Abstract
COVID-19 diagnosis is usually based on PCR test using radiological images, mainly chest Computed Tomography (CT) for the assessment of lung involvement by COVID-19. However, textual radiological reports also contain relevant information for determining the likelihood of presenting radiological signs of COVID-19 involving lungs. The development of COVID-19 automatic detection systems based on Natural Language Processing (NLP) techniques could provide a great help in supporting clinicians and detecting COVID-19 related disorders within radiological reports. In this paper we propose a text classification system based on the integration of different information sources. The system can be used to automatically predict whether or not a patient has radiological findings consistent with COVID-19 on the basis of radiological reports of chest CT. To carry out our experiments we use 295 radiological reports from chest CT studies provided by the ‘‘HT médica" clinic. All of them are radiological requests with suspicions of chest involvement by COVID-19. In order to train our text classification system we apply Machine Learning approaches and Named Entity Recognition. The system takes two sources of information as input: the text of the radiological report and COVID-19 related disorders extracted from SNOMED-CT. The best system is trained using SVM and the baseline results achieve 85% accuracy predicting lung involvement by COVID-19, which already offers competitive values that are difficult to overcome. Moreover, we apply mutual information in order to integrate the best quality information extracted from SNOMED-CT. In this way, we achieve around 90% accuracy improving the baseline results by 5 points.
Keywords: COVID-19, Radiological report, Text classification, Natural language processing, Named entity recognition
Highlights
-
•
Automatic detection of suspicious COVID-19 in Spanish radiology reports.
-
•
Natural Language Processing techniques to detect cases suspicious of COVID-19.
-
•
Automatic extraction of COVID-19 related disorders using SNOMED-CT.
-
•
Applying Named Entity Recognition to improve classification systems.
-
•
Using Machine Learning and feature representation in text classification task.
1. Introduction
The new coronavirus 2019 disease (COVID-19) is creating an important and urgent threat to global health. Since the outbreak in early December 2019 in Wuhan, Hubei Province, China, more than 180 countries contain a high number of infected people and the number is still rising. To mitigate the burden on the healthcare system, while providing the best possible care for patients, an efficient and effective diagnosis of the disease is needed [1].
Many efforts are being focusing on developing automated solutions to support medical experts in the early detection of the disease based on medical images. Prediction models that combine variables or features to estimate the risk of people becoming infected is helping clinicians to deal with the COVID-19 outbreak [2]. These models require innovative approaches that provide immediate and real-time results. In particular, the Named Entity Recognition (NER) task aims to detect mentions of interesting entities within unstructured textual reports which can help to refine the COVID-19 detection task [3].
Chest, and more specifically, lung involvement is by far the most common site of organ involvement in COVID-19. Together with patient's symptoms, the use of Ray-X and Computed Tomography (CT) are the commonest approaches to diagnosis and staging the severity of lung involvement by COVID-19. Several studies have tested the viability and specificity of chest CT for the COVID-19 disease being currently included in most clinical protocols for COVID-19 assessment. The most typical CT features of COVID-19 chest involvement are ground-glass opacities, usually bilateral with peripheral distribution and the presence of bronchovascular thickening or bronchiectasias within lesions. Atypical findings for COVID-19 are considered pleural effusion, lymphadenopaties, lung consolidations or cavitations [[4], [5], [6], [7]].
Nowadays, although radiologists and clinicians are able to accurately detect and characterize cases of COVID-19 based on chest CT examinations, their tasks are manual and time-consuming, especially when many cases need to be examined, making it necessary to automate support tools for medical specialists. As we referenced before, most efforts have been focused on developing supporting Artificial intelligence (AI) tools to help in the evaluation of medical images (mainly RX and CT) for the diagnosis of COVID-19. However, few studies have addressed this issue from the perspective of the clinical and radiological text-based reports. Disorder-related clinical or radiological text-based reports are a potential source of information about the likelihood of presenting signs of COVID-19 manifestations for each patient. In this paper, we propose an automatic system for extracting and analyzing disorders related to COVID-19 from CT radiological reports. We apply AI and Natural Language Processing (NLP) tools to automatically extract disorders. For our experiments we have used a corpus composed of 295 radiological reports suspicious of COVID-19 and labeled in binary form (whether the patient has the virus or not). This corpus contains chest CT scans and has been provided by the radiological clinic ‘‘HT médica".
The main goal of this paper is to study the impact of integrating external information from biomedical ontologies to improve the automatic detection of COVID-19 in textual radiological reports. However, other important motivations can be highlighted like for example the detection of unexpected findings related to COVID-19 and early notification of the cases, and monitoring the incidence and prevalence of COVID-19 in radiology units.
In addition, in order to detect disorders we use in our experiments the latest available Spanish version of SNOMED-CT as a source of knowledge (release date: 2020-04-30). This version is updated with the most recent concepts related to COVID-19. Disorders extracted from the text (e.g. bronchiectasis) are an important feature of the classification system. It should be noted that not all the disorders detected have the same relevance for achieving a correct classification. For this reason, we will use mutual information to distinguish those disorders that provide important information. In other word, a fracture does not contribute as much to detecting the COVID-19 as a bilateral pneumonia. Our system includes these important features by using different word representation vectors concatenated with the text of the radiological report. Finally, the final vector is included in the classification algorithm.
The remainder of the paper is structured as follows: in Section 2 we comment on some related studies. The dataset used and the automatic disorder extraction system are described in Section 3. In Section 4 we study the representation of the features. Machine learning approaches are shown in Section 5. The results are presented in Section 6. Finally, discussion and conclusions are presented in Sections 7 and 8 respectively.
2. Related work
In the current literature, many studies have been conducted on the automatic detection of COVID-19. Most of these studies have focused on the classification of CT images by using Machine Learning (ML) technologies due to their high capability of feature extraction.
On the one hand, traditional ML algorithms are beginning to be a key technology in the detection of the virus. Barstugan et al. [8] used Support Vector Machine (SVM) and features to label images as coronavirus and non-coronavirus (infected/non-infected). This study used 150 C T images for COVID-19 classification. On the other hand, deep learning has been growing in recent years, driven largely by increased computing power and the availability of massive new datasets. Using these algorithms, Butt et al. [9] classified CT images of COVID-19 into three classes: COVID-19, influenza-A viral pneumonia, and healthy cases. The dataset consisted of total 618 images and they achieved an 87.6% overall classification accuracy.
Given the importance of early prediction of COVID-19, there are many other studies related to deep learning and classification of CT images achieving competitive results [[10], [11], [12], [13], [14]]. Moreover, Wynants et al. [1] conducted a review and critical evaluation of published studies of predictive models for the diagnosis of COVID-19 in patients with suspicious of infection. These prediction models can be divided into three categories: models for the general population to predict the risk of being infected of COVID-19, models to support the diagnosis of COVID-19 in patients with suspicious of infection, and models to support the prognostication of patients with COVID-19. All models reported moderate to excellent predictive performance.
Concerning the treatment of textual information for detecting COVID-19, not very much research can be found. For example, some studies use NLP to automate the extraction of COVID-19-related discussion from social media [15] or to analyze the research literature on COVID-19 [[16], [17], [18]]. There are also studies on biomedical corpus available in English that have been explored to extract signs and symptoms from texts [13]. Moreover, some recent studies show the potential benefit of using NLP in the classification of textual radiological reports [19,20]. However, no specific research can be found on the automatic detection of possible cases of COVID-19 applying NLP technologies such as NER or information extraction.
