Abstract
Cell biology is moving from observing molecules to controlling them in real time, a critical step towards a mechanistic understanding of how cells work. Initially developed from light-gated ion channels to control neuron activity, optogenetics now describes any genetically encoded protein system designed to accomplish specific light-mediated tasks. Recent photosensitive switches employ many ingenious designs that bring spatial and temporal control within reach for almost any protein or pathway of interest. This next generation optogenetics includes light-controlled protein-protein interactions and shapeshifting photosensors, which in combination with live microscopy enable acute modulation and analysis of dynamic protein functions in living cells. We provide a brief overview of various types of optogenetic switches. We then discuss how diverse approaches have been employed to control cytoskeleton dynamics with light through Rho GTPase signaling, microtubule and actin assembly, mitotic spindle positioning and intracellular transport and highlight advantages and limitations of different experimental strategies.
Introduction
The 21st century has seen remarkable advances in light microscopy. Combined with improving labeling techniques, such as fluorescent proteins that just celebrated their 25th birthday, live-cell imaging has repeatedly demonstrated just how dynamic cells are. Cells continuously reorganize their shape and internal structure. Both are essential to build and operate complex multicellular organisms in which each cell type has unique functions reflected in their intracellular organization and dynamics. Forces driven by the pulling and pushing of the cell’s cytoskeleton underlie cell movement, identity and many pathological processes [1,2]. Consequently, polarized and meaningful cell dynamics depend greatly on the spatial and temporal control of protein activities and interactions downstream of extracellular signaling inputs as well as intracellular control circuits. Ideally, a cell biologist would be able to specifically, locally and acutely interfere with intracellular protein activities to perturb steady-state biochemistry and interrogate cell biological consequences, and the development of technologies to achieve this goal continues to be a driving force of cell biology discovery.
Cytoskeleton filament systems assemble, disassemble and reorganize within minutes, and molecular motors transport cargo at rates of tens of micrometers per minute. Thus, compared to these time scales, experimental approaches that rely solely on removing gene expression or inhibition of protein production by RNA or CRISPR interference are quite slow and not well suited to investigate rapid intracellular dynamics at a mechanistic level. By the time one observes a phenotype, cell and cytoskeleton dynamics have reached a new steady state, which complicates the interpretation of such experiments. Cells also frequently respond to genetic alterations with compensatory mechanisms and genetic approaches do not allow subcellular spatial control.
One experimental agent that, at least in non-plant cells, is orthogonal to most biological processes and that can be controlled with very high spatial and temporal accuracy is light. While high power lasers have been used to locally destroy and cut cellular structures, the first ‘optogenetics’ experiments involved expression of light-gated ion channels in neuronal cells to locally photostimulate nervous system circuits [3–5]. Nowadays, optogenetics more broadly describes any genetically encoded protein system designed to accomplish a specific light-mediated task. Many aspects of plant development and metabolism depend on the ability to sense light through photosensitive domains of phototropins, cryptochromes and phytochromes that contain different kinds of chromophores (Fig. 1) [6]. Photochemical reactions driven by photon absorption result in conformational changes that can be harnessed to accomplish