Abstract
Compartmentalization of metabolic enzymes through protein-protein interactions is an emerging mechanism for localizing and regulating metabolic activity. Self-assembly into linear filaments is a common strategy for cellular compartmentalization of enzymes. Polymerization is often driven by changes in the metabolic state of the cell, suggesting it is a strategy for shifting metabolic flux in response to cellular demand. While polymerization of metabolic enzymes is widespread, observed from bacteria to humans, we are just beginning to appreciate their role in regulating cellular metabolism. In most cases, one functional role of metabolic enzyme filaments is allosteric control of enzyme activity. Here, we highlight recent findings providing insight into the structural and functional significance of filamentation of metabolic enzymes in cells.
Introduction
Metabolic processes in the cytosol have traditionally been viewed as a disordered mixture of freely diffusing molecules and proteins. Compartmentalization into classical membrane-bound organelles has long been understood to play a central role in organizing metabolic activity. However, recent cellular, biochemical, and structural studies are driving a new appreciation that cellular metabolism is localized and controlled to a surprising degree by protein-protein interactions. In many different pathways, single or multiple enzymes assemble into cellular ultrastructures that are distinct from phase-separated structures like stress granules or P-bodies. Many self-assembling enzymes have punctate localization, but several dozen have have filamentous localization patterns that suggest a high degree of order.
The discovery that metabolic enzymes reversibly form filaments and other supramolecular complexes in response to nutrient availability suggests the formation of higher order structures is essential for their cellular function. High-throughput imaging screens of GFP-labeled libraries have enabled the identification of self-associating proteins in live cells. For example, recent studies in yeast have shown that approximately 100 proteins, including 60 metabolic enzymes, formed micrometer-sized assemblies, including many that dynamically and reversibly assembled depending on the metabolic state of the cell [1–4]. These results suggest self-association of metabolic enzymes is an important mechanism regulating cellular metabolic networks. However, in many cases the functional significance of self-associating metabolic enzymes is unknown. Filamentous enzymes have been identified from most major metabolic pathways, such as nucleotide, fatty acid, and glucose metabolism, and have been described in prokaryotic to eukaryotic organisms. These filament-forming enzymes are structurally diverse, often adopt various conformations, and consequently give rise to a variety of 3-dimensional filament architectures. This review focuses on recent structural, biochemical, and cell biological characterization of a selection of enzyme filaments that highlight principles of metabolic control by enzyme self-assembly.
Structural analysis of metabolic enzyme filaments
Filaments of a variety of metabolic enzymes have been reconstituted in vitro from purified protein. Importantly, this provides opportunities to study the direct effects of polymerization on enzyme function outside of the complex metabolic environment of the cell. Such studies with isolated protein make it possible to unambiguously identify, for example, the determinants for enzyme polymerization, as well as the effects of polymerization on enzyme kinetics. In a number of recent studies, cryo-electron microscopy has been used to determine the structures of enzyme filaments, providing insight into the molecular mechanisms of polymerization-based allosteric regulation. These studies have found a surprising diversity of mechanisms by which enzyme activity can be controlled in the filament, including conformational selection, increased cooperativity, and channeling of intermediates.
Many enzymes must undergo a series of conformational changes in order to catalyze their reactions; locking such enzymes in a single conformation within a filament therefore results in inhibition. For instance, hexokinases switch between an open and closed conformation during their catalytic cycles, and filaments of the yeast hexokinase glucokinase 1 (Glk1) inhibit activity by selectively stabilizing the closed conformation of the enzyme (Fig. 1A) [5]. In this manner, filament formation tends to build upon existing regulatory mechanisms by altering the conformational dynamics of the enzyme in the polymerized versus non-polymerized state. One might also imagine that polymerization might act to produce novel enzyme conformations, or to generate new regulatory sites at filament interfaces, but mechanisms like these have not yet been described for any enzyme filaments.
Figure 1. Filament formation has diverse effects on metabolic enzyme regulation.