On the other hand, ontologies have become an increasingly important component of biomedical studies, especifically in NER tasks. This is due to the fact that they provide researchers with common terminologies for presenting information in a structured way. Ontologies, terminologies and dictionaries such as UMLS [21], SNOMED-CT [22] and ICD-10 are the most popular. These ontologies make it possible to identify and extract relevant information from the biomedical literature such as fractures, abnormalities, disorders, findings, and so on [[23], [24], [25], [26]]. Zuccon et al. [27] use SNOMED-CT as a feature for the classification task of radiological texts.
Several classification methods integrate different features extracted from ontologies. However, the inclusion of a number of features does not always improve the final system. In fact, dimensionality is a major problem when using high dimensional features [28]. For this reason, several approaches have been proposed and used for dimensionality reduction [29]. Feature selection involves the election of a subset of the original, which is a widely used dimensionality reduction technique. Several studies have experimented with approaches to the selection of subsets of features [30,31]. They demonstrate that the use of an effectively selected subset of characteristics can achieve better performance than the use of the original set. In this context, many techniques have been used to reduce the dimensionality of features [[32], [33], [34]], more specifically in the biomedical domain [28,[35], [36], [37]].
In this study we address a classification task of radiological reports. This is a difficult task since all the reports are from CT studies with initial clinical suspicion of COVID-19 being labeled as consistent with COVID-19 and not consistent with COVID-19. To carry out this study, we first use an automatic entity recognizer to extract virus-related disorders using SNOMED-CT terminology. Next, we applied a method of dimensionality reduction in order not to take into account all the recognized disorders. Finally, the features and report are jointly entered into the ML algorithm to predict whether a patient has chest involvement by COVID-19 or not.
3. Materials and resources
3.1. Dataset
A real-life dataset for document classification is used to evaluate different document representation methods. The dataset is composed of 295 anonymous CT scan reports with suspicious of COVID-19 collected between April 3, 2020 and April 24, 2020. This clinical corpus has been provided by the radiological clinic ‘‘HT médica’‘. The dataset was annotated by radiology experts and all reports included in the corpus contain suspicious of COVID-19 making the automatic classification to detect whether a patient has the disease a hard challenge. In addition, the corpus is labeled in binary form: consistent with COVID-19 and not consistent with COVID-19.
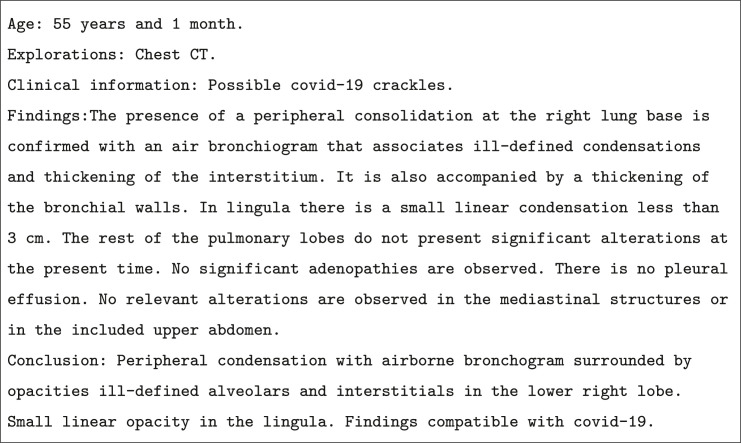
The corpus is written in Spanish and is anonymized to preserve patient identity. In addition, the radiology reports are divided into sections such as patient's age, examination performed (chest CT), clinical information, findings and conclusions. Fig. 1 displays an example of a radiology report of a patient with COVID-19.
Fig. 1.
Example of Spanish radiology report annotated with COVID-19. (See the English translation in Figure A.11 in Appendix A).
The study included 295 patients, 52.2% men and 47.8% women, aged 16 to 97 with a mean age of 56 years. Other corpus statistics are presented in Table 1 .
Table 1.
Dataset analysis.
| COVID-19 | Non-COVID-19 | |
|---|---|---|
| Number of documents | 158 | 137 |
| Vocabulary size | 2162 | 2017 |
| Avg. of sentence in the reports | 24.58 | 23.32 |
| Avg. of tokens in the reports | 161.11 | 147.97 |
3.2. Named Entity Recognition using SNOMED-CT
As we mentioned previously, one of the main purposes of this study is to automatically extract disorders related to COVID-19. In order to detect these disorders we use the BSB NER system for Spanish described in Ref. [38]. This system, among other things, uses information sources related to the biomedical domain such as UMLS, SNOMED-CT and ICD-10 to extract all entities in a determined text. To carry out the task of NER, the system develops a normalizing process in the text. BSB tries to match concepts with ontologies and terminologies in a way that returns the beginning and end of each entity.
Since SNOMED-CT offers semantic categories to easily extract concepts included in them, we decided to apply a filter to the BSB system to extract only the SNOMED-CT biomedical concepts included in the semantic type ‘‘disorder".1 Moreover, we also include an updated list2 of SNOMED-CT concepts related to SARS-CoV-2 not included in the April release version.
In addition, the SNOMED-CT National Reference Center for Spain has activated a contingency mechanism to respond to the challenge of having standardized and sufficiently accurate concepts for coding clinical and epidemiological records relating to Severe Acute Respiratory Syndrome (SARS) coronavirus.3 Therefore, it is considered a reference terminology with regard to COVID-19. The terminology describes that a disorder is ‘‘always and necessarily an abnormal clinical state".
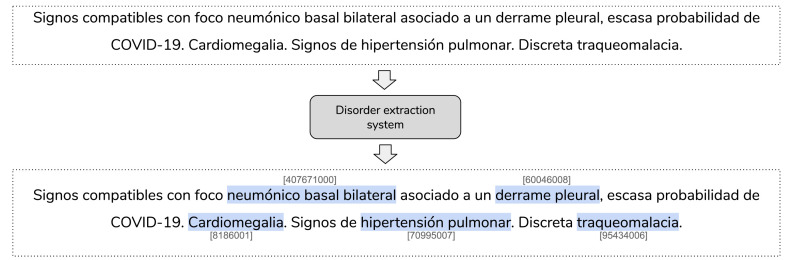
Fig. 2 shows an example of the process carried out by the entity extraction system using SNOMED-CT terminologies as the information source. The system recognizes ‘‘disorders” such as bilateral opacities in ground glass (opacidades en vidrio deslustrado), atypical pneumonia (neumonía atípica) and patchy infiltrate (infiltrado parcheado), among others.
Fig. 2.
Example of disorder extraction system using SNOMED-CT terminology. Translation: Signs compatible with bilateral basal pneumonic focus associated with pleural effusion, low probability of COVID-19. Cardiomegaly. Signs of pulmonary hypertension. Discrete tracheomalacia.
4. Features selection
Features selection is the process of finding a subset of characteristics from the original set of features forming patterns in a given dataset, according to the defined criterion [39]. Features selection plays an important role in text categorization because it can provide additional information to the ML algorithm for better classification.
