPAL, 5 mM EDTA) supplemented with EDTA-free Protease Inhibitor Cocktail and PhosStop (Sigma-Aldrich) for 10 min on cold ice. Lysates were centrifuged and supernatants were collected. Protein content was quantified and samples were prepared using NuPAGE antioxidant and LDS sample buffer (ThermoFisher) and heated for 10 min at 70°C. Each sample containing 10 μg of proteins were loaded into 12% NuPAGE Bis-Tris precast gels and transferred to PVDF membranes. The membranes were blocked with 5% milk in TBS and 0.1% Tween (TBST) and incubated with anti-HA (Y-11, sc-805, Santa Cruz Biotechnology) overnight at 4°C. Next day, the membrane was washed with TBST and incubated with goat anti-rabbit HRP secondary antibodies. The antigen-antibody complexes were visualized using a Clarity Western ECL kit (BIO-RAD). The membrane was stripped, and the western blot was repeated using β-actin primary antibody (sc-69879, Santa Cruz Biotechnology) to check for loading.
Apoptosis assay
Human ovarian cancer cell line SKOV3 was obtained from the American Type Culture Collection (ATCC; www.atcc.org) and cultured according to standard mammalian tissue culture protocols, and sterile technique in McCoy’s 5a modified medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin/100 μg/ml streptomycin. Cells were plated in 96-well plates at 1 × 104 cells/well and incubated to attach overnight. The following day, media was replenished with fresh media containing IncuCyte® Cytotox Green and cells were exposed to the different compounds in Table 3 (Figure 4C) at various concentrations. Subsequently, the plate was imaged by IncuCyte S3 live-cell analysis system and images were collected every 2 h for 48 h and data were analyzed using the IncuCyte S3 basic analyzer.
Plasma stability
Mouse plasma (GenTex: GTX73236) was diluted to 80% with 0.05 M PBS (pH 7.4) at 37°C. The reactions were initiated by the addition of the test compounds to 1 ml of preheated plasma solution to yield a final concentration of 200 μM (37°C in triplicate). Samples (50 μl) were subsequently taken at 0, 15, 30, 45, 60, 90 and 120 min and added to 200 μl acetonitrile (4°C) in order to deproteinize the plasma. After vortex mixing for 1 min and centrifugation at 4°C for 15 min at 14,000 rpm, the clear supernatants were analyzed by Mass Spec Analysis. The values represent the mean of three independent experiments.
Molecular modeling
Covalent docking of compounds in Figures 2 and 3, was obtained using Gold (Cambridge Crystallographic Data Center; www.ccdc.cam.ac.uk) and Protein Data Bank entry 2OPZ. The covalent models reported for compound 10 and wt-BIR3, Lys311Tyr BIR3, and Lys311His-BIR3 were prepared form the Protein Data Bank entry 2OPZ and modified using SYBYL-X 2.1.1 (Certara, Princeton, NJ), including energy minimizations. The table of content graphic figure was generated using Chimera (http://www.cgl.ucsf.edu/chimera; UCSF). The coordinates for compound 10 in complex with wt-BIR3 and the mutants are included as supplementary information.
DELFIA (Dissociation-Enhanced Lanthanide Fluorescence Immunoassay)
The assay was conducted using the same protocol as we recently reported33 measured using the VICTOR X5 microplate reader (PerkinElmer) with excitation and emission wavelengths of 340 and 615 nm, respectively. The final protein concentrations were 30 nM for each XIAP BIR3 mutant and the final antibody concentrations used were 22.2 ng/well. DELFIA assay buffer (PerkinElmer) was used to prepare the protein, peptide and antibody solutions and the incubations were done at room temperature and different times as indicated in Tables 1–3. All of the samples were normalized to 1% DMSO and reported as % inhibition. The IC50 values were calculated from dose-response curves by GraphPad Prism version 7 and the reported SE was derived from replicate measurements.
