ABSTRACT
Most research on the impact of the gut microbiome on animal nutrition is designed to identify the effects of single microbial taxa and single metabolites of microbial origin, without considering the potentially complex network of interactions among co-occurring microorganisms. Here, we investigated how different microbial associations and their fermentation products affect host nutrition, using Drosophila melanogaster colonized with three gut microorganisms (the bacteria Acetobacter fabarum and Lactobacillus brevis, and the yeast Hanseniaspora uvarum) in all seven possible combinations. Some microbial effects on host traits could be attributed to single taxa (e.g. yeast-mediated reduction of insect development time), while other effects were sex specific and driven by among-microbe interactions (e.g. male lipid content determined by interactions between the yeast and both bacteria). Parallel analysis of nutritional indices of microbe-free flies administered different microbial fermentation products (acetic acid, acetoin, ethanol and lactic acid) revealed a single consistent effect: that the lipid content of both male and female flies is reduced by acetic acid. This effect was recapitulated in male flies colonized with both yeast and A. fabarum, but not for any microbial treatment in females or males with other microbial complements. These data suggest that the effect of microbial fermentation products on host nutritional status is strongly context dependent, with respect to both the combination of associated microorganisms and host sex. Taken together, our findings demonstrate that among-microbe interactions can play a critically important role in determining the physiological outcome of host–microbiome interactions in Drosophila and, likely, in other animal hosts.
KEY WORDS: Acetobacter, Lipid content, Host–microbe interactions, Hanseniaspora, Lactobacillus, Microbial community
Summary: The impact of individual microorganisms in the Drosophila gut microbiome on insect nutrition, particularly lipid storage, is strongly dependent on both the presence of co-occurring microorganisms and host sex.
INTRODUCTION
The gut of many animals is colonized by a diverse community of microorganisms (collectively known as the microbiome) that can influence the health and fitness of the animal host. The gut microbiome contributes to various host functions, including the degradation of dietary constituents, synthesis of essential nutrients, modulation of host immunity and protection against pathogens (Herp et al., 2019; Hooper et al., 2012; Huang et al., 2015; Karasov et al., 2011; Read and Holmes, 2017; Rolhion and Chassaing, 2016; Sommer and Bäckhed, 2013; Thaiss et al., 2016; Wong et al., 2016). Numerous studies have linked specific microorganisms and microbial metabolites with host traits (e.g. Morton et al., 2019; Tripathi et al., 2018). However, it is increasingly recognized that the traits of individual microorganisms can be influenced by interactions with other members of the microbiome (Douglas, 2020; Kundu et al., 2019; Noecker et al., 2019). Theoretical models have predicted that among-microbe interactions, which range from beneficial to antagonistic and are widespread throughout microbial communities, can have varying levels of metabolic dependencies and cross-feeding of metabolites between taxa (Coyte et al., 2015; Freilich et al., 2011; Levy and Borenstein, 2013; Magnúsdóttir et al., 2017; Noecker et al., 2019; Zelezniak et al., 2015). These interactions can lead to the net production of metabolites that none of the individual taxa in the community are able to synthesize in isolation (Wintermute and Silver, 2010). As a result, data for associations between animals and single microbial taxa may not accurately predict how individual taxa perform in a complex community.
A valuable approach to investigate among-microbial interactions is to use in vitro and simplified gnotobiotic animal systems with pairs or small groups of microbial taxa. For example, some Bacteriodes spp. in the human gut degrade dietary polysaccharides and the simple carbohydrates liberated support the growth of co-occurring taxa and promote the release of metabolites that influence human health (Mahowald et al., 2009; Rakoff-nahoum et al., 2016; Rodriguez-Castaño et al., 2019). In the lower termite gut, folate-producing bacteria release 5-formyl-tetrahydrofolate and the spirochete Treponema primitia uses this metabolite as a co-factor to produce acetic acid, an energy source for the insect host (Graber and Breznak, 2005). In the honey bee gut microbiome, numerous candidate cross-feeding events have been identified between the bacteria Bifidobacterium spp., Gilliamella apicola and Snodgrassella alvi, resulting in improved digestion of the pollen diet and utilization of bacterial by-products by co-occurring taxa (Kešnerová et al., 2017; Zheng et al., 2019).
The focus of this study was the impact of among-microbe interactions on the nutritional physiology of the fruit fly Drosophila melanogaster, which is a fast-emerging model for gut–microbiome interactions (Broderick and Lemaitre, 2012; Douglas, 2019; Erkosar et al., 2013; Wong et al., 2016). Three taxonomic groups, the bacteria of the family Acetobacteraceae and order Lactobacillales, and the Saccharomycetales yeasts, are the dominant taxa in the Drosophila gut microbiome (Adair et al., 2018; Chandler et al., 2011, 2012; Quan and Eisen, 2018; Wong et al., 2011). To date, most research has focused either on the bacteria or yeast partners, with evidence that co-associations involving between two and five bacterial species can recapitulate the effects of conventional multi-species communities on certain traits under laboratory conditions (Gould et al., 2018; Newell and Douglas, 2014; Rohlfs and Kürschner, 2010). For example, D. melanogaster reared in co-association with pairs of Acetobacter and Lactobacillus species likely display cooperative metabolism and lower fly lipid content relative to microbe-free flies (Consuegra et al., 2020; Newell and Douglas, 2014; Sommer and Newell, 2019). Indications that interactions between bacteria and yeasts may be important determinants of Drosophila traits come largely from a single study (Fischer et al., 2017), which demonstrated that female flies prefer to both feed and lay eggs on substrate bearing a Saccharomyces–Acetobacter co-culture relative to mono-cultures and an Acetobacter strain unable to produce acetic acid. Furthermore, research on both Acetobacter–Lactobacillus interactions and Acetobacter–Saccharomyces interactions (Fischer et al., 2017; Sommer and Newell, 2019) has identified four microbial metabolites as candidate mediators of among-microbe and microbe–host interactions: acetic acid, acetoin, ethanol and lactic acid. Importantly, these four metabolites have also been implicated in the regulation of Drosophila metabolism, immunity and behavior (Devineni and Heberlein, 2013; Farine et al., 2017; Fry, 2014; Hang et al., 2014; Hoffmann and Parsons, 1984; Iatsenko et al., 2018; Kamareddine et al., 2018; Shin et al., 2011).
The specific goal of this study was to understand whether and how among-microbe interactions may influence Drosophila performance and nutrient allocation. We used three representative strains of gut microorganisms: two bacteria, Acetobacter fabarum and Lactobacillus brevis, widely used in Drosophila microbiome research (Dobson et al., 2015; Sommer and Newell, 2019; White et al., 2018); and the yeast Hanseniaspora uvarum, a prevalent species in wild Drosophila populations (Chandler et al., 2012; De Camargo and Phaff, 1957) that has also been used for laboratory experiments (Hoang et al., 2015; Murgier et al., 2019; Palanca et al., 2013; Scheidler et al., 2015; Solomon et al., 2019). We administered the microorganisms combinatorially to give associations with one, two or all three taxa. The Drosophila in these treatments are described as gnotobiotic, meaning that they have a defined complement of microorganisms. In parallel, we quantified the effect of four microbial fermentation products, acetic acid, acetoin, ethanol and lactic acid, on nutrient allocation in flies reared under microbiologically sterile conditions (known as axenic flies) and linked these results to the fermentation product profiles in the axenic and gnotobiotic flies. This study demonstrates how non-additive among-microbe effects can influence Drosophila performance and nutrition, and it establishes the contribution of individual microbial metabolites in the observed among-microbe interactions.
MATERIALS AND METHODS
Insects and microorganisms
The three microorganisms were: Acetobacter fabarum DsW54 (ACE), Lactobacillus brevis DmCS_003 (LAC) and Hanseniaspora uvarum FYH04C (YST). Details of their provenance and culture protocols are provided in Table 1.
Table 1.
Microbial strains
The stock culture of D. melanogaster strain Canton S (Wolbachia free) was maintained at 25°C with 50% relative humidity on a 12 h:12 h light:dark cycle on a yeast–glucose (Y–G) diet comprising 10% inactive brewer's yeast (MP Biomedicals), 10% glucose (Sigma) and 1.2% Drosophila type II agar (Apex) with preservatives 0.04% phosphoric acid (Fisher Scientific) and 0.42% propionic acid (Fisher Scientific).
Axenic flies were prepared by a standard protocol for egg dechorionation (Koyle et al., 2016). Briefly, flies from the stock culture were allowed to oviposit for 16–18 h on grape juice agar plates (Y–G diet with 1% Drosophila agar and ∼15% Welch's grape juice concentrate), and the deposited eggs were thrice washed in 0.6% hypochlorite solution for 5 min, followed by three rinses in sterile deionized water. Approximately 60 eggs were aseptically transferred to 50 ml sterile conical Falcon tubes (Globe Scientific, Inc.) containing 7.5 ml autoclaved Y–G diet without preservatives, and they were raised under standard culture conditions to generate axenic flies.
To produce gnotobiotic insects colonized with one, two or all three microbial strains, microorganisms were administered aseptically to Falcon tubes containing dechorionated eggs at a density of 5×106 cells (50 μl suspension of microorganism at 1×108 cells ml−1) in phosphate-buffered saline (PBS) (Cold Spring Harbor, 2018). For treatments containing multiple microbial species, the taxa were added in equal proportion to give the same total concentration. To prepare the microorganisms, a loop of frozen glycerol stock was streaked onto solid medium and, following 1–2 days of growth, a single colony was picked and grown in 5 ml broth to mid-logarithmic phase (see Table 1 for details). Cell densities were determined by optical density at 600 nm (OD600) on 96-well plates (Globe Scientific, Inc.) using an xMark Spectrophotometer (BioRad) and diluted to the required cell number based on calibration curves of OD600 against number of colony-forming units (CFUs) constructed for each strain (Table 1).
Experimental design
The eight treatments comprised axenic insects and seven gnotobiotic treatments of insects administered every combination of mono-, di- and tri-association with ACE, LAC and YST, with six replicates (i.e. Falcon tubes) per treatment. The insects were reared from dechorionated eggs under the standard culture conditions and scored twice daily (2 and 10 h post-dawn) for number of pupae and number of empty puparia to obtain time to pupation and eclosion, respectively. On the first day post-eclosion (dpe), the flies in each Falcon tube were aseptically transferred to a fresh Falcon tube containing autoclaved Y–G diet. At 5 dpe, the flies were anesthetized using CO2, separated by sex by visualizing morphological differences in genitalia, counted, and analyzed with one sample of each sex per Falcon tube for microbial load (two flies per sample) and nutritional indices (five flies per sample). In addition, ca. 20 mg (10–20 flies per sample) of each sex from two Falcon tubes per treatment were used for quantification of short chain fatty acids (SCFAs); the number of flies available for quantification varied with treatment. The mass of each sample used for nutritional indices and SCFA content was determined with a Mettler-Toledo balance, to an accuracy of 0.1 mg. All samples were flash frozen in liquid nitrogen and stored at −80°C. This experiment was replicated 3 times independently to generate a total of 18 replicates of males and females for each treatment.
Microbial load in flies
The abundance of each microbial strain per fly of each sex at 5 dpe was scored as number of CFUs. Two male flies and two female flies from each Falcon tube were surface sterilized in 500 μl 0.3% hypochlorite solution in a sterile 1.5 ml centrifuge tube with gentle shaking by hand, followed by two rinses in sterile PBS. The flies were transferred to a sterile 2 ml screw-cap microcentrifuge tube containing 100 µl sterile lysis matrix D beads (MP Biomedicals) and 200 µl sterile PBS, homogenized using a FastPrep-24 instrument at 4.0 m s−1 for 30 s, diluted with 800 µl sterile PBS, and then inoculated in duplicate onto the appropriate medium (Table 1) using a WASP-2 spiral plater (Microbiology International). A Protocol3 instrument (Microbiology International) was utilized to enumerate CFUs after 2–3 days of growth at 30°C, and the data were normalized to CFUs per fly. The detection limit was 10 CFUs per fly; this value was added for statistical analysis to eliminate zeros from microbial treatments in the dataset.
