ABSTRACT
High physical stability is required for the development of monoclonal antibodies (mAbs) into successful therapeutic products. Developability assays are used to predict physical stability issues such as high viscosity and poor conformational stability, but protein aggregation remains a challenging property to predict. Among different types of stresses, air–water and solid–liquid interfaces are well known to potentially trigger protein instability and induce aggregation. Yet, in contrast to the increasing number of developability assays to evaluate bulk properties, there is still a lack of experimental methods to evaluate antibody stability against interfaces. Here, we investigate the potential of a hydrophobic nanoparticle surface-mediated stress assay to assess the stability of mAbs during the early stages of development. We evaluate this surface-mediated accelerated stability assay on a rationally designed library of 14 variants of a humanized IgG4, featuring a broad span of solubility values and other developability properties. The assay could identify variants characterized by high instability against agitation in the presence of air–water interfaces. Remarkably, for the set of investigated molecules, we observe strong correlations between the extent of aggregation induced by the surface-mediated stress assay and other developability properties of the molecules, such as aggregation upon storage at 45°C, self-association (evaluated by affinity-capture self-interaction nanoparticle spectroscopy) and nonspecific interactions (estimated by cross-interaction chromatography, stand-up monolayer chromatography (SMAC), SMAC*). This highly controlled surface-mediated stress assay has the potential to complement and increase the ability of the current set of screening techniques to assess protein aggregation and developability potential of mAbs during the early stages of drug development.
Abbreviations:AC-SINS: Affinity-Capture Self-Interaction Nanoparticle Spectroscopy; AMS: Ammonium sulfate precipitation; ANS: 1-anilinonaphtalene-8-sulfonate; CIC: Cross-interaction chromatography; DLS: Dynamic light scattering; HIC: Hydrophobic interaction chromatography; HNSSA: Hydrophobic nanoparticles surface-stress assay; mAb: Monoclonal antibody; NP: Nanoparticle; SEC: Size exclusion chromatography; SMAC: Stand-up monolayer chromatography; WT: Wild type
KEYWORDS: antibodies, developability, interfaces, surfaces, aggregation, stability, formulation
Introduction
The successful development of biotherapeutics is not limited to the optimization of biological activity but also relies on the developability potential of drug candidates. This property is achieved by careful selection, optimization and design of biophysical properties such as nonspecificity, conformational stability, aggregation propensity and viscosity.1–3 Misbehavior of drug candidates in developability assays is associated with detrimental consequences for the successful development of bioprocesses and stable drug formulations, and can even compromise drug pharmacokinetics.4,5 A comprehensive study by Jain and coworkers2 demonstrated that a high developability potential of monoclonal antibodies (mAbs) appears to be a prerequisite for advancing monoclonal antibodies to the market.
The formation of protein aggregates during bioprocessing6,7 and upon long-term storage of drug product8,9 is considered as a critical quality attribute because it can alter drug potency and potentially induce immunogenicity.10 Consequently, the aggregation propensity of drug candidates is a critical developability parameter.
Many experimental assays have been proposed that attempt to predict protein aggregation propensity, either during bioprocessing or storage, from other surrogate properties, such as thermal stability,11–13 reversible self-interactions,14–23 nonspecific interactions with surfaces24,25 and other biomolecules,4,26–28 hydrophobic interactions,14 and solubility.29
Different computational approaches have also been developed to optimize protein sequences to increase intrinsic stability against partial unfolding and aggregation,30–34 by increasing repulsive interactions via increase of the protein net charge,35,36 predicting the second virial coefficient B2237,38 or identifying aggregation hot-spots or aggregation-prone patches to be re-designed (CamSol,39 SAP,40,41 TANGO,34 Aggrescan3D).32,42,43
Despite the extensive efforts to develop experimental and computational screening tools for the prediction of highly aggregation prone drug candidates during early stages, validated assays still remain to be identified.30,44 Accordingly, the prediction of long-term stability remains particularly challenging44–46 because proteins may follow several different aggregation pathways, which are highly specific to the combination of protein and buffer conditions under analysis.12,47–49 Moreover, most accelerated aggregation studies have been performed in the presence of stresses that may substantially differ from the real processing conditions.
Recent studies2,44,50,51 have assessed the developability potential of a large set of antibodies using numerous high-throughput methods. None of these methods individually correlated with aggregation upon storage under different formulations, underlining the pressing need to combine multiple biophysical properties in one single aggregation propensity “score” to predict protein long-term stability against aggregation in an accurate manner.44,50 Currently, this aggregation propensity “score” is evaluated based on a pool of computational and experimental methods that assess bulk properties of the molecules. However, it has now become clear that among the various driving-forces of aggregation, the presence of air–water,52–56 oil–liquid and solid–liquid interfaces57–59 can likely trigger protein instability and aggregation.53 Therefore, together with the current set of bulk properties, including protein interactions, thermal unfolding or aggregation upon thermal stress, the evaluation of the global developability score during the early stages of candidate selection and formulation development should also consider protein stability against interfaces. This information can complement the current set of developability assays and provide orthogonal data on interface-induced stability, therefore strengthening the current multi-variate combination of developability properties. Yet, in contrast to the large number of high-throughput assays to evaluate bulk properties, experimental methods to evaluate antibody stability against interfaces remain much less developed. Challenges include the need to simulate a variety of different interfaces with which the molecule can interact during bioprocessing, formulation and delivery, as well as the accurate control of surfaces during the assay under both stagnant and flow conditions.
To address these challenges, we recently developed an accelerated stability assay60 that provides a highly controlled surface-mediated driving force for aggregation based on polymeric nanoparticles.61 Due to the small size of these nanoparticles and the corresponding high surface-to-volume ratio, it is possible to accurately control the total area exposed to the protein solution and neglect the contribution of other interfaces of the test container, including the air–water interface. Moreover, the surface properties of our nanoparticles can be finely tuned by leveraging the flexibility of polymer chemistry. Although it is unrealistic to simulate all the different types of materials that the molecule might contact, the stability of proteins against different types of interfaces is mediated by a variety of interactions, which may be simplified and considered as a complex combination of a limited number of fundamental forces. With our assay, we can test molecules against a set of model surfaces, including hydrophobic as well as positively and negatively charged, providing a first estimation of the stability of the molecule against different interfaces.60 This assay requires only µg amounts of protein, is performed in a few hours and can be implemented with relatively high-throughout.
Here, we focus on hydrophobic nanoparticles and evaluate this assay on a library of 14 antibody variants previously designed to span a broad range of developability potential.62 We observe that the aggregation propensity measured by our hydrophobic surface-mediated aggregation assay correlated with other developability properties of the molecules, including solubility and aggregation at 45°C over several weeks. The highly controlled surface-mediated aggregation test is therefore of high value to complement stability and developability studies of therapeutic proteins during the early stages of their development. Together with the current set of methods, the assay has the potential to increase the ability to predict aggregation propensity with minimal experimental effort, in particular when libraries of variants of the same molecule need to be screened.
Results
Hydrophobic nanoparticles surface-mediated stress assay
The hydrophobic nanoparticle assay was implemented as previously described and characterized.60 In the following, we briefly describe its principle using our model IgG4.
Before exposing our model IgG4 to the hydrophobic surface stress, we tested the stability of the molecule in 10 mM HEPES, 10 mM MES, 10 mM NaCl at pH 6.6 at room temperature. After 24 h incubation, we did not observe any increase in size by dynamic light scattering (DLS) (Figure 1a), thereby confirming the monomeric state and stability of the IgG4 in this formulation condition. In contrast, the addition of hydrophobic nanoparticles in a 1:50 NP:mAb surface ratio to the same stable formulation led to the formation of larger species within a few minutes, as shown by DLS. The nanoparticle-protein aggregates were also imaged by optical microscopy, where the aggregated protein was stained by 1-anilinonaphtalene-8-sulfonate (ANS) fluorescence (Figure 1b). ANS is an extrinsic dye that shows strong fluorescence in hydrophobic environments, and can therefore report on the unfolding and aggregation of proteins (Figure S2).60 The Hb− nanoparticles are characterized in terms of size and charge in Table 1.60,61
Figure 1.
