ABSTRACT
C-terminus of HSC70-interacting protein (CHIP) encoded by the gene STUB1 is a co-chaperone and E3 ligase that acts as a key regulator of cellular protein homeostasis. Mutations in STUB1 cause autosomal recessive spinocerebellar ataxia type 16 (SCAR16) with widespread neurodegeneration manifesting as spastic-ataxic gait disorder, dementia and epilepsy. CHIP−/− mice display severe cerebellar atrophy, show high perinatal lethality and impaired heat stress tolerance. To decipher the pathomechanism underlying SCAR16, we investigated the heat shock response (HSR) in primary fibroblasts of three SCAR16 patients. We found impaired HSR induction and recovery compared to healthy controls. HSPA1A/B transcript levels (coding for HSP70) were reduced upon heat shock but HSP70 remained higher upon recovery in patient- compared to control-fibroblasts. As SCAR16 primarily affects the central nervous system we next investigated the HSR in cortical neurons (CNs) derived from induced pluripotent stem cells of SCAR16 patients. We found CNs of patients and controls to be surprisingly resistant to heat stress with high basal levels of HSP70 compared to fibroblasts. Although heat stress resulted in strong transcript level increases of many HSPs, this did not translate into higher HSP70 protein levels upon heat shock, independent of STUB1 mutations. Furthermore, STUB1(−/−) neurons generated by CRISPR/Cas9-mediated genome editing from an isogenic healthy control line showed a similar HSR to patients. Proteomic analysis of CNs showed dysfunctional protein (re)folding and higher basal oxidative stress levels in patients. Our results question the role of impaired HSR in SCAR16 neuropathology and highlight the need for careful selection of proper cell types for modeling human diseases.
KEY WORDS: CHIP/STUB1, Cortical neurons, CRISPR/Cas9, Heat shock response, Induced pluripotent stem cells, SCAR16
Summary: Cell viability, expression and immunocytochemical analyses reveal that mutations in STUB1 that cause SCAR16 impair the heat-shock response in patient-derived fibroblasts, but not in iPSC-derived cortical neurons.
INTRODUCTION
Mutations in STUB1 (STIP1 homology and U-box containing protein 1) cause early-onset autosomal recessive cerebellar ataxia type 16 (OMIM 615768), a movement disorder that is characterized by atrophy of the cerebellum, brainstem and spinal cord, leading to loss of muscle coordination, unsteady gait, impaired fine motor skills and slurred speech (Shi et al., 2013; Bettencourt et al., 2015). Recently, patients with STUB1 ataxia were shown to present a broader neurodegeneration with complex clinical phenotypes of cognitive impairment, epilepsy and hypogonadism in addition to spastic-ataxic movement disorder (Shi et al., 2013; Heimdal et al., 2014; Shi et al., 2014; Synofzik et al., 2014; Hayer et al., 2017).
STUB1 codes for the C-terminus of HSC70-interacting protein (CHIP), which plays an important role in protein quality control. Its two major functions are linked to two distinct structural domains: via its tetratricopeptide repeat (TPR) domain, CHIP acts as a co-chaperone of HSC70/HSP70 and HSP90 and inhibits ATPase and refolding activity (Ballinger et al., 1999); and via its U box domain, it acts as an E3 ligase tagging chaperone-bound substrates, as well as tagging other substrates with ubiquitin (Jiang et al., 2001; Murata et al., 2003). Degradation of those proteins or organelles occurs via the ubiquitin-proteasome system (UPS) or the autophagy-lysosome system (Zhang et al., 2005; Yao, 2010; Guo et al., 2015). Both mechanisms play a major role in protein quality control and sustain proper cellular homeostasis.
More recently, CHIP was identified as a regulator of many other processes, such as TFEB activity and thereby macroautophagy regulation (Guo et al., 2015; Sha et al., 2017), necroptosis (Seo et al., 2016; Tang et al., 2018), cAMP and AMPK signaling (Schisler et al., 2013; Rinaldi et al., 2019), oxidative metabolism (Ravi et al., 2018), chaperone-mediated autophagy (Ferreira et al., 2015) and neuronal preconditioning (Lizama et al., 2018). In addition to this, CHIP was shown to be cytoprotective in many forms of neurodegeneration by degrading inter alia α-synuclein (Shin et al., 2005; Tetzlaff et al., 2008), LRRK2 (Ding and Goldberg, 2009), APP and BACE1 (Kumar et al., 2007; Singh and Pati, 2015), as well as huntingtin (Jana et al., 2005; Miller et al., 2005). This leads to the assumption that CHIP might be a key player in neurodegeneration and a promising target for the treatment of many neurodegenerative diseases. Furthermore, CHIP was previously linked to cardiac and muscular disorders and several different types of cancers (reviewed by Joshi et al., 2016).
CHIP expression is highest in tissues with high metabolic activity, i.e. heart, skeletal muscle and brain (Ballinger et al., 1999). In mouse brains, CHIP is expressed primarily in neurons of the cerebellum, pons, medulla oblongata, hippocampus and cerebral cortex (Sahara et al., 2005; Anderson et al., 2010). CHIP−/− mice display severe cerebellar atrophy specifically in the Purkinje cell layer, with a distinct motor impairment phenotype. Furthermore, CHIP knockout mice were shown to have decreased stress tolerance and increased age-related phenotypes leading to decreased lifespan and a gonadal dysfunction (Dai et al., 2003; Min et al., 2008; Shi et al., 2014). Upon thermal challenge, 100% of CHIP−/− mice die during heat shock or shortly after (Dai et al., 2003), suggesting a key role for CHIP in stress response, and more specifically in heat shock response (HSR).
Environmental stressors such as heat, heavy metals and reactive oxygen species, but also pathophysiological stressors, such as protein aggregation, inflammation and tissue injury, can induce HSR, which is characterized by a rapid increase in the expression of heat shock proteins (reviewed by Richter et al., 2010). This induction of heat shock proteins is regulated by the key transcription factor heat shock factor 1 (HSF1). Under basal conditions, HSF1 is present in the cytoplasm in its inactive monomeric form, shielded by HSC70/HSP70 and HSP90 (Abravaya et al., 1991). Upon stress, the chaperones blocking HSF1 are recruited to the site of accumulating misfolded proteins, thereby releasing HSF1 (Santoro, 2000). HSF1 quickly trimerizes, undergoes post-translational modifications that modulate its activity and translocates to the nucleus (Sarge et al., 1993). Here, it binds to heat shock responsive elements, leading to the transcription of heat shock proteins, such as HSPA1A/B (encoding HSP70) and DNAJB1 (encoding HSP40) that act to refold or clear misfolded proteins and confer cytoprotection (reviewed by Richter et al., 2010). Other small HSPs (sHSPs), such as HSPB1, HSPB5 and HSPB8 assist by binding to unfolded proteins to prevent aggregation (Acunzo et al., 2012). Once cellular homeostasis is re-established, free HSPs inhibit HSF1 and attenuate the HSR (Abravaya et al., 1991; Baler et al., 1992; Vjestica et al., 2013), allowing a tight regulation of protein quality control during stress conditions.
CHIP was shown to directly interact with HSF1 upon stress induction, co-translocating from the cytoplasm to the nucleus and thereby increasing HSP70 expression in non-neuronal cell types, such as fibroblasts, human embryonic kidney (HEK) cells and retinal epithelium cells, with no or a reduced response in CHIP−/− or CHIPmut cells (Dai et al., 2003; Kim et al., 2005; Qian et al., 2006). Furthermore, CHIP expression is required for HSP70 turnover upon recovery (Qian et al., 2006). Dai et al. (2003) also showed that HSP70 expression upon heat shock in CHIP−/− mice was strongly reduced compared to wild type in most tissues, with an almost complete absence of HSP70 in brain, heart and spleen.
To understand the pathomechanism induced by mutant STUB1, we assessed the HSR in fibroblasts of three SCAR16 patients and three healthy controls and found an impaired HSR in patients. To reflect the central nervous system being the major focus of disease in SCAR16, we next investigated HSR in cortical neurons (CNs) generated from induced pluripotent stem cells (iPSCs). Although heat stress resulted in increased transcript levels of HSPs, this surprisingly did not translate into higher protein levels of HSP70. Proteomic analysis of patient and control CNs showed disturbances in protein folding, the ubiquitin system and oxidative stress response. Both approaches question the role of impaired HSR in SCAR16 neuropathology and highlight the need for the careful selection of proper cell types for disease modeling.
RESULTS
CHIP mutations do not alter viability of fibroblasts during prolonged heat stress
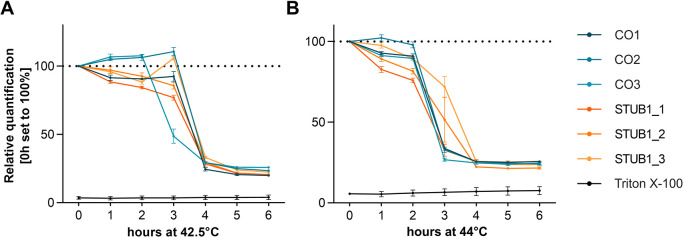
As CHIP has been linked to cell viability upon heat shock, we assessed the effect of prolonged heat stress on the viability of fibroblasts in three patients with STUB1 mutations (STUB1_1, STUB1_2 and STUB1_3) in comparison to three healthy controls (CO1, CO2, CO3) (for further details see Table S1). CHIP protein levels were confirmed to be reduced in all three patient-derived fibroblasts compared to the three healthy controls (Fig. S1A). For cell viability analysis, we exposed cells to either 42.5°C or 44°C for 6 h and quantified the signal with the CyQuant direct cell proliferation assay. Cell viability decreased rapidly between 3 and 4 h of prolonged heat stress at 42.5°C [mean±s.e.m. (%); 3 h heat shock, 86.67±9.16; and 4 h heat shock, 29.01±1.15] (Fig. 1A). At 44°C, cell death mostly occurred after 2 to 3 h of maintained heat shock (2 h heat shock, 87.53±3.16; 3 h heat shock, 41.5±6.95; and 4 h heat shock, 24.88±0.54) (Fig. 1B). No consistent difference was detected between patients and controls. We therefore could not determine an effect of STUB1 mutations on fibroblast viability upon prolonged heat stress.