To facilitate the automated counting, samples from di- and tri-associations were plated onto media supplemented with antimicrobial(s) that selectively suppressed one of the microorganisms: 100 µg ml−1 kanamycin sulfate (Sigma) to suppress ACE, 10 µg ml−1 ampicillin sodium salt (Sigma) to suppress LAC, 500 µg ml−1 methylparaben (Apex) under high CO2 on mMRS medium [modified De Man, Rogosa and Sharpe medium: 1.25% bacto-proteose peptone, 0.75% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% dipotassium hydrogen phosphate, 0.2% triammonium citrate, 0.02% magnesium sulfate heptahydrate, 0.005% manganese sulfate tetrahydrate and 1.2% agar (for plates only); all ingredients from Sigma, except bacto-proteose peptone from Becton Dickinson] to suppress YST (Fig. S1A–D). Preliminary experiments confirmed that each antimicrobial did not suppress growth of the non-target microorganisms (Fig. S1E), apart from methylparaben, which had a negative effect on ACE growth and was not used in plates selecting for ACE growth (Fig. S1B). For ACE and YST co-associations, colonies were differentiated by morphology: ACE has small orange/tan colonies and YST has large off-white colonies.
Negative controls comprised axenic flies plated onto mMRS and YPD [yeast–peptone–dextrose medium: 1% yeast extract, 2% bacto-peptone, 2% glucose and 1.5% agar (for plates only); all ingredients from Sigma, except bacto-peptone from Becton Dickinson] agar without antimicrobials and incubated at 30°C for 1 week. Individual replicates were discarded if microbial contamination was present on plates for the entire experiment. Microbial biomass (dry mass) was estimated following the method of Norland et al. (1987), using estimates of biovolume (ACE 1.1 μm3, LAC 1.7 μm3, YST 38.7 μm3) derived from published data (Cleenwerck et al., 2008; Gries and Ly, 2009; Cadez and Smith, 2011).
Nutritional indices
The lipid, carbohydrate and protein content of the flies was quantified by the procedure of Newell and Douglas (2014) as an index of fly nutritional status. Briefly, each fly sample was homogenized in 125 μl ice-cold TET buffer (10 mmol l−1 Tris pH 8, 1 mmol l−1 EDTA, 0.1% Triton X-100) and 100 μl lysis matrix D beads, using a FastPrep-24 at 4 m s−1 for 45 s. For protein analysis, 10 μl was removed from the homogenate and diluted 1:6 for males and 1:7 for females in ice-cold TET buffer. The remaining homogenate was incubated at 72°C for 30 min to inactivate endogenous enzymes. Samples were stored at −80°C prior to quantification.
Protein content was determined using the DC Protein Assay Kit (Bio-Rad) according to manufacturer's protocol, with a standard curve using 0–1.4 mg ml−1 bovine serum albumin. For lipid content, the triacylglyceride (TAG) content was determined by incubating 5 μl heat-inactivated homogenate with 37.5 μl lipase (20 U ml−1 in 20 mmol l−1 potassium phosphate, 20 mmol l−1 EDTA and 20 mmol l−1 magnesium chloride, pH 7.5; Sigma L9518), followed by assay of liberated glycerol, using the Free Glycerol Reagent (Sigma-Aldrich F6428) following the manufacturer's protocol with 0–3 mg ml−1 triolein equivalent glycerol for the standards. Absorbance attributed to endogenous free glycerol was subtracted from TAG for quantification. Glucose, trehalose and glycogen were assayed with the glucose kit (Sigma GAGO20) following the manufacturer's protocol, with 0–0.8 mg ml−1 glucose, 0–0.2 mg ml−1 trehalose and 0–0.2 mg ml−1 glycogen as standards. For trehalose and glycogen determination, 2.5 μl trehalase (1 U ml−1; Sigma T0167) and 5 μl amyloglucosidase (2 U ml−1 in 5 mmol l−1 acetic acid, 5 mmol l−1 sodium acetate; Sigma A7420), respectively, were added to samples and incubated for 1 h at 37°C prior to quantification of glucose. For trehalose measurements, 2 μl 1 mmol l−1 EDTA and 2 μl 0.2 mol l−1 sodium citrate dihydrate were added to samples and incubated for 10 min at 37°C prior to the addition of the enzyme. All samples and standards were assayed in duplicate, and the absorbance values of the two technical replicates were averaged. Final absolute quantifications for all nutritional indices were normalized to fly mass.
SCFA content
SCFAs were assayed using the propyl esterification derivatization method of Cai et al. (2017) on ca. 20 mg whole bodies of pooled flies for each sex, with quantification by an Agilent 7890A gas chromatograph coupled with an Agilent 5975 mass spectrometer (GC-MS, Agilent Technologies, Santa Clara, CA, USA) at the Penn State Metabolomics Facility, University Park, PA, USA. A targeted analysis was performed to quantify the following 12 SCFAs: acetate, propionate, butyrate, isobutyrate, isovalerate, valerate, 2-methylbutyrate, 2-methylpentanoate, 3-methylpentanoate, 2-methylhexanoate, 4-methylvalerate and heptanoate. Each fly sample was homogenized at 6500 rpm for 60 s (Precellys, Bertin Technologies, Rockville, MD, USA) in 1 ml 5 mmol l−1 NaOH, with 10 μg ml−1 caproic acid-6,6,6-d3 (internal standard) using 1.0 mm diameter zirconia/silica beads (BioSpec, Bartlesville, OK, USA), and centrifuged at 13,200 g for 20 min at 4°C; 500 μl of sample supernatant was added to 500 μl 1-propanol:pyridine (3:2 v/v), followed by addition of 100 μl propyl chloroformate (esterification reagent). Then, samples were vortexed for 1 min, with a subsequent 1 h incubation at 60°C. The derivatized samples were extracted with a two-step hexane extraction (300 μl+200 μl) following the procedure of Zheng et al. (2013), yielding 500 μl, which was transferred to a glass autosampler vial. A standard curve was generated for each analyte to quantify biological concentration (metabolite amount normalized to fly fresh mass in μmol g−1) of SCFAs. The data was range scaled to normalize SCFA titer across three experimental replicates; values were centered to the mean metabolite concentration and scaled by the range of metabolites quantified in each experimental replicate (van de Berg et al., 2006).
Dietary administration of fermentation products to flies
The effect of microbial fermentation products on fly nutritional indices was assayed using a modification of the CAFE method (Ja et al., 2007) to enable parallel quantification of food consumption in adult flies and remove effects of larval manipulation to the diet. The feeding chamber comprised a sterile 50 ml Falcon tube, containing a filter paper (∼25×50 mm) wetted with 0.5 ml sterile deionized water to maintain humidity. Calibrated capillary tubes (1–5 µl, Drummond Scientific Co.) were aseptically filled to 5 µl with a chemically defined liquid food. The diet followed the protocol of Piper et al. (2014) with the following modifications. First, the agar was omitted. Second, glucose was provided as sole sugar source, as in the Y–G diet used in this study, but at 5% (w/v) because 10% (the concentration in Y–G diet) was viscous and supported very low feeding rates in the CAFE system (unpublished data). Third, the cholesterol was dissolved in 2.5:1 mmol ratio of cyclodextrin to cholesterol (replacing ethanol) for improved solubilization (Christian et al., 1997). Finally, acetic acid buffer was replaced by citric acid buffer, with pH adjusted to 4.8 using 10 mol l−1 NaOH. Three capillaries were inserted into the lid of each Falcon tube. Pilot experiments confirmed that food consumption by the insects did not differ significantly between the liquid holidic diet and liquid meridic diet comprising 5% glucose and 5% yeast extract (Fig. S2A) with high insect survival (>80%; Fig. S2B) and small differences in nutrient allocation (Fig. S2C–E). We also compared fly traits on liquid and solid media (Fig. S2C–F), finding that flies on the liquid diets had lower or similar survival, TAG and glucose content compared with those on the solid diets, while mass per fly also did not vary significantly, apart from the elevated values for females on the solid meridic diet.
The experimental design of the metabolite administration assay tested one metabolite – acetic acid, acetoin, ethanol or lactic acid – at three treatment levels: 0, 0.15 and 0.3 mol l−1 based on concentrations used in Kim et al. (2018) for acetic acid effects on Drosophila, which was adopted for all four metabolites. The concentrations generally reflect published data on microbial production of these metabolites (Adler et al., 2014; Aranda-Díaz et al., 2020; Barata et al., 2012; Consuegra et al., 2020; Hall et al., 2018). Exceptionally, acetoin was administered at higher concentrations than reported in Drosophila cultures to enable comparison with the other metabolites in the experiment (Adler et al., 2014; Barata et al., 2011). Three independent experiments were conducted for each metabolite, with male and female flies in separate replicate chambers for each experiment. All the chambers of a given diet were stored together in a sterile air-tight Snapware 23-cup container (Corelle Brands, Chicago, IL, USA). To control for diet evaporation, one fly-free chamber was included in each container. The experiments used axenic flies that had been raised to 3 dpe, with six flies per chamber, incubated at 25°C on a 12 h:12 h light:dark cycle. Each chamber was scored daily for 4 days for the number of live insects and the volume of food in each capillary tube (quantified from the liquid height to an accuracy of 0.1 µl). Capillary tubes containing fresh diet were replaced daily. On day 4 of the experiment, all living flies from each feeding chamber were weighed, flash frozen in liquid nitrogen and stored at −80°C prior to quantification of the protein, glucose and TAG content as above, except protein samples were diluted 1:3 for males and 1:4 for females.
To calculate food consumption, the change in height in each of the three capillaries over the 24 h test period was determined, and then summed to obtain the total amount consumed. The change in height of the liquid column in the three capillaries in the negative control was also scored and subtracted from the experimental values to control for evaporation. To determine diet consumption per fly per day, the value per chamber was divided by the number of live flies at the end of each day (i.e. it is assumed that flies which died in each 24 h period did not feed over that period).
Statistics
All statistics were performed using R version 3.6.1 (http://www.R-project.org/) with α=0.05 as the cut-off for statistical significance of model predictors. An analysis of variance (ANOVA) was implemented to examine microbial abundance, relative insect numbers, fly mass, nutritional indices, feeding rates and SCFA content. All models used the ‘lmer’ function in the lme4 package (Bates et al., 2015) for mixed-effect models, except for relative insect numbers as no random effect was required. Residuals from each model were visually assessed for normality and homoscedasticity. See Table S1 for specific information on predictors and random effects included in models. Differences between fixed effects for the ANOVA models were determined by type II Wald F-test with Kenward–Roger degrees of freedom approximation using the ‘Anova’ function in the car package (Fox and Weisberg, 2019); type III method was used for feeding rates due to repeated measures. Either a post hoc Tukey's or Dunnett's test was implemented to discriminate between effects of predictors on response variables with the emmeans package (https://CRAN.R-project.org/package=emmeans). The contribution of random effects in the model was tested by an analysis of deviance using the ‘Anova’ function in the car package. The multi-model inference (MuMIn) package (https://CRAN.R-project.org/package=MuMIn) was used to calculate the marginal and conditional R2 values to assess model fit of fixed and random effects. For SCFA content analysis, effect sizes (ω2) for each predictor and interactions were calculated from ANOVA table results provided by lmerTest (Kuznetsova et al., 2017) to determine the degree of association for fixed effects. Treatments were considered to have an effect on metabolite concentrations when ω2≥0.01 (Cohen, 1988; Kirk, 1996). For the analysis of flies administered fermentation products, the best linear unbiased predictions were estimated for all response variables to obtain between-trial effect sizes with the ‘ranef’ function for the controls (without added metabolite) and were assessed for between-trial variation when significant results were found for analysis of deviance. Specifically, we simulated the posterior distributions (n=10,000) from each model using the ‘REsim’ function in the merTools package (https://CRAN.R-project.org/package=merTools).
Insect development time summary statistics (Kaplan–Meir method) were calculated using the ‘survfit’ function in the survival package (https://CRAN.R-project.org/package=survival). A Cox mixed-effect model was used to assess the impact of administered microorganisms on pupation and eclosion rates with the coxme package (https://CRAN.R-project.org/package=coxme). Microbial treatments were coded as a categorical fixed effect predictor, and replicate (i.e. each Falcon tube) was nested within each of the three experiments as a categorical random effect. Pairwise comparisons between all treatments were assessed by post hoc Tukey test using the multcomp package (Hothorn et al., 2008). An analysis of deviance was performed as ANOVA models.
