Abstract
Coronavirus infection causes diffuse alveolar damage leading to acute respiratory distress syndrome. The absence of ex vivo models of human alveolar epithelium is hindering an understanding of coronavirus disease 2019 (COVID-19) pathogenesis. Here, we report a feeder-free, scalable, chemically defined, and modular alveolosphere culture system for the propagation and differentiation of human alveolar type 2 cells/pneumocytes derived from primary lung tissue. Cultured pneumocytes express the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor angiotensin-converting enzyme receptor type-2 (ACE2) and can be infected with virus. Transcriptome and histological analysis of infected alveolospheres mirror features of COVID-19 lungs, including emergence of interferon (IFN)-mediated inflammatory responses, loss of surfactant proteins, and apoptosis. Treatment of alveolospheres with IFNs recapitulates features of virus infection, including cell death. In contrast, alveolospheres pretreated with low-dose IFNs show a reduction in viral replication, suggesting the prophylactic effectiveness of IFNs against SARS-CoV-2. Human stem cell-based alveolospheres, thus, provide novel insights into COVID-19 pathogenesis and can serve as a model for understanding human respiratory diseases.
Keywords: organoids, pneumocytes, SARS-CoV-2, interferons, surfactants, ACE2, ARDS, protease, respiratory cells, cytokine storm
Graphical Abstract

Highlights
-
•
Stroma-free long-term expansion and differentiation of adult human lung stem cells
-
•
AT2 response to SARS-CoV-2 infection mirrors features of COVID-19 lungs
-
•
Infected AT2s upregulate IFNs and apoptotic pathways and decrease surfactants
-
•
Low-dose IFN pre-treatment blocks SARS-CoV-2 replication in alveolospheres
Tata and colleagues report defined conditions for long-term expansion and differentiation of adult human primary alveolar stem cells. Cultured AT2s are conducive to SARS-CoV-2 infection and elicit transcriptome-wide changes that mirror COVID-19 histopathology, including upregulation of inflammatory responses, cell death, and downregulation of surfactant expression, leading to pneumocyte dysfunction.
Introduction
In coronavirus disease 2019 (COVID-19), lung disease is the primary cause for mortality. Histopathological analyses reveal widespread alveolar damage and pneumonia, which may eventually progress to acute respiratory distress syndrome (ARDS) (Bradley et al., 2020). This clinically challenging manifestation is accompanied by the production of multiple cytokines (“cytokine storm”), loss of parenchyma, immune infiltration, and fluid filled alveoli, all of which contribute to acute respiratory failure and eventual death (Huang et al., 2020a; Zhu et al., 2020). The causative agent of COVID-19 is the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus uses the same receptor—ACE2 (angiotensin-converting enzyme receptor type-2)—for entry into target cells as the closely related viruses SARS-CoV (2003) and NL-63 (Hoffmann et al., 2020). However, the unique clinical symptoms and increased transmissibility of SARS-CoV-2 suggest that it uses different mechanisms to both infect and evade host immune responses, including the production of type I and type III interferons (IFN-IIIs) (Hou et al., 2020; Huang et al., 2020a; Wu and McGoogan, 2020; Zhu et al., 2020). To develop safe and effective therapies for COVID-19, it is critically important to understand the cell-type-specific innate immune mechanisms triggered in response to viral entry and how they orchestrate adaptive immune responses. One way to achieve this goal is to infect target cells of the adult human lung with virus ex vivo and to follow molecular and cellular responses over time. Ideally, this approach must be performed under well-defined, modular conditions that can easily be adapted to high-throughput pharmaco-genomic screens for therapeutic discovery. We report here the results of this approach by using SARS-CoV-2 infection of 3D alveolosphere cultures of primary human alveolar epithelial type-2 cells (AT2s), the stem cells of the distal alveolar region.
Single-cell transcriptome profiling and immunolocalization studies showed that AT2s exhibit the highest enrichment of SARS-CoV-2 receptor ACE2, and its associated protease TMPRSS2, in the human distal lung (Hou et al., 2020; Muus et al., 2020; Sungnak et al., 2020; Ziegler et al., 2020). AT2s can both self-renew and differentiate into thin and flat gas exchanging alveolar epithelial type-1 cells (AT1s). In addition, they secrete surfactant proteins, namely, SFTPA and SFTPD, that promote alveolar patency but also can directly bind many viruses and other microbial pathogens to facilitate opsonization and phagocytosis (Crouch and Wright, 2001; McCormack and Whitsett, 2002). Therefore, AT2s play a key role in providing a first line of defense against viruses and in restoring cell numbers after injury. However, currently we do not know the nature of the pathways that are dysregulated in human AT2s in response to SARS-CoV-2 infection and how these pathways intersect with other forms of defense mechanisms. It is also unclear whether and how AT2s maintain stem cell characteristics while activating anti-viral defense mechanisms. Alveolosphere cultures derived from adult AT2s provide the opportunity to address these questions.
Numerous studies have demonstrated the potential of primary-tissue-derived organoids to serve as models for disease pathogenesis, organogenesis, and tissue repair (Drost and Clevers, 2018; Jacob et al., 2017; Lancaster and Huch, 2019; Lancaster and Knoblich, 2014; Neal et al., 2018; Yamamoto et al., 2017). For example, recent studies using intestinal organoids combined with SARS-CoV-2 infection revealed the infectability of intestinal epithelium and associated cellular responses (Lamers et al., 2020; Yang et al., 2020). In the case of the lung, AT2s have the ability to generate alveolospheres, which can proliferate and differentiate into AT1s (Barkauskas et al., 2013, 2017; Chung et al., 2018; Dye et al., 2015; Hogan and Tata, 2019; Katsura et al., 2019; Lancaster and Knoblich, 2014; Lee et al., 2013; Nikolić et al., 2018; Shiraishi et al., 2019a). However, current conditions require the co-culture of AT2s with PDGFRα+ fibroblasts isolated from the alveolar stem cell niche or lung endothelial cells isolated from fetal tissues (Barkauskas et al., 2017; Lancaster and Huch, 2019; McQualter et al., 2010). In addition, current culture media are poorly defined and contain unknown factors derived from fetal bovine serum (FBS) or calf serum and bovine pituitary extracts (Barkauskas et al., 2017). Such complex conditions do not provide a modular system in which AT2s can be either selectively expanded or differentiated into AT1 (Shiraishi et al., 2019a, 2019b; Weiner et al., 2019). Such defined conditions are needed to study cell-type-specific effects and for high-throughput pharmaco-genomic studies to discover drugs for treating diseases.
To overcome these challenges, we have developed chemically defined conditions for human AT2 expansion and differentiation in alveolosphere cultures. We demonstrate that SARS-CoV-2 infects and propagates in AT2s in these alveolospheres. Complementary assays were used to assess the transcriptome-wide changes in response to SARS-CoV-2 infection, and the results were directly compared with transcriptome data from COVID-19 patients. Furthermore, we show that viral infection induces the production of IFNs and that different types of IFNs affect AT2 behavior in alveolosphere culture.
Results
Establishment of Chemically Defined Conditions for Alveolosphere Cultures
The cellular composition and properties of 3D culture models are highly dependent on culture conditions (Barkauskas et al., 2017; Drost and Clevers, 2018; Hu et al., 2018; Huch et al., 2013; Neal et al., 2018; Peng et al., 2018; Velasco et al., 2019). Purified, lineage-labeled mouse AT2s can be grown as alveolospheres (Barkauskas et al., 2013, 2017; Katsura et al., 2019; Lee et al., 2013). However, current culture conditions use a complex medium containing many variable components (serum and bovine pituitary extract) and require the addition of lung-resident PDGFRα+ fibroblasts to support AT2 growth. We, therefore, established defined conditions for long-term propagation of AT2s. Initially we used mouse cells, and then subsequently similar conditions were adapted for human AT2 cultures. Briefly, we performed single-cell transcriptome analysis on AT2s grown in mouse tracheal epithelial cell (MTEC) media (Figures S1A and S1B) and then mined the single-cell RNA sequencing (scRNA-seq) data for ligand-receptor pairs differentially expressed in epithelial cells and fibroblasts (Figures S1C and S1D). Based on these data, we tested different combinations of ligands and small molecules, including interleukin-1β (IL-1β), a recently identified AT2 niche-derived molecule, in a basal medium to generate a serum-free feeder-free medium (SFFF) (Figures S1E–S1H; see Table S1 for the full composition of the medium) (Katsura et al., 2019). SFFF medium supports the growth of AT2s characterized by numerous mature lamellar bodies packed with surfactants (Figure S1I). Of note, although the addition of IL-1β enhanced alveolosphere size, it did not increase alveolosphere number. Recent studies identified that inhibition of bone morphogenetic protein (BMP) signaling prevents AT2 differentiation (Chung et al., 2018). Therefore, to promote a higher proportion of AT2s in alveolospheres, we supplemented SFFF medium with inhibitors of BMP signaling (Noggin and DMH1), to generate AT2 maintenance medium (AMM) (Figure S2A). In this medium, pneumocytes maintain AT2 identity and can be sub-cultured for over six passages (Figures S2B–S2F). Furthermore, we formulated a medium (AT2 differentiation medium [ADM]) that contains 10% FBS and stimulated AT2 differentiation, leading to a dramatic induction of cells expressing markers of AT1s (see Table S1 for the full composition of the media; Figures S2G and S2H).
Defined Culture Conditions for Human Alveolosphere Cultures
Next, we sought to establish SFFF culture conditions for human AT2s. We purified AT2s from healthy lungs using HTII-280 antibody (Gonzalez et al., 2010) and established alveolosphere cultures using SFFF medium with human-specific recombinant proteins in the absence of stromal cells (Table S2; Figure S3A). Anddition of IL-1β for the first 4–7 days of culture slightly enhanced the size and number of alveolospheres, although this effect varied in magnitude from donor to donor (Figures S3B–S3D). Therefore, we did not use IL-1β treatment for further experiments. Long-term sub-culture over 10 passages and sphere quantification (P6) confirmed the ability of SFFF medium to sustain long-term human AT2 self-renewal and maintenance of morphology across passages (Figures 1 A–1C; Figures S3E and S3F). Immunostaining for general lung epithelial cell (NKX2-1) and AT2-specific markers (SFTPC, SFTPB, HTII-280, and DC-LAMP) revealed that alveolospheres were composed solely of AT2s and that neither airway (SOX2, SCGB1A1, and TP63) nor AT1s (AGER) were present (Figures 1D–1F; Figures S3G and S3H). Quantitative RT-PCR (qRT-PCR) analyses for AT2 markers further corroborated these findings (Figure S3F).
Figure 1.
Establishment of Chemically Defined Human Lung Alveolosphere Culture System
(A) Schematic representation of human alveolosphere cultures and passaging in SFFF medium.
(B) Representative images of human alveolospheres from different passages. Scale bar: 100 μm.
(C) Quantification of the colony formation efficiency (CFE) of human alveolospheres at different passages.
(D) Immunostaining for SFTPC (green), SFTPB (red), and AGER (gray) (left panel) or SFTPB (green), HTII-280 (red), and DC-LAMP (gray) (right panel) at P1 and P3 human alveolospheres cultured in SFFF medium for 14 days.
(E) Immunostaining for SFTPC (green) and HTII-280 (red) in cells dissociated from alveolospheres at P2 (top) and P8 (bottom).
(F) Quantification of HTII-280+ SFTPC+ cells/total 4′,6-diamidino-2-phenylindole (DAPI)+ cells derived from alveolospheres dissociation from P2 (orange) and P8 (blue).
(G) Schematic representation of human AT2 to AT1 differentiation in alveolospheres. AT2s were cultured in SFFF medium for 10 days, followed by culture in ADM for 14 days.
(H) Immunostaining for SFTPC (green) and AGER (red) in human alveolospheres cultured under ADM condition for 14 days. Scale bars: 100 μm (B); 50 μm (D); 20 μm (E); 20 μm (H). DAPI (blue) shows nuclei in (D), (E), and (H). Data are presented as mean ± SEM.