Fig. 1.
Cell viability of fibroblasts upon heat stress is not impaired by dysfunctional CHIP. (A,B) Viability of fibroblasts (%) during prolonged heat stress at 42.5°C (A) and 44°C (B) for 6 h of three controls and three STUB1 patients. Data are mean±s.d. of triplicates. Triton X-100 was used as negative control for cell viability.
STUB1 mutations cause impaired heat shock response induction and recovery in fibroblasts
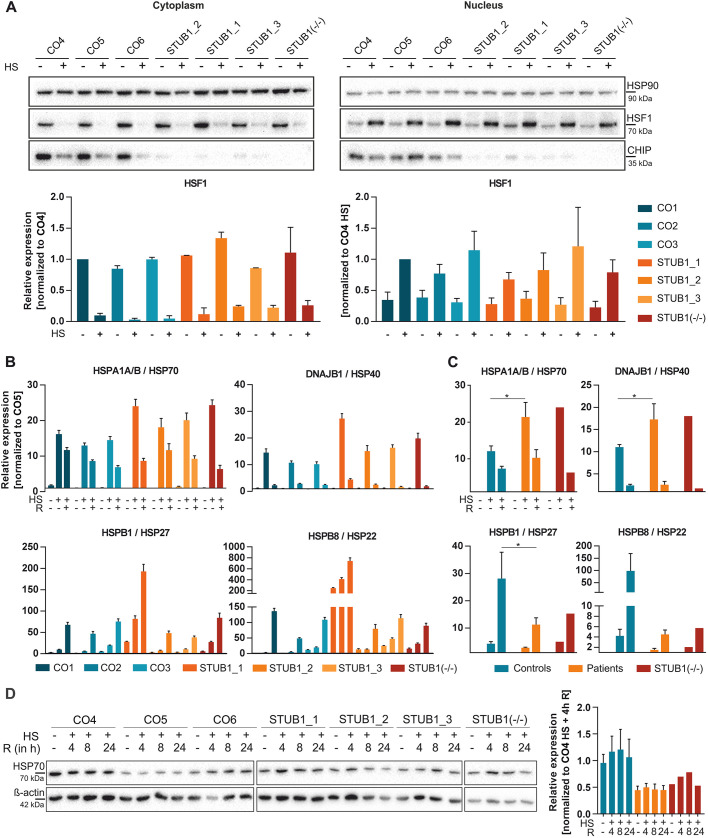
Previous reports have shown that CHIP plays a role in both the induction and recovery of the HSR in HEK and HeLa cells, as well as fibroblasts (Dai et al., 2003; Kim et al., 2005; Qian et al., 2006; Zhang et al., 2015). To test whether CHIP mutations lead to dysfunctional HSR, we analyzed HSF1 translocation upon heat shock, HSR induction by transcript analysis and protein expression analysis and HSR recovery by protein analysis in three STUB1 patient- and three healthy age- and gender-matched control-derived fibroblasts.
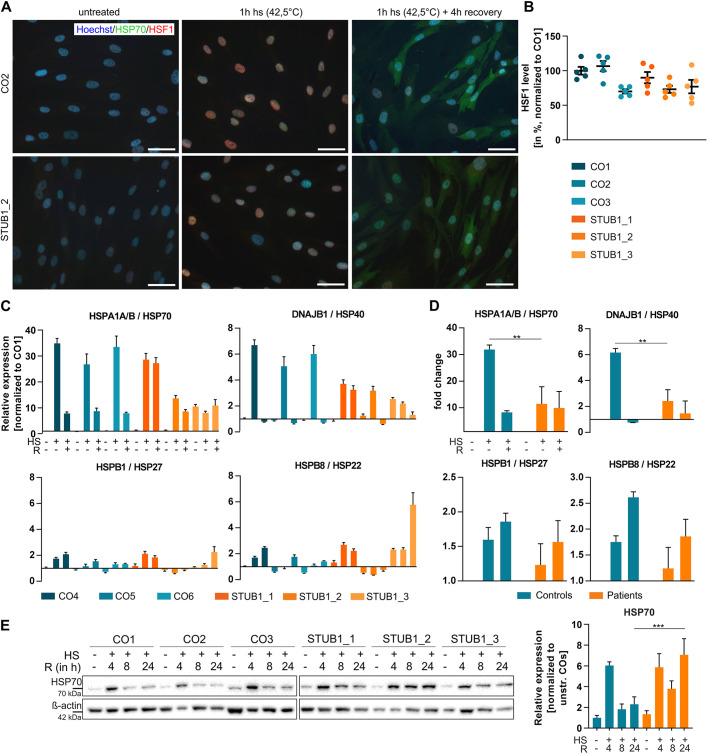
For the analysis of HSF1 translocation from cytoplasm to nucleus, we fixed cells after 1 h heat shock at 42.5°C and immunocytochemically stained for HSF1, HSP70 and with Hoechst 33258 (Fig. 2A). In all six cell lines (three patients, three controls), HSF1 and HSP70 proteins were barely detectable in unstressed conditions. Upon heat shock, HSF1 levels in the nuclei strongly increased and HSP70 accumulated in nuclear bodies. When 4 h of recovery at 37°C was added, HSF1 levels in the nuclei returned to baseline but cytosolic expression of HSP70 increased strongly. Representative images of two cell lines are shown in Fig. 2A. Upon quantification of nuclear HSF1 levels at 1 h of heat shock, we saw a trend towards lower HSF1 levels in patients compared to controls [mean±s.e.m. (%); controls, 92.3±5.0; and patients, 80.0±4.5; P=0.09] (Fig. 2B).
Fig. 2.
Mutant CHIP impairs HSR induction and recovery in fibroblasts. (A) Immunocytochemical staining of HSF1 (red), HSP70 (green) and with Hoechst (blue) in fibroblasts without stress, after heat shock (1 h, 42.5°C) and after recovery (4 h, 37°C). Exemplary images of one control and one patient are shown. Scale bars: 50 µm. (B) Quantification of HSF1 levels in nuclei of heat-shocked fibroblasts, given in percent and normalized to control line CO1. (C) Transcript analysis by qRT-PCR of HSPA1A/B, DNAJB1, HSPB1 and HSPB8 in unstressed, heat shocked (HS) and recovered (R) samples. Values are normalized to CO1 and the housekeeping genes GAPDH and TBP. n=3 replicates. (D) Pooled analysis of three controls and three patients. Transcript levels were normalized to the respective basal levels. (E) Western blot analysis of HSP70 protein levels in fibroblasts without stress, after 1 h heat shock and up to 24 h of recovery, with ß-actin as the loading control. One representative image out of three experiments is shown. Quantification is based on the normalization to unstressed controls. Three controls and three patients were pooled. Data are mean±s.e.m. **P<0.01, ***P<0.001, one-way ANOVA.
When transcripts of heat shock-related genes were analyzed, a strong increase of HSPA1A/B (coding for HSP70) and DNAJB1 (coding for HSP40) was observed after 1 h of heat shock (Fig. 2C,D). However, this induction was significantly lower in patients compared to controls [mean±s.e.m.; HSPA1A/B in controls, 31.89±1.39-fold; and in patients, 11.44±5.25-fold (P=0.004). DNAJB1 in controls, 6.17±0.25-fold; and in patients, 2.42 fold±0.71 (P=0.001)]. Levels in controls and patients decreased with an additional 4 h of recovery. HSPB1 (coding for HSP27) and HSPB8 (coding for HSP22) transcription slightly increased upon recovery [HSPB1 in controls, 1.86±0.1-fold; and in patients, 1.56±0.25-fold. HSPB8 in controls, 2.61±0.09-fold; and in patients, 1.86±0.27-fold (P=0.10)] (Fig. 2C,D). HSP90AA1 (HSP90), HSPA8 (HSC70) and HSPA5 (BiP/GRP78) were neither strongly altered upon heat shock or recovery nor different between patients and controls (Fig. S1B,C).
In terms of protein levels, we saw very low levels of HSP70 under basal conditions in all cell lines and a strong induction of HSP70 at 1 h heat shock plus 4 h recovery (mean±s.e.m.; unstressed controls, 1±0.23; 1 h heat shock plus 4 h recovery; controls, 6.03±0.36; unstressed patients, 1.67±0.33; 1 h heat shock plus 4 h recovery; patients, 7.34±1.1) (Fig. 2E). Induction was 6.03-fold in controls and 4.39-fold in patients (P=0.1). However, HSP70 turnover, and thus HSR recovery, differed in controls and patients with prolonged recovery times, with significantly higher HSP70 levels after 24 h recovery in patients (controls, 2.88±0.76; patients, 7.07±1.55; P=0.0004).
In summary, we were able to show that HSF1 translocates to the nucleus upon heat shock in fibroblasts, with a slight but not significant reduction caused by STUB1 mutations. This led to a significantly lower induction of HSPA1A/B and DNAJB1 in patients. HSP70 protein levels were strongly induced at 1 h heat shock plus 4 h recovery compared to unstressed levels in both patients and controls, but levels in patients remained high at 8 h and 24 h after heat shock, indicating an impaired HSR recovery and HSP70 turnover.
iPSC-derived CNs show typical morphology and express neuronal markers
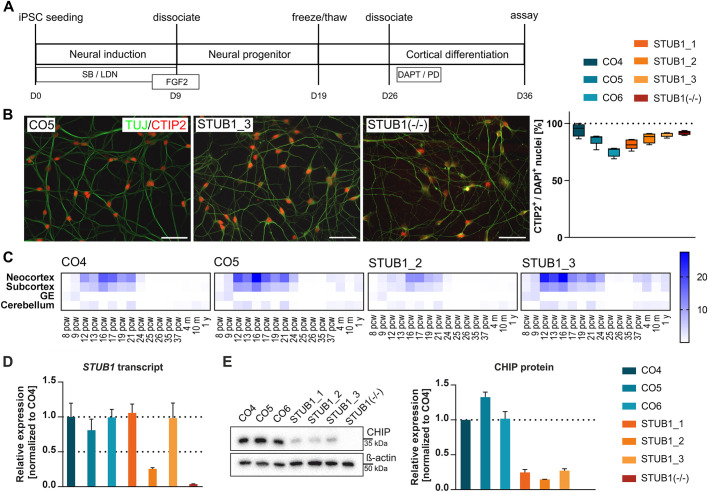
As SCAR16 primarily affects the central nervous system, we next investigated the HSR in CNs. For this, we generated iPSCs from three SCAR16 patients (STUB1_1, STUB1_2 and STUB1_3) and three gender-matched healthy controls (CO4, CO5 and CO6) (for further details see Table S1). Genomic integrity and pluripotency were confirmed for all cell lines [data not shown; for details see Schuster et al. (2018)]. Additionally, we generated a STUB1 knockout line STUB1(−/−) by CRISPR/Cas9-mediated genome editing of the isogenic control line CO5 (Schuster et al., 2019). iPSCs were differentiated into CNs according to a previously published protocol, with slight modifications (Shi et al., 2012; Rehbach et al., 2019) (Fig. 3A). After 36 days of differentiation, the cultures were highly homogeneous, with all cells being positive for ß-III-tubulin (TUJ) and with more than 75% of cells being positive for CTIP2 (also known as BCL11B) (mean±s.e.m., 86.7±1.3%), a cortical layer V marker (Fig. 3B). There was no significant difference in the percentage of CTIP2+ nuclei between STUB1 patients, knockout and controls.