optogenetic work (Fig. 1a). Optogenetic experimental strategies can be roughly categorized into techniques that alter the localization of a protein of interest resulting in activation or inactivation in a specific subcellular region or organelle, approaches that uncage a constitutively active protein, and maybe most relevant to understanding physiological function, techniques that locally inactivate a specific activity of a protein or protein complex. The light-oxygen-voltage (LOV2) domain of oat phototropin 1 may be the most versatile and has been used to build protein modules that either dimerize or dissociate upon blue light stimulation, or directly change protein conformation (Fig. 2). However, instead of reviewing the basic designs of optogenetic switches [7,8], we highlight examples of how optogenetic building blocks have been used to directly target cytoskeleton dynamics and function in living cells with high spatial and temporal accuracy (Table 1).
Figure 1. Comparison of plant-derived photosensitive domains.
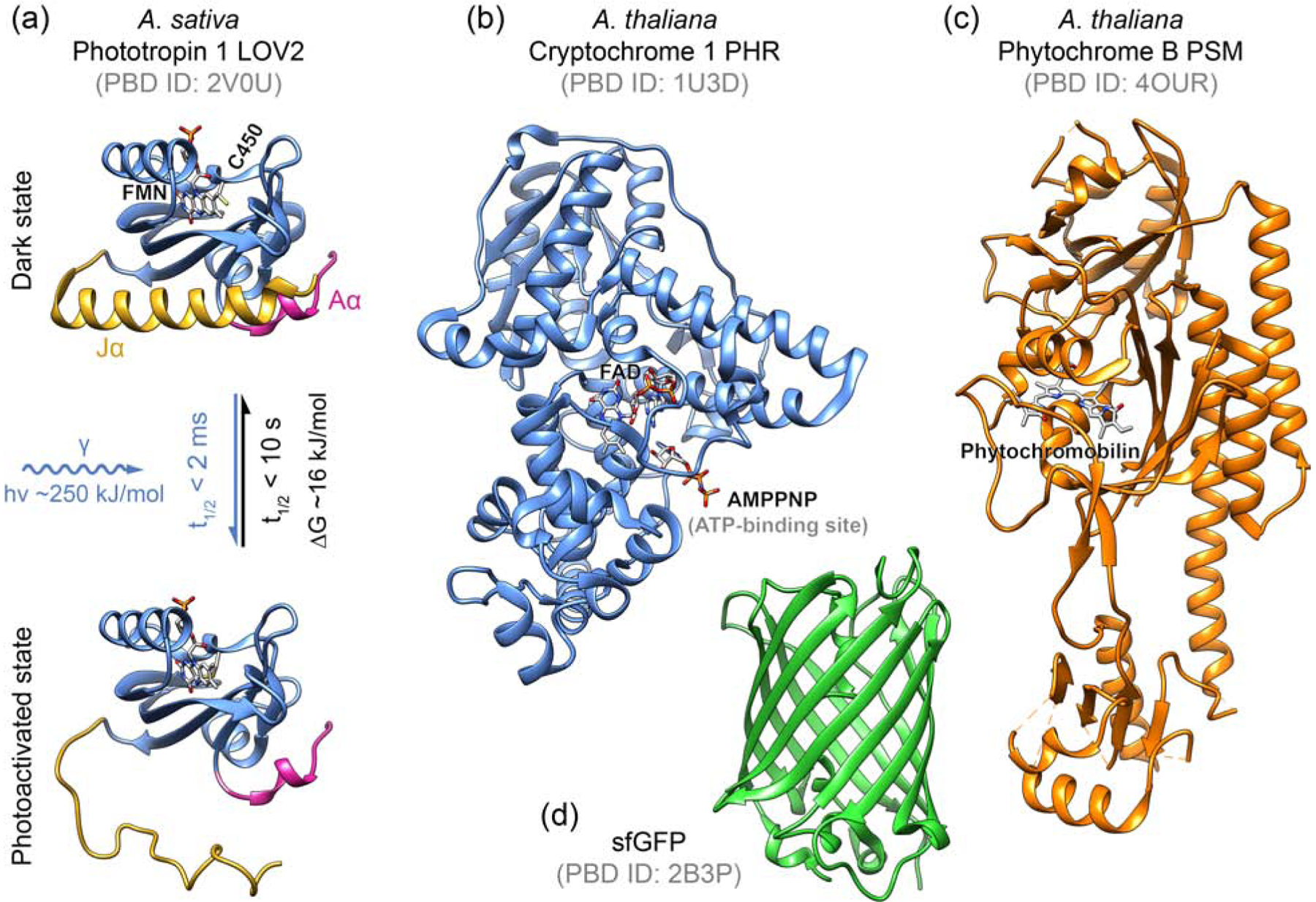
(a) The LOV2 domain is the smallest of the three major plant photosensors, and photochemistry and resulting structural changes have been characterized extensively [56]. Absorption of a blue light photon by the flavin mononucleotide (FMN) co-factor drives formation of a metastable covalent adduct with a conserved cysteine, which results in the undocking of the helices flanking the LOV2 domain core and unfolding of the C-terminal Jα helix [57–59]. LOV2 photoactivation is very fast [60,61], although the light response of LOV2-based optogenetic switches will always be substantially slower. The fungal blue light sensor VVD has a very similar overall fold [62]. (b) The photolyase homology region (PHR) of cryptochromes bind a flavin adenine dinucleotide (FAD) chromophore as well as ATP. Blue light induces cryptochrome dimerization although the underlying photochemistry is incompletely understood [63]. Shown is the crystal structure of cryptochrome Cry1 as a Cry2 structure is not available [64]. (c) The phytochrome B photosensory module (PSM) binds a plant-specific phytochromobilin (or phycocyanobilin) chromophore that in animal cells must be added or supplied by co-expression of the biosynthetic enzyme system [65,66], although bacterial phytochromes utilize a biliverdin (BV) chromophore that may be less limiting in animal cells [67]. Although not fully understood, rotation of the phytochromobilin pyrrole rings likely underlies PhyB photoconversion [68]. (d) sfGFP is shown for size comparison [69].
Figure 2. Overview of LOV2-domain based optogenetic strategies to modulate protein activities.
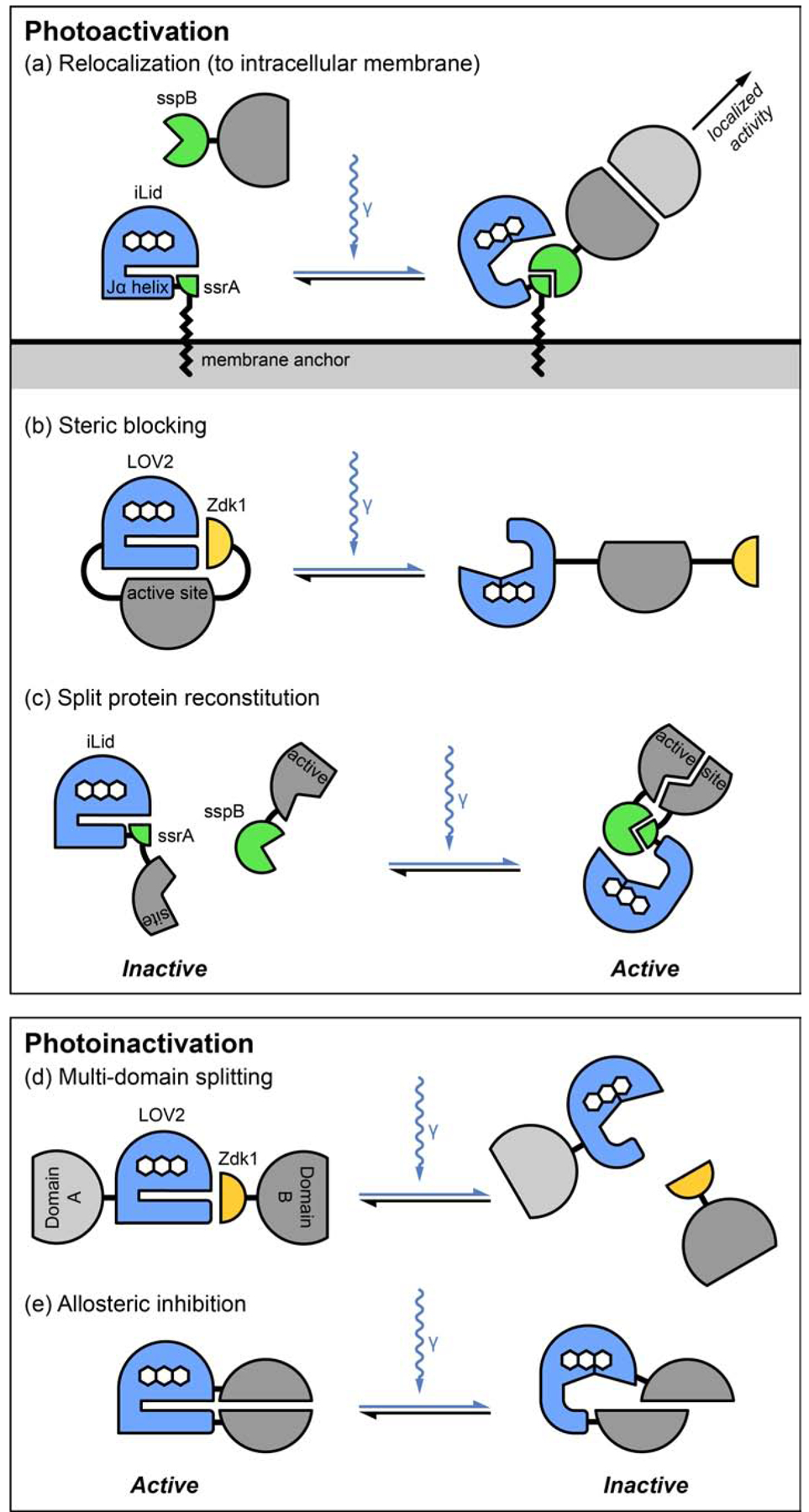
(a) Light-induced dimerization can recruit activating factors, for example Rho GEFs, to specific intracellular sites. As indicated in the main text this also works with other light-induced dimerizers such as Cry2/CIBN or PhyB/PIF. The same approach can also be used to sequester and thus inactivate proteins away from their normal site of action. (b) The light sensitive LOV2/Zdk1 protein interaction module can sterically block access to an active or binding site in a target protein in the dark. (c) In principle, the activity of split proteins can be reconstituted by bringing half-domains back together by light-mediated dimerization. However, because of the difficulty in designing such proteins, this approach has not yet been used much in optogenetics. (d) The light sensitive LOV2/Zdk1 module can be used to dissociate and thus disrupt the function of multi-domain proteins. (e) Conformational changes in photosensors can propagate allosteric inhibition into split target proteins. It should be noted that swapping a light-induced dimerizer, i.e. iLID/nano, for a light-sensitive interaction module, i.e. LOV2/Zdk1, would be expected to relatively easily reverse the directionality of many of these designs and convert photoactivation into photoinhibition or vice versa.