Supplementary Material
Acknowledgements
Financial support was obtained in part by the NIH, with grants CA168517 and NS107479 (to MP) and a City of Hope - UC Riverside Biomedical Research Initiative (CUBRI) grant (to MP), and UC CRCC CRN-18524906 and UCOP LFR-17-476732 grants to JJPP. MP holds the Daniel Hays Chair in Cancer Research at the School of Medicine at UCR. PU is a recipient of the 2017-2018 Pease Cancer Fellowship through the Division of Biomedical Sciences, School of Medicine at UCR. We thank Dr. J. Momper and his associates at the University of California San Diego, Drug Development Pipeline core facility for plasma stability data. HA-XIAP-BIR3 plasmid was a gift from Dr. Colin Duckett (Addgene plasmid # 25689). Molecular graphics were performed with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Our agents can be distributed in small amounts (1 - 5mg) for research purposes upon request and signing of a standard material transfer agreement.
Abbreviations used
- XIAP
X-linked Inhibitor of Apoptosis Protein
- BIR
baculovirus IAP repeat domains
- cIAP1
cellular Inhibitor of Apoptosis Protein 1
- DELFIA
Dissociation-Enhanced Lanthanide Fluorescent Immunoassay
- DMF
Dimethylformamide
- ivDde
1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3-methylbutyl
- HATU
1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- DIPEA
N,N-diisopropylethylamine
- AISF
4-(Acetylamino)phenyl]imidodisulfuryl difluoride
- DBU
1,8-Diazabicyclo[5.4.0]undec-7-ene
- THF
Tetrahydrofuran
- rt
room temperature
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website. Figure S1 contains the individual thermal shift data plots that generated the date reported in Tables 1–3. Figure S2 and Figure S3 report additional aqueous stability of selected compounds at neutral pH and in simulated gastric fluid (pH=1.5). Figure S4 reports SDS gel electrophoresis data with XIAP BIR3 mutant Lys311Ala in presence of various compounds. Figure S5 reports basic characterization of key compounds from Table 3. PDB files relative to the models prepared for compound 10 in complex with wt-BIR3, Lys311Tyr BIR3, and Lys311His BIR3 are also provided. A Molecular Formula Strings file is also available.
Notes: The authors declare no competing financial interest
References
- 1.Souers AJ; Leverson JD; Boghaert ER; Ackler SL; Catron ND; Chen J; Dayton BD; Ding H; Enschede SH; Fairbrother WJ; Huang DC; Hymowitz SG; Jin S; Khaw SL; Kovar PJ; Lam LT; Lee J; Maecker HL; Marsh KC; Mason KD; Mitten MJ; Nimmer PM; Oleksijew A; Park CH; Park CM; Phillips DC; Roberts AW; Sampath D; Seymour JF; Smith ML; Sullivan GM; Tahir SK; Tse C; Wendt MD; Xiao Y; Xue JC; Zhang H; Humerickhouse RA; Rosenberg SH; Elmore SW ABT-199, a Potent and Selective Bcl-2 Inhibitor, Achieves Antitumor Activity While Sparing Platelets. Nat Med 2013, 19, 202–208. [DOI] [PubMed] [Google Scholar]
- 2.Bruncko M; Oost TK; Belli BA; Ding H; Joseph MK; Kunzer A; Martineau D; McClellan WJ; Mitten M; Ng SC; Nimmer PM; Oltersdorf T; Park CM; Petros AM; Shoemaker AR; Song X; Wang X; Wendt MD; Zhang H; Fesik SW; Rosenberg SH; Elmore SW Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-Xl. J Med Chem 2007, 50, 641–662. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z; Sun C; Olejniczak ET; Meadows RP; Betz SF; Oost T; Herrmann J; Wu JC; Fesik SW Structural Basis for Binding of Smac/Diablo to the Xiap Bir3 Domain. Nature 2000, 408, 1004–1008. [DOI] [PubMed] [Google Scholar]