The two-sample Kolmogorov–Smirnov test was used to assess differences in the cumulative distribution function of each microbial treatment against the axenic fly treatment; a Bonferroni correction was used for multiple tests (i.e. α=0.05/7). The skewness and kurtosis of each treatment were calculated with the DescTools package (https://CRAN.R-project.org/package=DescTools); 95% confidence intervals [bias corrected and accelerated (BCa) method] were generated with 10,000 bootstrap replicates to enable comparison among treatments. D'Agostino and Anscombe tests were performed to test for skewness and kurtosis, respectively, for each treatment using the moments package (https://CRAN.R-project.org/package=moments); a Bonferroni correction was used for multiple tests (i.e. α=0.05/8).
To determine whether the elevated insect mortality in the treatments including YST reduced development time by selective death of slowly developing individuals, a re-sampling approach of the axenic eclosion data was implemented to impute the ‘missing’ flies of the YST treatment using the ‘sample’ function (replace=TRUE). The missing flies comprised 347 insects (29% of 1195 axenic flies observed) that were randomly selected from the axenic eclosion distribution for each simulation (n=5000). A Cox mixed-effect model was performed for each simulation to assess the proportion of significant results and changes in the hazard ratio. In addition, skewness and kurtosis were calculated for each simulation.
Nutritional indices were analyzed using several multivariate methods. First, principal component analysis (PCA) was implemented with a correlation matrix to visualize the nutritional status of each fly replicate for each sex separately using the vegan package (https://CRAN.R-project.org/package=vegan). TAG, glucose and trehalose content of each sex were square root-transformed to reduce right skew. Principal components were correlated with microbial abundance using the ‘envfit’ function, and the resulting vectors were plotted onto PCA plots. Second, a permutational multivariate ANOVA (PERMANOVA) was performed using autoscaled data to identify the influence of microbiota on nutritional status for each sex. Presence of microbes was coded as three binary fixed effect predictors with dummy codes using the ‘adonis’ function in vegan with Euclidean distances (999 permutations). Then, ordination plots of female and male nutritional status were correlated with a Procrustean analysis and significance was assessed using a Procrustean randomization test (‘protest’ function in vegan with 999 permutations). Finally, a structural equation model (SEM) was used to identify how microbiota influenced each nutritional index with the piecewiseSEM 2.0.2 package (Lefcheck, 2016). Mixed effect linear models were constructed for the piecewise SEM analysis to determine the relationship for microbial abundance of each taxon and each nutritional index (see Fig. S4A for links tested). Indirect effects in SEMs were assessed by multiplying significant standardized coefficients of the relationship between microbial abundance and the effect of individual taxa on a nutritional index.
Fly survival was investigated using a mixed effect logistic regression with the lme4 package using the ‘glmer’ function. Metabolite concentration and sex were included as categorical fixed effects. Total food consumed per fly was included as a continuous covariate in the CAFE experiments other than the method development experiments (feeding rate was not obtained for flies on solid media). Experimental replicate was included as categorical random effect with replicate (i.e. each Falcon tube) nested in experimental replicate for survival analysis to account for over-dispersion by using replicate as an observation level random effect. Wald's χ2 test was performed to examine the effect of predictors using the ‘Anova’ function. A post hoc Dunnett's test was used to compare each metabolite concentration with the control treatment for both sexes. An analysis of deviance was performed as ANOVA models.
A moderation analysis was performed to assess how each microorganism influenced the association between SCFA content and a given nutritional index. A mixed-effect multiple linear regression was performed for each sex separately with microbes included as three separate fixed effects with binary dummy codes and raw SCFA content as a continuous predictor with the ‘lmer’ function. Experimental replicate was included as a categorical random effect. An ANOVA effect test table was generated as for SCFA content analysis. A Bonferroni correction was implemented to control for multiple tests (i.e. α=0.05/30). A follow-up analysis was performed to examine the effect of ACE and YST presence on acetic acid titer and TAG content in male flies, and LAC predictor was removed from the analysis as it had no strong moderation effect on acetic acid titer. The model was performed as before, but a categorical random effect of treatment was nested within experimental replicate. The function ‘emtrends’ was used to assess the moderation effect of ACE presence on simple slopes for treatments with and without YST. An analysis of deviance and R2 values were performed as ANOVA models.
RESULTS
Microbial abundance
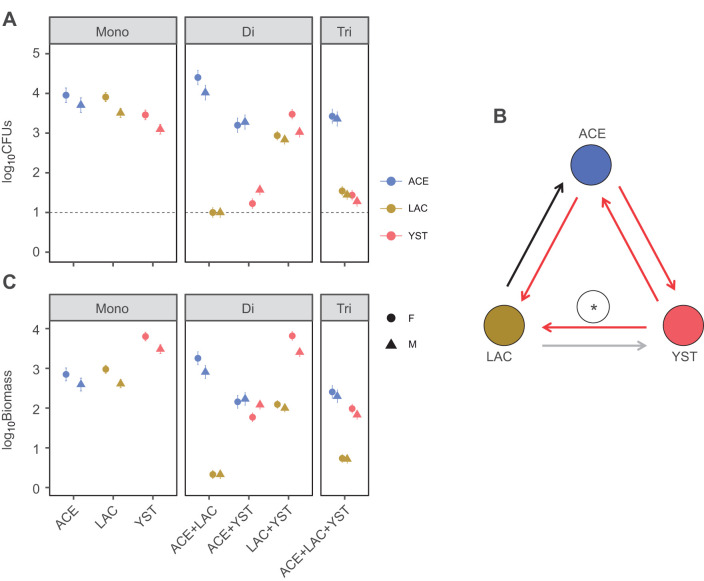
First, we quantified the microbial abundance in the flies at 5 dpe with the three microorganisms (Table 1) in each of the seven microbial treatments. The majority of flies sampled contained at least 10 cells of the microbial taxon that had been administered and no other microorganisms, except for the di-association treatment with ACE and LAC, where LAC was below the detection limit, i.e. ≤10 CFUs fly−1, for all replicates (Fig. 1A). No microorganisms were detected in the axenic flies. The among-treatment variation differed significantly by sex for total microbial abundance, with female flies harboring more microbial cells than males in all treatments except the ACE/YST di-association (Table S1A). The effects of co-association on abundance of individual taxa varied among the microorganisms and were predominantly negative (Fig. 1A,B). The abundance of both LAC and YST was reduced by co-association with ACE, and both ACE and LAC were also significantly suppressed by YST. These negative effects were also evident in the tri-association, although YST ameliorated the negative effect of ACE on LAC. Just one positive interaction was identified: ACE populations were increased in the di-association with LAC (Fig. 1A), as reported previously (Newell and Douglas, 2014). Although ACE tended to be the most abundant taxon in co-associations, YST (which is 17–26 times the estimated biomass per cell of ACE and LAC) attained a comparable or greater biomass than the bacteria in co-associations (Fig. 1C). In addition, the inclusion of total estimated microbial biomass increased model explanatory power from 40% in the total abundance model to 63% (Table S1A). This result reinforces the finding of Keebaugh et al. (2018) that microbial biomass indices improve the statistical power of analyses of microbial abundance in Drosophila, even though different conversion factors were adopted (empirical data for Escherichia coli and Saccharomyces cerevisiae in their study, and an allometric equation based on biovolume estimates in the literature for this study).
Fig. 1.
Abundance of microorganisms in flies. (A) Number of colony forming units (CFUs) per fly (means±s.e., n=18). The black line indicates the limit of detection per fly. The estimated marginal means and standard error from ANOVA analyses are plotted. Results from statistical analyses are provided in Table S1A. (B) Pairwise effects of co-association on microbial abundance (ACE, Acetobacter fabarum; LAC, Lactobacillus brevis; YST, Hanseniaspora uvarum). Arrows indicate co-association effects (black, positive; red, negative; and gray, null) and the yeast amelioration of the negative effect of ACE on LAC in the tri-association is shown by the asterisk. (C) Estimated biomass (pg) of microorganisms (means±s.e., n=18). Data in A and C are for female (F) and male (M) flies. Mono, mono-association; Di, di-association; Tri, tri-association.
Survival and development time of Drosophila
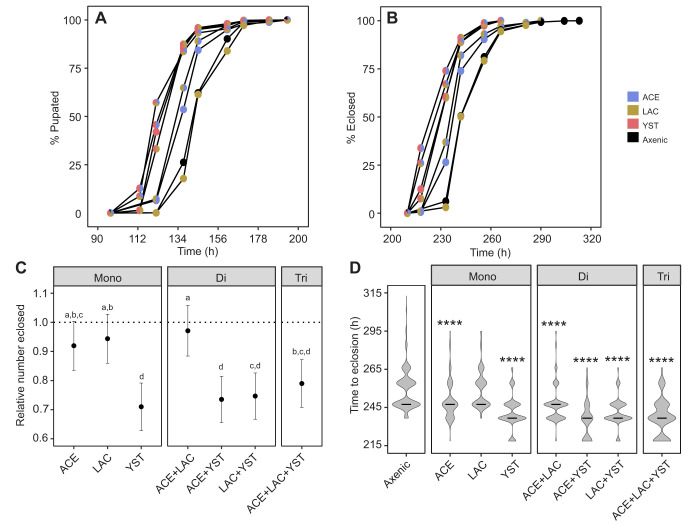
Our first analysis of Drosophila administered the seven microbial treatments (every combination of ACE, LAC and YST), together with axenic insects as a control, focused on insect performance. YST, irrespective of co-associating bacteria, significantly decreased development time to both pupation and eclosion compared with bacteria-only and axenic treatments (Fig. 2A,B and Table 2). ACE significantly decreased development time relative to the axenic treatment. LAC alone did not change development time relative to the axenic treatment, nor influence the effect of ACE in the di-association (Fig. 2A,B and Table 2). These results are broadly consistent with published evidence that axenic cultivation extends larval development time of Drosophila (e.g. Murgier et al., 2019; Newell and Douglas, 2014; Shin et al., 2011), although this developmental delay can be much reduced or undetectable on high nutrient diets (e.g. Storelli et al., 2011; Tefit and Leulier, 2017; Wong et al., 2014).
Fig. 2.
Development time of insects colonized with different microorganisms. (A,B) Time to pupation (A) and eclosion (B). Plots of Kaplan–Meier results, showing mean pupation and eclosion rates at the observed times (A, Cox mixed-effect model treatment effect: integrated χ29=2491.67, P<0.0001 with full summary provided in Table S1B; B, Cox mixed-effect model treatment effect: integrated χ29=2187.88, P<0.0001 with full summary displayed in Table S1B). (C) Number of insects eclosed relative to axenic treatment. The estimated marginal means and 95% confidence intervals (CI) from ANOVA analysis are plotted (n=18), along with letter rankings from post hoc Tukey tests (ANOVA treatment effect: F6,110=6.88, P=3.20×10−6, R2=0.27). The dashed line is the average value for axenic flies; CI that do not overlap with this line indicate a significant difference from axenic insects. (D) Violin plots for time to eclosion. The probability density function and median time (black bar) to eclosion are shown. The development time of each microbial treatment was compared with that of axenic insects by Kolmogorov–Smirnov tests (****P<0.0001; statistical analyses provided in Table S1C).
Table 2.
