Abstract
Despite evidence that SARS-CoV-2 infection is systemic in nature, there is little known about the effects that SARS-CoV-2 infection or exposure has on many host cell types, including primitive and mature hematopoietic cells. The hematopoietic system is responsible for giving rise to the very immune cells that defend against viral infection and is a source of hematopoietic stem cells (HSCs) and progenitor cells (HPCs) which are used for hematopoietic cell transplantation (HCT) to treat hematologic disorders, thus there is a strong need to understand how exposure to the virus may affect hematopoietic cell functions. We examined the expression of ACE2, to which SARS-CoV-2 Spike (S) protein binds to facilitate viral entry, in cord blood derived HSCs/HPCs and in peripheral blood derived immune cell subtypes. ACE2 is expressed in low numbers of immune cells, higher numbers of HPCs, and up to 65% of rigorously defined HSCs. We also examined effects of exposing HSCs/HPCs and immune cells to SARS-CoV-2 S protein ex vivo. HSCs and HPCs expand less effectively and have less functional colony forming capacity when grown with S protein, while peripheral blood monocytes upregulate CD14 expression and show distinct changes in size and granularity. That these effects are induced by recombinant S protein alone and not the infectious viral particle suggests that simple exposure to SARS-CoV-2 may impact HSCs/HPCs and immune cells via S protein interactions with the cells, regardless of whether they can be infected. These data have implications for immune response to SARS-CoV-2 and for HCT.
Graphical Abstract.
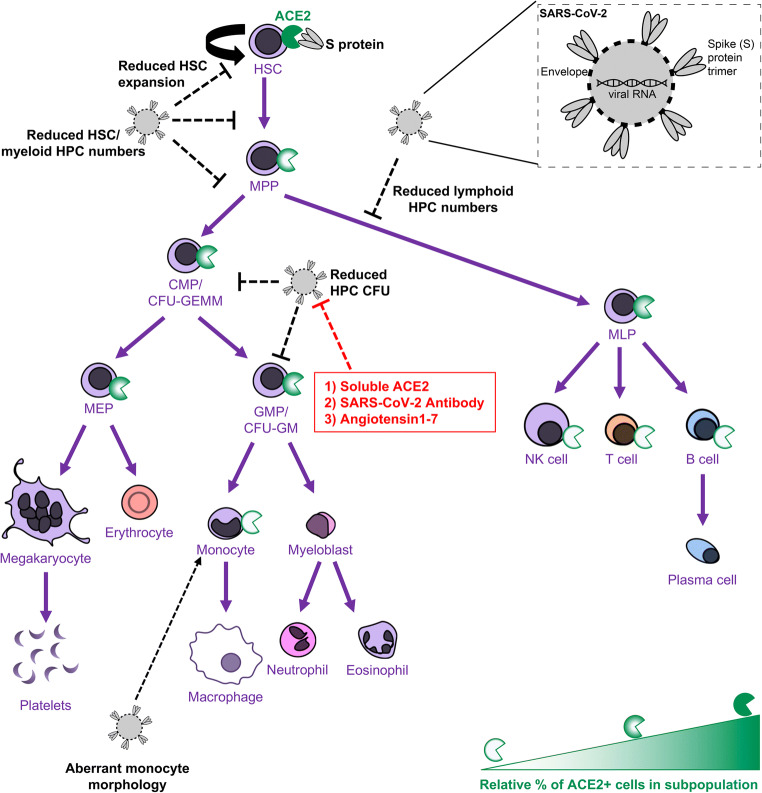
• Human HSCs, HPCs, and immune cells express ACE2 on the cell surface, making them potentially susceptible to SARS-CoV-2 infection.
• SARS-CoV-2 S protein, which binds to ACE2, induces defects in the colony forming capacity of human HPC and inhibits the expansion of HSC/HPC subpopulations ex vivo. These effects can be at least partially neutralized by treatment with SARS-CoV-2 targeting antibody, recombinant human ACE2, or Angiotensin1–7.
• S protein also induces aberrant morphological changes in peripheral blood derived monocytes ex vivo.
• Thus, there are many different manners in which SARS-CoV-2 virus may impact the functional hematopoietic system, which has important implications for hematological manifestations of COVID-19 (i.e. thrombocytopenia and lymphopenia), immune response, and hematopoietic stem cell transplant in the era of COVID-19.
Electronic supplementary material
The online version of this article (10.1007/s12015-020-10056-z) contains supplementary material, which is available to authorized users.
Keywords: SARS-CoV-2, COVID-19, Spike protein, ACE2, Hematopoietic stem and progenitor cells, Hematopoiesis, Immune cells, Cord blood, Hematopoietic cell expansion
Introduction
The devastating effects of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory coronavirus 2 (SARS-CoV-2) infection highlights the need to understand the virus’s effect on host tissues [1–3]. One of the alarming aspects of COVID-19 is the systemic nature of the disease and the paucity of information available regarding effects of infection on many different host cell types [4, 5]. Hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) have not been well studied in context of SARS-CoV-2 infection, though the impacts on these cell types could have important implications for immune response, hematologic complications [6], and stem cell based therapies, such as hematopoietic cell transplantation (HCT) [7–9]. While mature immune cells have been a current principle focus with regard to host responses, few if any studies have examined whether these immune cells are themselves susceptible to infection and what effects exposure to the virus may have directly on immune cells.
Coronaviruses are a family of pathogens historically not considered highly infective or deadly until the outbreak of severe acute respiratory syndrome (SARS) caused by the SARS coronavirus (SARS-CoV) in 2002 and the subsequent outbreak of Middle East respiratory syndrome (MERS) caused by the MERS coronavirus (MERS-CoV) in 2012. COVID-19 represents the third major outbreak of a disease caused by coronavirus infection in less than two decades, highlighting the need to better understand this family of viruses in order to address the current and future public health crises. SARS, MERS, and COVID-19 manifest primarily as respiratory syndromes, but growing evidence indicates that many different host cell types are infected or impacted by coronaviruses [4, 10, 11]. The coronavirus family is so named due to their crown-like appearance, a result of the spike (S) proteins that extend from the viral envelope and bind host cell membrane-bound proteins to facilitate viral entry to host cells by endocytosis [12]. The functional host receptor that binds SARS-CoV-2 is the cell surface glycoprotein, Angiotensin converting enzyme 2 (ACE2) [13]. In normal physiological context, ACE2 is a regulatory protein that converts the hormone Angiotensin II to Angiotensin 1–7 [14], which is a peptide involved in hypertension [15]. In this coronavirus infection, SARS-CoV-2 S protein binds to host cell ACE2, facilitating viral entry. Within the host cell, SARS-CoV-2 then replicates its viral machinery to propagate infection; this is known as a productive infection. A recent study demonstrated that SARS-CoV-2 can infect CD4+ T cells but does not actively replicate within the host T cell [16], thus SARS-CoV-2 may affect cells in different ways, including by both productive and abortive infection.
It is important to know how HSCs/HPCs and immune cells may be affected by SARS-CoV-2 infection because disruption to hematopoiesis or direct impacts on immune cells can lead to dysregulated immune responses and other hematologic complications. Large percentages of COVID-19 patients have presented with lymphopenia and thrombocytopenia, suggesting there are SARS-CoV-2 induced effects on primitive and/or mature blood cell populations [6, 17]. Further, HSCs/HPCs derived from bone marrow, mobilized peripheral blood, and cord blood (CB) are sources for hematopoietic cell transplantation (HCT) used to treat malignant and non-malignant hematologic disorders [18–21]. Thus, it is important to establish if SARS-CoV-2 exposure may impact HSCs/HPCs to be used for HCT, as this may in turn affect the efficacy of transplantation. If SARS-CoV-2 exposure may impact the efficacy of HCT, it will be critical to put in place stringent screening procedures for stem cell donation and banking of stem cell sources such as CB.