The input to the classification system is the text of the radiological report and the concepts extracted from SNOMED-CT, and therefore two independent representation models are created. These representation models are explained below.
4.1. Document representation
For the representation of the text included in the radiological report, we tried different methods of representation including word embeddings and TF-IDF [40]. TF-IDF is combination of two statistical techniques, Term Frequency (TF) and Inverse Document Frequency (IDF). The main benefit of the TF-IDF score is that its value increases with the corresponding number of times a word appears in the document, but is offset by the occurrence of the word in the collection of documents.
The TF-IDF is a Bag Of Words (BOW) weighting model used to give weights to the terms in a document collection by measuring how often a term is found within a document (TF), offset by how often the term is found within the entire collection (IDF). The BOW assumption is that a document can be considered as a feature vector where each element in the vector indicates the presence (or absence) of a word. The BOW model is the simplest approach used in NLP and text classification. In this model, a text (such as a word or sentence) is represented as the bag (multiset) of its keywords.
First, in the IDF calculation (Eq (1)) we use smooth to prevent zero divisions, specifically the constant ‘‘1″ is added to the numerator and denominator of the IDF as if an extra document was seen containing every term in the collection exactly once.
| (1) |
where:
N = total number of documents in the collection.
= number of documents containing t (term).
Afterwards, Eq. (2) shows the calculation of the TF-IDF used. TF-IDF incorporates local and global parameters, because it takes into consideration not only the isolated term but also the term within the document collection.
| (2) |
where:
= number of occurrences of t in d (document).
The use of this simple TF could lead to problems when we have a repeated term in a document, therefore, the TF of a document in a vector space is usually also normalized. In all our experiments we use the L2 normalization or Euclidean norm.
On the other hand, neural networks use an embedding layer as an input, which makes it possible to represent words and documents using a dense vector representation. In our case, we use FastText embeddings from SUC (Spanish Unannotated Corpora4 ) because they provide greater coverage for our vocabulary [41].
In this study, four different BOW methods are explored to represent the text of the radiology report:
-
(1)
TF-IDF.
-
(2)
TF-IDF by disabling the reweighting of the IDF.
-
(3)
TF-IDF with word-based n-grams model. This method uses not only unigrams but also bigrams and trigrams. For example, for the sentence ‘‘bilateral pleural effusion” we would have the following n-grams: ‘‘bilateral”, ‘‘pleural”, ‘‘effusion”, ‘‘bilateral pleural”, ‘‘pleural effusion”, and ‘‘bilateral pleural effusion”. With this method we can better capture the semantics using the proximity of the words and their occurrence in the document.
-
(4)
TF-IDF with word-based n-grams model and disabled IDF. This method uses unigrams, bigrams and trigrams and disables the IDF calculation.
-
(5)
FastText SUC.
4.2. SNOMED-based features representation
For the representation of the concepts extracted by the automatic entity detection system it is also necessary to use numerical vectors.
After conducting several tests with different representation vectors, we decided to use two types:
-
(1)
TF. This method consists of assigning the frequency of the SNOMED-CT concept (number of occurrences in the radiological report) in the representation vector.
-
(2)
Binary TF. In this case we use a binary vector in which 1 represents that the concept SNOMED-CT occurs in the report and 0 that it does not occur.
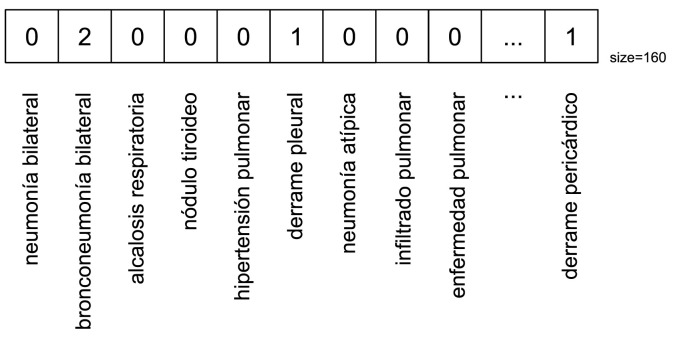
In order to clarify the concept representation vector, Fig. 3 displays an example where each position represents an SNOMED-CT concept. In the example of the vector, the second position in the vector refers to the concept of ‘‘bilateral bronchopneumonia” and it occurs twice in the radiological report. It is important to highlight that the NER system has recognized 164 different disorders in the COVID-19 corpus and for that reason the vector size is 164.
Fig. 3.
Example of SNOMED-CT concept representation vector using TF.
4.3. Mutual information and feature reduction
In this section we introduce the process carried out for the reduction of dimensionality of the features. Dimensionality reduction of the raw input variable space is an essential pre-processing step in the classification process. We apply dimensional reduction for two main reasons: computational cost and classification accuracy. It has been observed that added irrelevant features may actually degrade the performance of classifiers if the number of training samples is small relative to the number of features [34]. In order to reduce the dimensionality, we first need to use an approach that identifies the most relevant features to our classification problem and the less relevant ones. Later, we select the most informative features using the top percentiles. In other words, those disorders related to the COVID-19 that provide more information to the classification algorithm.
One of the most effective approaches for optimal feature extraction is based on Mutual Information (MI). MI is known to characterize the dependence between random variables beyond the second order moment (correlation) and can be used for multivariate selection, by choosing the features which jointly maximize the prediction given a set of previously selected features [42]. The function relies on non-parametric methods based on entropy estimation from k-nearest neighbors distances as described in Ref. [42,43].
We apply the MI scoring function to all previously extracted SNOMED-CT disorders. We found 164 different disorders named in the radiology reports. Table 2 summarizes the top 10 high-scoring features and the associated SNOMED-CT code. In this table we can appreciate disorders such as bilateral pneumonia, ground-glass opacities and bilateral bronchopneumonia are common disorders when the prognosis corresponds to COVID-19. All these disorders are usually related to pneumonia and infectious lung problems.
Table 2.
Top 10 high-scoring features of COVID-19 related disorders.
| # | SNOMED-CT code | Disorder |
|---|---|---|
| 1 | 407,671,000 | Bilateral pneumonia |
| 2 | 63,531,000,122,103 | Ground-glass opacities |
| 3 | 396,286,008 | Bilateral bronchopneumonia |
| 4 | 68,409,003 | Organized pneumonia |
| 5 | 63,521,000,122,101 | Patchy infiltrate |
| 6 | 95,436,008 | Lung consolidation |
| 7 | 101,401,000,119,103 | Pulmonary granuloma |
| 8 | 233,935,004 | Pulmonary thromboembolism |
| 9 | 59,282,003 | Pulmonary embolism |
| 10 | 63,541,000,122,106 | Interstitial Pneumonia |
On the contrary, the MI approach offers disorders that are not related to COVID-19 such as breast carcinoma, pericardial effusion and lung carcinoma. These disorders are common to detect in chest CT but are not related to the virus. For this reason we apply dimensional reduction in the features.
Selecting the optimal number of features to be included in the ML algorithm is an essential part of our experiments. For the experimentation we use the top 50th percentile of our features. Initially, we found 164 different disorders in the radiology reports. After selecting the top 50th percentile of the features, we will use 82 disorders (the 82 most informative disorders for our classification system).