- 4.Oltersdorf T; Elmore SW; Shoemaker AR; Armstrong RC; Augeri DJ; Belli BA; Bruncko M; Deckwerth TL; Dinges J; Hajduk PJ; Joseph MK; Kitada S; Korsmeyer SJ; Kunzer AR; Letai A; Li C; Mitten MJ; Nettesheim DG; Ng S; Nimmer PM; O’Connor JM; Oleksijew A; Petros AM; Reed JC; Shen W; Tahir SK; Thompson CB; Tomaselli KJ; Wang B; Wendt MD; Zhang H; Fesik SW; Rosenberg SH An Inhibitor of Bcl-2 Family Proteins Induces Regression of Solid Tumours. Nature 2005, 435, 677–681. [DOI] [PubMed] [Google Scholar]
- 5.Oost TK; Sun C; Armstrong RC; Al-Assaad AS; Betz SF; Deckwerth TL; Ding H; Elmore SW; Meadows RP; Olejniczak ET; Oleksijew A; Oltersdorf T; Rosenberg SH; Shoemaker AR; Tomaselli KJ; Zou H; Fesik SW Discovery of Potent Antagonists of the Antiapoptotic Protein Xiap for the Treatment of Cancer. J Med Chem 2004, 47, 4417–4426. [DOI] [PubMed] [Google Scholar]
- 6.Tse C; Shoemaker AR; Adickes J; Anderson MG; Chen J; Jin S; Johnson EF; Marsh KC; Mitten MJ; Nimmer P; Roberts L; Tahir SK; Xiao Y; Yang X; Zhang H; Fesik S; Rosenberg SH; Elmore SW ABT-263: A Potent and Orally Bioavailable Bcl-2 Family Inhibitor. Cancer Res 2008, 68, 3421–3428. [DOI] [PubMed] [Google Scholar]
- 7.Deveraux QL; Reed JC Iap Family Proteins--Suppressors of Apoptosis. Genes Dev 1999, 13, 239–252. [DOI] [PubMed] [Google Scholar]
- 8.Salvesen GS; Duckett CS Iap Proteins: Blocking the Road to Death’s Door. Nat Rev Mol Cell Biol 2002, 3, 401–410. [DOI] [PubMed] [Google Scholar]
- 9.Cong H; Xu L; Wu Y; Qu Z; Bian T; Zhang W; Xing C; Zhuang C Inhibitor of Apoptosis Protein (Iap) Antagonists in Anticancer Agent Discovery: Current Status and Perspectives. J Med Chem 2019, article ASAP doi: 10.1021/acs.jmedchem.8b01668. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y; Rich RL; Myszka DG; Wu H Requirement of Both the Second and Third Bir Domains for the Relief of X-Linked Inhibitor of Apoptosis Protein (Xiap)-Mediated Caspase Inhibition by Smac. J Biol Chem 2003, 278, 49517–49522. [DOI] [PubMed] [Google Scholar]
- 11.Wu G; Chai J; Suber TL; Wu JW; Du C; Wang X; Shi Y Structural Basis of Iap Recognition by Smac/Diablo. Nature 2000, 408, 1008–1012. [DOI] [PubMed] [Google Scholar]
- 12.Samuel T; Welsh K; Lober T; Togo SH; Zapata JM; Reed JC Distinct Bir Domains of Ciap1 Mediate Binding to and Ubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 2 and Second Mitochondrial Activator of Caspases. J Biol Chem 2006, 281, 1080–1090. [DOI] [PubMed] [Google Scholar]
- 13.Cai Q; Sun H; Peng Y; Lu J; Nikolovska-Coleska Z; McEachern D; Liu L; Qiu S; Yang CY; Miller R; Yi H; Zhang T; Sun D; Kang S; Guo M; Leopold L; Yang D; Wang S A Potent and Orally Active Antagonist (Sm-406/at-406) of Multiple Inhibitor of Apoptosis Proteins (Iaps) in Clinical Development for Cancer Treatment. J Med Chem 2011, 54, 2714–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen F; Alicke B; Elliott LO; Flygare JA; Goncharov T; Keteltas SF; Franklin MC; Frankovitz S; Stephan JP; Tsui V; Vucic