Summary statistics for average and median development time
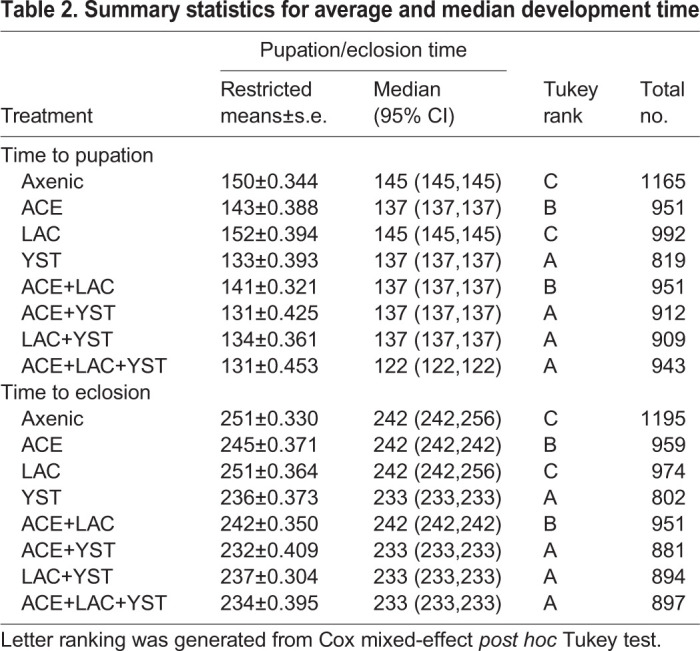
The total number of pupae and flies was reduced by 15–29% in YST-bearing Drosophila compared with axenic insects (Fig. S3A; Fig. 2C, respectively). This result suggests that the presence of YST reduces the larval population prior to late third instar when larvae wander to pupate. None of the bacterial associations (mono- or di-association) significantly affected the number of insects pupated or eclosed relative to the axenic treatment and the bacteria also did not alter the effect of YST in co-associations (Fig. 2C; Fig. S3A,B and Table S1C).
The finding that YST reduced both development time and number of surviving insects raised the possibility that the treatment may have disproportionately increased mortality of slowly developing insects, thereby artifactually inflating the development rate. This hypothesis was supported by the finding that the presence of YST decreased skewness for time to eclosion compared with the axenic or bacterial treatments (Fig. 2D). To address this issue, we conducted 5000 simulations in which time to eclosion for insects bearing YST was supplemented with values randomly drawn from the dataset for axenic insects. The YST median development time was shifted to the axenic time in these simulations (Fig. S3C), indicating that the elevated developmental rate of insects with YST could be explained by disproportionate mortality of slowly developing individuals.
Nutritional indices
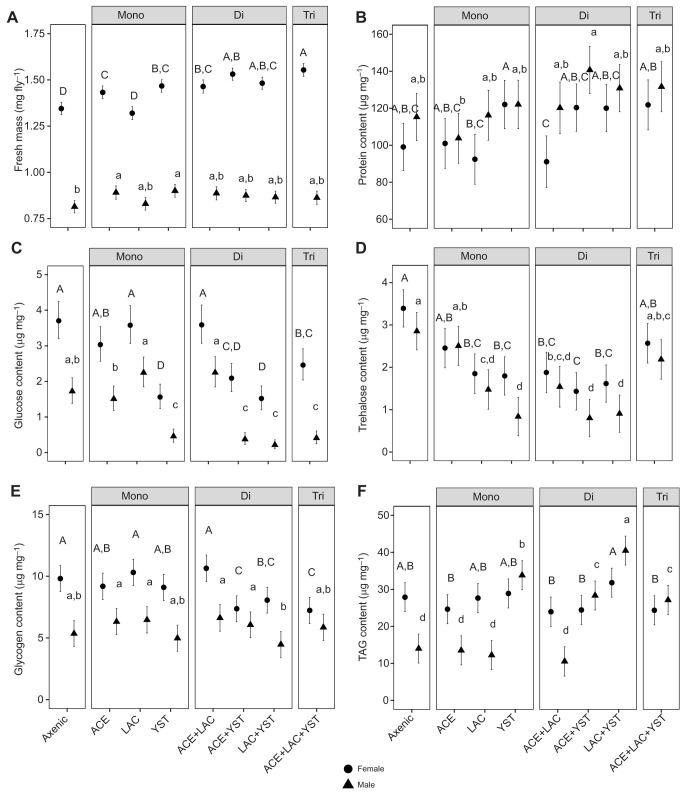
The mass and four of the five nutritional indices (glucose, glycogen, trehalose, TAG) of the flies at 5 dpe varied significantly with the interaction term between microbial treatment and sex; the interaction term for protein content was not significant, although the main effects of sex and microbial treatment were significant (Fig. 3; Table S1D).
Fig. 3.
Individual nutritional indices in flies colonized with different microorganisms. (A) Average fresh mass. (B) Protein content. (C) Glucose content. (D) Trehalose content. (E) Glycogen content. (F) Triacylglyceride (TAG) content. The estimated marginal mean and 95% CI are plotted from each ANOVA model (n=18). Letters indicate post hoc Tukey test results with female- and male-specific comparisons indicated by capital and lowercase letters, respectively. A full description of statistical tests is given in Table S1D.
Female flies bearing microbes weighed more on average than axenic flies (except LAC-associated flies) and bacterial co-associations with YST significantly increased female fly mass, while male flies showed little variation in mass across all treatments (Fig. 3A). Further inspection of the data revealed that males had a significantly lower TAG content than females in both axenic flies and flies bearing bacteria, as has also been observed for Drosophila in routine culture (Jehrke et al., 2018; Wong et al., 2014), while the stereotypical difference in TAG content between the sexes was ameliorated in the presence of YST (Fig. 3F).
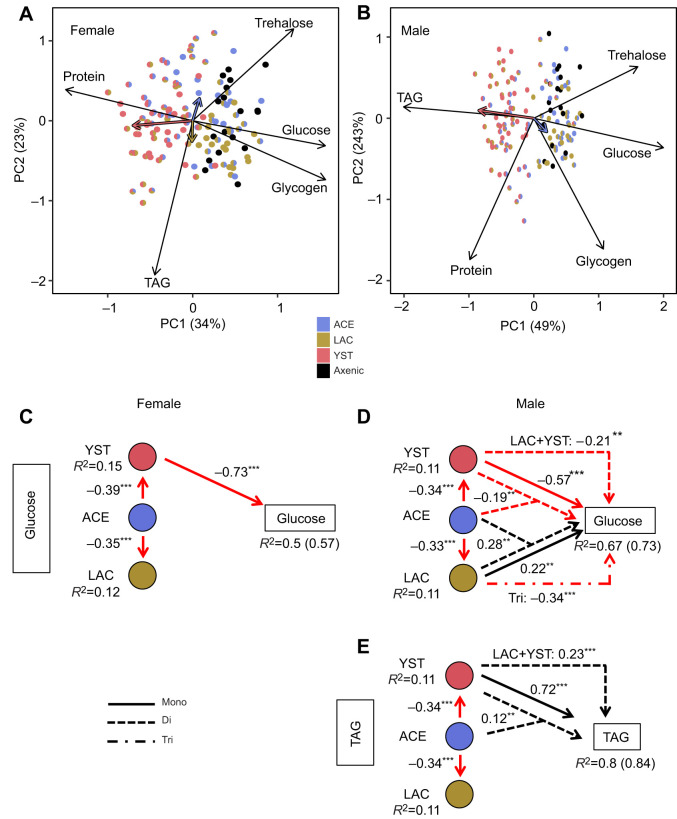
To investigate the overall nutritional status of the flies further, we applied PCA (Fig. 4A,B). For both sexes, ordination plots significantly correlated with each other as determined by Procrustes analysis (m2=0.681, r=0.565, P=0.001). YST was a strong separator on the first principal component axis (PC1) for both males and females, but other effects differed between the sexes. YST-containing treatments were associated with investment in protein content for females, but in TAG content for males, while bacteria-only and axenic treatments increased carbohydrate content (glucose, trehalose and glycogen) for both sexes.
Fig. 4.
Nutritional status of flies colonized with different microorganisms. Principal component analyses (PCA) for (A) females and (B) males. Black arrows indicate loading indices and colored vectors correspond to correlation of microbial abundance (log10-transformed CFUs fly−1+1) with PCA axes. (C–E) Significant results from structural equation models are shown for female glucose (C) and for male glucose (D) and TAG (E) content. Red and black arrows indicate negative and positive associations, respectively. The standardized coefficient for each significant association and marginal R2 values for all response variables are shown, with conditional R2 values shown in parentheses if needed. *P<0.05, **P<0.01, ***P<0.001. Full structural equation model test results are provided in Table S1E and Fig. S4A.
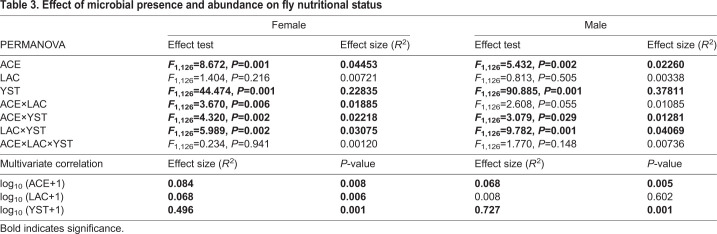
Multivariate correlation of PC1 and PC2 with microbial abundance detected significant effects of all three microbial taxa on the nutritional status of the insects, apart from LAC in males; the effect size of YST was substantially greater than that of the bacteria (Table 3). The PERMANOVA full factorial analysis (Table 3) further showed that the effect of YST was significantly influenced by co-association with either ACE or LAC in both sexes. A significant interaction between the two bacteria, ACE and LAC, was also evident for females.
Table 3.
Effect of microbial presence and abundance on fly nutritional status
We then implemented structural equation modeling to investigate the contributions of the different microbial taxa and their abundance on each nutritional index (Fig. S4A,B). The most pronounced effects of microbial composition were obtained for fly glucose and TAG content (marginal R2>0.5 in at least one sex), while the explanatory power for protein content was particularly weak (marginal R2=0.08–0.2) (Fig. S4B). The model outputs for glucose and TAG content are shown in Fig. 4. For female flies, YST negatively impacted glucose content, while ACE indirectly promoted glucose content by lowering the YST population (standardized coefficient=0.28) (Fig. 4C). The relationships were more complex in males. YST alone reduced the glucose content but increased the TAG content, and both of these effects were dampened by co-association with ACE or LAC. In addition, LAC promoted male glucose content, both alone and in co-association with ACE (Fig. 4D,E).
Nutritional indices of flies administered microbial fermentation metabolites
We hypothesized that the effects of microorganisms on the nutritional indices of the flies were mediated, at least partly, by by-products of microbial metabolism that alter nutrient allocation in the fly (see Introduction). We tested the hypothesis by feeding adult flies on chemically defined liquid diet, administered via capillary tubes and supplemented individually with acetic acid, acetoin, ethanol or lactic acid. The flies feeding from the capillary tubes increased daily consumption per fly over the full 4 days of the experiment for female flies, and over the first 2 days followed by stable feeding rates between days 3 and 4 for male flies (Fig. S5A,B). On average 85% and 82% of female and male flies, respectively, survived during the 4 day study period with significant differences by concentration for all metabolites, except acetoin, and a sex-specific difference in survival for lactic acid (Fig. S5C and Table S1F). Generally, the indices scored did not vary significantly between the controls (i.e. flies on fermentation metabolite-free diets) for the three independent experiments per fermentation metabolite or between the experiments on different fermentation metabolites. Exceptionally, among-experiment variation was obtained for food consumption in the acetoin control and glucose content in the ethanol control (Fig. S5C); and the mean glucose content of male flies in the acetoin control was significantly greater than in the lactic acid control (Table S1F).
The effect of metabolite concentration (0, 0.15 and 0.3 mol l−1) and fly sex was evaluated for TAG, glucose and protein content, with fly mass and food consumption included in each model as covariates to discriminate between the effects of each metabolite and associated covariates on nutritional status. Acetic acid significantly reduced both TAG and protein content of the flies, with significant effects for both concentrations tested (0.15 and 0.3 mol l−1) for females and for one concentration for males (Fig. 5; Table S1F). This result could not be attributed to effects of acetic acid on food consumption (the volume of diet ingested, used as a covariate in the analysis, was not significant; Table S1F) but TAG content was positively associated with fly mass in females for both experiments (Fig. 5; Table S1F). The other three metabolites had no significant effect, apart from a change in glucose content of females administered 0.3 mol l−1 acetoin and lactic acid and a small reduction in female protein content at 0.3 mol l−1 lactic acid (Table S1F).
Fig. 5.