Fig. 3.
Characterization of iPSC-derived neurons. (A) Schematic representation of the experimental procedure of the differentiation of iPSCs into iPSC-derived CNs. SB, SB431542; LDN, LDN193189; PD, PD0325901. (B) Immunocytochemical stainings of CNs on D36 for TUJ (green) and CTIP2 (red). Scale bars: 50 µm. Exemplary stainings of three lines are shown. Quantification of the percentage of CTIP2+/DAPI+ cells in controls, patients and STUB1(−/−) was performed for four to five fields per cell line. Values are given as boxplots showing minimum to maximum, with the mean indicated by a line. (C) Transcript expression in CNs match the best expression found in neocortical tissue at postconception week 16. Heatmaps were produced for two controls (CO4 and CO5) and two patients (STUB1_2 and STUB1_3) by Wilcoxon rank-sum comparisons of CN transcripts to the BrainSpan atlas, and results are shown as –log 10 P-values of significant differences. All four generated lines display a similar expression pattern. (D) Transcript level of STUB1 in three controls, three patients and the homozygous knockout line. Levels are normalized to CO4. Dotted lines indicate full transcript level and 50% reduced transcript level. (E) CHIP protein expression level was analyzed by western blotting. One representative blot is shown. Bands are quantified densitometrically and normalized to ß-actin and CO4. Data are mean±s.e.m. n=3. GE, ganglionic eminence; iPSC, induced pluripotent stem cells; pcw, postconception week.
To assess the spatial and temporal identity of the generated CNs and to exclude differences in differentiation potential and state, we compared transcript analysis data of biological triplicates of CO4, CO5, STUB1_2 and STUB1_3 with transcript data from the developing human brain [BrainSpan atlas (www.brainspan.org) data from fetal and early childhood postmortem tissue up to 1 year of age]. The gene signature of both patient and control iPSC-derived CNs most significantly matched the co-expressed gene sets in the neocortex and the subcortex at postconception week (pcw) 12 to 21, with the strongest correlation at pcw 16 (Fig. 3C). The four analyzed cell lines display only slight differences in the expression patterns independent of STUB1 mutations and show the same temporospatial pattern.
In generated CNs, we quantified STUB1 transcript levels and CHIP protein expression levels. In terms of transcript levels, patients STUB1_1 and STUB1_3 showed no difference to the controls (Fig. 3D), whereas at the protein level, CHIP expression was reduced in the CNs of both patients to 25% of control levels (Fig. 3E). STUB1_2 neurons showed a reduced STUB1 transcript level to 25% of controls (Fig. 3D), resulting in an even lower protein level of 15% compared to wild type (Fig. 3E). STUB1(−/−) showed a transcript level of 3% compared to controls (Fig. 3D), and no detectable protein (Fig. 3E), confirming the homozygous knockout of STUB1.
CHIP mutations do not alter cell viability during prolonged heat stress in CNs
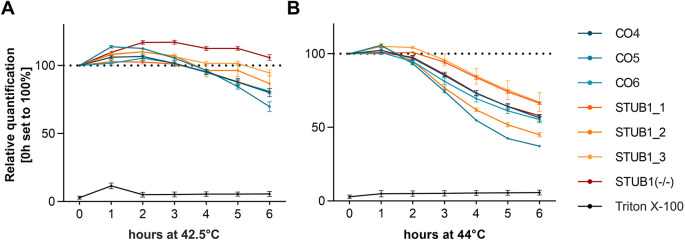
To examine the effect of heat shock on the viability of CNs, we exposed CNs of three controls, three STUB1 patients and STUB1(−/−) to prolonged heat stress at either 42.5°C or 44°C for 6 h. Interestingly, signal intensity of the CyQuant direct cell proliferation assay slightly increased in all cell lines at 42.5°C and only reached levels below baseline after more than 4 h of heat stress. After 6 h of heat stress, a mean value of 85.5±11.1% of viable cells was reached (Fig. 4A). At 44°C, lethality of CNs was higher, reaching a mean value of 55.0±10.3% of viable cells after 6 h of heat stress (Fig. 4B). No consistent difference in cell viability was observable between controls, patients and the STUB1(−/−) line.
Fig. 4.
Viability of CNs upon heat shock is not impaired by dysfunctional CHIP. (A,B) Cell viability (%) during prolonged heat shock at 42.5°C (A) and 44°C (B) was investigated in three controls, three STUB1 patients and a STUB1(−/−) line. Data are mean±s.d. of triplicates. Triton X-100 was used as negative control for cell viability.
Mutations in STUB1 do not impair the heat shock response in CNs
We next investigated the HSR in CNs after 1 h of heat shock at 42.5°C. To assess nuclear translocation of HSF1, we performed subcellular fractionation of the nucleus and cytoplasm. Proper separation of nuclei from the cytoplasmic fraction was verified by the presence and absence of histone 3 (H3), respectively (Fig. S2A). In unstressed conditions, HSF1 was highly expressed in the cytosol and weakly expressed in the nucleus. Upon heat shock, HSF1 was hyperphosphorylated, as seen by a mass shift, and was translocated to the nucleus (Fig. 5A), as almost no HSF1 remained in the cytosol after heat shock. HSF1 translocation and expression levels were not altered by mutant STUB1 (Fig. 5A). CHIP levels were shown to diminish after heat shock in the cytosol in controls but did not increase drastically in the nucleus.
Fig. 5.
Mutant CHIP does not impair the HSR in iPSC-derived CNs. (A) Cytoplasmic and nuclear fractions of HSP90, HSF1 and CHIP before and after heat shock (heat shock, 1 h, 42.5°C) in CNs of three controls, three patients and STUB1(−/−). HSP90 and HSF1 levels were quantified densitometrically and normalized to HSP90, and to CO5/CO5 heat shock (HS). (B) Transcript analysis of HSPA1A/B, DNAJB1, HSPB1 and HSPB8 was performed by qRT-PCR. Values are normalized to CO5 and the housekeeping genes GAPDH and TBP. Each bar represents a triplicate. Data are mean±s.e.m. (C) Fold change of HSPs compared to baseline. Transcript levels of B were pooled for controls and patients. (D) Western blot analysis of HSP70 levels of unstressed and heat-shocked CNs with various times of recovery (R, in hours). HSP70 levels were quantified densitometrically, normalized to ß-actin and pooled for controls and patients. n=3 western blots were performed as technical replicates. Data are mean±s.e.m. *P<0.05; one-way ANOVA.
In terms of transcript levels, heat strongly induced the transcription of HSPA1A/B (encoding HSP70) and DNAJB1 (encoding HSP40) (Fig. 5B,C). This induction was significantly higher in patients and STUB1(−/−) compared to controls [mean±s.e.m.; HSPA1A/B in controls, 12.06±1.18-fold; in patients, 21.38±3.25-fold (P=0.017); and in STUB1(−/−), 24-fold (P=0.03). DNAJB1 in controls, 11.06±0.46 fold; in patients, 17.28±2.9-fold (P=0.038); and in STUB1(−/−), 18-fold (P=0.1)]. After 4 h of recovery, levels decreased in all lines again [HSPA1A/B in controls, 7.31±0.54-fold; in patients, 10.26±1.81-fold; and in STUB1(−/−), 6.3-fold. DNAJB1 in controls, 2.40±0.22-fold; in patients, 2.58±0.61-fold, and in STUB1(−/−): 1.7-fold]. HSPB1 (encoding HSP27) and HSPB8 (encoding HSP22) transcription slightly increased upon heat shock by two to fourfold but highly increased after 4 h recovery at 37°C [HSPB1 in controls, 28.16±7.86-fold; in patients, 11.25±2.05-fold (P=0.037); and in STUB1(−/−), 15.3-fold (P=0.3). HSPB8 in controls, 98.5±57.65-fold; in patients, 4.5±0.7-fold (P=0.11); and in STUB1(−/−), 5.7-fold (P=0.3)]. The increase of HSPB1 was significantly higher in controls compared to patients. However, variability for HSPB1 and HSPB8 was very high between lines, probably because of low basal expression levels. HSP90AA1 (HSP90), HSPA8 (HSC70) and HSPA5 (BiP/GRP78) were neither strongly altered upon heat shock or recovery nor different between patients and controls (Fig. S2B,C).
Unexpectedly, in terms of protein levels, HSP70 expression under basal conditions was already very high in CNs, with slight variations between lines and independent of STUB1 mutations (Fig. S2D). We barely saw an increase of HSP70 after heat shock and at various times of recovery [increase after 1 h heat shock plus 4 h recovery normalized to baseline (mean±s.e.m.) of controls, 1.27±0.3-fold; patients, 1.12±0.15-fold; STUB1(−/−), 1.25-fold]. However, when absolute HSP70 levels were compared, we saw slightly lower levels in patients compared to controls [unstressed HSP70 level of controls, 1±0.16; patients, 0.47±0.07 (P=0.27); and STUB1(−/−), 0.58. Heat shock (1 h) plus 4 h recovery HSP70 level of controls, 1.22±0.29; patients, 0.52±0.07 (P=0.25); and STUB1(−/−), 0.73] (Fig. 5D).