To induce the differentiation of AT2s, we first tested differentiation medium containing 10% bovine serum, but there were few or no AGER+ (AT1) cells in alveolospheres (Figure S3I). We therefore switched to human serum and found that this change induced a robust expression of the AT1 marker AGER that was co-incident with a decrease in SFTPC (Figures 1G and 1H; Figure S3J). Significantly, the AGER+ cells show the large, thin, and flat morphology characteristic of type-1 pneumocytes in vivo (Figure 1H). Collectively, these data indicate that our newly developed SFFF culture conditions facilitated the long-term expansion of primary human AT2s in the absence of feeder cells and that the addition of human serum stimulated the cells to differentiate into AT1s.
Alveolosphere-Derived AT2s Express Viral Receptors and Are Permissive to SARS-CoV-2 Infection
Recent studies have indicated that SARS-CoV-2 receptor ACE2 and a key protease, TMPRSS2, which is needed for proteolytic cleavage of the viral spike protein, are expressed in AT2s (Hoffmann et al., 2020; Hou et al., 2020; Muus et al., 2020; Sungnak et al., 2020; Ziegler et al., 2020). We therefore assessed the expression and localization of ACE2 and TMPRSS2 in pneumocytes derived from alveolospheres cultured in SFFF media or ADM that contain AT2s only or a mixture of AT2 and AT1s, respectively. Immunostaining in combination with a well-known apical marker of AT2 (HTII-280), polarity marker ZO1, and membrane marker EpCAM showed that ACE2 is localized at the apical surface (similar to HTII-280 and ZO1), whereas TMPRSS2 is enriched at the basal side of AT2s (Figures 2 A–2C). We then quantified the number of AT2s that express ACE2 and TMPRSS2 on single-cell preparations of alveolospheres. Our data revealed that 40% of AT2s express ACE2, whereas about 80% are positive for TMPRSS2 (Figures 2D–2F). We did not find ACE2 and TMPRSS expression in differentiated (AGER+) AT1s, a finding consistent with prior single-cell transcriptome as well as immunolabeling assays on human lungs (Figure 2G; Hou et al., 2020; Muus et al., 2020).
Figure 2.
Alveolosphere-Derived AT2s Express Viral Receptors and Are Permissive to SARS-CoV-2 Infection
(A) Immunostaining for ACE2 (green) (left panel) and TMPRSS2 (green) (right panel) with HTII-280 (red) and AGER (gray).
(B) Co-staining for epithelial cell membrane marker EPCAM (green) with TMPRSS2 (red) and ACE2 (gray).
(C) Co-staining for ACE2 (red) (left panel) and TMPRSS2 (red) (right panel) with polarity marker ZO1 (green) and HTII-280 (gray).
(D) Co-immunostaining for ACE2 (green) (left panel) and TMPRSS2 (green) (right panel) with HTII-280 (red) in AT2s dissociated from P5-cultured alveolospheres.
(E) Co-immunostaining for ACE2 (green), HTII-280 (red), and TMPRSS2 (gray) in AT2s from alveolospheres.
(F) Quantification of percent ACE2+/total HTII-280+ cells (left) and percent TMPRSS2+/total HTII-280+ cells.
(G) Immunostaining for ACE2 (red) (left panel) and TMPRSS2 (red) (left panel) with HTII-280 (green) and AGER (gray) in alveolospheres cultured in ADM for 14 days.
(H) Schematic representation for SARS-CoV-2-GFP infection in human alveolospheres. AT2s were cultured on Matrigel-coated plates in SFFF medium for 10–12 days, followed by infection with SARS-CoV-2 virus and RNA isolation or histological analysis after different time points.
(I) Representative wide-field microscopy images from control and SARS-CoV-2-GFP-infected human lung alveolospheres.
(J) Quantification of low-infected (1–10 GFP+ cells) and high-infected (10 or more GFP+ cells) alveolospheres 24, 48, and 72 h post SARS-CoV-2GFP virus infection.
(K) Viral titers were measured by plaque assays using media collected from lung alveolosphere cultures at 24, 48, and 72 h post-infection.
(L) SARS-CoV-2 negative-strand-specific reverse transcription followed by qRT-PCR targeting two different genomic loci (1202–1363 and 848–981) in mock- (blue) and SARS-CoV-2-infected human alveolospheres at 72 h post-infection. Asterisks show p < 0.05. Scale bars: 30 μm (A, B, and C); 20 μm (D); and 20 μm (F). White box in merged image indicates region of single-channel images. All quantification data are presented as mean ± SEM.
To test whether SARS-CoV-2 can infect alveolosphere-derived AT2s, we used a recently developed reverse-engineered SARS-CoV-2 virus harboring a GFP-fusion protein (Hou et al., 2020). Human alveolospheres were cultured on a Matrigel surface in SFFF media (lacking IL-1β) for 10–12 days, incubated with SARS-CoV-2-GFP for 2 h, washed with phosphate-buffered saline (PBS) to remove residual viral particles, and then collected for analysis over 72 h. GFP was detected as early as 48 hours post-infection (hpi) in virus-exposed but not in control alveolospheres (Figures 2H and 2I). Quantification of GFP-expressing cells at 24, 48, and 72 hpi revealed a gradual decrease in the number of GFP+ cells (Figure 2J). Consistent with this finding, plaque-forming assays using culture supernatants revealed that viral release peaks at 24 h but later declined (Figure 2K). This observation was consistent across cells from three different donors. Of note, we observed a significant number of viral particles immediately after infection despite numerous washes with PBS. This result was likely due to the entrapment of virus in the Matrigel. Nevertheless, the viral titer increased at 24 hpi, demonstrating that SARS-CoV-2 productively replicates in AT2s (Figure 2K). qRT-PCR further revealed the presence of viral RNA in SARS-CoV-2-infected cells compared to controls (Figure S4A). To further confirm virus replication, we performed qRT-PCR using primers that specifically recognize the minus strand of the virus. Indeed, we observed viral replication in alveolosphere cultures (Figure 2L).
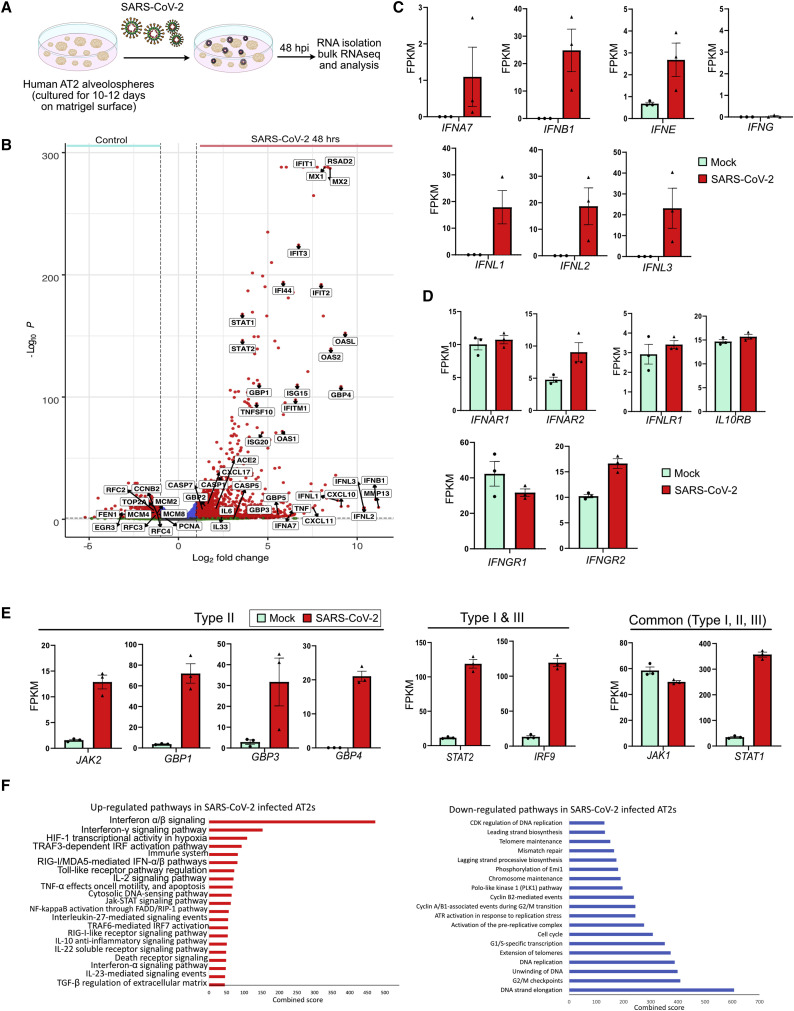
AT2s Activate IFN and Inflammatory Pathways in Response to SARS-CoV-2 Infection
To gain insights into the response of AT2s to SARS-CoV-2 (wild type), we performed unbiased genome-wide transcriptome profiling on alveolospheres cultures 48 h after infection (Figure 3 A). Of all the sequenced reads, viral transcripts accounted for 4.7%, indicating that virus was propagating in AT2s (Figure S4B). Previous studies have shown that in response to viral infection, target cells typically produce type I IFNs (IFN-Is) and IFN-IIIs (α/β and λ, respectively) that subsequently activate targets of transcription factors IRF, STAT1/2, and NF-κB including, IFN-stimulated genes (ISGs), inflammatory chemokines, and cytokines that go on to exert anti-viral defense mechanisms (Barrat et al., 2019). It was therefore significant that a differential gene expression analysis of infected versus uninfected alveolospheres revealed an enrichment of transcripts related to general viral response genes, including multiple IFNs and their targets (Figures 3A–3F). Specifically, SARS-CoV-2-infected AT2s were enriched for transcripts of IFN-Is (IFNA7, IFNB1, and IFNE) as well as IFN-IIIs (IFNL1, IFNL2, and IFNL3) but not type II IFN (IFNG) ligands (Figures 3B and 3C; Figure S4C). Receptors for IFN-Is (IFNAR1 and IFNAR2), type II IFNs (IFNGR1 and IFNGR2) and IFN-IIIs (IFNLR1 and IL10RB) were expressed in control AT2s, and a modest increase was found for IFNAR2 and IFNGR2 after SARS-CoV-2 infection (Figures 3B and 3D; Platanias, 2005; Syedbasha and Egli, 2017). These data indicate that in response to SARS-CoV-2 infection, AT2s produce IFN-I and IFN-III ligands, which can potentially act by either by autocrine or paracrine (neighboring AT2s) mechanisms to activate their cognate receptors. Indeed, a large number of IFN target genes including ISGs, IFN-induced protein-coding genes (IFIs), and IFN-induced protein with tetratricopeptide repeats-coding genes (IFITs), were upregulated in SARS-CoV-2-infected AT2s (Figures 3B and 3E). Additionally, key transcription factors STAT1 and STAT2 that are known to be components of the signaling pathways downstream of IFN receptors were also upregulated in infected AT2s.
Figure 3.
Transcriptome Profiling Revealed Enrichment of IFN, Inflammatory, and Cell Death Pathways in SARS-CoV-2-Infected Pneumocytes
(A) Schematic for SARS-CoV-2-GFP infection in human alveolospheres. AT2s cultured in SFFF medium were infected with SARS-CoV-2 virus followed by RNA isolation at 48 h after infection.
(B) Volcano plot showing upregulated (right) and downregulated (left) genes in alveolospheres cultured in SFFF infected with SARS-CoV-2. DESeq2 was used to perform statistical analysis.
(C–E) Expression levels of listed genes in mock- (green) and SARS-CoV-2-infected (red) human alveolospheres detected by bulk RNA-seq. IFN ligands (C), receptors (D), as well as downstream targets (E), are shown. Data are presented as fragments per kilobase million (FPKM) mean ± SEM.
(F) Pathway enrichment analysis of upregulated (left, red) and downregulated (right, blue) genes in SARS-CoV-2-infected alveolospheres. Scale shows the combined score obtained from BioPlanet database through Enrichr.