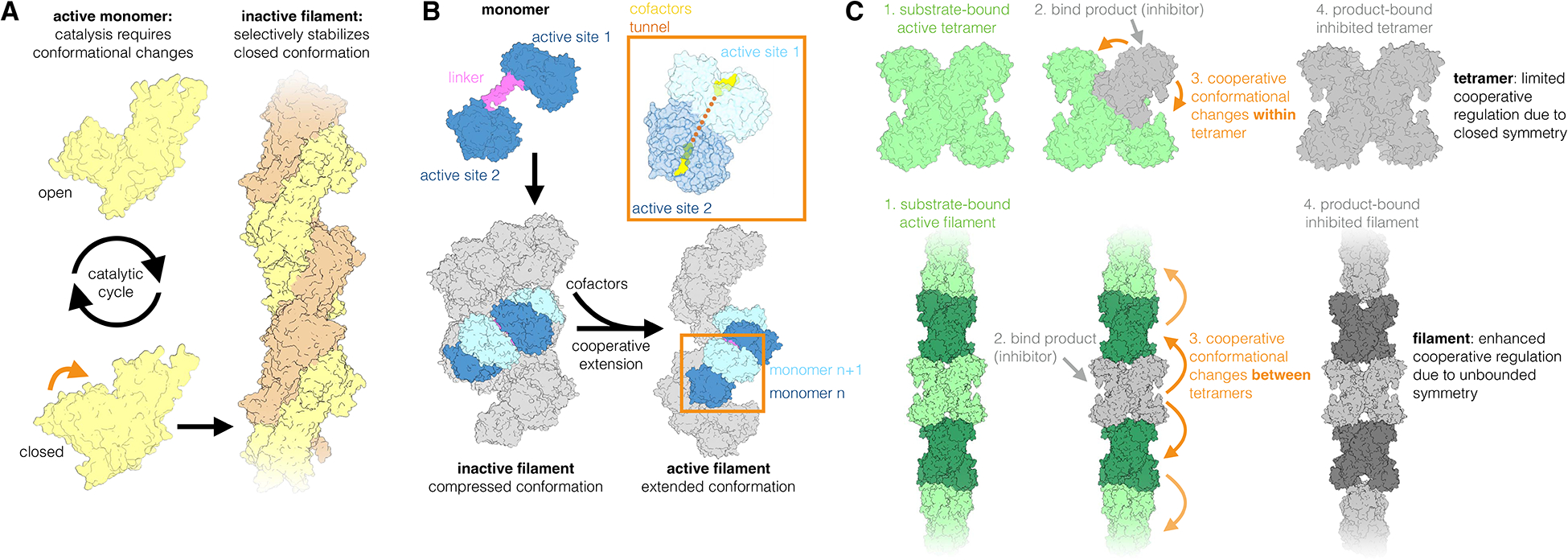
(A) Yeast glucokinase 1 (Glk1) assembles into inactive filaments. Glk1 forms two-stranded filaments that selectively stabilize the closed conformation, inhibiting the enzyme by preventing conformational cycling. (B) Filament formation is essential to the activity of bacterial alcohol-aldehyde dehydrogenase (AdhE). The two AdhE active sites are in separate domains connected by a linker (pink). AdhE assembles into compressed filaments, which cooperatively extend into active filaments upon addition of cofactors. Two adjacent monomers are colored within each filament (monomer n; blue and monomer n+1; light blue), with the remainder of the filament in grey. In the active filament a tunnel connects sequential active sites, allowing efficient transfer of reaction intermediates. (C) Human CTP synthase 2 (CTPS2) filaments enable highly cooperative regulation. CTPS2 forms tetramers, which switch from an active to an inactive conformation upon binding their product, a feedback inhibitor. In the tetramer cooperative inhibition is limited to four protomers, while polymerization expands cooperativity by linking the conformational state of many tetramers, producing ultrasensitive regulation.
Remarkably, a number of enzymes have been shown to assemble into multiple filament forms with unique 3-dimensional architectures, depending on their ligand-binding state. Perhaps the most striking example of this is seen in filaments of human acetyl-CoA carboxylase (ACC1), which forms two distinct filaments: In the presence of its allosteric regulator citrate, ACC1 assembles into single-stranded filaments with increased activity. By contrast, ACC1 forms inactive, double-stranded filaments when bound to the C-terminal domain of the breast cancer type 1 susceptibility protein [6]. In other cases, enzyme filaments are much more conformationally plastic, comprising enzyme subunits in multiple, dynamic conformations. For instance, human Inosine Monophosphate Dehydrogenase 2 (IMPDH2) filaments can simultaneously accommodate enzymes in a range of active and inactive conformations [7,8]. IMPDH2 adopts a characteristic “compressed” conformation upon binding its downstream feedback inhibitor GTP. When incorporated into a filament, IMPDH2 resists this compression, allowing it to remain active under conditions where the free enzyme would otherwise be inhibited [8]. Similarly, the bifunctional bacterial enzyme aldehyde-alcohol dehydrogenase (AdhE) forms compressed filaments that adopt an extended conformation upon addition of cofactors NAD+ and Fe2+ (Fig. 1B). Polymerization is critical to AdhE activity, with the extended filaments likely representing the active form of the enzyme [9,10]. In the case of AdhE, enzyme assembly allows for direct channeling of the acetaldehyde intermediate between active sites, providing a mechanism for increased flux.