While the primary disease presentation for coronaviruses is a productive infection of the lungs [2, 22], SARS-CoV and MERS-CoV also affect the hematopoietic system via either a productive or abortive infection that leads to aberrant cytokine responses and in some cases induces apoptosis of immune cells [10, 11, 23–25]. Further, a recent study demonstrated that a subpopulation of HSCs express the ACE2 receptor and that exposure to SARS-CoV-2 S protein leads to upregulation of inflammatory response genes in these cells [26]. It is important to evaluate how SARS-CoV-2 exposure may affect the efficacy of HCT and the implications that may have for CB banking and bone marrow or mobilized peripheral blood donation [7, 9]. Here, we confirm expression of ACE2 in subpopulations of HSCs and examine a full panel of primitive and lineage committed progenitor cells as well as fully mature immune cells for ACE2 expression. The expression of ACE2 on subsets of these cells indicates potential susceptibility to interactions with SARS-CoV-2 S protein. We also demonstrate marked suppressive effects on HPC functional colony forming cell capacity, reduced expansion of HSCs/HPCs, and physiological changes induced in peripheral blood (PB) cells when cells are grown in the presence of recombinant SARS-CoV-2 S protein. These phenotypic changes induced by recombinant S protein alone suggests that exposure of HSCs/HPCs and immune cells to SARS-CoV-2 may impact primitive and mature hematopoietic cells regardless of its ability to infect them via host cell interactions with S protein.
Materials and Methods
Umbilical Cord Blood (CB) Processing
Umbilical cord blood units were obtained from the Cleveland Cord Blood Center (CCBC), Cleveland, OH, USA, collected through CCBC’s Volunteer Donating Communities in Cleveland, OH, Atlanta, GA and San Francisco, CA, and were processed as previously described [27–29] within 48 h of collection. Cord blood units were washed with PBS to remove preservatives, layered on Ficoll-Paque Plus (GE Healthcare, Chicago, IL, USA), spun at 1600 RPM for 30 min, and low-density cells were collected from the intermediate layer. For RT-qPCR, low density cells were lineage depleted using the lineage depletion kit and then enriched for CD34+ cells using CD34 MicroBead Kit (Milltenyi Biotec, Bergisch Gladbach, Germany). Lineage positive cells were also collected by eluting from the lineage depletion column. High density blood cells were also collected from the pellet produced by the gradient spin. For expansion assays and flow cytometry analysis, low density cells were enriched for CD34+ cells, using two CD34 MicroBead Kit columns for increased purity (up to 95% purity). High density granulocytes were isolated by lysing red blood cells in RBC lysis buffer (Biolegend, San Diego, CA, USA) for 15 min.
Peripheral Blood (PB) Processing
Whole PB was obtained from healthy donors, layered on Ficoll-Paque Plus, spun at 1600 RPM for 30 min, and low-density cells were collected from the intermediate layer.
Flow Cytometry Analysis
Cells were stained according to the immunophenotypes identified in Table S1 using the antibodies listed in Table S2. Cells were blocked with human FC receptor block, incubated with stain for 30 min at 4 °C, washed twice, and subjected to flow cytometry on a BD LSR II or a BD LSRFortessa flow cytometer. ACE2+ gates were set on each individual subpopulation using isotype controls. Data were analyzed using FlowJo v10.6.2 (BD, Franklin Lakes, NJ, USA).
RT-qPCR
RNA was harvested from 5 × 105-1 × 106 cells using Qiagen RNeasy Micro Plus Kit. cDNA was synthesized using SuperScriptIV (ThermoFisher, Waltham, MA, USA). RT-qPCR was run on an Applied Biosystems (Foster City, CA, USA) QuantStudio 6 using Fast SYBR Green Master Mix (Thermo Fisher). Primers used were: ACE2_forward- 5’ CATTGGAGCAAGTGTTGGATCTT 3′; ACE2_reverse- 5’ GAGCTAATGCATGCCATTCTCA 3″; GAPDH_forward- 5’ ACCCACTCCTCCACCTTTGAC 3′; GAPDH_reverse-5’ TGTTGCTGTAGCCAAATTCGTT 3′; TMPRSS2_forward- 5’ TCACACCAGCCATGATCTGT 3′; TMPRSS2_reverse- 5’ TATCCCCTATCAGCCACCAG 3′.
SDS-PAGE/ Western Blotting
Protein from 5 × 105 cells was harvested by directly lysing cells in 2x Laemmli Sample Buffer (BioRad Laboratories, Hercules, CA, USA) +/- 5% 2-mercaptoethanol. Lysed cells were boiled for 5 min at 95 °C and spun at maximum speed to pellet insoluble debris. Supernatants were run on 7% or 10% SDS-PAGE Mini-Protean TGX gels (BioRad) at 200 V for 30-45 min. Proteins were transferred to 0.22 μm nitrocellulose membrane (ThermoFisher). Membranes were blocked with 5% milk, immunoblotted with 1:1000 dilution of primary antibody, washed, incubated with 1:2500 dilution of hrp conjugated secondary antibodies, washed, and developed using SuperSignal Picoluminescent substrate (ThermoFisher) and a BioRad ChemiDoc Touch. Primary antibodies used were goat anti-hACE2 (R&D AF933); rabbit anti-TMPRSS2 (Abcam ab109131, Cambridge, UK); mouse anti-GAPDH (ThermoFisher AM4300).
Colony Forming Unit (CFU) and Expansion Assays
All colony forming unit assays were plated in 1% v/v methylcellulose, with 30% v/v FBS, 2 mM Glutamine, 2 × 10−5 M 2-Mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), 1 U/mL Epogen (EPO) (Amgen Inc., Thousand Oaks, CA, USA), 10 ng/mL recombinant human (rhu) Granulocyte-macrophage colony stimulating factor (GM-CSF), 10 ng/mL rhu Interleukin-3 (IL-3), 50 ng/mL rhu Stem cell factor (SCF) (R&D Systems, Minneapolis, MN, USA). To test direct effects of S protein on HPC colony formation capacity, 300 CD34+ enriched cells were plated in triplicate with varying doses (10 ng/mL, 50 ng/mL, 250 ng/mL, or 1000 ng/mL) of recombinant SARS-CoV-2 S protein (ACRO Biosystems SPN-C52H8, Newark, DE, USA) or PBS control. To test the effects of S protein on HSC/HPC ex vivo expansion, expansion assays were performed as previously described [18, 30, 31]. 100,000–200,000 CD34+ enriched cells were plated in liquid culture media in the presence or absence of 1 μg/mL recombinant S protein with growth factors for expansion: RPMI1640 + 10% FBS + 100 ng/mL rhu Thrombopoietin (TPO), rhuSCF, rhu Fms-like tyrosine kinase 3 ligand (FLT3L) (R&D Systems). Cells were expanded for 7 days in a humidified 5% O2, 5% CO2, 37 °C incubator, then 1 × 106 cells were stained for flow cytometry analysis and 500 cells were plated in triplicate in methylcellulose for HPC CFU assay. All CFU assays were grown in a humidified 5% O2, 5% CO2, 37 °C incubator for 12 days and were scored to distinguish colony forming unit- granulocyte, macrophage (CFU-GM) and colony forming unit- granulocyte, erythroid, macrophage, megakaryocyte (CFU-GEMM) colonies. This combination does not pick up Burst-forming unit- erythroid (BFU-E) colonies, as previously described [32].