Pathway analysis revealed all three classes of IFN targets were upregulated, but the most prominent were IFN-I and IFN-III signaling. Despite the absence of type II IFN ligands (IFNG), we observed a significant upregulation of canonical targets of IFNγ-response mediators in SARS-CoV-2-infected AT2s (Figures 3B and 3E). This finding suggests that there is a significant overlap of downstream targets and cross-talk between different classes of IFN pathways, as described previously (Barrat et al., 2019; Bartee et al., 2008). Other prominent upregulated genes include chemokines (CXCL10, CXCL11, and CXCL17) and programmed cell-death-related genes (TNFSF10, CASP1, CASP4, CASP5, and CASP7) (Figure 3B; Figures S4D and S4E). In contrast, we observed a significant downregulation of transcripts associated with DNA replication and cell cycle (PCNA, TOP2A, MCM2, and CCNB2) in infected AT2s (Figure 3B; Figure S4F). Selected targets were validated using independent qRT-PCR assays at early (48 h) and late (120 h) time points post-infection (Figure S5A). Taken together, transcriptome analysis revealed a significant upregulation of IFN and inflammatory and cell death signaling, juxtaposed to the downregulation of proliferation-related transcripts, in alveolosphere-derived AT2s in response to SARS-CoV-2.
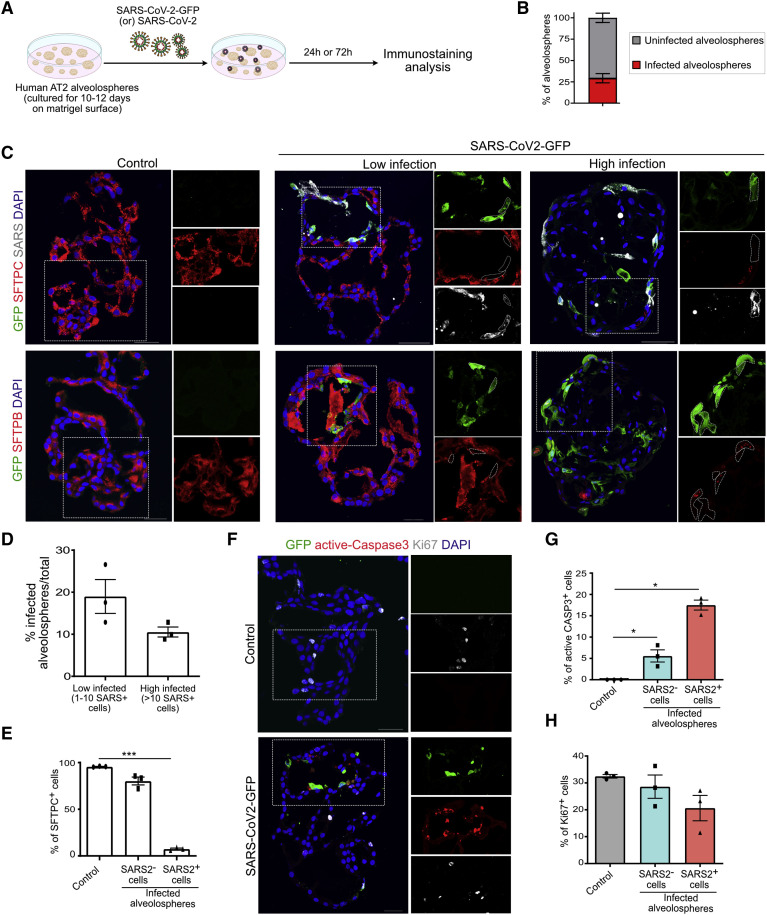
SARS-CoV-2 Infection Induces Loss of Surfactants and Pneumocyte Death
To gain further insights into how primary AT2s respond early to SARS-CoV-2 infection, we analyzed cellular changes in alveolospheres by using immunohistochemistry (Figure 4 A). Quantification of infected alveolospheres revealed that 29.22% are SARS+ (Figure 4B). Immunostaining revealed the co-expression of GFP and SARS-CoV-2 spike protein in infected alveolospheres (Figure 4C). We found variation in the number of GFP+ cells in each alveolosphere. Therefore, we broadly categorized alveolospheres into low (1–10 cells) and high (>10) groups, depending on the number of SARS+ cells in each alveolosphere (Figures 4C and 4D). Next, analyses for AT2 markers, including SFTPC, SFTPB, and HTII-280, revealed a dramatic loss or decrease in the expression of surfactant proteins SFTPC and SFTPB in infected cells (GFP+ or SARS+) but not in control alveolospheres (Figures 4C and 4E; Figure S5B). Of note, HTII-280 expression was unchanged (Figure S5C). The loss of surfactant protein expression was more apparent in highly infected alveolospheres (Figure S5B). Some of the GFP+ cells showed a slightly elongated morphology, resembling that of AT1s, but immunostaining for AT1 markers revealed that infected cells did not differentiate into AT1s (Figure S5D). These data are in accordance with our scRNA-seq data that AT2s downregulate surfactant expression in response to SARS-CoV-2 infection.
Figure 4.
SARS-CoV-2 Infection Induces Loss of Surfactants and AT2 Death
(A) Schematic for SARS-CoV-2-GFP infection in human alveolospheres. Alveolospheres were cultured in SFFF medium, infected with SARS-CoV-2 virus, and collected for histological analysis.
(B) Quantification of the percentage of SARS-CoV-2-infected alveolospheres.
(C) Immunostaining for GFP (green), SFTPC (red), and SARS (gray) (top panel) and GFP (green) and SFTPB (red) (bottom panel) in control, “low,” and “high” SARS-CoV-2-GFP-infected human lung alveolospheres at 72 h post-infection. Scale bar: 50 μm.
(D) Quantification of low-infected (1–10 SARS-CoV-2+ cells) and high-infected (10 or more SARS-CoV-2+ cells) alveolospheres.
(E) Quantification of SFTPC+ cells in uninfected control and SARS− and SARS+ cells in virus-infected alveolospheres.
(F) Immunostaining for GFP (green) in combination with the apoptotic marker active caspase 3 (red) and proliferation marker Ki67 (gray) in control and SARS-CoV-2-GFP-infected alveolospheres. Scale bar: 30 μm.
(G and H) Quantification of active caspase-3 (CASP3)+ (G) and Ki67+ (H) cells in uninfected control (gray), SARS-CoV-2− cells (blue), and SARS-CoV-2+ cells in infected alveolospheres. The white box in the merged image indicates the region of single-channel images. DAPI stains nuclei (blue). All quantification data are presented as mean ± SEM.
Histopathological evidence suggests that there is a loss of alveolar parenchyma in COVID-19 lungs (Bradley et al., 2020; Huang et al., 2020a). To test whether SARS-CoV-2 infection induces cell death, we performed immunostaining for active caspase-3, a marker for apoptotic cells. Apoptotic cells were found in alveolospheres exposed to virus but not in controls, suggesting that AT2s undergo cell death in response to SARS-CoV-2 infection (Figure 4F). Significantly, we observed cell death in both SARS+ and SARS− cells suggesting a paracrine mechanism inducing cell death in uninfected neighboring cells (Figure 4G). Furthermore, immunostaining for Ki67, a marker for proliferating cells revealed no apparent difference in overall cell replication in virus-exposed alveolospheres compared to controls (Figure 4H). Taken together, these data show that SARS-CoV-2 infection induces the downregulation of surfactant proteins and an increase in cell death in AT2s by both cell autonomous and non-autonomous mechanisms.
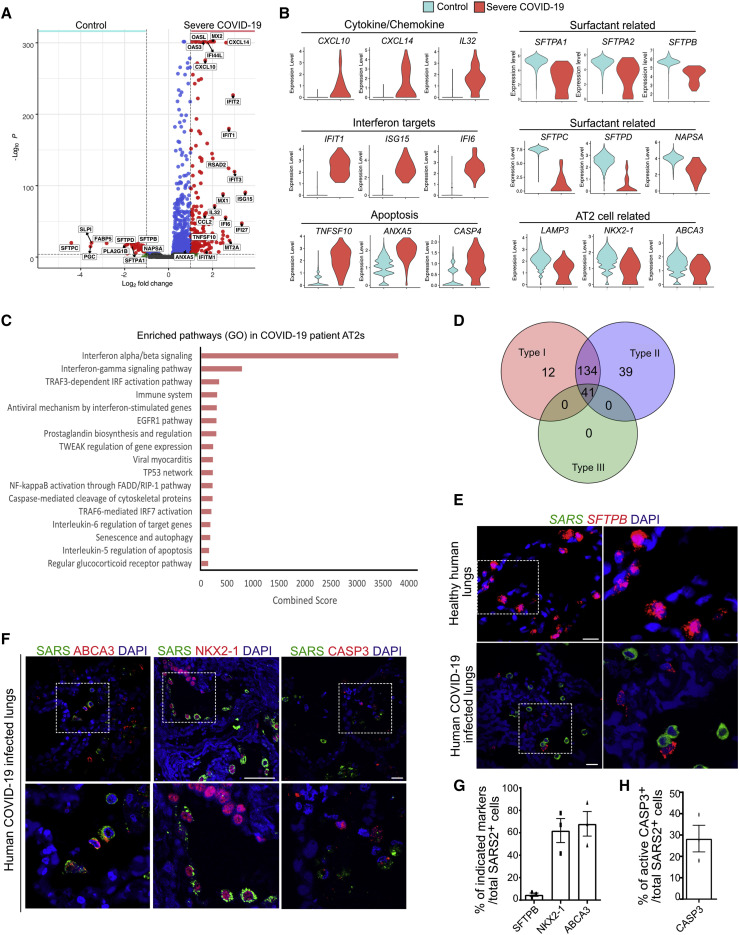
Transcriptome-wide Similarities in AT2s from SARS-CoV-2-Infected Alveolospheres and COVID-19 Lungs
To directly compare SARS-CoV-2-induced responses in AT2s in alveolospheres to changes seen in COVID-19 lungs, we used a publicly available scRNA-seq dataset from bronchoalveolar lavage fluid (BALF) obtained from six severe COVID-19 patients (Bost et al., 2020; Liao et al., 2020). First, we compared the gene expression profiles of AT2s from COVID-19 patient lungs with AT2s from healthy lungs (Figures S6A–S6D). We found a significant upregulation of chemokines (CXCL10, CXCL14, and IL32), IFN targets (IFIT1, ISG15, and IFI6), and cell-death-pathway (TNFSF10, ANXA5, and CASP4) -related transcripts in COVID-19 patient AT2s (Figures 5A and 5B). Intriguingly, surfactant genes, including SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD, as well as NAPSA, a gene product that catalyzes the processing of the pro-form of surfactant proteins into mature proteins, were significantly downregulated in COVID-19 patient AT2s, whereas changes in other AT2 markers were minimal and insignificant (Figures 5A and 5B). Pathway analysis revealed a significant enrichment for IFN-I and type II IFN signaling, inflammatory programs, and cell death pathways in COVID-19 AT2s (Figures 5C and 5D). We then made a direct comparison of transcripts between AT2s from SARS-CoV-2-infected ex vivo cultures and COVID-19 patient lungs. This comparison revealed a striking similarity in upregulated transcripts (Figure S6E). These included an upregulation of chemokines and cytokines, including IFN ligands and their targets, indicating that AT2s derived from alveolospheres respond similarly to AT2s from human lungs after SARS-CoV-2 infection (Figures S6E and S6F). We then extended these findings to COVID-19 lungs and uncovered that SFTPB but not other AT2s markers, NKX2-1 or ABCA3, were downregulated in SARS+ cells (Figures 5E–5G). Similar to alveolospheres, we observed active caspase-3 in SARS+ cells in COVID-19 human lungs (Figure 5H).
Figure 5.
Transcriptome-wide Similarities in AT2s from SARS-CoV-2-Infected Alveolospheres and COVID-19 Lungs
(A) Volcano plot shows specific genes enriched in AT2s in bronchioalveolar lavage fluid from severe COVID-19 patients (right) and AT2s isolated from healthy lungs (control) (left). Wilcoxon rank-sum test was used for the statistical analysis.