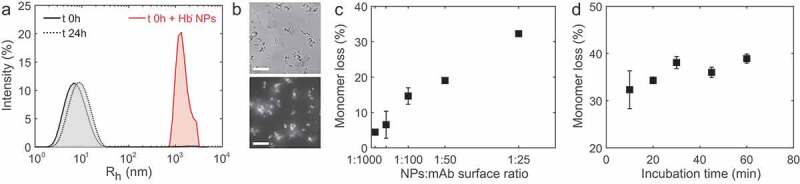
Surface-induced aggregation of IgG4 wild type in the hydrophobic nanoparticle surface-mediated aggregation assay. (a) Size distribution of our model IgG4 at the initial time point in absence and presence of hydrophobic nanoparticles, which trigger the formation of micron size aggregate in a time scale of few seconds. The IgG4 sample is stable in absence of hydrophobic nanoparticles. Incubation was performed at room temperature in 10 mM NaCl, 10 mM HEPES, 10 mM MES buffer at pH 6.6. (b) Bright field (top) and fluorescence (bottom) microscopy images of aggregates stained by ANS. Scale bar is 50 μm. (c) The monomer loss scales linearly with the hydrophobic surface introduced by the nanoparticles (results shown were taken after 30 min incubation), and (d) is constant after 30 min stagnant incubation at room temperature (ratio NPs:mAb 1:50).
Table 1.
Characterization of the hydrophobic nanoparticles used in the HNSSA, in terms of size and charge. Variations refer to standard deviations of mean values measured with nanoparticles produced in five independent production batches
| Nanoparticles | Diameter (nm) | PDI | ζ-potential (mV) |
|---|---|---|---|
| Hydrophobic (Hb−) | 75 ± 5 | 0.05 ± 0.01 | −38 ± 7 |
We quantified the monomer loss by UV absorbance after separating the nanoparticles and the aggregates from the monomer by centrifugation (Figure S1). The monomeric state of the supernatant was confirmed by DLS and size exclusion chromatography (SEC) (Figure S3). Increasing nanoparticle concentration increases the extent of aggregation, thereby confirming that the total amount of surface introduced into the system is the driving force for protein destabilization (Figure 1c). We note that the ratio between the total surface of the nanoparticles and the surface of the air–water interface varies between approximately 10 to 100 fold, and the contribution of the air–water interface can therefore be neglected.
The aggregation process reaches a plateau after 30 min incubation (Figure 1d). In all experiments described below, we, therefore, evaluated the monomer conversion after 30 min incubation.
The hydrophobic nanoparticle surface-mediated aggregation assay correlates with agitation stress at the air–water interface
We tested the hydrophobic nanoparticle surface-mediated aggregation assay (HNSSA) as surface-mediated stress assay by comparing the monomer loss measured by the HNSSA with the values obtained in a typical agitation stress assay in the presence of an air–water interface, which is a notorious major source of protein destabilization.25,59,63–67 To this aim, we used an antibody library with variants featuring a range of solubility values (Table S1) that were computationally designed using the solubility predictor CamSol method.62 This algorithm predicts the intrinsic solubility of proteins based on a linear combination of specific physicochemical properties derived from the amino acid sequence, such as hydrophobicity, charge at neutral pH, α-helix and β-sheet propensities, which can be further corrected in a structural context.[38] Low intrinsic CamSol scores correspond to poor solubility, whereas higher values indicate increased variant solubility. The antibody library used here differs slightly from that used in a previous report,62 as we excluded two variants due to poor available protein yields. We incubated the variants in glass vials under shaking at 1400 rpm at room temperature in presence of an air–water interface.
In the following, the Pearson correlation coefficient (rp) and Spearman’s rank-order correlation coefficient (rs) were used to correlate the outcome of different assays. The Pearson coefficient reports for the linearity in a correlation between two variables, with a value of 1 or −1 for purely direct or reverse linear relationships, respectively. The Spearman non-parametric coefficient describes the monotonicity (direction) of the correlation, with a value of 1 for a positive correlation and −1 for a negative correlation throughout the dataset. A p-value is associated with these coefficients, as they are obtained from a statistical test that compares correlation against the alternative hypothesis of no correlation.
Both the HNSSA and the agitation assay were able to identify the variants with the most pronounced difference in interface stability, indicating that our hydrophobic nanoparticles can effectively recognize molecules with particularly high/low stability in the presence of air–water interfaces. A generally fair correlation (rp = 0.68; p ≤ 0.01 and rs = 0.54; p = 0.05; exclusion of mAbs 11 and 16 results in coefficients of rp = 0.62; p = 0.03 and rs = 0.27; p > 0.1) was obtained between the HNSSA and the conventional agitation assay (Figure 2, Figure S4). Deviations in the correlation may be due to the absence of hydrodynamic mixing in the HNSSA, as mechanical agitation and shear stresses have been shown to act synergistically with surfaces in enhancing protein destabilization.55,56,61,68–70 Moreover, the molecular mechanisms underlying protein aggregation on a solid interface are likely different from the air/water interface.
Figure 2.
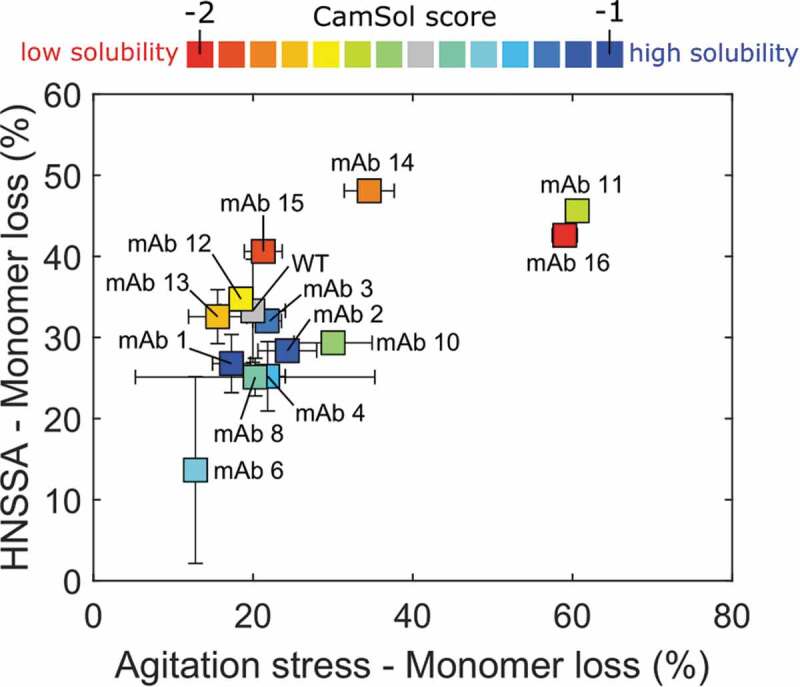
Correlation between the monomer loss induced by our HNSSA (1:50 surface ratio NPs:mAb) and an agitation stress assay performed with an air headspace in glass vials incubated at 1400 rpm for 1.5 h. The variants were formulated in both assays at 0.50 mg/mL in 20 mM HEPES, 10 mM NaCl at pH 7.6. The HNSSA and agitation stress assays were performed in duplicates. The different points correspond to the indicated variants, the color gradient from red to blue reflects the CamSol solubility score, from low to high solubility, respectively. The color coding is preserved throughout the presented work. Error bars correspond to one standard deviation.
We note that the nanoparticle-based assay can also be used to screen for stabilizing formulation conditions and measure the effect of excipients and buffer components. For instance, in a recent work, we applied the HNSSA to investigate the effects of surfactants and determine the optimal concentration of Tween 80 to protect the mAb in solution from surface-induced instability.60
While the assay provides a quantitative way to rank the relative surface-stability of different molecules, the interpretation of the method to determine an acceptable threshold of stability is unavoidably subjected to a certain level of arbitrariness. For instance, within a set of variants, a molecule could be considered unstable if its value differs more than a certain percentage from the average. The exact value of this percentage will depend on how conservative the choice should be and will be specific on the context. Application of the method to an increasing number of molecules and conditions will likely provide an estimation of this percentage in the future.
Accelerated stability correlates with the HNSSA for the antibody variant library
We next investigated the correlation of our surface-mediated stress assay with accelerated stability under moderate thermal stress. To this aim, we performed two independent storage stability studies, during which the variants were incubated at low ionic strength for 6 weeks at a moderate thermal stress at 45°C to promote aggregation, which was evaluated by SEC. During the first replicate study, we observed evaporation for samples of mAb 1, which was therefore excluded. In the second replicate run, mAb 12 could not be measured because of the low availability of material due to poor expression.
Upon aggregation, the monomeric variants formed soluble and insoluble aggregates, as well as fragments. The latter species represented on average 13% and 3% by mass after 6-week incubation for each independent replicate study, respectively (Table S2), and did not affect aggregation.