Table 1:
Overview of optogenetic switches to control cytoskeleton dynamics.
| Photosensor (Chromophore) | Peak excitation wavelength | Mechanism | Description | |
|---|---|---|---|---|
| Rho-GTPase signaling | ||||
| PA-Rac1 | LOV2 (FMN) | On: 450 nm Off: dark |
steric inhibition | Release of LO2V-mediated inhibition of Rac1 (Q61L) GEF-binding [9] |
| Photoactivated Cdc42 and Rac1 GEFs | PhyB/PIF (PCB) | On: 664 nm Off: 725 nm |
recruitment | Activation of Rho-GTPase signaling through GEF membrane recruitment [14] |
| Photoactivated Cdc42 GEF | BphP1/PpsR2 (BV) | On: 740 nm Off: 650 nm |
recruitment | Activation of Rho-GTPase signaling through GEF membrane recruitment [67] |
| Photoactivated Cdc42 and Rac1 GEFs | iLID/nano (FMN) | On: 450 nm Off: dark |
recruitment | Activation of Rho-GTPase signaling through GEF membrane recruitment [18] |
| PR_GEF | TULIPs (FMN) | On: 450 nm Off: dark |
recruitment | RhoA activation through GEF membrane recruitment [20–22] |
| optoGEF-RhoA | Cry2/CIBN (FAD) | On: 450 nm (dark) Off: dark (450 nm) |
Recruitment (sequestration) | RhoA activation (inactivation) through GEF membrane (mitochondria) recruitment [23] |
| opto-PI3K | PhyB/PIF (PCB) | On: 664 nm Off: 725 nm |
recruitment | Activation of Rho-GTPase signaling through PI3K membrane recruitment [15] |
| Photoactivated PI3K | Magnets (FAD) | On: 450 nm Off: dark |
recruitment | Membrane recruitment of PI3K activity [19] |
| LOVTRAP GEF (and Rho GTPases) | LOV2/Zdks (FMN) | On: 450 nm Off: dark |
sequestration | Rac1 GEF (or dominant active RhoA, Rac1) release from outer mitochondria membrane [41] |
| PI-Rho GTPases (GEFs) | LOV2 (FMN) | On: dark Off: 450 nm |
Allosteric inhibition | LOV2 insertion in surface loops propagating allosteric conformational inactivation [47] |
| Cytoskeleton dynamics | ||||
| SxIP-iLID | iLID/nano (FMN) | On: 450 nm Off: dark |
association | Recruitment to growing microtubule plus ends [32] |
| n-EB1 | LOV2/Zdk1 (FMN) | On: dark Off: 450 nm |
domain splitting | Dissociation of the EB1 microtubule-binding and +TIP adaptor domains [45] |
| Z-lock cofilin (aTAT) | LOV2/Zdks (FMN) | On: 450 nm Off: dark |
steric inhibition | Uncaging of f-actin severing (microtubule acetylation) through LOV2/Zdk dissociation [42] |
| NuMA localization | iLID/nano (FMN) TULIPs (FMN) | On: 450 nm Off: dark |
recruitment | Localization of the spindle positioning factor NUMA to specific cortical sites [33,34] |
| Optogenetic ‘knocksideways’ | iLID/nano (FMN) | On: dark Off: 450 nm |
sequestration | TACC3 or KIF18A removal from mitotic spindles by outer mitochondria membrane recruitment [35] |
| Clustering | Cry2olig (FAD) | On: dark (450 nm) Off: 450 nm (dark) |
oligomerization | Inactivation (activation) of endocytosis (actin assembly) through crosslinking / clustering [39] |
| Integrin (receptor) clustering | Cry2 CLICR (FAD) | On: 450 nm Off: dark |
oligomerization | Receptor activation through cell membrane associated clustering [70] |
| OptoIntegrin | PhyB/PIF (PCB) | On: 664 nm Off: 725 nm |
association | Extracellular modulation of cell-matrix interaction [71] |
| Cargo transport | ||||
| Motor recruitment | TULIPs (FMN) | On: 450 nm Off: dark |
association | Recruitment of kinesin, dynein, and myosin motor domains to different organelles [26,27,29] |
| Motor recruitment | PhyB/PIF (PCB) | On: 664 nm Off: 725 nm |
association | Recruitment of kinesin and dynein motor domains to different organelles [30] |
| Motor recruitment | CRY2/CIBN (FAD) | On: 450 nm Off: dark |
association | Recruitment of kinesin and dynein motor domains to different organelles [72] |
| Motor processivity | VVD (FAD) | On: 450 nm Off: dark |
dimerization | Kinesin motor homodimerization [29] |
| Motor gear shifting | LOV2 (FMN) | On: 450 nm Off: dark |
allosteric | Control of myosin and kinesin speed/directionality through lever arm conformation changes [50] |
Light-controlled Rho GTPase signaling.
Many local cytoskeleton activities such as f-actin and microtubule dynamics during cell migration are controlled by low molecular weight Rho GTPases. It is thus no surprise that initial optogenetic approaches directed toward cytoskeleton dynamics in living cells targeted Rho GTPase activities either directly or indirectly. In one of the first demonstrations of direct optogenetic control of Rho GTPase signaling in cells, Wu et al linked a LOV2 domain to the N-terminus of a dominant active GTPase-deficient mutant of Rac1(Q61L) with the hypothesis that in the dark LOV2 sterically blocks access of downstream effectors to the Rac1 binding site [9]. Indeed, local blue light exposure of photoactivatable PA-Rac1 stimulates lamellipodia f-actin polymerization and can direct cell migration in vitro [9,10] and in vivo [11,12]. However, further analysis of the PA-Rac1 photoactivation mechanism showed that it also relies on a serendipitously introduced Ca2+ binding site between the LOV2 and Rac1 moieties [13], highlighting the difficulty of rational photoswitch design and explaining why this approach did not work for other Rho GTPases.