In summary, we were able to show that HSF1 translocates to the nucleus upon heat shock in CNs and that this leads to the induction of HSPA1A/B and DNAJB1 transcripts, which surprisingly did not translate to increased HSP70 protein levels. Induction of HSPA1A/B and DNAJB1 was significantly higher in patients and STUB1(−/−) compared to controls. Furthermore, we barely saw an induction of HSP70 on protein level in both controls and patients.
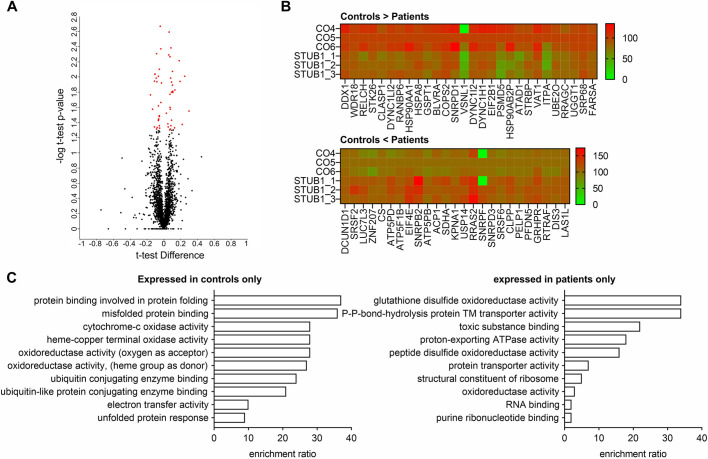
Proteome analysis of CNs reveals impaired protein folding and ubiquitination in SCAR16 patients
Proteomic analysis based on mass spectrometry (MS) with liquid chromatography (LC)/MS-MS and label-free quantification (LFQ) was applied to identify quantitative differences in proteome-wide protein levels between CNs of controls and patients (n=3 for both biological groups). We identified 53 proteins with significantly altered levels (Fig. 6A); 28 proteins were increased (Fig. 6B, upper panel) and 25 proteins were reduced in controls compared to patients (Fig. 6B, lower panel). Additionally, 185 proteins were exclusively expressed in at least two control cell lines but not in any patient cell line (Table S3), and 228 proteins were exclusively expressed in at least two patient cell lines but not in any control cell line (Table S4). Based on WebGestalt analysis (Liao et al., 2019) of proteins that were exclusively expressed in at least two controls, gene ontology (GO) terms associated with protein folding and refolding, and the ubiquitin system, were highly enriched (Fig. 6C, left panel). In contrast, GO terms associated with oxidative stress coping were enriched in patients only (Fig. 6C, right panel). In terms of specific interaction partners of CHIP, we found differences between controls and patients for some proteins, including PSEN1/2, IGF1R, TRAF2 and TRAF6 (Fig. S3).
Fig. 6.
Proteome analysis of CNs from three controls and three STUB1 patients revealed protein (re)folding disturbances and increased oxidative stress in patients. (A) The volcano plot illustrates significantly differentially expressed proteins in patients versus controls. The –log 10 two-tailed unpaired Student's t-test P-value is plotted against the t-test difference in terms of controls versus patients. The significance threshold is set to P=0.05/1.30 (−log 10). (B) Heatmap displaying significantly dysregulated proteins, for proteins downregulated (upper panel) and upregulated in patients (lower panel). Values are given as log 10 LFQ intensity. Control line CO5 is set to 100%. (C) GO analysis with WebGestalt of proteins expressed in at least two of three controls but not in patients (left panel), and in at least two of three patients but not in controls (right panel). Proteins were subjected to GO classification in terms of molecular function. A threshold of five proteins per classification was set. Enrichment ratios of GO terms are shown.
DISCUSSION
Proteotoxic stress and a compromised HSR are associated with many neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, Huntington's disease and amyotrophic lateral sclerosis (reviewed by San Gil et al., 2017). This suggests an innate susceptibility of neurons to stress, homeostatic changes and disturbances in protein quality control, thereby highlighting the significance of chaperones in neuroprotection.
In this study, we investigated the pathological effect of STUB1 mutations causing SCAR16 on the HSR. Analyzing patient-derived fibroblasts, we confirmed an impaired HSR induction on transcript level and an impaired HSR recovery on protein level. Furthermore, we developed a human iPSC-based SCAR16 disease model with CNs as the affected cell type. Despite differences in heat-inducible transcript levels in STUB1-mutant CNs, we could not detect an impaired heat shock induction or recovery at the protein level in CNs of STUB1 patients.
In a rodent model of SCAR16, CHIP−/− mice exhibit decreased stress tolerance, pronounced heat sensitivity, increased oxidative stress and lethality upon heat shock (Dai et al., 2003; Min et al., 2008). In vitro, CHIP knockdown in mesodermal cell lines caused reduced cell viability upon prolonged heat stress and impaired induction and recovery of the HSR (Dai et al., 2003; Kim et al., 2005; Qian et al., 2006; Zhang et al., 2015).
In human SCAR16 cell models, we found a striking difference in heat stress tolerance between cell types. Although fewer than 30% of fibroblasts survived 4 h of heat shock at 42.5°C, more than 95% of neurons were still viable. This might indicate a higher stress vulnerability of fibroblasts and a stronger response to this toxic stimulus. Although Dai et al. (2003) reported 50% lethality of murine CHIP(−/−) fibroblasts after 60 min at 42°C, our patient-derived fibroblasts did not show relevant lethality after 60 min of heat shock at 42.5°C. This difference might be due to low levels of remaining CHIP in patient fibroblasts with residual activity that potentially rescues the drastic toxic effect seen in CHIP knockout fibroblasts. The better stress coping of CNs could be a result of the immaturity of the CNs as efficiency of the proteostasis network strongly declines with terminal differentiation (Vonk et al., 2020) and with aging (Morishima et al., 2008).
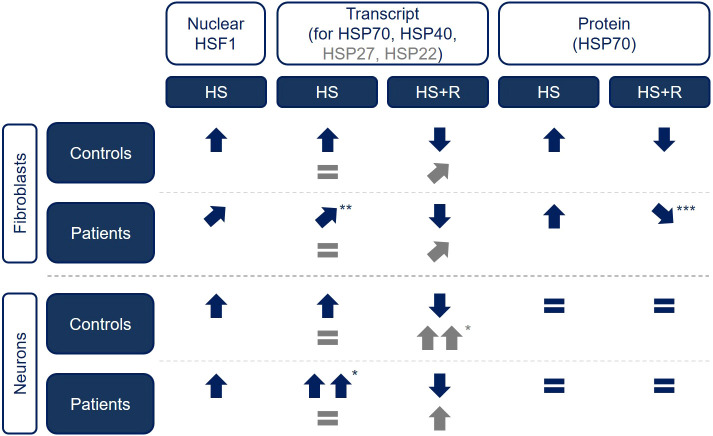
Our data imply that STUB1 mutations, and thereby dysfunctional CHIP, do not affect the cell viability negatively upon heat shock in fibroblasts and CNs. We next evaluated the induction of HSR and its recovery in patient- and healthy control-derived fibroblasts. Using immunocytochemical analysis of heat shocked cells, we showed that HSF1 translocated to the nucleus in patients and controls, with a trend towards lower levels in controls. Furthermore, when different transcript levels in fibroblasts were analyzed, we observed a lower increase in HSPA1A/B and DNAJB1 transcript levels upon heat shock in patients compared to controls. This might be caused by slightly lower nuclear HSF1 levels and/or by differential post-translational modifications (PTMs) of HSF1, as PTMs were reported to strongly alter the activity of HSF1 (reviewed by Gomez-Pastor et al., 2018). Surprisingly, at the protein level, we did not see reduced HSP70 levels in patients compared to controls after a 4 h recovery from heat shock as would be expected from the lower transcript levels, but instead we observed a six to sevenfold increase compared to unstressed levels in both biological groups. Yet, we see an impaired HSR recovery, as indicated by the remaining high levels of HSP70 after prolonged recovery (8 h and 24 h), in line with results presented by Qian et al. (2006). This leads to the conclusion that either HSP70 ubiquitin tagging is reduced in CHIP-mutant patient cells or that the amount of misfolded proteins in patients is simply higher, requiring a prolonged high expression of HSPs in those cells, or a combination of both. The maintained HSP70 protein expression in SCAR16 patient-derived cells might also lead to the observed similar cell viability upon prolonged heat shock. In conclusion, we confirm an impaired induction and recovery of the HSR caused by dysfunctional CHIP, which is the first such observation in patient-derived cells (summarized in Fig. 7).
Fig. 7.
Comparative overview of heat shock-related changes in control and SCAR16 patient fibroblasts and CNs. Experimental results of control- and patient-derived fibroblasts (Fig. 2) and control- and patient iPSC-derived CNs (Fig. 5) are summarized. *P<0.05, **P<0.01, ***P<0.001. ↓, reduction of level; ↑, increase of level; = no change in level. Comparison is based on unstressed condition for heat shock, and on heat shock for heat shock and recovery. HS, heat shock; HS+R, heat shock and recovery.
As SCAR16 primarily causes degeneration of the central nervous system, we next sought to analyze the HSR in patient neurons. We therefore generated patient-specific iPSCs from fibroblasts and differentiated those, together with gender-matched controls, into CNs as a disease-relevant cell type. This provides a disease model with endogenous protein levels and the patient's own genetic background. Although mutations in STUB1_1 and STUB1_3 do not alter STUB1 transcript levels, protein stability is strongly reduced by the mutations, leading to a lower CHIP level (Heimdal et al., 2014; Ronnebaum et al., 2014; Pakdaman et al., 2017; Kanack et al., 2018; Shi et al., 2018; Madrigal et al., 2019). STUB1_2 shows reduced transcript and protein levels.