CTP synthase (CTPS) highlights the diversity of mechanisms by which filament formation regulates enzyme activity. The cellular phenomenon of CTPS polymerization is broadly conserved across kingdoms, but surprisingly polymerization leads to dramatically different functional outcomes in bacteria and in humans, and even regulatory differences between isoforms of the human enzyme. CTPS undergoes a conserved active-inactive conformational cycle controlled by substrate and product binding. E. coli CTPS forms inhibited filaments composed of the enzyme in the product-bound, inactive conformation, while human CTPS1 behaves in essentially the opposite manner, forming hyper-active filaments composed of enzyme in the substrate-bound conformation [11]. The second human CTPS isoform, CTPS2, assembles into filaments in either the active or inactive conformation, but unlike IMPDH2, does not appear to form filaments composed of enzyme in mixed conformations. Rather, entire CTPS2 filaments dynamically switch between the active and inactive conformation in response to changes in levels of substrate and product. CTPS2 filaments therefore link the conformational-and thus activity-state of many subunits within a filament, resulting in highly cooperative regulation (Fig. 1C) [12]. One can compare this to more canonical examples of cooperativity, such as the hemoglobin tetramer where the coupling of conformational states is restricted to four subunits by a closed symmetry; theoretically unbounded, linear polymers can couple the conformational state of potentially thousands of enzyme subunits. This could allow, for example, a more rapid inactivation of enzyme in response to accumulation of a feedback inhibitor. Similar models of “conformational spread” have previously been used to describe the propagation of conformational changes across large-scale 2-dimensional arrays of membrane proteins, which also produce ultrasensitive regulation [13–15].
Importantly, E. coli and human CTPS polymerize via entirely different filament contact sites, producing filaments with distinct structures [11,12]. This may suggest that the importance of enzyme polymerization lies primarily in the integration of regulatory information across many subunits of the same enzyme, rather than the emergent 3-dimensional architecture of the filaments themselves. A recent study revealed that many proteins are poised to evolve the ability to form polymers; single point mutations on the surfaces of various E. coli proteins were sufficient to cause them to self-assemble into homomeric filaments [16]. Polymerization into filaments may therefore provide a relatively simple, easy to evolve mechanism for enhancing the regulation of metabolic enzymes.
Cellular functions and dynamics
While a number of metabolic enzymes have been shown to assemble into filaments or micrometer-sized structures, in many cases the functional and regulatory significance of this phenomena on cellular metabolism is unknown. Proposed roles of filament formation have been discussed in detail in recent reviews [17–20]. We briefly discuss the potential cellular functions of filament formation, focusing on recent publications.
Consistent with the biochemical effects observed in vitro, filament formation is likely a mechanism to control enzymatic activity in the more complex environment of the cell. A majority of filamentous metabolic enzymes detailed to date assemble in the active conformation [5], suggesting that filament formation could increase pathway flux under conditions of high metabolic demand. For enzymes that assemble into multiple filament forms, such as IMPDH2 [8], filament assembly could be a mechanism to increase cooperativity and reduce feedback inhibition of enzyme activity, buffering the activity against transient changes in metabolite concentrations. Alternatively, the reversible sequestering of metabolic enzymes by filament formation is a mechanism to decrease the flux of metabolites through the pathway. This has been observed for bacterial CTPS, where the enzyme assembles into filaments of stacked tetramers held in an inactive conformation in high cellular levels of CTP [21]. In periods of low CTP levels in the cell, CTPS can be rapidly reactivated upon release of tetramers from the filaments. A second example is yeast Glk1, which forms inactive filaments upon glucose addition that disassemble upon glucose removal (Fig. 1A) [5]. Filamentation of Glk1 has been proposed to buffer the level of active enzyme, thus setting a maximal rate of glucose phosphorylation. Consistent with this hypothesis, cells expressing filament-incompetent Glk1 display decreased fitness.