For the more insoluble antibodies, protein precipitates were visually observed in some samples. These insoluble aggregates did not enter the SEC column and led to an overall mass loss indicated by a decrease of the total area under the chromatograms.
We observed a certain degree of variability between the two replicate studies in terms of absolute amounts of aggregates and fragments formed (see Tables S2, S3 and S4). However, the relative ranking of the variants was very consistent across the two replicate studies. Therefore, we decided to use the variant ranking instead of the absolute amount of aggregates. The variants were ranked depending on the relative extent of aggregation compared to the wild type (WT), with a high rank (>0) corresponding to a high amount of aggregates formed upon incubation, and a low rank (<0) corresponding to a lower aggregation extent.
After 6-weeks incubation, the ranking in the soluble and insoluble aggregate of the variants correlated with the predicted solubility, as measured by SEC and the CamSol score, respectively (Figure 3a) (rp = 0.76 and rs = 0.76; p ≤ 0.01 for soluble aggregates, and rp = −0.82 and rs = −0.83; p ≤ 0.01 for insoluble aggregates). The ranking in soluble aggregation of the variants correlated inversely with the ranking in insoluble aggregate formation (rp = −0.93 and rs = −0.94; p ≤ 0.01). In other words, upon incubation, variants predicted to be more soluble preferably form soluble aggregates, whereas less soluble variants tend to form insoluble aggregates (Figure 3b). In contrast, the aggregation extent weakly correlated to the thermal unfolding of the antigen-binding fragment (Fab) domains of the variants (Figure S6).
Figure 3.
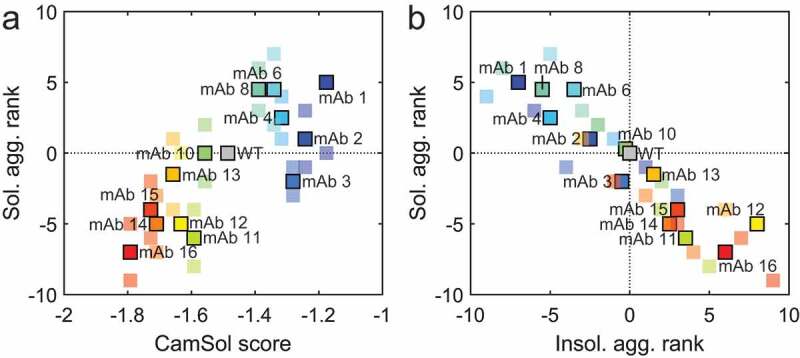
Behavior of the variant library relative to the WT in accelerated thermal stability studies (6 weeks incubation at 45°C). (a) The variant ranking in terms of soluble aggregate formation relative to the WT correlates directly to the CamSol solubility score. (b) Correlation between the variant rakings of soluble aggregates against insoluble aggregates. Soluble variants tend to form soluble aggregates, while insoluble variants tend to form insoluble aggregates. In (a) and (b), a high rank value (>0) corresponds to high amounts of aggregates formed compared to the WT, while a low rank value (<0) corresponds to a lower amount of aggregates formed compared to the WT. The bright and big marker points correspond to the average rankings between the two replicate incubations, while small markers correspond to the individual ranking obtained for each replicate incubation. The samples were prepared as quintuples (run 1) and triplicates (run 2) and incubated in 20 mM HEPES, 10 mM NaCl at pH 7.6.
We correlated these results with our HNSSA at a surface stress of 1:50 NPs:mAb. Remarkably, the ranking and the monomer loss in the HNSSA correlate inversely with the ranking of the soluble aggregate formation (rp = −0.94, rs = −0.92; p ≤ 0.01) (Figure 4a, d), and directly with the ranking of the insoluble aggregate formation (rp = 0.84, rs = 0.87; p ≤ 0.01) (Figure 4b, d). A direct correlation is also obtained between the HNSSA ranking and the total aggregation ranking (considering both soluble and insoluble aggregation) (rp = 0.83, rs = 0.84; p ≤ 0.01) (Figure 4c, d). The correlation between the monomer loss in the HNSSA and the aggregation extent for each individual replicate study is shown in Figure S7.
Figure 4.
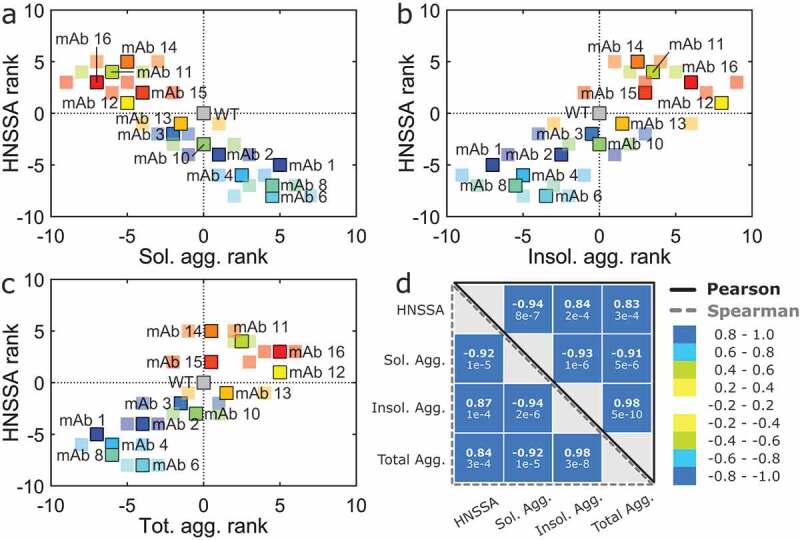
Variants ranking in the HNSSA compared to the ranking in the accelerated thermal stability. The variants ranking in the HNSSA correlates (a) inversely with the soluble aggregates ranking in the accelerated thermal stress assay, (b) directly with the ranking of the amount of insoluble aggregates and (c) directly with the ranking of the total amount of aggregates formed (soluble and insoluble) evaluated by SEC. (d) Aggregation and solubility scores obtained with the monomer loss in the HNSSA (not the rankings) and in the thermal stability at 45°C. Bold numbers indicate the Pearson and Spearman rank-order correlation coefficients, while the values indicated below correspond to the respective p-values. High (>0) and low (<0) rank values correspond respectively to higher and lower amounts of aggregates formed compared to the WT. The bright and larger marker points correspond to the average rankings between two replicate incubations, while small markers correspond to the individual ranking obtained for each replicate incubation. The samples were prepared in 20 mM HEPES, 10 mM NaCl at pH 7.6 and incubated in two runs for 6 weeks at 45 °C. The HNSSA was performed in duplicates, and the nanoparticles introduced in a 1:50 NPs:mAb ratio.
HNSSA correlation with other experimental developability assays
The previous results show that the HNSSA provides orthogonal information on interface-induced stability and can strengthen the current multi-variate combination of developability properties to predict aggregation. To further test the relationship between the HNSSA and other properties of the antibodies, we used eight developability assays recently applied to the variant library62 to measure hydrophobicity, thermal stability, solubility, self-association, in vivo clearance and nonspecificity (Table S3). In the correlations shown below, the monomer loss evaluated by the HNSSA was correlated to the different developability assays.
We first experimentally determined the solubility of the different variants by ammonium sulfate (AMS) precipitation, which probes the propensity of the proteins to precipitate upon progressive sequestration of water by the salt.62 The HNSSA probes for hydrophobic and charged surface interactions. The correlation between AMS precipitation and our HNSSA (Figure 5) was not significant (rp = −0.43 and rs = −0.47; p ≥ 0.1).
Figure 5.
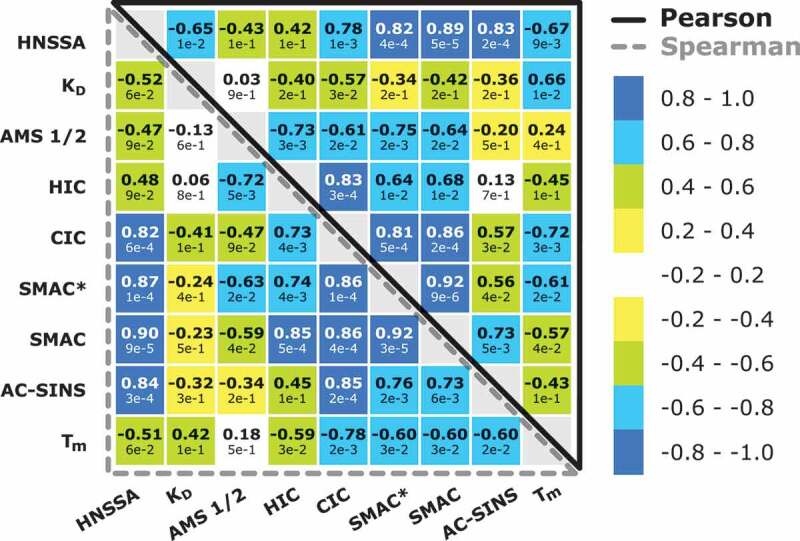
Developabilixty score matrix showing the Pearson’s and Spearman’s correlation rank coefficient of the HNSSA and other developability assays. Bold numbers indicate the Pearson and Spearman rank-order correlation coefficients, while the values indicated below correspond to the respective p-values.