D; Wong H; Fairbrother WJ Orally Bioavailable Antagonists of Inhibitor of Apoptosis Proteins Based on an Azabicyclooctane Scaffold. J Med Chem 2009, 52, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 15 <j/>15.Flygare JA; Beresini M; Budha N; Chan H; Chan IT; Cheeti S; Cohen F; Deshayes K; Doerner K; Eckhardt SG; Elliott LO; Feng B; Franklin MC; Reisner SF; Gazzard L; Halladay J; Hymowitz SG; La H; LoRusso P; Maurer B; Murray L; Plise E; Quan C; Stephan JP; Young SG; Tom J; Tsui V; Um J; Varfolomeev E; Vucic D; Wagner AJ; Wallweber HJ; Wang L; Ware J; Wen Z; Wong H; Wong JM; Wong M; Wong S; Yu R; Zobel K; Fairbrother WJ Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (Iap) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152). J Med Chem 2012, 55, 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaither A; Porter D; Yao Y; Borawski J; Yang G; Donovan J; Sage D; Slisz J; Tran M; Straub C; Ramsey T; Iourgenko V; Huang A; Chen Y; Schlegel R; Labow M; Fawell S; Sellers WR; Zawel L A Smac Mimetic Rescue Screen Reveals Roles for Inhibitor of Apoptosis Proteins in Tumor Necrosis Factor-Alpha Signaling. Cancer Res 2007, 67, 11493–11498. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto K; Saito B; Miyamoto N; Oguro Y; Tomita D; Shiokawa Z; Asano M; Kakei H; Taya N; Kawasaki M; Sumi H; Yabuki M; Iwai K; Yoshida S; Yoshimatsu M; Aoyama K; Kosugi Y; Kojima T; Morishita N; Dougan DR; Snell GP; Imamura S; Ishikawa T Design and Synthesis of Potent Inhibitor of Apoptosis (Iap) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]Pyrazine Scaffold as a Novel Proline Mimetic. J Med Chem 2013, 56, 1228–1246. [DOI] [PubMed] [Google Scholar]
- 18.Li L; Thomas RM; Suzuki H; De Brabander JK; Wang X; Harran PG A Small Molecule Smac Mimic Potentiates Trail- and Tnfalpha-Mediated Cell Death. Science 2004, 305, 1471–1474. [DOI] [PubMed] [Google Scholar]
- 19.Ndubaku C; Varfolomeev E; Wang L; Zobel K; Lau K; Elliott LO; Maurer B; Fedorova AV; Dynek JN; Koehler M; Hymowitz SG; Tsui V; Deshayes K; Fairbrother WJ; Flygare JA; Vucic D Antagonism of C-Iap and Xiap Proteins Is Required for Efficient Induction of Cell Death by Small-Molecule Iap Antagonists. ACS Chem Biol 2009, 4, 557–566. [DOI] [PubMed] [Google Scholar]
- 20.Peng Y; Sun H; Nikolovska-Coleska Z; Qiu S; Yang CY; Lu J; Cai Q; Yi H; Kang S; Yang D; Wang S Potent, Orally Bioavailable Diazabicyclic Small-Molecule Mimetics of Second Mitochondria-Derived Activator of Caspases. J Med Chem 2008, 51, 8158–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng R; Sun H; Liu L; Lu J; McEachern D; Wang G; Wen J; Min P; Du Z; Lu H; Kang S; Guo M; Yang D; Wang S A Potent Bivalent Smac Mimetic (Sm-1200) Achieving Rapid, Complete, and Durable Tumor Regression in Mice. J Med Chem 2013, 56, 3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun H; Lu J; Liu L; Yi H; Qiu S; Yang CY; Deschamps JR; Wang S Nonpeptidic and Potent Small-Molecule Inhibitors of Ciap-1/2 and Xiap Proteins. J Med Chem 2010, 53, 6361–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H; Nikolovska-Coleska Z; Lu J; Meagher JL; Yang CY; Qiu S; Tomita Y; Ueda Y; Jiang S; Krajewski K; Roller PP; Stuckey JA; Wang S Design, Synthesis, and Characterization