Filament formation can also allow for increased catalytic efficiency by substrate channeling, the facilitated transfer of the metabolic product of one enzyme to the next without release of the metabolite into the bulk cytosol. Substrate channeling can occur between the active sites of bifunctional enzymes that catalyze sequential steps of the metabolic pathway, such as CTPS where ammonium passes from the glutamine hydrolysis and amidoligase active sites [22]. Channeling can also occur between two or more individual, sequential metabolic enzymes of a pathway. Theoretical and experimental data have demonstrated that these enzymes do not have to physically interact as the proximity of consecutive enzymes is sufficient to increase catalytic efficiency by increased local concentration of intermediates [23]. Additionally, it can be a mechanism to sequester toxic byproducts, preventing their release into the cytosol. For example, bacterial AdhE catalyzes consecutive steps converting acetyl-CoA to ethanol. In the first step, acetyl-CoA is converted to a toxic intermediate, acetaldehyde, by the aldehyde dehydrogenase domain followed by its conversion to ethanol by the alcohol dehydrogenase domain. Within the AdhE filament, a tunnel is formed allowing acetaldehyde to be transported to the alcohol dehydrogenase domain without being released into the cytoplasm (Fig. 1B)[10].
The dynamics and regulation of filament formation is critically important to understand their functions in cell physiology. CTPS and IMPDH2 catalyze rate-limiting steps in the de novo synthesis of pyrimidine and purine nucleotides, respectively. Both enzymes form cytoplasmic and nuclear filamentous assemblies, termed ‘cytoophidia’ or ‘rods and rings’, in cells and tissues [24–26]. The physiological roles of these assemblies are incompletely understood but are thought to reduce feedback inhibition of enzyme activity, increasing the production of de novo nucleotide synthesis in response to proliferative signals [8]. Treatment of cells with the glutamine analog 6-diazo-5-oxo-L-norleucine (DON), an inhibitor of both purine and pyrimidine biosynthesis, induces the formation of both CTPS and IMPDH2 assemblies. While both IMPDH2 and CTPS form dynamic assemblies, unexpectedly, they were observed to co-assemble in HeLa cervical adenocarcinoma cells [27]. The filaments were intertwined with but distinct from one another, suggesting the involvement of an additional factor to tether the filaments together. These observations raise the possibility that the mixed filaments represent a mechanism to coordinate the regulation of nucleotide metabolic pathways.
A second example of dynamic regulation of filament formation in cells is enzymes in the glycolytic pathway. Glycolytic enzymes from yeast to human cells have been shown to assemble into filaments or micrometer-sized assemblies. In S. cerevisiae, six of the ten enzymes in the glycolytic pathway have been shown to coalesce [2,4,5]. However, different cellular stresses appear to differentially regulate their clustering. Alterations in nutrient availability induced S. cerevisiae glycolytic enzymes to form independent assemblies of undetermined function [2]. Conversely, in response to hypoxia glycolytic enzymes co-cluster and are thought to represent glycolytic metabolons, functioning to increase glycolytic flux during times of metabolic demand [29]. Similarly, several glycolytic enzymes in C. elegans neurons clustered in micrometer-sized assemblies in response to hypoxic stress, suggesting the formation of a metabolon to support the energetic demands of synaptic transmission [28]. In mammalian cells, the liver isoform of the glycolytic enzyme phosphofructokinase-1 (PFKL), which catalyzes the rate-limiting step committing glucose to breakdown, assembles into elongated filaments of stacked tetramers in vitro [28–32] and dynamic punctae less than 200 nanometers in size in MTLn3 rat mammary adenocarcinoma cells [28]. A small subset of these PFKL punctae docked at or near the plasma membrane, where we hypothesize PFKL particles interact with transmembrane ion transport proteins to provide localized ATP for ion homeostasis. Upon stimulation of cells with extracellular citrate, PFKL forms micrometer-sized assemblies of unknown function. However, the ability of PFKL to form filaments is required for its recruitment (unpublished) suggests that these represent a form of filamentous PFKL in cells. PFKL also assembles into micrometer-sized complexes in HepG2 human hepatocarcinoma cells under hypoxic stress [29] and in HeLa and Hs578T human triple negative breast cancer cells, where it colocalizes with other rate-limiting glycolytic and gluconeogenic enzymes [30]. The significance of these findings on cellular metabolism and glycolytic flux is an area of current inquiry.