5. Automatic classification methodology
ML provides an effective way to automate the analysis and diagnosis for medical reports. This approach can potentially reduce the workload of radiologists. In this section we will briefly detail the models and algorithms used in our study and then show the results obtained in the following section.
5.1. Traditional machine learning
After testing several traditional ML algorithms, we decided to show the experiments using SVM because we have obtained the best results with this simple ML algorithm. SVM transfers features into space where it can better classify features with kernel functions [44,45]. SVM-based approaches can handle large feature spaces with excellent classification accuracy. This technique is the first kernel bases learning algorithm and the most commonly used kernel are linear, polynomial, Radial Basis Function (RBF) and sigmoid. Linear kernel function was used in our system with the parameter C = 1. The parameter C controls the trade-off between frequency of error and complexity of decision rule.
5.2. Deep learning
Deep learning is a subfield of machine learning that has been growing in recent years, driven largely by increased computing power and the availability of massive new datasets. The objective of deep learning is to capture non-linear patterns in data by adding layers of parameters to the model. In our study, we explore three techniques associated with deep learning: LSTM, BiLSTM and CNN.
Hochreiter and Schmidhuber [46] showed a variation of a recurrent neural network named Long Short Term Memory network (LSTM) that with a special hidden unit acting like a memory cell plus a gradient-based back-propagation technique makes it possible to selectively retain relevant information from previous step, while the input sequence is being parsed element by element [47]. Afterwards, Bi-directional Long Short-Term Memory (BiLSTM) is an extension of traditional LSTM that can improve model performance on sequence classification problems [48]. The main goal of BiLSTM is to split the neurons of a regular LSTM into two directions, one for positive time direction (forward states), and another for negative time direction (backward states). On the other hand, Convolutional Neural Network (CNN) uses layers with convulsion filters that are applied to local features [49]. Finally, we also tested a basic Artificial Neural Network (ANN) with different word representations. This network is structured in a sequential mode and composed of dense layers.
We have tested these networks with different hyperparameter values and the ones that performed the best are presented below. For the hyperparameter optimization we have tried the following parameters and values: size of layers: [50, 100, 150], batch size: [8, 32, 64], dropout rate: [0.25, 0.5], activation: [relu, tahn].
After conducting the hyperparameter optimization, we have used 100 neurons in the case of LSTM, BiLSTM and ANN, and 50 neurons for CNN. A batch size of 8 for BiLSTM, CNN and ANN, and 16 for LSTM was employed. For all networks, a max pooling layer was appended to the model and a dense layer of size 50 with Rectified Linear Unit (ReLU) activation added. After this, we applied a dropout function to help prevent overfitting. Specifically, we have used a dropout rate of 0.25. This was followed by a dense layer with the sigmoid activation function in order to produce the desired binary output. For the optimizer, we leverage the adam optimizer which performs well for NLP data.
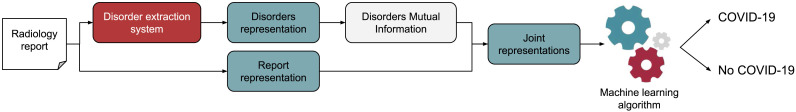
Finally, the proposed architecture is shown in Fig. 4 . As we can see, the first step consists of disorders entity recognition within the radiological report. The disorders are represented using different types of BOW. Mutual information is used on the disorders to reduce the dimensionality of the BOW vector obtaining those SNOMED-CT concepts most important for the virus classification. In addition, the text of the radiological report is also represented using a BOW vector. The representation vectors are concatenated and introduced into the ML algorithm to predict whether the patient contains the COVID-19 infection or not consistent with COVID-19 called non-COVID-19.
Fig. 4.
Machine learning system architecture for COVID-19 detection.
6. Results and analysis
We investigate the performance of the proposed methods using the suspected corpus of COVID-19 and compare the results obtained with the well known feature extraction methods proposed: TD-IDF, TF, MI-based feature extraction method and word embeddings for neural networks.
In order to present the results and make an analysis of them, we have divided this section into four subsections including the evaluation metrics used to evaluate the systems, the results obtained, an analysis of the features used, and finally an error analysis showing the confusion matrix.
6.1. Evaluation metrics
Five statistical parameters such as precision, recall, accuracy, F1-score and Matthews Correlation Coefficient (MCC) are applied to determine the evaluation of the proposed approach using the macro-average method. The equations of the different metrics are described below:
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where TP, TN, FP and FN mean true positive, true negative, false positive and false negative, respectively.
6.2. Experimental result
To evaluate the performance of the classification model, a 10-fold cross-validation method was used.
Table 3 shows the general results in terms of accuracy using the SVM classifier. The rows of the table display the representation technique for the radiology report (see Section.4.1), while the columns of the table show the method chosen to represent the disorders related to COVID-19. These methods are: baseline (without SNOMED-CT features), TF, TF + MI (TF with reduced dimensionality), TF-bin (binary TF) and TF-bin + MI (binary TF with reduced dimensionality).
Table 3.
Summary of results obtained Acc. with different features representations using the SVM algorithm.
| Document representation | SNOMED-CT representation |
||||
|---|---|---|---|---|---|
| Baseline (%) | TF (%) | TF + MI (%) | TF-bin (%) | TF-bin + MI (%) | |
| TF-IDF | 85.08 | 83.39 | 87.10 | 85.02 | 89.15 |
| TF-IDF disable IDF | 84.90 | 81.69 | 87.39 | 82.71 | 87.12 |
| TF-IDF n-grams | 84.88 | 81.02 | 86.42 | 81.36 | 86.78 |
| TF-IDF n-grams disable IDF | 85.42 | 81.36 | 87.14 | 82.36 | 87.46 |
Since the corpus contains suspicious reports of COVID-19 the base systems already reach high values, around 85% in detecting the virus. This means that all reports contain information about the virus but the algorithm is able to detect whether the patient definitely has COVID-19. This baseline obtains high values that are difficult to surpass.
The results show that the use of all the extracted features (164 disorders related to COVID-19) does not improve the baseline. For example, using as document representation TF-IDF and n-grams, the baseline obtained 85.08% of accuracy, and by adding the vector of disorder features the accuracy decreases to 83.39% and 85.02% (TF and binary TF, respectively).
However, the reduction in dimensionality using the recognized SNOMED-CT disorders surpasses the baseline values. In this case, instead of using 164 disorders we used the 82 most important and representative ones. This indicates that applying few but good features increases the accuracy of the classifier.
All the results applying MI improve the baseline. For instance, using TF-IDF to represent the radiology report and binary TF with MI to represent the disorders, we achieve our best result (89.15% of accuracy).
To better clarify the results obtained previously, we provide Table 4 . In this table we can see the results according to each class (COVID-19 and Non-COVID-19). In addition, we also show the measure macro-average. With this table we try to show the improvement over the baseline (without features) by using and integrating binary TF and MI, reducing the dimensionality of the feature vector. In terms of precision, recall, F1, AUC and MCC in all scenarios the integration of disorders improves the baseline.
Table 4.