A possibly more adaptable method to locally control Rho GTPase activity is through optogenetic localization of their guanosine exchange factors (GEFs). Levskaya et al achieved this first through light-induced heterodimerization of the phytochrome PhyB photosensory domain with its downstream transcription factor effector PIF [14]. Localized membrane recruitment of the PIF-tagged Rac1 GEF Tiam results in local lamellipodia protrusion similar to PA-Rac1 photoactivation. Moving up one step in the signaling cascade that controls Rac1 activity, PhyB/PIF-mediated phosphoinositide 3-kinase (PI3K) recruitment by localized red light exposure locally produces phosphatidylinositol (3,4,5)-trisphosphate (PIP3) [15]. In contrast to PA-Rac1, this only generates a transient increase in Rac1 activity, which is still sufficient to direct cell migration.
Because the PhyB/PIF system can be difficult to implement (Fig. 1; Box 1), blue light activated dimerization modules [16–19] have more recently been employed to localize Rho GEFs (Fig. 2a). For example, using a LOV2-derived dimerization system in which a PDZ domain binding peptide is inaccessible at the C-terminus of the dark-state LOV2 Jα helix, local RhoA activation through light-induced recruitment of a RhoA GEF DH domain induces f-actin contractility and cleavage furrow formation anywhere on the anaphase cell cortex [20]. The Gardel lab then localized RhoA GEFs in interphase cells to generate local actomyosin contractility either on stress fibers [21] or epithelial cell-cell junctions [22]. Combined with quantitative physical models of the underlying cytoskeleton these studies highlight the strength of optogenetics to dissect cell dynamics, for example by showing how alternating RhoA-mediated contractility and membrane internalization shortens epithelial cell-cell junctions. Similarly, Valon et al use the cryptochrome Cry2/CIB1N dimerization module to locally recruit RhoA ArhGEF11 to the membrane and induce actomyosin contractility [23], which induces cell migration away from the site of optogenetic stimulation by generating rear contractility and retraction [24]. Extending optogenetic RhoA activation to morphogenetic processes at the multicellular level, Cry2/CIB1N-mediated photoactivated membrane-binding of RhoGEF2 can generate surprisingly geometrically precise epithelial folding patterns in early Drosophila embryos [25].
Box 1: Limitations of optogenetic experiments.
The ‘dark side’ of photoactivation.
Diffraction sets a physical lower boundary to the spatial precision with which an area of interest can be exposed to light in a microscope, but photochemistry kinetics further limit the accuracy of intracellular photoactivation. Blue light sensitive domains revert to the dark state through thermal decay, and the rate at which this occurs is critical for how sharp of a photoactivation boundary can be generated. Slow dark state reversal allows photoactivated molecules to diffuse further, thus broadening the boundary between activated and not activated regions. The half-life of the LOV2 photoactivated state is around 5–10 seconds, sufficient to generate intracellular gradients of freely diffusible molecules (Fig. 3) [45]. Photoactivated Cry2 is substantially more stable, with a half-life of minutes, which can still be fast enough to generate gradients if diffusion is confined, for example at the cell membrane or an organelle. Interestingly, a recent comparison with iLID/nano and Cry2/CIBN finds that Magnets, a dimerization system based on the fungal blue light photoreceptor VVD [19], produced the best spatial confinement, and the authors speculate that this is because both parts of the Magnet dimerizer need to be photoactivated restricting interaction to the light-exposed region [73]. In contrast to these blue light sensitive systems, PhyB photoconverts between two relatively stable states either by red or infrared light. Thus, while longer wavelengths may be less damaging to cells and penetrate deeper into biological specimens, precise spatial control of the PhyB/PIF interaction requires complementary 650 nm and 750 nm illumination patterns. However, even then the PhyB/PIF dissociation rate is in the order of seconds. This is comparable to the dissociation rate of π-EB1 with a fast dark reversal LOV2(I427V) variant suggesting similar spatial accuracy [14,45]. An important trade-off for high spatial control, however, is that fast dark state reversal requires frequent (1 Hz or faster) exposure to photoactivation light, which even at low intensities can accumulate significant phototoxicity over longer time periods and may also contribute to temperature increases of the specimen. Another point to consider is that dark is never truly dark as the inactive and activated states of photosensors exist in a chemical equilibrium. Even in the dark, an estimated ~1.5% of LOV2 molecules are in the activated state with the Jα helix dissociated, which increases ~60-fold in response to blue light [59]. This limits the dynamic range of individual LOV2 molecules although this can likely be increased by dimerization or other multivalent interactions.
Lights, camera, and overlapping action spectra:
Due to overlapping excitation spectra, it can be challenging to avoid unintended photoactivation when combining optogenetics and fluorescence microscopy. Blue light sensitive domains such as LOV2 or Cry2 are only compatible with imaging of red or far red fluorescent proteins. Even 515 nm, a frequently used YFP excitation line will photoactivate LOV2 [51]. Thus, combining FRET-based protein activity sensors with LOV2 optogenetics requires FRET between new near-infrared fluorescent proteins [74]. Combining fluorescence microscopy with PhyB/PIF red light sensitive optogenetics is possibly even more problematic, because the PhyB action spectrum spans almost the entire visible light range [75]. Both blue and green light cause PhyB photoconversion, and frequent 750 nm light exposure is required to avoid accumulation of the stable PhyB photoactivated state that has a half-life of hours [30]. It is noteworthy that both PhyB isomers have absorbance minima around 515 nm, making YFP perhaps the most suitable choice for fluorescence microscopy with PhyB/PIF. Lastly, one should not forget that animal cells also express cryptochromes. Although it remains unknown if vertebrate cryptochromes are light-sensitive in a physiological context (in insects they certainly are) [76], this highlights that appropriate blue light controls in the absence of the designed photoswitch should be incorporated in any optogenetic experiment. Unfortunately, this is not always current standard practice in the field.