Neutralizing the Effects of S Protein
We tested the ability of different agents to neutralize the effects of S protein. To test whether direct effects of S protein on HPC colony formation capacity can be neutralized, 350 CD34+ enriched cells were plated in triplicate with 250 ng recombinant SARS-CoV-2 S protein alone, 250 ng SARS-CoV-2 antibody (Sino Biological 40,591-MM43, Beijing, China) alone, 250 ng soluble rhu ACE2 (Sigma Aldrich) alone, 250 ng Angiotensin1–7 (Sigma Aldrich) alone, 250 ng recombinant S protein pre-incubated at room temperature for 30 min with 250 ng SARS-CoV-2 antibody, 250 ng recombinant S protein pre-incubated at 37 °C for 30 min with 250 ng soluble rhu ACE2 (Sigma Aldrich), or 250 ng recombinant S protein with 250 ng Angiotensin1–7 (Sigma Aldrich). To test the whether the effects of S protein on HSC/HPC ex vivo expansion could be neutralized, 100,000 CD34+ enriched cells were plated in liquid culture media with growth factors and with 1 μg/mL recombinant SARS-CoV-2 S protein alone, 1 μg/mL SARS-CoV-2 antibody (Sino Biological 40,591-MM43, Beijing, China) alone, 1 μg/mL soluble rhu ACE2 (Sigma Aldrich) alone, 1 μg/mL Angiotensin1–7 (Sigma Aldrich) alone, 1 μg/mL recombinant S protein pre-incubated at room temperature for 30 min with 1 μg/mL SARS-CoV-2 antibody, 1 μg/mL recombinant S protein pre-incubated at 37 °C for 30 min with 1 μg/mL soluble rhu ACE2 (Sigma Aldrich), or 1 μg/mL recombinant S protein with 1 μg/mL Angiotensin1–7 (Sigma Aldrich). Cells were expanded for 7 days in a humidified 5% O2, 5% CO2, 37 °C incubator, then 1 × 106 cells were stained for flow cytometry analysis and 500 cells were plated in triplicate in methylcellulose for HPC CFU assay.
Statistical Analysis
For assays consisting of only two test conditions (i.e., control versus S protein), data were analyzed using paired t-tests including matching for the same cord blood unit or same peripheral blood donor. Where multiple t-tests were performed for a given experiment, multiple testing correction was performed, and adjusted P-value was reported. For assays consisting of more than two test conditions (i.e., control versus S protein versus input) including dose-dependence assays, generalized linear modeling was performed using a model that corrects for sample variability from different cord blood units or peripheral blood donations followed by ANOVA and TukeyHSD post-hoc testing comparing all conditions to all other conditions. Reported P-values are the results of the TukeyHSD post-hoc tests. This data was analyzed in R version 3.6.3 and the code used was as follows: lm = aov(counts ~ treatment+samplenumber); anova(lm); TukeyHSD(lm). For all CFU assays, the average counts of technical triplicates was used as the value for biological replicates and n was determined based off of the number of biological replicates.
Results
Hematopoietic Stem and Progenitor Cells Express Cell Surface ACE2
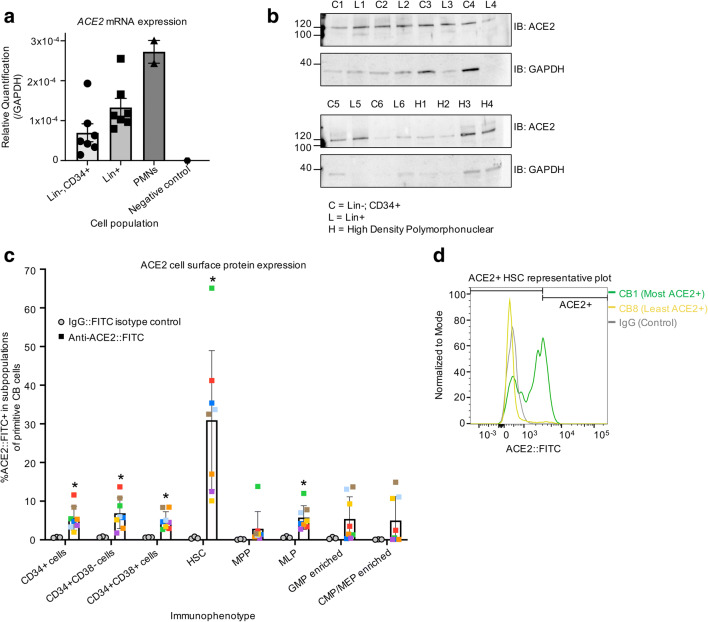
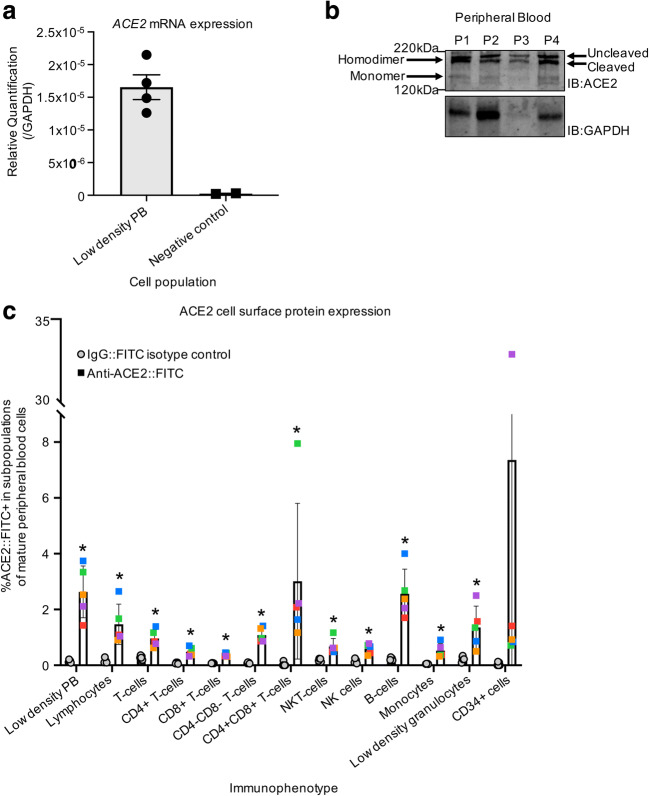
We examined whether CD34+ primitive CB hematopoietic cells express ACE2 and are thus potentially susceptible to SARS-CoV-2. RNA was harvested from lineage depleted (lin-) CD34+ enriched cells (enriched for HSCs/HPCs), from lineage enriched (lin+) cells (enriched for more mature blood cells), and from high density polymorphonuclear leukocytes (PMN). RT-qPCR demonstrates that ACE2 mRNA is expressed in all three of these cell populations (Fig. 1a). Protein harvested and subjected to SDS-PAGE followed by western blotting demonstrates that ACE2 protein is also present in these three cell populations (Fig. 1b). Cells were stained and analyzed by FACS to determine the cell surface expression of ACE2 on rigorously immunophenotypically defined subpopulations of HSCs/HPCs (Fig. S1, Table S1 and S2). ACE2 was expressed on 3.3–11.6% of CD34+ cells, including 10.1–65.1% of rigorously purified HSCs (CD34 + CD38-CD45RA-CD49f + CD90+); 0.4–13.8% of multipotent progenitor cells (MPPs; CD34 + CD38-CD45RA-CD49f-CD90-); and 2.7–12% of multipotent lymphoid progenitor cells (MLPs; CD34 + CD38-CD45RA + CD10+) (Fig. 1c). ACE2 expression was observed on the cell surface of 0.1–14.9% of cells enriched for common myeloid progenitors/ megakaryocyte-erythroid progenitors (CMPs/MEPs; CD34 + CD38 + CD10-CD45RA-) and 0.3–13.7% of cells enriched for granulocyte-macrophage progenitors (GMPs; CD34 + CD38 + CD10-CD45RA+) (Fig. 1c). This suggests that HSCs have the highest subpopulation of ACE2 expressing cells, making them potentially the most susceptible hematopoietic cells to an ACE2 dependent mechanism of SARS-CoV-2 infection or impact on host cells. However, the percentage of cell surface ACE2+ cells and level of ACE2 expression in these cells varied greatly by sample within all subpopulations of cells, and particularly in HSCs (Fig. 1d).
Fig. 1.