Analyzing the HSR, we could show that the translocation of HSF1 from the cytoplasm to the nucleus upon heat shock occurs invariable of CHIP functionality, suggesting no direct impact of CHIP mutations on trimerization of HSF1 or its translocation in CNs. CHIP levels declined upon heat shock in the cytoplasm, but did not correspondingly increase in the nucleus. Potentially, nuclear CHIP levels did already return to baseline in our experimental setup with 1 h heat shock, as Anderson and colleagues showed a strong accumulation in the nucleus only 5-10 min after heat shock and a decrease back to baseline within 30-60 mins (Anderson et al., 2010).
When transcript levels of HSPs were analyzed, we observed a higher increase in HSPA1A/B and DNAJB1 upon heat shock in patient neurons compared to controls, independent of nuclear HSF1 levels. This might be caused by differential PTMs of HSF1 as described above. Higher HSPA1A/B and DNAJB1 transcription might also be caused by ‘preconditioning’ to stress and thermotolerance in patients: levels of misfolded proteins are presumably higher in CHIP mutant cells, causing mild proteotoxic stress and an induction of the HSR before the heat stimulus which potentiates the HSR upon subsequent stress exposure (Burdon, 1987). Stress preconditioning might only play a role in CNs and not in fibroblasts as non-dividing cells are more vulnerable to proteotoxic stress in general.
Transcript levels of the sHSPs HSPB1 (coding for HSP27) and HSPB8 (coding for HSP22) were strongly increased during the recovery steps from heat shock, indicating two stages of transcript induction with an early induction of HSPA1A/B and DNAJB1 and, unexpectedly, a delayed induction of HSPB1 and HSPB8. HSP27 and HSP22 bind to unfolded proteins, prevent their aggregation and protect cells from toxicity caused by aggregates. Both sHSPs also enable stress resistance and inhibit apoptosis (Acunzo et al., 2012). This might explain the higher levels in controls compared to patients and delayed induction. Interestingly, mutations in both HSPB1 and HSPB8 induce neurodegeneration, highlighting the importance of both sHSPs in the stress response in neurons in particular.
Unexpectedly, at the protein level, we neither detected a prominent induction of HSP70 protein nor a difference in HSP70 level upon recovery. HSP70 levels at 4 h recovery after heat stress were only 1.1- to 1.2-fold higher than at its basal levels, compared to six to sevenfold higher levels in fibroblasts. These differences cannot be explained by varying CHIP expression in these cell types, as the basal CHIP protein expression levels in fibroblasts and CNs were comparable (Fig. S4), but there might be several other reasons: (1) an impaired HSR in the brain might be relevant and disease-causing but not in neurons; (2) alterations of the HSR that were seen in patient-derived fibroblasts but not in CNs are not disease-relevant, as the diseased cell type does not show any changes; or (3) the low HSR is an in vitro artifact of the monoculture and the culture conditions, as the basal HSP70 level is already very high. As persistently high levels of HSP70 are detrimental to cells (Feder et al., 1992; Volloch and Sherman, 1999), an additional induction of HSP70 upon stress might be prevented in iPSC-derived CNs. Considering the role of CHIP in this process, we observed only slightly lower HSP70 protein levels in patients but no difference in fold changes. Furthermore, the comparison of STUB1(−/−) with its isogenic control revealed very similar results to the comparison of patients and controls. This confirms our findings in patient lines and supports the limited effect of CHIP on the HSR in CNs (summarized in Fig. 7).
Although the HSR is evolutionarily well conserved, tissue and cell variability of the chaperone in humans is prominent: Hageman and Kampinga (2009) analyzed the temporal and the spatial expression pattern of 45 tissues using the UniGene database and observed strong tissue specificity for the expression of HSPH, HSPA and DNAJ isoforms. Notably, the expression of HSPA1A is highly variable between tissues and developmental stages (embryoid body to adult). Focusing on the brain, histological analyses after heat shock in rodents showed the strongest HSP70 induction in the dentate gyrus, hypothalamus and cerebellum (Blake et al., 1990; Li et al., 1992). Of interest, a comparison of HSR in neurons and glia in vitro revealed a higher induction of HSP70 in glial cells compared to neurons (Nishimura et al., 1991; Marcuccilli et al., 1996; Vogel et al., 1997; Batulan et al., 2003). This might be caused by differential co-chaperone levels: HSPBP1 expression in neurons is higher than in astrocytes and is directly linked to HSR inhibition (Zhao et al., 2017); and HSPB1/HSP27 levels are oppositely correlated (Satoh and Kim, 1995). However, although HSP27 levels do not increase upon heat shock in astrocytes, this increase is observed in neurons (Satoh and Kim, 1995). Other HSPs, such as DNAJB2A (HSP70 family) and HSJ1a/b (HSP40 family), were reported to be highly enriched in neuronal tissue (Cheetham et al., 1992; Chapple and Cheetham, 2003). In our study, we additionally observed a strong cell specificity of HSPB1 and HSPB8, with higher expression under basal conditions in fibroblasts, but higher induction upon recovery of heat stress in neurons. This might point towards an important role for both sHSPs in stress coping in CNs.
For a broader picture, proteomic analysis of control and patient CNs was performed. Comparisons point towards aberrations in protein folding and the ubiquitin system, as well as an increased oxidative stress level in unstressed patient cells. This is in line with reports that show that mutations alter the ability of CHIP to ubiquitinate its substrates (Heimdal et al., 2014; Ronnebaum et al., 2014; Pakdaman et al., 2017; Kanack et al., 2018; Shi et al., 2018). However, with more than 200 E3 ligases encoded in the human body, substrates are not exclusively ubiquitinated by one E3 ligase; for example, parkin and CHIP act redundantly on some substrates (Morishima et al., 2008). Furthermore, keeping in mind the function of CHIP in tagging misfolded proteins with ubiquitin for degradation, the importance of CHIP might only show up in a state with a high abundancy of misfolded proteins.
In summary, our results question the role of impaired HSR in SCAR16 neuropathology and point to protein degradation and oxidative stress as potentially more critical factors in the pathogenesis of CHIP-related neurodegeneration. By demonstrating major differences in the HSR of fibroblasts and induced CNs of SCAR16 patients, this study highlights the need for the careful selection of proper cell types for modeling human diseases.
MATERIALS AND METHODS
Patients
Three patients with genetically confirmed compound heterozygous or homozygous mutations in STUB1 were included in this study. Patient STUB1_1 had a homozygous c.367C>G; p.L123V mutation, whereas patients STUB1_2 and STUB1_3 carried the compound heterozygous mutations c.355C>T; p.R119*; c.880A>T, p.I294T and c.433A>C, p.K145Q; c.728C>T, p.P243L, respectively (Hayer et al., 2017). All patients were severely affected by SCAR16 with ataxia (3/3), spasticity (3/3), epilepsy (2/3), dementia (2/3) and hypogonadism (1/3). The controls were age- and gender-matched for fibroblast analyses and gender-matched for iPSC-derived neuronal analyses. Detailed information on study participants and cell lines is provided in Table S1. The study was approved by the Ethics Committee of the University of Tübingen (vote 598/2011BO1). Informed written consent was obtained from all participants or their legal guardian.
Fibroblast cultivation
Human dermal fibroblasts were obtained from skin biopsies and maintained at 37°C, 5% CO2 and 100% relative humidity in fibroblast medium consisting of Dulbecco's modified eagle medium (DMEM) (Merck Millipore) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) in cell culture flasks. Upon reaching high confluency, cells were split by washing with PBS followed by 5 min trypsination and passaging into new flasks, or seeding at a defined density depending on the assay (20,000 cells/cm2 for immunocytochemical analysis and 30,000 cells/cm2 for protein analysis).
Reprogramming of fibroblasts to iPSCs
The generation of iPSCs from fibroblasts was performed according to a previously published protocol (Okita et al., 2011) with minor modifications. In brief, 105 human dermal fibroblasts were nucleofected with 1 µg of each episomal plasmid [pCXLE-hUL, pCXLE-hSK and pCXLE-hOCT4 (Addgene numbers 27080, 27078 and 27076, respectively)] with Nucleofector 2b (Lonza). Fibroblasts were cultivated in fibroblast medium before adding 2 ng/ml FGF2 (Peprotech) on day 2. The following day, medium was changed to Essential 8 (E8) medium [DMEM/F12 (Life Technologies), 64 mg/l L-ascorbic acid 2-phosphate magnesium (Sigma-Aldrich), 1% insulin-transferrin-selenium-supplement (100×) (Life Technologies), 10 ng/ml FGF2, 2 ng/ml TGFß1 (Peprotech) and 100 ng/ml heparin (Sigma Aldrich)] with 100 μM NaB (Sigma-Aldrich). After 3 to 4 weeks, with a change of medium every other day, colonies were manually picked and expanded onto Matrigel-coated plates (Corning) in a feeder-free system in E8 medium. The iPSCs were frozen in E8 medium with 40% KnockOut serum replacement (Life Technologies), 10% dimethyl sulfoxide (Sigma-Aldrich) and 10 μM Y-27632 (Selleckchem). Genomic integrity was confirmed by excluding plasmid integration and performing whole-genome single nucleotide polymorphism genotyping [Infinium OmniExpressExome-8-BeadChip (Illumina) or CytoScan HD technology (Affymetrix), and copy number analysis (CNVPartition plug-in (Illumina)], and resequencing of the mutation sites. Pluripotency was confirmed by ALP expression, immunocytochemical analysis of pluripotency markers, transcript analysis of pluripotency genes and embryoid-body-based differentiation of iPSCs into cells of all three germ layers. For detailed descriptions see Schuster et al. (2018).
Targeted STUB1 knockout with CRISPR/Cas9
To generate the STUB1 knockout line STUB1(−/−), we targeted exons 2 and 3 of STUB1 in a dual-cleavage approach to enhance the efficiency of knockout generation. CRISPR-RNAs (crRNAs) (Integrated DNA Technologies) were designed using the CRISPOR web tool (Haeussler et al., 2016). The iPSCs of control line CO5 were nucleofected with two crRNA-ATTO550-tracrRNA (Integrated DNA Technologies) ribonucleoprotein complexes with Cas9 protein (Integrated DNA Technologies), followed by fluorescence-activated cell sorting of ATTO550+ cells with a Sony Cell Sorter SH800Z, single cell seeding and manual picking and expansion of clones. Homozygous or heterozygous knockout was validated by PCR analysis and Sanger sequencing. The top five exonic off-target effects, as predicted by CRISPOR for each cleavage site, were excluded by Sanger sequencing in the isogenic control, as well as the generated clones. For detailed experimental setup and characterization of STUB1(−/−) see Schuster et al. (2019).