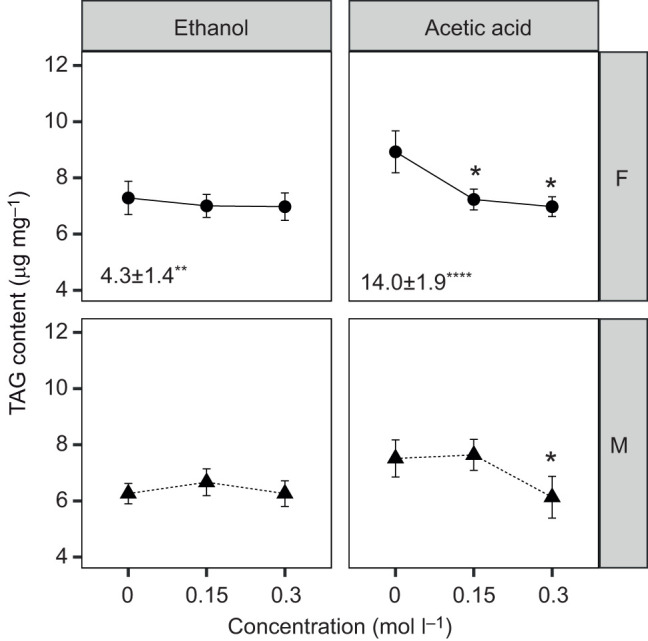
TAG content of axenic flies administered microbial fermentation compounds. The effect of ethanol and acetic acid on TAG content is indicated with raw means±s.e. (n=18). *P<0.05 for post hoc Dunnett's test from each ANOVA model. The covariate slopes and s.e. for mass per fly from each ANOVA model are included at the bottom of each panel when significant; the volume of diet consumed by the flies was non-significant for all experiments (*P<0.05, **P<0.01, ****P<0.0001). Statistical analyses are provided in Table S1F and ANOVA model plots are included in the extended version of Fig. S5 (Dryad: https://datadryad.org/stash/share/7vqNjVDRg5yTuB_fdVIxYE-Tlly4ky9al5RdySr37TI).
To investigate the link between acetic acid and microbial effects on fly TAG and protein content, we quantified the acetic acid content of the flies colonized with different microbial taxa. As described below, we extended our analysis to other SCFAs, as a check for the possible contribution of other SCFAs to this effect.
SCFA content and association with host nutrient allocation
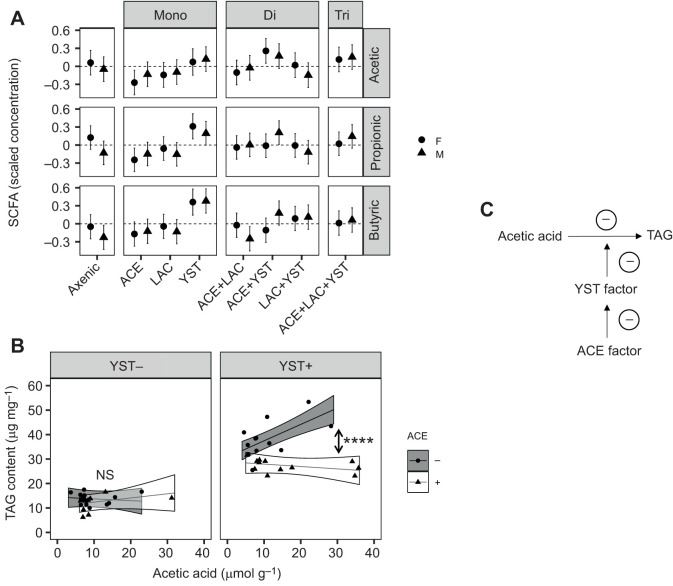
Of the 12 SCFAs tested (see Materials and Methods), just three compounds were detected in the adult flies: acetic acid, butyric acid and propionic acid, which are also the three dominant SCFAs found in the mammalian intestine (Cummings et al., 2004). The normalized content of all three SCFAs in Drosophila (Fig. 6A) was significantly increased in flies bearing YST with differences by sex in butyric acid (Fig. 6A; Table S1G). For acetic acid, this effect was increased by the YST×ACE interaction and also varied with both LAC and sex (Table S1G). ACE also significantly affected propionic and butyric acid content, although the magnitude of this effect varied with sex and LAC (Fig. 6A; Table S1G).
Fig. 6.
Short chain fatty acid (SCFA) content of Drosophila colonized with different microorganisms and the association between SCFA acetic acid and TAG content. (A) SCFA profile of flies. The dashed horizontal line indicates the mean concentration across all treatments for each SCFA (acetic, propionic and butyric acid). The estimated marginal means and 95% CI from ANOVA analyses are plotted (n=6). CI that do not overlap with zero indicate a significant effect of microbial treatment on SCFA concentration. Statistical analyses are provided in Table S1G. (B) Moderation analysis of microbial effect on TAG content by acetic acid titer. The data are the linear prediction for male fly TAG content by acetic acid titer for the presence (+) and absence (−) of ACE and YST in all combinations from the moderation analysis with the results averaged over LAC treatments. The ribbon around the line represents the CI with symbols displaying individual replicates sampled. A post hoc linear contrast was performed comparing the effect of the presence of ACE on the regression slopes with and without YST. Asterisks indicate statistical significance (****P<0.0001); NS, not significant. (C) Proposed ACE and YST factors influencing the relationship between acetic acid and TAG (see Discussion for details).
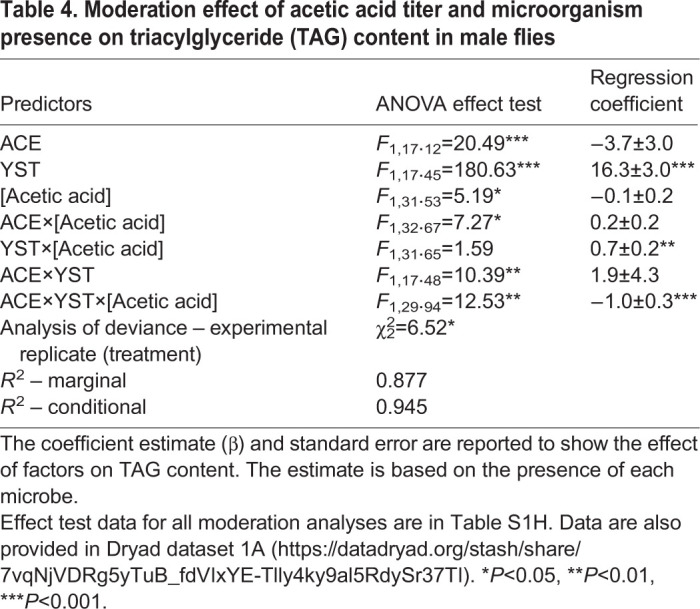
A moderation analysis was performed to investigate how the microorganisms influenced the association between SCFA content and each nutritional index. A significant relationship was obtained between one SCFA, acetic acid, and one nutritional index, TAG, in male flies only (Table S1H). Specifically, the association between TAG and acetic acid content varied significantly with the presence of both ACE and YST (Table 4), with a correlated increase in acetic acid titer and decrease in TAG content of flies bearing ACE and YST (Fig. 6B). This finding suggests that a YST factor may modulate the effect of acetic acid on TAG content observed in Fig. 5, while an ACE factor is capable of reducing the YST factor to decrease TAG content as acetic acid titer increases (Fig. 6C).
Table 4.
Moderation effect of acetic acid titer and microorganism presence on triacylglyceride (TAG) content in male flies
DISCUSSION
This study on the interaction between gut microorganisms and their Drosophila host yielded two key results. First, the impact of individual bacterial and yeast taxa on the performance and nutritional status of Drosophila is strongly influenced by both the presence of other microorganisms and host sex. Second, the relationship between a key microbial fermentation product, acetic acid, and fly lipid content is strongly dependent on both microbiota composition and sex. Here, we address the processes that may contribute to the interactive effects of the bacteria and yeast, focusing the last part of the Discussion specifically on the role of acetic acid. Our interpretation of the results is informed by two important aspects of the Drosophila–microbiome system. (1) Viable microbial cells are shed via the feces onto the diet (Fink et al., 2013; Inamine, et al., 2018), and these shed cells both are available for re-ingestion (i.e. fecal–oral cycling) and can proliferate in the food, thereby altering the nutritional composition of the food consumed by the insects (Broderick and Lemaitre, 2012; Huang and Douglas, 2015; Martino et al., 2018; Wong et al., 2015). In this study, we focused on microbial populations in the insects, while recognizing that some microbial effects on the nutritional status of Drosophila may be mediated by diet-associated microorganisms. (2) Various studies indicate that the effect of gut microorganisms on the nutritional status of Drosophila can be influenced by post-ingestive processes, including nutrient assimilation across the gut wall and nutrient allocation by the insect (e.g. Shin et al., 2011; Storelli et al., 2011; Kamareddine et al., 2018), and this is the primary focus of this study. Nevertheless, we recognize that the nutritional physiology of Drosophila can be influenced by microbial effects on feeding rates (e.g. Huang and Douglas, 2015; Wong et al., 2014, 2017; Yamada et al., 2015), and that this important facet of the Drosophila–microbiome interaction remains to be investigated systematically.
Many of the among-microbe interactions and sex-specific effects of the microorganisms involved the YST treatment (an isolate of H. uvarum from wild Drosophila). Previous studies on gut bacteria in Drosophila and other animals have shown that most among-bacteria interactions reduce bacterial abundance, i.e. they are negative interactions (Coyte and Rakoff-Nahoum, 2019; Faust et al., 2018; Gould et al., 2018; Newell and Douglas, 2014; Venturelli et al., 2018). This study extends this generality to a yeast, including evidence for both mutual reduction in numbers of live YST and ACE in flies colonized with both taxa and a negative effect of YST on LAC abundance (Fig. 1). The basis for these effects in the Drosophila system remains to be determined but may include competition for nutrients in the food and Drosophila gut, as well as toxicity of certain metabolic by-products released from the different microorganisms (Dashko et al., 2014; Hibbing et al., 2010; Mitri and Foster, 2013). In some instances, however, the cross-feeding of metabolic by-products between microbial taxa (Fischer et al., 2017) can promote microbial abundance in Drosophila. For example, the LAC-dependent promotion of ACE abundance (Consuegra et al., 2020; Newell and Douglas, 2014; this study) is likely mediated by LAC-derived fermentation metabolites (e.g. lactic acid) (Consuegra et al., 2020; Sommer and Newell, 2019). In addition, the effect of YST on LAC abundance may be due to YST-derived small metabolites as observed in other yeast–Lactobacillus systems (Ponomarova et al., 2017).
The effect of YST on Drosophila performance was highly significant and independent of the presence of bacteria. The pattern of reduced survival but increased developmental rate to adulthood of Drosophila with YST obtained in our study has also been reported by Murgier et al. (2019), who compared pre-adult performance of Drosophila associated with a different strain of the same yeast species (H. uvarum) to a treatment comprising dried S. cerevisiae routinely used to maintain Drosophila cultures. Our dataset is compatible with the interpretation that YST killed slowly developing larvae (Fig. 2D; Fig. S3C). Recognizing that yeasts release a wide range of fermentation metabolites and other compounds (Arguello et al., 2013; Bueno et al., 2019; Halbfeld et al., 2014; Jolly et al., 2014; Krause et al., 2018; Rossouw et al., 2008), we hypothesize that one or more of the YST products accumulate in the diet to levels that are toxic for the slower-developing larval insects. YST may, in this way, exert very strong selection for rapid Drosophila development time. A high priority for future research is to identify and investigate the mode of action of the putative insecticidal products of YST.