Subpopulations of cord blood derived HSCs/HPCs express cell surface ACE2. (a/b) RT-qPCR to test for ACE2 mRNA expression (a) and SDS-PAGE followed by western blot with indicated antibodies to test for ACE2 protein expression (b) in CB lineage enriched (L = Lin+) cells; low density CB lineage depleted and CD34+ enriched cells (C = Lin-CD34+), and CB high density polymorphonuclear cells (H=PMN). ACE2 expression is shown relative to GAPDH expression. Matching numbers in labels indicate samples that came from the same cord blood unit. (c) Low density cord blood CD34+ enriched cells were stained with fluorochrome conjugated antibodies and analyzed with flow cytometry to define the indicated immunophenotypes and determine ACE2 expression on these subpopulations. ACE2+ gate was defined using rabbit IgG isotype control. Matched colors of points indicate the same cord blood unit. ACE2 staining n = 8, IgG control n = 3; stats: t-tests comparing FITC+ % in ACE2 staining versus IgG staining, corrected for multiple testing. *P < 0.05 (d) Histogram shows representative ACE2 staining for HSCs from two cord blood units with the highest and lowest number of ACE2+ cells. Also shown is rabbit IgG isotype control
HSC/HPC Colony Formation Is Inhibited by SARS-CoV-2 S Protein
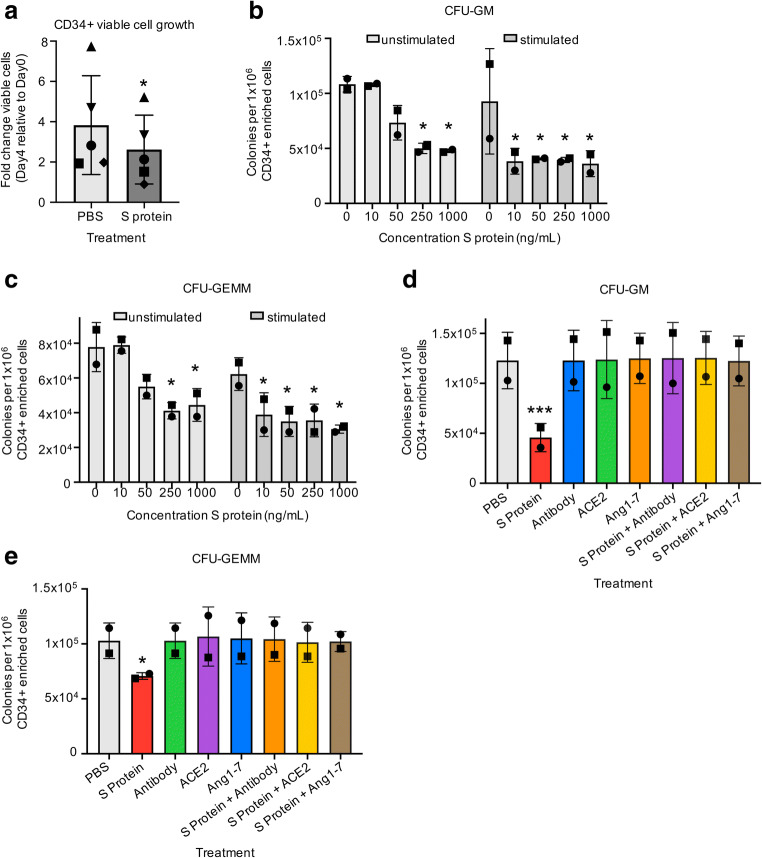
To examine the functional impact of hematopoietic cell exposure to SARS-CoV-2, we incubated CB derived CD34+ enriched cells with full-length recombinant S protein. We first examined whether S protein induced changes in viable cell growth of CD34+ cells in the presence of stimulating growth factors (rhuTPO/rhuSCF/rhuFLT3L). We observed significant changes in total viable cell number with cells grown in the absence of S protein exhibiting an average 3.8-fold increase in viable cells grown in the presence of S protein exhibiting an average 2.6-fold increase in viable cells after 4 days incubation in stimulating growth conditions (Fig. 2a). We next examined whether S protein exposure affects the functional HPC colony forming capacity of CD34+ cells using either freshly harvested CD34+ cells or CD34+ cells that we stimulated overnight with stimulating growth factors to put the cells into cycle. We plated these cells in semi-solid methylcellulose in the presence of serum and growth factors and with varying doses of S protein in HPC CFU assays. Plates were scored for granulocyte-macrophage progenitors (CFU-GM) and multipotential progenitors (CFU-GEMM). S protein exposure induced dose-dependent decreases in CFU-GM and CFU-GEMM colonies in unstimulated cells at 50–250 ng/mL with significantly decreased colony formation at 250 ng/mL and 1000 ng/mL doses compared to the control group (0 ng/mL unstimulated cells) (Fig. 2b, c). Stimulated cells trended toward a stronger response to S protein, with significantly decreased CFU-GM and CFU-GEMM colonies compared to the control group (0 ng/mL unstimulated cells) at all evaluated doses, including the lowest dose (10 ng/mL), indicating that cycling CD34+ cells may be more sensitive to S protein exposure (Fig. 2b, c). To test whether the direct effects on functional HPC colony formation induced by S protein can be neutralized, we utilized three different methods. First, we pre-incubated S protein with a SARS-CoV-2 Spike neutralizing mouse monoclonal antibody (Sino Biological 40,591-MM43). This antibody was determined in development to inhibit the infection of ACE2 expressing human cells by SARS-CoV-2 Spike pseudovirus. Next, we included the peptide fragment Angiotensin1–7 (Ang1–7) with S protein addition without pre-incubation. Ang1–7 is an important product of ACE2 that counters inflammatory responses triggered by infection [26, 33] and has been used ex vivo to reverse the inflammatory cascade induced by S protein in primitive stem cells. Finally, we pre-incubated S protein with soluble rhu ACE2, which can bind to S protein [13] and prevents its mediation of SARS-CoV-2 pseudovirus entry into ACE2 expressing human cells [34, 35]. All three of these conditions were tested separately to determine whether S protein effects on HPC CFU were neutralized in the presence of serum and growth factors. CFU-GM (Fig. 2d) and CFU-GEMM (Fig. 2e) numbers were rescued by all three neutralizing agents, indicating the observed effects are caused directly by the S protein and reveals three ways to neutralize these direct effects.
Fig. 2.