As a measure of colloidal stability and propensity to aggregation, we next determined the diffusion interaction parameter KD of each variant (Figure 5), which is directly related to the second virial coefficient B22.71 KD has been correlated with antibody solubility,72 antibody solution viscosity73 and aggregation propensity.74 All determined KD values of the variants were negative and below −5.34 mL/g, suggesting that protein attraction was prevailing.75,76 A non-significant correlation was observed between the HNSSA and KD values (rp = −0.65; p ≥ 0.01 and rs = −0.52; p ≥ 0.05). In fact, it has been reported that KD may be a poor predictor of aggregation behavior in scenarios where hydrophobic interactions may become the governing forces in solution and outbalance electrostatic interactions.77
We next applied hydrophobic interaction chromatography (HIC) to determine protein hydrophobicity, which has been associated with aggregation,41 viscosity and in vivo clearance.78 The principle of the assay relies on modulating the interactions between mAbs and a hydrophobic stationary phase through the application of a salt gradient. Hydrophobic mAbs show longer retention times compared to less hydrophobic mAbs at increasing salt concentrations.79 We observed that the measured variant hydrophobicities correlate with their aggregation propensities under thermally stressed conditions, measured from the total amount of aggregates determined by SEC (rp = 0.70, rs = 0.70; p < 0.01, and rp = 0.75; p < 0.01, rs = 0.54; p = 0.06 in the first and second replicate runs, respectively). However, when comparing the HNSSA and HIC, the determined correlation was non-significant (rp = 0.42, rs = 0.48; p ≥ 0.1, Figure 5). This poor correlation might arise from the different hydrophobic materials used in both assays, and the fact that the hydrophobic nanoparticles used in the HNSSA carry a residual negative charge, which might play a role in the aggregation of the different variants. Moreover, we have observed that the nanoparticles not only trigger the adsorption of the protein on their surface but are also responsible for further aggregation events, which are not probed for with HIC.60
Next, we determined the unfolding temperature of the Fab domain of the mAbs under equal buffer and concentration conditions (pH 7.6, 20 mM HEPES, 10 mM NaCl, 1 mg/mL) as the accelerated temperature studies mentioned above. A low correlation was determined between the HNSSA and Tm (rp = −0.67, rs = −0.51; p < 0.1, Figure 5, Table S1). These results corroborate the weak link between thermal stability and other developability assays, as previously reported.2,62
After characterizing solubility, aggregation propensity, hydrophobicity and thermal stability, we measured nonspecific protein interactions by several methods. Cross-interaction chromatography (CIC,80) measures nonspecific interactions between the mAb under analysis and polyclonal antibodies immobilized on a column. Insoluble mAbs have been shown to cross-interact more frequently with immobilized polyclonal antibodies, resulting in a longer retention time on the column. CIC has been correlated with solubility experiments80 and in vivo clearance.81 The correlation between the HNSSA and CIC is fairly high (rp = 0.78, rs = 0.82; p ≤ 0.01, Figure 5), suggesting that the HNSSA captures nonspecific interactions as well as solubility to a certain extent.
In addition to CIC, we applied stand-up monolayer adsorption chromatography (SMAC) and SMAC* to evaluate the nonspecific interactions of mAbs with different chromatographic surfaces.14,62 Retention times are inversely related to the colloidal stability of mAbs, meaning that mAbs prone to precipitation or aggregation exhibit longer retention times on the column. Of all developability assays, these two assays correlated the highest with the HNSSA (SMAC*: rp = 0.82, rs = 0.87; p ≤ 0.01, SMAC: rp = 0.89, rs = 0.90; p ≤ 0.01, Figure 5).
In addition, high agreement between affinity-capture self-interaction nanoparticle spectroscopy (AC-SINS), a high-throughput assay for antibody self-association,20 and the HNSSA was determined (rp = 0.83, rs = 0.84, p ≤ 0.01, Figure 5).
Overall, several developability assays highly correlated with our HNSSA, particularly with methods probing for self-association (AC-SINS) and nonspecific interactions (CIC, SMAC and SMAC*), as summarized in Figure 5.
Correlation of the HNSSA with in silico developability assays
We next investigated the performance of the HNSSA compared to in silico predictors for globular protein aggregation and solubility (Figure 6). As mentioned in previous sections, the variants prepared in this work were selected based on their sequence-based (intrinsic) CamSol score, which scores the variants based on a linear combination of specific physicochemical properties derived from the amino acid sequence, such as hydrophobicity, charge at neutral pH, α-helix and β-sheet propensities [38]. A fairly good correlation could be observed between the HNSSA and the CamSol score (rp = −0.73 and rs = −0.75; p ≤ 0.01). Another predictor of protein aggregation propensity, SAP,82 is based on hydrophobic surface patches determined by structural analysis and short molecular dynamic simulations. The structural SAP score correlated with the HNSSA to a similar extent as CamSol (rp = −0.73 and rs = −0.75; p ≤ 0.01). The method based on the Developabilty Index (DI) is an extension of the SAP algorithm and considers not only hydrophobic contributions to aggregation but also electrostatic forces.83 A comparably high correlation between DI and the HNSSA was found as the other predictors (rp = −0.76 and rs = −0.73; p ≤ 0.01) when using the β-index of 0.0498 at 40°C.83 The average aggregation score of the Aggrescan3D method42 correlated to a lesser extent with the HNSSA (rp = 0.63 and rs = 0.68; p > 0.01), but highly with CamSol (rp = −0.88 and rs = −0.93; p ≤ 0.001) and SAP (rp = 0.93 and rs = 0.89; p ≤ 0.001). Finally, the protein solubility predictor Protein-Sol84 did not correlate with the HNSSA (rp = −0.39 and rs = −0.31; p > 0.1). The highest correlation of Protein-Sol was determined with Aggrescan3D (rp = −0.80 and rs = −0.59; p ≤ 0.03).
Figure 6.
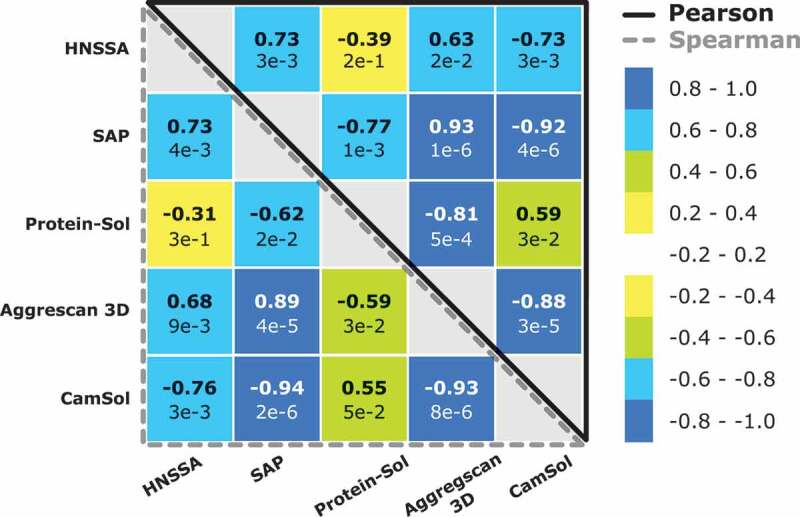
In silico predictors score matrix showing the Pearson’s and Spearman’s correlation rank coefficients between the HNSSA, the CamSol score and three other common in silico predictors. Bold numbers indicate the Pearson and Spearman rank-order correlation coefficients, while the values indicated below correspond to the respective p-values.
In conclusion, the HNSSA correlates to a certain extent with in silico predictions of solubility and aggregation. In particular, with the current antibody dataset, the HNSSA correlates to the highest amount with the in silico predictor SAP probing for hydrophobicity, the related DI index and the CamSol solubility score.