Our analysis of the nutritional indices of 5 day old adult flies revealed markedly different effects of YST on nutrient allocation in male and female Drosophila, with significantly increased protein content of females and TAG (lipid) content of males (Figs 3 and 4). The female response is fully in keeping with published evidence that females derive protein from dietary yeast for ovary maturation and egg production, underpinned at the molecular level by heightened expression of yolk protein genes for vitellogenesis (Bownes et al., 1988; Roy et al., 2018; Terashima and Bownes, 2004). The male response to YST comprised the loss of the stereotypical lower lipid content in male than female flies that is exhibited by Drosophila in a range of rearing conditions (Jehrke et al., 2018; Schwasinger-Schmidt et al., 2012; Wong et al., 2014), and also by axenic flies and Drosophila bearing bacteria (Fig. 3A). The effect of YST on TAG in male flies matches the effect of eliminating the function of the Drosophila lipase gene brummer in neurons and somatic cells of the testes (Wat et al., 2020). The evidence that brummer expression in these organs plays an important role in global lipid homeostasis (Wat et al., 2020) raises the possibility that the nutritional consequences of YST in male flies involves a system-level change in the regulation of lipid metabolism via a brummer-dependent process. In addition, changes in feeding behavior and dietary nutrient composition may provide insight into the microbe-mediated effects on nutrient allocation.
Whatever the mechanistic basis of the YST-mediated increase in TAG of male flies, this effect is ameliorated by co-colonization with ACE (Figs 3F and 4E). Importantly, aspects of the interactive effect of YST and ACE on male TAG content did not match well to published evidence that various Acetobacter strains reduce fly TAG content (Chaston et al., 2014; Newell and Douglas, 2014; Shin et al., 2011). Contrary to these previous studies, flies colonized with ACE in this study did not have significantly lower TAG content than axenic flies. The likely basis for this discrepancy is a difference in experimental design. Acetobacter growing in the food consume dietary glucose, thereby reducing the availability of glucose substrate for TAG synthesis by Drosophila (Huang and Douglas, 2015). Dietary glucose would be less depleted for the 5 day old flies analyzed in this study than in previous work because we transferred the newly eclosed flies to fresh diet, while published studies reared the Drosophila with the Acetobacter inoculum from eggs without refreshing the diet at adulthood. A further discrepancy is the finding from our structural equation modeling that the effects of microorganisms on fly TAG content could not be predicted from microbial population size, contrary to the published finding of a negative correlation between fly TAG content and Acetobacter load (Chaston et al., 2014). This difference likely arises from the inclusion of YST in our analyses.
Insight into the basis of the interactive effect of ACE and YST on fly TAG content comes from consideration of the response of adult Drosophila to microbial fermentation products. Our demonstration that dietary acetic acid significantly reduces TAG content of axenic flies in both sexes is consistent with previous studies, which have, additionally, shown that acetic acid-mediated TAG reduction is mediated by enhanced antimicrobial peptide production in the gut and systemic insulin signaling (Kamareddine et al., 2018; Shin et al., 2011).
Our study demonstrates, further, that the relationship between acetic acid titer and Drosophila TAG content is strongly influenced by the presence and composition of the gut microbiota. The first issue is the microbial source of acetic acid. To date, acetic acid in the Drosophila system has been identified as the product of aerobic fermentation by Acetobacteraceae, a metabolic trait that is displayed when the ethanol substrate is cross-fed from yeasts or heterofermentative lactobacilli (Fischer et al., 2017; Shin et al., 2011; Sommer and Newell, 2019). However, this interpretation may be incomplete because, although (as predicted) fly acetic acid levels are greatest in the ACE+YST treatments, they are also significantly elevated in YST-only flies compared with axenic flies. We hypothesize that YST may be a net producer of acetic acid, as demonstrated for various yeasts (Jolly et al., 2014), including a different strain of the same species, H. uvarum (Bueno et al., 2019). A further potential source of metabolic complexity is the production of acetic acid by various heterofermentative lactobacilli (Adler et al., 2013; Oude Elferink et al., 2001), although we obtained no indication of this effect from the acetic acid content of flies bearing the heterofermentative LAC used in this study.
Taken together, these considerations lead to the apparently paradoxical conclusion that male flies bearing YST have elevated TAG levels, despite their high acetic acid titer. This paradox can be resolved by invoking a YST-derived factor that suppresses the metabolic response of male flies to acetic acid, and the reversal of this effect by an ACE factor that suppresses the YST factor (Fig. 6C). Priorities for future research are twofold: to establish the identity of the putative YST and ACE factors; and to investigate how the putative YST factor may interact with the IMD (immune deficiency) and insulin signaling pathways (Kamareddine et al., 2018; Shin et al., 2011) that mediate TAG reduction by acetic acid and the brummer-mediated regulation of TAG levels in males.
Our analysis of the SCFA profiles detected propionic acid and butyric acid in Drosophila, with elevated titers in YST-bearing flies. Although we obtained no significant relationship between these SCFAs and Drosophila nutritional indices, these microbe-derived SCFAs are important effectors of gut microbiome–host interactions in mammals (Den Besten et al., 2013; Gentile and Weir, 2018), including transgenerational effects on energy homeostasis (Kimura et al., 2020). In addition, propionic acid has been identified as an appetite stimulant for Drosophila larvae under nutrient stress (Depetris-Chauvin et al., 2017). Considering the evolutionary conservation of many aspects of gut microbiome interactions and metabolism across the animal kingdom (Douglas, 2019; Musselman and Kühnlein, 2018), we cannot exclude the possibility that these SCFAs may influence Drosophila metabolism at different developmental stages, on different diets or over longer time scales than used in this study.
We conclude by considering how this study contributes to our understanding of the central role of gut microorganisms and the microbial metabolite acetic acid as determinants of Drosophila lipid content. Although most research on this topic has been conducted on Drosophila associated with a single bacterial strain (e.g. Chaston et al., 2014; Ma et al., 2019; Shin et al., 2011), there is growing interest in the effects of among-microbe interactions on various fly traits (Aranda-Díaz et al., 2020; Consuegra et al., 2020; Fischer et al., 2017; Gould et al., 2018; Judd et al., 2018; Sommer and Newell, 2019). Here, we demonstrate that the impact of a microbial community on metabolism-related traits, especially lipid content, cannot be predicted reliably from the study of mono-associations because the effect of individual microorganisms is strongly influenced by other microorganisms, especially yeast–bacterial interactions, and by host sex. There is growing evidence that this complexity is not peculiar to Drosophila but applies to other animals, including humans (Bolnick et al., 2014; Haro et al., 2016; Markle et al., 2013; Weger et al., 2019). Because Drosophila is superbly amenable to large experiments, including combinatorial designs of microbial treatments (Gould et al., 2018; this study), it is an excellent system to investigate the fundamental processes underlying gut microbiome–host interactions.
Supplementary Material
Acknowledgements
We thank Dr Lynn Johnson (Cornell Statistical Consulting Unit) and Dr Mary Centrella for statistical advice as well as Dr Greg Loeb, Dr Michael Sheehan and Dr John Chaston for helpful feedback on the manuscript. Arturo Vera-Ponce de León collected the Drosophila and isolated the yeast strain identified in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.G.M., A.D.P., A.E.D.; Methodology: J.G.M., G.P., J.C., A.D.P., A.E.D.; Investigation: J.G.M., G.P., J.C.; Writing - original draft: J.G.M., A.E.D.; Writing - review & editing: J.G.M., G.P., J.C., A.D.P., A.E.D.; Supervision: A.D.P., A.E.D.; Project administration: A.D.P., A.E.D.; Funding acquisition: A.D.P., A.E.D.
Funding
This research was funded by a National Institutes of Health grant R01GM095372 to A.E.D. and in part through the Pennsylvania Department of Health using Tobacco CURE funds to A.D.P. J.G.M. was supported by the Dean's Excellence Diversity Fellowship and Sarkaria Insect Physiology and Toxicology Fellowship from Cornell University. Deposited in PMC for release after 12 months.
Data availability
All the data presented in the main text, as well as an extended version of Fig. S5, are available from the Dryad digital repository (McMullen et al., 2020): dryad.ngf1vhhrj
Supplementary information
Supplementary information available online at https://jeb.biologists.org/lookup/doi/10.1242/jeb.227843.supplemental
References
- Adair K. L., Wilson M., Bost A. and Douglas A. E. (2018). Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. ISME J. 12, 959-972. 10.1038/s41396-017-0020-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler P., Bolten C. J., Dohnt K., Hansen C. E. and Wittmann C. (2013). Core fluxome and metafluxome of lactic acid bacteria under simulated cocoa pulp fermentation conditions. Appl. Environ. Microbiol. 79, 5670-5681. 10.1128/AEM.01483-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler P., Frey L. J., Berger A., Bolten C. J., Hansen C. E. and Wittmann C. (2014). The key to acetate: metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation-simulating conditions. Appl. Environ. Microbiol. 80, 4702-4716. 10.1128/AEM.01048-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Díaz A., Obadia B., Dodge R., Thomsen T., Hallberg Z. F., Güvener Z. T., Ludington W. B. and Huang K. C. (2020). Bacterial interspecies interactions modulate pH-mediated antibiotic tolerance. eLife 9, e51493 10.7554/eLife.51493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello J. R., Sellanes C., Lou Y. R. and Raguso R. A. (2013). Can yeast (S. cerevisiae) metabolic volatiles provide polymorphic signaling? PLoS ONE 8, e70219 10.1371/journal.pone.0070219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata A., Campo E., Malfeito-Ferreira M., Loureiro V. and Ferreira V. (2011). Analytical and sensorial characteriation of the aroma of wines produced with sour rotten grapes using GC-O and GC-MS: identifiation of key aroma compounds. J. Agric. Food Chem. 59, 2543-2553. 10.1021/jf104141f [DOI] [PubMed] [Google Scholar]
- Barata A., Santos S. C., Malfeito-Ferreira M. and Loureiro V. (2012). New insights into the ecological interaction between grape berry microorganisms and Drosophila flies during development of sour rot. Microb. Ecol. 64, 416-430. 10.1007/s00248-012-0041-y [DOI] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B. and Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1-48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bolnick D. I., Snowberg L. K., Hirsch P. E., Lauber C. L., Org E., Parks B., Lusis A. J., Knight R., Caporaso J. G. and Svanbäck R. (2014). Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat. Commun. 5, 4500 10.1038/ncomms5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M., Scott A. and Shirras A. (1988). Dietary components modulate yolk protein gene transcription in Drosophila melanogaster. Development 103, 119-128. [DOI] [PubMed] [Google Scholar]
- Broderick N. A. and Lemaitre B. (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307-321. 10.4161/gmic.19896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno E., Martin K. R., Raguso R. A., McMullen J. G. II, Hesler S. P., Loeb G. M. and Douglas A. E. (2019). Response of wild spotted wing Drosophila (Drosophila suzukii) to microbial volatiles. J. Chem. Ecol. 46, 688-698. 10.1007/s10886-019-01139-4 [DOI] [PubMed] [Google Scholar]
- Cadez N. and Smith M. T. (2011). Hanseniaspora Zikes (1912). In The Yeasts (ed. Kurtzman C. P., Fell J. W. and Boekhout T.), pp. 421-434. Amsterdam, Netherlands: Elsevier B.V. [Google Scholar]
- Cai J., Zhang J., Tian Y., Zhang L., Hatzakis E., Krausz K. W., Smith P. B., Gonzalez F. J. and Patterson A. D. (2017). Orthogonal comparison of GC−MS and 1 H NMR spectroscopy for short chain-fatty acid quantitation. Anal. Chem. 89, 7900-7906. 10.1021/acs.analchem.7b00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. A., Lang J. M., Bhatnagar S. and Eisen J. A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7, e1002272 10.1371/journal.pgen.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. A., Eisen J. A. and Kopp A. (2012). Yeast communities of diverse Drosophila species: comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 78, 7327-7336. 10.1128/AEM.01741-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J. M., Newell P. D. and Douglas A. E. (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 5, 1-12. 10.1128/mBio.01631-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian A. E., Haynes M. P., Phillips M. C. and Rothblat G. H. (1997). Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid Res. 38, 2264-2272. [PubMed] [Google Scholar]