Deeper summary of the SVM algorithm and performance improvement using different representations of documents and features.
| Doc. Repr. | SNOMED-CT Repr. | COVID-19 |
Non-COVID-19 |
Macro-avg |
AUC (%) | MCC (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P (%) | R (%) | F1 (%) | P (%) | R (%) | F1 (%) | P (%) | R (%) | F1 (%) | ||||
| TF-IDF | Baseline | 84.34 | 88.61 | 86.42 | 86.05 | 81.02 | 83.46 | 85.19 | 84.81 | 84.94 | 84.81 | 70.01 |
| TF-bin + MI | 87.50 | 93.04 | 90.18 | 91.34 | 84.67 | 87.88 | 89.42 | 88.85 | 89.03 | 88.95 | 78.27 | |
| TF-IDF | Baseline | 85.19 | 87.34 | 86.25 | 84.96 | 82.48 | 83.70 | 85.07 | 84.91 | 84.98 | 84.91 | 69.99 |
| disable IDF | TF-bin + MI | 84.88 | 92.41 | 88.48 | 90.24 | 81.02 | 85.38 | 87.56 | 86.81 | 87.03 | 86.71 | 74.27 |
| TF-IDF n-grams | Baseline | 83.53 | 89.87 | 86.59 | 87.20 | 79.56 | 83.21 | 85.36 | 84.72 | 84.90 | 84.72 | 70.08 |
| TF-bin + MI | 86.50 | 89.24 | 87.85 | 87.12 | 83.94 | 85.50 | 86.81 | 86.59 | 86.68 | 86.59 | 73.40 | |
| TF-IDF n-grams | Baseline | 84.43 | 89.24 | 86.77 | 86.72 | 81.02 | 83.77 | 85.57 | 85.13 | 85.27 | 85.13 | 70.70 |
| disable IDF | TF-bin + MI | 85.80 | 91.77 | 88.69 | 89.68 | 82.48 | 85.93 | 87.74 | 87.13 | 87.31 | 87.13 | 74.87 |
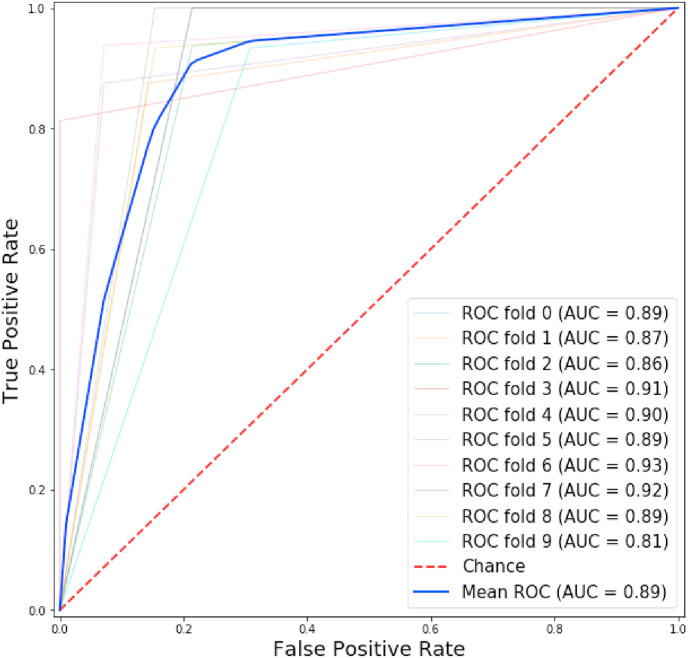
On the other hand, AUC (Area under the ROC curve) measures the entire two-dimensional area underneath the entire Receiver Operating Characteristic (ROC) curve. The ROC curve is a common tool used with binary classifiers in machine learning methods. ROC curves have been frequently used in the biomedical domain [50,51] because it provides an effective approach for characterising the performance of classifiers in terms of sensitivity to specificity. Fig. 5 shows the ROC curve with the AUC values of each 10-fold cross-validation using the best system presented before (SVM classifier, text representation with TF-IDF and SNOMED represented with binary TF and mutual information). This figure displays that each folder obtains different AUC, reaching a maximum of 0.93 of AUC and a minimum of 0.81. Finally, the mean obtained is 0.89 of AUC.
Fig. 5.
Cross-validation ROC curve using SNOMED-CT disorders detected.
Additionally, in this section we present a basic comparison of the different neural networks using the same dataset as mentioned above and 10-fold cross-validation. Since neural networks are currently widely used in different areas of the NLP, we would like to show how they work in this particular domain by detecting suspected cases of COVID-19.
Table 5 summarizes the best results obtained after testing different hyperparameters of neural networks and word representations. As we can appreciate, in all cases the neural networks achieve better results with the incorporation of features using mutual information on COVID-19 related disorders. The best result without using extra features (baseline) has been obtained with CNN and using FastText SUC embeddings reaching 84.03% accuracy. On the other hand, the best result adding SNOMED features has been obtained by the basic ANN with TF-IDF n-grams and disable IDF as document representation and binary TF with MI to represent the features. In this scenario, the system achieves 84.75% accuracy. In our particular case, we can conclude that the neuronal networks are far from reaching the best system proposed in Table 3 in which 89.15% accuracy is achieved.
Table 5.
Results using different neural networks and document presentations detecting suspicious cases of COVID-19.
| Model | Doc. Repr. | SNOMED-CT Repr. | P (%) | R (%) | F1 (%) | Acc.(%) | AUC (%) | MCC (%) |
|---|---|---|---|---|---|---|---|---|
| Basic ANN | TF-IDF n-grams | Baseline | 79.57 | 81.05 | 79.55 | 81.49 | 81.05 | 63.23 |
| disable IDF | TF-bin + MI | 85.66 | 85.23 | 85.28 | 85.54 | 85.24 | 70.90 | |
| CNN | FastText SUC | Baseline | 84.17 | 84.52 | 83.66 | 84.03 | 84.52 | 68.67 |
| TF-bin + MI | 85.17 | 84.96 | 84.47 | 84.75 | 84.96 | 70.11 | ||
| LSTM | FastText SUC | Baseline | 80.22 | 77.45 | 75.77 | 76.59 | 77.45 | 57.46 |
| TF-bin + MI | 82.66 | 81.01 | 79.11 | 79.69 | 81.01 | 63.51 | ||
| BiLSTM | FastText SUC | Baseline | 78.01 | 74.40 | 71.63 | 73.00 | 74.40 | 52.01 |
| TF-bin + MI | 78.92 | 76.45 | 75.41 | 76.62 | 76.45 | 55.09 |
6.3. Improving the system by applying MI and feature reduction
In this section we present how to improve the use of dimensionality reduction using MI. To accomplish this task, we designed Fig. 6 . This figure has been created using the best system obtained in the previous results (SVM algorithm). For the representation of the radiological report we use the TF-IDF method. In order to represent the SNOMED-CT features related to the COVID-19 we use the binary TF method.
Fig. 6.