Discussion
Here, we describe an accelerated surface-stress assay probing for protein stability against aggregation based on hydrophobic nanoparticles. This assay provides a highly controlled amount of surface and induces the aggregation of different antibodies within a few hours, requiring a minimal amount of material. Remarkably, by analyzing a series of different protein variants, we have demonstrated that the stability ranking obtained with our HNSSA correlates with other developability properties of the molecules, including stability at interfaces in the presence of agitation stress and aggregation over several weeks.
Although the validity of the assay over a broader range of molecules and formulation conditions remains to be tested, this work represents a first step toward demonstrating the potential of the nanoparticle-based assay as an accelerated interface-stress screening tool. This assay can fill the current gap in standardized assays to probe interface-induced instability, providing orthogonal information and complementing the current set of bulk techniques to evaluate protein developability during early-stage candidate selection. Moreover, the simplicity of the assay allows further development into high-throughput configurations, such as plate or microfluidic assays.60,85,86 Finally, the strategy can be expanded in the future to different nanoparticles characterized by different surface properties (such as negative or positive charge).60 This combination of different surface properties will provide further flexibility to mimic different types of interfaces over a larger range of pH values, providing a good overview of the stability of the molecules against a broad range of different surfaces and devices.
Materials and methods
Model monoclonal antibody and variants library
The humanized anti-trinitrophenyl monoclonal IgG4 antibody (145.2 kDa) used in this study was provided by Novo Nordisk A/S. A single amino acid substitution within the free hinge region (S241P) was introduced to avoid the formation of half-mAb, which is a common problem for IgG4 antibodies87 (See primary sequence in Supplementary Information). The solubility library was previously designed and characterized in a previous study62 via CIC, HIC, SMAC, Stand-up Monolayer Chromatography* (SMAC*) and AMS. Information about variant expression and purification can be found in Supplementary Information.
Hydrophobic nanoparticles synthesis
Hydrophobic nanoparticles were prepared according to a previously described protocol.61 In brief, batch emulsion polymerization was used, with 2.5 g of hydrophobic monomer (either BA, EHA) and 75 mg of sodium dodecyl sulfate in 45 mL water. Before the initiation of the reaction, the system was set under an inert atmosphere by purging a nitrogen flux for 20 min at room temperature. The mix temperature was then raised to 70°C by means of an oil bath equipped with a thermocouple, after which the radical initiator, potassium persulfate (30 mg dissolved in 2.5 mL of water), was added. The reaction was stopped after 4 h. The resulting dispersion was cleaned with ion exchange resins (Dowex Marathon M3, Sigma-Aldrich) to remove the surfactant as well as other possible electrolytes. The effectiveness of the surfactant removal step was verified by measuring the electric conductivity of the dispersion before and after the ion exchange (data not shown). The initiator left a weak negative charge on the nanoparticles necessary for particle stabilization.
The hydrophobic nanoparticle stability under different buffer conditions was tested by monitoring increases in size with DLS under 30 min incubation in stagnant conditions (data not shown).
Characterization of the hydrophobic nanoparticles
The nanoparticles stock solid content after purification was measured by thermogravimetric analysis. Their hydrodynamic radius and ζ-potential were evaluated on a Zetasizer Nano (Malvern, U.K.) working in backscattering mode at a fixed angle of 173° and a laser source operating at 633 nm. The hydrodynamic radius of the species in solution was obtained by DLS according to Equation (1) after obtaining the diffusion coefficient using the method of cumulants:
The ζ-potential was measured in Milli-Q water (Milli-Q Synergy Water Purification System, Merck Millipore, MA, USA), based on Equation (2):
where μ represents the electrophoretic mobility μ of the species in solution measured based on the instrument’s laser Doppler effect, ε the dielectric constant and η the medium viscosity.
Hydrophobic nanoparticle surface-mediated aggregation assay
Protein samples were buffer exchanged in 30 mM NaCl (Fisher Scientific, U.K), 10 mM HEPES (Sigma Aldrich, Germany) and pH 6.6 using spin filters with a 30 kDa cutoff polyethersulfone membrane (Vivaspin 500, Sartorius, U.K.). The concentration after buffer exchange was at least 10 mg/mL. The samples were then diluted in the desired buffer at 1.0 mg/mL. The HNSSA protocol is depicted in Figure S1. Two replicate samples containing the desired amount of nanoparticle surface (either 1:1000 or 1:50 of NPs:mAb surface ratio) were prepared in each corresponding buffer. The protein and nanoparticle surfaces were computed based on their hydrodynamic radius measured by DLS, assuming a spherical shape. The mAb sample and the nanoparticle sample were then mixed 1:1 v/v, yielding a final protein concentration of 0.50 mg/mL, and incubated for 30 min at stagnant conditions and room temperature. Three control samples were also prepared by mixing the mAb sample and a buffer sample in a 1:1 v/v ratio without nanoparticles. After aggregation, monomer separation from the aggregated species was achieved by adding MgCl2 to a final concentration of 50 mM to destabilize the colloids in solution, followed by centrifugation for 3 h at 17200 g. One of the control samples was kept and evaluated without a centrifugation step to estimate the monomer loss resulting from protein adsorption on the test tube walls due to centrifugation only and to potential destabilization induced by the addition of MgCl2. To verify the efficient removal of all aggregates, the size distribution in the supernatant was measured by DLS, performed on a Zetasizer instrument (Malvern, U.K.) working in backscattering mode at 173° with a laser source at 633 nm. Two replicate and two control samples were acquired for each investigated buffer condition.
The monomeric state of the supernatants was further investigated by SEC. In brief, 100 μL of supernatant was injected at 0.75 mL/min in a Superdex 200 10/300 GL column branched on an Agilent 1100 series HPLC unit (Santa Clara, CA, USA). The mobile phase consisted in a 200 mM L-Arginine (Sigma Life Science, USA) and 100 mM sodium phosphate (Sigma-Aldrich, Germany) at pH 6.5. Chromatograms were collected at 280 nm and analyzed on the Agilent ChemStation software (Santa Clara, CA, USA).
Alternatively, the monomer concentration was measured by UV absorption at 280 nm (NanoDrop Lite, Thermo-Fisher Scientific, USA).
Aggregate formation was also monitored qualitatively by DLS and imaged by optical microscopy under bright field on a Ti2-U inverted microscope (Nikon, Switzerland) equipped with a camera (Zyla sCMOS 4.2P-CL10, Andor, U.K.) and a 60x oil immersion objective. The microscope was also equipped with an LED light source emitting at 365 nm (Omicron Laserage Laserprodukte GmbH, Germany), and a DAPI UC BP filter set (352 − 402 nm/417 − 477 nm, AHF Analysentechnik AG, Germany) for the acquisition of images in fluorescence of the aggregates stained with ANS (25 μM).
Agitation stress assay
The agitation stress assays were performed in 5 mL borosilicate glass vials (Fiolax, Schott AG, Switzerland), incubating the samples for 1.5 h on a thermomixer (Eppendorf ThermoMixer comfort) operating at 1400 rpm and 25°C. Four hundred microliter of the antibody formulations (0.50 mg/mL in 20 mM HEPES, 10 mM NaCl at pH 7.6) was prepared per glass vial, leaving an air headspace. Two replicates were acquired for each condition, and the monomer loss was evaluated based on control unshaken vials. The monomeric protein was recovered after 3 h centrifugation at 17 200 g immediately after agitation, and the supernatant concentration was measured by UV absorption at 280 nm on the NanoDrop.
Dynamic light scattering for determination of the self-interaction parameter KD
The average hydrodynamic radius (Rh) was determined by extrapolating measurements of Rh at high protein concentrations to zero protein concentration to avoid the effects of mAb self-association or repulsion on Rh determination.74 Experiments were conducted using DLS using a DynaPro plate reader (Wyatt Technology). Twenty microliter of protein samples of 4, 8, 12, 16 and 20 mg/ml in 20 mM HEPES and 10 mM NaCl, pH 8, was measured in 384-well clear bottom plates (Corning) and the hydrodynamic radius was determined by extrapolating to zero concentration. Forty acquisitions were recorded for 5 seconds at 25°C with 0.5 and 1000 nm as low and high cutoff values, respectively. Data processing and calculation of diffusion coefficients were performed using Dynamics (version: 7.5.0.17; Wyatt Technology). The KD values were determined by a linear fit of the measured (mutual) diffusion coefficients (Dm) as a function of protein concentration:
Differential scanning fluorimetry
Thermal unfolding was measured in high sensitivity capillaries using a Prometheus NT.48 instrument (NanoTemper Technologies). Before the analysis, variant samples were buffer exchanged to 20 mM HEPES, 10 mM NaCl and pH 7.6 and 8.2, respectively. The antibody variants were subjected to a linear thermal ramp (1°C/min, from 20°C to 95°C) and 20% laser excitation intensity. Fluorescence at 350 and 330 nm was collected and the midpoints of the thermal unfolding reaction (Tm) were determined from the first derivative of the fluorescence ratio (F350/F330). Data processing was performed using PR control (version: 1.11; NanoTemper Technologies).