Cord blood HSCs/HPCs exhibit reduced colony forming capacity in the presence of SARS-CoV-2 S protein. (a) CD34+ enriched cells were plated at 100,000 cells/mL in media with stimulating growth factors (rhuTPO/rhuSCF/rhuFLT3L) and with 1 μg/mL recombinant S protein or PBS control and grown for 4 days in 5% O2 and 5% CO2 at 37 °C. Viable cells were counted using a hemocytometer and Trypan Blue viability stain. n = 5, stats: paired t-test, different symbols indicate the same cord blood unit in different treatment conditions. (b-c) CD34+ enriched cells were either taken for direct plating (unstimulated) or were grown for 24 h with stimulating growth factors (stimulated). 300 CD34+ enriched cells were plated in triplicate with the indicated doses of recombinant S protein or PBS control in 1% v/v methylcellulose with growth factors and serum and grown for 12 days in 5% O2 and 5% CO2 at 37 °C. CFU/1x106cells were calculated. Stats: general linearized modeling including stimulated and unstimulated cells at varying doses in the same model followed by ANOVA with TukeyHSD post hoc tests. Significance codes indicate comparison of sample to 0 ng/mL (PBS) control in unstimulated cells. (d-e) 350 freshly harvested CD34+ cells were plated in triplicate in 1% v/v methylcellulose with serum and growth factors and with PBS control, 250 ng/mL recombinant S protein alone, 250 ng/mL S protein pre-incubated with 250 ng/mL SARS-CoV-2 neutralizing antibody (Antibody), or 250 ng/mL S protein with 250 ng/mL Angiotensin1–7 (Ang1–7) and grown for 12 days in 5% O2 and 5% CO2 at 37 °C. Total CFU/1x106cells were calculated. Stats: general linearized modeling followed by ANOVA with TukeyHSD post hoc tests, matched symbols indicate the same cord blood unit in different treatment conditions. Shown are significance codes comparing all treatment levels to PBS control. *P < 0.05, ***P < 0.0005
HSC/HPC Expansion Is Significantly Inhibited by Exposure to SARS-CoV-2 S Protein
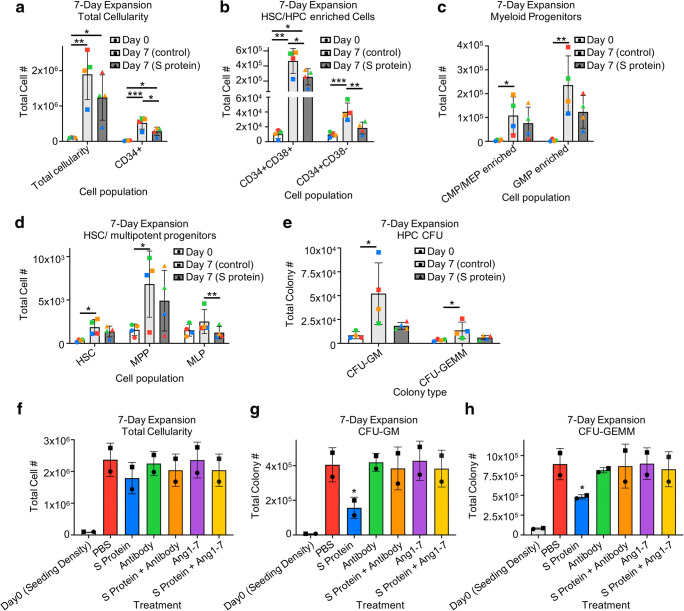
We evaluated effects of S protein on ex vivo expansion of CD34+ enriched cells in the presence of stimulating growth factors. Cells were analyzed for immunophenotype by flow cytometry and total number of cell populations per well were calculated. Cells incubated with/without S protein demonstrated respective average expansions of 11.9-fold/18.3-fold increases in total cellularity,13.8-fold/26.4-fold increases in CD34+ cells (Fig. 3a), 1.9-fold/4.3-fold increases in CD34 + CD38- cells enriched for HSCs and more primitive progenitors, and 73.7-fold/166.5-fold increases in CD34 + CD38+ cells enriched for mature progenitors (Fig. 3b). We observed respective expansions of 18.4-fold/29.3-fold change in CMP/MEP enriched cells, 73.7-fold/166.5-fold increases in GMP enriched cells (Fig. 3c), 4.1-fold/6.2-fold increases in HSCs and 2.8-fold/4.0-fold change in MPP (Fig. 3d). Incubation with S protein induced an average 0.86-fold decrease in numbers of MLP, while MLP in the absence of S protein exhibited a modest 1.7-fold expansion (Fig. 3d). Thus, expansion of HSCs, MPPs, CMPs/MEPs, and GMPs were all reduced in the presence of S protein. While MLPs did not show significant expansion in any condition, there were significantly more total MLP grown without S protein compared to with S protein. To determine effects on expansion of functional HPC, cells were expanded for 7 days in the presence of stimulating growth factors, and in the presence or absence of S protein, washed to remove residual unbound S protein, and plated in HPC CFU assays in methylcellulose containing FBS and stimulating growth factors. Cells that were expanded in the absence of S protein exhibited significant expansion of CFU-GM/ CFU-GEMM (13.3-fold/4-fold average expansion), while cells grown in the presence of S protein did not exhibit significant expansion in total CFU-GM/CFU-GEMM (4.5-fold/1.8-fold average expansion) (Fig. 3e). This indicates that ex vivo exposure to SARS-CoV-2 S protein affects the expansion and numbers of HSCs/HPCs. We also examined whether the S protein effects on expansion of HSCs/HPCs and functional HPC CFU can be neutralized. Pre-incubation of S protein with SARS-CoV-2 neutralizing antibody showed a trend of rescuing expansion of several rigorously defined hematopoietic populations, including total CD34+ cells, HSCs, and CMPs/MEPs, and significantly enhanced expansion of GMPs compared to S protein treatment alone (Fig. S2a). The addition of Ang1–7 to the expansion media partially rescued the expansion of CMP/MEP and significantly enhanced expansion of GMPs compared to S protein treatment alone, with little effect on more primitive immunophenotypes (Fig. S2b). Interestingly, both pre-incubation with the neutralizing antibody and the addition of Ang1–7 to the expansion media showed a trend of rescuing the expansion of total cellularity (Fig. 3f) and fully rescued the expansion of CFU-GM (Fig. 3g) and CFU-GEMM (Fig. 3h). These data indicate that the observed effects on expansion are due directly to exposure to the recombinant S protein and provides insight into at least partially neutralizing these effects.
Fig. 3.
Cord blood HSCs/HPCs exhibit reduced expansion in the presence of SARS-CoV-2 S protein. (a-h) CD34+ enriched cells were plated at 100,000–200,000 cells/mL in media with stimulating growth factors and with PBS control, 1 μg/mL recombinant S protein alone, 1 μg/mL S protein pre-incubated with 1 μg/mL SARS-CoV-2 neutralizing antibody (Antibody), 1 μg/mL S protein pre-incubated with 1 μg/mL rhu ACE2, or 1 μg/mL S protein with 1 μg/mL Angiotensin1–7 (Ang1–7) and grown for 7 days in 5% O2 and 5% CO2 at 37 °C. (a-d). Cells were then analyzed by flow cytometry for the indicated cell populations and total cell numbers were calculated or (e-h) 350–500 CD34+ cells were plated in triplicate in 1% v/v methylcellulose with serum and growth factors and grown for 12 days in 5% O2 and 5% CO2 at 37 °C. Total CFU were calculated. n = 4/2 for (a-e)/(f-h), stats: generalized linear modelling followed by ANOVA with TukeyHSD post hoc tests, matched colors of points for 3a-e and matched symbols for points for 3f-h indicate the same cord blood unit in different treatment conditions. For (f-h) significance codes shown are for the comparison of the indicated treatment PBS control. *P < 0.05, **P < 0.005,***P < 0.0005
Immune Cell Subpopulations Express Low Levels of Cell Surface ACE2
To determine whether immune cells themselves are susceptible to infection, we isolated low density mononuclear cells from PB of healthy donors. RNA was harvested from pooled PB and subjected to RT-qPCR, revealing that ACE2 is expressed at the mRNA level in these cells (Fig. 4a). Protein was harvested from pooled PB and run on SDS-PAGE followed by immunoblotting with an antibody against ACE2 in non-reducing conditions, revealing that ACE2 protein is detectable in PB and that ACE2 runs at the predicted molecular weight of the ACE2 homodimer as well as the ACE2 monomer; further, there are two distinct bands visible on the western blot, possibly indicating that ACE2 is expressed as both its full-length and cleaved isoforms in blood cells (Fig. 4b) [36]. We determined cell surface ACE2 expression on specific populations of immune cells by flow cytometry analysis (Table S1, example gating strategy Fig. S3). ACE2 is expressed on 1.4–3.7% of low density PB cells, 0.9–2.6% of size-defined lymphocytes, 0.5–2.5% of size-defined low-density granulocytes, and 0.3–0.9% of CD14+ monocytes. Examining more rigorously defined subpopulations of immune cells, ACE2 is expressed on 1.7–4% of CD19+ B-cells, 0.6–1.4% of CD3+ T-cells, 0.3–0.8% of CD3-CD56+ Natural Killer (NK) cells, and 0.5–1.1% of CD3 + CD56+ NK T-cells (Fig. 4c). Specific subpopulations of T-cells also exhibit low numbers of ACE2+ cells, with ACE2 cell surface expression found on 0.3–0.7% of CD4+ T-cells, 0.3–0.4% of CD8+ T-cells, 0.8–1.4% of CD4-CD8- T-cells, and 2–8% of CD4 + CD8+ T-cells (Fig. 4c). PB CD34+ cells, which are enriched for HPC, have an ACE2+ population of 0.7–1.4%, with one outlier sample exhibiting cell surface ACE2 expression in over 30% of PB derived CD34+ (Fig. 4c). This may be a technical artifact or may be indicative of population variability of ACE2 expression on PB derived HPC. Together, these data indicate that ACE2, which is thought to be the receptor protein primarily responsible for SARS-CoV-2 infection, is expressed on the cell surface of low numbers of PB immune cells.