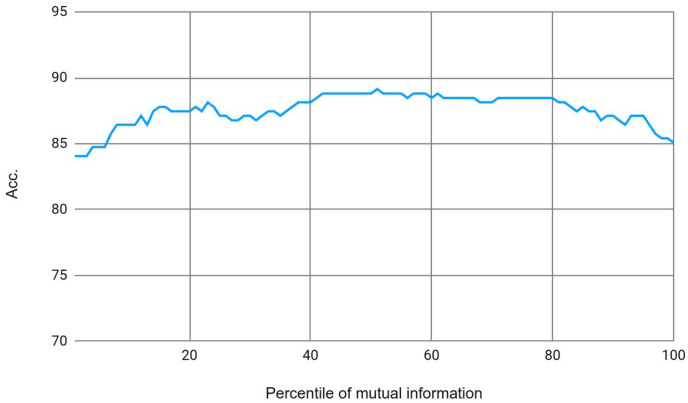
Results in terms of accuracy according to the selected percentile of MI.
On the Y-axis the figure shows the accuracy obtained by the classification system. X-axis shows the percentile used, in other words, how many disorders to use and include in the system.
Initially we had 164 COVID-19 related disorders, but as we have seen from the results in Table 3, using all 164 disorders does not improve the classification; instead using the 50th percentile improves the baseline, achieving up to 89.15% accuracy.
This figure shows the use of different percentiles and the results obtained by the system. The 50th percentile was the decision chosen since it obtained the best score, and with the 50th percentile the feature vector was reduced to 82 disorders. These 82 disorders were the features that provided the most extra information to the classifier for detecting whether a patient had the virus or not.
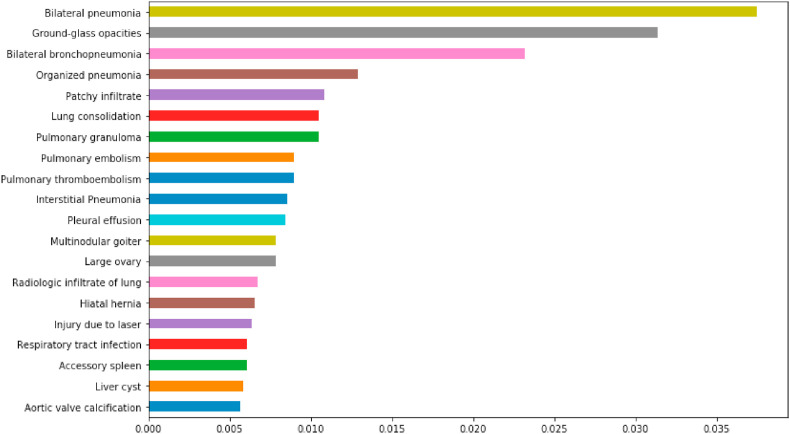
Finally, Fig. 7 shows the SNOMED-CT concepts obtained by applying MI and taking into account the 50th percentile. This figure shows the 20 disorders that are most representative for the classification system and its value by using MI. We do not show the 82 disorders for reasons of space.
Fig. 7.
The 20 most representative SNOMED-CT disorders detected to enhance COVID-19 text classification.
6.4. Error analysis
A good error analysis shows something about why a given method is effective or ineffective for a problem. Basically, error analysis involves examining the errors committed by a system.
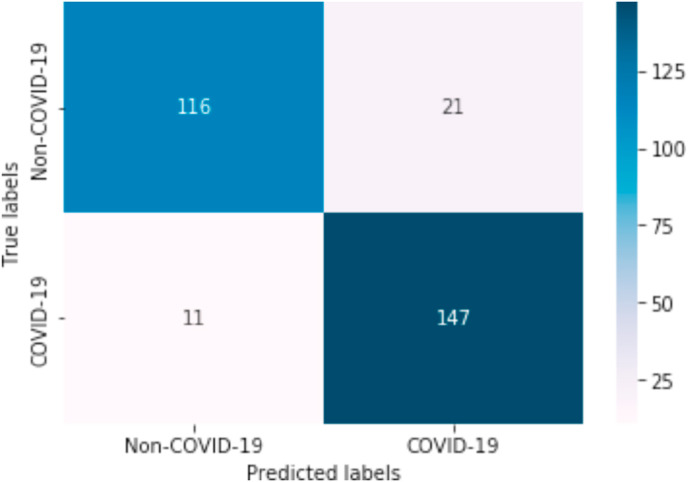
Fig. 8 shows the confusion matrix obtained with the best experiment using SVM algorithm with TF-IDF for document representation and the binary TF method to represent the SNOMED-CT features related to the COVID-19. In this image we can observe that of 295 radiology reports, the model does not classify well 10.85% (32 documents). On the other hand, the system labels 263 documents correctly. In addition, the matrix presents the number of TP, TN, FP and FN. In our particular case the system returns 147 TP, 116 TN, 11 FN and 21 FP which means that the method is very successful.
Fig. 8.
Confusion matrix using SNOMED-CT disorders detected.
To better understand the system errors, one example of false positive is shown in Fig. 9 and one of false negative is shown in Fig. 10 .
Fig. 9.
False positive radiology report in Spanish for a patient with no COVID-19. (See the English translation in Figure A.12 in Appendix A).
Fig. 10.
False negative radiology report in Spanish for a patient with COVID-19. (See the English translation in Figure A.13 in Appendix A).
In Fig. 9, the system detected that the patient had COVID-19 because in the report words were found like: activar protocolo COVID (activate COVID protocol), infiltrados (infiltrates), engrosamientos bronquiales (bronchial thickenings) or neumonía viral (viral pneumonia). All these words are closely related to the virus although some of them are terms of denial and this causes confusion in the classification algorithm.
The false negative shown in Fig. 10 shows the examination of a patient with COVID-19 but was wrongly detected by our system. In this examination we find words like: no se observan (not observed) and no presentan (not present). In addition, it also includes some adjectives that minimize the importance of findings such as pequeña (small) and no significativas (not significant). In this example we also see that the expert used words like consolidation and condensation instead of infiltration which may have caused the system not to detect and classify it incorrectly. In conclusion, these errors suggest improvements in the system for future work.
7. Discussion
COVID-19 has spread widely around the world since the first case was detected at the end of 2019. Early diagnosis of the disease is important for treatment and patient isolation in order to prevent the spread of the virus.
Most studies are based on chest CT images [8,11,13,14]. Regarding textual analysis, other studies use NLP to automate the extraction of COVID-19-related discussion from social media [15] or to analyze the research literature on COVID-19 [[16], [17], [18]]. There are also studies on biomedical corpus available in English that have been explored to extract signs and symptoms in texts [13]. Since these studies use the existing literature, we propose a novel method focused on NLP using a dataset based on chest CT examinations to detect and characterize actual cases of COVID-19 suspicion in patients.
This is an experimental study that uses terminology related to the biomedical domain to extract relevant information. Specifically, we used the most relevant disorders as features in order to help the classification system predict more effectively. On the one hand, we use as baseline radiological reports from chest CT studies, and on the other hand, we use the MI method to extract those relevant features and incorporate them into the classification algorithm.
In order to extract the disorders, we use an automatic entity extraction system created in previous studies. The source of information used has been SNOMED-CT in its latest version and an updated list of concepts related to COVID-19 proposed by SNOMED-CT. All the disorders recognized by the system were not considered equally important. For this reason, we apply a feature reduction method based on its importance in detecting COVID-19.