Accelerated temperature stress aggregation of variants library
The variant library was buffer exchanged to 20 mM HEPES, 10 mM NaCl, pH 7.6 in Millipore Amicon ultracentrifugation (Merck) devices with a molecular cutoff of 50 kDa. The final protein concentration of 1 mg/ml was determined using A280 nm absorbance (Dropsense, Trinean). We performed two studies preparing samples for the first study in triplicates and for the second study as quintuples. Samples were pipetted into 12 × 32 mm glass screw neck vials (quick thread, LectraBond cap, PTFE/silicone septa P/N 186000384 c) and parafilm was wrapped around the lid. Samples were incubated for 6 weeks at 45°C. To test whether evaporation was occurring during incubation, samples were weighed at time point 0 and after incubation. After incubation, samples were gently centrifuged at 2000 rpm for 1 min to spin down possible condensate on the glass vial. SEC-HPLC analysis was performed by injecting 20 µg into a TSK G3000 SWXL SEC Column (5 µm, 7.8 × 300 mm; Tosoh Bioscience) assembled on an HPLC system (1200 model, Agilent Technologies). Eluent consisted of 122 mM Na2HPO4, 78 mM NaH2PO4, 300 mM NaCl and 4% 2-propanol at pH 6.8. The flow rate was set to 0.8 mL/min of 100% eluent at a column temperature of 28°C for 24 min. 20 µg of protein was injected on the column and detection occurred at 215 nm and 280 nm. Data processing was performed using ChemStation (Agilent). Monomer, soluble aggregates and fragments peak areas were determined. Total peak areas were also calculated to check for possible evaporation or precipitation, which would result in a large increase in peak area or decrease, respectively.
The aggregation extent was evaluated according to the following equations, which derive from the law of conservation of mass:
where the fraction of soluble aggregates formed between time zero (Aagg, t0) and a defined time lapse t (Aagg, t) is determined from the area under the chromatogram corresponding to species larger than the monomer; the insoluble aggregates were quantified as the total mass loss observed upon incubation, defined as the difference between the initial total area under the curve At0, and the final one At. The total amount of aggregates has been calculated by considering the initial and final areas of both the monomer (M) and the fragments (F), indicated as AM+F. The correlation between the amount of soluble and insoluble aggregates in the two replicate studies is shown in Figure S5.
Supplementary Material
Acknowledgments
We want to thank Laila Sakhnini from Novo Nordisk A/S for valuable discussions. A.M.W.P. was funded by Innovation Fund Denmark and the STAR program at Novo Nordisk A/S. We acknowledge ETH Zurich for financial support.
Funding Statement
This work was supported by the Innovation Fund Denmark and the STAR program at Novo Nordisk A/S [--].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Jarasch A, Koll H, Regula JT, Bader M, Papadimitriou A, Kettenberger H.. Developability assessment during the selection of novel therapeutic antibodies. J Pharm Sci. 2015;104(6):1885–12. doi: 10.1002/jps.24430. [DOI] [PubMed] [Google Scholar]
- 2.Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Nat Acad Sci. 2017;114(5):944. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zurdo J. Developability assessment as an early de-risking tool for biopharmaceutical development. Pharm Bioprocess. 2018;1(1):29–50. doi: 10.4155/pbp.13.3. [DOI] [Google Scholar]
- 4.Avery LB, Wade J, Wang M, Tam A, King A, Piche-Nicholas N, Kavosi MS, Penn S, Cirelli D, Kurz JC, et al. Establishing in vitro in vivo correlations to screen monoclonal antibodies for physicochemical properties related to favorable human pharmacokinetics. mAbs. 2018;10(2):244–55. doi: 10.1080/19420862.2017.1417718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson CL, Devine PWA, Phillips JJ, Higazi DR, Lloyd C, Popovic B, Arnold J, Buchanan A, Lewis A, Goodman J, et al. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci Rep. 2016;6(1):38644. doi: 10.1038/srep38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arosio P, Rima S, Morbidelli M. Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at low pH: from oligomers to larger aggregates. Pharm Res. 2013;30(3):641–54. doi: 10.1007/s11095-012-0885-3. [DOI] [PubMed] [Google Scholar]
- 7.Vázquez-Rey M, Lang DA. Aggregates in monoclonal antibody manufacturing processes. Biotechnol Bioeng. 2011;108(7):1494–508. doi: 10.1002/bit.23155. [DOI] [PubMed] [Google Scholar]
- 8.Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm Res. 2003;20(9):1325–36. doi: 10.1023/A:1025771421906. [DOI] [PubMed] [Google Scholar]
- 9.Das TK, Nema S. Protein particulate issues in biologics development. Am Pharm Rev. 2008;(May 2008):52–57. [Google Scholar]
- 10.Filipe V, Jiskoot W, Basmeleh AH, Halim A, Schellekens H, Brinks V. Immunogenicity of different stressed IgG monoclonal antibody formulations in immune tolerant transgenic mice. mAbs. 2012;4(6):740–52. doi: 10.4161/mabs.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brader ML, Estey T, Bai S, Alston RW, Lucas KK, Lantz S, Landsman P, Maloney KM. Examination of thermal unfolding and aggregation profiles of a series of developable therapeutic monoclonal antibodies. Mol Pharm. 2015;12(4):1005–17. doi: 10.1021/mp400666b. [DOI] [PubMed] [Google Scholar]
- 12.Weiss WF, Young TM, Roberts CJ. Principles, approaches, and challenges for predicting protein aggregation rates and shelf life. J Pharm Sci. 2009;98(4):1246–77. doi: 10.1002/jps.21521. [DOI] [PubMed] [Google Scholar]
- 13.Sakhnini LI, Greisen PJ, Wiberg C, Bozoky Z, Lund S, Wolf Perez A-M, Karkov HS, Huus K, Hansen -J-J, Bülow L, et al. Improving the developability of an antigen binding fragment by aspartate substitutions. Biochemistry. 2019;58(24):2750–59. doi: 10.1021/acs.biochem.9b00251. [DOI] [PubMed] [Google Scholar]
- 14.Kohli N, Jain N, Geddie ML, Razlog M, Xu L, Lugovskoy AA. A novel screening method to assess developability of antibody-like molecules. mAbs. 2015;7(4):752–58. doi: 10.1080/19420862.2015.1048410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam ME, Geng SB, Bender C, Ludwig SD, Linden L, Hoet R, Tessier PM. Biophysical and sequence-based methods for identifying monovalent and bivalent antibodies with high colloidal stability. Mol Pharm. 2018;15(1):150–63. doi: 10.1021/acs.molpharmaceut.7b00779. [DOI] [PubMed] [Google Scholar]
- 16.Estep P, Caffry I, Yu Y, Sun T, Cao Y, Lynaugh H, Jain T, Vásquez M, Tessier PM, Xu Y, et al. An alternative assay to hydrophobic interaction chromatography for high-throughput characterization of monoclonal antibodies. mAbs. 2015;7(3):553–61. doi: 10.1080/19420862.2015.1016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng SB, Wittekind M, Vigil A, Tessier PM. Measurements of monoclonal antibody self-association are correlated with complex biophysical properties. Mol Pharm. 2016;13(5):1636–45. doi: 10.1021/acs.molpharmaceut.6b00071. [DOI] [PubMed] [Google Scholar]