Optogenetic re-localization to modulate cytoskeleton dynamics
Photoactivated recruitment can also localize protein activities other than Rho GTPases. For example, the Kapitein group has used TULIPs, a LOV2-based heterodimerization module, to link organelle cargoes to kinesin, dynein and myosin motor domains and demonstrated light-induced redistribution of peroxisomes, endosomes and mitochondria [26,27], and in vitro control of microtubule gliding [28]. However, all variations of localization-mediated photoactivation require expression of engineered constructs that become dominant active under illumination conditions, but still compete with endogenous activities and may also have unintended consequences in the dark. For example, kinesin motor activity in the absence of cargo-binding can still affect microtubule network organization. To improve dynamic range and specificity of light-activated transport systems, the Kapitein group most recently introduced a two-step system in which both cargo binding and motor processivity are blue light-activated [29]. Blue light can also be combined with red light-sensitive PhyB/PIF dimerization to independently control trafficking and distribution of different sets of organelles [30] although crosstalk challenges between channels remain (Box 1). In vitro, blue light-activated multimerization of kinesin motors can drive complex self-organization of dynamic microtubule arrays to analyze and direct the non-equilibrium dynamic properties of such assemblies [31]. Utilizing a similar recruitment strategy, Adikes et al invented a system to localize proteins to growing microtubule ends through iLID/nano-mediated dimerization with the microtubule plus-end adaptor EB1 and show that light-induced microtubule plus end crosslinking with f-actin slows microtubule polymerization [32].
In the so far only example of spatial optogenetic control of a structural mitotic spindle protein, two independent research teams used different optimized LOV2-based dimerization modules to dissect the molecular mechanism of dynein-mediated spindle positioning in either mammalian cells or C. elegans embryos [33,34]. Both teams find that localized recruitment of NuMA to the cell cortex is sufficient to reconstitute a functional cortical dynein motor complex that can drive spindle positioning. Interestingly, in both systems light-mediated localization of the dynein motor itself does not work possibly because the long coiled-coil NuMA molecule is required for the correct cortical dynein anchoring geometry. It should be noted that spatially precise photoactivation is difficult in mitosis because of the small size of mammalian metaphase spindles. Immobilization at the cell cortex is important here, as slow dark recovery rates present challenges in achieving sharp photoactivation boundaries of a cytoplasmic freely diffusible protein.
In a variation of the pharmacological ‘knocksideways’ approach, optogenetic recruitment to the cell cortex or mitochondria can also acutely remove proteins from a site of action, for example from mitotic spindles [35,36]. However, efficient sequestration will greatly depend on relative affinities and binding kinetics of the physiological binding interaction. In addition, this or similar inactivation methods trapping proteins in the nucleus [37] or in Cry2/CIBN oligomer clusters [38,39] do not allow subcellular spatial control, and the effect of Cry2 oligomerization likely clustering many hundreds of molecules is difficult to predict. Nevertheless, replacing endogenous Delta with a Cry2-tagged variant that is inactivated by blue light-induced clustering in the cell membrane has recently been implemented to dissect temporal Notch signaling dynamics in Drosophila [40].
Direct photoswitching of protein activity
The above examples highlight a powerful and easily implemented optogenetic strategy that utilizes light-induced dimerization to recruit protein activities to ectopic sites where they were not before. Currently, only one optogenetic system exists that functions in the opposite direction. LOVTRAP relies on small engineered peptides, Zdks, that bind the LOV2 domain dark-state with high affinity [41]. Thus, in contrast to light-induced dimerization modules, blue light results in reversible dissociation of the LOV2/Zdk heterodimer. While the LOVTRAP module was initially also used to release proteins of interest, such as the Rac1 GEF Vav2 from mitochondria [41], the Hahn lab recently proposed that the LOV2/Zdk interaction could directly cage protein active sites (Fig. 2b) [42]. For example, blue light uncaging the f-actin binding and thus filament severing activity of Z-lock cofilin can enhance actin monomer recycling to promote leading edge polymerization. However, successful implementation of this Z-lock idea depends on fine-tuning of the relative binding affinities of the LOV2/Zdk module and the binding interaction to be disrupted, as well as on the overall active site geometry in relation to the linkers connecting LOV2 and Zdk [43]. A similar uncaging strategy employs light-sensitive oligomerization of the photoactivatable DRONPA fluorescent protein to locally activate membrane-bound Cdc42 [44].
While the vast majority of optogenetic approaches activate a protein of interest, in order to understand physiological intracellular protein functions in space and time, different methods are needed to acutely inactivate proteins. We recently utilized the LOV2/Zdk1 interaction to disrupt the function of EB1 by blue light [45]. EB1 is a dimer with an N-terminal calponin homology domain that recognizes growing microtubule plus ends and a C-terminal domain that recruits numerous other +TIP proteins to growing microtubule ends [46]. By inserting the LOV2/Zdk1 module in the disordered linker between these two domains, we demonstrated reversible light-induced dissociation of the +TIP complex and inhibition of microtubule polymerization with high spatial and temporal accuracy. In addition, migrating cells in which endogenous EB1 was replaced with this photo-inactivated π-EB1 turned away from local blue light exposure indicating that +TIP complex function is required for persistent directional cell movement. We posit that this strategy could be used to render many other multidomain proteins light sensitive (Fig. 2d). To adapt this idea to more complex cell systems, we are currently developing genome editing to directly insert light sensitive LOV2/Zdk1 modules into multi-domain proteins and have been able to replace endogenous EB1 with the light-sensitive version in one step in various human cell types (Fig. 3).
Figure 3. Example of a spatiotemporal optogenetics experiment with subcellular resolution.