Fig. 4.
Low numbers of peripheral blood derived immune cells express cell surface ACE2. (a) RT-qPCR to test for ACE2 mRNA expression in low density PB cells. ACE2 expression is shown relative to GAPDH expression. (b) Protein was harvested from low density PB cells and subjected to SDS-PAGE in non-reducing conditions followed by western blot with the indicated antibodies (IB = immunoblot). (c) Low density PB cells were stained with fluorochrome conjugated antibodies and analyzed with flow cytometry to define the indicated immunophenotypes and determine ACE2 expression on these subpopulations. ACE2+ gate was defined using rabbit IgG isotype control. Matched colors of points indicate the same peripheral blood donor. ACE2 staining n = 5, IgG control n = 5; stats: t-tests comparing FITC+ % in ACE2 staining versus IgG staining, corrected for multiple testing. *P < 0.05
Peripheral Blood Cells Respond to Exposure to the SARS-CoV-2 S Protein
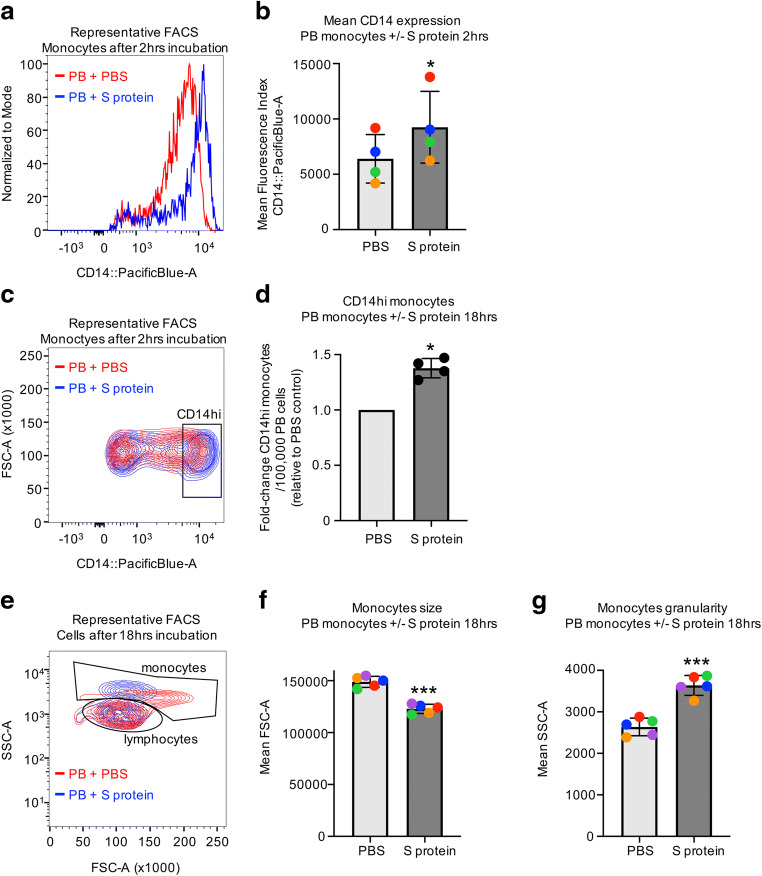
To examine effects of mature blood cell exposure to SARS-CoV-2, low density PB cells were incubated with full length recombinant SARS-CoV-2 S protein. Short-term 2 h incubation with S protein in the absence of serum leads to significant increases in CD14 staining by flow cytometry (Fig. 5a, b) in forward and side scatter defined monocytes. Additionally, there is an accumulation of more CD14hi monocytes (Fig. 5c), with an average of 1.38-fold more per 1 × 105 total cells relative to controls (Fig. 5d), though there are no significant differences in number of total CD14+ monocytes (data not shown). Thus, short-term exposure of PB to S protein induces increased CD14 expression. We examined what effects S protein exposure has on PB cells when they are cultured longer in the presence of serum. Overnight (18 h) incubation with S protein leads to distinct changes in the morphology of PB cells (Fig. 5e). Flow cytometry analysis of cells shows that cells in in the monocyte size distribution become smaller (decrease in forward scatter) and more granular (increase in side scatter) compared to cells incubated with PBS (Fig. 5f). This shift in morphology may be indicative of cell death, suggesting that PB monocytes grown ex vivo in the presence of S protein may eventually undergo apoptosis or necrosis.
Fig. 5.
Peripheral blood cells respond ex vivo to exposure to SARS-CoV-2 S protein. (a-d) Low density PB was incubated for 2 h with 1 μg/mL SARS-CoV-2 S recombinant S protein or PBS control. (a) Representative histogram from 1 PB sample treated with S protein or PBS showing CD14 expression of forward and side scatter gated monocytes and (b) mean fluorescence intensities for CD14 staining. n = 4, stats: paired t-test, different colors of points indicate PBs from the same donor. (c) Representative contour plot from 1 PB sample treated with S protein or PBS showing CD14 expression of forward and side scatter gated monocytes, the gating strategy for CD14hi cells and (d) difference in CD14hi monocytes represented as a fold-change of S protein treated cells relative to PBS treated cells. n = 4, stats: paired t-test performed on total numbers of CD14hi monocytes. (e-g) Low density PB was incubated for 18 h with serum in the presence of 1 μg/mL SARS-CoV-2 S recombinant S protein or PBS control. (e) Representative contour plot from 1 PB sample showing all cells excluding debris and the monocyte gate used. (f) Mean values for forward scatter (FSC) and (g) side scatter (SSC) of monocytes from PB samples after incubation with S protein or PBS. n = 5, stats = paired t-test, different colors of points indicate PBs from the same donor. FACS = fluorescence activated flow cytometry. *P < 0.05, ***P < 0.0005
Discussion
Our results reveal previously unknown effects of SARS-CoV-2 S protein exposure on HSCs, HPCs, and immune cells. ACE2, the cell surface receptor protein to which SARS-CoV-2 S protein binds and is currently considered the principal factor required for viral entry to host cells, is expressed at low levels (<5%) on PB immune cells, including T-cells, B-cells, NK cells, NKT cells, and monocytes. Despite this low number of immune cells that are susceptible to ACE2 dependent mechanisms of infection, ex vivo exposure to SARS-CoV-2 S protein leads to distinct morphological changes in PB monocytes. Most primitive and lineage committed HPC derived from CB also include low numbers of ACE2+ cells (<15%). Interestingly, HSCs consistently exhibit the highest percentage of ACE2 expressing cells when compared to HPCs and mature blood cells, though there is variability in ACE2 expressing populations from sample to sample. Importantly, ex vivo exposure of HSCs/HPCs to SARS-CoV-2 S protein alters their functional proliferative/expansion characteristics.
Thus many hematologic manifestations of COVID-19 reported may be due to direct SARS-CoV-2 impact on HSCs/HPCs. Two common hematologic complications associated with COVID-19 are thrombocytopenia and lymphopenia [4, 6, 17]. Interestingly, we observed significantly decreased capacity for the development of MLP when HSCs/HPCs are treated ex vivo with SARS-CoV-2 S protein. MLP are progenitors that differentiate into T-cells, B-cells and NK cells, so dampening the ability of HSCs and MPPs to differentiate to lymphoid primed cells may partly explain the lower numbers of lymphocytes observed in COVID-19 patients. Further, we also observed decreased numbers of CFU-GM and CFU-GEMM when CD34+ cells are grown ex vivo in the presence of S protein, as well as reduced expansion of CMP/MEP/CFU-GEMM/CFU-GM when CD34+ cells are expanded in the presence of S protein. Thus, it is possible that the observed decreases in cells that are precursors to megakaryocytes may partially explain the reduction in circulating platelets in COVID-19 patients. Given that HSCs exhibit a higher subpopulation of ACE2+ cells than other primitive and mature hematopoietic cells, it is also possible that these observations are a result of SARS-CoV-2 impact on HSCs themselves. Because of the large variability that we see in ACE2 expressing HSC numbers, it is possible that the clinical variability associated with SARS-CoV-2 may be in part explained by differing immune responses due to differential susceptibility of HSCs to SARS-CoV-2 infection/interactions. Further, we observed that monocytes exhibit morphology changes that may be indicative of cell death after incubation with the S protein. This suggests that more mature immune cells may be directly affected by SARS-CoV-2 exposure, which could also explain the aforementioned hematologic complications.