Neuronal differentiation
To model SCAR16 in vitro, we generated neurons of cortical layer V and VI according to a published protocol (Shi et al., 2012; Rehbach et al., 2019) with modifications. In brief, iPSCs were dissociated with 0.02% EDTA (Carl Roth) in PBS (Sigma-Aldrich) and seeded at a density of 3×105/cm2 in E8 medium supplemented with 10 µM Y-27632. The following day, the medium was replaced by neural induction medium [1:1 N2/B27, 500 nM LDN-193189 (Th. Geyer) and 10 µM SB431542 (Sigma-Aldrich)], which, from there onwards, was changed every day. The neural induction medium was supplemented with 20 ng/ml FGF2 on day 8. The next day, cultures were split by detachment with Accutase (Sigma-Aldrich) for 10 min and seeded in N2/B27 medium with 20 ng/ml FGF2 and 10 µM Y-27632 onto Matrigel-coated six-well plates. N2/B27 medium with 20 ng/ml FGF2 was added on the next day. From day 11 onward, cells were cultured in N2/B27 medium with a change of medium every other day. Neural precursors were dissociated and frozen on day 19. For maturation, frozen precursors were thawed and seeded in N2/B27 medium with 10 µM Y-27632, with a change of medium every other day. On day 26, cells were detached with Accutase and reseeded at the desired assay density (for immunocytochemical staining, 1×105/cm2; and for protein and RNA isolation, 4-6×105/cm2) on poly-l-ornithine (Sigma-Aldrich) and Matrigel-coated wells. On day 27 and 29, the medium was changed to N2/B27 supplemented with 10 µM PD0325901 (Tocris) and 10 µM DAPT (Sigma-Aldrich). From day 31 and up to the analysis on day 36, the cells were cultured in N2/B27 medium with medium changes every other day. For heat shock experiments, cells were exposed to 42.5°C with prewarmed medium for 1 h, with direct harvesting of cells for RNA isolation and protein fractionation, and an additional 4, 8 or 24 h recovery at 37°C for protein analysis.
Immunocytochemistry and image analysis
Cells were washed with PBS followed by fixation with 4% paraformaldehyde (Merck Millipore) and subsequent washing. Fixed cells were permeabilized and blocked with 5% bovine serum albumin (Sigma-Aldrich) in PBS with 0.1% Triton X-100 (Carl Roth). Afterwards, cells were stained with anti-β-III-tubulin (TUJ, mouse, 1:1000, T8660, Sigma-Aldrich), anti-CTIP2 (rat, 1:200, ab18465, Abcam), anti-HSF1 (rabbit, 1:500, 4356T, Cell Signaling Technology) and/or anti-HSP70 (mouse, 1:500, ADI-SPA-810, Enzo Life Sciences), all followed by labeling with Alexa Fluor-conjugated secondary antibodies (Invitrogen). Hoechst 33258 staining (1:10,000, H1398, Invitrogen) was used to counterstain for nuclei. Coverslips were mounted with Dako mounting solution (Agilent Dako) onto microscope slides. Four or five random fields per coverslip per cell line were used for quantification. Images were acquired using an Observer Z1 fluorescence microscope (Zeiss), and exposure time was kept constant. Quantifications were conducted using the cell counter plug-in in FiJi (Schindelin et al., 2012) and threshold mask settings in ImageJ (https://imagej.nih.gov/ij/).
Transcript analysis by qRT-PCR
For RNA isolation, cells were scraped off in RLT buffer (Qiagen) and lysed in the well. RNA was isolated with an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. A total of 500 ng RNA was reverse-transcribed into cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. For real-time PCR, 3 µl of 1.25 ng/µl cDNA was mixed with 2 µl of 2 µM primer pairs and 5 µl of SYBR Green Select Master Mix (Applied Biosystems). Primers are listed in Table S2. The qRT-PCR program was as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 1 s, 60°C for 30 s and 72°C for 5 s, and subsequently 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. The specificity of PCR products was confirmed by melting curve analysis. Real-time PCR amplifications were performed on a Viia7 Real-Time PCR System (Applied Biosystems) and primers were run in triplicate. The housekeeping genes GAPDH and TBP were amplified to standardize the amount of sample cDNA. Analysis was performed with QuantStudio Software V1.3 (Thermo Fisher Scientific).
Cell viability analysis
Fibroblasts were seeded at a density of 12.5×104/cm2 on 96-well plates in triplicate. CNs were seeded at a density of 3×105/cm2 on D33 on 96-well plates coated with poly-l-ornithine and Matrigel in quadruplicate. To assess cell viability upon heat shock, the CyQuant direct cell proliferation assay (Thermo Fisher Scientific) was used according to the manufacturer's instructions. In brief, CQD nucleic acid stain and CQD background suppressor dye were diluted in PBS and added to the cells, followed by 1 h incubation at 37°C. Triton X-100 (1%) was used as a negative control for cell viability. The plate reader SpectraMax M (Molecular Devices) was set to 42.5°C or 44°C, and plates were measured for 6 h with excitation/emission at 485/525 nm.
Protein isolation
Pellets of primary fibroblasts or CNs were lysed in RIPA buffer (Sigma-Aldrich) containing 1×cOmplete protease inhibitor cocktail (PI) (Roche) for 45 min on a rotator at 4°C. Cell debris was pelleted at 15,800 g at 4°C for 30 min. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
Subcellular fractionation
To separate nuclei from cytoplasm in CNs, an Abcam protocol was applied with slight modifications. In brief, cells were scraped off on ice in fractionation buffer [20 mM HEPES (pH 7.4) (Carl Roth), 10 mM KCl (Carl Roth), 2 mM MgCl2 (Merck Millipore), 1 mM EDTA (Carl Roth), 1 mM EGTA (AppliChem), 1 mM dithiothreitol (AppliChem) and 1×PI] and incubated for 15 min on ice. Samples were then passed through a 27-gauge needle (Sigma-Aldrich) ten times, then incubated on ice for 20 min. Samples were centrifuged at 750 g for 5 min at 4°C and supernatant containing cytoplasm was transferred to a fresh tube. One quarter of the total amount of 5×RIPA buffer [750 mM NaCl (VWR), 5% Igepal CA-630 (Sigma-Aldrich), 2.5% Sodium deoxycholate (Carl Roth), 0.5% SDS (Sigma-Aldrich) and 250 mM Tris (pH 8.0) (AppliChem)] with 1×PI was added. The nuclear pellet was washed with fractionation buffer followed by ten more passes through a 27-gauge needle and 10 min centrifugation at 750 g at 4°C. The supernatant was discarded, the pellet was resuspended in 1×RIPA buffer plus PI and sonicated to shear genomic DNA. Protein isolation was performed according to the procedure described above.
Western blotting and densitometric analysis
Protein (10-30 µg) was eluted in 5×pink buffer (Thermo Fisher Scientific) at 95°C. Samples were separated on 10% polyacrylamide gels in 1×NuPAGE MOPS SDS running buffer (Novex) and transferred onto a Hybond-P polyvinylidene difluoride membrane (Merck Millipore). Membranes were blocked in 5% milk in TBS-T, incubated overnight with primary antibodies [ß-actin (mouse, 1:20,000, A5441, Thermo Fisher Scientific), CHIP (rabbit, 1:10,000, ab134064, Abcam), H3 (horseradish peroxidase (HRP)-tagged, 1:100,000, ab21054, Abcam), HSF1 (rabbit, 1:1000, 4356T, Cell Signaling Technology), HSP70 (mouse, 1:50,000, ADI-SPA-810, Enzo Life Sciences), HSP90 (mouse, 1:100,000, ADI-SPA-831-050, Enzo Life Sciences)] in Western Blocking Reagent (Roche) at 4°C, followed by three washes with TBS-T and incubation with HRP-conjugated secondary antibodies (Jackson ImmunoResearch) for 1 h at room temperature. Proteins were visualized using the Immobilon Western chemiluminescent HRP substrate (Merck Millipore). Bands were quantified with ImageJ and normalized to respective loading controls.
RNA Sequencing analysis
Isolated RNA at a concentration of 20 ng/µl was further purified with a TruSeq mRNA v2Kit (polyA, Illumina). Biological triplicates were sequenced on a HiSeq2500 using paired-end chemistry and 2×125 cycles. The sequence depth was ∼100 million reads per sample. Runs were performed by deCODE Genetics.
To assess the differentiation state of generated CNs, cross-platform comparisons with spatiotemporal data from the developing human brain of the BrainSpan atlas were performed by pairwise comparison of generated CNs with each BrainSpan atlas sample. By ranking gene expression for each pairwise comparison, rank difference values for all genes were used to calculate Spearman’s rank correlation coefficients. A Wilcoxon rank-sum test was applied if a category of interest (neocortex, subcortex, ganglionic eminence or cerebellum) had significantly higher Spearman’s correlation coefficients than the background of all paired correlations. –Log 10 P-values of significant differences of CNs and spatiotemporal brain data were shown in heat maps.
Mass spectrometry
Equal amounts of samples were purified using SDS PAGE (Invitrogen). Coomassie blue-stained gel pieces were excised and in-gel digested using trypsin as described previously (Borchert et al., 2010). Extracted peptides were desalted using C18 StageTips and subjected to LC/MS-MS analysis, performed on an Easy nano-LC (Thermo Fisher Scientific) coupled to an LTQ Orbitrap Elite (Thermo Fisher Scientific) as described previously (Franz-Wachtel et al., 2012). Equal amounts of the peptide mixtures were injected onto the column in high-performance LC (HPLC) solvent A (0.1% formic acid) at a flow rate of 500 nl/min and subsequently eluted with a 127 min segmented gradient of 5–33-50-90% of HPLC solvent B (80% acetonitrile in 0.1% formic acid) at a flow rate of 200 nl/min. Using an Orbitrap Elite, the 15 most intense precursor ions were sequentially fragmented in each scan cycle. In all measurements, sequenced precursor masses were excluded from further selection for 60 s. The target values were 5000 charges for MS/MS fragmentation and 106 charges for the MS scan. CNs of three controls (CO4, CO5 and CO6) and three STUB1 patients (STUB1_1, STUB1_2 and STUB1_3) were analyzed.