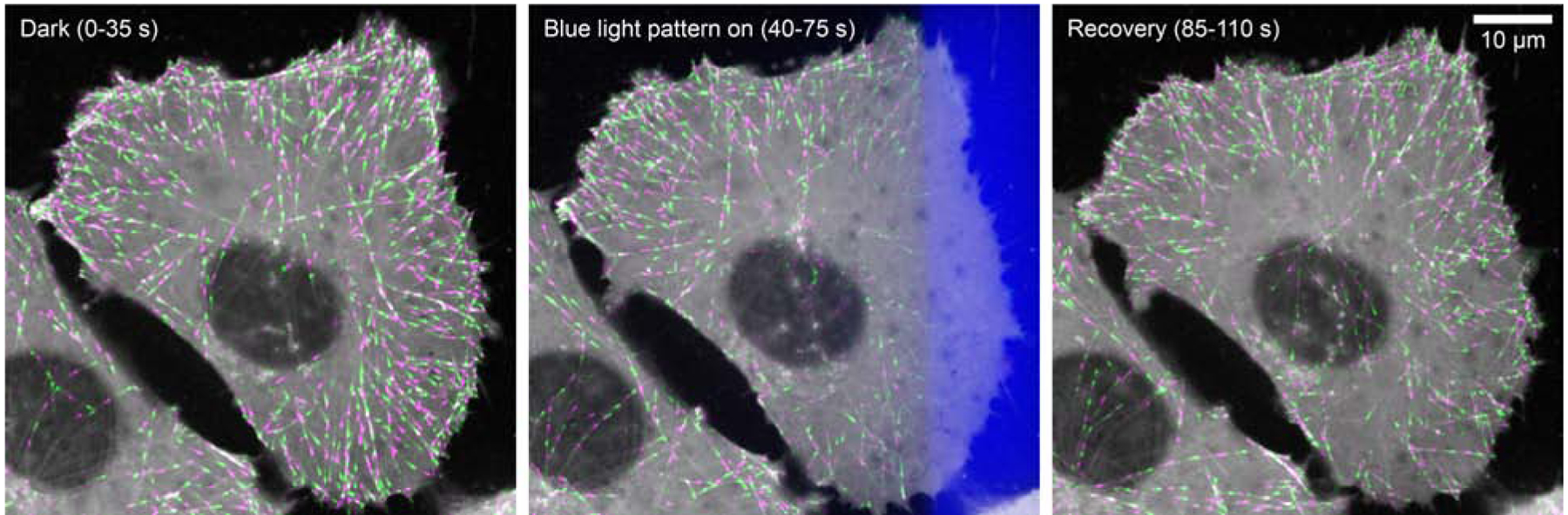
Shown is a human tissue culture cell in which a photosensitive LOV2/Zdk1 protein-protein interaction module was inserted into the endogenous EB1 gene by CRISPR/Cas9 genome editing, which replaces the wild-type EB1 protein with the light sensitive π-EB1 variant in one step. The images show an overlay of the mCherry-tagged π-EB1 C-terminal half on growing microtubule ends from short time-lapse sequences (8 images every 5 seconds) in alternating green and magenta before, during and after localized 470 nm blue light exposure. The light-exposed region is indicated by the blue overlay in the middle panel. Note the gradient of π-EB1 dissociation near the edge of the light-exposed region that results from diffusion of photoactivated molecules. Sharper boundaries can be achieved with LOV2 domain variants with faster dark recovery rates [45].
An even more direct way to render proteins light-sensitive is through direct insertion of a LOV2 domain into exposed surface loops predicted to propagate allosteric conformational changes (Fig. 2e). Dagliyan et al test this idea for several different signaling molecules including the Rac1 GEF Vav2 and show that in this case local Vav2 photoinactivation results in cell retraction from the site of blue light exposure [47,48]. A related approach is to design split proteins in which inactive but properly folded subdomains are brought back together through a light-induced dimerizer to reconstitute protein activity (Fig. 2c). New computational methods to predict split protein designs, which has traditionally been very difficult and may not always be possible, may open this strategy to optogenetics [49]. LOV2 conformation changes have also been used to alter lever arm geometries in both kinesin and myosin motors that can alter speed and directionality as a function of blue light exposure [50]. However, these light-controlled motors have not yet been employed inside cells.
Summary and outlook
Light control of specific intracellular protein activities has only been around for a relatively short time, but optogenetic approaches are already casting new light on many dynamic cell processes. Many basic optogenetic experiments can be easily and inexpensively implemented on standard fluorescent light microscopes, but spatial control requires additional equipment to achieve localized light exposure such as laser-scanning or digital micromirror device illumination systems [51] (Box 1). Still, current photoactivation technologies are relatively static, and to gain more precise dynamic control of fast intracellular processes in cells or organisms, it will be necessary to develop feedback between microscopy and on-the-fly image analysis to direct photoactivation light patterns. It will also be interesting to see how optogenetics can break the diffraction barrier. Stimulated emission depletion (STED) reduces the point spread function diameter with a surrounding donut beam of inactivation light and recent work suggests STED nanoscale control of channelrhodopsins [52]. Similarly, STED optogenetics may be possible with phytochrome photoconversion between stable states driven by different wavelengths, but this has not yet been tested [53]. Bioluminescence resonance energy transfer between luciferase and blue light sensors [54] may provide yet another opportunity for nanoscale control by restricting activation to where the luciferase (i.e. the light source) and the photosensor are sufficiently close. However, spatial resolution will still be limited by the photo-and biochemical characteristics of the optogenetic switch (Box 1). In any case, it seems a safe bet that as with fluorescent proteins there are unusual photosensors in the wild that we do not yet know about, with complementary or more desirable characteristics than what is available now, for example narrower action spectra or faster reversion kinetics. Such properties might also be gained through further photosensor engineering or development of synthetic cofactors. The emerging field of ‘photopharmacology’ that uses synthetic photoswitches further broadens possible experimental approaches [55]. In summary, light-controlled proteins are an important addition to the cell biologist’s toolkit. Being a relatively young field, however, many properties of optogenetic systems remain poorly characterized, and experimentalists need to be aware of the demands and limitations of their specific experiment design. Nevertheless, the future of optogenetics surely looks bright.
Acknowledgements
This work was supported by NIH grants R01 GM094819, R01 NS107480, R21 CA224194 and S10 RR026758 to T.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bodor DL, Pönisch W, Endres RG, Paluch EK: Of Cell Shapes and Motion: The Physical Basis of Animal Cell Migration. Dev Cell 2020, 52:550–562. [DOI] [PubMed] [Google Scholar]
- 2.Elting MW, Suresh P, Dumont S: The Spindle: Integrating Architecture and Mechanics across Scales. Trends Cell Biol 2018, 28:896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K: Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005, 8:1263–1268. [DOI] [PubMed] [Google Scholar]
- 4.Zemelman BV., Lee GA, Ng M, Miesenböck G: Selective photostimulation of genetically chARGed neurons. Neuron 2002, 33:15–22. [DOI] [PubMed] [Google Scholar]
- 5.Hegemann P, Nagel G: From channelrhodopsins to optogenetics. EMBO Mol Med 2013, 5:173–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Gelderen K, Kang C, Pierik R: Light signaling, root development, and plasticity. Plant Physiol 2018, 176:1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losi A, Gardner KH, Möglich A: Blue-Light Receptors for Optogenetics. Chem Rev 2018, 118:10659–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klewer L, Wu YW: Light-Induced Dimerization Approaches to Control Cellular Processes. Chem - A Eur J 2019, 25:12452–12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM: A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato T, Kawai K, Egami Y, Kakehi Y, Araki N: Rac1-dependent lamellipodial motility in prostate cancer PC-3 cells revealed by optogenetic control of Rac1 activity. PLoS One 2014, 9:e97749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, He L, Wu YI, Hahn KM, Montell DJ: Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 2010, 12:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A: Differential Regulation of Protrusion and Polarity by PI(3)K during Neutrophil Motility in Live Zebrafish. Dev Cell 2010, 18:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler A, Barends TRM, Udvarhelyi A, Lenherr-Frey D, Lomb L, Menzel A, Schlichting I: Structural details of light activation of the LOV2-based photoswitch PA-Rac1. ACS Chem Biol 2015, 10:502–509. [DOI] [PubMed] [Google Scholar]
- 14.Levskaya A, Weiner OD, Lim WA, Voigt CA: Spatiotemporal control of cell signalling using a light- switchable protein interaction. Nature 2009, 461:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graziano BR, Gong D, Anderson KE, Pipathsouk A, Goldberg AR, Weiner OD: A module for Rac temporal signal integration revealed with optogenetics. J Cell Biol 2017, 216:2515–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Spatial and temporal optogenetics to dissect signalling circuit dynamics underlying Rac1mediated cell migration.