These data also provide interesting speculative insight into mechanisms that may be at play in initiating a SARS-CoV-2 induced cytokine storm syndrome (CSS). CSS is an aberrant overactivation of cytokine signaling that leads to a toxic excessive immune response [37–40]. There is evidence that monocytes play a key role in COVID-19 induced CSS [38, 41]. We demonstrated that ex vivo exposure to SARS-CoV-2 S protein leads to an initial increase in CD14hi monocytes. Interestingly, CD14hi cells are increased in abundance and are associated with increased production of inflammatory cytokines in COVID-19 patients in the early recovery stage [40]. We also observed that longer incubation ex vivo with S protein results in marked size reduction and increase in granularity of the total monocyte population, which may be indicative of monocyte cell death. There are two aspects of this study to note with respect to our findings. First, we reported a very low number of immune cells expressing the ACE2 receptor protein that is typically associated with facilitating host cell interactions with SARS-CoV-2. This includes monocytes, of which <1% are ACE2+. This implies that a separate host cell receptor may be interacting with SARS-CoV-2 S protein, underscoring the need to screen for additional host receptors that may facilitate infection. Indeed, it has been postulated that this and other coronaviruses such as SARS-CoV and MERS-CoV can infect host cells via S protein binding to cell surface receptors that are not their “primary” receptor [42–46]. Second, in this study, PB cells and HSCs/HPCs are only being exposed to SARS-CoV-2 protein, not the full viral particle, which indicates that SARS-CoV-2 may impact cells regardless of whether viral entry actually occurs in these cells. Perhaps SARS-CoV-2 S protein induces or blocks signaling in these cells by binding ACE2 or other cell surface receptors. Thus, immune cells may be directly affected by SARS-CoV-2 exposure by interaction with viral proteins and not simply as a result of normal host cell responses to infection. Understanding intracellular effects of SARS-CoV-2 S protein on HSCs/HPCs is a next step in experimental evaluation.
TMPRSS2 is a cell surface protease that has demonstrated catalytic activity against SARS-CoV-2 S protein and ACE2 [13, 36] and may play a role in SARS-CoV-2 infection [13]. During the course of these studies, we also explored TMPRSS2 expression in low density PB and CB CD34+ enriched cells, but we did not detect its expression by RT-qPCR or western blot (data not shown). This may indicate it is not expressed in these cells or it is just expressed on a low percentage of a specific subpopulation of cells and is therefore below our limit of detection. Early studies focusing on SARS-CoV-2 indicated proximal cell surface TMPRSS2 protein is required to cleave S protein into two subunits, S1 and S2, priming the virus for host entry [13]. However, a recent study demonstrated that virus pseudotyped using SARS-CoV-2 S protein can infect cells expressing ACE2 regardless of TMPRSS2 expression, and that infection is not enhanced by expression of TMPRSS2 [47]. Thus, the occurrence of TMPRSS2 expression may not immediately rule out that a specific type of cell can be impacted by interaction with the virus. This is important to consider while evaluating host tissues for susceptibility to viral infection and warrants further studies.
Importantly, we also examined several methods which are proposed to inhibit SARS-CoV-2 infection in the context of neutralizing S protein impacts on HSCs/HPCs. We observed that many of the effects induced by the S protein on hematopoietic cells ex vivo can be neutralized by antibodies targeting the S protein, by treatment with soluble recombinant human ACE2, or by treatment with Angiotensin1–7. Current and past studies have proposed and demonstrated the feasibility of using similar therapeutic interventions for the treatment of SARS-CoV-2 infection [26, 33, 34, 48]. Future studies should explore whether these agents may be used therapeutically to specifically target the effects on the hematopoietic system induced by SARS-CoV-2 exposure, (i.e. to prevent or reverse thrombocytopenia or lymphopenia in COVID-19 patients).
A limitation of our studies is that we performed our functional assays using recombinant S protein instead of live virus. It will be important to determine answers to several outstanding questions to fully understand the impact of SARS-CoV-2 and potentially other coronaviruses on the hematopoietic and immune systems so that we may better understand how to treat this particular facet of the disease: 1) Are HSCs/HPCs and immune cells directly impacted by exposure to live SARS-CoV-2? 2) Are HSCs/HPCs and immune cells simply impacted by exposure to SARS-CoV-2, perhaps by an aberrant induction of signaling pathways, or are these cells actually infected? If they are infected, what form does the infection take (i.e., is it an abortive infection that does not lead to active replication of the virus, thus accounting for the low viral titer found in blood samples?) 3) How long does the virus remain in or interactive with cells? Can HSCs/HPCs and immune cells actually act as harbor for dormant SARS-CoV-2 particles, allowing for a re-emergence of symptoms after the patient initially recovers? 4) Can hematologic manifestations of COVID-19 be treated by directly targeting the mechanisms by which HSCs/HPCs are impacted by SARS-CoV-2, and 5) what are the implications for HCT by bone marrow, CB, or mobilized PB that may have been exposed to SARS-CoV-2?
While the world awaits much-anticipated vaccines to SARS-CoV-2, it is critical for the scientific community to fully assess all host tissues that may be impacted by infection and to explore short- and long-term implications for infections in the context of these tissues. We have demonstrated that specific subpopulations of HSCs/HPCs and immune cells express ACE2 receptor, which may make them vulnerable to infection/interaction with SARS-CoV-2. Importantly, our data along with the recent report by Ratacjzak, et al. [26] suggests that HSCs may be particularly susceptible, and this vulnerability may vary greatly from patient to patient, which may partly explain variability in patient responses that has been reported. We have also shown that exposure to just one small piece of the SARS-CoV-2 viral particle, S protein, can induce both phenotypic and functional changes in HSCs/HPCs as well as aberrant changes in morphology in PB monocytes, suggesting that even absent a true infection of these cells, SARS-CoV-2 may impact the functional ability of these cells, thus affecting the host response to infection.
Electronic supplementary material
(PDF 317 kb)
Funding
This work was supported by US Public Health Service Grants R35HL139599 and U54DK106846 to H.E.B. and T32DK007519 to J.R. (PI H.E.B.). The authors have no conflicts to declare.
Compliance with Ethical Standards
Comment on Ethics
Informed consent was obtained for all donors and normal donor human peripheral blood (PB) was collected in accordance with Indiana University School of Medicine’s Institutional Review Board (Study #1011002987).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
James Ropa, Email: jropa@iu.edu.