In relation to the terms detected, pulmonary embolism and pulmonary thromboembolism are important disorders automatically detected by the NER system. The available biological and clinical data raise concerns about unsuspected pulmonary embolism, and these studies shows that patients with COVID-19 are at risk of acute pulmonary embolism and pulmonary thromboembolism [[52], [53], [54], [55]]. Since we have added a list of virus-related terms proposed by SNOMED-CT, the NER system has recognized four of them and classified them among the 40 best for predicting COVID-19 (three of these terms are included in the top 10). These terms are: bilateral interstitial pneumonia, interstitial pneumonia, patchy infiltrate and ground-glass opacities.
This study was carried out using 295 radiological reports provided by ‘‘HT médica". All of these reports contain cases of COVID-19 suspicion and are labeled as COVID-19 (consistent with COVID-19) and not COVID-19 (not consistent with COVID-19). Since all the reports are cases of suspicion, the classification task can be considered a difficult challenge.
Our results show that by adding extra information to the classifier, it improves the base case. Our best result was obtained using TF-IDF for the representation of the CT exam, TF in binary form to represent the concepts of SNOMED-CT and dimensionality reduction taking into account 82 virus-related disorders. The ML classifier that presented the best results was SVM achieving 89.15% accuracy.
We find some limitations during the development of the NLP system and its validation: the number of CT chest exams provided was limited, so the number of disorders detected was as well. In addition, there are automatically recognized disorders that are only loosely related to COVID-19 so they do not add extra information to the classification system.
Although the main goal of our paper is the integration of disorders extracted from SNOMED-CT in order to develop a system for detecting suspicious cases of COVID-19 in textual radiological reports, there are also other interesting motivations that arise from our study:
-
•
Detection of unexpected findings related to COVID-19 in patients who attend the clinic for another reason not related to the virus.
-
•
Monitoring the incidence and prevalence of COVID-19 in radiology or clinical units through radiological reports, using these to detect new outbreaks of the disease.
-
•
Early notification of COVID-19 cases.
-
•
Retrospective search for COVID-19 findings in patients with chest CT in the months prior to the pandemic.
8. Conclusion
A robust ML model is developed using automatically extracted radiological findings consistent with COVID-19 in chest CT reports. These results demonstrate that a traditional ML approach has the ability to predict the presence of the virus in a radiological examination. To improve the approach, a NER system was used to extract COVID-19 related disorders and include them as additional information to the algorithm.
The best system proposed (SVM) is potentially efficient, quality and cost effective obtaining 89.15% accuracy. For this reason, this system will be used in real scenarios by radiologists as a decision support tool for detecting suspicious cases of COVID-19.
In future work, we are planning to study the impact of negation in our systems because it is demonstrated that the correct treatment of negation cues such as ‘‘without” or ‘‘not” is paramount when we work with clinical textual information, e.g. no se visualiza derrame pleural (no pleural effusion visualized). In addition, and since our system will be put into practice in the ‘‘HT médica", we will enrich the corpus with new training samples because the clinic experts will continue to evaluate future radiology reports for detection of suspicious cases of COVID-19. Using these new examinations we will perform more in-depth analysis comparing different classification models [56,57]. The system will be retrained on new studies and it is expected that the accuracy will be increased. Other ontologies and terminologies related to the biomedical domain will be explored in future work, as well as other semantic types (findings, procedures and body structure, among others) will be included in the system in order to detect COVID-19 in radiological reports. Finally, the processing of acronyms such as COPD (chronic obstructive pulmonary disease or EPOC in Spanish) or LSI (lódulo superior izquierdo in Spanish) is another important task in the biomedical domain that could be interesting to study. As future work we plan to use both the description and the acronym in clinical documents in order to see their impact on the final system.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
This work has been partially supported by the LIVING-LANG project [RTI2018-094653-B-C21] of the Spanish Government and the Fondo Europeo de Desarrollo Regional (FEDER).
Footnotes
Appendix A. Translation cases
Fig. A.11.
Example of English radiology report annotated with COVID-19.
Fig. A.12.
False positive radiology report in English for a patient with no COVID-19.
Fig. A.13.
False negative radiology report in English for a patient with COVID-19.
References
- 1.Wynants L., Van Calster B., Bonten M.M., Collins G.S., Debray T.P., De Vos M., Haller M.C., Heinze G., Moons K.G., Riley R.D. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. Br. Med. J. 2020:369. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arabi Y.M., Murthy S., Webb S. COVID-19: a novel coronavirus and a novel challenge for critical care. Intensive Care Med. 2020:1–4. doi: 10.1007/s00134-020-05955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goulart R.R.V., de Lima V.L.S., Xavier C.C. A systematic review of named entity recognition in biomedical texts. J. Braz. Comput. Soc. 2011;17(2):103–116. [Google Scholar]
- 4.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am. J. Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez-Oro R., Nuez J.T., Martínez-Sanz G. English Ed. Medicina Clinica; 2020. Radiological Findings for Diagnosis of SARS-CoV-2 Pneumonia (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., Revel M.P. Diagnostic and interventional imaging; 2020. COVID-19 Pneumonia: a Review of Typical CT Findings and Differential Diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanne J.P. 2020. Chest CT Findings in 2019 Novel Coronavirus (2019-nCoV) Infections from Wuhan, China: Key Points for the Radiologist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barstugan M., Ozkaya U., Ozturk S. 2020. Coronavirus (Covid-19) Classification Using Ct Images by Machine Learning Methods. arXiv preprint arXiv:200309424. [Google Scholar]
- 9.Butt C., Gill J., Chun D., Babu B.A. Applied Intelligence; 2020. Deep learning system to screen coronavirus disease 2019 pneumonia; p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shi Y. 2020. Lung Infection Quantification of Covid-19 in Ct Images with Deep Learning. arXiv preprint arXiv:200304655. [Google Scholar]
- 11.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:200310849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Wong A. 2020. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. arXiv preprint arXiv:200309871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelodar H., Wang Y., Orji R., Huang H. 2020. Deep Sentiment Classification and Topic Discovery on Novel Coronavirus or Covid-19 Online Discussions: Nlp Using Lstm Recurrent Neural Network Approach. arXiv preprint arXiv:200411695. [DOI] [PubMed] [Google Scholar]
- 16.Lopez C.E., Vasu M., Gallemore C. 2020. Nderstanding the Perception of COVID-19 Policies by Mining a Multilanguage Twitter Dataset. arXiv preprint arXiv:200310359. [Google Scholar]