- 16.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL: Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 2010, 7:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M: TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat Methods 2012, 9:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B: Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A 2015, 112:112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawano F, Suzuki H, Furuya A, Sato M: Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun 2015, 6:6256. [DOI] [PubMed] [Google Scholar]
- 20.Wagner E, Glotzer M: Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J Cell Biol 2016, 213:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M, Gardel ML: Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat Commun 2017, 8:15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavanaugh KE, Staddon MF, Munro E, Banerjee S, Gardel ML: RhoA Mediates Epithelial Cell Shape Changes via Mechanosensitive Endocytosis. Dev Cell 2020, 52:152–166.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Unique combination of optogenetics and quantitative modeling to test how RhoA-mediated contractility controls epithelial cell-cell junction dynamics.
- 23.Valon L, Marín-Llauradó A, Wyatt T, Charras G, Trepat X: Optogenetic control of cellular forces and mechanotransduction. Nat Commun 2017, 8:14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennig K, Wang I, Moreau P, Valon L, DeBeco S, Coppey M, Miroshnikova YA, Albiges-Rizo C, Favard C, Voituriez R, et al. : Stick-slip dynamics of cell adhesion triggers spontaneous symmetry breaking and directional migration of mesenchymal cells on one-dimensional lines. Sci Adv 2020, 6:eaau5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izquierdo E, Quinkler T, De Renzis S: Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat Commun 2018, 9:2366. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Tissue level optogenetics shows that RhoA activation is sufficient to drive apical contraction and epithelial folding in early Drosophila development.
- 26.Van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC: Optogenetic control of organelle transport and positioning. Nature 2015, 518:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harterink M, Van Bergeijk P, Allier C, De Haan B, Van Den Heuvel S, Hoogenraad CC, Kapitein LC: Light-controlled intracellular transport in Caenorhabditis elegans. Curr Biol 2016, 26:R153–R154. [DOI] [PubMed] [Google Scholar]
- 28.Tas RP, Chen CY, Katrukha EA, Vleugel M, Kok M, Dogterom M, Akhmanova A, Kapitein LC: Guided by Light: Optical Control of Microtubule Gliding Assays. Nano Lett 2018, 18:7524–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nijenhuis W, van Grinsven MMP, Kapitein LC: An optimized toolbox for the optogenetic control of intracellular transport. J Cell Biol 2020, doi: 10.1083/jcb.201907149. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Presents innovative approaches for improved control of blue light-activated intracellular transport systems using iLID/nano and VVD dimerization.
- 30.Adrian M, Nijenhuis W, Hoogstraaten RI, Willems J, Kapitein LC: A Phytochrome-Derived Photoswitch for Intracellular Transport. ACS Synth Biol 2017, 6:1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First demonstration of simultaneous blue and red light sensitive optogenetics in the same cells to control transport of different sets of organelles independently.
- 31.Ross TD, Lee HJ, Qu Z, Banks RA, Phillips R, Thomson M: Controlling organization and forces in active matter through optically defined boundaries. Nature 2019, 572:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Analysis of in vitro non-equilibrium microtubule network dynamics generated by spatially controlled kinesin motor oligomerization.
- 32.Adikes RC, Hallett RA, Saway BF, Kuhlman B, Slep KC: Control of microtubule dynamics using an optogenetic microtubule plus end-F-actin cross-linker. J Cell Biol 2018, 217:779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okumura M, Natsume T, Kanemaki MT, Kiyomitsu T: Dynein–dynactin–NuMA clusters generate cortical spindle-pulling forces as a multiarm ensemble. Elife 2018, 7:e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• In one of the few examples of local optogenetics in mitotic cells this study and ref [34] demonstrate control of spindle positioning by light-mediated NuMA recruitment to the mitotic cell cortex.
- 34.Fielmich LE, Schmidt R, Dickinson DJ, Goldstein B, Akhmanova A, Van Den Heuvel S: Optogenetic dissection of mitotic spindle positioning in vivo. Elife 2018, 7:e38198. [DOI] [PMC free article] [PubMed] [Google Scholar]; • See note to ref [33].
- 35.Milas A, Jagrić M, Martinčić J, Tolić IM: Optogenetic reversible knocksideways, laser ablation, and photoactivation on the mitotic spindle in human cells. In Methods in Cell Biology. 2018:191–215. [DOI] [PubMed] [Google Scholar]
- 36.Jagrić M, Risteski P, Martinčić J, Milas A, Tolić IM: Optogenetic control of PRC1 reveals its role in kinetochore alignment at the metaphase plate. bioRxiv 2019, doi: 10.1101/865394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooikaas PJ, Martin M, Mühlethaler T, Kuijntjes GJ, Peeters CAE, Katrukha EA, Ferrari L, Stucchi R, Verhagen DGF, Van Riel WE, et al. : MAP7 family proteins regulate kinesin-1 recruitment and activation. J Cell Biol 2019, 218:1298–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Do Heo W: Reversible protein inactivation by optogenetic trapping in cells. Nat Methods 2014, 11:633–636. [DOI] [PubMed] [Google Scholar]
- 39.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL: An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 2014, 5:4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan R, Necakov A, Trylinski M, Harish RK, Krueger D, Esposito E, Schweisguth F, Neveu P, De Renzis S: Optogenetic inhibition of Delta reveals digital Notch signalling output during tissue differentiation. EMBO Rep 2019, 20:e47999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM: LOVTRAP: An optogenetic system for photoinduced protein dissociation. Nat Methods 2016, 13:755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone OJ, Pankow N, Liu B, Sharma VP, Eddy RJ, Wang H, Putz AT, Teets FD, Kuhlman B, Condeelis JS, et al. : Optogenetic control of cofilin and αTAT in living cells using Z-lock. Nat Chem Biol 2019, 15:1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teets FD, Watanabe T, Hahn KM, Kuhlman B: A Computational Protocol for Regulating Protein Binding Reactions with a Light-Sensitive Protein Dimer. J Mol Biol 2020, doi: 10.1016/j.jmb.2019.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou XX, Chung HK, Lam AJ, Lin MZ: Optical control of protein activity by fluorescent protein domains. Science 2012, 338:810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Haren J, Charafeddine RA, Ettinger A, Wang H, Hahn KM, Wittmann T: Local control of intracellular microtubule dynamics by EB1 photodissociation. Nat Cell Biol 2018, 20:252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• By photosensitive dissociation of the two functional domains of EB1, this study shows local and reversible inhibition of microtubule growth and consequences on cell migration.