(B) Violin plots show gene expression of cytokines and chemokines (CXCL10, CXCL14, and IL32), interferon targets (IFIT1, ISG15, and IFI6), apoptosis targets (TNFSF10, ANXA5, and CASP4), surfactant-related targets (SFTPC, SFTPD, and NAPSA), and AT2-related targets (LAMP3, NKX2-1, and ABCA3) in AT2s derived from control and severe COVID-19 patient lungs.
(C) Pathway enrichment analysis shows signaling pathways enriched in AT2s derived from severe COVID-19 patients. Scale shows combined score obtained from BioPlanet database through Enrichr.
(D) Venn diagram shows enrichment of upregulated transcripts associated with different IFN pathways in AT2s derived from COVID-19 human lungs.
(E) RNA in situ hybridization for SARS-CoV-2 (green) and SFTPB (red) on control and COVID-19 lung sections.
(F) Co-immunostaining for SARS-CoV-2 (green), ABCA3 (red) (left); SARS-CoV2 (green) and NKX2-1 (red) (middle); and active caspase-3 (CASP3) (red) (right) on COVID-19 lung sections. DAPI stains nuclei. Scale bars: 20 μm and 60 μm (NKX2-1).
(G) Quantification of SFTPB+ cells, NKX2-1+ cells, and ABCA3+ cells in total SARS-CoV-2+ cells.
(H) Quantification of active CASP3+ cells in total SARS-CoV-2+ cells. Data are presented as mean ± SEM.
AT2s Respond to Exogenous IFNs and Recapitulate Features Associated with SARS-CoV-2 Infection
Our transcriptome analysis revealed a striking similarity in IFN signatures in AT2s from alveolospheres and human lungs after SARS-CoV-2 infection. Previous studies have shown that IFNs induce cellular changes in a context-dependent manner. For example, IFNα and IFNβ provide protective effects in response to influenza virus infection in the lungs, whereas IFNγ induces apoptosis in intestinal cells in response to chronic inflammation (Koerner et al., 2007; Takashima et al., 2019). To test the direct effects of IFNs on AT2s, we treated alveolospheres with purified recombinant IFNα, IFNβ, and IFNγ in SFFF media and cultured them for 72 h (Figure 6 A). First, we observed detached cells in all treatments, with a maximal ∼3-fold increased effect in IFNγ-treated alveolospheres (Figure 6B). Immunostaining for active caspase-3 revealed a significant induction of cell death in response to all IFN treatments, with a maximal effect with IFNγ (Figures 6C and 6D; Figure S7A). In contrast, we observed a significant reduction in cell proliferation in IFNβ and IFNγ treatments, as revealed by immunostaining for Ki67, a marker for cell proliferation (Figures 6E and 6F). Significantly, immunostaining revealed a reduction of SFTPB expression in alveolospheres treated with all IFNs compared with that of controls (Figure 6E). A similar trend was observed for SFTPC and SFTPB transcripts, as assessed by qRT-PCR (Figures S7B and S7C). These data are in accordance with transcriptome data from AT2 alveolospheres after SARS-CoV-2 infection (Figure 3B). Of note, treatment with IFNα, IFNβ, and IFNγ significantly enhanced the levels of ACE2, but not TMPRSS2 transcripts, which is in line with results of previous studies in other cell types (Hou et al., 2020; Ziegler et al., 2020; Figures S7D and S7E). A similar trend was observed in SARS-CoV-2-infected cells, suggesting a positive loop that involves IFNs and ACE2 that subsequently amplifies SARS-CoV-2 infection (Figure S7F).
Figure 6.
IFN Treatment Recapitulates Features of SARS-CoV-2 Infection Including Cell Death and Loss of Surfactants in Alveolosphere-Derived AT2s
(A) Schematic of experimental design. Human lung alveolospheres were treated with IFNα, IFNβ or IFNγ for 72 h.
(B) Representative images of control and IFNα-, IFNβ-, and IFNγ-treated human lung alveolospheres.
(C) Immunostaining for active-caspase-3 (green), HTII-280 (red), and SOX2 (gray) in control and IFN-treated alveolospheres. DAPI stains nuclei (blue). Scale bar: 30 μm.
(D) Quantification of active caspase-3+ cells in total DAPI+ (per alveolosphere) cells in control and IFN-treated human alveolospheres.
(E) Immunostaining for SFTPB (green), Ki67 (red), and AGER (gray) in controls and IFNα-, IFNβ-, or IFNγ-treated human alveolospheres. DAPI stains nuclei (blue). Scale bar: 30 μm.
(F) Quantification of Ki67+ cells in total DAPI+ cells in control and IFN-treated human alveolospheres. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(G) Schematic of IFNs or IFN inhibitor treatment followed by SARS-CoV-2 infection.
(H) Viral titers in control (gray), ruxolitinib-treated (orange), IFNα-treated (blue), and IFNγ-treated (green) cultures were determined by plaque assay using media collected from alveolosphere cultures at 24 and 48 h post-infection. Data are presented as mean ± SEM.
Pre-treatment with IFNs Reduces SARS-CoV-2 Replication in Alveolospheres
Recent studies suggested that pre-treatment with IFNs reduced SARS-CoV-2 replication in Calu-3 and Vero-2 cells. We then tested the effect of pre-treatment of alveolospheres with IFNs before viral infection (Clementi et al., 2020; Felgenhauer et al., 2020) because our above data from IFN treatments alone led to an increase in AT2 death. Therefore, we pre-treated alveolospheres with a lower dose of IFNα and IFNγ (10 ng) for 18 h prior to viral infection (Figure 6G). Subsequent plaque-forming assays at 24 hpi and 48 hpi revealed that pre-treatment with IFNs significantly reduced the viral titers in alveolospheres (Figure 6H). In addition, we also tested the effect of IFN signaling inhibition on viral replication. For this test, we pretreated alveolospheres with ruxolitinib, an inhibitor of IFN signaling, for 18 h and continued treatment following viral infection (Figure 6G). Plaque-forming assays revealed an increase in the viral replication (Figure 6H). Taken together, these data suggest that pre-treatment with IFNs gives a prophylactic effect, whereas IFN inhibition promotes viral replication.
Discussion
Here, we used scRNA-seq-guided AT2-fibroblast interactome maps to develop and optimize the first chemically defined, serum-free, and stroma-free alveolosphere culture conditions for mouse and human AT2 expansion, maintenance, and differentiation. Our newly developed alveolosphere conditions facilitate long-term passaging and large-scale expansion, features conducive to infection by viruses and potentially other pathogens, as well as large-scale biochemical assays. Furthermore, our alveolosphere culture conditions are well suited for high-throughput, pharmaco-genomic screening to identify both anti-viral drugs and pathways that control cell fate choices in certain pathophysiological conditions.
Using alveolosphere cultures, we demonstrate that AT2s express a SARS-CoV-2 receptor, ACE2, and are sensitive to virus infection. Transcriptome profiling further revealed the emergence of an “inflammatory state” in which AT2s activated the expression of numerous IFNs, cytokines, chemokines, and cell-death-related genes at later times post-infection. These data are consistent with earlier studies showing delayed host innate immune responses after SARS-CoV (2003) infection, until later times (Menachery et al., 2014), but also underscore the need for kinetic analyses of host responses at different times after infection. Both transcriptome and immunohistochemical analysis revealed a downregulation of surfactant proteins in SARS-CoV-2-infected alveolospheres. Interestingly, these findings are in line with previous observations of surfactant protein downregulation after influenza injury (Kebaabetswe et al., 2015). The loss of surfactant proteins has adverse repercussions for the host, as surfactants are essential for preventing alveolar collapse and for controlling both innate and adaptive immune responses (Crouch and Wright, 2001; McCormack and Whitsett, 2002). Our finding that the type-II IFN pathway is activated in AT2s ex vivo is somewhat surprising, as typically it is the IFN-I and IFN-III pathways that are activated in cells by viral infection (Barrat et al., 2019; Bartee et al., 2008). Significantly, these unexpected findings from alveolosphere-derived AT2s mirror responses in AT2s from COVID-19 patient lungs, further supporting the relevance of alveolosphere-derived AT2 for SARS-CoV-2 studies. Our study further provided evidence that pre-treatment with IFNs shows prophylactic effectiveness in alveolospheres. Future studies will reveal whether such a prophylactic effect can provide resistance to viral propagation in vivo.
There are several reasons why AT2s grown in alveolosphere cultures are preferred over the currently used cell lines, such as Calu-3, A549, Vero, and H1299. For example, A549 cells derived from a human lung adenocarcinoma have been widely used as surrogates for alveolar epithelial cells in viral infection studies (Hoffmann et al., 2020; Letko et al., 2020; Osada et al., 2014; Ujie et al., 2019). However, the A549 cell line lacks the cardinal features of lung epithelial cells, including the ability to form epithelial tight junctions; they also harbor numerous genetic alterations (Mason and Williams, 1980; Osada et al., 2014). More importantly, A549 cells do not express the SARS-CoV-2 receptor ACE2, and viral infection studies rely on the ectopic expression of this receptor (Blanco-Melo et al., 2020). Accordingly, transformed cell lines do not faithfully recapitulate the native lung epithelial cells (Mason and Williams, 1980). In contrast, AT2-based alveolospheres are highly polarized epithelial structures that retain molecular and morphological features and maintain the ability to differentiate into AT1s under suitable conditions.
Other models, including AT2s derived from directed differentiation of induced pluripotent stem cells (iPSCs), can serve as alternative models for ex vivo studies (Jacob et al., 2017; Yamamoto et al., 2017). Indeed, recent studies using primary stem cells and iPSC-derived 3D cultures from lung, brain, kidney, and intestine have provided insights into SARS-CoV-2 cell tropism, viral replication kinetics, factors that promote viral entry, and the associated cellular responses (Huang et al., 2020b; Jacob et al., 2020; Lamers et al., 2020; Monteil et al., 2020; Ramani et al., 2020; Yang et al., 2020). Furthermore, such models have been useful for the discovery of anti-viral drugs as well as preclinical humanized ex vivo models to test the efficacy of existing therapeutically relevant molecules (Monteil et al., 2020; Yang et al., 2020). Consistent with our data, a recent study using iPSC-derived AT2s (iAT2s) uncovered epithelial intrinsic inflammatory responses, including IFN pathways, after SARS-CoV-2 infection (Huang et al., 2020b). Despite these similarities, primary lung stem cell-derived models and iPSC-derived iAT2 models have their unique advantages and disadvantages. For example, although iPSC-derived models are more conducive to genetic modulation, they may not be suitable for studying age-specific phenomena. This is particularly important in the case of severe COVID-19, as this disease is more severe in aged than in younger populations and the cellular responses to infection may differ in adolescents compared to the elderly (Muus et al., 2020; Ziegler et al., 2020). Nevertheless, human stem cell-based models will be very valuable for rapid and scalable disease modeling and drug discovery.
One limitation of alveolosphere models in general is that they lack the complete cellular complexity of native tissues. However, such simplified models also offer several advantages. First, as has already been demonstrated here, they serve as an excellent model to study intrinsic cellular mechanisms that are triggered in response to viruses and other pathogens. For example, our study identified numerous IFN ligands and target genes that are activated by AT2s in response to SARS-CoV-2 infection. Second, our study found that AT2s activate type II IFN signaling in the absence of IFNG ligands, suggesting the possibility of unknown non-canonical type II ligands. As discussed above, currently used cancer cell lines, e.g., Vero cells and A549 cells, are defective in their responses to certain IFN pathways (Blanco-Melo et al., 2020; Desmyter et al., 1968; Osada et al., 2014) and, therefore, are not suitable for studying IFN signaling. As we demonstrated here, alveolosphere-derived AT2s express receptors and downstream components needed for IFN signal transduction. Therefore, alveolosphere-derived AT2s offer a faithful model that recapitulates in vivo cellular contexts for studying and identifying components that are either specific or shared between different classes of IFN signaling. Third, although not tested here, alveolosphere-derived AT2s can be combined with other cell types, e.g., specific immune cell subsets, to study the interactions between AT2s and immune cells in the presence and absence of pathogens, such as SARS-CoV-2. Indeed, such efforts were made in the context of tumor-immune cell interactions (Neal et al., 2018).