of a Potent, Nonpeptide, Cell-Permeable, Bivalent Smac Mimetic That Concurrently Targets both the Bir2 and Bir3 Domains in Xiap. J Am Chem Soc 2007, 129, 15279–15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H; Nikolovska-Coleska Z; Lu J; Qiu S; Yang CY; Gao W; Meagher J; Stuckey J; Wang S Design, Synthesis, and Evaluation of a Potent, Cell-Permeable, Conformationally Constrained Second Mitochondria Derived Activator of Caspase (Smac) Mimetic. J Med Chem 2006, 49, 7916–7920. [DOI] [PubMed] [Google Scholar]
- 25.Sun H; Nikolovska-Coleska Z; Yang CY; Xu L; Tomita Y; Krajewski K; Roller PP; Wang S Structure-Based Design, Synthesis, and Evaluation of Conformationally Constrained Mimetics of the Second Mitochondria-Derived Activator of Caspase That Target the X-Linked Inhibitor of Apoptosis Protein/Caspase-9 Interaction Site. J Med Chem 2004, 47, 4147–4150. [DOI] [PubMed] [Google Scholar]
- 26.Sun W; Nikolovska-Coleska Z; Qin D; Sun H; Yang CY; Bai L; Qiu S; Wang Y; Ma D; Wang S Design, Synthesis, and Evaluation of Potent, Nonpeptidic Mimetics of Second Mitochondria-Derived Activator of Caspases. J Med Chem 2009, 52, 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S Design of Small-Molecule Smac Mimetics as Iap Antagonists. Curr Top Microbiol Immunol 2011, 348, 89–113. [DOI] [PubMed] [Google Scholar]
- 28.Zobel K; Wang L; Varfolomeev E; Franklin MC; Elliott LO; Wallweber HJ; Okawa DC; Flygare JA; Vucic D; Fairbrother WJ; Deshayes K Design, Synthesis, and Biological Activity of a Potent Smac Mimetic That Sensitizes Cancer Cells to Apoptosis by Antagonizing Iaps. ACS Chem Biol 2006, 1, 525–533. [DOI] [PubMed] [Google Scholar]
- 29.Hennessy EJ; Saeh JC; Sha L; MacIntyre T; Wang H; Larsen NA; Aquila BM; Ferguson AD; Laing NM; Omer CA Discovery of Aminopiperidine-Based Smac Mimetics as Iap Antagonists. Bioorg Med Chem Lett 2012, 22, 1690–1694. [DOI] [PubMed] [Google Scholar]
- 30.Huang JW; Zhang Z; Wu B; Cellitti JF; Zhang X; Dahl R; Shiau CW; Welsh K; Emdadi A; Stebbins JL; Reed JC; Pellecchia M Fragment-Based Design of Small Molecule X-Linked Inhibitor of Apoptosis Protein Inhibitors. J Med Chem 2008, 51, 7111–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barile E; Marconi GD; De SK; Baggio C; Gambini L; Salem AF; Kashyap MK; Castro JE; Kipps TJ; Pellecchia M hBfl-1/hNOXA Interaction Studies Provide New Insights on the Role of Bfl-1 in Cancer Cell Resistance and for the Design of Novel Anticancer Agents. ACS Chem Biol 2017, 12, 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stebbins JL; Santelli E; Feng Y; De SK; Purves A; Motamedchaboki K; Wu B; Ronai ZA; Liddington RC; Pellecchia M Structure-Based Design of Covalent Siah Inhibitors. Chem Biol 2013, 20, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baggio C; Gambini L; Udompholkul P; Salem AF; Aronson A; Dona A; Troadec E; Pichiorri F; Pellecchia M Design of Potent Pan-Iap and Lys-Covalent Xiap Selective Inhibitors Using a Thermodynamics Driven Approach. J Med Chem 2018, 61, 6350–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonsdale R; Ward RA Structure-Based Design of Targeted Covalent Inhibitors. Chem Soc Rev 2018, 47, 3816–3830. [DOI] [PubMed] [Google Scholar]