Conclusions and future perspectives
A growing list of metabolic enzymes have been discovered to form filaments in vitro and in micrometer-sized assemblies in cells. Future research will provide insight into why metabolic enzymes have evolved the ability to form filaments and how filament formation contributes to the metabolic regulation of the cell. One potential way in which filamentation of metabolic enzymes may be exploited is for the treatment of diseases in which dysregulation of metabolic enzymes and pathways contributes to their initiation and pathogenesis. As filamentation of metabolic enzymes is important for the activity and allosteric regulation it raises the possibility that pharmaceuticals can be designed to enhance or inhibit filament formation as a mechanism to specifically control enzyme activity. This has been successfully achieved for glutaminase C (GAC). GAC converts glutamine to glutamate and ammonia and its expression is positively correlated with malignancy in cancers [32]. GAC self-assembles into helical, double-stranded oligomers that are catalytically active [33]. The GAC inhibitor bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide (BPTES) locks GAC into an inactive tetrameric confirmation, inhibiting filament formation and decreasing the rate of cancer cell proliferation. Future research will reveal if this strategy is a specific and effective method for targeting other filament forming enzymes.
Table 1.
Functional consequences of polymerization for some metabolic enzymes
| Enzyme | Species | Biochemical function | Allosteric effect |
|---|---|---|---|
| CTPS1 | human | increased activity | stabilize active conformation |
| CTPS2 | human | increased cooperativity | couple structural transitions among protomers |
| CTPS | E. coli | decreased activity | stabilize inactive conformation |
| IMPDH2 | human | reduced sensitivity to inhibitors | stabilize conformation with low affinity for inhibitor |
| Glk1 | yeast | decreased activity | stabilize inactive conformation |
| AdhE | E. coli | increased activity | substrate channeling |
| ACC1 | human | increased activity | stabilize active conformation |
| ACC1-BRCA1 | human | decreased activity | stabilize inactive conformation |
Acknowledgements
This work was supported by National Institutes of Health grant 5R01GM118396-04 and West Virginia University start-up funding (BAW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Noree C et al. (2010) Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol 190, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noree C et al. (2019) A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. [DOI] [PMC free article] [PubMed] [Google Scholar]; * In a large-scale screen of GFP-labeled proteins in yeast, the authors demonstrate that sixty metabolic enzymes form micrometer-sized assemblies, some of which dynamically assembled depending on the metabolic state of the cell. In certain pathways, multiple metabolic enzymes were observed to aggregate. Further, the ability to assemble was enriched in enzymes that catalyze branch point reactions in several metabolic pathways.
- 3.Narayanaswamy R et al. (2009) Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. U.S.A 106, 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Q-J et al. (2016) Filamentation of Metabolic Enzymes in Saccharomyces cerevisiae. J Genet Genomics 43, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoddard PR et al. (2020) Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science 367, 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Glucokinase catalyzes the ATP-dependent phosphorylation of glucose. The authors show that yeast glucokinase (Glk1) forms two-stranded filaments in which Glk1 is inactive. In cells, Glk1 forms filaments upon glucose addition, which disassemble upon glucose removal. The authors propose that filamentation of Glk1 is a mechanism regulating glucose phosphorylation by buffering the levels of active enzyme, thus setting the maximal rate of phosphorylation. This paper is an excellent example of structural biology informing cell biology.
- 6.Hunkeler M et al. (2018) Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558, 470–474 [DOI] [PubMed] [Google Scholar]; ** Acetyl-CoA carboxylase (ACC1) catalyzes the ATP-dependent carboxylation of acetyl-CoA, a rate-limiting step in acetyl-CoA biosynthesis. The authors determined structures of ACC1 filaments in active and inactive conformations. This paper highlights the diversity of filament architectures that can be produced by a single enzyme.
- 7.Anthony SA et al. (2017) Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson MC and Kollman JM (2020) Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. eLife 9, e53243. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Inosine monophosphate dehydrogenase (IMPDH) catalyzes the rate-limiting step in guanine nucleotide biosynthesis. This study reveals that human IMPDH2 forms filaments in a variety of conformations, and that polymerization reduces the sensitivity of IMPDH2 to feedback inhibition by GTP. Importantly, this likely reflects a physiological role for IMPDH2 filaments in proliferation and the immune response, where there is an increased demand for guanine nucleotides.
- 9.Kim G et al. (2019) Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity. Nat Commun 10, 4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pony P et al. (2020) Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation. Nature Communications 11, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]; * Aldehyde-alcohol dehydrogenase (AdhE) is a bifunctional enzyme involved in bacterial fermentation, catalyzing consecutive enzymatic reactions converting acetyl-CoA to ethanol. This paper reports the structure of helical AdhE filaments, termed spirosomes, where the catalytic sites were topologically separated. Spirosomes underwent a conformational change in the presence of cofactors, suggesting filamentation regulates catalytic activity.