In conclusion, we provide chemically defined and modular culture conditions for the selective expansion and differentiation of AT2s that retain the cardinal features of AT2s, including the ability to self-renew, produce surfactants, and differentiate into AT1s. SARS-CoV-2 infection of the defined AT2 alveolosphere system revealed an induction of autocrine and presumed paracrine IFN signaling and inflammatory pathways that blocked surfactant production and proliferation and induced cell death. This newly developed alveolosphere model offers a unique system for studying SARS-CoV-2 infection and developing effective therapies for COVID-19 and other respiratory diseases.
Limitations of Study
Human lung tissue specimens used for generating alveolospheres came from healthy donors from different age groups. Our study has not taken into account the differences in tissue composition or cell behavior among different age groups. Future studies will need to consider these variables in studying the properties of AT2 cells in 3D cultures. Similarly, the affect age on viral infection and the associated responses were not accounted in our study. Although the AT2 expansion medium is completely defined, AT2 differentiation medium currently uses human serum. Development of a defined AT2 differentiation medium will be valuable for genetic and pharmacological screenings and will most likely advance the use of alveolospheres for cell-based therapies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal anti-Prosurfactant protein C | Millipore | Cat# ab3786, RRID:AB_91588 |
| Anti-ABCA3 | Seven Hills Bioreagents | Cat# WMAB-17G524 |
| Monoclonal Rat anti-RAGE/AGER | R&D systems | Cat# MAB1179, RRID:AB_2289349 |
| Polyclonal Goat Human RAGE/AGER | R&D systems | Cat# AF1145, RRID:AB_354628 |
| Rat Monoclonal anti-Ki67 | Thermo Fisher Scientific | Cat# 14-5698-82, RRID: AB_10854564 |
| Mouse Monoclonal anti-HTII-280 | Terrace Biotech | Cat# TB-27AHT2-280 |
| Rat monoclonal anti-DC-Lamp | Novus Biologicals | Cat# DDX0191P RRID: AB_2827532 |
| Rabbit Polyclonal anti-SFTPB | Thermo Fisher Scientific | Cat# PA5-42000 RRID: AB_2609628 |
| Mouse Monoclonal anti-SFTPB | Thermo Fisher Scientific | MA1-204 RRID: AB_2633311 |
| Rabbit polyclonal anti-ACE2 | Sino biological | Cat# 10108-H31H |
| Mouse Monoclonal anti-ACE2 | R&D systems | Cat# MAB933-SP RRID: AB_2223153 |
| Mouse Monoclonal anti-TMPRSS2 | Santa Cruz Biotechnology | Cat# sc-515727 RRID: AB_10863728 |
| Rabbit monoclonal anti-TMPRSS2 | Abcam | Cat# ab109131 RRID: AB_10863728 |
| Mouse Monoclonal anti- SARS-CoV/ SARS-CoV-2 | Genetex | Cat# GTX632604 |
| Rabbit polyclonal anti- SARS-CoV/ Coronavirus Nucleocapsid | Thermo Fisher Scientific | Cat# PA1-41098 RRID: AB_1087200 |
| Rabbit monoclonal anti- Active Caspase-3 | BD Biosciences | Cat# 559565 RRID: AB_397274 |
| Rabbit polyclonal anti-p63 | Genetex | Cat# GTX102425 RRID: AB_1952344 |
| Goat-anti-SOX2 | R&D systems | Cat# AF2018 RRID: AB_355110 |
| Rabbit monoclonal Anti-TTF1/NKX2-1 | Abcam | Cat# ab76013 RRID: AB_1310784 |
| Goat polyclonal Anti-Scgb1a1 | Santa Cruz Biotechnology | Cat# sc-9772 RRID: AB_2238819 |
| Mouse Monoclonal Anti-Human EPCAM-488 | BioLegend | Cat# 324209 RRID: AB_756083 |
| Rabbit Polyclonal ZO-1 | Abcam | Cat# ab216880 |
| Mouse SARS antiserum | Hou et al., 2020 | N/A |
| Alexa Flour 488 goat anti-mouse IgM | Thermo Fisher Scientific | Cat# 10680 RRID: AB_10892893 |
| Alexa Fluor 546 goat anti-hamster IgG | Thermo Fisher Scientific | Cat# A21111, RRID: AB_2535760 |
| Alexa Fluor 594 goat anti-mouse IgG1 | Thermo Fisher Scientific | Cat# A21125, RRID:AB_2535767 |
| Alexa Fluor 594 donkey anti-goat IgG | Thermo Fisher Scientific | Cat# A11058, RRID:AB_2534105 |
| Alexa Fluor 647 goat anti-mouse IgG1 | Thermo Fisher Scientific | Cat# A21240, RRID:AB_2535809 |
| Alexa Fluor 647 goat anti-rabbit IgG | Thermo Fisher Scientific | Cat# A21245, RRID:AB_2535813 |
| Alexa Fluor 647 donkey anti-mouse IgG | Thermo Fisher Scientific | Cat# A31571, RRID:AB_162542 |
| Mouse-CD31 MicroBeads | Miltenyi Biotec | Cat# 130-097-418 RRID:AB_2814657 |
| Mouse-CD45 MicroBeads | Miltenyi Biotec | Cat# 130-052-301 |
| Human-CD326 (EpCAM) microbeads | Miltenyi Biotec | Cat# 130-061-101 RRID:AB_2832928 |
| Human TruStain FcX | Biolegend | Cat# 422032 |
| Mouse monoclonal CD326 (EpCAM) | Thermo Fisher Scientific | Cat#14-5791-81 |
| Rat Monoclonal PE anti-mouse CD140a | Biolegend | Cat# 135905 |
| RNAscope® Probe- Hs-SFTPB-C2 | ACD | Cat# 544251-C2 |
| RNAscope® Probe- V-nCoV2019-S | ACD | Cat# 848561 |
| Bacterial and Virus Strains | ||
| icSARS-CoV-2-GFP | Hou et al., 2020 | GenBank: MT461670 |
| SARS-CoV-2 USA-WA1/2020 | (kind gift of Greg Sempowski, Duke University, US) | N/A |
| Biological Samples | ||
| Human lung tissue | The University of North Carolina at Chapel Hill or Yale University | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SB431542 | Abcam | Cat# Ab120163 |
| CHIR99021 | Tocris | Cat# 4423 |
| BIRB796 | Tocris | Cat# 5989 |
| DMH-1 | Tocris | Cat# 41-261-0 |
| Heparin | Sigma-Aldrich | Cat# H3149 |
| N-Acetyl-L-cysteine | Sigma-Aldrich | Cat# A9165 |
| Human serum | Sigma-Aldrich | Cat# H4522 |
| Human EGF | GIBCO | Cat# PHG0313 |
| Mouse FGF7 | R&D systems | Cat# 5028-KG-025 |
| Mouse FGF10 | R&D systems | Cat# 751004 |
| Y27632 2HCl | Selleckchem | Cat# S1049 |
| B-27 Supplement (50X) | Thermo Fisher Scientific | Cat# 17504044 |
| N-2 Supplement (100X) | Thermo Fisher Scientific | Cat# 17502048 |
| Insulin-Transferrin-Selenium (100X) | Thermo Fisher Scientific | Cat# 41400-045 |
| Glutamax | Thermo Fisher Scientific | Cat# 35050061 |
| HEPES (1M) | Thermo Fisher Scientific | Cat# 15630080 |
| Antibiotic-Antimycotic (100X) | Thermo Fisher Scientific | Cat# A5955-100ML |
| MEM | GIBCO | Cat # 11095 |
| Penicillin/Streptomycin | GIBCO | Cat # 15140 |
| MEM NEAA | GIBCO | Cat# 11140 |
| Human FGF10 | BioLegend | Cat# 559304 |
| Mouse IL-1β | BioLegend | Cat# 575104 |
| Human IL-1β | BioLegend | Cat# 579404 |
| Mouse TNFα | BioLegend | Cat# 575204 |
| Mouse Noggin | Peprotech | Cat# 250-38 |
| Human FGF10 | BioLegend | Cat# 559304 |
| Human IFNβ | Peprotech | Cat# 300-02BC |
| Human IFNα | BioLegend | Cat# 592704 |
| Human IFNγ | BioLegend | Cat# 570204 |
| Ruxolitinib | Cayman | Cat# 11609 |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Dispase | Corning | Cat# 354235 |
| DNase I | Thermo Fisher Scientific | Cat# 10104159001 |
| Collagenase type I | GIBCO | Cat# 17100-017 |
| Matrigel | Corning | Cat# 354230 |
| Lysotracker | Thermo Fisher Scientific | Cat# L7526 |
| Accutase | Sigma-Aldrich | Cat# A6964 |
| Citrate Buffer, pH 6.0 (10X) | Sigma-Aldrich | Cat# C9999 |
| Fluoromount-G, with DAPI | Thermo Fisher Scientific | Cat# 00-4959 |
| PBS | GIBCO | Cat# 20012027 |
| Sodium bicarbonate | Sigma-Aldrich | Cat# S5761 |
| Sodium hydroxide | Sigma-Aldrich | Cat# S8045 |
| Terra PCR Direct Polymerase | Takara | Cat# 639271 |
| TrypLE Select Enzyme | Thermo Fisher Scientific | Cat# #12563029 |
| TRIzol LS Reagent #11588616 | Invitrogen | Cat# 11588616 |
| Direct-zol RNA MicroPrep kit | Zymoresearch | Cat# 11-330M |
| SuperScript III | Thermo Fisher Scientific | Cat# 18080400 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | New England BioLabs | Cat# E7490 |
| NEBNext Ultra II RNA Library Prep Kit for Illumina | New England BioLabs | Cat# E7770 |
| Thermo Scientific Maxima Reverse Transcriptase (200 U/μL) |
Thermo Fisher Scientific | Cat# EP0742 |
| Exonuclease I | New England BioLabs | Cat# M0293 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat# FC-131-1096 |
| Glutaraldehyde | Sigma | Cat# G5882 |
| Sodium cacodylate buffer pH 7.4 | Electron Microscopy Sciences | Cat# 11652 |
| Uranyl acetate | Electron Microscopy Sciences | Cat# 22400 |
| Tannic Acid | Sigma | Cat# 403040 |
| Osmium tetroxide | Electron Microscopy Sciences | Cat# 19180 |
| Propylene oxide | Polysciences | Cat# 00236-1 |
| EMbed 812 | Electron Microscopy Sciences | Cat# 14120 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE141634, 152586 |
| Experimental Models: Cell Lines | ||
| N/A | N/A | |
| Experimental Models: Organisms/Strains | ||
| Sftpctm1(cre/ERT2)Blh | Rock et al., 2011 | N/A |
| Rosa26R-CAG-lsl-tdTomato | Arenkiel et al., 2011 | N/A |
| Oligonucleotides | ||
| See Table S3 | N/A | N/A |
| Recombinant DNA | ||
| N/A | N/A | N/A |
| Software and Algorithms | ||
| dropSeqPipe v0.3 | N/A | https://hoohm.github.io/dropSeqPipe |
| Seurat v3.0.6 | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| R v3.6.3 | N/A | https://www.r-project.org/ |
| R Studio v1.2.5033 | N/A | https://rstudio.com/ |
| FastQC v0.11.9 | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Cutadapt v1.18 | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Trimmomatic v0.39 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| HISAT 2.2.0 | http://daehwankimlab.github.io/hisat2/ | |
| SAMtools v1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Subread v2.0.1 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| EnhancedVolcano v1.5.4 | N/A | https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html |
| Enrichr | Kuleshov et al., 2016 | https://maayanlab.cloud/Enrichr/ |
| GraphPad Prism 7 | Graph Pad | https://www.graphpad.com |
| FIJI | NIH | https://fiji.sc |
| Other | ||
| Previously published datasets reanalyzed in this study | GEO | GSE145926, and GSE135893 |
Resource Availability
Lead Contact
Further information and requests for resource/reagents should be directed to and will be fulfilled by the Lead Contact, Purushothama Rao Tata (pt93@duke.edu).