- 35.Pettinger J; Jones K; Cheeseman MD Lysine-Targeting Covalent Inhibitors. Angew Chem Int Ed Engl 2017, 56, 15200–15209. [DOI] [PubMed] [Google Scholar]
- 36.Narayanan A; Jones LH Sulfonyl Fluorides as Privileged Warheads in Chemical Biology. Chem Sci 2015, 6, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H; Mukherjee P; Liu R; Evrard E; Wang D; Humphrey JM; Butler TW; Hoth LR; Sperry JB; Sakata SK; Helal CJ; Am Ende CW Introduction of a Crystalline, Shelf-Stable Reagent for the Synthesis of Sulfur(VI) Fluorides. Org Lett 2018, 20, 812–815. [DOI] [PubMed] [Google Scholar]
- 38.Dong J; Krasnova L; Finn MG; Sharpless KB Sulfur(VI) Fluoride Exchange (Sufex): Another Good Reaction for Click Chemistry. Angew Chem Int Ed Engl 2014, 53, 9430–9448. [DOI] [PubMed] [Google Scholar]
- 39.Baggio C; Udompholkul P; Barile E; Pellecchia M Enthalpy-Based Screening of Focused Combinatorial Libraries for the Identification of Potent and Selective Ligands. ACS Chem Biol 2017, 12, 2981–2989. [DOI] [PubMed] [Google Scholar]
- 40.Lewis J; Burstein E; Reffey SB; Bratton SB; Roberts AB; Duckett CS Uncoupling of the Signaling and Caspase-Inhibitory Properties of X-Linked Inhibitor of Apoptosis. J Biol Chem 2004, 279, 9023–9029. [DOI] [PubMed] [Google Scholar]
- 41.Li X; Chen W; Zeng W; Wan C; Duan S; Jiang S Microrna-137 Promotes Apoptosis in Ovarian Cancer Cells Via the Regulation of Xiap. Br J Cancer 2017, 116, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Favalli N; Bassi G; Scheuermann J; Neri D DNA-Encoded Chemical Libraries - Achievements and Remaining Challenges. FEBS Lett 2018, 592, 2168–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favalli N; Biendl S; Hartmann M; Piazzi J; Sladojevich F; Graslund S; Brown PJ; Nareoja K; Schuler H; Scheuermann J; Franzini R; Neri D A DNA-Encoded Library of Chemical Compounds Based on Common Scaffolding Structures Reveals the Impact of Ligand Geometry on Protein Recognition. ChemMedChem 2018, 13, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neri D; Lerner RA DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu Rev Biochem 2018, 87, 479–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaikuad A; Koch P; Laufer SA; Knapp S The Cysteinome of Protein Kinases as a Target in Drug Development. Angew Chem Int Ed Engl 2018, 57, 4372–4385. [DOI] [PubMed] [Google Scholar]
- 46.Wu S; Luo Howard H; Wang H; Zhao W; Hu Q; Yang Y Cysteinome: The First Comprehensive Database for Proteins with Targetable Cysteine and Their Covalent Inhibitors. Biochem Biophys Res Commun 2016, 478, 1268–1273. [DOI] [PubMed] [Google Scholar]
- 47.Liu Q; Sabnis Y; Zhao Z; Zhang T; Buhrlage SJ; Jones LH; Gray NS Developing Irreversible Inhibitors of the Protein Kinase Cysteinome. Chem Biol 2013, 20, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Q; Ouyang X; Wan X; Gajiwala KS; Kath JC; Jones LH; Burlingame AL; Taunton J Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J Am Chem Soc 2017, 139, 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anscombe E; Meschini E; Mora-Vidal R; Martin MP; Staunton D; Geitmann M; Danielson UH; Stanley WA; Wang LZ; Reuillon T; Golding BT; Cano C; Newell DR; Noble ME; Wedge SR; Endicott JA; Griffin RJ Identification and Characterization of an Irreversible Inhibitor of Cdk2. Chem Biol 2015, 22, 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akcay G; Belmonte MA; Aquila B; Chuaqui C; Hird AW; Lamb ML; Rawlins PB; Su N; Tentarelli S; Grimster NP; Su Q Inhibition of Mcl-1 through Covalent Modification of a Noncatalytic Lysine Side Chain. Nat Chem Biol 2016, 12, 931–936. [DOI] [PubMed] [Google Scholar]