- 18.Geng Steven B, Cheung JK, Narasimhan C, Shameem M, Tessier PM. Improving monoclonal antibody selection and engineering using measurements of colloidal protein interactions. J Pharm Sci. 2014;103(11):3356–63. doi: 10.1002/jps.24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayaraman J, Wu J, Brunelle MC, Cruz AMM, Goldberg DS, Lobo B, Shah A, Tessier PM. Plasmonic measurements of monoclonal antibody self-association using self-interaction nanoparticle spectroscopy. Biotechnol Bioeng. 2014;111(8):1513–20. doi: 10.1002/bit.25221. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Caffry I, Wu J, Geng SB, Jain T, Sun T, Reid F, Cao Y, Estep P, Yu Y, et al. High-throughput screening for developability during early-stage antibody discovery using self-interaction nanoparticle spectroscopy. mAbs. 2014;6(2):483–92. doi: 10.4161/mabs.27431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessier PM, Wu J, Dickinson CD. Emerging methods for identifying monoclonal antibodies with low propensity to self-associate during the early discovery process. Expert Opin Drug Deliv. 2014;11(4):461–65. doi: 10.1517/17425247.2014.876989. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann M, Winzer M, Weber C, Gieseler H. Prediction of protein aggregation in high concentration protein solutions utilizing protein-protein interactions determined by low volume static light scattering. J Pharm Sci. 2016;105(6):1819–28. doi: 10.1016/j.xphs.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 23.Wälchli R, Fanizzi F, Massant J, Arosio P. Relationship of PEG-induced precipitation with protein-protein interactions and aggregation rates of high concentration mAb formulations at 5 °C. Eur J Pharm Biopharm. 2020;151:53–60. doi: 10.1016/j.ejpb.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs SA, Wu S-J, Feng Y, Bethea D, O’Neil KT. Cross-interaction chromatography: a rapid method to identify highly soluble monoclonal antibody candidates. Pharm Res. 2010;27(1):65. [DOI] [PubMed] [Google Scholar]
- 25.Höger K, Mathes J, Frieß W. IgG1 adsorption to siliconized glass vials—influence of pH, ionic strength, and nonionic surfactants. J Pharm Sci. 2015;104(1):34–43. doi: 10.1002/jps.24239. [DOI] [PubMed] [Google Scholar]
- 26.Hotzel I, Theil F-P, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4(6):753–60. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wardemann HS, Yurasov A, Schaefer JW, Young EM, Nussenzweig MC , Predominant Autoantibody Production by Early Human B Cell Precursors. Science. 2003;301(5638):1374. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu T-Y, Torrey J, Thomas J, Bobrowicz P, Vasquez M, et al. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26(10):663–70. doi: 10.1093/protein/gzt047. [DOI] [PubMed] [Google Scholar]
- 29.Banks DD, Latypov RF, Ketchem RR, Woodard J, Scavezze JL, Siska CC, Razinkov VI. Native-state solubility and transfer free energy as predictive tools for selecting excipients to include in protein formulation development studies. J Pharm Sci. 2012;101(8):2720–32. doi: 10.1002/jps.23219. [DOI] [PubMed] [Google Scholar]
- 30.Meric G, Robinson AS, Roberts CJ. Driving forces for nonnative protein aggregation and approaches to predict aggregation-prone regions. Annu Rev Chem Biomol Eng. 2017;8(1):139–59. doi: 10.1146/annurev-chembioeng-060816-101404. [DOI] [PubMed] [Google Scholar]
- 31.Cellmer T, Bratko D, Prausnitz JM, Blanch HW. Protein aggregation in silico. Trends Biotechnol. 2007;25(6):254–61. doi: 10.1016/j.tibtech.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trainor K, Broom A, Meiering EM. Exploring the relationships between protein sequence, structure and solubility. Curr Opin Struct Biol. 2017;42:136–46. doi: 10.1016/j.sbi.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Costanzo JA, O’Brien CJ, Tiller K, Tamargo E, Robinson AS, Roberts CJ, Fernandez EJ. Conformational stability as a design target to control protein aggregation. Protein Eng Des Sel. 2014;27(5):157–67. doi: 10.1093/protein/gzu008. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 35.Miklos AE, Kluwe C, Der B, Pai S, Sircar A, Hughes R, Berrondo M, Xu J, Codrea V, Buckley P, et al. Structure-based design of supercharged, highly thermoresistant antibodies. Chem Biol. 2012;19(4):449–55. doi: 10.1016/j.chembiol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien CJ, Blanco MA, Costanzo JA, Enterline M, Fernandez EJ, Robinson AS, Roberts CJ. Modulating non-native aggregation and electrostatic protein–protein interactions with computationally designed single-point mutations. Protein Eng Des Sel. 2016;29(6):231–43. doi: 10.1093/protein/gzw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco MA, Sahin E, Robinson AS, Roberts CJ. Coarse-grained model for colloidal protein interactions, B 22, and protein cluster formation. J Phys Chem B. 2013;117(50):16013–28. doi: 10.1021/jp409300j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long WF, Labute P. Calibrative approaches to protein solubility modeling of a mutant series using physicochemical descriptors. J Comput Aided Mol Des. 2010;24(11):907–16. doi: 10.1007/s10822-010-9383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sormanni P, Aprile FA, Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427(2):478–90. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Prediction of aggregation prone regions of therapeutic proteins. J Phys Chem B. 2010;114(19):6614–24. doi: 10.1021/jp911706q. [DOI] [PubMed] [Google Scholar]
- 41.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Nat Acad Sci. 2009;106(29):11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambrano R, Jamroz M, Szczasiuk A, Pujols J, Kmiecik S, Ventura S. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015;43(Web Server issue):W306–W313. doi: 10.1093/nar/gkv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conchillo-Sole O, de Groot NS, Avilés FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentiluomo L, Svilenov HL, Augustijn D, El Bialy I, Greco ML, Kulakova A, Indrakumar S, Mahapatra S, Morales MM, Pohl C, et al. Advancing therapeutic protein discovery and development through comprehensive computational and biophysical characterization. Mol Pharm. 2020;17(2):426–40. doi: 10.1021/acs.molpharmaceut.9b00852. [DOI] [PubMed] [Google Scholar]
- 45.Hofmann M, Gieseler H. Predictive screening tools used in high-concentration protein formulation development. J Pharm Sci. 2018;107(3):772–77. doi: 10.1016/j.xphs.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Bajaj H, Sharma VK, Badkar A, Zeng D, Nema S, Kalonia DS. Protein structural conformation and not second virial coefficient relates to long-term irreversible aggregation of a monoclonal antibody and ovalbumin in solution. Pharm Res. 2006;23(6):1382. doi: 10.1007/s11095-006-0018-y. [DOI] [PubMed] [Google Scholar]
- 47.Roberts D, Keeling R, Tracka M, van der Walle CF, Uddin S, Warwicker J, Curtis R. Specific ion and buffer effects on protein–protein interactions of a monoclonal antibody. Mol Pharm. 2015;12(1):179–93. doi: 10.1021/mp500533c. [DOI] [PubMed] [Google Scholar]
- 48.Hawe A, Wiggenhorn M, van de Weert M, Garbe JHO, Mahler H-C, Jiskoot W. Forced degradation of therapeutic proteins. J Pharm Sci. 2012;101(3):895–913. doi: 10.1002/jps.22812. [DOI] [PubMed] [Google Scholar]
- 49.Wälchli R, Vermeire P-J, Massant J, Arosio P. Accelerated aggregation studies of monoclonal antibodies: considerations for storage stability. J Pharm Sci. 2020;109(1):595–602. doi: 10.1016/j.xphs.2019.10.048. [DOI] [PubMed] [Google Scholar]
- 50.Gentiluomo L, Roessner D, Frieß W. Application of machine learning to predict monomer retention of therapeutic proteins after long term storage. Int J Pharm. 2020;577:119039. doi: 10.1016/j.ijpharm.2020.119039. [DOI] [PubMed] [Google Scholar]
- 51.Klijn ME, Hubbuch J. Correlating multidimensional short-term empirical protein properties to long-term protein physical stability data via empirical phase diagrams. Int J Pharm. 2019;560:166–74. doi: 10.1016/j.ijpharm.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Wiesbauer J, Prassl R, Nidetzky B. Renewal of the air–water interface as a critical system parameter of protein stability: aggregation of the human growth hormone and its prevention by surface-active compounds. Langmuir. 2013;29(49):15240–50. doi: 10.1021/la4028223. [DOI] [PubMed] [Google Scholar]
- 53.Koepf E, Eisele S, Schroeder R, Brezesinski G, Friess W. Notorious but not understood: how liquid-air interfacial stress triggers protein aggregation. Int J Pharm. 2018;537(1):202–12. doi: 10.1016/j.ijpharm.2017.12.043. [DOI] [PubMed] [Google Scholar]