Materials Availability
This study did not generate new unique materials.
Data and Code Availability
RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GEO: GSE141634 (MTEC alveolosphere scRNA-seq) and GEO: GSE152586 (Bulk RNA-seq from SARS-CoV-2 infected alveolospheres). Previously published sequencing data that were re-analyzed here are available under accession code GEO: GSE145926 (scRNA-seq data from severe COVID-19 patient lungs) and GEO: GSE135893 (scRNA-seq data from control lungs). All analysis code is available from corresponding author upon request.
Experimental Model and Subject Details
Animal Studies
Mouse strains used in this study were maintained in the C57BL/6 background, including Sftpctm1(cre/ERT2)Blh(Sftpc-CreER) (Rock et al., 2011) and Rosa26R-CAG-lsl-tdTomato (Arenkiel et al., 2011) mice were described previously. For lineage labeling, two doses of Tamoxifen (0.2 mg/g body weight, Sigma-Aldrich) was given via Intraperitoneal (IP). All Animal experiments were approved by the Duke University Institutional Animal care and Use Committee. All animals were handled in accordance with the NIH and AAALAC guidelines for humane care and use of laboratory animals. Mice used for experiments were not involved in previous procedures and were drug or test naive. Mice were kept in a SPF breeding area under specified pathogen free conditions. Cages, bedding, food and water were autoclaved. Animals were maintained on the same 12h light/dark cycle and monitored daily by caretakers or researchers. Health monitoring did not reveal any infections in the past 18 months.
Human lung tissue
Excised sub transplant-quality human lung tissues from donors without preexisting chronic lung diseases were obtained from the Marsico Lung Institute at the University of North Carolina at Chapel Hill under the University of North Carolina Biomedical Institutional Review Board-approved protocols (#03-1396) and from Yale University (#0901004626). Informed consent was obtained from all participants where necessary.
Human COVID-19 infected subjects
Tissue blocks or cut sections obtained from three COVID-19 autopsy lungs were obtained from Drs. Ross. E. Zumwalt (University of New Mexico, Albuquerque, NM) and Edana Stroberg (Office of the Chief Medical Examiner, Oklahoma City, OK). Donor demographics are described below. The paraffin blocks were cut to produce 5 μm serial sections for immunofluorescence.
Donor 1. 40-year-old, male. Medical history: Diabetes mellitus. Clinical course: This donor had upper respiratory infection (URI) symptoms three days before he was found dead at home. No intubation occurred. Postmortem testing of the lung was positive for SARS-CoV-2.
Donor 2. 77-year-old, male. Medical history: Acute pancreatitis, acute cholecystitis, and splenectomy. Clinical course: This donor was transferred to the ER because of fever and respiratory distress six days before death. He was dead shortly after arrival. A swab from the nasal cavity and the postmortem lung were positive for SARS-CoV-2.
Donor 3. 91-year-old, female. Medical history: Coronary artery disease, hyperlipidemia, and hypertension. Clinical course: This donor was transferred to the ER because of URI symptoms, hypoxia, weakness, and shortness of breath. A nasal swab was positive for SARS-CoV-2. She was treated with high flow nasal canula, but died from acute pneumonia due to SARS-CoV-2 complicated by an acute hypoxic respiratory failure.
Method Details
Mouse lung tissue dissociation
Lung dissociation and FACS were performed as described previously (Chung et al., 2018). Briefly, lungs were intratracheally inflated with 1ml of enzyme solution containing Dispase (5 U/ml), DNase I (0.33U/ml) and Collagenase type I (450 U/ml) in DMEM/F12. Separated lung lobes were diced and incubated with 3ml enzyme solution for 30min at 37°C with rotation. The reaction was quenched with an equal amount of DMEM/F12+10% FBS medium and filtered through a 100μm strainer. The cell pellet was resuspended in red blood cell lysis buffer (100μM EDTA, 10mM KHCO3, 155mM NH4Cl) for 5min, washed with DMEM/F12 containing 10% FBS and filtered through a 40μm strainer. Total cells were centrifuged at 450 g for 5min at 4°C and the cell pellet was processed for AT2 isolation by FACS.
Human lung tissue dissociation
Human lung dissociation was as described previously (Zacharias et al., 2018). Briefly, pleura was removed and remaining human lung tissue (approximately 2g) washed with PBS containing 1% Antibiotic-Antimycotic and cut into small pieces. Visible small airways and blood vessels were carefully removed to avoid clogging. Then samples were digested with 30 mL of enzyme mixture (Collagenase type I: 1.68 mg/ml, Dispase: 5U/ml, DNase: 10U/ml) at 37°C for 1h with rotation. The cells were filtered through a 100μm strainer and rinsed with 15ml DMEM/F12+10% FBS medium through the strainer. The supernatant was removed after centrifugation at 450 g for 10min and the cell pellet was resuspended in red blood cell lysis buffer for 10min, washed with DMEM/F12 containing 10% FBS and filtered through a 40μm strainer. Total cells were centrifuged at 450 g for 5 min at 4°C and the cell pellet was processed for AT2 isolation.
Isolation of human and mouse AT2s
AT2s were isolated by Magnetic-activated cell sorting (MACS) or Fluorescence-activated cell sorting (FACS) based protocols. For mouse AT2 isolation the total lung cell pellet was resuspended in MACS buffer (1x PBS, pH 7.2, 1% BSA, and 2mM EDTA). CD31/CD45 positive cells were depleted using MACS beads according to the manufacturer’s instructions. After CD31/CD45 depletion AT2s were sorted based on TdTomato reporter and for AT2s without a reporter, cells were stained using the following antibodies: EpCAM/CD326, PDGFRα/CD140a and Lysotracker as described previously (Katsura et al., 2019). For isolation of human AT2s, approximately 2-10 million total lung cells were resuspended in MACS buffer and incubated with Human TruStain FcX for 15min at 4°C followed by HTII-280 (1:60 dilution) antibody for 1h at 4°C. The cells were washed twice with MACS buffer and then incubated with anti-mouse IgM microbeads for 15min at 4°C. The cells were loaded into the LS column and labeled cells collected magnetically. For FACS based purification of human AT2s, the total lung cell pellet was resuspended in MACS buffer. Cells were positively selected for the EpCAM population using CD326 (EpCAM) microbeads according to the manufacturer’s instructions. CD326 selected cells were stained with HTII-280 and LysoTracker at 37°C for 25min followed by secondary Alexa anti-mouse IgM-488 for 10min at 37°C. Sorting was performed using a FACS Vantage SE and SONY SH800S.
Alveolosphere culture
Mouse conventional Alveolosphere culture (using MTEC medium) was performed as described previously (Barkauskas et al., 2013). Briefly, FACS sorted lineage labeled AT2s (1-3 × 103) from Sftpc-CreER; R26R-lsl-tdTomato mice and PDGFRα+ (5 × 104) cells were resuspended in MTEC/Plus or serum free medium and mixed with an equal volume of growth factor-reduced Matrigel.
For feeder free culture, AT2s (1-3 × 103) were resuspended in serum free medium and mixed with an equal amount of Matrigel. For drop culture, 3 drops of 50μl of cells-medium/Matrigel mixture were plated in each well of a 6-well plate. The medium was changed every other day. For detailed SFFF and AMM media composition see Table S1. For human alveolosphere culture, HTII-280+ human AT2s (1-3 × 103) were resuspended in serum free medium and mixed with an equal amount of Matrigel and plated in 6 well plates. For detailed mouse and human SFFF media composition, see Tables S1 and S2.
Alveolosphere passaging
Mouse alveolosphere passaging experiment was performed in AMM medium, composition as described above. Briefly, FACS sorted mouse AT2s (2 × 103) were resuspended in AMM medium and mixed with an equal volume of Matrigel. 3 drops of 50 μl of cells-medium/Matrigel mixture were plated in each well of a 6-well plate for each biological replicate (n = 3). For every passage mouse IL-1β (10ng/ml) was added for the first 4 days and subsequently, the media was replaced with AMM without IL-1β. The medium was changed every three days. Mouse alveolosphere were passaged every 10 days. For human alveolosphere passages, AT2s (3 × 103) were resuspended in SFFF medium and mixed with an equal volume of Matrigel. 3 drops of 50μl of cells-medium/Matrigel mixture were plated in each well of a 6-well plate for each donor (n = 3). Alveolospheres were passaged every 10-14 days.
AT2 differentiation
For detailed mouse and human AT2-Differentiation medium (ADM) composition see Tables S1 and S2, respectively. For differentiation, mouse alveolospheres were cultured in AMM for 10 days were switched to AT2-differentiation medium followed by culture for an additional 7 days, except where stated otherwise. For differentiation, human alveolospheres cultured in SFFF media for 10 days were switched to ADM and cultured for an additional 12-15 days, except where stated otherwise. The medium was changed every three days. Human AT2-Differentiation medium contains human serum instead of FBS.
Cell culture and SARS-CoV-2 Virus Propagation
Vero E6 cells were the kind gift of Greg Grey and were maintained at 37°C and 5% CO2 in MEM supplemented with 1% Penicillin/Streptomycin, 10% FBS, 1mM Sodium Pyruvate, and 1x MEM NEAA. To grow stocks of SARS-CoV-2, Vero E6 cells were inoculated with BEI isolate SARS-CoV-2 USA-WA1/2020 (kind gift of Greg Sempowski) at a MOI 0.01 in viral growth media (MEM supplemented with 1% Pen/Strep, 2% FBS, 1mM Sodium Pyruvate, and 1x MEM NEAA). After 1h incubation at 37°C, viral growth media was added to bring the final volume to 30 ml. Virus was collected after 72 h and titered on Vero E6 cells using 2x MEM and 0.7% Oxoid Agar and standard procedures. Plaques were stained with crystal violet and counted.
Alveolosphere infection experiment for bulk RNaseq and qPCR studies
To infect alveolosphere cultures, cells were washed with 1 mL PBS then virus was added to cells at a MOI of 1. Virus and cells were incubated for 3.5 hours at 37°C after which virus was removed and cell culture media was added. Infection proceeded for 48 or 120 hours and then alveolospheres were washed with PBS, dissociated as described above. Finally, alveolosphere derived cells were stored in Trizol and stored at −80°C.
Infection of AT2 alveolospheres with SARS-CoV-2
Human alveolosphere cultures were briefly washed twice with 500μl 1X PBS. SARS-CoV-2-GFP (icSARS-CoV-2-GFP virus was described previously (Hou et al., 2020). Briefly, seven cDNA fragments covering the entire SARS-CoV-2 WA1 genome were amplified by RT-PCR using PrimeSTAR GXL HiFi DNA polymerase (TaKaRa). Junctions between each fragment contain non-palindromic sites BsaI (GGTCTCNˆNNNN) or BsmBI (CGTCTCNˆNNNN) with unique four-nucleotide cohesive ends. Fragment E and F contain two BsmBI sites at both termini, while other fragments harbor BsaI sites at the junction. Each fragment was cloned into high-copy vector pUC57 and verified by Sanger sequencing. A silent mutation T15102A was introduced into a conserved region in nsp12 in plasmid D as a genetic marker. GFP was inserted by replacing the ORF7 gene. Cultures were then inoculated with 200μl of 1x107 PFU/ml of icSARS-CoV-2-GFP virus (Hou et al., 2020) or 200μl of 1X PBS for mock cultures. Alveolospheres were allowed to incubate at 37°C supplemented with 5% CO2 for 2h. Following incubation, the inoculum was removed, and alveolosphere cultures were washed three times with 500μl 1X PBS. 1mL of SFFF media was added to each culture. Alveolospheres were incubated at 37°C for 72h, with samples taken every 24h during infection. To sample, 100μl of media was removed. Equal volumes of fresh media were then added to the cultures to replace the sampled volume. Viral titers were ultimately determined after 72h by plaque assay on Vero E6 cells (USAMRIID). Viral plaques were visualized by neutral red staining after 3 days (Hou et al., 2020). For histological analysis alveolospheres were fixed for 7 days in 10% formalin solution followed by 3 washes in PBS.