Hal E. Broxmeyer, Email: hbroxmey@iupui.edu
References
- 1.Feng W, Zong W, Wang F, Ju S. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review. Molecular Cancer. 2020;19(1):100. doi: 10.1186/s12943-020-01218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JF-W, Yuan S, Kok KH, To KKW, Chu H, Yang J, Xing F, Liu J, Yip CCY, Poon RWS, Tsoi HW, Lo SKF, Chan KH, Poon VKM, Chan WM, Ip JD, Cai JP, Cheng VCC, Chen H, Hui CKM, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J’, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. Extrapulmonary manifestations of COVID-19. Nature Medicine. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 5.Mallapaty S. Mini organs reveal how the coronavirus ravages the body. Nature. 2020;583(7814):15–16. doi: 10.1038/d41586-020-01864-x. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Zhang R, He G. Hematological findings in coronavirus disease 2019: Indications of progression of disease. Annals of Hematology. 2020;99(7):1421–1428. doi: 10.1007/s00277-020-04103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broxmeyer HE, Parker GC. Impact of COVID-19 and future emerging viruses on hematopoietic cell transplantation and other cellular therapies. Stem Cells and Development. 2020;29(10):625–626. doi: 10.1089/scd.2020.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratajczak MZ. How to “detronize” virus in crown – Questions to current stem cell therapies. Stem Cell Reviews and Reports. 2020;16(3):425–426. doi: 10.1007/s12015-020-09978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dholaria B, Savani BN. How do we plan hematopoietic cell transplant and cellular therapy with the looming COVID-19 threat? British Journal of Haematology. 2020;189(2):239–240. doi: 10.1111/bjh.16597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tynell J, Westenius V, Rönkkö E, Munster VJ, Melén K, Österlund P, Julkunen I. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. The Journal of General Virology. 2016;97(2):344–355. doi: 10.1099/jgv.0.000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Chu H, Li C, Wong BHY, Cheng ZS, Poon VKM, Sun T, Lau CCY, Wong KKY, Chan JYW, Chan JFW, To KKW, Chan KH, Zheng BJ, Yuen KY. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: Implications for pathogenesis. The Journal of Infectious Diseases. 2014;209(9):1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang F, Yang J, Zhang Y, Dong M, Wang S, Zhang Q, Liu FF, Zhang K, Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1-7: Novel therapeutic targets. Nature Reviews. Cardiology. 2014;11(7):413–426. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner AJ, Hiscox JA, Hooper NM. ACE2: From vasopeptidase to SARS virus receptor. Trends in Pharmacological Sciences. 2004;25(6):291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee A, Nasir JA, Budylowski P, Yip L, Aftanas P, Christie N, Ghalami A, Baid K, Raphenya AR, Hirota JA, Miller MS, McGeer AJ, Ostrowski M, Kozak RA, McArthur AG, Mossman K, Mubareka S. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases. 2020;26(9):2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan W-J, et al. Clinical characteristics of coronavirus disease 2019 in China. The New England Journal of Medicine. 2020;382(18):1709–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Guo B, Capitano M, Broxmeyer HE (2019). Past, present, and future efforts to enhance the efficacy of cord blood hematopoietic cell transplantation. [version 1; peer review: 3 approved]. F1000Res, 8. [DOI] [PMC free article] [PubMed]
- 19.Huang X, Broxmeyer HE. Progress towards improving homing and engraftment of hematopoietic stem cells for clinical transplantation. Current Opinion in Hematology. 2019;26(4):266–272. doi: 10.1097/MOH.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 20.Broxmeyer HE. Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfusion and Apheresis Science. 2016;54(3):364–372. doi: 10.1016/j.transci.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broxmeyer HE, Milano F, Cord Blood Association (2019). Cord Blood Transplantation: State of the Science. Available at: https://www.cb-association.org/state-of-the-science [Accessed August 21, 2020].
- 22.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung CY, Poon LLM, Ng IHY, Luk W, Sia SF, Wu MHS, Chan KH, Yuen KY, Gordon S, Guan Y, Peiris JSM. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: Possible relevance to pathogenesis. Journal of Virology. 2005;79(12):7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law HKW, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JSM, Lau YL. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Zhou J, Wong BHY, Li C, Chan JFW, Cheng ZS, Yang D, Wang D, Lee ACY, Li C, Yeung ML, Cai JP, Chan IHY, Ho WK, To KKW, Zheng BJ, Yao Y, Qin C, Yuen KY. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. The Journal of Infectious Diseases. 2016;213(6):904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratajczak MZ, et al. (2020). SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45- precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 Inflammasome. Stem Cell Reviews and Reports, 1-12 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 27.Huang X, Guo B, Liu S, Wan J, Broxmeyer HE. Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment. Nature Communications. 2018;9(1):2741. doi: 10.1038/s41467-018-05178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farag SS, Srivastava S, Messina-Graham S, Schwartz J, Robertson MJ, Abonour R, Cornetta K, Wood L, Secrest A, Strother RM, Jones DR, Broxmeyer HE. In vivo DPP-4 inhibition to enhance engraftment of single-unit cord blood transplants in adults with hematological malignancies. Stem Cells and Development. 2013;22(7):1007–1015. doi: 10.1089/scd.2012.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broxmeyer HE, Hoggatt J, O’Leary HA, Mantel C, Chitteti BR, Cooper S, Messina-Graham S, Hangoc G, Farag S, Rohrabaugh SL, Ou X, Speth J, Pelus LM, Srour EF, Campbell TB. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nature Medicine. 2012;18(12):1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo B, Huang X, Lee MR, Lee SA, Broxmeyer HE. Antagonism of PPAR-γ signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nature Medicine. 2018;24(3):360–367. doi: 10.1038/nm.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broxmeyer HE, Srour EF, Hangoc G, Cooper S, Anderson SA, Bodine DM. High-efficiency recovery of functional hematopoietic progenitor and stem cells from human cord blood cryopreserved for 15 years. Proc Natl Acad Sci USA. 2003;100(2):645–650. doi: 10.1073/pnas.0237086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broxmeyer HE, Hangoc G, Cooper S, Ribeiro RC, Graves V, Yoder M, Wagner J, Vadhan-Raj S, Benninger L, Rubinstein P. Growth characteristics and expansion of human umbilical cord blood and estimation of its potential for transplantation in adults. Proc Natl Acad Sci USA. 1992;89(9):4109–4113. doi: 10.1073/pnas.89.9.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magalhaes, G. S., da Rodrigues-Machado, M., G., Motta-Santos, D., Campagnole-Santos, M. J., & RAS, S. (2020). Activation of Ang-(1-7)/mas receptor is a possible strategy to treat coronavirus (SARS-CoV-2). Infection Frontiers in Physiology, 11, 730. [DOI] [PMC free article] [PubMed]
- 34.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alhenc-Gelas F, Drueke TB. Blockade of SARS-CoV-2 infection by recombinant soluble ACE2. Kidney International. 2020;97(6):1091–1093. doi: 10.1016/j.kint.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. Journal of Virology. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta P, McAuley D, Brown M, Sanchez E, Tattersall RS, Manson JJ, HLH Across Speciality Collaboration, UK COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pence BD. Severe COVID-19 and aging: Are monocytes the key? Geroscience. 2020;42(4):1051–1061. doi: 10.1007/s11357-020-00213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. The Journal of Allergy and Clinical Immunology. 2020;146(1):119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen W, Su W, Tang H, le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Dong L, Cui X, Miao Y, Wang D, Dong J, Xiao C, Chen W, Wang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ognibene A, Lorubbio M, Magliocca P, Tripodo E, Vaggelli G, Iannelli G, Feri M, Scala R, Tartaglia AP, Galano A, Pancrazzi A, Tacconi D. Elevated monocyte distribution width in COVID-19 patients: The contribution of the novel sepsis indicator. Clinica Chimica Acta. 2020;509:22–24. doi: 10.1016/j.cca.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, et al. Hematological findings in SARS patients and possible mechanisms (review) International Journal of Molecular Medicine. 2004;14(2):311–315. [PubMed] [Google Scholar]
- 43.Yu X-J, Luo C, Lin JC, Hao P, He YY, Guo ZM, Qin L, Su J, Liu BS, Huang Y, Nan P, Li CS, Xiong B, Luo XM, Zhao GP, Pei G, Chen KX, Shen X, Shen JH, Zou JP, He WZ, Shi TL, Zhong Y, Jiang HL, Li YX. Putative hAPN receptor binding sites in SARS_CoV spike protein. Acta Pharmacologica Sinica. 2003;24(6):481–488. [PubMed] [Google Scholar]
- 44.Chan C-M, Chu H, Wang Y, Wong BHY, Zhao X, Zhou J, Yang D, Leung SP, Chan JFW, Yeung ML, Yan J, Lu G, Gao GF, Yuen KY. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome coronavirus. Journal of Virology. 2016;90(20):9114–9127. doi: 10.1128/JVI.01133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ropa J, Broxmeyer HE. An expanded role for dipeptidyl peptidase 4 in cell regulation. Current Opinion in Hematology. 2020;27(4):215–224. doi: 10.1097/MOH.0000000000000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 317 kb)