- 11.Lynch EM et al. (2017) Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol 24, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]; * CTP synthase (CTPS) is a critical enzyme in nucleotide metabolism, catalyzing the rate-limiting step in CTP biosynthesis. This study reveals diversity in the effects of polymerization on CTPS activity, demonstrating that while bacterial CTPS filaments are inhibited, filaments of human CTP synthase 1 (CTPS1) have increased activity. It is also shown that human and bacterial CTPS filaments assemble via entirely different contact sites, leading to distinct filament architectures.
- 12.Lynch EM and Kollman JM (2020) Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat. Struct. Mol. Biol 27, 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study reveals that the second human CTPS isoform, CTPS2, forms both active and inactive filaments. CTPS2 filaments can switch between the active and inactive filament forms in response to changes in substrate and product levels. Because the conformational states of many CTPS2 subunits within a filament are linked, this produces highly cooperative regulation. This ultrasensitive mechanism of regulation may aid in understanding the function of other enzyme filaments which can alternate between multiple conformations.
- 13.Bray D and Duke T (2004) Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu Rev Biophys Biomol Struct 33, 53–73 [DOI] [PubMed] [Google Scholar]
- 14.Sourjik V and Berg HC (2004) Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 [DOI] [PubMed] [Google Scholar]
- 15.Marx SO et al. (1998) Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors). Science 281, 818–821 [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Seisdedos H et al. (2017) Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244–247 [DOI] [PubMed] [Google Scholar]
- 17.Aughey GN and Liu J-L (2015) Metabolic regulation via enzyme filamentation. Crit. Rev. Biochem. Mol. Biol 51, 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park CK and Horton NC (2019) Structures, functions, and mechanisms of filament forming enzymes: a renaissance of enzyme filamentation. Biophys Rev 11, 927–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Seisdedos H et al. (2019) Infinite Assembly of Folded Proteins in Evolution, Disease, and Engineering. Angew. Chem. Int. Ed. Engl 58, 5514–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connell JD et al. (2012) Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu. Rev. Cell Dev. Biol 28, 89–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry RM et al. (2014) Large-scale filament formation inhibits the activity of CTP synthetase. Elife 3, e03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endrizzi JA et al. (2004) Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry 43, 6447–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellana M et al. (2014) Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol 32, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carcamo WC et al. (2011) Induction of Cytoplasmic Rods and Rings Structures by Inhibition of the CTP and GTP Synthetic Pathway in Mammalian Cells. PLOS ONE 6, e29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K et al. (2011) Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J Genet Genomics 38, 391–402 [DOI] [PubMed] [Google Scholar]
- 26.Gou K-M et al. (2014) CTP synthase forms cytoophidia in the cytoplasm and nucleus. Experimental Cell Research 323, 242–253 [DOI] [PubMed] [Google Scholar]
- 27.Chang C-C et al. (2018) Interfilament interaction between IMPDH and CTPS cytoophidia. FEBS J. 285, 3753–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang S et al. (2016) Glycolytic Enzymes Localize to Synapses under Energy Stress to Support Synaptic Function. Neuron 90, 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb BA et al. (2017) The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J Cell Biol 216, 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]; * Phosphofructokinase-1 catalyzes a rate-limiting step in glycolysis, the phosphorylation of fructose 6-phosphate to fructose 1,6-bisphosphate. This study demonstrates that the liver isoform of phosphofructokinase-1 (PFKL) assembled into filaments of stacked tetramers in a substrate-dependent manner. When expressed in cells, PFKL formed dynamic punctae that docked proximal to the plasma membrane and formed micrometer-sized assemblies in response to exposure to extracellular citrate.
- 30.Jin M et al. (2017) Glycolytic Enzymes Coalesce in G Bodies under Hypoxic Stress. Cell Reports 20, 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohnhorst CL et al. (2017) Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem 292, 9191–9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L and Cui H (2015) Targeting Glutamine Induces Apoptosis: A Cancer Therapy Approach. Int J Mol Sci 16, 22830–22855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreira APS et al. (2013) Active glutaminase C self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J. Biol. Chem 288, 28009–28020 [DOI] [PMC free article] [PubMed] [Google Scholar]