MS data processing
The data were processed using MaxQuant software suite v.1.5.2.8 (Cox and Mann, 2008). The database search was performed with MaxQuant, using the Andromeda search engine (Cox et al., 2011). MS/MS spectra were searched against a target-decoy Uniprot database consisting of 95,972 protein entries from Homo sapiens and 245 commonly observed contaminants. Full specificity was required for trypsin; up to two missed cleavages were allowed. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine and acetylation of the N-terminus were set as variable modifications. Initial mass tolerance was set to 4.5 ppm for precursor ions and 0.5 Da for fragment ions. Peptide, protein and modification site identifications were reported at a false discovery rate of 0.01, estimated by the target/decoy approach (Elias and Gygi, 2007). The label-free algorithm was enabled, as was the ‘match between runs’ option (Luber et al., 2010). LFQ protein intensities from the MaxQuant data output were used for relative protein quantification. Downstream bioinformatical analysis (two-sample t-tests and volcano plots) was performed using the Perseus software package, version 1.5.0.15. The data were filtered for contaminants, reverse and only identified by site entries.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (version 8). One-way ANOVA or a two-tailed unpaired Student's t-test was applied. Statistical analysis was restricted to inter-biological comparison for the same experimental condition. P-values were corrected for multiple comparisons and P<0.05 was considered statistically significant. Unless indicated otherwise, all data are shown as mean±s.e.m.
Supplementary Material
Acknowledgements
We are grateful to Jonathan Schisler for the recommendation of the specific CHIP antibody. We acknowledge Melanie Kraft and Yvonne Schelling for their excellent technical assistance. L.S. and M.S. are members of the European Reference Network for Rare Neurological Diseases (Project ID 739510).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., S.H., L.S.; Methodology: S.S., S.H., L.S.; Validation: S.S., E.H.; Formal analysis: S.S., A.V., J.A.; Investigation: S.S., E.H., A.V., J.A.; Resources: M.S., L.S.; Writing - original draft: S.S., S.H., L.S.; Writing - review & editing: S.S., E.H., A.V., J.A., M.S., S.O., B.M., S.H., L.S.; Visualization: S.S.; Supervision: S.S., S.O., B.M., S.H., L.S.; Project administration: S.S., S.H., L.S.; Funding acquisition: L.S.
Funding
This work was supported by the Bundesministerium für Bildung und Forschung within the frame of E-Rare (01GM1905A to S.H. and L.S., and 01GM1907A to L.S.).
Data availability
RNA-Seq raw data available upon request to the corresponding author.
Supplementary information
Supplementary information available online at https://dmm.biologists.org/lookup/doi/10.1242/dmm.045096.supplemental
References
- Abravaya K., Phillips B. and Morimoto R. I. (1991). Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperatures. Genes Dev. 5, 2117-2127. 10.1101/gad.5.11.2117 [DOI] [PubMed] [Google Scholar]
- Acunzo J., Katsogiannou M. and Rocchi P. (2012). Small heat shock proteins HSP27 (HspB1), alphaB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int. J. Biochem. Cell Biol. 44, 1622-1631. 10.1016/j.biocel.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Anderson L. G., Meeker R. B., Poulton W. E. and Huang D. Y. (2010). Brain distribution of carboxy terminus of Hsc70-interacting protein (CHIP) and its nuclear translocation in cultured cortical neurons following heat stress or oxygen-glucose deprivation. Cell Stress Chaperones 15, 487-495. 10.1007/s12192-009-0162-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler R., Welch W. J. and Voellmy R. (1992). Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 117, 1151-1159. 10.1083/jcb.117.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L.-Y. and Patterson C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535-4545. 10.1128/MCB.19.6.4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batulan Z., Shinder G. A., Minotti S., He B. P., Doroudchi M. M., Nalbantoglu J., Strong M. J. and Durham H. D. (2003). High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J. Neurosci. 23, 5789-5798. 10.1523/JNEUROSCI.23-13-05789.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt C., de Yébenes J. G., López-Sendón J. L., Shomroni O., Zhang X., Qian S.-B., Bakker I. M. C., Heetveld S., Ros R., Quintáns B. et al. (2015). Clinical and neuropathological features of spastic ataxia in a spanish family with novel compound heterozygous mutations in STUB1. Cerebellum 14, 378-381. 10.1007/s12311-014-0643-7 [DOI] [PubMed] [Google Scholar]
- Blake M. J., Nowak T. S. Jr. and Holbrook N. J. (1990). In vivo hyperthermia induces expression of HSP70 mRNA in brain regions controlling the neuroendocrine response to stress. Brain Res. Mol. Brain Res. 8, 89-92. 10.1016/0169-328X(90)90014-5 [DOI] [PubMed] [Google Scholar]
- Borchert N., Dieterich C., Krug K., Schutz W., Jung S., Nordheim A., Sommer R. J. and Macek B. (2010). Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837-846. 10.1101/gr.103119.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon R. H. (1987). Thermotolerance and the heat shock proteins. Symp. Soc. Exp. Biol. 41, 269-283. [PubMed] [Google Scholar]
- Chapple J. P. and Cheetham M. E. (2003). The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J. Biol. Chem. 278, 19087-19094. 10.1074/jbc.M212349200 [DOI] [PubMed] [Google Scholar]
- Cheetham M. E., Brion J. P. and Anderton B. H. (1992). Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem. J. 284, 469-476. 10.1042/bj2840469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367-1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V. and Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794-1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L. et al. (2003). CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22, 5446-5458. 10.1093/emboj/cdg529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. and Goldberg M. S. (2009). Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS ONE 4, e5949 10.1371/journal.pone.0005949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E. and Gygi S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207-214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Feder J. H., Rossi J. M., Solomon J., Solomon N. and Lindquist S. (1992). The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 6, 1402-1413. 10.1101/gad.6.8.1402 [DOI] [PubMed] [Google Scholar]
- Ferreira J. V., Soares A. R., Ramalho J. S., Pereira P. and Girao H. (2015). K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci. Rep. 5, 10210 10.1038/srep10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz-Wachtel M., Eisler S. A., Krug K., Wahl S., Carpy A., Nordheim A., Pfizenmaier K., Hausser A. and Macek B. (2012). Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol. Cell. Proteomics 11, 160-170. 10.1074/mcp.M111.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R., Burchfiel E. T. and Thiele D. J. (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4-19. 10.1038/nrm.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Ying Z., Wang H., Chen D., Gao F., Ren H. and Wang G. (2015). Regulation of autophagic flux by CHIP. Neurosci. Bull. 31, 469-479. 10.1007/s12264-015-1543-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J. and Kampinga H. H. (2009). Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 14, 1-21. 10.1007/s12192-008-0060-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer S. N., Deconinck T., Bender B., Smets K., Zuchner S., Reich S., Schöls L., Schüle R., De Jonghe P., Baets J. et al. (2017). STUB1/CHIP mutations cause Gordon Holmes syndrome as part of a widespread multisystemic neurodegeneration: evidence from four novel mutations. Orphanet J. Rare Dis. 12, 31 10.1186/s13023-017-0580-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimdal K., Sanchez-Guixé M., Aukrust I., Bollerslev J., Bruland O., Jablonski G. E., Erichsen A. K., Gude E., Koht J. A., Erdal S. et al. (2014). STUB1 mutations in autosomal recessive ataxias - evidence for mutation-specific clinical heterogeneity. Orphanet J. Rare Dis. 9, 146 10.1186/s13023-014-0146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana N. R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K. and Nukina N. (2005). Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 280, 11635-11640. 10.1074/jbc.M412042200 [DOI] [PubMed] [Google Scholar]
- Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J. and Patterson C. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938-42944. 10.1074/jbc.M101968200 [DOI] [PubMed] [Google Scholar]
- Joshi V., Amanullah A., Upadhyay A., Mishra R., Kumar A. and Mishra A. (2016). A decade of boon or burden: what has the CHIP ever done for cellular protein quality control mechanism implicated in neurodegeneration and aging? Front. Mol. Neurosci. 9, 93 10.3389/fnmol.2016.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanack A. J., Newsom O. J. and Scaglione K. M. (2018). Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP. J. Biol. Chem. 293, 2735-2743. 10.1074/jbc.RA117.000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-A., Yoon J.-H., Kim D.-K., Kim S.-G. and Ahn S.-G. (2005). CHIP interacts with heat shock factor 1 during heat stress. FEBS Lett. 579, 6559-6563. 10.1016/j.febslet.2005.10.043 [DOI] [PubMed] [Google Scholar]
- Kumar P., Ambasta R. K., Veereshwarayya V., Rosen K. M., Kosik K. S., Band H., Mestril R., Patterson C. and Querfurth H. W. (2007). CHIP and HSPs interact with beta-APP in a proteasome-dependent manner and influence Abeta metabolism. Hum. Mol. Genet. 16, 848-864. 10.1093/hmg/ddm030 [DOI] [PubMed] [Google Scholar]