- 46.van Haren J, Wittmann T: Microtubule Plus End Dynamics − Do We Know How Microtubules Grow? BioEssays 2019, 41:e1800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, Dokholyan NV., Hahn KM: Engineering extrinsic disorder to control protein activity in living cells. Science 2016, 354:1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dagliyan O, Dokholyan NV., Hahn KM: Engineering proteins for allosteric control by light or ligands. Nat Protoc 2019, 14:1863–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagliyan O, Krokhotin A, Ozkan-Dagliyan I, Deiters A, Der CJ, Hahn KM, Dokholyan NV.: Computational design of chemogenetic and optogenetic split proteins. Nat Commun 2018, 9:4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura M, Chen L, Howes SC, Schindler TD, Nogales E, Bryant Z: Remote control of myosin and kinesin motors using light-activated gearshifting. Nat Nanotechnol 2014, 9:693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Haren J, Adachi LS, Wittmann T: Optogenetic Control of Microtubule Dynamics In Cytoskeleton Dynamics: Methods and Protocols. Edited by Maiato H. Springer US; 2020:211–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahlberg MA, Ramakrishnan C, Willig KI, Boyden ES, Deisseroth K, Dean C: Investigating the feasibility of channelrhodopsin variants for nanoscale optogenetics. Neurophotonics 2019, 6:015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziegler T, Möglich A: Photoreceptor engineering. Front Mol Biosci 2015, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim CK, Cho KF, Kim MW, Ting AY: Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. Elife 2019, 8:e43826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hüll K, Morstein J, Trauner D: In Vivo Photopharmacology. Chem Rev 2018, 118:10710–10747. [DOI] [PubMed] [Google Scholar]
- 56.Christie JM: Phototropin Blue-Light Receptors. Annu Rev Plant Biol 2007, 58:21–45. [DOI] [PubMed] [Google Scholar]
- 57.Harper SM, Neil LC, Gardner KH: Structural basis of a phototropin light switch. Science 2003, 301:1541–1544. [DOI] [PubMed] [Google Scholar]
- 58.Halavaty AS, Moffat K: N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 2007, 46:14001–14009. [DOI] [PubMed] [Google Scholar]
- 59.Yao X, Rosen MK, Gardner KH: Estimation of the available free energy in a LOV2-Jα photoswitch. Nat Chem Biol 2008, 4:491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eitoku T, Nakasone Y, Matsuoka D, Tokutomi S, Terazima M: Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. J Am Chem Soc 2005, doi: 10.1021/ja052523i. [DOI] [PubMed] [Google Scholar]
- 61.Konold PE, Mathes T, Weienborn J, Groot ML, Hegemann P, Kennis JTM: Unfolding of the C-Terminal Jα Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. J Phys Chem Lett 2016, 7:3472–3476. [DOI] [PubMed] [Google Scholar]
- 62.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR: Conformational switching in the fungal light sensor vivid. Science 2007, 316:1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Zuo Z, Wang X, Liu Q, Gu L, Oka Y, Lin C: Beyond the photocycle — how cryptochromes regulate photoresponses in plants? Curr Opin Plant Biol 2018, 45:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J: Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A 2004, 101:12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uda Y, Goto Y, Oda S, Kohchi T, Matsuda M, Aoki K: Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc Natl Acad Sci U S A 2017, 114:11962–11967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kyriakakis P, Catanho M, Hoffner N, Thavarajah W, Hu VJ, Chao SS, Hsu A, Pham V, Naghavian L, Dozier LE, et al. : Biosynthesis of Orthogonal Molecules Using Ferredoxin and Ferredoxin-NADP+ Reductase Systems Enables Genetically Encoded PhyB Optogenetics. ACS Synth Biol 2018, 7:706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaberniuk A, Shemetov AA, Verkhusha VV.: A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods 2016, 13:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD: Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci U S A 2014, 111:10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS: Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 2006, 24:79–88. [DOI] [PubMed] [Google Scholar]
- 70.Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, Schaffer DV.: Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat Commun 2015, 6:6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baaske J, Mühlhäuser WWD, Yousefi OS, Zanner S, Radziwill G, Hörner M, Schamel WWA, Weber W: Optogenetic control of integrin-matrix interaction. Commun Biol 2019, 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duan L, Che D, Zhang K, Ong Q, Guo S, Cui B: Optogenetic control of molecular motors and organelle distributions in cells. Chem Biol 2015, 22:671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benedetti L, Barentine AES, Messa M, Wheeler H, Bewersdorf J, De Camilli P: Light-activated protein interaction with high spatial subcellular confinement. Proc Natl Acad Sci U S A 2018, 115:E2238–E2245. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper provides the most detailed comparison to date of the performance of the three main blue light sensitive dimerizers in live cells.
- 74.Shcherbakova DM, Cox Cammer N, Huisman TM, Verkhusha VV., Hodgson L: Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET article. Nat Chem Biol 2018, 14:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sethe Burgie E, Bussell AN, Lye SH, Wang T, Hu W, McLoughlin KE, Weber EL, Li H, Vierstra RD: Photosensing and Thermosensing by Phytochrome B Require Both Proximal and Distal Allosteric Features within the Dimeric Photoreceptor. Sci Rep 2017, 7:13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michael AK, Fribourgh JL, Van Gelder RN, Partch CL: Animal Cryptochromes: Divergent Roles in Light Perception, Circadian Timekeeping and Beyond. Photochem Photobiol 2017, 93:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]