- 17.Hu Y., Chen M., Wang Q., Zhu Y., Wang B., Li S., Xu Y., Zhang Y., Liu M., Wang Y. 2020. From SARS to COVID-19: A Bibliometric Study on Emerging Infectious Diseases with Natural Language Processing Technologies. [Google Scholar]
- 18.Müller M., Salathé M., Kummervold P.E. 2020. Covid-Twitter-Bert: A Natural Language Processing Model to Analyse COVID-19 Content on Twitter. arXiv preprint arXiv:200507503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Úbeda P.L., Díaz-Galiano M.C., Lopez L.A.U., Martín-Valdivia M.T., Martín-Noguerol T., Luna A. Proceedings of the LREC 2020 Workshop on Multilingual Biomedical Text Processing. MultiligualBIO 2020; 2020. Transfer learning applied to text classification in Spanish radiological reports; pp. 29–32. [Google Scholar]
- 20.López-Úbeda P., Díaz-Galiano M.C., Noguerol T.M., Ureña-López A., Martín-Valdivia M.T., Luna A. Detection of unexpected findings in radiology reports: a comparative study of machine learning approaches. Expert Syst. Appl. 2020:113647. [Google Scholar]
- 21.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(suppl_1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spackman K.A., Campbell K.E., Côté R.A., Snomed R.T. Proceedings of the AMIA Annual Fall Symposium. American Medical Informatics Association; 1997. A reference terminology for health care; p. 640. [PMC free article] [PubMed] [Google Scholar]
- 23.Bada M. Biomedical Literature Mining. Springer; 2014. Mapping of biomedical text to concepts of lexicons, terminologies, and ontologies; pp. 33–45. [DOI] [PubMed] [Google Scholar]
- 24.López-Ubeda P., Dıaz-Galiano M.C., Martın-Valdivia M.T., Urena-López L.A. Sinai en tass 2018 task 3. clasificando acciones y conceptos con umls en medline. Proceedings of TASS. 2018:2172. [Google Scholar]
- 25.López-Úbeda P., Díaz-Galiano M.C., Martín-Valdivia M.T., López L.A.U. CLEF (Working Notes) 2018. Machine learning to detect ICD10 codes in causes of death. [Google Scholar]
- 26.Úbeda P.L., Galiano M.C.D., Lopez L.A.U., Martín-Valdivia M.T. Proceedings of the 5th Workshop on BioNLP Open Shared Tasks. vols. 115–120. 2019. Using Snomed to recognize and index chemical and drug mentions. [Google Scholar]
- 27.Zuccon G., Wagholikar A.S., Nguyen A.N., Butt L., Chu K., Martin S., Greenslade J. Automatic classification of free-text radiology reports to identify limb fractures using machine learning and the snomed ct ontology. AMIA Summits on Translational Science Proceedings. 2013;2013:300. [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S.K., Sarkar S., Mitra P. Feature selection techniques for maximum entropy based biomedical named entity recognition. J. Biomed. Inf. 2009;42(5):905–911. doi: 10.1016/j.jbi.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Fodor I.K. A Survey of Dimension Reduction Techniques. Tech. Rep.; Lawrence Livermore National Lab.; CA (US): 2002. [Google Scholar]
- 30.Forman G. An extensive empirical study of feature selection metrics for text classification. J. Mach. Learn. Res. 2003;3(Mar):1289–1305. [Google Scholar]
- 31.Chen Y., Li Y., Cheng X.Q., Guo L. International Conference on Information Security and Cryptology. Springer; 2006. Survey and taxonomy of feature selection algorithms in intrusion detection system; pp. 153–167. [Google Scholar]
- 32.MacKay D.J., Mac Kay D.J. Cambridge university press; 2003. Information Theory, Inference and Learning Algorithms. [Google Scholar]
- 33.Vergara J.R., Estévez P.A. A review of feature selection methods based on mutual information. Neural Comput. Appl. 2014;24(1):175–186. [Google Scholar]
- 34.Shadvar A. 2012. Dimension Reduction by Mutual Information Feature Extraction. arXiv preprint arXiv:12073394. [Google Scholar]
- 35.Batet M., Sánchez D., Valls A. An ontology-based measure to compute semantic similarity in biomedicine. J. Biomed. Inf. 2011;44(1):118–125. doi: 10.1016/j.jbi.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Zare M., Pahl C., Nilashi M., Salim N., Ibrahim O. A review of semantic similarity measures in biomedical domain using SNOMED-CT. J of Soft Computi Decis Support Syst. 2015;2(6):1–13. [Google Scholar]
- 37.Groza T., Verspoor K. Assessing the impact of case sensitivity and term information gain on biomedical concept recognition. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0119091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López-Úbeda P., Díaz-Galiano M.C., Montejo-Ráez A., Martín-Valdivia M.T., Ureña-López L.A. An integrated approach to biomedical term identification systems. Appl. Sci. 2020;10(5):1726. [Google Scholar]
- 39.Cios K.J., Pedrycz W., Swiniarski R.W., Kurgan L.A. Springer Science & Business Media; 2007. Data Mining: a Knowledge Discovery Approach. [Google Scholar]
- 40.Salton G., Buckley C. Term-weighting approaches in automatic text retrieval. Inf. Process. Manag. 1988;24(5):513–523. [Google Scholar]
- 41.Bojanowski P., Grave E., Joulin A., Mikolov T. Enriching word vectors with subword information. Trans Assoc Computational Linguistics. 2017;5:135–146. [Google Scholar]
- 42.Kraskov A., Stögbauer H., Grassberger P. Estimating mutual information. Phys. Rev. 2004;69(6) doi: 10.1103/PhysRevE.69.066138. [DOI] [PubMed] [Google Scholar]
- 43.Ross B.C. Mutual information between discrete and continuous data sets. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0087357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vapnik V. Springer science & business media; 2013. The Nature of Statistical Learning Theory. [Google Scholar]
- 45.Yuan Y., Huang T. A polynomial smooth support vector machine for classification. In: Li X., Wang S., Dong Z.Y., editors. Advanced Data Mining and Applications. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. pp. 157–164. [Google Scholar]
- 46.Hochreiter S., Schmidhuber J. Advances in Neural Information Processing Systems. 1997. LSTM can solve hard long time lag problems; pp. 473–479. [Google Scholar]
- 47.Feng Y., Teh H.S., Cai Y. Deep learning for chest radiology: a review. Current Radiology Reports. 2019;7(8):24. [Google Scholar]
- 48.Schuster M., Paliwal K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997;45(11):2673–2681. [Google Scholar]
- 49.Kim Y. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) Association for Computational Linguistics; Doha, Qatar: 2014. Convolutional neural networks for sentence classification; pp. 1746–1751.https://www.aclweb.org/anthology/D14-1181 URL. [DOI] [Google Scholar]
- 50.Lasko T.A., Bhagwat J.G., Zou K.H., Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. J. Biomed. Inf. 2005;38(5):404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Zweig M.H., Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 52.Bompard F., Monnier H., Saab I., Tordjman M., Abdoul H., Fournier L., Sanchez O., Lorut C., Chassagnon G., Revel M.p. Pulmonary embolism in patients with Covid-19 pneumonia. European Respiratory Journal. 2020;56(1):2001365. doi: 10.1183/13993003.01365-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deshpande C. Thromboembolic findings in COVID-19 autopsies: pulmonary thrombosis or embolism? Ann. Intern. Med. 2020;173(5):394–395. doi: 10.7326/M20-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rotzinger D., Beigelman-Aubry C., von Garnier C., Qanadli S. Thrombosis research; 2020. Pulmonary Embolism in Patients with COVID-19: Time to Change the Paradigm of Computed Tomography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaminetzky M., Moore W., Fansiwala K., Babb J.S., Kaminetzky D., Horwitz L.I., McGuinness G., Knoll A., Ko J.P. Pulmonary embolism on CTPA in COVID-19 patients. Radiology: Cardiothoracic Imaging. 2020;2(4) doi: 10.1148/ryct.2020200308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calvo B., Santafé Rodrigo G. scmamp: statistical comparison of multiple algorithms in multiple problems. The R Journal. 2016;8/1 Aug 2016. [Google Scholar]
- 57.Demšar J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006;7(Jan):1–30. [Google Scholar]