- Cleenwerck I., Gonzalez Á., Camu N., Engelbeen K., De Vos P. and De Vuyst L. (2008). Acetobacter fabarum sp. nov., an acetic acid bacterium from a Ghanaian cocoa bean heap fermentation. Int. J. Syst. Evol. Microbiol. 58, 2180-2185. 10.1099/ijs.0.65778-0 [DOI] [PubMed] [Google Scholar]
- Cohen J. (1988). Statistical Power Analysis for the Behavioral Sciences. 2nd edn Hillsdale, NJ: Lawrence Earlbaum Associates. [Google Scholar]
- Cold Spring Harbor (2018). PBS (pH 7.4). Cold Spring Harb. Protoc. 10.1101/pdb.rec099085 [DOI] [Google Scholar]
- Consuegra J., Grenier T., Akherraz H., Rahioui I., Gervais H., da Silva P. Leulier F. (2020). Metabolic cooperation among commensal bacteria supports Drosophila juvenile growth under nutrient stress. ISCIENCE 23, 101232 10.1016/j.isci.2020.101232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K. Z. and Rakoff-Nahoum S. (2019). Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538-R544. 10.1016/j.cub.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K. Z., Schluter J. and Foster K. R. (2015). The ecology of the microbiome: Networks, competition, and stability. Science 350, 663-666. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- Cummings J. H., Rombeau J. L. and Sakata T. (2004). Physiological and Clinical Aspects of Short-Chain Fatty Acids. 1st edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- Dashko S., Zhou N., Compagno C. and Piškur J. (2014). Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 14, 826-832. 10.1111/1567-1364.12161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camargo R. and Phaff H. J. (1957). Yeasts occurring in Drosophila flies and in fermenting tomato fruits in Northern California. J. Food Sci. 22, 367-372. 10.1111/j.1365-2621.1957.tb17024.x [DOI] [Google Scholar]
- Den Besten G., van Eunen K., Groen A. K., Venema K., Reijngoud D.-J. and Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325-2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A., Galagovsky D., Chevalier C., Maniere G. and Grosjean Y. (2017). Olfactory detection of a bacterial short-chain fatty acid acts as an orexigenic signal in Drosophila melanogaster larvae. Sci. Rep. 7, 1-14. 10.1038/s41598-017-14589-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni A. V. and Heberlein U. (2013). The evolution of Drosophila melanogaster as a model for alcohol research. Annu. Rev. Neurosci. 36, 121-138. 10.1146/annurev-neuro-062012-170256 [DOI] [PubMed] [Google Scholar]
- Dobson A. J., Chaston J. M., Newell P. D., Donahue L., Hermann S. L., Sannino D. R., Westmiller S., Wong A. C.-N., Clark A. G., Lazzaro B. P. et al. (2015). Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat. Commun. 6, 6312 10.1038/ncomms7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. E. (2019). Simple animal models for microbiome research. Nat. Rev. Microbiol. 17, 764-775. 10.1038/s41579-019-0242-1 [DOI] [PubMed] [Google Scholar]
- Douglas A. E. (2020). The microbial exometabolome: ecological resource and architect of microbial communities. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190250 10.1098/rstb.2019.0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkosar B., Storelli G., Defaye A. and Leulier F. (2013). Host-intestinal microbiota mutualism: “learning on the fly”. Cell Host Microbe 13, 8-14. 10.1016/j.chom.2012.12.004 [DOI] [PubMed] [Google Scholar]
- Farine J.-P., Habbachi W., Cortot J., Roche S. and Ferveur J.-F. (2017). Maternally-transmitted microbiota affects odor emission and preference in Drosophila larva. Sci. Rep. 7, 1-10. 10.1038/s41598-017-04922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K., Bauchinger F., Laroche B., de Buyl S., Lahti L., Washburne A. D., Gonze D. and Widder S. (2018). Signatures of ecological processes in microbial community time series. Microbiome 6, 1-13. 10.1186/s40168-018-0496-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink C., Staubach F., Kuenzel S., Baines J. F. and Roeder T. (2013). Noninvasive analysis of microbiome dynamics in the fruit fly Drosophila melanogaster. Appl. Environ. Microbiol. 79, 6984-6988. 10.1128/AEM.01903-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C., Trautman E. P., Crawford J. M., Stabb E. V., Handelsman J. and Broderick N. A. (2017). Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. Elife 6, e18855 10.7554/eLife.18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. and Weisberg S. (2019). An {R} Companion to Applied Regression. 3rd edn SAGE Publications. [Google Scholar]
- Freilich S., Zarecki R., Eilam O., Segal E. S., Henry C. S., Kupiec M., Gophna U., Sharan R. and Ruppin E. (2011). Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2, 589 10.1038/ncomms1597 [DOI] [PubMed] [Google Scholar]
- Fry J. D. (2014). Mechanisms of naturally evolved ethanol resistance in Drosophila melanogaster. J. Exp. Biol. 217, 3996-4003. 10.1242/jeb.110510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile C. L. and Weir T. L. (2018). The gut microbiota at the intersection of diet and human health. Science 362, 776-780. 10.1126/science.aau5812 [DOI] [PubMed] [Google Scholar]
- Gould A. L., Zhang V., Lamberti L., Jones E. W., Obadia B., Korasidis N., Gavryushkin A., Carlson J. M., Beerenwinkel N. and Ludington W. B. (2018). Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA 115, E11951-E11960. 10.1073/pnas.1809349115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J. R. and Breznak J. A. (2005). Folate cross-feeding supports symbiotic homoacetogenic spirochetes. Appl. Environ. Microbiol. 71, 1883-1889. 10.1128/AEM.71.4.1883-1889.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gries O. and Ly T. (2009). Volume three, the firmicutes. In Bergey's Manula of Systematic Bacteriology (ed. Vos P. De., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H. and Whitman W. B.), pp. 203-208. Heidelberg, Germany: Springer Berlin Heidelberg. [Google Scholar]
- Halbfeld C., Ebert B. E. and Blank L. M. (2014). Multi-capillary column-ion mobility spectrometry of volatile metabolites emitted by Saccharomyces cerevisiae. Metabolites 4, 751-774. 10.3390/metabo4030751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. E., Loeb G. M., Cadle-Davidson L., Evans K. J. and Wilcox W. F. (2018). Grape sour rot: a four-way interaction involving the host, yeast, acetic acid bacteria, and insects. Phytolpathol 108, 1429-1442. 10.1094/PHYTO-03-18-0098-R [DOI] [PubMed] [Google Scholar]
- Hang S., Purdy A. E., Robins W. P., Wang Z., Mandal M., Chang S., Mekalanos J. J. and Watnick P. I. (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592-604. 10.1016/j.chom.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro C., Rangel-Zúñiga O. A., Alcalá-Díaz J. F., Gómez-Delgado F., Pérez-Martínez P., Delgado-Lista J., Quintana-Navarro G. M., Landa B. B., Navas-Cortés J. A., Tena-Sempere M. et al. (2016). Intestinal microbiota is influenced by gender and body mass index. PLoS ONE 11, 1-16. 10.1371/journal.pone.0154090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herp S., Brugiroux S., Garzetti D., Ring D., Jochum L. M., Beutler M., Eberl C., Hussain S., Walter S., Gerlach R. G. et al. (2019). Mucispirillum schaedleri antagonizes Salmonella virulence to protect mice against colitis. Cell Host Microbe 25, 681-694.e8. 10.1016/j.chom.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Hibbing M. E., Fuqua C., Parsek M. R. and Peterson S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15-25. 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D., Kopp A. and Chandler J. A. (2015). Interactions between Drosophila and its natural yeast symbionts—Is Saccharomyces cerevisiae a good model for studying the fly-yeast relationship? PeerJ 3, e1116 10.7717/peerj.1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A. and Parsons P. A. (1984). Olfactory response and resource utilization in Drosophila: interspecific comparisons. Biol. J. Linn. Soc. 22, 43-53. 10.1111/j.1095-8312.1984.tb00798.x [DOI] [Google Scholar]
- Hooper L. V., Littman D. R. and Macpherson A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268-1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T., Bretz F. and Westfall P. (2008). Simultaneous inference in general parametric models. Biometrical J. 50, 346-363. 10.1002/bimj.200810425 [DOI] [PubMed] [Google Scholar]
- Huang J.-H. and Douglas A. E. (2015). Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol. Lett. 11, 20150469 10.1098/rsbl.2015.0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-H., Jing X. and Douglas A. E. (2015). The multi-tasking gut epithelium of insects. Insect Biochem. Mol. Biol. 67, 1-6. 10.1016/j.ibmb.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatsenko I., Boquete J.-P. and Lemaitre B. (2018). Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity 49, 929-942. 10.1016/j.immuni.2018.09.017 [DOI] [PubMed] [Google Scholar]
- Inamine H., Ellner S. P., Newell P. D., Luo Y., Buchon N. and Douglas A. E. (2018). Spatio-temporally heterogeneous population dynamics of gut bacteria inferred from fecal-time series data. mBio 9, e01453-e01417. 10.1128/mBio.01453-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja W. W., Carvalho G. B., Mak E. M., de la Rosa N. N., Fang A. Y., Liong J. C., Brummel T. and Benzer S. (2007). Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA 104, 8253-8256. 10.1073/pnas.0702726104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehrke L., Stewart F. A., Droste A. and Beller M. (2018). The impact of genome variation and diet on the metabolic phenotype and microbiome composition of Drosophila melanogaster. Sci. Rep. 8, 1-15. 10.1038/s41598-018-24542-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly N. P., Varela C. and Pretorius I. S. (2014). Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215-237. 10.1111/1567-1364.12111 [DOI] [PubMed] [Google Scholar]
- Judd A. M., Matthews M. K., Hughes R., Veloz M., Sexton C. E. and Chaston J. M. (2018). Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Appl. Environ. Microbiol. 84, e00662-e00618. 10.1128/AEM.00662-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L., Robins W. P., Berkey C. D., Mekalanos J. J. and Watnick P. I. (2018). The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab. 28, 449-462.e5. 10.1016/j.cmet.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov W. H., Martínez del Rio C. and Caviedes-Vidal E. (2011). Ecological physiology of diet and digestive systems. Annu. Rev. Physiol. 73, 69-93. 10.1146/annurev-physiol-012110-142152 [DOI] [PubMed] [Google Scholar]
- Keebaugh E. S., Yamada R., Obadia B., Ludington W. B. and Ja W. W. (2018). Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 4, 247-259. 10.1016/j.isci.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kešnerová L., Mars R. A. T., Ellegaard K. M., Troilo M., Sauer U. and Engel P. (2017). Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, 1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G., Huang J. H., McMullen J. G. II, Newell P. D. and Douglas A. E. (2018). Physiolgoical responses of insects to microbial fermentation products: Insights from the interactions between Drosophila and acetic acid. J. Insect Physiol. 106, 13-19. 10.1016/j.jinsphys.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Miyamoto J., Ohue-Kitano R., Watanabe K., Yamada T., Onuki M., Aoki R., Isobe Y., Kashihara D., Inoue D. et al. (2020). Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 367, eaaw8429 10.1126/science.aaw8429 [DOI] [PubMed] [Google Scholar]
- Kirk R. E. (1996). Practical significance: a concept whose time has come. Educ. Psychol. Meas. 56, 746-759. 10.1177/0013164496056005002 [DOI] [Google Scholar]
- Koyle M. L., Veloz M., Judd A. M., Wong A. C. N., Newell P. D., Douglas A. E. and Chaston J. M. (2016). Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J. Vis. Exp. e54219 10.3791/54219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause D. J., Kominek J., Opulente D. A., Shen X. X., Zhou X., Langdon Q. K., DeVirgilio J., Hulfachor A. B., Kurtzman C. P., Rokas A. et al. (2018). Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl. Acad. Sci. USA 115, 11030-11035. 10.1073/pnas.1806268115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P., Manna B., Majumder S. and Ghosh A. (2019). Species-wide metabolic interaction network for understanding natural lignocellulose digestion in termite gut microbiota. Sci. Rep. 9, 1-13. 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P. B. and Christensen R. H. B. (2017). lmerTest Package: Tests in linear mixed effects models. J. Stat. Softw. 82, 1-26. 10.18637/jss.v082.i13 [DOI] [Google Scholar]
- Lefcheck J. S. (2016). piecewiseSEM: Piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573-579. 10.1111/2041-210X.12512 [DOI] [Google Scholar]