- 54.Koepf E, Schroeder R, Brezesinski G, Friess W. The film tells the story: physical-chemical characteristics of IgG at the liquid-air interface. Eur J Pharm Biopharm. 2017;119:396–407. doi: 10.1016/j.ejpb.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Rudiuk S, Cohen-Tannoudji L, Huille S, Tribet C. Importance of the dynamics of adsorption and of a transient interfacial stress on the formation of aggregates of IgG antibodies. Soft Matter. 2012;8(9):2651–61. doi: 10.1039/c2sm07017k. [DOI] [Google Scholar]
- 56.Shieh IC, Patel AR. Predicting the agitation-induced aggregation of monoclonal antibodies using surface tensiometry. Mol Pharm. 2015;12(9):3184–93. doi: 10.1021/acs.molpharmaceut.5b00089. [DOI] [PubMed] [Google Scholar]
- 57.Nakanishi K, Sakiyama T, Imamura K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J Biosci Bioeng. 2001;91(3):233–44. doi: 10.1016/S1389-1723(01)80127-4. [DOI] [PubMed] [Google Scholar]
- 58.Roach P, Farrar D, Perry CC. Interpretation of protein adsorption: surface-induced conformational changes. J Am Chem Soc. 2005;127(22):8168–73. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 59.Bee JS, Chiu D, Sawicki S, Stevenson JL, Chatterjee K, Freund E, Carpenter JF, Randolph TW. Monoclonal antibody interactions with micro- and nanoparticles: adsorption, aggregation, and accelerated stress studies. J Pharm Sci. 2009;98(9):3218–38. doi: 10.1002/jps.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kopp MRG, Capasso Palmiero U, Arosio P. A nanoparticle-based assay to evaluate surface-induced antibody instability. Mol Pharm. 2020;17(3):909–18. doi: 10.1021/acs.molpharmaceut.9b01168. [DOI] [PubMed] [Google Scholar]
- 61.Grigolato F, Colombo C, Ferrari R, Rezabkova L, Arosio P. Mechanistic origin of the combined effect of surfaces and mechanical agitation on amyloid formation. ACS Nano. 2017;11(11):11358–67. doi: 10.1021/acsnano.7b05895. [DOI] [PubMed] [Google Scholar]
- 62.Adriana-Michelle WP, Pietro S, Jonathan SA, Laila IS, Ileana R-L, Jais RB, Annette JG, De M Leonardo, Daniel EO, Michele V, Nikolai L.. In vitro and in silico assessment of the developability of a designed monoclonal antibody library. mAbs. 2019;11(2):388–400, DOI: 10.1080/19420862.2018.1556082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahler H‐C, Fischer S, Randolph TW, Carpenter JF.. Protein Aggregation and Particle Formation: Effects of Formulation, Interfaces, and Drug Product Manufacturing Operations. In Wang W, Roberts CJ, editors. Aggregation of Therapeutic Proteins. 2010. doi: 10.1002/9780470769829.ch7 [DOI] [Google Scholar]
- 64.Krayukhina E, Tsumoto K, Uchiyama S, Fukui K. Effects of syringe material and silicone oil lubrication on the stability of pharmaceutical proteins. J Pharm Sci. 2015;104(2):527–35. doi: 10.1002/jps.24184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerwin BA. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: structure and degradation pathways. J Pharm Sci. 2007;97(8):2924–35. doi: 10.1002/jps.21190. [DOI] [PubMed] [Google Scholar]
- 66.Kueltzo LA, Wang WEI, Randolph TW, Carpenter JF. Effects of solution conditions, processing parameters, and container materials on aggregation of a monoclonal antibody during freeze-thawing. J Pharm Sci. 2008;97(5):1801–12. doi: 10.1002/jps.21110. [DOI] [PubMed] [Google Scholar]
- 67.Basu P, Thirumangalathu R, Randolph TW, Carpenter JF. IgG1 aggregation and particle formation induced by silicone–water interfaces on siliconized borosilicate glass beads: a model for siliconized primary containers. J Pharm Sci. 2013;102(3):852–65. doi: 10.1002/jps.23434. [DOI] [PubMed] [Google Scholar]
- 68.Maa Y-F, Hsu CC. Protein denaturation by combined effect of shear and air-liquid interface. Biotechnol Bioeng. 2000;54(6):503–12. doi:. [DOI] [PubMed] [Google Scholar]
- 69.Mehta SB, Lewus R, Bee JS, Randolph TW, Carpenter JF. Gelation of a monoclonal antibody at the silicone oil–water interface and subsequent rupture of the interfacial gel results in aggregation and particle formation. J Pharm Sci. 2015;104(4):1282–90. doi: 10.1002/jps.24358. [DOI] [PubMed] [Google Scholar]
- 70.Grigolato F, Arosio P. Synergistic effects of flow and interfaces on antibody aggregation. Biotechnol Bioeng. 2020;117(2):417–28. doi: 10.1002/bit.27212. [DOI] [PubMed] [Google Scholar]
- 71.Some D, Hitchner E, Ferullo J. Characterizing protein-protein interactions via static light scattering: nonspecific interactions. Am Biotechnol Lab. 2009;27:16–20. [Google Scholar]
- 72.Pindrus M, Shire SJ, Kelley RF, Demeule B, Wong R, Xu Y, Yadav S. Solubility challenges in high concentration monoclonal antibody formulations: relationship with amino acid sequence and intermolecular interactions. Mol Pharm. 2015;12(11):3896–907. doi: 10.1021/acs.molpharmaceut.5b00336. [DOI] [PubMed] [Google Scholar]
- 73.Connolly BD, Petry C, Yadav S, Demeule B, Ciaccio N, Moore JR, Shire S, Gokarn Y. Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103(1):69–78. doi: 10.1016/j.bpj.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saluja A, Fesinmeyer RM, Hogan S, Brems DN, Gokarn YR. Diffusion and sedimentation interaction parameters for measuring the second virial coefficient and their utility as predictors of protein aggregation. Biophys J. 2010;99(8):2657–65. doi: 10.1016/j.bpj.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yadav S, Shire SJ, Kalonia DS. Viscosity behavior of high-concentration monoclonal antibody solutions: correlation with interaction parameter and electroviscous effects. J Pharm Sci. 2011;101(3):998–1011. doi: 10.1002/jps.22831. [DOI] [PubMed] [Google Scholar]
- 76.Yadav S, Shire SJ, Kalonia DS. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J Pharm Sci. 2010;99(12):4812–29. doi: 10.1002/jps.22190. [DOI] [PubMed] [Google Scholar]
- 77.Kumar V, Dixit N, Zhou L, Fraunhofer W. Impact of short range hydrophobic interactions and long range electrostatic forces on the aggregation kinetics of a monoclonal antibody and a dual-variable domain immunoglobulin at low and high concentrations. Int J Pharm. 2011;421(1):82–93. doi: 10.1016/j.ijpharm.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 78.Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I, et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111(52):18601–06. doi: 10.1073/pnas.1421779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haverick M, Mengisen S, Shameem M, Ambrogelly A. Separation of mAbs molecular variants by analytical hydrophobic interaction chromatography HPLC: overview and applications. mAbs. 2014;6(4):852–58. doi: 10.4161/mabs.28693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacobs SA, Wu S-J, Feng Y, Bethea D, O’Neil KT. Cross-interaction chromatography: a rapid method to identify highly soluble monoclonal antibody candidates. Pharm Res. 2010;27(1):65–71. [DOI] [PubMed] [Google Scholar]
- 81.Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vásquez M, Wittrup KD, et al. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015;7(4):770–77. doi: 10.1080/19420862.2015.1043503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106(29):11937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lauer TM, Agrawal NJ, Chennamsetty N, Egodage K, Helk B, Trout BL. Developability index: a rapid in silico tool for the screening of antibody aggregation propensity. J Pharm Sci. 2012;101(1):102–15. doi: 10.1002/jps.22758. [DOI] [PubMed] [Google Scholar]
- 84.Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kopp MRG, Arosio P. Microfluidic approaches for the characterization of therapeutic proteins. J Pharm Sci. 2018;107(5):1228–36. doi: 10.1016/j.xphs.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 86.Charmet J, Arosio P, Knowles TPJ. Microfluidics for protein biophysics. J Mol Biol. 2018;430(5):565–80. doi: 10.1016/j.jmb.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 87.Angal S, King DJ, Bodmer MW, Turner A, Lawson ADG, Roberts G, Pedley B, Adair JR. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol. 1993;30(1):105–08. doi: 10.1016/0161-5890(93)90432-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