- Li Y., Chopp M., Yoshida Y. and Levine S. R. (1992). Distribution of 72-kDa heat-shock protein in rat brain after hyperthermia. Acta Neuropathol. 84, 94-99. 10.1007/BF00427221 [DOI] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E. J., Shi Z. and Zhang B. (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199-W205. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizama B. N., Palubinsky A. M., Raveendran V. A., Moore A. M., Federspiel J. D., Codreanu S. G., Liebler D. C. and McLaughlin B. (2018). Neuronal Preconditioning Requires the Mitophagic Activity of C-terminus of HSC70-Interacting Protein. J. Neurosci. 38, 6825-6840. 10.1523/JNEUROSCI.0699-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M. et al. (2010). Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279-289. 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Madrigal S. C., McNeil Z., Sanchez-Hodge R., Shi C.-H., Patterson C., Scaglione K. M. and Schisler J. C. (2019). Changes in protein function underlie the disease spectrum in patients with CHIP mutations. J. Biol. Chem. 294, 19236-19245. 10.1074/jbc.RA119.011173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcuccilli C. J., Mathur S. K., Morimoto R. I. and Miller R. J. (1996). Regulatory differences in the stress response of hippocampal neurons and glial cells after heat shock. J. Neurosci. 16, 478-485. 10.1523/JNEUROSCI.16-02-00478.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. M., Nelson R. F., Gouvion C. M., Williams A., Rodriguez-Lebron E., Harper S. Q., Davidson B. L., Rebagliati M. R. and Paulson H. L. (2005). CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 25, 9152-9161. 10.1523/JNEUROSCI.3001-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.-N., Whaley R. A., Sharpless N. E., Lockyer P., Portbury A. L. and Patterson C. (2008). CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28, 4018-4025. 10.1128/MCB.00296-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y., Wang A. M., Yu Z., Pratt W. B., Osawa Y. and Lieberman A. P. (2008). CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum. Mol. Genet. 17, 3942-3952. 10.1093/hmg/ddn296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S., Chiba T. and Tanaka K. (2003). CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int. J. Biochem. Cell Biol. 35, 572-578. 10.1016/S1357-2725(02)00394-1 [DOI] [PubMed] [Google Scholar]
- Nishimura R. N., Dwyer B. E., Clegg K., Cole R. and de Vellis J. (1991). Comparison of the heat shock response in cultured cortical neurons and astrocytes. Brain Res. Mol. Brain Res. 9, 39-45. 10.1016/0169-328X(91)90128-K [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. et al. (2011). A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409-412. 10.1038/nmeth.1591 [DOI] [PubMed] [Google Scholar]
- Pakdaman Y., Sanchez-Guixe M., Kleppe R., Erdal S., Bustad H. J., Bjorkhaug L., Haugarvoll K., Tzoulis C., Heimdal K., Knappskog P. M. et al. (2017). In vitro characterization of six STUB1 variants in spinocerebellar ataxia 16 reveals altered structural properties for the encoded CHIP proteins. Biosci. Rep. 37, BSR20170251 10.1042/BSR20170251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S. B., McDonough H., Boellmann F., Cyr D. M. and Patterson C. (2006). CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440, 551-555. 10.1038/nature04600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi S., Parry T. L., Willis M. S., Lockyer P., Patterson C., Bain J. R., Stevens R. D., Ilkayeva O. R., Newgard C. B. and Schisler J. C. (2018). Adverse effects of fenofibrate in mice deficient in the protein quality control regulator, CHIP. J. Cardiovasc. Dev. Dis. 5, 43 10.3390/jcdd5030043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbach K., Kesavan J., Hauser S., Ritzenhofen S., Jungverdorben J., Schüle R., Schöls L., Peitz M. and Brüstle O. (2019). Multiparametric rapid screening of neuronal process pathology for drug target identification in HSP patient-specific neurons. Sci. Rep. 9, 9615 10.1038/s41598-019-45246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K., Haslbeck M. and Buchner J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253-266. 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Delle Donne R., Catalanotti B., Torres-Quesada O., Enzler F., Moraca F., Nisticò R., Chiuso F., Piccinin S., Bachmann V. et al. (2019). Feedback inhibition of cAMP effector signaling by a chaperone-assisted ubiquitin system. Nat. Commun. 10, 2572 10.1038/s41467-019-10037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnebaum S. M., Patterson C. and Schisler J. C. (2014). Emerging evidence of coding mutations in the ubiquitin-proteasome system associated with cerebellar ataxias. Hum. Genome Var. 1, 14018 10.1038/hgv.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara N., Murayama M., Mizoroki T., Urushitani M., Imai Y., Takahashi R., Murata S., Tanaka K. and Takashima A. (2005). In vivo evidence of CHIP up-regulation attenuating tau aggregation. J. Neurochem. 94, 1254-1263. 10.1111/j.1471-4159.2005.03272.x [DOI] [PubMed] [Google Scholar]
- San Gil R., Ooi L., Yerbury J. J. and Ecroyd H. (2017). The heat shock response in neurons and astroglia and its role in neurodegenerative diseases. Mol. Neurodegener. 12, 65 10.1186/s13024-017-0208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M. G. (2000). Heat shock factors and the control of the stress response. Biochem. Pharmacol. 59, 55-63. 10.1016/S0006-2952(99)00299-3 [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Murphy S. P. and Morimoto R. I. (1993). Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13, 1392-1407. 10.1128/MCB.13.3.1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J.-I. and Kim S. U. (1995). Differential expression of heat shock protein HSP27 in human neurons and glial cells in culture. J. Neurosci. Res. 41, 805-818. 10.1002/jnr.490410611 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisler J. C., Rubel C. E., Zhang C., Lockyer P., Cyr D. M. and Patterson C. (2013). CHIP protects against cardiac pressure overload through regulation of AMPK. J. Clin. Invest. 123, 3588-3599. 10.1172/JCI69080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S., Saravanakumar S., Schöls L. and Hauser S. (2019). Generation of a homozygous CRISPR/Cas9-mediated knockout human iPSC line for the STUB1 locus. Stem Cell Res. 34, 101378 10.1016/j.scr.2018.101378 [DOI] [PubMed] [Google Scholar]
- Schuster S., Schelling Y., Synofzik M., Höflinger P., Schols L. and Hauser S. (2018). Establishment of STUB1/CHIP mutant induced pluripotent stem cells (iPSCs) from a patient with Gordon Holmes syndrome/SCAR16. Stem Cell Res. 29, 166-169. 10.1016/j.scr.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Seo J., Lee E.-W., Sung H., Seong D., Dondelinger Y., Shin J., Jeong M., Lee H.-K., Kim J.-H., Han S. Y. et al. (2016). CHIP controls necroptosis through ubiquitylation- and lysosome-dependent degradation of RIPK3. Nat. Cell Biol. 18, 291-302. 10.1038/ncb3314 [DOI] [PubMed] [Google Scholar]
- Sha Y., Rao L., Settembre C., Ballabio A. and Eissa N. T. (2017). STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 36, 2544-2552. 10.15252/embj.201796699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P. and Livesey F. J. (2012). Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 7, 1836-1846. 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang J., Li J.-D., Ren H., Guan W., He M., Yan W., Zhou Y., Hu Z., Zhang J. et al. (2013). Identification of CHIP as a novel causative gene for autosomal recessive cerebellar ataxia. PLoS ONE 8, e81884 10.1371/journal.pone.0081884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-H., Schisler J. C., Rubel C. E., Tan S., Song B., McDonough H., Xu L., Portbury A. L., Mao C.-Y., True C. et al. (2014). Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP. Hum. Mol. Genet. 23, 1013-1024. 10.1093/hmg/ddt497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-H., Rubel C., Soss S. E., Sanchez-Hodge R., Zhang S., Madrigal S. C., Ravi S., McDonough H., Page R. C., Chazin W. J. et al. (2018). Disrupted structure and aberrant function of CHIP mediates the loss of motor and cognitive function in preclinical models of SCAR16. PLoS Genet. 14, e1007664 10.1371/journal.pgen.1007664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Klucken J., Patterson C., Hyman B. T. and McLean P. J. (2005). The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J. Biol. Chem. 280, 23727-23734. 10.1074/jbc.M503326200 [DOI] [PubMed] [Google Scholar]
- Singh A. K. and Pati U. (2015). CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of beta-secretase. Aging Cell 14, 595-604. 10.1111/acel.12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M., Schüle R., Schulze M., Gburek-Augustat J., Schweizer R., Schirmacher A., Krägeloh-Mann I., Gonzalez M., Young P., Züchner S. et al. (2014). Phenotype and frequency of STUB1 mutations: next-generation screenings in Caucasian ataxia and spastic paraplegia cohorts. Orphanet J. Rare Dis. 9, 57 10.1186/1750-1172-9-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.-B., Li Y.-S., Li S.-H., Cheng Y., Zhang S., Luo H.-Y., Mao C.-Y., Hu Z.-W., Schisler J. C., Shi C.-H. et al. (2018). Anisomycin prevents OGD-induced necroptosis by regulating the E3 ligase CHIP. Sci. Rep. 8, 6379 10.1038/s41598-018-24414-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff J. E., Putcha P., Outeiro T. F., Ivanov A., Berezovska O., Hyman B. T. and McLean P. J. (2008). CHIP targets toxic α-Synuclein oligomers for degradation. J. Biol. Chem. 283, 17962-17968. 10.1074/jbc.M802283200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A., Zhang D., Liu J. and Oliferenko S. (2013). Hsp70-Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1-driven heat stress-associated transcription. PLoS Genet. 9, e1003886 10.1371/journal.pgen.1003886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P., Dux E. and Wiessner C. (1997). Effect of heat shock on neuronal cultures: importance of protein synthesis and HSP72 induction for induced tolerance and survival. Metab. Brain Dis. 12, 203-217. 10.1007/BF02674613 [DOI] [PubMed] [Google Scholar]
- Volloch V. Z. and Sherman M. Y. (1999). Oncogenic potential of Hsp72. Oncogene 18, 3648-3651. 10.1038/sj.onc.1202525 [DOI] [PubMed] [Google Scholar]
- Vonk W. I. M., Rainbolt T. K., Dolan P. T., Webb A. E., Brunet A. and Frydman J. (2020). Differentiation drives widespread rewiring of the neural stem cell chaperone network. Mol. Cell 78, 329-345.e9. 10.1016/j.molcel.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.-P. (2010). The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer 1, 779-786. 10.1177/1947601910383277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C. and Pearl L. H. (2005). Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525-538. 10.1016/j.molcel.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Zhang W., Liu Z., Bao X., Qin Y., Taylor A., Shang F. and Wu M. (2015). CHIP knockdown reduced heat shock response and protein quality control capacity in lens epithelial cells. Curr. Mol. Med. 15, 652-662. 10.2174/1566524015666150831131636 [DOI] [PubMed] [Google Scholar]
- Zhao T., Hong Y., Yin P., Li S. and Li X.-J. (2017). Differential HspBP1 expression accounts for the greater vulnerability of neurons than astrocytes to misfolded proteins. Proc. Natl. Acad. Sci. USA 114, E7803-E7811. 10.1073/pnas.1710549114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.