- Levy R. and Borenstein E. (2013). Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 110, 12804-12809. 10.1073/pnas.1300926110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Bou-Sleiman M., Joncour P., Indelicato C. E., Frochaux M., Braman V., Litovchenko M., Storelli G., Deplancke B. and Leulier F. (2019). Commensal gut bacteria buffer the impact of host genetic variants on Drosophila developmental traits under nutritional stress. iScience 19, 436-447. 10.1016/j.isci.2019.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D. A., Bauer E., Noronha A., Greenhalgh K., Jäger C., Baginska J., Wilmes P. et al. (2017). Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81-89. 10.1038/nbt.3703 [DOI] [PubMed] [Google Scholar]
- Mahowald M. A., Rey F. E., Seedorf H., Turnbaugh P. J., Fulton R. S., Wollam A., Shah N., Wang C., Magrini V., Wilson R. K. et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 106, 5859-5864. 10.1073/pnas.0901529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J. G. M., Frank D. N., Mortin-Toth S., Robertson C. E., Feazel L. M., Rolle-Kampczyk U., Von Bergen M., McCoy K. D., Macpherson A. J. and Danska J. S. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084-1088. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- Martino M. E., Joncour P., Leenay R., Gervais H., Shah M., Hughes S., Gillet B., Beisel C. and Leulier F. (2018). Bacterial adaptation to the host's diet is a key evolutionary force shaping Drosophila-Lactobacillus symbiosis. Cell Host Microbe 24, 109-119. 10.1016/j.chom.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen J. G. II, Peters-Schulze G., Cai J., Patterson A. D. and Douglas A. E. (2020). Data from: How gut microbiome interactions affect nutritional traits of Drosophila melanogaster, v3, Dryad, Dataset. 10.5061/dryad.ngf1vhhrj [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S. and Foster K. R. (2013). The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 47, 247-273. 10.1146/annurev-genet-111212-133307 [DOI] [PubMed] [Google Scholar]
- Morton J. T., Aksenov A. A., Nothias L. F., Foulds J. R., Quinn R. A., Badri M. H., Swenson T. L., Van Goethem M. W., Northen T. R., Vazquez-Baeza Y. et al. (2019). Learning representations of microbe–metabolite interactions. Nat. Methods 16, 1306-1314. 10.1038/s41592-019-0616-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgier J., Everaerts C., Farine J. P. and Ferveur J. F. (2019). Live yeast in juvenile diet induces species-specific effects on Drosophila adult behaviour and fitness. Sci. Rep. 9, 1-12. 10.1038/s41598-019-45140-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman L. P. and Kühnlein R. P. (2018). Drosophila as a model to study obesity and metabolic disease. J. Exp. Biol. 221, jeb163881 10.1242/jeb.163881 [DOI] [PubMed] [Google Scholar]
- Newell P. D. and Douglas A. E. (2014). Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80, 788-796. 10.1128/AEM.02742-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Chaston J. M., Wang Y., Winans N. J., Sannino D. R., Wong A. C. N., Dobson A. J., Kagle J. and Douglas A. E. (2014). In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Front. Microbiol. 5, 576 10.3389/fmicb.2014.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker C., Chiu H.-C., McNally C. P. and Borenstein E. (2019). Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies. mSystems 4, e00579-e00519. 10.1128/mSystems.00579-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norland S., Heldal M. and Tumyr O. (1987). On the relation between dry matter and volume of bacteria. Microb. Ecol. 13, 95-101. 10.1007/BF02011246 [DOI] [PubMed] [Google Scholar]
- Oude Elferink S. J. W. H., Krooneman J., Jan C., Spoelstra S. F., Faber F., Elferink S. O., Krooneman J. and Spoelstra S. F. (2001). Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 67, 125-132. 10.1128/AEM.67.1.125-132.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanca L., Gaskett A. C., Günther C. S., Newcomb R. D. and Goddard M. R. (2013). Quantifying variation in the ability of yeasts to attract Drosophila melanogaster. PLoS ONE 8, e75332 10.1371/journal.pone.0075332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Blanc E., Leitão-Gonçalves R., Yang M., He X., Linford N. J., Hoddinott M. P., Hopfen C., Soultoukis G. A., Niemeyer C. et al. (2014). A holidic medium for Drosophila melanogaster. Nat. Methods 11, 100-105. 10.1038/nmeth.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarova O., Gabrielli N., Sévin D. C., Mülleder M., Zirngibl K., Bulyha K., Andrejev S., Kafkia E., Typas A., Sauer U. et al. (2017). Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345-357.e6. 10.1016/j.cels.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan A. S. and Eisen M. B. (2018). The ecology of the Drosophila-yeast mutualism in wineries. PLoS ONE 13, 1-25. 10.1371/journal.pone.0196440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Foster K. R. and Comstock L. E. (2016). The evolution of coooperation within the gut microbiota. Nature 533, 255-259. 10.1038/nature17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M. N. and Holmes A. J. (2017). Towards an integrative understanding of diet-host-gut microbiome interactions. Front. Immunol. 8, 1-9. 10.3389/fimmu.2017.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Castaño G. P., Dorris M. R., Liu X., Bolling B. W., Acosta-Gonzalez A. and Rey F. E. (2019). Bacteroides thetaiotaomicron starch utilization promotes quercetin degradation and butyrate production by Eubacterium ramulus. Front. Microbiol. 10, 1-8. 10.3389/fmicb.2019.01145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs M. and Kürschner L. (2010). Saprophagous insect larvae, Drosophila melanogaster, profit from increased species richness in beneficial microbes. J. Appl. Entomol. 134, 667-671. 10.1111/j.1439-0418.2009.01458.x [DOI] [Google Scholar]
- Rolhion N. and Chassaing B. (2016). When pathogenic bacteria meet the intestinal microbiota. Philos. Trans. R. Soc. B Biol. Sci. 371, 20150504 10.1098/rstb.2015.0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D., Næs T. and Bauer F. F. (2008). Linking gene regulation and the exo-metabolome: A comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9, 1-18. 10.1186/1471-2164-9-530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Saha T. T., Zou Z. and Raikhel A. S. (2018). Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 63, 489-511. 10.1146/annurev-ento-020117-043258 [DOI] [PubMed] [Google Scholar]
- Scheidler N. H., Liu C., Hamby K. A., Zalom F. G. and Syed Z. (2015). Volatile codes: Correlation of olfactory signals and reception in Drosophila-yeast chemical communication. Sci. Rep. 5, 1-13. 10.1038/srep14059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwasinger-Schmidt T. E., Kachman S. D. and Harshman L. G. (2012). Evolution of starvation resistance in Drosophila melanogaster: measurement of direct and correlated responses to artificial selection. J. Evol. Biol. 25, 378-387. 10.1111/j.1420-9101.2011.02428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. C., Kim S.-H., You H., Kim B., Kim A. C., Lee K.-A., Yoon J.-H., Ryu J.-H. and Lee W.-J. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670-674. 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- Solomon G. M., Dodangoda H., McCarthy-Walker T. T., Ntim-Gyakari R. R. and Newell P. D. (2019). The microbiota of Drosophila suzukii influences the larval development of Drosophila melanogaster. PeerJ 19, e8097 10.7717/peerj.8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F. and Bäckhed F. (2013). The gut microbiota - masters of host development and physiology. Nat. Rev. Microbiol. 11, 227-238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Sommer A. J. and Newell P. D. (2019). Metabolic basis for mutualism between gut bacteria and its impact on the Drosophila melanogaster host. Appl. Environ. Microbiol. 85, 1-15. 10.1128/AEM.01882-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli G., Defaye A., Erkosar B., Hols P., Royet J. and Leulier F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient signaling. Cell Metab. 14, 403-414. 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Tefit M. A. and Leulier F. (2017). Lactobacillus plantarum favors the early emergence of fit and fertile adult Drosophila under chronic undernutrition. J. Exp. Biol. 220, 900-907. 10.1101/080549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima J. and Bownes M. (2004). Translating available food into the number of eggs laid by Drosophila melanogaster. Genetics 167, 1711-1719. 10.1534/genetics.103.024323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss C. A., Zmora N., Levy M. and Elinav E. (2016). The microbiome and innate immunity. Nature 535, 65-74. 10.1038/nature18847 [DOI] [PubMed] [Google Scholar]
- Tripathi A., Marotz C., Gonzalez A., Vázquez-Baeza Y., Song S. J., Bouslimani A., McDonald D., Zhu Q., Sanders J. G., Smarr L. et al. (2018). Are microbiome studies ready for hypothesis-driven research? Curr. Opin. Microbiol. 44, 61-69. 10.1016/j.mib.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Berg R. A., Hoefsloot H. C. J., Westerhuis J. A., Smilde A. K. and van der Werf M. J. (2006). Centering, scaling, and transformations: imporving the biological informaiton content of metabolomics data. BMC Genomics 7, 142 10.1186/1471-2164-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli O. S., Carr A. V., Fisher G., Hsu R. H., Lau R., Bowen B. P., Hromada S., Northen T. and Arkin A. P. (2018). Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, 1-19. 10.15252/msb.20178157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat L. W., Chao C., Bartlett R., Buchanan J. L., Millington J. W., Chih H. J., Chowdhury Z. S., Biswas P., Huang V., Shin L. J. et al. (2020). A role for triglyceride lipase brummer in the regulation of sex differences in Drosophila fat storage and breakdown. PLoS Biol. 18, e3000595 10.1371/journal.pbio.3000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger B. D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., Foata F., Berger B., Balvay A., Foussier A. et al. (2019). The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 29, 362-382. 10.1016/j.cmet.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. and Taylor J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (ed. Innis M. A., Gelfand D. H., Sninsky J. J. and White T. J.), pp. 315-322. New York, NY: Academic Press Inc. [Google Scholar]
- White M. K., Matthews M. K., Hughes R. C., Sommer A. J., Griffitts J. S., Newell P. D. and Chaston J. M. (2018). A metagenome-wide association study and arrayed mutant library confirm Acetobacter lipopolysaccharide genes are necessary for association with Drosophila melanogaster. G3 Genes, Genomes, Genet. 8, 1119-1127. 10.1534/g3.117.300530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans N. J., Walter A., Chouaia B., Chaston J. M., Douglas A. E. and Newell P. D. (2017). A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol. Ecol. 26, 4536-4550. 10.1111/mec.14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute E. H. and Silver P. A. (2010). Emergent cooperation in microbial metabolism. Mol. Syst. Biol. 6, 1-7. 10.1038/msb.2010.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C. N., Ng P. and Douglas A. E. (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889-1900. 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C.-N., Dobson A. J. and Douglas A. E. (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 217, 1894-1901. 10.1242/jeb.101725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C.-N., Luo Y., Jing X., Franzenburg S., Bost A. and Douglas A. E. (2015). The host as driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl. Environ. Microbiol. 81, 6232-6240. 10.1128/AEM.01442-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C. N., Vanhove A. S. and Watnick P. I. (2016). The interplay between intestinal bacteria and host metabolism in health and disease: lessons from Drosophila melanogaster. Dis. Model. Mech. 9, 271-281. 10.1242/dmm.023408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C.-N., Wang Q.-P., Morimoto J., Senior A. M., ihoreau M., Neeley G. G., Simpson S. J. and Ponton F. (2017). Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr. Biol. 27, 2397-2406. 10.1016/j.cub.2017.07.022 [DOI] [PubMed] [Google Scholar]
- Yamada R., Deshpande S. A., Bruce K. D., Mak E. M. and Ja W. W. (2015). Microbes promote amino acid harvest to resuce undernutrition in Drosophila. Cell Reports 10, 865-872. 10.1016/j.celrep.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezniak A., Andrejev S., Ponomarova O., Mende D. R., Bork P. and Patil K. R. (2015). Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 112, 6449-6454. 10.1073/pnas.1421834112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Qiu Y., Zhong W., Baxter S., Su M., Li Q., Xie G., Ore B. M., Qiao S., Spencer M. D. et al. (2013). A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9, 818-827. 10.1007/s11306-013-0500-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Perreau J., Elijah Powell J., Han B., Zhang Z., Kwong W. K., Tringe S. G. and Moran N. A. (2019). Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 116, 25909-25916. 10.1073/pnas.1916224116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.