Interferon treatment
For interferon and cytokine treatment experiment, Human AT2s (2.5 × 104) from P2 or P3 passage were cultured on the surface of matrigel. Prior to the plating of cells 12 well plates were precoated with matrigel (1:1 matrigel and SFFFM mix) for 30min. AT2s were grown in SFFFM without IL-1β for 7 to 10 days to allow the formation of alveolospheres. Alveolospheres were treated with 20ng/ml interferons (IFNα, IFNβ, IFNγ) for 12h or 72h for RNA isolation and quantitative PCR. For histological analysis, Alveolospheres were treated with indicated interferons for 72h. Human alveolosphere cultures were pretreated with 10ng IFNα or 10ng IFNγ for 18h prior to virus infection. For IFN inhibition studies, alveolospheres were treated with 1μM Ruxolitinib throughout the culture time.
Alveolospheres fixation and sectioning
Alveolospheres were fixed with 4% paraformaldehyde (PFA) at 4°C for 2h or at room temperature for 1h, respectively. Submersion cultures of alveolospheres were first immersed in 1% low melting agarose (Sigma) and fixed with 4% PFA at room temperature for 30 min. For OCT frozen blocks, alveolospheres were washed with PBS, embedded in OCT and cryosectioned (8-10μm). For paraffin blocks, samples were dehydrated, embedded in paraffin and sectioned at 7μm.
Immunostaining on COVID-19 lungs
Immunohistochemical staining was performed on COVID-19 autopsy lung sections according to a protocol as previously described (Hou et al., 2020). Briefly, paraffin-embedded sections were baked at 60 °C for 2–4 hours and deparaffinized with xylene (2 changes × 5 min) and graded ethanol (100% 2 × 5 min, 95% 1 × 5 min, 70% 1 × 5 min). After rehydration, quenching of endogenous peroxidase was performed with 0.5% hydrogen peroxide in methanol for 15 min. Antigen retrieval was performed by boiling the slides in 0.1 M sodium citrate pH 6.0 (3 cycles with microwave settings: 100% power for 6.5 min, 60% for 6 min, and 60% for 6 min, refilling the Coplin jars with distilled water after each cycle). After cooling and rinsing with distilled water, slides washed in PBS, and blocked with 4% normal donkey serum, for an hour at RT. Primary antibody (NKX2-1:1:500, ABCA3: 1:1000, SARS-CoV-2 nucleocapsid: 1:500, Anti-SARS mouse antiserum: 1:4000, cleaved-CASP3: 1:200) were diluted in 4% normal donkey serum in PBST and incubated overnight at 4 °C. Mouse and rabbit gamma globulin was used as an isotype control at the same concentration as the primary antibody. Sections were washed in PBST, and secondary antibodies were applied for 1h at RT. After washing in PBST, the Vector® TrueVIEW Autofluorescence Quenching Kit (Vector laboratories) was used to reduce background staining, and glass coverslips were placed over tissue sections with the ProLong Gold Antifade Reagent with DAPI (Invitrogen).
Immunostaining on alveolospheres
Paraffin sections were first dewaxed and rehydrated before antigen retrieval. OCT section were defrosted and washed with PBS. Antigen retrieval was performed using 10mM sodium citrate buffer in either an antigen retrieval system (Electron Microscopy Science) or water bath (90°C for 15 min), or 0.05% Trypsin (Sigma-Aldrich) treatment for 5 min at room temperature. Sections were washed with PBS, permeabilized in PBST (0.1% Triton X-100 in PBS), and incubated with 1% BSA and 0.1% Triton X-100 in PBS for 30 min at room temperature followed by primary antibodies at 4°C overnight. Sections were then washed 3 times in PBST, incubated with secondary antibodies in blocking buffer for 1h at room temperature, washed with PBST 3 times, and mounted using Fluor G reagent with DAPI. Primary antibodies were as follows: Prosurfactant protein C (1:500), RAGE/AGER (Rat, 1:500), human RAGE/AGER (Gt, 1:500), DC-Lamp (1:250), SFTPB (Rb, 1:500), SFTPB (Ms, 1:500), ACE2 (Rb, 1:500), ACE2 (Ms, 1:500), TMPRSS2 (Rb, 1:500), TMPRSS2 (Ms, 1:250), SARS (Rb, 1:500), SARS (Ms, 1:500), Active Caspase-3 (1:500), NKX2-1(1:500), SCGB1A1 (1:250), TP63 (1:500), HTII-280 (1:250), Ki67 (1:250), ABCA3 (1:250), EPCAM (1:500), ZO1 (1:250), SOX2 (1:500).
For quantifying the stainings on near single cell suspensions, Alveolosphere bubbles were dissociated using TrypLE Select Enzyme at 37°C for 15min. Matrigel was disrupted by vigorous pipetting. Alveolosphere derived cells were then plated on matrigel precoated (5%–10% Matrigel for 30min) coverslips or chamber slides for 2-3h. Cells were then fixed in 4% paraformaldehyde.
RNA in situ hybridization
RNA-ISH was performed on paraffin-embedded 5 μm tissue sections of COVID-19 autopsy lungs according to the manufacturer’s instructions (Advanced Cell Diagnostics). Sections were deparaffinized with xylene (2 changes × 5 min) and 100% ethanol (2 changes × 1 min), and then incubated with hydrogen peroxide for 10 min, followed by target retrieval in boiling water for 15 min, and incubation with Protease Plus (Advanced Cell Diagnostics) for 15 min at 40°C. Slides were hybridized with custom probes at 40°C for 2 hours, and signals were amplified according to the manufacturer’s instructions.
Image acquisition, processing and quantification
For Alveolosphere number and size quantitation images were obtained at 1.25x objective, all other phase contrast images were taken at 10x or 20x objective using Zeiss Axiovert 200 microscope. Alveolosphere numbers and sizes were quantified using FIJI ImageJ software. Human Alveolosphere numbers and sizes (> 300 μm in perimeter) were counted on day 15, except where stated otherwise. Microscope. Scale bar 1 mm, except where stated otherwise. Confocal images were collected using Olympus Confocal Microscope FV3000 using a 20X or 40X or 60x objective.
Electron microscopy
Sample preparation for electron microscopy was performed as described previously (Jacob et al., 2017). Alveolospheres were fixed for 3h in 2.5% glutaraldehyde in 0.1M cacodylate buffer pH 7.4 at room temperature. The sample was then washed in 0.1M cacodylate buffer three times for 10min each, post-fixed in 1% Tannic Acid in 0.1M cacodylate buffer for 5min at room temperature and washed again three times in 0.1M cacodylate buffer. Alveolospheres were post-fixed overnight in 1% osmium tetroxide in 0.1M cacodylate buffer in the dark at 4°C. Samples were washed three times in 0.1N acetate buffer for 10 min and block stained in 1% Uranyl acetate for 1h at room temperature. Next, the sample was dehydrated through acetone on ice: 70%, 80%, 90%, 100% for 10min each, and then incubated with propylene oxide at room temperature for 15min. The sample was changed into EMbed 812 for 3h at room temperature followed by fresh Embed 812 and left overnight at room temperature, after which it was embedded in freshly prepared EMbed 812 and polymerized overnight at 60°C. Sections were prepared at 70 nm and grids were stained in 1% aqueous Uranyl Acetate for 5 min at room temperature followed by lead citrate for 2.5min at room temperature. Sections on grids were imaged on FEI Tecnai G2 Twin at a magnification of 2200x and 14500x.
RNA isolation and qRT-PCR
For RNA isolation, Alveolospheres were dissociated into single-cell suspension using TrypLE Select Enzyme at 37°C for 10min. The cell pellet was resuspended in 300μl of TRIzol LS Reagent Total RNA was extracted using the Direct-zol RNA MicroPrep kit according to the manufacturer’s instructions with DNase I treatment. Reverse transcription was performed using SuperScript III with random hexamer or negative-strand specific RT primer. 600ng of purified total RNA was used from each sample. The specificity of negative strand RT primer was confirmed by using viral genomic RNA purified from viral supernatants as this contains only positive strand. Negative strand RNA was detected at high levels in our infected organoids (Ct value for primer- 1202-1363, 24.76 ± 0.73; primer- 848-981, 24.79 ± 0.80) compared to RNA isolated from viral inoculum (Ct value > 29). Quantitative RT-PCR assays were performed using StepOnePlus system (Applied Biosystems) with PowerUp SYBR Green Master Mix. The relative quantities of mRNA for all target genes were determined using the standard curve method. Target-gene transcripts in each sample were normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers used are listed in Table S3.
Bulk RNA sequencing and differential gene expression analysis
Purified RNA (1 μg) from each sample was enriched for Poly-A RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, #E7490). Libraries were prepared using NEBNext Ultra II RNA Library Prep Kit for Illumina (New England BioLabs, #E7770). Paired-end sequencing (150 bp for each read) was performed using HiSeq X with at least 15 million reads for each sample. Quality of sequenced reads were assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). PolyA/T tails were trimmed using Cutadapt (Martin, 2011). Adaptor sequences were trimmed and reads shorter than 24 bp were trimmed using Trimmomatic (Bolger et al., 2014). Reads were mapped to the reference genomes of human (hg38) and SARS-CoV-2 (wuhCor1) obtained from UCSC using Hisat2 (Kim et al., 2019) with default setting. Duplicate reads were removed using SAMtools (Li et al., 2009). Fragment numbers were counted using the featureCounts option of SUBREAD (Liao et al., 2014). Normalization and extraction of differentially expressed genes (DEGs) between control and treatments were performed using an R package, DESeq2 (Love et al., 2014).
Droplet-based single-cell RNA sequencing (Drop-seq)
Alveolospheres embedded in Matrigel were incubated with Accutase at 37°C for 20min followed by incubation with 0.25% trypsin-EDTA at 37°C for 10min. Trypsin was inactivated using DMEM/F-12 Ham supplemented with 10% FBS and cells were then resuspended in PBS supplemented with 0.01% BSA. After filtration through 40μm strainer cells at a concentration of 100 cells/μl were run through microfluidic channels at 3,000 μl/h, together with mRNA capture beads at 3,000 μl/h and droplet-generation oil at 13,000 μl/h. DNA polymerase for pre-amplification step (1 cycle of 95°C for 3 min, 15-17 cycles of 98°C for 15 s, 65°C for 30 s, 68°C for 4 min and 1 cycle of 72°C for 10 min) was replaced by Terra PCR Direct Polymerase. The remaining steps were performed as described in the original Drop-seq protocol (Macosko et al., 2015). Libraries were sequenced (150-bp paired end) using HiSeq X.
Computational analysis for Drop-seq
The FASTQ files were processed using dropSeqPipe v0.3 (https://hoohm.github.io/dropSeqPipe) and mapped on the GRCm38 genome with annotation version 91. Unique molecular identifier (UMI) counts were then further analyzed using an R package Seurat v3.0.6 (Stuart et al., 2019). UMI counts were normalized using SCTransform v0.2 (Hafemeister and Satija, 2019). Principle components which are significant based on Jackstraw plots were used for generating t-SNE plots. After excluding duplets, specific cell clusters were identified based on enrichment for Sftpc, Sftpa1, Sftpa2, Sftpb, Lamp3, Abca3, Hopx, Ager, Akap5, Epcam, Vim, Pdgfra, Ptprc, Pecam1 and Mki67 in tSNE plot.