- 51.Hoppmann C; Wang L Proximity-Enabled Bioreactivity to Generate Covalent Peptide Inhibitors of P53-Mdm4. Chem Commun (Camb) 2016, 52, 5140–5143. [DOI] [PubMed] [Google Scholar]
- 52.Ashley SL; Sisson TH; Wheaton AK; Kim KK; Wilke CA; Ajayi IO; Subbotina N; Wang S; Duckett CS; Moore BB; Horowitz JC Targeting Inhibitor of Apoptosis Proteins Protects from Bleomycin-Induced Lung Fibrosis. Am J Respir Cell Mol Biol 2016, 54, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baggio C; Cerofolini L; Fragai M; Luchinat C; Pellecchia M HTS by NMR for the Identification of Potent and Selective Inhibitors of Metalloenzymes. ACS Med Chem Lett 2018, 9, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bottini A; Wu B; Barile E; De SK; Leone M; Pellecchia M High-Throughput Screening (HTS) by NMR Guided Identification of Novel Agents Targeting the Protein Docking Domain of Yoph. ChemMedChem 2016, 11, 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu B; Barile E; De SK; Wei J; Purves A; Pellecchia M High-Throughput Screening by Nuclear Magnetic Resonance (HTS by NMR) for the Identification of Ppis Antagonists. Curr Top Med Chem 2015, 15, 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barile E; Pellecchia M NMR-Based Approaches for the Identification and Optimization of Inhibitors of Protein-Protein Interactions. Chem Rev 2014, 114, 4749–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu B; Zhang Z; Noberini R; Barile E; Giulianotti M; Pinilla C; Houghten RA; Pasquale EB; Pellecchia M Hts by Nmr of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery. Chem Biol 2013, 20, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pellecchia M; Sem DS; Wuthrich K NMR in Drug Discovery. Nat Rev Drug Discov 2002, 1, 211–219. [DOI] [PubMed] [Google Scholar]
- 59.Martin-Gago P; Olsen CA Arylfluorosulfate-Based Electrophiles for Covalent Protein Labeling: A New Addition to the Arsenal. Angew Chem Int Ed Engl 2019, 58, 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X; van Veldhoven JPD; Offringa J; Kuiper BJ; Lenselink EB; Heitman LH; van der Es D; AP IJ Development of Covalent Ligands for G Protein-Coupled Receptors: A Case for the Human Adenosine A3 Receptor. J Med Chem 2019, 62, 3539–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fadeyi OO; Hoth LR; Choi C; Feng X; Gopalsamy A; Hett EC; Kyne RE Jr.; Robinson RP; Jones LH Covalent Enzyme Inhibition through Fluorosulfate Modification of a Noncatalytic Serine Residue. ACS Chem Biol 2017, 12, 2015–2020. [DOI] [PubMed] [Google Scholar]
- 62.Giuntini S; Balducci E; Cerofolini L; Ravera E; Fragai M; Berti F; Luchinat C Characterization of the Conjugation Pattern in Large Polysaccharide-Protein Conjugates by NMR Spectroscopy. Angew Chem Int Ed Engl 2017, 56, 14997–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