Computational analysis for single-cell RNA sequencing of COVID-19 patient lungs
Publicly available single-cell RNA-seq dataset of six severe COVID-19 patient lungs (GEO: GSE145926; Bost et al., 2020) and control lungs (GEO: GSE135893; Habermann et al., 2019) were obtained from Gene Expression Omnibus (GEO). EpCAM-positive epithelial cell cluster in the severe COVID-19 patient lungs was further clustered based on LAMP3, ABCA3, KRT5, KRT15, DNAH1, FOXJ1, SCGB3A1 and SCGB1A1. AT2s that have ≧ 1 UMI count of LAMP3, NKX2-1 and ABCA3 were utilized for comparison between severe COVID-19 patient lungs and control lungs. UMI counts were normalized and regressed to percentage of mitochondrial genes using SCTransform. Enriched genes in severe COVID-19 patient and control lungs were extracted using FindMarkers and shown in volcano plot drawn by R package Enhanced Volcano v1.5.4 Genes that have ≧ 2 log2 fold change were used as input for Enrichr (Kuleshov et al., 2016) query to get enriched signaling pathways through database - BioPlanet.
Quantification and Statistical Analysis
Statistical methods relevant to each figure are outlined in the figure legend. Sample size was not predetermined. Data are presented as means with standard error (SEM) to indicate the variation within each experiment. Statistics analysis was performed in Excel, Prism and R. A two-tailed Student’s t test was used for the comparison between two experimental conditions. For experiments with more than two conditions, statistical significance was calculated by ANOVA followed by the Tukey-HSD, Steel-Dwass or Dunnett’s test for multiple comparisons. We used Shapiro-Wilk test to test whether data are normally distributed and used Wilcoxon rank sum test for the comparison between two conditions that showed non-normal distributions.
Acknowledgments
We thank Brigid Hogan for advice and critical reading of the manuscript. We thank Peiying Shan (Yale University) and Randell lab members (University of North Carolina at Chapel Hill) for human lung preparation, the Duke Cancer Institute Flow Cytometry Shared Resource for cell sorting, the Duke University Light Microscopy Core Facility for imaging equipment and consultation, and the Duke Compute Cluster for help with computing sequencing data. This work was performed in part at the Duke University Shared Materials Instrumentation Facility, a member of the North Carolina Research Triangle Nanotechnology Network, which is supported by the National Science Foundation (grant ECCS-1542015) as part of the National Nanotechnology Coordinated Infrastructure. SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281 was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH. Biocontainment work was performed in the Duke Regional Biocontainment Laboratory, which received partial support for construction from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (UC6-AI058607). Y.K. is a fellow of the Japan Society for the Promotion of Science Overseas Research. V.S. is supported by a fellowship from Regeneration Next Initiative at Duke University. A.K. is supported by a medical scientist training program fellowship from NHLBI/NIH (F30HL143911). S.H.R. is supported by Cystic Fibrosis Foundation grant BOUCHE15R0 and NIH grant DK065988. This work was supported by a generous gift from the Chan Zuckerberg Foundation and NIH grants AI132178 and AI149644 to R.S.B. This work was supported by NHLBI/NIH (R00HL127181, R01HL146557, and R01HL153375), NIGMS (R21GM1311279), and funds from Regeneration NeXT and Kaganov-MEDx Pulmonary Research Initiative at Duke University to P.R.T. This work was partially supported by a grant from the United Therapeutics Corporation (to P.R.T.). P.R.T. is a Whitehead Scholar at Duke University.
Author Contributions
H.K. designed and optimized alveolosphere cultures, analyzed the data, and edited manuscript. V.S. designed and optimized alveolosphere cultures and co-wrote the manuscript. A.T. performed alveolosphere cultures, histology, and immunostainings; analyzed the data; and co-wrote the manuscript. Y.K. designed and performed scRNA-seq, bulk RNA-seq, RT-PCR, and computational analysis and co-wrote the manuscript. C.E.E., E.J.F., and B.E.H. performed viral infections. A.K. provided reagents. T.A. and Y.M. performed stainings and quantification on COVID-19 lung sections. P.J.L. and S.H.R. provided human lung tissues. R.C.B., N.S.H., and R.S.B. contributed reagents and recombinant viruses. P.R.T. designed, conceived, and supervised the work and co-wrote the manuscript. All authors reviewed and edited the manuscript.
Declaration of Interests
A patent application (PCT/US20/53158) related to this work has been filed. H.K. and P.R.T. are listed as co-inventors on this application. P.R.T. serves as a consultant for Cellarity and Surrozen.
Published: October 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2020.10.005.
Supplemental Information
References
- Arenkiel B.R., Hasegawa H., Yi J.J., Larsen R.S., Wallace M.L., Philpot B.D., Wang F., Ehlers M.D. Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PLoS One. 2011;6:e29423. doi: 10.1371/journal.pone.0029423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L.M. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C.E., Chung M.-I., Fioret B., Gao X., Katsura H., Hogan B.L.M. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat F.J., Crow M.K., Ivashkiv L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019;20:1574–1583. doi: 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E., Mohamed M.R., McFadden G. Tumor necrosis factor and interferon: cytokines in harmony. Curr. Opin. Microbiol. 2008;11:378–383. doi: 10.1016/j.mib.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R.A., tenOever B.R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020 doi: 10.1101/2020.03.24.004655. [DOI] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Marshall D., Lacy J.M., Williams T. Histopathology and Ultrastructural Findings of Fatal COVID-19 Infections. medRxiv. 2020 doi: 10.1101/2020.04.17.20058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.-I., Bujnis M., Barkauskas C.E., Kobayashi Y., Hogan B.L.M. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145:dev163014. doi: 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Ferrarese R., Criscuolo E., Diotti R.A., Castelli M., Scagnolari C., Burioni R., Antonelli G., Clementi M., Mancini N. Interferon-β-1a Inhibition of Severe Acute Respiratory Syndrome-Coronavirus 2 In Vitro When Administered After Virus Infection. J. Infect. Dis. 2020;222:722–725. doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E., Wright J.R. Surfactant proteins a and d and pulmonary host defense. Annu. Rev. Physiol. 2001;63:521–554. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A.H., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgenhauer U., Schoen A., Gad H.H., Hartmann R., Schaubmar A.R., Failing K., Drosten C., Weber F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R.F., Allen L., Gonzales L., Ballard P.L., Dobbs L.G. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J. Histochem. Cytochem. 2010;58:891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A.C., Gutierrez A.J., Bui L.T., Yahn S.L., Winters N.I., Calvi C.L., Peter L., Chung M.-I., Taylor C.J., Jetter C. Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. bioRxiv. 2019 doi: 10.1101/753806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv. 2019 doi: 10.1101/576827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B., Tata P.R. Cellular organization and biology of the respiratory system. Nat. Cell Biol. 2019 doi: 10.1038/s41556-019-0357-7. [DOI] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Gehart H., Artegiani B., LÖpez-Iglesias C., Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell. 2018;175:1591–1606.e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.-D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell. 2020 doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S.W., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.-L., Weaver T.E., Vedaie M., Hurley K. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell. 2017;21:472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.-K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell. 2020 doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Kobayashi Y., Tata P.R., Hogan B.L.M. IL-1 and TNFα Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Reports. 2019;12:657–666. doi: 10.1016/j.stemcr.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaabetswe L.P., Haick A.K., Gritsenko M.A., Fillmore T.L., Chu R.K., Purvine S.O., Webb-Robertson B.-J., Matzke M.M., Smith R.D., Waters K.M. Proteomic analysis reveals down-regulation of surfactant protein B in murine type II pneumocytes infected with influenza A virus. Virology. 2015;483:96–107. doi: 10.1016/j.virol.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner I., Kochs G., Kalinke U., Weiss S., Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J. Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Huch M. Disease modelling in human organoids. Dis. Model. Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Kim J., Gludish D., Roach R.R., Saunders A.H., Barrios J., Woo A.J., Chen H., Conner D.A., Fujiwara Y. Surfactant protein-C chromatin-bound green fluorescence protein reporter mice reveal heterogeneity of surfactant protein C-expressing lung cells. Am. J. Respir. Cell Mol. Biol. 2013;48:288–298. doi: 10.1165/rcmb.2011-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- Mason R.J., Williams M.C. Phospholipid composition and ultrastructure of A549 cells and other cultured pulmonary epithelial cells of presumed type II cell origin. Biochim. Biophys. Acta. 1980;617:36–50. doi: 10.1016/0005-2760(80)90222-2. [DOI] [PubMed] [Google Scholar]
- McCormack F.X., Whitsett J.A. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J. Clin. Invest. 2002;109:707–712. doi: 10.1172/JCI15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter J.L., Yuen K., Williams B., Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc. Natl. Acad. Sci. USA. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schäfer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5:e01174. doi: 10.1128/mBio.01174-14. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C., Luecken M.D., Eraslan G., Waghray A., Heimberg G., Sikkema L., Kobayashi Y., Vaishnav E.D., Subramanian A., Smilie C. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020 2020.04.19.049254. [Google Scholar]
- Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.-H., Salahudeen A.A., Smith A.R. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175:1972–1988.e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolić M.Z., Sun D., Rawlins E.L. Human lung development: recent progress and new challenges. Development. 2018;145:dev163485. doi: 10.1242/dev.163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.C., Logan C.Y., Fish M., Anbarchian T., Aguisanda F., Álvarez-Varela A., Wu P., Jin Y., Zhu J., Li B. Inflammatory Cytokine TNFα Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell. 2018;175:1607–1619.e15. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., Hogan B.L.M. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K., Shichino S., Ueha S., Nakajima T., Hashimoto S., Yamazaki S., Matsushima K. Mesenchymal-Epithelial Interactome Analysis Reveals Essential Factors Required for Fibroblast-Free Alveolosphere Formation. iScience. 2019;11:318–333. doi: 10.1016/j.isci.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K., Nakajima T., Shichino S., Deshimaru S., Matsushima K., Ueha S. In vitro expansion of endogenous human alveolar epithelial type II cells in fibroblast-free spheroid culture. Biochem. Biophys. Res. Commun. 2019;515:579–585. doi: 10.1016/j.bbrc.2019.05.187. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syedbasha M., Egli A. Interferon Lambda: Modulating Immunity in Infectious Diseases. Front. Immunol. 2017;8:119. doi: 10.3389/fimmu.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S., Martin M.L., Jansen S.A., Fu Y., Bos J., Chandra D., O’Connor M.H., Mertelsmann A.M., Vinci P., Kuttiyara J. T cell-derived interferon-γ programs stem cell death in immune-mediated intestinal damage. Sci. Immunol. 2019;4:eaay8556. doi: 10.1126/sciimmunol.aay8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujie M., Takada K., Kiso M., Sakai-Tagawa Y., Ito M., Nakamura K., Watanabe S., Imai M., Kawaoka Y. Long-term culture of human lung adenocarcinoma A549 cells enhances the replication of human influenza A viruses. J. Gen. Virol. 2019;100:1345–1349. doi: 10.1099/jgv.0.001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A.I., Jackson S.R., Zhao G., Quansah K.K., Farshchian J.N., Neupauer K.M., Littauer E.Q., Paris A.J., Liberti D.C., Scott Worthen G. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen. Med. 2019;4:17. doi: 10.1038/s41536-019-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Gotoh S., Korogi Y., Seki M., Konishi S., Ikeo S., Sone N., Nagasaki T., Matsumoto H., Muro S. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W.J., Frank D.B., Zepp J.A., Morley M.P., Alkhaleel F.A., Kong J., Zhou S., Cantu E., Morrisey E.E. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GEO: GSE141634 (MTEC alveolosphere scRNA-seq) and GEO: GSE152586 (Bulk RNA-seq from SARS-CoV-2 infected alveolospheres). Previously published sequencing data that were re-analyzed here are available under accession code GEO: GSE145926 (scRNA-seq data from severe COVID-19 patient lungs) and GEO: GSE135893 (scRNA-seq data from control lungs). All analysis code is available from corresponding author upon request.